Abstract
In eukaryotes, the nucleolus is the site of ribosome biosynthesis, an essential process in all cells. While human ribosome assembly is largely evolutionarily conserved, many of the regulatory details underlying its control and function have not yet been well-defined. The nucleolar protein RSL24D1 was originally identified as a factor important for 60S ribosomal subunit biogenesis. In addition, the PeBoW (BOP1–PES1–WDR12) complex has been well-defined as required for pre-28S rRNA processing and cell proliferation. In this study, we show that RSL24D1 depletion impairs both pre-ribosomal RNA (pre-rRNA) transcription and mature 28S rRNA production, leading to decreased protein synthesis and p53 stabilization in human cells. Surprisingly, each of the PeBoW complex members is also required for pre-rRNA transcription. We demonstrate that RSL24D1 and WDR12 coimmunoprecipitate with the RNA polymerase I subunit, RPA194, and regulate its steady-state levels. These results uncover the dual role of RSL24D1 and the PeBoW complex in multiple steps of ribosome biogenesis, and provide evidence implicating large ribosomal subunit biogenesis factors in pre-rRNA transcription control.
Keywords: 60S large ribosomal subunit, nucleolus, pre-rRNA processing, rRNA transcription, ribosome biogenesis
INTRODUCTION
Ribosome biogenesis (Ribi) is a highly conserved and essential process and among eukaryotes has been most thoroughly investigated in baker's yeast, Saccharomyces cerevisiae (S. cerevisiae) (Woolford and Baserga 2013). The two ribosomal subunits, large 60S (LSU) and small 40S (SSU), are assembled independently in the nucleolus and join in the cytoplasm to translate mRNAs to proteins. The process begins with RNA polymerase I (RNAPI) driving transcription of the ribosomal DNA (rDNA) to produce the 47S pre-ribosomal RNA (pre-rRNA) transcript in humans. The transcript is subsequently processed and modified by a wide range of trans-acting assembly factors to produce the 18S, 5.8S, and 28S mature rRNAs that form the SSU (18S) and LSU (5.8S, 28S, and RNAPIII-transcribed 5S), respectively (Aubert et al. 2018; Bohnsack and Bohnsack 2019). As rDNA transcription is the rate-limiting step of ribosome biosynthesis which underlies cell growth and proliferation, this process is under tight regulation to ensure cellular homeostasis (Laferte et al. 2006; Chedin et al. 2007; Kopp et al. 2007).
Pre-rRNA processing and ribosome assembly require the hierarchical association and dissociation of a large number of assembly factors. Some of these factors are required for the formation of both ribosomal subunits, while others are required for the synthesis of one of the two subunits (Bassler and Hurt 2019). Moreover, a group of factors known collectively as the transcription U3 small nucleolar RNA associated proteins (t-UTPs), a subcomplex of the SSU processome, are required for the early pre-rRNA transcription steps in eukaryotic cells in addition to their functional role in the processing of the 18S rRNA (Gallagher et al. 2004; Krogan et al. 2004; Prieto and McStay 2007). The t-UTPs associate with the SSU processome to coordinate this action (Krogan et al. 2004). While SSU processome components exhibit dual-functional roles in transcription and processing (Phipps et al. 2011), it remains largely unknown whether there are nucleolar ribosome assembly factors required for LSU biogenesis that play a similar role in RNAPI transcription.
Among LSU factors, it is well established that the PeBoW (PES1–BOP1–WDR12) complex is required for LSU biogenesis in eukaryotes. It is a conserved complex from yeast (Nop7–Erb1–Ytm1) that is required for internal transcribed spacer sequence 2 (ITS2) processing (Holzel et al. 2005). As an extension of this, PeBoW has been shown to be required for cell-cycle progression and cellular proliferation by stimulating ribosome biogenesis (Strezoska et al. 2002; Lapik et al. 2004; Holzel et al. 2005; Grimm et al. 2006). Overexpression of PeBoW proteins has been associated with several cancers (Killian et al. 2006; Fan et al. 2018; Yin et al. 2018; Li et al. 2020, 2022; Mi et al. 2021; Vellky et al. 2021). Nonetheless, PeBoW's function in other critical steps of ribosome biogenesis, such as RNAPI transcription, has not been studied to date.
RSL24D1 is an evolutionarily conserved protein previously studied in yeast with more recent work beginning to elucidate its role in human cells. The S. cerevisiae Rlp24 protein (ortholog of mammalian RSL24D1) is required for 27SB pre-rRNA processing to 5.8S and 25S rRNAs via ITS2 cleavage to produce the 60S ribosomal subunit (Saveanu et al. 2003). Rlp24 associates with both early and late stage pre-60S particles in the nucleolus and cytoplasm, respectively, and is replaced by the homologous ribosomal protein eL24 via the AAA–ATPase Drg1 (Kappel et al. 2012). In humans, RSL24D1 expression in tumor educated platelets has been negatively correlated with early-stage cancer progression and its up-regulation has been associated with familial hypercholesterolemia (Li et al. 2015; Ge et al. 2021). Our laboratory's previous RNAi screening campaign identified RSL24D1 as a factor important for the synthesis of ribosomes in human breast epithelial MCF10A cells (Farley-Barnes et al. 2018). More recently, RSL24D1 was also a hit in a screen for factors involved in 60S subunit assembly in HeLa cells (Dorner et al. 2022). However, the molecular mechanisms of how RSL24D1 participates in ribosome assembly in metazoan cells have not yet been fully understood.
In this study we establish mammalian RSL24D1 as a critical factor for LSU production. It is required for a normal nucleolar number in MCF10A cells, a highly predictive indicator of a function in Ribi (Farley-Barnes et al. 2018; Ogawa et al. 2021). More specifically, we confirm that RSL24D1 is required for cleavage at site 4 in ITS2 of pre-rRNAs to produce the 60S large subunit. Unexpectedly, we uncover a previously unidentified role for the LSU maturation factors, RSL24D1 and the PeBoW complex, in maintaining RPA194 protein levels and therefore important for sustaining RNAPI transcription. This newfound function is likely in part attributed to the identification of an interaction between RSL24D1 and WDR12 with RPA194 by coimmunoprecipitation. These results support the critical role for RSL24D1 in rRNA synthesis and reveal a link between LSU biogenesis and RNAPI transcription regulation.
RESULTS
RSL24D1 is required to maintain nucleolar number and viability in MCF10A human breast epithelial cells
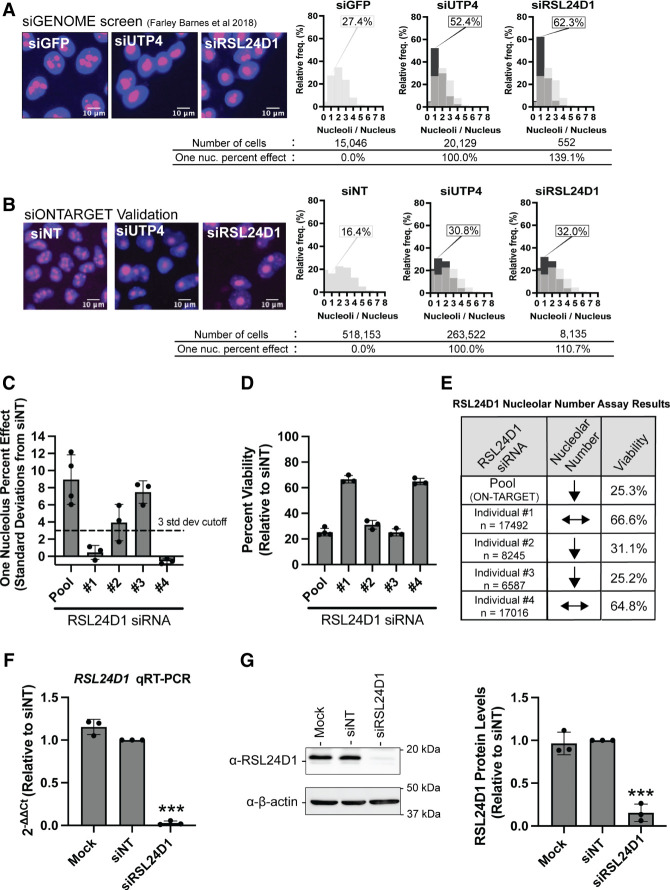
Previously, we performed a genome-wide, phenotypic RNAi screen to uncover novel protein regulators of nucleolar number using a library of siGENOME siRNAs (Horizon Discovery) (Farley-Barnes et al. 2018; Ogawa et al. 2021). The screen was performed in human breast epithelial MCF10A cells, a near-diploid noncancerous cell line, which contain an average of 2–3 nucleoli per cell nucleus. Proteins whose depletion led to a significant change in nucleolar number, either a decrease to one or an increase to five or more, were identified as candidate novel regulators of Ribi. RSL24D1 depletion caused a striking decrease in nucleolar number (139.1% effect) relative to the siGFP negative control (0% effect) and siUTP4 positive control (100% effect) (Fig. 1A). This surpassed the extremely stringent screen cutoff used of greater than three standard deviations from the mean percent effect for the entire screening population (>122% effect) (Farley-Barnes et al. 2018). Indeed, siRSL24D1 treatment produced a larger percentage of one-nucleolus harboring MCF10A cells (62.3%) than the positive control siUTP4 (52.4%) (Fig. 1A). These results, consistent with another RNAi screening effort to identify 60S assembly factors (Dorner et al. 2022), strengthened our hypothesis that RSL24D1's role in making ribosomes is conserved from yeast to humans.
FIGURE 1.
