Abstract
BACKGROUND:
Cardiac regeneration after injury is limited by the low proliferative capacity of adult mammalian cardiomyocytes (CMs). However, certain animals readily regenerate lost myocardium via a process involving dedifferentiation, which unlocks their proliferative capacities.
METHODS:
We bred mice with inducible, CM-specific expression of the Yamanaka factors, enabling adult CM reprogramming and dedifferentiation in vivo.
RESULTS:
Two days following induction, adult CMs presented a dedifferentiated phenotype and increased proliferation in vivo. Microarray analysis revealed that upregulation of ketogenesis was central to this process. Adenovirus-driven HMGCS2 overexpression induced ketogenesis in adult CMs and recapitulated CM dedifferentiation and proliferation observed during partial reprogramming. This same phenomenon was found to occur after myocardial infarction, specifically in the border zone tissue, and HMGCS2 knockout mice showed impaired cardiac function and response to injury. Finally, we showed that exogenous HMGCS2 rescues cardiac function following ischemic injury.
CONCLUSIONS:
Our data demonstrate the importance of HMGCS2-induced ketogenesis as a means to regulate metabolic response to CM injury, thus allowing cell dedifferentiation and proliferation as a regenerative response.
Keywords: HMGCS2, Ketogenesis, Reprogramming, Dedifferentiation, Proliferation
Introduction
The adult human heart is incapable of replacing damaged or lost tissue due to the low proliferation rate of adult cardiomyocytes (CMs).1 Yet, certain animals such as zebrafish, newts, and neonatal mice, exhibit robust regeneration of the myocardium after injury as a consequence of the dedifferentiation and proliferation of surviving CMs.2–5 A variety of approaches have been described to induce proliferation of adult CMs, including the delivery of growth factors or by gene manipulation strategies,6–9 but robust and fully-translational regeneration of the adult mammalian heart has not been achieved.7 In injured zebrafish hearts, some preexisting CMs undergo dedifferentiation followed by proliferation, which allows for complete cardiac regeneration.10 CM dedifferentiation is manifest by reactivation of embryonic cardiac gene expression and disassembly of contractile sarcomeres.2, 11, 12 The exact mechanisms responsible for triggering CM dedifferentiation are un-known. We hypothesized that if adult mammalian CMs were induced to dedifferentiate, this may enable CMs to initiate proliferation in response to relevant signals, thus allowing for adult heart regeneration in mammals.
Metabolic flexibility is essential for the heart to adapt to various changes in the microenvironment,13 and changes in metabolism and substrate utilization are well-demonstrated in CMs during development and following injury. Proliferative fetal CMs favor glycolysis to generate ATP during cardiac development; however, soon after birth, CMs begin to utilize primarily aerobic fatty acid (FA) metabolism. During the same time period, neonatal human CMs rapidly lose their proliferative ability,14 and as the heart enlarges through childhood, rod-shaped CMs undergo hypertrophy, rather than hyperplasia. When injured by hypoxic stress, CMs enlarge due to pathological hypertrophy and their sarcomeric structures become disorganized. During this process, they also regain a small amount of proliferative ability along with a metabolic switch to glycolysis.15 This suggests that CM metabolism, dedifferentiation, and proliferation are intrinsically linked. Yet, in adult mammals this adaptive response is not strong enough for complete cardiac regeneration after injury. Therefore, we hypothesized that exploring metabolic reprogramming may yield exploitable methods for inducing adult CM proliferation following injury.
As discovered by Dr. Shinya Yamanaka, somatic cells can be reprogrammed by forced expression of OCT4, SOX2, KLF4, and c-MYC (OSKM) into induced pluripotent stem cells (iPSCs) which have characteristics similar to those of embryonic stem cells.16 The reprogramming process involves several cellular changes including the induction of proliferation, morphological alterations, and a metabolic switch.17, 18 Recently, short-term expression of OSKM in adult CMs in vivo was shown to induce their dedifferentiation and reprogramming, enabling reentry of mammalian CMs into the cell cycle, thus facilitating heart regeneration.19 These data support our hypothesis that reprogramming-induced dedifferentiation is a feasible strategy for promoting cardiac regeneration. However, the mechanisms underlying these changes have not been fully explored. In addition, reprogramming has limitations as a regeneration approach, such as risks of tumorigenesis and loss of cellular epigenetic memory. Therefore, we aimed to identify potential downstream targets which may independently allow for inducing adult CM dedifferentiation and enable proliferation. Here, we show that HMGCS2-induced ketogenesis leads to metabolic switch in adult CMs during early reprogramming. This metabolic adaptation would increase adult CM dedifferentiation to facilitate cardiac regeneration after injury.
Methods
All data and materials are available within the article. Detailed methods are described in the Supplemental Material.
Animal Studies
All animal experiments were conducted in accordance with the Guides for the Use and Care of Laboratory Animals (ARRIVE guidelines), and all of the animal protocols have been approved by the Experimental Animal Committee, Academia Sinica, Taiwan.
Data Availability
Datasets are deposited and publicly available in the GEO database and the accession number is GSE169035.
Statistical Analysis
The statistical data were performed using GraphPad Prism and shown as mean ± standard error of the mean (SEM). Unpaired Studenťs t-test and two-way ANOVA were applied for statistical comparisons, and a P-value of < 0.05 was considered significant.
