Abstract
Congenital myasthenic syndrome (CMS) is a group of 32 disorders involving genetic dysfunction at the neuromuscular junction resulting in skeletal muscle weakness that worsens with physical activity. Precise diagnosis and molecular subtype identification are critical for treatment as medication for one subtype may exacerbate disease in another (Engel et al., Lancet Neurol 14: 420 [2015]; Finsterer, Orphanet J Rare Dis 14: 57 [2019]; Prior and Ghosh, J Child Neurol 36: 610 [2021]). The SNAP25-related CMS subtype (congenital myasthenic syndrome 18, CMS18; MIM #616330) is a rare disorder characterized by muscle fatigability, delayed psychomotor development, and ataxia. Herein, we performed rapid whole-genome sequencing (rWGS) on a critically ill newborn leading to the discovery of an unreported pathogenic de novo SNAP25 c.529C > T; p.Gln177Ter variant. In this report, we present a novel case of CMS18 with complex neonatal consequence. This discovery offers unique insight into the extent of phenotypic severity in CMS18, expands the reported SNAP25 variant phenotype, and paves a foundation for personalized management for CMS18.
Keywords: arthrogryposis multiplex congenita, hydrops fetalis, polyhydramnios, talipes cavus equinovarus
INTRODUCTION
The prevalence of congenital myasthenic syndrome (CMS) in northern Europeans, as estimated by a 2014 U.K. study was 9.2 cases/million in children under the age of 18 (Parr et al. 2014). This is likely an underestimate given the diagnostic limitations of genomic sequencing accessibility. CMS disorders are characterized by fatigable, transient, or permanent muscular weakness in the absence of myasthenia gravis–associated autoantibodies. The majority of CMS disorders occur because of defects in postsynaptic proteins associated with the acetylcholine receptor (Finsterer 2019). There is no standardized therapy because of rarity of cases, but most CMS subtypes respond to acetylcholinesterase inhibitors, which prevent degradation of acetylcholine in the synaptic cleft, and 3,4-diaminopyridine, which increases the amount of acetylcholine released into the synapse. Because these same therapies may exacerbate clinical manifestations in certain subtypes of CMS, it is important to establish a genetic diagnosis prior to initiating treatment (Engel et al. 2015; Finsterer 2019; Prior and Ghosh 2021). As the clinical presentation of CMS is highly variable depending on subtype and variant pathogenicity, prognosis predictions are currently difficult to establish. Clinical findings may range from mild exercise intolerance to disabling weakness and respiratory insufficiency (Abicht et al. 2003). However, although CMS profoundly affects the respiratory system, only the most severe cases require intubation and rarely does it lead to neonatal crisis (Finsterer 2019).
SNAP25 was first isolated and characterized in small rodents by Oyler et al. (1989) where it was localized to synaptic membranes in neurons found in the limbic and neocortical brain regions. As a member of the SNARE (soluble N-ethylmaleimide-sensitive factor) complex which mediates presynaptic vesicle fusion, SNAP25 is responsible for SNARE complex membrane stabilization and conformational change following protein–protein interactions at synapse junctions (Karmakar et al. 2019). Vesicle priming depends on the SNARE complex formation, which involves interactions of the α-helical domains of SNAP25, syntaxin, and synaptobrevin. Vesicle fusion occurs following calcium-triggered interactions between synaptotagmin and SNAP25 (Zhang et al. 2002; Zhou et al. 2015, 2017). Pathogenic variants in components of the SNARE complex have been associated with autism, intellectual disability, epilepsy, and movement disorders (Tang 2021).
Mice homozygous for a Snap25 null mutation exhibited an embryonic lethal phenotype and subsequent neuronal cell culture studies indicated that evoked but not spontaneous neurotransmitter release was eliminated in Snap25−/− lines, suggesting that SNAP25 is essential for Ca2+-triggered synaptic transmission (Washbourne et al. 2002). In humans, disease associated with SNAP25 follows an autosomal dominant inheritance pattern with the majority of de novo SNAP25 variants linked to early-onset developmental and epileptic encephalopathy (DEE). These patients display a variety of phenotypes that include developmental delay, seizures, and various neurological abnormalities such as muscular hypotonia and ataxia (Rohena et al. 2015; Hamdan et al. 2017; Fukuda et al. 2018; Klöckner et al. 2021). In these patients, a broad spectrum of seizure activity is noted with median age of onset for seizure activity reported to be 12 mo (Klöckner et al. 2021). To our knowledge, human variants in SNAP25 have not previously been linked to early neonatal demise.