RSL24D1 depletion reduces nucleolar number and cell viability in MCF10A cells. (A) RSL24D1 depletion using siGENOME siRNAs decreases nucleolar number. (Left) Representative images of nuclei stained with Hoechst (blue) and nucleoli stained with an anti-fibrillarin antibody (pink). siGFP was used as a negative control (two to three nucleoli/nucleus) and siUTP4 was used as a positive control (one nucleolus/nucleus). (Right) Histograms of the relative frequency of nucleoli per nucleus are shown for the controls and siRSL24D1. There is a decrease in nuclei with two to three nucleoli for positive controls and RSL24D1-depleted cells. Light gray bars show the nucleolar number distribution for the siGFP negative control, and black bars show the nucleolar number distribution for the indicated positive control or siRSL24D1. Total number of cells analyzed, percentage of cells harboring one nucleolus, and one nucleolus (one nuc.) percent effect for each treatment are indicated. (B) RSL24D1 depletion using ON-TARGETplus siRNAs decreases nucleolar number. Panels as above, except nontargeting (siNT) was used as a negative control. (C) siRNA pool and two of four individual RSL24D1 siRNAs (#2 and #3) decrease nucleolar number in MCF10A cells. Oligonucleotide deconvolution of the ON-TARGETplus siRNA pool was used to confirm the activity of individual siRNAs targeting RSL24D1 in the assay for nucleolar number. The graph indicates mean ± SD, the n = 3–4 separate experiments (indicated), and the dotted line is the cutoff of three SD from nontargeting (siNT) negative control. (D) siRNA pool and two of four individual RSL24D1 siRNAs (#2 and #3) reduce MCF10A cell viability over 50%. Quantitation of number of cells based on Hoechst staining, negative control (siNT) set at 100%. Graph indicates mean ± SD, n = 3–4 separate experiments. (E) Nucleolar number assay and cell viability results correlate for siRNA pool and individual siRNAs targeting RSL24D1. n = total number of cells analyzed for the three to four separate experiments completed from (B–D). Down arrow indicates a greater than three standard deviations from nontargeting (siNT) negative control one nucleolus percent effect, horizontal arrows indicate less than three standard deviations from siNT. Mean percent viability reported from D. (F) qRT-PCR confirmation of RSL24D1 knockdown in MCF10A cells. 2−ΔΔCt values, relative to a nontargeting (siNT) negative control and 7SL internal control primer, show knockdown of RSL24D1 by qRT-PCR using the indicated siRNAs. Graph indicates mean ± SD, n = 3 biological replicates. Data were analyzed by Student's t-test followed by multiple testing P-value correction (two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli) where (***) P ≤ 0.001. (G) Western blot confirmation of RSL24D1 knockdown in MCF10A cells. Mock and nontargeting (siNT) siRNAs are shown as negative controls. (Left) Representative western blot images using α-RSL24D1 and α-β-actin antibodies. (Right) Quantitation of RSL24D1 levels is reported relative to siNT and normalized to the β-actin loading control. Graph indicates mean ± SD, n = 3 biological replicates. Data were analyzed by one-way ANOVA with Dunnett's multiple comparisons test where (****) P ≤ 0.0001; ns = not significant.
To minimize the inclusion of off-target effect induced hits, we rescreened RSL24D1 using ON-TARGETplus siRNA reagents (siONT; Horizon Discovery; Supplemental Data S1), which are designed to reduce siRNA off-target effects (Jackson et al. 2006). We found that RSL24D1 depletion using a siONT pool reproduces the one-nucleolus phenotype (110.7% effect, 32.0% one-nucleolus cells) relative to the nontargeting (siNT) negative control (0% effect, 16.4% one-nucleolus cells) and the siUTP4 positive control (100% effect, 30.8% one nucleolus-cells) (Fig. 1B). Since this was a targeted validation of our previous siGENOME screen results, we used a cutoff threshold of producing a one-nucleolus percent effect greater than three standard deviations from the negative control siNT (>39.1% effect) for each of the four independent screen repetitions.
We performed deconvolution of the RSL24D1 siRNA pool to further validate the role of RSL24D1 in maintaining normal nucleolar number. The individual siRNAs from the siONT pool of four siRNAs were evaluated for their effects on nucleolar number and cell viability in RSL24D1-depleted cells. We found that two of the four individual siRNAs significantly reduced the number of nucleoli from 2–3 to only one, passing our cutoff criteria of producing an average one-nucleolus percent effect greater than three standard deviations from the negative control siNT for three independent experiments (Fig. 1C; Supplemental Table S1; Supplemental Fig. S1). Additionally, the siONT treatments that produced the one-nucleolus phenotype correlated with a greater reduction in cell viability (Fig. 1D,E). A correlation between the presence of one nucleolus upon RSL24D1 depletion and cell viability is expected if RSL24D1 is required for RiBi, an essential cellular process. Thus, we hypothesized that the reduction in nucleolar number upon RSL24D1 depletion was the result of defective RiBi.
As an important step to further confirm our observations with siRNAs targeting RSL24D1, we measured RSL24D1 mRNA levels and RSL24D1 protein levels after siONT pool treatment. Both mRNA levels, as measured by qRT-PCR, and protein levels, as measured by western blotting, were greatly reduced, indicating robust knockdown using this method (Fig. 1F,G).
Changes in nucleolar number and morphology have been connected with cancer pathology and prognosis for over 100 yr (Derenzini et al. 2017). Based on our result that RSL24D1 is necessary for maintenance of nucleolar number, we examined RSL24D1’s mRNA expression levels in Genotype-Tissue Expression (GTEx) unmatched normal and The Cancer Genome Atlas (TCGA) matched normal and tumor samples (Goldman et al. 2020). Although RSL24D1 expression is negatively correlated with cancer progression in tumor educated platelets (Ge et al. 2021), this is not what we observed for RSL24D1’s expression in tumor compared to normal tissue. RSL24D1 was significantly higher expressed in all tumor samples, including breast tumor, compared to normal tissue (Supplemental Fig. S2). RSL24D1’s increased expression in breast cancer specifically highlights the relevance of its expected role in RiBi in MCF10A cells.
RSL24D1 functions in pre-28S rRNA processing
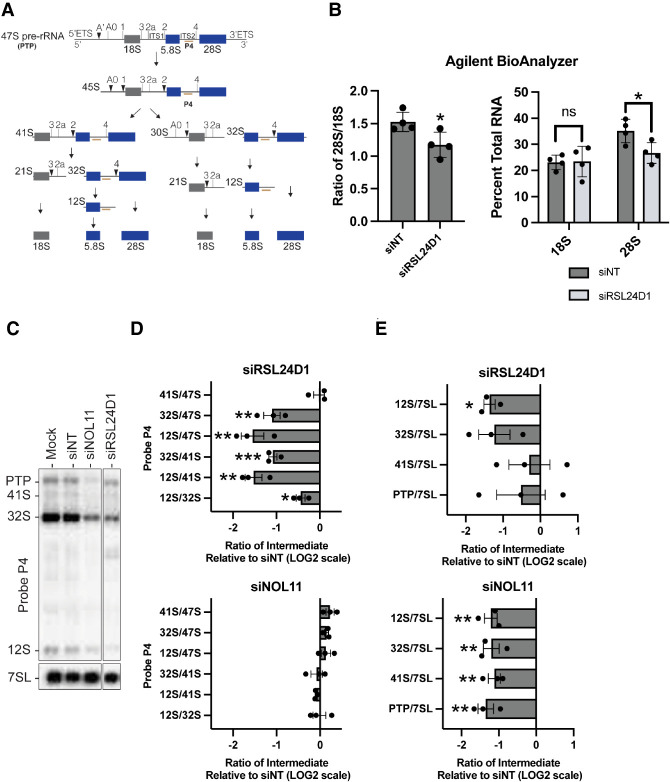
RSL24D1 was identified in a previous targeted RNAi depletion screen as a factor potentially involved in LSU processing and assembly in HeLa cells and 60S biogenesis in mouse embryonic stem cells (Durand et al. 2021; Dorner et al. 2022). Moreover, the RSL24D1 yeast ortholog, Rlp24, has been shown to be important for large subunit pre-rRNA processing (Saveanu et al. 2003). A key step during large subunit synthesis is the cleavage of ITS2 from maturing pre-60S subunits. In the 47S primary transcript precursor (PTP), ITS2 separates the 5.8S and 28S rRNAs and is eventually removed from the larger pre-rRNA precursor (Fig. 2A).
FIGURE 2.
RSL24D1 functions in pre-rRNA processing of the large ribosomal subunit. (A) Mammalian pre-rRNA processing schematic. The probe P4 hybridizes to the internal transcribed spacer (ITS2) sequence of the 47S pre-rRNA precursor which undergoes a series of cleavage steps to yield the small subunit 18S rRNA (gray) and the large subunit 5.8S and 28S rRNAs (blue). (B) RSL24D1 depletion in MCF10A cells reduces the levels of the mature 28S rRNA by Agilent BioAnalyzer. Nontargeting (siNT) was used as a negative control. (Left) Ratio of 28S/18S mature rRNAs. (Right) Percent of total RNA for both 18S and 28S mature rRNAs. Graphs indicate mean ± SD, n = 4 biological replicates. Data were analyzed by Student's t-test in where (*) P ≤ 0.05. (C–E) RSL24D1 is required for ITS2 processing. (C) Representative northern blots measuring steady-state levels of pre-rRNA intermediates when RSL24D1 is depleted in MCF10A cells. A probe for the 7SL RNA was used as a loading control. Negative controls were mock (no siRNA) and nontargeting (siNT), and siNOL11 was used as a positive control. (D) Defects in processing were determined by analyzing precursor-product relationships using ratio analysis of multiple precursors (RAMP) (Wang et al. 2014) and were reported relative to nontargeting (siNT) negative control. Graphs indicate mean ± SEM, n = 3 biological replicates. Data were analyzed by two-way ANOVA where (***) P ≤ 0.001, (**) P ≤ 0.01, and (*) P ≤ 0.05. (E) Changes in relative levels of pre-rRNA precursors were measured relative to nontargeting (siNT) negative control and normalized to 7SL internal loading control. Graphs indicate mean ± SEM, n = 3 biological replicates. Data were analyzed by two-way ANOVA where (**) P ≤ 0.01 and (*) P ≤ 0.05.
Because the expected pre-rRNA processing defects indicate aberrant LSU biogenesis, we sought to determine the extent to which RSL24D1 depletion by siONT pools affects the production of the mature 28S rRNA. Agilent BioAnalyzer quantitation shows a decrease in the ratio of mature 28S to 18S levels (Fig. 2B, left) in siRSL24D1 compared to the negative control, siNT treated MCF10A cells. This decrease in the 28S to 18S rRNA ratio is caused by a decrease in 28S rRNA levels specifically, while 18S levels do not change (Fig. 2B, right).
To further refine RSL24D1's role in LSU biogenesis, we probed the extent to which pre-rRNA processing was disrupted upon depletion of RSL24D1 using siONT pools in MCF10A cells by northern blot analysis, using the ITS2 probe P4 to detect defects in LSU biogenesis (Fig. 2A). Mock (no siRNA) and siNT were used as negative controls and siNOL11 was used as a positive control. After 72 h of siRNA knockdown, RSL24D1 depletion resulted in an apparent decrease in the 32S and 12S pre-rRNAs relative to the mock and siNT negative controls (Fig. 2C). These results concur with previous northern blot results where two of three siRNAs against RSL24D1 mRNA effected the same reduction in 12S pre-rRNA (Tafforeau et al. 2013). Quantitation of the ratios of each intermediate relative to its precursor in the processing pathway by the ratio analysis of multiple precursors (RAMP) method (Wang et al. 2014) revealed that the 12S/32S, 12S/41S, and 12S/PTP ratios were all significantly decreased after depletion of RSL24D1 (Fig. 2D). Furthermore, overall 12S levels were also significantly reduced (Fig. 2E). Although depletion of NOL11 led to a reduction in all pre-rRNA precursors probed (Fig. 2E), NOL11 depletion did not decrease the 12S/32S, 12S/41S, and 12S/PTP ratios, consistent with its role in pre-rRNA transcription and pre-18S rRNA processing (Fig. 2D; Freed et al. 2012). These results confirm that RSL24D1 is required for pre-LSU rRNA processing at site 4 in ITS2 and for maintaining levels of the 28S rRNA.