Results
In vivo CM-reprogramming induces metabolic switch, CM dedifferentiation and increased CM proliferation
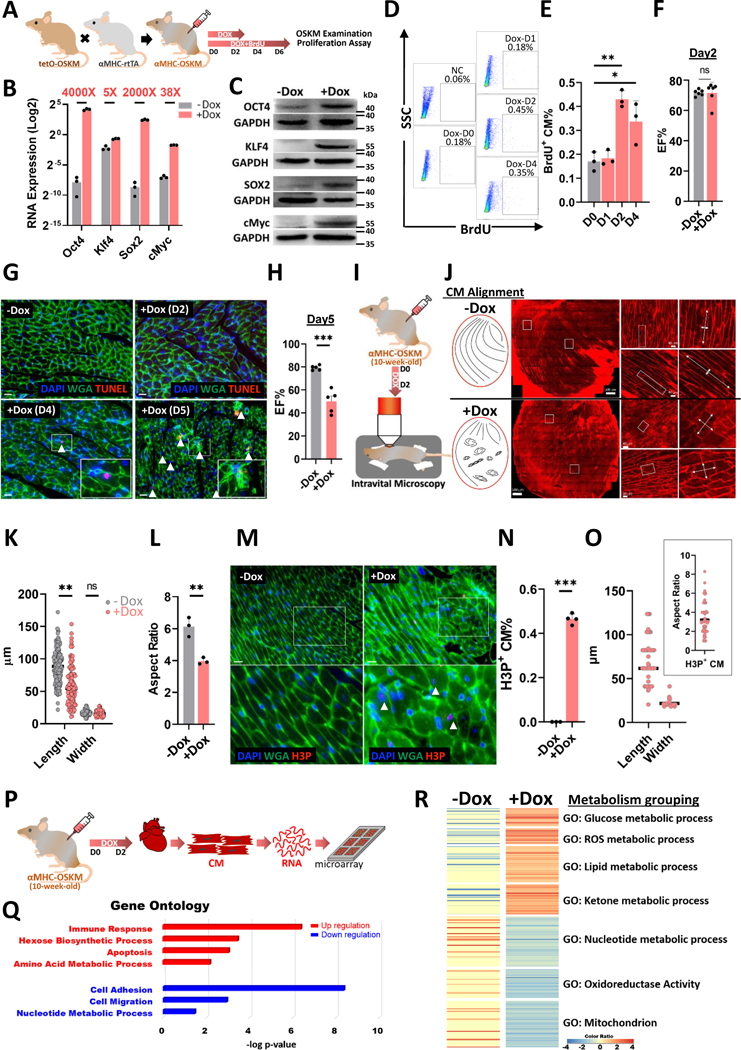
In order to examine the process of adult CM reprogramming in vivo, transgenic mice were bred to overexpress mouse OSKM specifically in adult CMs after doxycycline induction, as shown in Figure 1A. Figure 1B and 1C show successful induction of OSKM mRNA and protein expression in isolated transgenic, adult CMs after doxycycline treatment for 2 days. The substantial levels of OCR4 and SOX2 were detected without doxycycline treatment indicating a certain leakiness of the genetic system in these CM-specific reprogramming mice. Importantly, this high level of induction was detected mainly in CMs, and not in non-CMs in the heart, or other tissues isolated from doxycycline-treated mice (Figure S1A). We noted an increase of Oct4 expression in the lung and kidney, which may contribute toward the lethality of prolonged reprogramming induction in vivo. Tracking the degree of CM proliferation, a three-fold increase in BrdU+ CMs was found 2 days following doxycycline administration (Figure 1D and 1E). This proliferative response of adult CMs was highest at reprogramming day 2 compared to day 1 and 4. Gross heart function at reprogramming day 2 was measured by echocardiography and showed no difference in ejection fraction % (EF%) (Figure 1F). Six days of doxycycline treatment was lethal. Examining the heart tissue after five days of doxycycline we found more aggregated apoptotic cells (TUNEL+ population) in reprogramming hearts compared to the control (Figure 1G). At the same timepoint, the mice had a significantly lower EF% by echocardiography (Figure 1H). Thus, we concluded that continuous induction of OSKM in adult CMs severely impairs heart function, leading to death. On this basis, reprogramming day 2 was selected as the key time point for our further analyses. Using intravital microscopy (Figure 1I) to investigate the isolated whole hearts with membrane potential dye (Di-2-ANEPEQ) staining, we found that CMs in control (Ctrl) mice were normally aligned, but regions of poorly-aligned CMs were observed in the doxycycline-treated mice (Figure 1J). In addition, the in-vivo morphology of reprogramming CMs maintained their normal width but became shorter, leading to a rounder cell aspect ratio than control CMs (Figure 1K and 1L). By recording each contraction of the Ctrl or reprogramming hearts in vivo, we observed areas of disorganized or nonaligned contraction, consistent with the structural disruptions (Figure S2). Furthermore, heart tissue sectioning was performed to examine the relationship between CM alignment (WGA staining) and CM proliferation (H3P staining). We confirmed that doxycycline-induced hearts contained more proliferative CMs, and these same cells displayed a rounder morphology (around 50–60 μm in length and an aspect ratio of approximately 3) with poorer cell alignment (Figure 1M–1O). Lastly, CMs were isolated from the hearts of mice treated for 2 days with PBS or doxycycline, and RNA was extracted and subjected to microarray analysis (Figure 1P). Gene Ontology categorization showed that metabolism-related gene expression was significantly changed in the reprogramming CMs compared to the Ctrl CMs at reprogramming day 2 (Figure 1Q). The gene expression changes included the up-regulation of glucose and amino acid metabolism and down-regulation of nucleotide metabolism. Similar trends were shown in heat map analysis (Figure 1R); ketone metabolism-related gene expression was up-regulated and aerobic respiration-related genes were down-regulated in the adult reprogramming CMs compared to the Ctrl CMs. Taken together, these data show that temporary CM reprogramming resulted in dedifferentiation with changes in cell morphology, proliferation, and expression of genes associated with metabolism.
Figure 1. In vivo CM-reprogramming induces metabolic switch, CM dedifferentiation and increased CM proliferation.