In 2014, CMS18 was first described in a patient with a mutation specifically impacting SNAP25B. The Ile67Asn variant was found to disrupt the helical nature of the protein, affecting its interactions with other members of the SNARE complex, and was determined to be pathogenic by a dominant negative effect based on cell culture transfection studies (Shen et al. 2014). In the reported patient, disease manifestation was characterized by arthrogryposis, myasthenia, cortical hyperexcitability, ataxia, and intellectual disability. This patient was cyanotic at birth and recovered with oxygen therapy. At the time of the report the patient was 11 yr of age.
Herein, we present a case of severe SNAP25-related CMS that resulted in early neonatal demise in the sixth day of life because of respiratory failure following compassionate withdrawal of life-sustaining care. The de novo nonsense variant was identified as a part of the Utah NeoSeq Project (Fig. 1A) via interdisciplinary collaboration between the Utah Center for Genetic Discovery (UCGD) and ARUP Laboratories, which integrates multiple variant calling and analysis methods to identify genetic etiologies for undiagnosed conditions. The patient was enrolled into the research study following presentation of arthrogryposis multiplex congenita. A concurrent clinical standard arthrogryposes gene panel was negative.
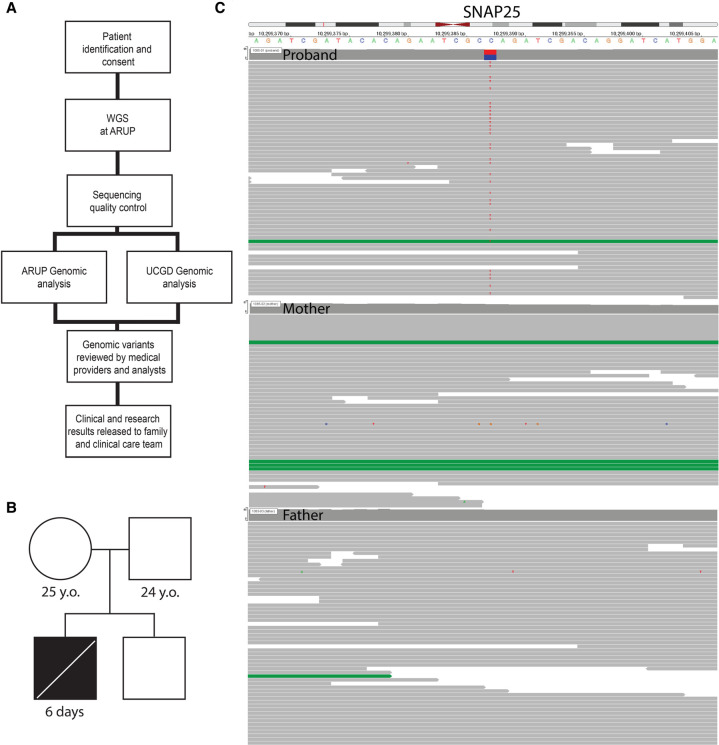
Figure 1.
NeoSeq workflow schematic, proband pedigree, and alignment data. (A) Schematic of NeoSeq protocol workflow: candidate patient identification and consent, whole-genome sequencing (WGS) at ARUP, UCGD and ARUP genomic analyses, sequencing quality control, review of genomic variant candidates, clinical and research results released to family and clinical care team. (B) Proband pedigree featuring unaffected mother, father, and brother. (C) Alignment of the SNAP25 gene for proband, mother, and father reveals heterozygous de novo mutation Chr 20 (GRCh38): g.10299389 C > T.