RSL24D1 is predicted to functionally associate with the PeBoW complex in human cells
To gain further biological insight into the role of RSL24D1, we used the human protein complex map (hu.MAP 2.0) to predict functional associations of RSL24D1 (Drew et al. 2021). hu.MAP 2.0 incorporates information from several recently published protein interaction and affinity purification mass spectrometry data sets to predict and catalog mammalian protein complexes. Of the medium-to-high confidence interacting proteins identified, the PeBoW complex (BOP1, PES1, WDR12) was predicted to interact with RSL24D1 (Table 1; Supplemental Table S2).
TABLE 1.
RSL24D1 is predicted to interact with PeBoW in human cells
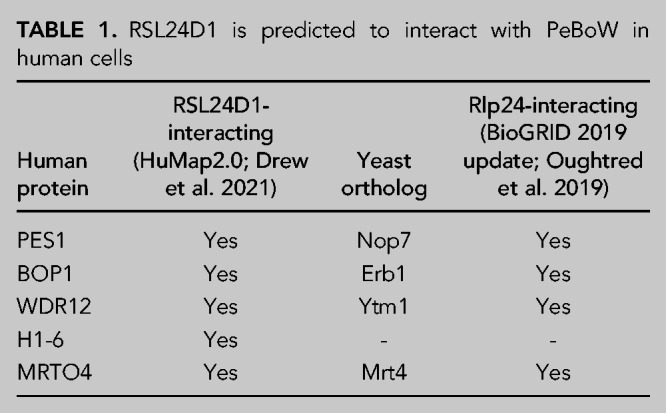
Interestingly, the yeast complex orthologous to PeBoW (Nop7 complex) containing factors Nop7, Erb1, and Ytm1 has been shown to interact with Rlp24, the yeast ortholog of RSL24D1, as constituents of pre-60S ribosomes (Table 1; Supplemental Table S2; Saveanu et al. 2003; Kater et al. 2017; Poll et al. 2021). While these interactions have been documented by various affinity capture experiments (Supplemental Table S2), a recent cross-link mass spectrometry study of yeast pre-60S ribosomes still found no evidence of direct interactions between Rlp24 and the Nop7 complex (Sailer et al. 2022). Furthermore, to our knowledge, there have been no documented human pre-60S structures containing both RSL24D1 and PeBoW. Thus, based on this unexplored predicted functional association between RSL24D1 and PeBoW in humans, we decided to further interrogate RSL24D1 and PeBoW's role in human RiBi together.
RSL24D1 and PeBoW regulate RNA polymerase I levels through their association with RPA194
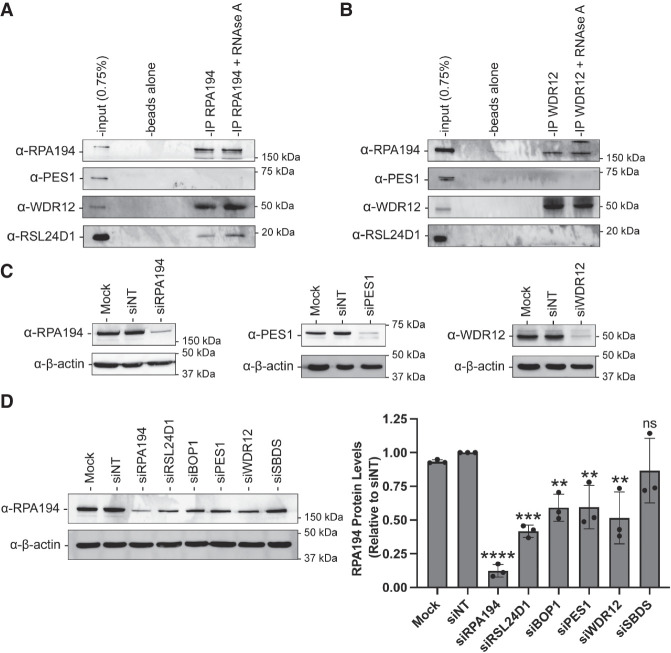
Previously, Sorino and coworkers performed immunoprecipitation of RPA194, the largest subunit of the RNAPI holoenzyme, followed by mass spectrometry. They identified RSL24D1 and WDR12 of the PeBoW complex as members of the RPA194 interactome (Sorino et al. 2020). These LSU maturation factors were among many confirmed RPA194 interacting proteins detected in their analysis, including NOL11 and UBF (Panov et al. 2006; Freed et al. 2012). To validate whether RSL24D1 and WDR12 interact or are in complex with RPA194, we performed coimmunoprecipitation experiments followed by western blotting. Intriguingly, antibodies against RPA194, but not unconjugated beads, coimmunoprecipitated endogenous RSL24D1 and WDR12 from MCF10A whole cell extracts (Fig. 3A). However, endogenous PES1 was not coimmunoprecipitated. Treatment of extracts with RNase A did not abrogate these interactions (Fig. 3A). The reciprocal coimmunoprecipitation with WDR12 also revealed that WDR12 interacts with RPA194 and that this interaction is not abrogated with RNase A treatment (Fig. 3B). In contrast, WDR12 did not coimmunoprecipitate either RSL24D1 or PES1, a member of the PeBoW complex. While we do not understand the lack of coimmunoprecipitation of WDR12 with its complex partner, PES1, this suggests that there may be a separate pool of WDR12 outside of the PeBoW complex that associates with RNAPI. Furthermore, we have not been able to confirm that RSL24D1 coimmunoprecipitates with PeBoW as predicted by hu.MAP 2.0 (Supplemental Table S2). The RPA194 and WDR12 antibodies used for coimmunoprecipitations and the PES1 antibody used for blotting were tested for specificity by western blotting after siRNA depletion. All antibodies were shown to be specific based on an observed decrease in the expected band after siRNA treatment (Fig. 3C).
FIGURE 3.
RSL24D1 and WDR12 associate with RPA194 and, along with BOP1 and PES1, regulate its levels. (A) MCF10A whole cell extracts were immunoprecipitated with α-RPA194 antibody with/without RNase A treatment. Input corresponds to 0.75% of the whole cell extract (WCE) used for immunoprecipitation. Unconjugated protein A beads were used as a negative control. RPA194, PES1, WDR12, and RSL24D1 were detected by western blotting with specific antibodies. Representative western blot images shown are one of two biological replicates. (B) Panel as above, except MCF10A whole cell extracts were immunoprecipitated with α-WDR12 antibodies. (C) α-RPA194, α-WDR12 antibodies for immunoprecipitation and α-PES1 antibody for blotting are specific. Western blot of MCF10A cell lysate using α-RPA194 (left), α-PES1 (middle), and α-WDR12 (right) antibodies. α-β-actin was used as an internal loading control. Mock and nontargeting (siNT) were used as negative controls, siRPA194, siPES1, and siWDR12 were used as positive controls to confirm a reduction in levels of their respective expected bands. (D) Depletion of RSL24D1 and PeBoW reduce RPA194 protein levels in MCF10A cells. Cells were treated with the indicated siRNAs for 72 h and total protein from whole cell extracts was harvested. Mock, nontargeting (siNT), and SBDS siRNAs are shown as negative controls. (Left) Representative western blot images using α-RSL24D1 and α-β-actin antibodies. (Right) Quantitation of RPA194 levels is reported relative to siNT and normalized to the β-actin loading control. Graph indicates mean ± SD, n = 3 biological replicates. Data were analyzed by one-way ANOVA with Dunnett's multiple comparisons test where (****) P ≤ 0.0001; (**) P ≤ 0.01; ns = not significant.
This finding led us to ask whether RSL24D1 and PeBoW could also influence RiBi at the level of pre-rRNA transcription in addition to their regulatory role in pre-LSU processing steps. First, due to the observed association of RSL24D1 and WDR12 with RPA194, we tested for changes in RPA194 protein levels after RSL24D1 and PeBoW complex member depletion. Mock (no siRNA), siNT, and siSBDS were used as negative controls, while siRPA194 was used as a positive control. siRNA depletion of RSL24D1, WDR12, and the two other PeBoW members, BOP1 and PES1, led to significant decreases in RPA194 levels in MCF10A cells (Fig. 3D). However, depletion of the Shwachman–Bodian–Diamond Syndrome protein (SBDS), an additional negative control, that functions in LSU biogenesis in downstream maturation steps outside of the nucleus (Warren 2018), did not have any effects on RPA194 levels. This indicates that it is most likely not a feedback mechanism of impaired later steps of RiBi that results in reductions in RPA194 levels.
For this reason, we have included SBDS depletion as a negative control in subsequent experiments. siRNAs targeting PeBoW members and SBDS were validated by qRT-PCR which revealed significant depletion of all targeted mRNA transcripts (Supplemental Fig. S3). Taken together, these results are consistent with a role for RSL24D1 and PeBoW complex members in maintaining adequate levels of the RPA194 protein in human cells.