A, The experimental design for investigating adult CM reprogramming in vivo. B, RNA expression level and induction level of OSKM in adult CMs after inducing OSKM reprogramming for 2 days. C, Protein expression level and induction level of OSKM in adult CMs after inducing OSKM reprogramming for 2 days. D, Flow cytometry analysis of isolated proliferative CMs through BrdU tracking in CM-reprogramming mice after OSKM induction. E, Percentage of proliferative CMs at each CM-reprogramming day determined by flow cytometry. F, Heart function measured by echocardiography in control or CM-reprogramming hearts after doxycycline treatment for 2 days. G, TUNEL staining of heart sections isolated from CM-OSKM mice after doxycycline treatment at the different time points. H, Heart function measured by echocardiography in control or CM-reprogramming hearts after doxycycline treatment for 5 days. I, Schematic diagram of intravital imaging protocol used for live investigating CM-reprogramming hearts after PBS or OSKM induction in vivo for 2 days. J, Investigation of CM alignment in the whole CM-reprogramming hearts by intravital microscopy after PBS or OSKM induction in vivo for 2 days. K, The morphology of CMs in CM-reprogramming hearts determined by length and width in intravital imaging data after PBS or OSKM induction in vivo for 2 days. Each dot represents one CM in one Ctrl or reprogramming heart. L, Aspect ratio determined by length-to-width ratio of each CM-reprogramming mouse in intravital imaging data after PBS or OSKM induction specifically in CMs in vivo for 2 days. Each dot represents one mouse sample. M, Immunofluorescence staining of heart tissue sections showing morphology of proliferative CMs through H3P and WGA staining on CM-reprogramming hearts after PBS or OSKM induction for 2 days. Arrow heads represented H3P+ proliferative CMs. Scale bars were 50 μm. N, Percentage of proliferative CM percentage (H3P+ %) in the heart tissue sections of CM-reprogramming hearts after PBS or OSKM induction for 2 days. O, The morphology of H3P+ CMs in three CM-reprogramming hearts determined by length, width, and aspect ratio in heart tissue sections after OSKM induction in vivo for 2 days. Each dot represents one CM in one Ctrl or reprogramming heart. P, The experimental design for discovering the detail mechanism for adult CM reprogramming at day 2 by microarray analysis. Q, Gene ontology analysis of gene expressional changes in adult CMs after PBS or OSKM induction for 2 days in vivo. R, Heat map showing metabolism-related gene expressional changes in adult CMs after PBS or OSKM induction for 2 days in vivo.
Cardiac-specific ketogenesis creates a systemic and specific metabolic switch along with mitochondrial changes, inducing CM dedifferentiation at reprogramming day 2
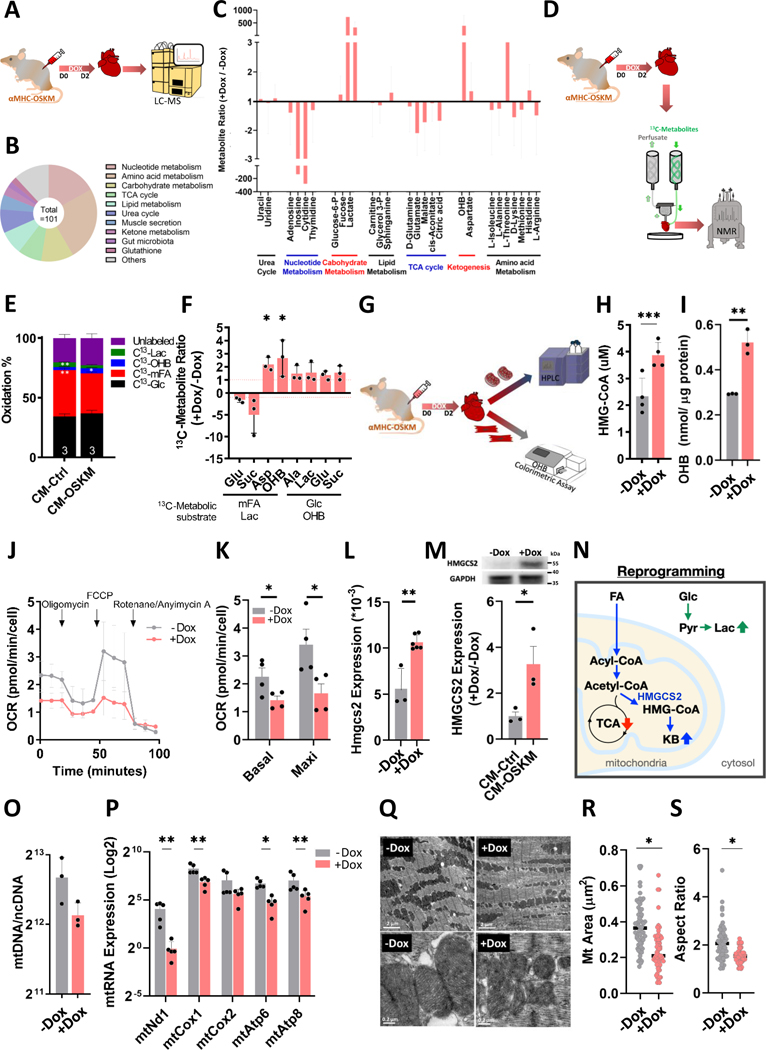
Since a metabolic switch appears implicated in adult CM dedifferentiation, we decided to clarify any rearrangement of metabolic pathways in adult CMs during the reprogramming process. First, the metabolites of Ctrl and CM-reprogramming heart lysates were analyzed by liquid chromatography-mass spectrometry (LC-MS), identifying 101 metabolites in both groups (Figure 2A and 2B). Grouping these hits revealed that glucose and ketone body metabolism-related metabolites were up-regulated in CM-reprogramming hearts (Figure 2C), in agreement with gene expression changes identified by microarray. On the contrary, tri-carboxylic acid (TCA) cycle and nucleotide metabolism-related metabolites were down-regulated in CM-reprogramming hearts (Figure 2C and 1R). In order to avoid influence by intermediate products derived from other tissues, an ex vivo heart system was set up and carbon NMR was used to detect the 13C-metabolites produced only from the exogenous addition of labeled substrates (Figure 2D).20 In NMR analysis, mixed fatty acids (mFA), which are the primary fuel for adult cardiac aerobic respiration, were decreased in the reprogramming hearts compared to the Ctrl hearts (Figure 2E). Although glucose and ketones slightly increased for ATP generation, the total percentage of aerobic respiration, derived from perfused 13C-metabolites, was decreased in the reprogramming hearts (Figure 2E and Figure S4). In addition, the amounts of Lactate and Alanine were 1.5 times higher in reprogramming hearts than in the Ctrl hearts, indicating that glycolysis was increased in the hearts two days following OSKM induction (Figure 2F). Interestingly, β-hydroxybutyrate (OHB) was 2 times higher in the reprogramming hearts than in the Ctrl hearts, indicating that ketogenesis