RESULTS
Clinical Presentation and Family History
The proband was a newborn male delivered at 35 wk gestation via emergent cesarean section because of severe polyhydramnios discovered on prenatal ultrasound. His weight was 2280 g, length was 40 cm, and occipital frontal circumference (OFC) was 36 cm. APGAR scores were 1, 6, 5, and 7 at 1, 5, 10, and 15 min, respectively. Upon delivery, he required intubation following ineffective respiratory effort and the need for positive pressure ventilation. Consistent with reported findings of disorders of the neuromuscular junction, prenatal ultrasound was remarkable for multiple fetal anomalies including severe polyhydramnios, multiple upper extremity joint contractures consistent with arthrogryposis, micrognathia, right clubfoot, left rocker-bottom foot, diffuse skin thickening, and hypomobility. Fetal growth was appropriate with slightly increased biparietal diameter. Physical exam confirmed ultrasound findings and additionally noted findings of undescended testicles, cleft palate, petechiae, hypotonia, and limited reactivity to stimuli. Echocardiogram and head ultrasound were normal. No seizure activity or epileptic spasms were noted on neurologic exam; however, neither a magnetic resonance imaging (MRI) nor electroencephalogram (EEG) were performed because of the critical condition of the patient. Autopsy reports describe multiple findings consistent with arthrogryposis (bilateral contractures of upper extremities with antecubital webbing, clinodactyly, bilateral hips, and knees stiffly extended with outward rotation of hips, left rocker-bottom foot and extreme right clubfoot, reduced skeletal muscle with mild fiber size variation and osteopenia, micrognathia with cleft soft palate, downslanted palpebral fissures and epicanthal folds, wide nasal root), fluid collections (pleural effusions, pericardial fluid, peritoneal fluid), evidence of acute hypoxic stress (increased extramedullary hematopoiesis, scattered hemorrhages), and dilated bilateral renal pelvises. The patient was the second child of nonconsanguineous parents. The mother is a healthy 25-yr old G2P1101. The father is a healthy 24-yr old. The sibling is a healthy male (Fig. 1B).
Genomic Analyses
Initial analysis included noninvasive prenatal testing for fetal aneuploidy and prenatal chromosomal single-nucleotide polymorphism (SNP) microarray (Reveal), which were reported as a normal male. After delivery and initial assessment, a standard arthrogryposes gene panel was ordered (Blueprint Genetics Arthrogryposes). Because of the infant's critical condition, the parents of the proband were consented under the University of Utah NeoSeq research protocol and rWGS was performed for the proband and parents. Overall, whole-genome sequencing (WGS) achieved ≥45-fold median coverage for all three sequenced individuals (Supplemental Table S2). Single-nucleotide variant (SNV) analysis methods from both UCGD and ARUP converged to identify a de novo stop-gain mutation: c.529C > T, p.Q177X in the SNAP25 gene (Table 1; Fig. 1C). The variant is predicted to result in a truncated protein product with loss of the terminal 30 residues and a large portion of the second α-helical domain (Fig. 2A). The variant is absent from the Genome Aggregation Database (gnomAD v2 and v3). Under ACMG/AMP guidelines, this variant meets criteria for pathogenicity: nonsense variant (PVS1_Strong), de novo paternity and maternity confirmed (PS2), and absence in population databases (PM2) (Richards et al. 2015). In this instance, the predicted lack of nonsense-mediated decay does not nullify the PVS1_Strong classification because the variant is located in a region critical to protein function (Abou Tayoun et al. 2018).
Table 1.
Variant table
| Gene | Chromosome (GRCh37) | HGVS cDNA reference | HGVS protein reference | Variant type | Predicted effect | dbSNP ID | Genotype |
|---|---|---|---|---|---|---|---|
| SNAP25 | Chr 20:g.10280037C > T | NM_001322908.2(SNAP25):c.529C > T | p.(Gln177*) | Substitution | Premature termination | n/a | Heterozygous |
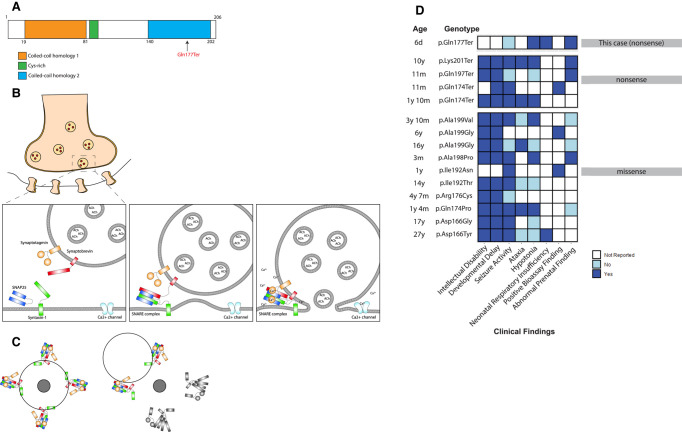
Figure 2.