RSL24D1 and the PeBoW complex are required for RNAPI promoter activity
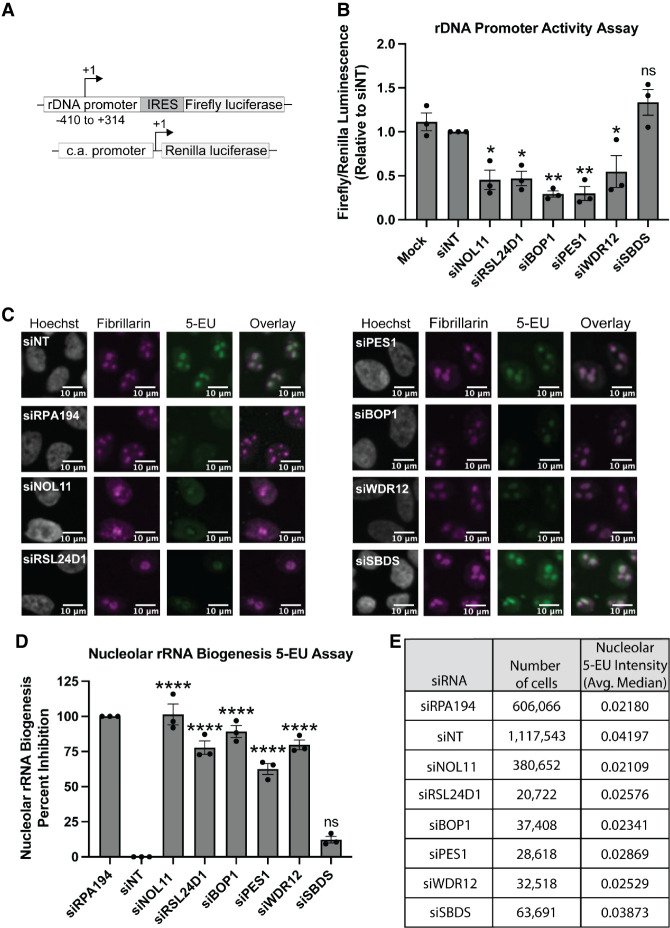
Since RPA194 is essential for RNAPI transcription of the pre-rRNA, we predicted that RSL24D1 and PeBoW would also be critical for RNAPI transcription. We assessed pre-rRNA transcription by RNAPI using a dual-luciferase reporter assay. In this system, the pHrD-IRES-Luc plasmid, which contains the firefly luciferase gene under the control of the human rDNA promoter, is cotransfected with a control plasmid containing the Renilla luciferase gene under the control of a constitutively active promoter (Fig. 4A; Ghoshal et al. 2004; Farley-Barnes et al. 2018). We assayed transcription in RSL24D1-, PES1-, BOP1-, or WDR12- siRNA depleted MCF10A cells where mock (no siRNA), siNT and siSBDS were used as negative controls, while siNOL11 was used as a positive control. Remarkably, depletion of RSL24D1 or individual PeBoW members revealed reduced rDNA promoter activity and thus decreased pre-rRNA transcription (Fig. 4B). This result was further corroborated by in cellula pulse labeling with the uridine analog 5-ethynyl uridine (5-EU) and the analysis of its incorporation into nascent nucleolar rRNA through immunofluorescent staining as in (Bryant et al. 2022). Strikingly, cells depleted of RSL24D1 or PeBoW components exhibited a strong reduction in nucleolar rRNA biogenesis relative to siNT and siSBDS negative control cells (Fig. 4C–E). This is consistent with previous results following depletion of Ribi factors required for pre-rRNA processing and transcription (Bryant et al. 2022) and almost to the same extent as siRPA194 and siNOL11 positive controls. Taken together, these results indicate that RSL24D1 and PeBoW modulate rDNA transcription, linking LSU processing factors to RNAPI transcription regulation.
FIGURE 4.
RSL24D1 and PeBoW complex members are required for optimal rDNA transcription. (A) Schematic of Firefly (pHrD-IRES-Luc) and Renilla luciferase plasmids for the rDNA promoter activity reporter system (Ghoshal et al. 2004); c.a. promoter: constitutively active promoter. (B) Depletion of RSL24D1 or PeBoW complex members leads to reduced rDNA promoter activity. MCF10A cells treated with the indicated siRNAs were transfected with reporter plasmids expressing Firefly and Renilla luciferase. Luminescence was quantified as a ratio in which Firefly gene expression, controlled by the human rDNA promoter, was normalized to Renilla gene expression, controlled by a constitutive promoter, and reported relative to nontargeting siRNA (siNT). Mock, nontargeting (siNT), and siSBDS were used as negative controls and siNOL11 was used as a positive control. Quantitation of results were reported relative to siNT. Graph indicates mean ± SEM, n = 3 biological replicates. Data were analyzed by one-way ANOVA with Dunnett's multiple comparisons test where (**) P ≤ 0.01 and (*) P ≤ 0.05; ns = not significant. (C) Nascent nucleolar RNA levels are reduced after siRNA depletion of RSL24D1 and individual PeBoW complex members by 5-EU visualization of nucleolar rRNA biogenesis. Seventy-two hours after siRNA transfection, MCF10A cells were supplemented with 1 mM 5-EU for 1 h before fixation. Representative images shown of fixed cells stained for DNA (Hoechst) and the nucleolar protein fibrillarin (FBL), and click chemistry was performed to conjugate AF488 azide to labeled nascent RNA (5-EU). (D) Quantitation of the results in C. Nucleolar rRNA biogenesis was quantified in MCF10A cells treated with the indicated siRNAs, where strongly reduced 5-EU signal corresponds to RNAPI inhibition (Bryant et al. 2022). The negative control nontargeting siRNA (siNT) (n = 16 wells per replicate) was set at 0% inhibition and the positive control siRPA194 (n = 16 wells per replicate) was set at 100% inhibition. siNOL11 and siSBDS were additional positive and negative controls, respectively (n = 1 well per replicate). Quantitation of results were reported relative to siNT. Graph indicates mean ± SD, n = 3 biological replicates. Data were analyzed by one-way ANOVA with Dunnett's multiple comparisons test where (****) P ≤ 0.0001; ns = not significant. (E) Summary data table of the quantitation in D. Total number of cells analyzed (sum of three independent repetitions) for each siRNA treatment. Average of the median nucleolar 5-EU intensity raw value from three independent repetitions for each siRNA treatment.
RSL24D1 is required for sustained protein synthesis
Since we discovered that RSL24D1 is required for pre-rRNA transcription and processing in human cells, we tested the extent to which RSL24D1 depletion impacts downstream processes, specifically global mRNA translation. We utilized a puromycin incorporation assay that labels nascent polypeptides as a proxy to monitor protein synthesis (Schmidt et al. 2009; Farley-Barnes et al. 2018). PeBoW, specifically BOP1, and SBDS depletion have been previously shown to disrupt protein synthesis using this assay (Sondalle et al. 2019; Wu et al. 2019). Treatment with 1 µM puromycin in RSL24D1-depleted MCF10A cells for 1 h followed by western blotting for incorporated puromycin shows significantly decreased protein synthesis relative to a siNT control (Fig. 5A,B). Mock (1 µM puromycin with no siRNA) and Mock at a half-dose of puromycin (0.5 µM) were used as negative controls whereas siNOL11 was a positive control. Our results connect RSL24D1's role in pre-rRNA transcription and large subunit maturation to a downstream significant reduction in ribosome function upon its depletion.
FIGURE 5.
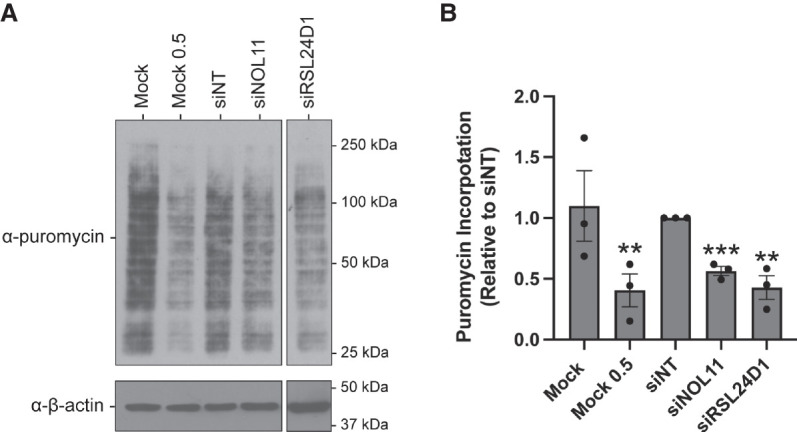
RSL24D1 depletion reduces global protein synthesis. (A) MCF10A cells were treated with 1 µM puromycin for 1 h following 72 h depletion with the indicated siRNAs. Representative western blot images using α-puromycin antibody, and α-β-actin antibody was used as a loading control. Mock (1 µM) and a nontargeting siRNA (siNT) were used as negative controls and siNOL11 was used as a positive control. Mock 0.5 µM indicates no siRNA and half the concentration of puromycin. β-actin was used as a loading control. (B) Quantitation of results in A were reported relative to siNT and normalized to the β-actin loading control. Graph indicates mean ± SEM, n = 3 biological replicates. Data were analyzed by Student's t-test followed by multiple testing P-value correction (two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli) where (***) P ≤ 0.001 and (**) P ≤ 0.01; ns = not significant.
RSL24D1 depletion induces the nucleolar stress response
Nucleolar stress denotes a key cellular response to drugs and environmental insults including impaired RiBi due to RNAPI transcription repression (Rubbi and Milner 2003; Lindstrom et al. 2018). When disrupted, the nucleolus can signal stress by activating the p53 pathway to initiate cell cycle arrest and apoptosis. Previously, PeBoW-complex members have been shown to induce the nucleolar stress response pathway when depleted (Holzel et al. 2005; Grimm et al. 2006). Likewise, we tested whether nucleolar perturbations described in Figure 1 upon RSL24D1 depletion were linked to concomitant p53 stabilization by western blotting for p53 levels in MCF10A cells depleted of RSL24D1. Indeed, knockdown of RSL24D1 induced p53 accumulation relative to the mock (no siRNA) and siNT negative controls (Fig. 6A). p53 stabilization following RSL24D1 knockdown was also observed in a recent genome-wide high-throughput screen conducted in A549 cells (Hannan et al. 2021). We further blotted for p53 transcriptional target gene p21 levels to orthogonally validate p53 activation and found these levels to be significantly increased relative to the mock (no siRNA) and siNT negative controls (Fig. 6B). Additionally, increased p21 transcript levels were observed by qRT-PCR in MCF10A cells (Fig. 6C). These findings show that RSL24D1 depletion increases p53 and p21 levels, likely as a result of nucleolar stress response induction upon impaired RiBi, as has been observed previously upon inhibition of pre-60S biogenesis (Sun et al. 2010; Fumagalli et al. 2012; Daftuar et al. 2013) or RNAPI activity (Bywater et al. 2012; Peltonen et al. 2014; Fu et al. 2017).
FIGURE 6.