is increased (Figure 2F). Several techniques were further utilized to confirm ketogenesis induction (Figure 2G). The main intermediate product of ketogenesis is HMG-CoA. Therefore, we isolated mitochondria and confirmed HMG-CoA amount was 2 times higher in the reprogramming hearts than in the Ctrl hearts (Figure 2H). The end product of ketogenesis, OHB, was measured >1.5 times higher in the reprogramming CMs than Ctrl CMs by an OHB colorimetric assay kit (Figure 2I). In order to control for potential metabolic effects of doxycycline, we performed the same metabolic profiling on C57BL/6J (normal) mice treated with doxycycline for 2 days. The levels of each 13C-metabolite were unchanged in Dox-treated normal hearts compared to PBS-treated CM-OSKM mice (Figure S5A). In addition, neither HMGCS2 expression nor OHB levels were significantly different (Figure S5B and S5C). Using Seahorse assay we found that the oxygen consumption rate (OCR) was lower in the adult reprogramming CMs than in the Ctrl CMs (Figure 2J and 2K). HMGCS2, the rate-limiting enzyme of ketogenesis, was significantly increased at both the RNA and protein levels (Figure 2L and 2M). A schematic summary of these changes is shown in Figure 2N. Several metabolic pathways such as ketogenesis and aerobic respiration occur in mitochondria, and changes of OCR are often accompanied by mitochondrial differences. Therefore, we assessed CM mitochondria presenting that mitochondrial copy numbers were lower and RNA expression was significantly lower in the reprogramming CMs compared to the Ctrl CMs (Figure 2O and 2P). Transmission electron microscopy (TEM) revealed that mitochondrial area and aspect ratio were both significantly decreased in the reprogramming hearts (Figure 2Q–2S). Mitochondrial fission has been reported to be related to proliferative induction through post-translational phosphorylation of DRP-1 on serine 616.21 Indeed, DRP-1 serine 616 phosphorylation was higher in reprogramming CMs compared to the Ctrl CMs (Figure S6A), while DRP-1 on serine 637 showed no difference (Figure S6B). In summary, these data indicate that during CM reprogramming by OSKM induction, a metabolic switch occurs, including increased ketogenesis and glycolysis with deceased aerobic respiration and altered mitochondrial structure and function. This switch occurs in synchrony with the induction of CM proliferation.
Figure 2. Cardiac-specific ketogenesis creates a systemic and specific metabolic switch along with mitochondrial changes, inducing CM dedifferentiation at reprogramming day 2.
A, The experimental design for metabolic profiling using LC-MS analysis. B, Hits detected by LC-MS analysis especially in both control and CM-reprogramming hearts. C, Grouping of metabolic hits detected by LC-MS analysis in control or CM-reprogramming hearts. D, The experimental design for metabolic profiling using a working heart system perfused with 13C-metabolites, detected by NMR. E, Oxidation percentage of control and CM-reprogramming hearts measured by 13C-glutamate level derived from different 13C-metabolic substrates through NMR analysis. F, Specific 13C-metabolites of control and CM-reprogramming hearts detected by NMR. G, The experimental design for measuring ketogenesis in the control or CM-reprogramming hearts. H, HMG-CoA level detected by HPLC in the isolated mitochondria from control or CM-reprogramming hearts. I, OHB level measured by OHB colorimetric assay in the isolated CMs from control or CM-reprogramming mice. J, OCR detected by Seahorse analysis in the isolated CMs from control or CM-reprogramming mice. K, Quantification of basal and maximal OCRs in the control or reprogramming CMs isolated from PBS or OSKM-treated hearts. L, RNA expression of Hmgcs2 normalized by Gapdh in CMs isolated from control or OSKM-treated mice. M, Protein expression of HMGCS2 in CMs isolated from control or OSKM-treated mice. N, Schematic diagram showing metabolic switch in adult CMs after OSKM induction for 2 days. O, Mitochondrial copy numbers detected by mtDNA through real-time PCR in control or reprogramming CMs isolated from PBS or OSKM-treated hearts. P, Mitochondrial RNA expression detected by real-time PCR in control or reprogramming CMs isolated from PBS or OSKM treated hearts. These RNA expressions were normalized by Gapdh. Q, Mitochondrial structure examined by TEM in isolated control or CM-reprogramming hearts. R, Mitochondrial size in isolated control or CM-reprogramming hearts, determined by TEM. S, Aspect ratio of mitochondrial length-to-width in isolated control or CM-reprogramming, determined by TEM.