SNAP25 structural domains with proband variant, SNARE complex–mediated vesicle docking and fusion, proposed dominant-negative mechanism schematic, and the coiled-coil homology 2 (CCH2) domain variant patient phenotypes. (A) Proband variant with respect to SNAP25 structural domains. The variant falls within the CCH2 domain. (B) Sequential diagram depicting SNARE complex–mediated evoked neurotransmitter release at the presynaptic membrane. V-SNARE: synaptobrevin (red) and T-SNAREs: SNAP25 (blue) and Syntaxin-1 (green) combine to form SNARE complex in N-ethylmaleimide sensitive factor (NSF)-mediated priming stage. Calcium influx triggers synaptotagmin (orange) calcium binding cytoplasmic domains (C2A and C2B) with SNARE complex, which result in vesicle fusion with presynaptic membrane. (C) Multiple SNARE complexes meet to traffic a single vesicle (black circle outline) toward a presynaptic membrane target fusion point (dark gray circle). If half of the SNARE complexes are disrupted (gray), the vesicle will not converge at the target fusion point providing an explanation of how a dominant-negative mechanism may occur. (D) Variants in the CCH2 domain displayed with age at time of report, phenotype and clinical findings. White indicates information was not reported, light blue indicates phenotype was reported as absent and dark blue indicates phenotype was reported as present. ClinVar accession numbers: Asp166Tyr (VCV001285522), p.Asp166Gly (VCV001285523), Gln174Pro (VCV001285524), p.Arg176Cys (VCV001683899), p.Ile192Thr (VCV001285525), p.Arg198Pro (VCV001066155), p.Ala199Gly-16y (VCV001285526), p.Ala199Val (VCV001065446), p.Gln174Ter-11m (VCV001285529), p.Gln197Ter (VCV000986340), p.Lys201Ter (VCV001285534). Other: p.Ile192Asn (PMID: 33147442), p.Ala199Gly-6y (PMID: 33147442), p.Gln174Ter-1y10m (PMID: 33299146).
Following whole-genome analysis with high priority given to genes associated with human phenotype ontology (HPO) terms, this variant emerged as the most likely candidate in this case due to the predicted pathogenicity and strong clinical overlap. The testing turn-around time was 7 d.
The clinical standard arthrogryposes panel resulted as negative 3 wk after collection as SNAP25 was not included in the panel (Supplemental Table S1).
DISCUSSION
This study is the first reported case of CMS18 resulting from the Q177X de novo variant. This variant would not have been identified through commercial gene panel testing, highlighting the utility of whole genome and exome sequencing in critically ill newborns. Although our patient does not appear to exhibit some of the common phenotypes in SNAP25-associated disease, it is possible that neurologic findings such as seizure activity, intellectual disability, and developmental delay would have been identified later in life or could have been identified on EEG. The severity of the CMS18 phenotype reported here is likely due to the pathogenic nature of the variant type consisting of protein truncation within a critical binding region in the coiled-coil homology 2 (CCH2) domain.
Without functional studies the pathogenic mechanism cannot be known, however we theorize that the variant is most likely pathogenic by a dominant-negative mechanism. Because the variant is located within the last 50 bp of the penultimate exon, it is not predicted to result in nonsense-mediated decay of the transcript, and therefore a truncated protein is likely produced (Richards et al. 2015). We hypothesize that disrupting interactions of the CCH2 domain with other members of the SNARE complex and calcium sensing domains of synaptotagmin results in unraveling of the SNARE complex and impaired vesicle fusion with the synaptic membrane (Fig. 2B; Gerona et al. 2000; Zhang et al. 2002, de Wit et al. 2009; Zhou et al. 2015, 2017). Moreover, because SNARE complexes operate cooperatively, several complexes are required for efficient vesicle docking and fusion (Mohrmann et al. 2010). Ablation of approximately half of the complexes, as seen in a heterozygous truncating variant, could impact overall vesicle positioning and fusion and would explain a dominant-negative effect (Fig. 2C).
The hypothesis for a dominant-negative mechanism is supported by the work of Klöckner et al. (2021) who found that nonsense variants in the terminal exon of SNAP25 displayed more severe phenotypes compared with splice variants in the second and third exon. Further, Alten et al. (2021) performed functional studies that confirmed that expression of one of these variants (Q174X) in wild-type neuronal cultures exerted a strong dominant-negative effect. Because our variant also results in the absence of similar critical carboxy-terminal residues, we predict that results of functional studies for Q177X would be similar to those of the Q174X variant, which deserves further research attention. Of note, among truncating variants within the CCH2 domain, there exists considerable variation in scope and severity of clinical phenotypes (Fig. 2D). It is also notable that Snap25+/− heterozygous mice have a relatively mild phenotype with no significant impairments, suggesting haploinsufficiency may not be adequate to explain the extent of disease pathophysiology seen in our patient and others with carboxy-terminal truncating variants (Alten et al. 2021). Potential explanations for the range of phenotypic variation within SNAP25-associated disease include genetic modifier effects, position-specific differential penetrance, differences in expressivity, or differences in ability to affect both evoked and spontaneous vesicle release (Alten et al. 2021). To better define the pathogenic effect of these variants, there is a need for in vitro functional analyses to compare to the patient phenotypic spectrum.