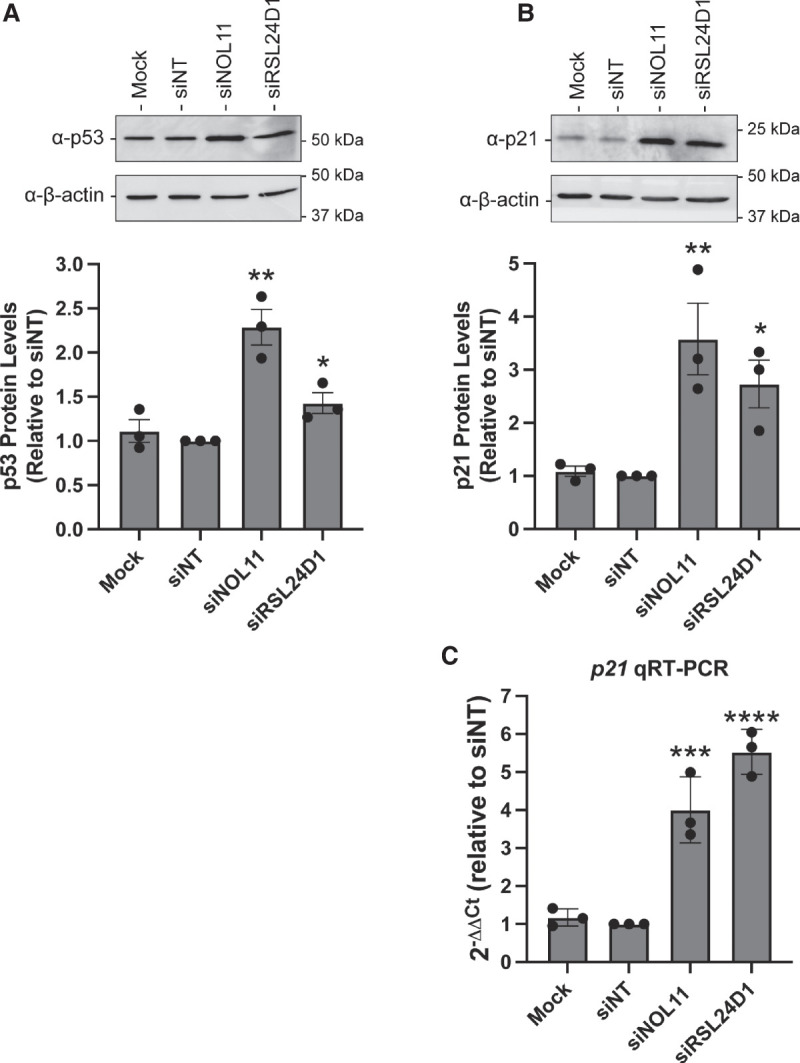
RSL24D1 depletion induces the nucleolar stress response. (A) RSL24D1 depletion increases p53 protein levels. MCF10A cells were treated with the indicated siRNAs for 72 h and total protein from whole cell extracts was harvested. Mock and nontargeting siRNA (siNT) were used as negative controls and siNOL11 was used as a positive control. (Top) Representative western blot images using α-p53 antibody, and α-β-actin antibody was used as a loading control. (Bottom) Quantitation of p53 levels is reported relative to siNT and normalized to the β-actin loading control. Graph indicates mean ± SD, n = 3 biological replicates. Data were analyzed by Student's t-test followed by multiple testing P-value correction (two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli) where (*) P ≤ 0.05, (**) P ≤ 0.01, (***) P ≤ 0.001, (****) P ≤ 0.0001. (B) RSL24D1 depletion increases p21 protein levels in MCF10A cells. Panel as above, except western blotting was performed using the α-p21 antibody. (C) RSL24D1 depletion increases p21 mRNA levels in MCF10A cells. qRT-PCR was performed and quantified using the 2−ΔΔCt method, relative to a siNT control and 7SL internal control primer. Nontargeting siRNA (siNT) and siNOL11 were used as negative and positive controls, respectively. Graph indicates mean ± SD, n = 3 biological replicates. Data were analyzed by Student's t-test followed by multiple testing P-value correction (two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli) where (***) P ≤ 0.001, (****) P ≤ 0.0001.
DISCUSSION
Here, we report the functional analysis of the putative RiBi assembly factor RSL24D1. This protein was previously identified to be important for 60S biogenesis (Durand et al. 2021; Dorner et al. 2022), and consistent with these reports, we demonstrate that RNAi mediated depletion of RSL24D1 specifically inhibits the late pre-rRNA processing steps required for 60S subunit formation. Moreover, repression of this pathway by RSL24D1 depletion leads to a global reduction in mRNA translation. We show that disruption of these processes after knockdown of RSL24D1 is associated with a reduction in nucleolar number (Farley-Barnes et al. 2018) and concomitant activation of the nucleolar stress response pathway. Remarkably, we uncover an unexpected role for RSL24D1 and the PeBoW complex for the efficient production of pre-rRNA by RNAPI, the first step of ribosome biogenesis. We demonstrate that RSL24D1 and PeBoW are positive regulators of rDNA promoter activity and RPA194 stability. We provide a likely mechanism through our confirmation of RSL24D1 and WDR12 association with RPA194, the largest subunit of the RNAPI enzyme complex.
In the present study, we provide strong evidence that LSU-associating assembly factors can also serve as dual-function factors in pre-rRNA transcription and processing. Several assembly factors are known to associate with the nascent pre-rRNA transcript to facilitate RNAPI recruitment and initiation. A subgroup of these factors, the t-UTPs, assemble synchronously to promote rDNA transcription and stimulate the formation of the SSU processome for the processing of the pre-40S subunit (Gallagher et al. 2004; Krogan et al. 2004; Prieto and McStay 2007). The results presented here are supported by previous studies that have identified nucleolar proteins that regulate both pre-rRNA transcription and pre-LSU processing, including splicing factor HTATSF1 and DNA repair protein FANCI (Corsini et al. 2018; Sondalle et al. 2019). We extend this framework to include the LSU factors RSL24D1 and PeBoW complex members (PES1–BOP1–WDR12) whose depletion leads to decreased RPA194 levels, down-regulated rDNA promoter activity, strongly decreased nucleolar rRNA biogenesis consistent with roles in pre-rRNA processing and transcription, and impaired pre-LSU processing.
Our results point toward both direct and potentially indirect mechanisms of RNAPI regulation by RSL24D1 and PeBoW. Consistent with the interactome results in Sorino et al. (2020), we observed an association of WDR12 and RSL24D1 with RPA194 by coimmunoprecipitation. Although a reduction in rDNA transcription and RPA194 levels extended to depletion of other PeBoW components, we did not find an association between PES1 and RPA194. This lack of association agrees with previously published PES1 and BOP1 interactomes, where RPA194 was not detected in either (Kellner et al. 2015). Interestingly, PES1 has been identified to be an interacting partner of upstream binding transcription factor (UBTF), which is required for the recruitment of the preinitiation complex along the rDNA promoter (Huttlin et al. 2017). Furthermore, all three PeBoW members interact with SIRT7, a positive regulator of RNAPI (Ford et al. 2006; Tsai et al. 2012).
Another potential explanation for PeBoW members all causing a reduction in rDNA transcription when individually depleted may be due to the interdependence of their steady state protein levels. Previous work has shown that depletion of individual PeBoW members results in a reduction in protein levels of both of the other complex members in human cells (Rohrmoser et al. 2007; Mi et al. 2021). Further studies investigating the crosstalk between these protein interactions and their influence on RNAPI transcription activation will be able to untangle the precise mechanism through which RSL24D1, PeBoW complex members, and maybe even other LSU maturation factors regulate this process.
Surprisingly, we were unable to coimmunoprecipitate PES1 with WDR12. This result was unexpected, as the presence of both proteins is well-established within the PeBoW complex (Holzel et al. 2005; Grimm et al. 2006; Rohrmoser et al. 2007). One potential explanation is that a large pool of endogenous WDR12 outside of the PeBoW complex led to our inability to detect the PES1–WDR12 association by coimmunoprecipitation. Although we validated WDR12 and RSL24D1's association with RPA194, further investigation will be critical to provide insight into the exact physical associations between RSL24D1, PeBoW, and RNAPI in human cells.
While RSL24D1 and PeBoW gene mutations are not yet known to be implicated in the molecular pathogenesis of ribosomopathies, the diseases of making ribosomes, several were shown to be linked to mutations in genes encoding LSU biogenesis factors. For example, mutations in SBDS are associated with Schwachman–Diamond syndrome (Boocock et al. 2003) and defects in RBM28 cause alopecia, neurological defects, and endocrinopathy (ANE) syndrome (Nousbeck et al. 2008; McCann et al. 2016; Bryant et al. 2021). Intriguingly, RSL24D1 was recently reported to be required for murine embryonic stem cell proliferation (Durand et al. 2021) and BOP1 in Xenopus anterior development (Gartner et al. 2022). Previously, it has been shown that PES1 is required for zebrafish and mouse embryonic development (Allende et al. 1996; Lerch-Gaggl et al. 2002) and more specifically Xenopus neural crest cell migration (Gessert et al. 2007). The cranial cartilage defects observed in Xenopus upon loss of BOP1 (Gartner et al. 2022) and PES1 (Gessert et al. 2007) are hallmarks of ribosomopathies (Farley-Barnes et al. 2019). It will be of interest to further dissect the roles of RSL24D1 and PeBoW in ribosome biogenesis as it relates to embryonic development in vertebrates. The prevalence of disorders arising from mutations in genes encoding assembly factors of the large subunit highlight the importance of faithful subunit processing for the steady production of ribosomes to maintain cellular homeostasis.
MATERIALS AND METHODS
Publicly available expression data sets
Genotype-Tissue Expression (GTEx) unmatched normal and The Cancer Genome Atlas (TCGA) matched normal and tumor expression data sets were obtained through the Xena platform (https://xena.ucsc.edu/) (Goldman et al. 2020). RNA-seq by Expectation-Maximization (RSEM) LOG2 fold expression levels for RSL24D1 were subtracted from the mean of the overall normal and tumor tissues combined for graphical visualization.
Cell culture and media
MCF10A cells (ATCC CRL-10317) were subcultured in Dulbecco's modified Eagles’ medium/nutrient mixture F-12 (Gibco 1130-032) containing horse serum (Gibco 16050), 10 µg/mL insulin (Sigma I1882), 0.5 µg/mL hydrocortisone (Sigma H0135), 100 ng/mL cholera toxin (Sigma C8052), and 20 ng/mL epidermal growth factor (Peprotech AF-100-15). Cells were maintained at 37°C, in a humidified atmosphere with 5% CO2. For the high-throughput nucleolar number and 5-EU screens, 3000 cells/well were reverse transfected into 384-well plates on day 0. For the dual-luciferase reporter assay, 75,000 cells/well were seeded into 12-well plates on day 0. For RNA or protein isolation, 100,000 cells/well were seeded into six-well plates on day 0.