HMGCS2 induction in adult hearts induces ketogenesis leading to a metabolic switch in an oxygen-dependent manner
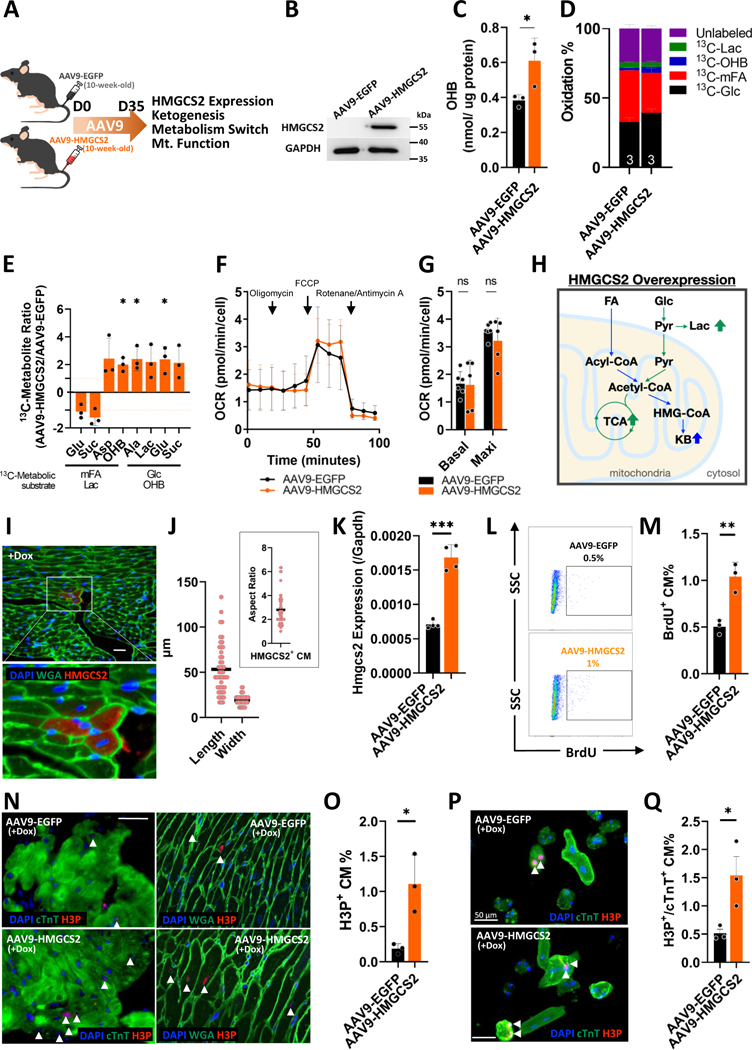
We determined to examine whether ketogenesis is the initiator of these metabolic changes in the heart. In vivo overexpression of HMGCS2 in normal adult mouse hearts was established through AAV9 infection to test if its overexpression would lead to the same metabolic changes found in reprogramming hearts (Figure 3A). AAV9-mediated HMGCS2 induction was sustained for approximately 2 months and the expression peaked 5 weeks after injection (Figure S7A). Transduction efficiency was ~88% by measuring EGFP+ cells using flow cytometry (Figure S7B). After AAV9 infection for 5 weeks, HMGCS2 expression and the amount of OHB were both confirmed to be increased in the isolated adult CMs from AAV9-HMGCS2 mice compared to AAV9-EGFP (Ctrl) mice (Figure 3B and 3C). Metabolic profiles of Ctrl and HMGCS2-overexpressing hearts were analyzed that ketogenesis was induced and aerobic FA metabolism was decreased in the HMGCS2-overexpressing hearts (Figure 3D and 3E). This was in agreement with what we observed in the OSKM-induced adult CMs. However, under normal oxygen conditions, energy demand could be supplied from aerobic glucose and OHB metabolism (Figure 3D and 3E). Glycolysis was also increased, likely to compensate for decreased ATP production via aerobic FA metabolism (Figure 3E). Seahorse analysis showed that OCR was not different in HMGCS2-overexpressing CMs compared to Ctrls (Figure 3F and 3G). In accordance with the finding that the total oxidation rate was not changed in the HMGCS2-overexpressing hearts, mitochondrial DNA and RNA expression, as well as mitochondrial structure, did not differ between Ctrl and HMGCS2-overexpressing hearts (Figure S8A–S8E). In addition, heart function was similar between Ctrl and HMGCS2-overexpressing hearts. Therefore, we conclude that cardiac HMGCS2 overexpression increases ketogenesis which in turn decreases aerobic FA metabolism and increases glycolysis and aerobic glucose metabolism, together maintaining oxygen consumption and overall energy production (Figure 3H). Although cardiac ketogenesis competitively reducing aerobic FA metabolism was shown in reprogramming hearts and cardiac HMGCS2-overexpressing hearts, the following metabolic outcome would differ based on environmental adaptation (Figure 2N and 3H). This implies that cardiac ketogenesis may lead to a “reprogramming metabolism” allowing adult CM dedifferentiation under temporary induction of Yamanaka’s factors. In order to test this hypothesis, cardiac HMGCS2 overexpression was performed on adult CM-OSKM mice to confirm if this overexpression would increase CM proliferative ability while inducing reprogramming. First, we examined the morphology of highly-HMGCS2-expressing CMs from CM-OSKM mice after OSKM induction for 2 days. These cells displayed a rounder morphology (50–60 μm in length and aspect ratio of approximately 3) similar to those we found in reprogramming CMs (Figure 3I and 3J). After HMGCS2 overexpression resulting from doxycycline treatment, the hearts showed ~2-fold higher Hmgcs2 RNA expression compared to the doxycycline treatment only or HMGCS2 overexpression only (Figure 3K). Through BrdU labeling of isolated adult CMs, flow cytometric analysis showed a two-fold increased BrdU+ population compared to the Ctrl reprogramming hearts (Figure 3L and 3M). Heart tissue sectioning was also performed and showed 2-fold increase in the H3P+ CM population in HMGCS2-overexpressing reprogramming hearts compared to the control reprogramming hearts (Figure 3N and 3O). These proliferative cells were also confirmed as HMGCS2+ CMs (Figure S9). However, the discrepancy that lower H3P+ CM population detected in AAV9-EGFP group than the +Dox group shown in Figure 1M may be due to the different mouse strains we used for AAV injection. Furthermore, the reprogramming induced CMs were isolated for in vitro culture and showed 3 times more H3P+/cTnT+ CMs in HMGCS2-overexpressing reprogramming hearts compared to the control reprogramming hearts (Figure 3P and 3Q). To further investigate whether HMGCS2 is necessary for the pro-proliferative effects of OSKM expression, trigenic mice with CM-specific Dox-inducible OSKM expression in HMGCS2+/− mice were bred. After two days of doxycycline and BrdU administration, adult CMs isolated from HMGCS2 heterozygous mutant CMs had a 2.5 times lower BrdU+ CM % than the Ctrl CMs (Figure S10A and S10B). In addition, adult CMs were isolated for in vitro culture and the proliferative H3P+/cTnT+ CM % was 3.5 times lower in HMGCS2 heterozygous mutant mice compared to the control. These proliferative CMs showed a rounder morphology than the non-proliferative CMs (Figure S10C and S10D). In conclusion, the results in Figure 3 show that HMGCS2 overexpression by itself induces ketogenesis but does not trigger proliferation of CMs. However, in CM-OSKM-induced hearts, HMGCS2 overexpression enhanced proliferation compared to the Ctrl.