Collectively, the study presented herein more precisely defines the CMS18 disorder and paves the foundation for future functional and structural analyses in the SNAP25 SNAREopathy.
METHODS
Genomic DNA Extraction and Sequencing
Genomic DNA was extracted from the proband umbilical cord blood and parental venous samples using the Chemagic Magnetic Separation Module I kit (PerkinElmer). DNA was then quantified with a broad-range double-stranded assay kit on a Qubit 1.0 fluorometer (ThermoFisher Scientific). Whole-genome libraries were prepared using the Illumina DNA Prep workflow (Illumina 20018704). This prep uses tagmentation and five cycles of PCR to produce dual-indexed, paired-end libraries. The libraries were sized and quantified using the Agilent 4200 TapeStation (Agilent) before being diluted and pooled into equimolar ratios. After a final dilution to 1.6 nM, the pool was spiked with 1% PhiX bacteriophage DNA as a sequencing control (Illumina). The final pool was then denatured and loaded on a NovaSeq 6000 sequencing platform for 2 × 150-bp paired-end sequencing on an S1 flow cell (Ilumina).
Sequence reads were generated using bcl2fastq (v2.20) and transferred with a custom script.
Analysis
The Utah NeoSeq pipeline performs alt-aware alignment and variant calling against the GRCh38 build of the human reference genome with alt and decoy contigs. BWA–MEM was used for alignment and the Sentieon suite (https://support.sentieon.com) was used for SNV and indel identification and joint genotyping, as adapted from the GATK “Best Practices” workflow (Li 2013; Caetano-Anolles 2022).
SNVs and short insertions/deletions (indels) were analyzed using Slivar, VAAST, Phevor, and VIQ (Kennedy et al. 2014; Singleton et al. 2014; Pedersen et al. 2021). Structural variants were called and analyzed using RUFUS, Smoove, and Manta (Chen et al. 2016; Feusier et al. 2019; Pedersen et al. 2020). Short tandem repeat (STR) expansions at known disease-associated loci were analyzed using gangSTR and STRling (Mousavi et al. 2019; Dashnow et al. 2021). SNVs were then selected and prioritized by mode of inheritance and by predicted deleteriousness, conservation, allele frequency, ClinVar classification, and known associations to assigned HPO terms (Table 2). Additionally in this case, genes from four different commercial arthrogryposis panels were searched (GeneDx, Blueprint Genetics, Fulgent, Prevention Genetics) (Supplemental Table S1).
Table 2.
HPO terms and associated genes
| Name | HP identifier |
|---|---|
| Arthrogryposis multiplex congenita | HP:0002804 |
| Hydrops fetalis | HP:0001789 |
| Micrognathia | HP:0000347 |
| Polyhydramnios | HP:0001561 |
| Talipes equinovarus | HP:0001762 |
| Fetal pyelectasis | HP:0010945 |
| Genes with >4 HPO matches | ACTA1, CHRNG, COL2A1, DOK7, FLNB, GBA, HBA1, HBA2, HSPG2, KAT6B, MUSK, MYH3, MYPN, NEK9, RAPSN, SLC26A2, TPM2, TPM3 |
| Genes with >3 HPO matches | ACTA1, ADGRG6, AGRN, AMER1, ARVCF, BICD2, CFL2, CHAT, CHRNA1, CHRNG, CHST14, CNTNAP1, COL13A1, COL1A2, COL2A1, COMT, DHCR24, DOK7, DYNC2H1, EBP, ECEL1, ERBB3, ERCC5, FAT4, FBN2, FIG4, FLNB, GBA, GBE1, GLE1, GP1BB, HACD1, HBA1, HBA2, HIRA, HRAS, HSD17B4, HSPG2, HYLS1, ITGA7, JMJD1C, KAT6B, KBTBD13, KIAA0586, KLHL41, LBR, LGI4, LMNA, LMOD3, MAFB, MAP3K20, MUSK, MYH3, MYH8, MYL2, MYO9A, MYOD1, MYPN, NALCN, NEB, NEK9, PHGDH, PI4KA, PIEZO1, PIEZO2, PIGA, PLXND1, PSAT1, PTH1R, RAPSN, REV3L, RREB1, RSPO2, RYR1, SEC24C, SELENON, SLC18A3, SLC25A1, SLC26A2, SLC5A7, SNAP25, SOX9, SYT2, TAPT1, TBX1, TMCO1, TNNI2, TNNT3, TPM2, TPM3, TRAIP, TRIP11, UFD1, VAC14, VAMP1, WDR34, WDR35, WDR60, WNT7A, ZC4H2, ZMPSTE24 |
HPO terms that were used in whole-genome sequencing analysis alongside genes associated with >4 and >3 HPO terms.