RNAi
All siRNAs were purchased from Horizon Discovery Biosciences (Supplemental Data S1). siGENOME SMARTpool siRNAs were used in the original screen (Farley-Barnes et al. 2018). For the 5-EU secondary screen validation of RSL24D1, PeBoW, and SBDS, ON-TARGETplus pools were used except for the NOL11 positive control which used the siGENOME SMARTpool siRNAs. For screen validation by deconvolution, the individual siONT set of four siRNAs that comprised the pool was used. The ON-TARGETplus pools were used in the remaining functional analysis. siRNA transfection was performed using Lipofectamine RNAiMAX Transfection Reagent (Invitrogen 13778150) as described in Ogawa et al. (2021). For the high-throughput screens, cells were reverse transfected into 384-well plates on day 0. siNT, siUTP4, siRPA194, and siNOL11 controls were added to 16 wells per plate, while our experimental samples were added to one well each for each independent screen repetition. For other assays, cells were transfected 24 h after plating. For siONT deconvolution, an individual siRNA targeting RSL24D1 was considered validated if it produced a mean one-nucleolus percent effect greater than or equal to +3 SD above the siNT mean, using the siNT SD.
5-EU incorporation assay for nucleolar rRNA biogenesis
Following 72 h of siRNA depletion, MCF10A cells were treated with 1 mM 5-ethynyl uridine (5-EU; Click Chemistry Tools 1261) for 1 h to label nascent RNA as in (Bryant et al. 2022). Briefly, cells were washed with PBS, fixed with 1% paraformaldehyde (Electron Microscopy Sciences 15710-S) in PBS for 20 min, and permeabilized with 0.5% (vol/vol) Triton X-100 in PBS for 5 min. Cells were blocked with 10% (vol/vol) FBS (MilliporeSigma F0926) in PBS for 1 h at room temperature. Nucleoli were stained with 72B9 primary anti-fibrillarin antibody (Reimer et al. 1987), 1:250 for 2 h at room temperature, followed by secondary AlexaFluor 647 goat anti-mouse immunoglobulin G (1:1000, Invitrogen A-21235) with Hoechst 33342 dye (1:3000) for nuclei detection for 1 h at room temperature. 5-EU incorporation was visualized by performing the following click reaction in PBS: CuSO4:5H2O (0.5 mg/mL resuspended in water, Acros Organics 197730010), ascorbic acid (20 mg/mL freshly resuspended in water, Alfa Aesar A15613), and AF488 azide (0.5 µM resuspended in DMSO, Click Chemistry Tools 1275) for 30 min at room temperature. Cells were then soaked with Hoechst 33342 (1:3000) for 30 min to dissociate excess azide dye. Cell images were acquired using IN Cell 2200 imaging system (GE Healthcare), and analysis was performed using a custom CellProfiler pipeline (Bryant et al. 2022).
Dual-luciferase reporter assay for pre-rRNA transcription
Following 48 h of siRNA depletion, MCF10A cells were cotransfected with 1000 ng of pHrD-IRES-Luc (Ghoshal et al. 2004) and 0.1 ng of a Renilla internal control plasmid using Lipofectamine 3000 (Thermo Fisher Scientific L3000015). Twenty-four h after plasmid transfection, cells were harvested and luminescence was measured using the Dual-luciferase Reporter Assay System (Promega E1910) following the manufacturer's instructions with a GloMax 20/20 luminometer (Promega). The ratio of pHrD-IRES-luciferase/Renilla activity was calculated to control for transfection efficiency and normalized to the nontargeting control.
Northern blots
After 72 h of siRNA-mediated depletion in MCF10A cells, total cellular RNA was extracted with TRIzol (Life Technologies 5596018) according to the manufacturer's protocol. For each sample, 4 µg of total RNA was resolved on a denaturing 1% agarose/1.25% formaldehyde gel using Tri/Tri buffer (Mansour and Pestov 2013) and transferred to a Hybond-XL membrane (GE Healthcare RPN 303S). Blots were hybridized to radiolabeled DNA oligonucleotide probes (P4: 5′-CGGGAACTCGGCCCGAGCCGGCTCTCTCTTTCCCTCTCCG-3′; 7SL: 5′-TGCTCCGTTTCCGACCTGGGCCGGTTCACCCCTCCTT-3′) and detected by phosphorimager as described previously (Pestov et al. 2008).
qRT-PCR analysis
After 72 h of siRNA-mediated depletion, RNA was extracted using TRIzol (Life Technologies 5596018) as per the manufacturer's instructions. cDNA preparation was performed using the iScript gDNA Clear cDNA Synthesis Kit (Bio-Rad 1725035), and qPCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad 1725121). All A260/230 values were above 1.7 prior to cDNA synthesis. Amplification of the 7SL RNA was used as an internal control, and analysis was completed using the comparative CT method (ΔΔCT). Bio-Rad PrimePCR Assay gene-specific primers were used to test RSL24D1 mRNA levels (Bio-Rad 10025636; RSL24D1, qHsaCID0021318) and the primers for BOP1, PES1, SBDS, WDR12, 7SL are available in Supplemental Data S2. Melt curves were performed for each sample to verify the amplification of a single product. Three biological replicates, each with three technical replicates, were measured.
Western blots
Following 72 h of siRNA depletion, cells were collected, and total protein was harvested and prepared as in (Farley-Barnes et al. 2018). A Bradford assay (Bio-Rad 5000006) was used to quantify amount of total protein. An amount of 25–50 µg of total protein was separated by SDS-PAGE and transferred to a PVDF membrane (Bio-Rad 1620177). Membranes were blocked for 1 h in 5% milk in 1× PBST (or 5% BSA in 1× PBST for western blots of immunoprecipitations) and incubated overnight with the specified primary antibodies at 4°C. Proteins were detected with the following antibodies: α-RSL24D1 (dilution 1:1000; Proteintech 25190-1-AP), α-RPA194 (dilution 1:1000; Santa Cruz sc-48385), α-WDR12 (1:1000; Bethyl Laboratories A302-650A), α-PES1 (1:1000; Bethyl Laboratories A300-902A), α-p53 (dilution 1:5000; Santa Cruz sc-126), α-p21 (dilution 1:400, Santa Cruz sc-6246), α-puromycin (dilution 1:10,000; Kerafast EQ0001), and α-β-actin (dilution 1:30,000; Sigma Aldrich A1978). The western blots were developed with enhanced chemiluminescence reagents (Thermo Scientific 34096). Images were acquired by digital imaging using the Bio-Rad ChemiDoc Imaging System. Images were quantified with ImageJ software.
Puromycin labeling assay
Global protein translation was assessed by treating cells with puromycin to label nascent polypeptide chains as in (Schmidt et al. 2009). Total cellular extracts were then analyzed by western blot using an anti-puromycin antibody (Kerafast EQ0001) at a 1:10,000 dilution. Quantitation was performed using ImageJ.
Coimmunoprecipitation
Protein A agarose beads (Cell Signaling Technologies 9863S) were washed and incubated in NET150 buffer (20 mM Tris HCl pH. 7.5, 150 mM NaCl, 0.05% NP-40) with 20 µg of the indicated antibodies overnight at 4°C with nutation. Harvested MCF10A cells were washed with PBS and incubated on ice for 10 min in NET150 buffer (with the addition of 1× protease inhibitors and 4 mM NEM). Total cell extracts were obtained by sonication and cleared by centrifugation at 16,000g for 10 min at 4°C after lysis. Supernatants were incubated with either antibody-bound or unconjugated Protein A beads for 2 h at 4°C with nutation. RNase A treated extracts were treated with 20 µg/mL RNase A (AMRESCO E866). After beads were washed five times with NET150, immunocomplexes were eluted in 2× Laemmli buffer and resolved on a 12% acrylamide gel. Western blotting was performed as described above.
Statistical analyses
All statistical analyses were performed in GraphPad Prism 8.2.1 (GraphPad Software) using the tests described in the figure legends.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank current and former members of the Baserga laboratory for helpful discussions and insights in the preparation of this manuscript. This work was supported by the following grants from the National Institutes of Health (NIH): 1R35GM131687 (to S.J.B.), 1F31DE030332 (to M.A.M.), and T32GM007223 (to C.J.B., M.A.M., and S.J.B.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079149.122.
Freely available online through the RNA Open Access option.
MEET THE FIRST AUTHORS
Amber F. Buhagiar.

Mason A. McCool.

Meet the First Author(s) is a new editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. Mason A. McCool and Amber F. Buhagiar are co-first authors of this paper, “Human pre-60S assembly factors link rRNA transcription to pre-rRNA processing.” Mason is a PhD candidate in the laboratory of Dr. Susan Baserga at Yale University (Molecular Biophysics and Biochemistry Department, MB&B). Amber is a first-year PhD student at the Albert Einstein College of Medicine (AECOM). Amber was a post-baccalaureate researcher in the Baserga Laboratory from 2018–2021. The main focus of the Baserga Laboratory is to discover and understand novel regulatory mechanisms of human ribosome biogenesis and how dysregulation of this process can lead to disease, namely cancer and ribosomopathies.
What are the major results described in your paper and how do they impact this branch of the field?
The major result of our paper is the discovery of an additional function of established large ribosomal subunit (LSU) assembly factors, RSL24D1 and the PeBoW complex (PES1, BOP1, WDR12), in the transcription of the ribosomal RNA by RNA polymerase I (RNAPI). While there are many known small ribosomal subunit (SSU) biogenesis factors that also regulate RNAPI transcription, it is exciting to observe regulation of this process from factors that lead to the formation of the LSU as well. This opens up the idea that even more factors can have dual roles in RNAPI transcription and LSU biogenesis, more tightly interconnecting these two steps of ribosome biogenesis than previously thought.
What led you to study RNA or this aspect of RNA science?
MAM: Starting graduate school, I was undecided on what area(s) of biochemistry that I wanted to pursue in my research. After completing a rotation in Susan Baserga's laboratory, it was the collaborative environment and friendly members there that sparked my initial interest in RNA biology. In addition to the people, I was drawn to the area of ribosome biogenesis specifically because of its relevance to human health and disease. I was happy to pursue a path of basic science research on an essential cellular process that could lead to downstream impacts in developing better treatments for human disease, specifically cancer.
AFB: After completing my undergraduate studies, I wanted to gain additional laboratory training in molecular biology before applying to graduate programs. I became interested in studying ribosome biogenesis because of the strong emphasis on basic science research that it would accompany, and because of its clinical relevance. After moving to Yale, it didn't take long for me to fall in love with the science and I developed a big heart for the tight-knit RNA community there.
What are some of the landmark moments that provoked your interest in science or your development as a scientist?
MAM: It has probably been landmark people more than moments that inspired my interest in science. First, my high school chemistry teacher, Mr. Kirian, inspired me with his enthusiasm in class and recommended me for a volunteer research assistant position in a university laboratory while I was at Findlay High School (OH) that got me started on my journey in research. During my undergraduate studies at Kenyon College, I was fortunate to have a close relationship with my professors in the classroom and research laboratory. The entire chemistry faculty at Kenyon, specifically Dr. John Hofferberth and Dr. Kerry Rouhier, were extremely supportive. They continually encouraged me to reach my full potential, which eventually led me to apply to graduate school to continue my path in science.