Figure 3. HMGCS2 induction in adult hearts induces ketogenesis leading to a metabolic switch in an oxygen-dependent manner.
A, Experimental design for HMGCS2 overexpression in vivo through AAV9 infection. B, Protein expression of HMGCS2 detected by western-blot in AAV9-EGFP or AAV9-HMGCS2 infected CMs. C, OHB level measured by colorimetric assay in the isolated CMs from AAV9-EGFP or AAV9-HMGCS2 infected hearts. D, Oxidation percentage of AAV9-EGFP or AAV9-HMGCS2 infected hearts measured by 13C-glutamate level derived from different 13C-metabolic substrates though NMR analysis. E, Specific 13C-metabolites of AAV9-EGFP or AAV9-HMGCS2 infected hearts detected by NMR. F, OCR detected by Seahorse analysis in the isolated CMs from AAV9-EGFP or AAV9-HMGCS2 infected mice. G, Quantification of basal and maximal OCRs in the AAV9-EGFP or AAV9-HMGCS2 infected CMs. H, The representative diagram of metabolic switch in adult CMs after AAV9-EGFP or AAV9-HMGCS2 infection. I, Immunofluorescence staining of heart tissue sections showing morphology of highly HMGCS2 expressing CMs through HMGCS2 and WGA staining on CM-reprogramming hearts after OSKM induction for 2 days. Scale bar was 50 μm. J, The morphology of highly HMGCS2-expressing CMs in three CM-reprogramming hearts determined by length, width, and aspect ratio in the heart tissue sections after OSKM induction in vivo for 2 days. Each dot represents one CM in 3 reprogramming hearts. K, RNA expression of Hmgcs2 normalized by Gapdh in CMs isolated from control or OSKM-treated mice after HMGCS2 overexpression. L, The percentage of isolated proliferative CM measured by flow cytometry through BrdU tracking in control or HMGCS2 overexpressed CM-reprogramming mice. M, The statistics of flow cytometric data representing the percentage of proliferative CMs in control or HMGCS2 overexpressed CM-reprogramming mice. N, Immunofluorescence of heart tissue sections to present morphology of proliferative CMs through H3P and WGA staining on control or HMGCS2 overexpressed CM-reprogramming hearts. Arrow Heads represented H3P+ proliferative CMs. Scale bar were 50 μm. O, Quantification of proliferative CM percentage (H3P+%) in the heart tissue sections of control or HMGCS2 overexpressed CM-reprogramming hearts. P, Immunofluorescence of isolated CMs showing morphology of proliferative CMs through H3P and cTnT staining from control or HMGCS2 overexpressed CM-reprogramming hearts. Scale bar was 50 μm. Q, Proliferative CM percentage (H3P+/cTnT+ %) in the isolated CMs from control or HMGCS2 overexpressed CM-reprogramming hearts.
CM-specific HMGCS2 knockout mice present a metabolic switch along with impaired mitochondria leading to a pathological hypertrophy
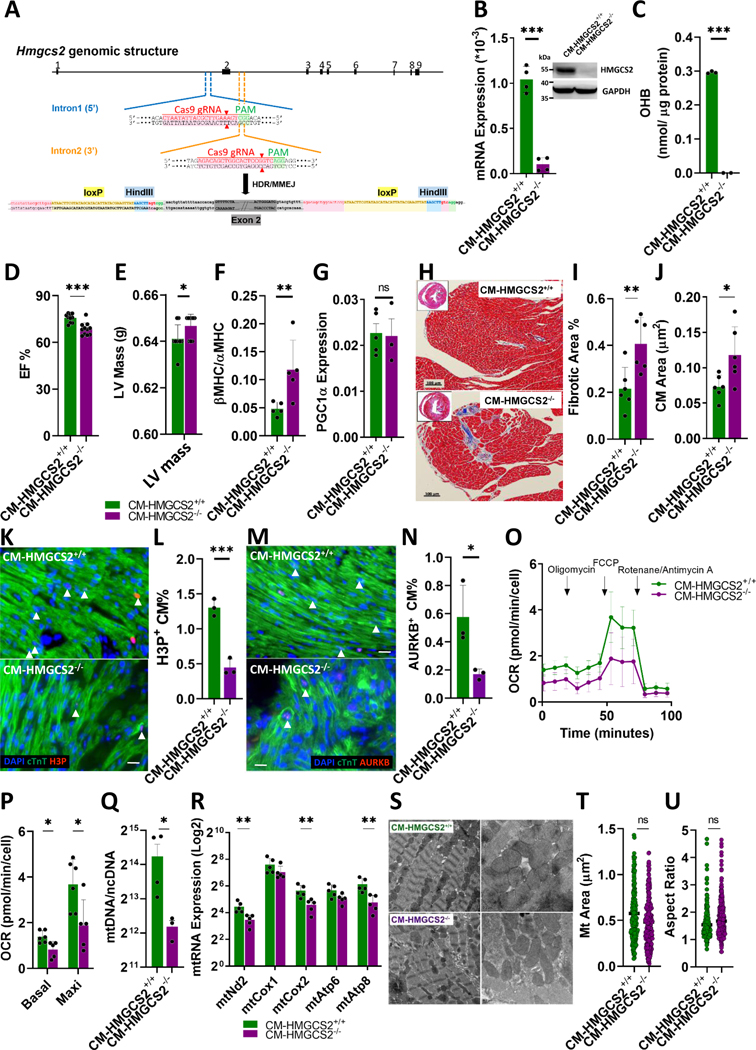
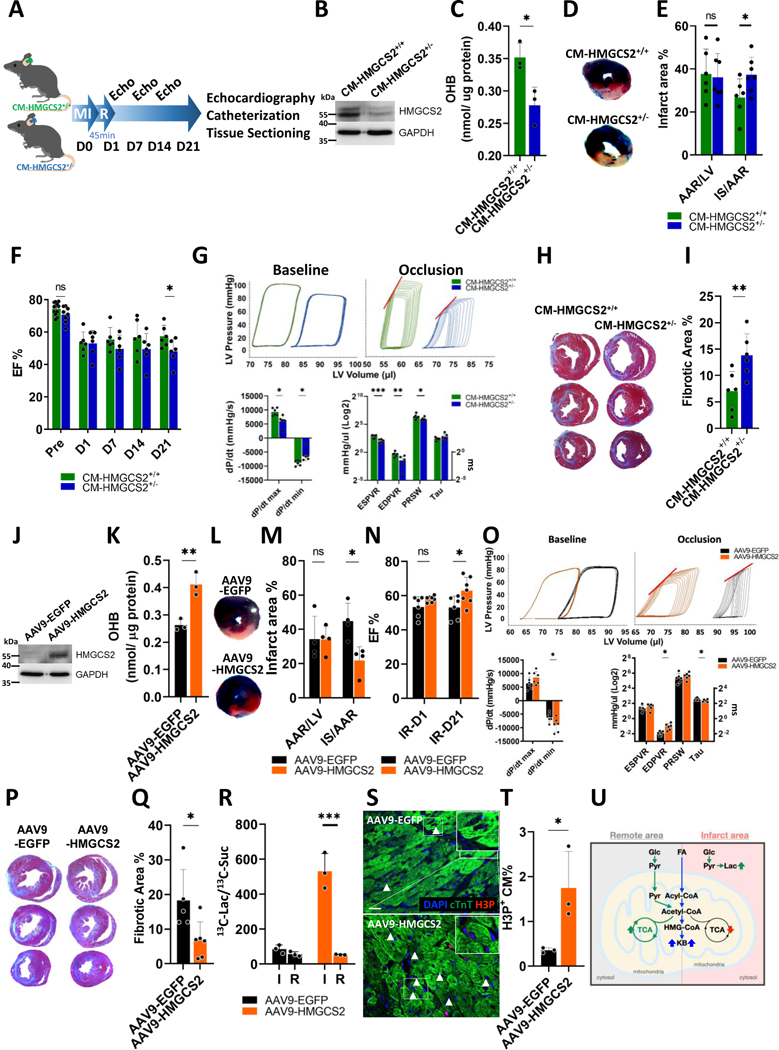