ADDITIONAL INFORMATION
Data Deposition and Access
The Utah NeoSeq Project consent allows for case level genomic and phenotypic data for approved uses, but not public distribution. The sequencing data has been deposited in the NCBI database of Genotypes and Phenotypes (dpGaP) and variant information is available in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession number VCV001712269.1.
Ethics Statement
This project (Utah NeoSeq) was approved by the University of Utah Institutional Review Board (IRB_00125940). Written informed consent was obtained from all participants in the study. Written consent was obtained from the parents or legal guardians of any participant under the age of 16.
Acknowledgments
We acknowledge all of the members of the Utah NeoSeq Project Team who make this research possible: Jessica Comstock, Colin Maguire, Mudsar Ahmad, Hunter Best, Lorenzo Botto, Luca Brunelli, Ashley Bunker, Russell Butterfield, Janice Byrne, John Carey, Devin Close, Nodira Codell, Michael Cormier, Jacob Durtschi, Josue Flores-Daboub, Luaiva Floyd, Eric Fredrickson, Makenzie Fulmer, Bushra Gorsi, Edgar Hernandez, Teresa Janecki, Yuan Ji, Ashley Joseph Jason Kunisaki, Gordon Lemmon, Tracey Lewis, Manndi Loertscher, Nicola Longo, Luke Maese, Kelsey Nicholson, Brendan O'Fallon, David Pattison, Christian Paxton, Brandy Petersen, Bennet Peterson, Lucilla Pizzo, Aaron Quinlan, Paul Rindler, Joshua Schiffman, Chelsea Solorzano, Jainy Thomas, Martin Tristani-Firouzi, Hunter Underhill, Gabor Marth, Matt Velinder, David Viskochil, William Watkins, Joseph Worden, Mark Yandell, and Jian Zhao.
Author Contributions
H.M.R. and T.W. drafted the initial manuscript. R.M., B.M., S.E.B., P.B-T., A.F., T.J.N., S.R., C.H., C.M., R.P., J.L.B., B.J.S., and S.M.J. conceptualized and designed the study. H.M.R., T.W., R.M., B.M., S.E.B., P.B.-T., A.F., T.J.N., S.R., C.H., C.M., R.P., B.O., J.L.B., B.J.S., and S.M.J. analyzed and interpreted the data. B.M., S.E.B., A.F., T.J.N., and C.H. established the analytical strategy. K.N. performed the sample processing, extracting, and sequencing experiments. D.B. collected patient samples. R.P., D.B., and S.M.J. recruited patients. R.M., B.M., S.E.B., P.B.-T., A.F., T.J.N., S.R., C.H., C.M., K.N., D.B., R.P., B.O., S.M., J.L.B., B.J.S., and S.M.J. reviewed and revised the manuscript. All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
Funding
The Utah NeoSeq Project is supported by the University of Utah Center for Genomic Medicine, ARUP Laboratories, the Ben B. and Iris M. Margolis Foundation, and the R. Harold Burton Foundation. Sequence alignment, variant calling, and variant interpretation analyses were performed at the Utah Center for Genetic Discovery Core facility, part of the Health Sciences Center Cores at the University of Utah. This work utilized resources and support from the Center for High Performance Computing at the University of Utah. The computational resources used were partially funded by the National Institutes of Health (NIH) Shared Instrumentation Grant 1S10OD021644-01A1.
Competing Interest Statement
Barry Moore has a financial relationship with Fabric Genomics, which provided web-based analysis tools used in this study.