AFB: My research journey was initiated the summer before my senior year at Brown University, when I pursued an internship with Dr. James Shorter at UPenn studying components of the protein homeostasis network. This was the first time I stepped foot in a research laboratory during college and the Shorter laboratory welcomed me with open arms! As a biomedical engineering major, I was trained to view the physical world through the lens of design, so naturally I approached the cell in terms of subsystems and subcomponents and how the integration of those networks “fit together.” This frame of reference allowed me to recognize and immediately appreciate the sophistication underpinning cellular organization. I figured out right away that I loved laboratory work and wanted to make a career out of it.
If you were able to give one piece of advice to your younger self, what would that be?
MAM: In life and in research, you can't control or always anticipate the results. However, you can control your process, how you react, and hopefully make a nice discussion section out of it.
What are your subsequent near- or long-term career plans?
AFB: During my PhD at AECOM, I intend to continue tackling exciting questions in molecular biology. In the long term, I hope to apply my training in either industry or academia and commit my career to mentoring the next generation of aspiring scientists and physicians, especially women from underrepresented backgrounds.
How did you decide to work together as co-first authors?
Most of the work in the Baserga laboratory consists of individual laboratory members following up with mechanistic assays on novel ribosome biogenesis factors identified through previously published high-throughput screens. However, laboratory members are continually lending a helping hand in others’ projects as well. The collaboration was initiated after Amber's original exciting results on the hit she was studying, RSL24D1, and the PeBoW complex pointed to a surprising role in pre-rRNA transcription by RNA polymerase I when Mason was able to confirm those results using a 5-EU assay developed in the laboratory. Amber's work at the bench continued until her transition to AECOM for her graduate studies. After, Mason took over the experimental work and the collaboration together continued through the writing and editing process, to ensure we could share these discoveries even though we were no longer in the same location.
REFERENCES
- Allende ML, Amsterdam A, Becker T, Kawakami K, Gaiano N, Hopkins N. 1996. Insertional mutagenesis in zebrafish identifies two novel genes, pescadillo and dead eye, essential for embryonic development. Genes Dev 10: 3141–3155. 10.1101/gad.10.24.3141 [DOI] [PubMed] [Google Scholar]
- Aubert M, O'Donohue MF, Lebaron S, Gleizes PE. 2018. Pre-ribosomal RNA processing in human cells: from mechanisms to congenital diseases. Biomolecules 8: 123. 10.3390/biom8040123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler J, Hurt E. 2019. Eukaryotic ribosome assembly. Annu Rev Biochem 88: 281–306. 10.1146/annurev-biochem-013118-110817 [DOI] [PubMed] [Google Scholar]
- Bohnsack KE, Bohnsack MT. 2019. Uncovering the assembly pathway of human ribosomes and its emerging links to disease. EMBO J 38: e100278. 10.15252/embj.2018100278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boocock GR, Morrison JA, Popovic M, Richards N, Ellis L, Durie PR, Rommens JM. 2003. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet 33: 97–101. 10.1038/ng1062 [DOI] [PubMed] [Google Scholar]
- Bryant CJ, Lorea CF, de Almeida HL Jr, Weinert L, Vedolin L, Pinto EVF, Baserga SJ. 2021. Biallelic splicing variants in the nucleolar 60S assembly factor RBM28 cause the ribosomopathy ANE syndrome. Proc Natl Acad Sci 118: e2017777118. 10.1073/pnas.2017777118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant CJ, McCool MA, Abriola L, Surovtseva YV, Baserga SJ. 2022. A high-throughput assay for directly monitoring nucleolar rRNA biogenesis. Open Biol 12: 210305. 10.1098/rsob.210305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, Wall M, Cluse L, Drygin D, Anderes K, et al. 2012. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell 22: 51–65. 10.1016/j.ccr.2012.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chedin S, Laferte A, Hoang T, Lafontaine DL, Riva M, Carles C. 2007. Is ribosome synthesis controlled by Pol I transcription? Cell Cycle 6: 11–15. 10.4161/cc.6.1.3649 [DOI] [PubMed] [Google Scholar]
- Corsini NS, Peer AM, Moeseneder P, Roiuk M, Burkard TR, Theussl HC, Moll I, Knoblich JA. 2018. Coordinated control of mRNA and rRNA processing controls embryonic stem cell pluripotency and differentiation. Cell Stem Cell 22: 543–558.e512. 10.1016/j.stem.2018.03.002 [DOI] [PubMed] [Google Scholar]
- Daftuar L, Zhu Y, Jacq X, Prives C. 2013. Ribosomal proteins RPL37, RPS15 and RPS20 regulate the Mdm2-p53-MdmX network. PLoS One 8: e68667. 10.1371/journal.pone.0068667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derenzini M, Montanaro L, Trere D. 2017. Ribosome biogenesis and cancer. Acta Histochem 119: 190–197. 10.1016/j.acthis.2017.01.009 [DOI] [PubMed] [Google Scholar]
- Dorner K, Badertscher L, Horvath B, Hollandi R, Molnar C, Fuhrer T, Meier R, Sarazova M, van den Heuvel J, Zamboni N, et al. 2022. Genome-wide RNAi screen identifies novel players in human 60S subunit biogenesis including key enzymes of polyamine metabolism. Nucleic Acids Res 50: 2872–2888. 10.1093/nar/gkac072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew K, Wallingford JB, Marcotte EM. 2021. hu.MAP 2.0: integration of over 15,000 proteomic experiments builds a global compendium of human multiprotein assemblies. Mol Syst Biol 17: e10016. 10.15252/msb.202010016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S, Bruelle M, Bourdelais F, Bennychen B, Blin-Gonthier J, Isaac C, Huyghe A, Seyve A, Vanbelle C, Meyronet D, et al. 2021. RSL24D1 sustains steady-state ribosome biogenesis and pluripotency translational programs in embryonic stem cells. bioRxiv 10.1101/2021.05.27.443845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan P, Wang B, Meng Z, Zhao J, Jin X. 2018. PES1 is transcriptionally regulated by BRD4 and promotes cell proliferation and glycolysis in hepatocellular carcinoma. Int J Biochem Cell Biol 104: 1–8. 10.1016/j.biocel.2018.08.014 [DOI] [PubMed] [Google Scholar]
- Farley-Barnes KI, McCann KL, Ogawa LM, Merkel J, Surovtseva YV, Baserga SJ. 2018. Diverse regulators of human ribosome biogenesis discovered by changes in nucleolar number. Cell Rep 22: 1923–1934. 10.1016/j.celrep.2018.01.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley-Barnes KI, Ogawa LM, Baserga SJ. 2019. Ribosomopathies: old concepts, new controversies. Trends Genet 35: 754–767. 10.1016/j.tig.2019.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. 2006. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev 20: 1075–1080. 10.1101/gad.1399706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EF, Prieto JL, McCann KL, McStay B, Baserga SJ. 2012. NOL11, implicated in the pathogenesis of North American Indian childhood cirrhosis, is required for pre-rRNA transcription and processing. PLoS Genet 8: e1002892. 10.1371/journal.pgen.1002892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Xu L, Qi L, Tian H, Yi D, Yu Y, Liu S, Li S, Xu Y, Wang C. 2017. BMH-21 inhibits viability and induces apoptosis by p53-dependent nucleolar stress responses in SKOV3 ovarian cancer cells. Oncol Rep 38: 859–865. 10.3892/or.2017.5750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S, Ivanenkov VV, Teng T, Thomas G. 2012. Suprainduction of p53 by disruption of 40S and 60S ribosome biogenesis leads to the activation of a novel G2/M checkpoint. Genes Dev 26: 1028–1040. 10.1101/gad.189951.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ. 2004. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev 18: 2506–2517. 10.1101/gad.1226604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner C, Messmer A, Dietmann P, Kuhl M, Kuhl SJ. 2022. Functions of block of proliferation 1 during anterior development in Xenopus laevis. PLoS One 17: e0273507. 10.1371/journal.pone.0273507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Yuan L, Cheng B, Dai K. 2021. Identification of seven tumor-educated platelets RNAs for cancer diagnosis. J Clin Lab Anal 35: e23791. 10.1002/jcla.23791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessert S, Maurus D, Rossner A, Kuhl M. 2007. Pescadillo is required for Xenopus laevis eye development and neural crest migration. Dev Biol 310: 99–112. 10.1016/j.ydbio.2007.07.037 [DOI] [PubMed] [Google Scholar]
- Ghoshal K, Majumder S, Datta J, Motiwala T, Bai S, Sharma SM, Frankel W, Jacob ST. 2004. Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. J Biol Chem 279: 6783–6793. 10.1074/jbc.M309393200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, et al. 2020. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 38: 675–678. 10.1038/s41587-020-0546-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm T, Holzel M, Rohrmoser M, Harasim T, Malamoussi A, Gruber-Eber A, Kremmer E, Eick D. 2006. Dominant-negative Pes1 mutants inhibit ribosomal RNA processing and cell proliferation via incorporation into the PeBoW-complex. Nucleic Acids Res 34: 3030–3043. 10.1093/nar/gkl378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan KM, Soo P, Wong MS, Lee JK, Hein N, Poh P, Wysoke KD, Williams TD, Montellese C, Smith LK, et al. 2022. Nuclear stabilization of p53 requires a functional nucleolar surveillance pathway. Cell Rep 41: 111571. 10.1016/j.celrep.2022.111571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzel M, Rohrmoser M, Schlee M, Grimm T, Harasim T, Malamoussi A, Gruber-Eber A, Kremmer E, Hiddemann W, Bornkamm GW, et al. 2005. Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J Cell Biol 170: 367–378. 10.1083/jcb.200501141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP, Parzen H, et al. 2017. Architecture of the human interactome defines protein communities and disease networks. Nature 545: 505–509. 10.1038/nature22366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et al. 2006. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA 12: 1197–1205. 10.1261/rna.30706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappel L, Loibl M, Zisser G, Klein I, Fruhmann G, Gruber C, Unterweger S, Rechberger G, Pertschy B, Bergler H. 2012. Rlp24 activates the AAA-ATPase Drg1 to initiate cytoplasmic pre-60S maturation. J Cell Biol 199: 771–782. 10.1083/jcb.201205021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater L, Thoms M, Barrio-Garcia C, Cheng J, Ismail S, Ahmed YL, Bange G, Kressler D, Berninghausen O, Sinning I, et al. 2017. Visualizing the assembly pathway of nucleolar pre-60S ribosomes. Cell 171: 1599–1610.e1514. 10.1016/j.cell.2017.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner M, Rohrmoser M, Forne I, Voss K, Burger K, Muhl B, Gruber-Eber A, Kremmer E, Imhof A, Eick D. 2015. DEAD-box helicase DDX27 regulates 3' end formation of ribosomal 47S RNA and stably associates with the PeBoW-complex. Exp Cell Res 334: 146–159. 10.1016/j.yexcr.2015.03.017 [DOI] [PubMed] [Google Scholar]