CM-specific HMGCS2 knockout mice were established to investigate the effects on cardiac metabolism. Conditional HMGCS2 knockout mice (floxP-HMGCS2) were bred by inserting loxP sites flanking exon 2 of HMGCS2 in the genome (Figure 4A). Through cross-breeding with αMHC promoter-driven CRE mice, CM-specific knockout mice (CM-HMGCS2−/−) were established (Figure S11A). These mice showed loss of HMGCS2 RNA and protein expression in isolated CMs even when we loaded more total protein (40 μg) (Figure 4B). OHB was undetectable in adult CM-HMGCS2−/− CMs (Figure 4C). Cardiac function was then measured by echocardiography and catheterization. EF% was significantly decreased in the CM-HMGCS2−/− mice (Figure 4D), and left ventricular pressure-volume indices revealed impaired heart function in the knockout mice compared to the Ctrl mice (Figure S11B). The dP/dt values were higher in CM-HMGCS2+/+ mice compared to CM-HMGCS2−/− mice under basal conditions (Figure S11B). During occlusion, CM-HMGCS2+/+ hearts showed higher ESPVR, EDPVR, and PRSW values and lower Tau values than HMGCS2−/− hearts (Figure S10B). The left ventricular (LV) mass was found to be higher in the CM-HMGCS2−/− mice compared to Ctrl mice (Figure 4E). The hypertrophic marker by the ratio of βMHC to αMHC expression showed increased 2-fold in the CM-HMGCS2−/− mice (Figure 4F), while PGC1α expression was not changed indicating the presence of pathological hypertrophy in the CM-HMGCS2−/− mice (Figure 4G). Through trichrome staining of adult mouse heart tissue sections, the percentage of fibrotic areas and CM size were both significantly increased in the CM-HMGCS2−/− mice compared to the CM-HMGCS2+/+ mice (Figure 4H–4J). Gradual decreases of HMGCS2 expression were detected with embryonic hearts showing much higher HMGCS2 expression than adult hearts (Figure S11C). In addition, the proliferative ability of CMs isolated from newborn CM-HMGCS2−/− mice was also examined, showing a significantly lower H3P+ and AURKB+ CM% compared to the Ctrl mice (Figure 4K–4N). Upon reaching adulthood, the number of rod-shaped CMs in CM-HMGCS2−/− mice were reduced by approximately half compared to Ctrl mice (Figure S11D). Thus, we hypothesize that CM-HMGCS2−/− mice generated less total CMs during development and hypertrophy may be used as a compensatory mechanism to maintain sufficient heart growth during adolescence. The metabolic profile of adult CM-HMGCS2−/− hearts was analyzed, revealing that glucose became the principal substrate for aerobic respiration (Figure S11E) and that glycolysis also increased, as shown by increased Lactate (Figure S11F). Ketogenesis was also reduced, shown by decreased OHB (Figure S10F). The OCR was also measured and found to be significantly decreased in CMs isolated from CM-HMGCS2−/− mice (Figure 4O and 4P). Accordingly, total mitochondrial DNA copies and the RNA expression of key mitochondrial genes were lower than those in CM-HMGCS2+/+ mice (Figure 4Q and 4R). TEM was used to find both misalignment and altered mitochondrial size and aspect ratio of CM-HMGCS2−/− mice compared to CM-HMGCS2+/+ mice (Figure 4S–4U). These data indicate that reduction of HMGCS2 expression in the CMs leads to the rearrangement of metabolic pathways along with impaired mitochondria, resulting in pathological hypertrophy and damaged heart function.
Figure 4. CM-specific HMGCS2 knockout mice present a metabolic switch along with impaired mitochondria leading to a pathological hypertrophy.