Referees
Xin-Ming Shen
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Abicht A, Müller JS, Lochmüller H. (2003) Congenital myasthenic syndromes overview. In GeneReviews® (ed. Adam MP, Mirzaa GM, Pagon RA, et al. ). University of Washington, Seattle. [PubMed] [Google Scholar]
- Abou Tayoun AN, Pesaran T, DiStefano MT, Oza A, Rehm HL, Biesecker LG, Harrison SM; ClinGen Sequence Variant Interpretation Working Group (ClinGen SVI). 2018. Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum Mutat 39: 1517–1524. 10.1002/humu.23626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alten B, Zhou Q, Shin OH, Esquivies L, Lin PY, White KI, Sun R, Chung WK, Monteggia LM, Brunger AT, et al. 2021. Role of aberrant spontaneous neurotransmission in SNAP25-associated encephalopathies. Neuron 109: 59–72.e5. 10.1016/j.neuron.2020.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano-Anolles D. 2022. Somatic short variant discovery (SNVs + Indels). GATK. https://gatk.broadinstitute.org/hc/en-us/articles/360035894731-Somatic-short-variant-discovery-SNVs-Indels- [Google Scholar]
- Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, Cox AJ, Kruglyak S, Saunders CT. 2016. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32: 1220–1222. 10.1093/bioinformatics/btv710 [DOI] [PubMed] [Google Scholar]
- Dashnow H, Pedersen BS, Hiatt L, Brown J, Beecroft SJ, Ravenscroft G, LaCroix AJ, Lamont P, Roxburgh RH, Rodrigues MJ, et al. 2021. STRling: a k-mer counting approach that detects short tandem repeat expansions at known and novel loci. bioRxiv 10.1101/2021.11.18.469113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit H, Walter AM, Milosevic I, Gulyás-Kovács A, Riedel D, Sørensen JB, Verhage M. 2009. Synaptotagmin-1 docks secretory vesicles to syntaxin-1/SNAP-25 acceptor complexes. Cell 138: 935–946. 10.1016/j.cell.2009.07.027 [DOI] [PubMed] [Google Scholar]
- Engel AG, Shen XM, Selcen D, Sine SM. 2015. Congenital myasthenic syndromes: pathogenesis, diagnosis, and treatment. Lancet Neurol 14: 420–434. 10.1016/S1474-4422(14)70201-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feusier J, Watkins WS, Thomas J, Farrell A, Witherspoon DJ, Baird L, Ha H, Xing J, Jorde LB. 2019. Pedigree-based estimation of human mobile element retrotransposition rates. Genome Res 29: 1567–1577. 10.1101/gr.247965.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsterer J. 2019. Congenital myasthenic syndromes. Orphanet J Rare Dis 14: 57. 10.1186/s13023-019-1025-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda H, Imagawa E, Hamanaka K, Fujita A, Mitsuhashi S, Miyatake S, Mizuguchi T, Takata A, Miyake N, Kramer U, et al. 2018. A novel missense SNAP25b mutation in two affected siblings from an Israeli family showing seizures and cerebellar ataxia. J Hum Genet 63: 673–676. 10.1038/s10038-018-0421-3 [DOI] [PubMed] [Google Scholar]
- GATK best practice workflows. Available from: https://gatk.broadinstitute.org/hc/en-us/sections/360007226651-Best-Practices-Workflows
- Gerona RR, Larsen EC, Kowalchyk JA, Martin TF. 2000. The C terminus of SNAP25 is essential for Ca2+-dependent binding of synaptotagmin to SNARE complexes. J Biol Chem 275: 6328–6336. 10.1074/jbc.275.9.6328 [DOI] [PubMed] [Google Scholar]
- Hamdan FF, Myers CT, Cossette P, Lemay P, Spiegelman D, Laporte AD, Nassif C, Diallo O, Monlong J, Cadieux-Dion M, et al. 2017. High rate of recurrent de novo mutations in developmental and epileptic encephalopathies. Am J Hum Genet 101: 664–685. 10.1016/j.ajhg.2017.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmakar S, Sharma LG, Roy A, Patel A, Pandey LM. 2019. Neuronal SNARE complex: a protein folding system with intricate protein-protein interactions, and its common neuropathological hallmark, SNAP25. Neurochem Int 122: 196–207. 10.1016/j.neuint.2018.12.001 [DOI] [PubMed] [Google Scholar]
- Kennedy B, Kronenberg Z, Hu H, Moore B, Flygare S, Reese MG, Jorde LB, Yandell M, Huff C. 2014. Using VAAST to identify disease-associated variants in next-generation sequencing data. Curr Protoc Hum Genet 81: 6.14.1–6.14.25. 10.1002/0471142905.hg0614s81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöckner C, Sticht H, Zacher P, Popp B, Babcock HE, Bakker DP, Barwick K, Bonfert MV, Bönnemann CG, Brilstra EH, et al. 2021. De novo variants in SNAP25 cause an early-onset developmental and epileptic encephalopathy. Genet Med 23: 653–660. 10.1038/s41436-020-01020-w [DOI] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM., arxiv:1303.3997 [q-bio.GN].