- Killian A, Sarafan-Vasseur N, Sesboue R, Le Pessot F, Blanchard F, Lamy A, Laurent M, Flaman JM, Frebourg T. 2006. Contribution of the BOP1 gene, located on 8q24, to colorectal tumorigenesis. Genes Chromosomes Cancer 45: 874–881. 10.1002/gcc.20351 [DOI] [PubMed] [Google Scholar]
- Kopp K, Gasiorowski JZ, Chen D, Gilmore R, Norton JT, Wang C, Leary DJ, Chan EK, Dean DA, Huang S. 2007. Pol I transcription and pre-rRNA processing are coordinated in a transcription-dependent manner in mammalian cells. Mol Biol Cell 18: 394–403. 10.1091/mbc.e06-03-0249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol Cell 13: 225–239. 10.1016/s1097-2765(04)00003-6 [DOI] [PubMed] [Google Scholar]
- Laferte A, Favry E, Sentenac A, Riva M, Carles C, Chedin S. 2006. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev 20: 2030–2040. 10.1101/gad.386106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapik YR, Fernandes CJ, Lau LF, Pestov DG. 2004. Physical and functional interaction between Pes1 and Bop1 in mammalian ribosome biogenesis. Mol Cell 15: 17–29. 10.1016/j.molcel.2004.05.020 [DOI] [PubMed] [Google Scholar]
- Lerch-Gaggl A, Haque J, Li J, Ning G, Traktman P, Duncan SA. 2002. Pescadillo is essential for nucleolar assembly, ribosome biogenesis, and mammalian cell proliferation. J Biol Chem 277: 45347–45355. 10.1074/jbc.M208338200 [DOI] [PubMed] [Google Scholar]
- Li G, Wu XJ, Kong XQ, Wang L, Jin X. 2015. Cytochrome c oxidase subunit VIIb as a potential target in familial hypercholesterolemia by bioinformatical analysis. Eur Rev Med Pharmacol Sci 19: 4139–4145. [PubMed] [Google Scholar]
- Li JL, Chen C, Chen W, Zhao LF, Xu XK, Li Y, Yuan HY, Lin JR, Pan JP, Jin BL, et al. 2020. Integrative genomic analyses identify WDR12 as a novel oncogene involved in glioblastoma. J Cell Physiol 235: 7344–7355. 10.1002/jcp.29635 [DOI] [PubMed] [Google Scholar]
- Li YZ, Zhang C, Pei JP, Zhang WC, Zhang CD, Dai DQ. 2022. The functional role of Pescadillo ribosomal biogenesis factor 1 in cancer. J Cancer 13: 268–277. 10.7150/jca.58982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom MS, Jurada D, Bursac S, Orsolic I, Bartek J, Volarevic S. 2018. Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene 37: 2351–2366. 10.1038/s41388-017-0121-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour FH, Pestov DG. 2013. Separation of long RNA by agarose-formaldehyde gel electrophoresis. Anal Biochem 441: 18–20. 10.1016/j.ab.2013.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann KL, Teramoto T, Zhang J, Tanaka Hall TM, Baserga SJ. 2016. The molecular basis for ANE syndrome revealed by the large ribosomal subunit processome interactome. Elife 5: e16381. 10.7554/eLife.16381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi L, Qi Q, Ran H, Chen L, Li D, Xiao D, Wu J, Cai Y, Zhang S, Li Y, et al. 2021. Suppression of ribosome biogenesis by targeting WD repeat domain 12 (WDR12) inhibits glioma stem-like cell growth. Front Oncol 11: 751792. 10.3389/fonc.2021.751792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousbeck J, Spiegel R, Ishida-Yamamoto A, Indelman M, Shani-Adir A, Adir N, Lipkin E, Bercovici S, Geiger D, van Steensel MA, et al. 2008. Alopecia, neurological defects, and endocrinopathy syndrome caused by decreased expression of RBM28, a nucleolar protein associated with ribosome biogenesis. Am J Hum Genet 82: 1114–1121. 10.1016/j.ajhg.2008.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa LM, Buhagiar AF, Abriola L, Leland BA, Surovtseva YV, Baserga SJ. 2021. Increased numbers of nucleoli in a genome-wide RNAi screen reveal proteins that link the cell cycle to RNA polymerase I transcription. Mol Biol Cell 32: 956–973. 10.1091/mbc.E20-10-0670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oughtred R, Stark C, Breitkreutz BJ, Rust J, Boucher L, Chang C, Kolas N, O'Donnell L, Leung G, McAdam R, et al. 2019. The BioGRID interaction database: 2019 update. Nucleic Acids Res 47: D529–D541. 10.1093/nar/gky1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panov KI, Friedrich JK, Russell J, Zomerdijk JC. 2006. UBF activates RNA polymerase I transcription by stimulating promoter escape. EMBO J 25: 3310–3322. 10.1038/sj.emboj.7601221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltonen K, Colis L, Liu H, Jaamaa S, Zhang Z, Af Hallstrom T, Moore HM, Sirajuddin P, Laiho M. 2014. Small molecule BMH-compounds that inhibit RNA polymerase I and cause nucleolar stress. Mol Cancer Ther 13: 2537–2546. 10.1158/1535-7163.MCT-14-0256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestov DG, Lapik YR, Lau LF. 2008. Assays for ribosomal RNA processing and ribosome assembly. Curr Protoc Cell Biol Chapter 22: Unit 22 11. 10.1002/0471143030.cb2211s39 [DOI] [PubMed] [Google Scholar]
- Phipps KR, Charette J, Baserga SJ. 2011. The small subunit processome in ribosome biogenesis-progress and prospects. Wiley Interdiscip Rev RNA 2: 1–21. 10.1002/wrna.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poll G, Pilsl M, Griesenbeck J, Tschochner H, Milkereit P. 2021. Analysis of subunit folding contribution of three yeast large ribosomal subunit proteins required for stabilisation and processing of intermediate nuclear rRNA precursors. PLoS One 16: e0252497. 10.1371/journal.pone.0252497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto JL, McStay B. 2007. Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev 21: 2041–2054. 10.1101/gad.436707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer G, Pollard KM, Penning CA, Ochs RL, Lischwe MA, Busch H, Tan EM. 1987. Monoclonal autoantibody from a (New Zealand black x New Zealand white)F1 mouse and some human scleroderma sera target an MTr 34,000 nucleolar protein of the U3 RNP particle. Arthritis Rheum 30: 793–800. 10.1002/art.1780300709 [DOI] [PubMed] [Google Scholar]
- Rohrmoser M, Holzel M, Grimm T, Malamoussi A, Harasim T, Orban M, Pfisterer I, Gruber-Eber A, Kremmer E, Eick D. 2007. Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol Cell Biol 27: 3682–3694. 10.1128/MCB.00172-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi CP, Milner J. 2003. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J 22: 6068–6077. 10.1093/emboj/cdg579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailer C, Jansen J, Sekulski K, Cruz VE, Erzberger JP, Stengel F. 2022. A comprehensive landscape of 60S ribosome biogenesis factors. Cell Rep 38: 110353. 10.1016/j.celrep.2022.110353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu C, Namane A, Gleizes PE, Lebreton A, Rousselle JC, Noaillac-Depeyre J, Gas N, Jacquier A, Fromont-Racine M. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol Cell Biol 23: 4449–4460. 10.1128/MCB.23.13.4449-4460.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, Pierre P. 2009. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6: 275–277. 10.1038/nmeth.1314 [DOI] [PubMed] [Google Scholar]
- Sondalle SB, Longerich S, Ogawa LM, Sung P, Baserga SJ. 2019. Fanconi anemia protein FANCI functions in ribosome biogenesis. Proc Natl Acad Sci 116: 2561–2570. 10.1073/pnas.1811557116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorino C, Catena V, Bruno T, De Nicola F, Scalera S, Bossi G, Fabretti F, Mano M, De Smaele E, Fanciulli M, et al. 2020. Che-1/AATF binds to RNA polymerase I machinery and sustains ribosomal RNA gene transcription. Nucleic Acids Res 48: 5891–5906. 10.1093/nar/gkaa344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strezoska Z, Pestov DG, Lau LF. 2002. Functional inactivation of the mouse nucleolar protein Bop1 inhibits multiple steps in pre-rRNA processing and blocks cell cycle progression. J Biol Chem 277: 29617–29625. 10.1074/jbc.M204381200 [DOI] [PubMed] [Google Scholar]
- Sun XX, Wang YG, Xirodimas DP, Dai MS. 2010. Perturbation of 60 S ribosomal biogenesis results in ribosomal protein L5- and L11-dependent p53 activation. J Biol Chem 285: 25812–25821. 10.1074/jbc.M109.098442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DL. 2013. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol Cell 51: 539–551. 10.1016/j.molcel.2013.08.011 [DOI] [PubMed] [Google Scholar]
- Tsai YC, Greco TM, Boonmee A, Miteva Y, Cristea IM. 2012. Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol Cell Proteomics 11: 60–76. 10.1074/mcp.A111.015156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellky JE, Ricke EA, Huang W, Ricke WA. 2021. Expression, localization, and function of the nucleolar protein BOP1 in prostate cancer progression. Am J Pathol 191: 168–179. 10.1016/j.ajpath.2020.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Anikin L, Pestov DG. 2014. Two orthogonal cleavages separate subunit RNAs in mouse ribosome biogenesis. Nucleic Acids Res 42: 11180–11191. 10.1093/nar/gku787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren AJ. 2018. Molecular basis of the human ribosomopathy Shwachman-Diamond syndrome. Adv Biol Regul 67: 109–127. 10.1016/j.jbior.2017.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford JL Jr, Baserga SJ. 2013. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195: 643–681. 10.1534/genetics.113.153197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Hong J, Wang Z, Hu J, Chen R, Hu Z, Li B, Hu X, Zhang Z, Ruan Y. 2019. Abnormal ribosome biogenesis partly induced p53-dependent aortic medial smooth muscle cell apoptosis and oxidative stress. Oxid Med Cell Longev 2019: 7064319. 10.1155/2019/7064319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Zhou L, Zhan R, Zhang Q, Li M. 2018. Identification of WDR12 as a novel oncogene involved in hepatocellular carcinoma propagation. Cancer Manag Res 10: 3985–3993. 10.2147/CMAR.S176268 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.