- Mohrmann R, de Wit H, Verhage M, Neher E, Sørensen JB. 2010. Fast vesicle fusion in living cells requires at least three SNARE complexes. Science 330: 502–505. 10.1126/science.1193134 [DOI] [PubMed] [Google Scholar]
- Mousavi N, Shleizer-Burko S, Yanicky R, Gymrek M. 2019. Profiling the genome-wide landscape of tandem repeat expansions. Nucl Acids Res 47: e90. 10.1093/nar.gkz501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyler GA, Higgins GA, Hart RA, Battenberg E, Billingsley M, Bloom FE, Wilson MC. 1989. The identification of a novel synaptosomal-associated protein, SNAP-25, differentially expressed by neuronal subpopulations. J Cell Biol 109: 3039–3052. 10.1083/jcb.109.6.3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr JR, Andrew MJ, Finnis M, Beeson D, Vincent A, Jayawant S. 2014. How common is childhood myasthenia? The UK incidence and prevalence of autoimmune and congenital myasthenia. Arch Dis Child 99: 539–542. 10.1136/archdischild-2013-304788 [DOI] [PubMed] [Google Scholar]
- Pedersen BS, Layer R, Quinlan AR. 2020. smoove: structural-variant calling and genotyping with existing tools (Version 0.2.8) [Computer software] https://github.com/brentp/smoove.
- Pedersen BS, Brown JM, Dashnow H, Wallace AD, Velinder M, Tristani-Firouzi M, Schiffman JD, Tvrdik T, Mao R, Best DH, et al. 2021. Effective variant filtering and expected candidate variant yield in studies of rare human disease. NPJ Genom Med 6: 60. 10.1038/s41525-021-00227-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior DE, Ghosh PS. 2021. Congenital myasthenic syndrome from a single center: phenotypic and genotypic features. J Child Neurol 36: 610–617. 10.1177/0883073820987755 [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. 2015. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17: 405–424. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohena L, Neidich J, Truitt Cho M, Gonzalez KD, Tang S, Devinsky O, Chung WK. 2015. Mutation in SNAP25 as a novel genetic cause of epilepsy and intellectual disability. Rare Dis 1: e26314. 10.4161/rdis.26314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen XM, Selcen D, Brengman J, Engel AG. 2014. Mutant SNAP25B causes myasthenia, cortical hyperexcitability, ataxia, and intellectual disability. Neurology 83: 2247–2255. 10.1212/WNL.0000000000001079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MV, Guthery SL, Voelkerding KV, Chen K, Kennedy B, Margraf RL, Durtschi J, Eilbeck K, Reese MG, Jorde LB, et al. 2014. Phevor combines multiple biomedical ontologies for accurate identification of disease-causing alleles in single individuals and small nuclear families. Am J Hum Genet 94: 599–610. 10.1016/j.ajhg.2014.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang BL. 2021. SNAREs and developmental disorders. J Cell Physiol 236: 2482–2504. 10.1002/jcp.30067 [DOI] [PubMed] [Google Scholar]
- Washbourne P, Thompson PM, Carta M, Costa ET, Mathews JR, Lopez-Benditó G, Molnár Z, Becher MW, Valenzuela CF, Partridge LD, et al. 2002. Genetic ablation of the t-SNARE SNAP-25 distinguishes mechanisms of neuroexocytosis. Nat Neurosci 5: 19–26. 10.1038/nn783 [DOI] [PubMed] [Google Scholar]
- Zhang X, Kim-Miller MJ, Fukuda M, Kowalchyk JA, Martin TF. 2002. Ca2+-dependent synaptotagmin binding to SNAP-25 is essential for Ca2+-triggered exocytosis. Neuron 34: 599–611. 10.1016/S0896-6273(02)00671-2 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Lai Y, Bacaj T, Zhao M, Lyubimov AY, Uervirojnangkoorn M, Zeldin OB, Brewster AS, Sauter NK, Cohen AE, et al. 2015. Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis. Nature 525: 62–67. 10.1038/nature14975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Zhou P, Wang AL, Wu D, Zhao M, Südhof TC, Brunger AT. 2017. The primed SNARE-complexin-synaptotagmin complex for neuronal exocytosis. Nature 548: 420–425. 10.1038/nature23484 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Utah NeoSeq Project consent allows for case level genomic and phenotypic data for approved uses, but not public distribution. The sequencing data has been deposited in the NCBI database of Genotypes and Phenotypes (dpGaP) and variant information is available in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession number VCV001712269.1.