A, Schematic diagram showing establishment of CM-specific HMGCS2 knockout mice. B, RNA and protein expression of HMGCS2 detected by real-time PCR and western-blot in isolated CMs from 10-week-old CM-HMGCS2+/+ or CM-HMGCS2−/− mice. 40 μg protein was loaded for examination. RNA expression was normalized by Gapdh. C, OHB level measured by OHB colorimetric assay in isolated CMs from 10-week-old CM-HMGCS2+/+ or CM-HMGCS2−/− mice. D, Ejection fraction of 10-week-old CM-HMGCS2+/+ or CM-HMGCS2−/− hearts measured by echocardiography. E, Left ventricular mass of 10-week-old CM-HMGCS2+/+ or CM-HMGCS2−/− hearts measured by echocardiography. F, RNA expression of hypertrophic marker βMHC and αMHC measured by real-time PCR in the isolated CMs from 10-week-old CM-HMGCS2+/+ or CM-HMGCS2−/− mice. RNA expression was normalized by Gapdh. G, RNA expression of PGC1α measured by real-time PCR in the isolated CMs from 10-week-old CM-HMGCS2+/+ or CM-HMGCS2−/− mice. H, The fibrotic area in 10-week-old CM-HMGCS2+/+ or CM-HMGCS2−/− hearts shown by Masson Trichrome staining of heart tissue sections. I, Quantification of fibrotic percentage in 10-week-old CM-HMGCS2+/+ or CM-HMGCS2−/− hearts measured by Masson Trichrome Staining. J, Quantification of CM size in 10-week-old CM-HMGCS2+/+ or CM-HMGCS2−/− hearts measured by Masson Trichrome Staining. K, Immunofluorescence of heart tissue sections to present morphology of proliferative CMs through H3P and cTnT staining on 1-day-old control or CM-HMGCS2−/− hearts. Arrow Heads represented H3P+/cTnT+ proliferative CMs. Scale bars were 50 μm. L, Quantification of proliferative CM percentage (H3P+%) in the heart tissue sections of 1-day-old control or CM-HMGCS2−/− hearts. M, Immunofluorescence of heart tissue sections showing morphology of proliferative CMs through AURKB and cTnT staining on 1-day-old control or CM-HMGCS2−/− hearts. Arrow Heads represented AURKB+/cTnT+ proliferative CMs. Scale bars were 50 μm. N, The statistics of proliferative CM percentage (AURKB+%) in the heart tissue sections of 1-day-old control or CM-HMGCS2−/− hearts. O, OCR detected by Seahorse analysis in the isolated CMs from CM-HMGCS2+/+ or CM-HMGCS2−/− mice. P, Quantification of basal and maximal OCRs in the isolated CMs from 10-week-old CM-HMGCS2+/+ or CM-HMGCS2−/− mice. Q, Mitochondrial copy numbers detected by mtDNA through real-time PCR in the isolated CMs from 10-week-old CM-HMGCS2+/+ or CM-HMGCS2−/− mice. R, Mitochondrial RNA expression detected by real-time PCR in adult CMs isolated from 10-week-old CM-HMGCS2+/+ or CM-HMGCS2−/− mice. RNA expressions were normalized by Gapdh. S, Mitochondrial structure examined by TEM in 10-week-old CM-HMGCS2+/+ or CM-HMGCS2−/− hearts. T, Mitochondrial size of isolated 10-week-old CM-HMGCS2+/+ or CM-HMGCS2−/− hearts in TEM data. U, The aspect ratio of mitochondrial length-to-width in isolated 10-week-old CM-HMGCS2+/+ or CM-HMGCS2−/− hearts in TEM data.
Loss of CM-specific HMGCS2 expression worsens heart function after cardiac ischemia reperfusion (cI/R) injury and can be rescued by exogenous HMGCS2 expression
Although HMGCS2 expression is low in the adult heart, its role in development and maintenance of heart function is clearly shown in Figure 4. Next, we decided to investigate any role of HMGCS2 in cardiac protection or regeneration, since injured hearts may create a similar environment as OSKM-induced reprogramming hearts. Both hearts showed significant upregulation of dedifferentiation markers Myh7 and Nkx2.5 (Figure S12A), and Hmgcs2 was highly upregulated in CMs after inducing OSKM in adult CM of MI mice (Figure S12B). Besides, HMGCS2 expression was reported directly regulated by MYC protein.22 CM-HMGCS2−/− mice displayed impaired heart function even without cardiac injury (Figure 4). However, CM-HMGCS2+/− mice, confirmed to have half the amount of HMGCS2 expression, still produced OHB equal to the control mice and maintained normal heart function (Figure S13A–S13C). Therefore, HMGCS2+/− mice were utilized to investigate the role of HMGCS2 in cardiac regeneration following cI/R injury (Figure 5A). HMGCS2 expression and OHB were both significantly lower in the CM-HMGCS2+/− mice one day after performing cI/R surgery (Figure 5B and 5C) which was the peak of HMGCS2 up-regulation in CM-HMGCS2+/+ mice (Figure S12C). One day after cI/R, CM-HMGCS2+/− mice showed a significantly larger infarct area% than CM-HMGCS2+/+ mice (Figure 5D and 5E). EF% was reduced in both CM-HMGCS2+/+ and CM-HMGCS2+/− mice one day following cI/R injury. Weekly analysis revealed that EF% slightly decreased over time in CM-HMGCS2+/− mice, whereas CM-HMGCS2+/+ mice showed a small increase in EF% due to innate recovery. At D21 this difference was statistically significant (Figure 5F). Heart function was further characterized by catheterization at D21 post-injury and all indices indicated poorer heart function in CM-HMGCS2+/− mice than CM-HMGCS2+/+ mice (Figure 5G). The percentage of fibrotic area was also higher in the CM-HMGCS2+/− mice compared to the CM-HMGCS2+/+ mice at D21 post-cI/R (Figure 5H and 5I). Next, we examined whether exogenous induction of HMGCS2 expression into CM-HMGCS2+/− mice may rescue cardiac function after cI/R injury. After exogenous HMGCS2 induction by AAV9 for 5 weeks, HMGCS2 expression and OHB amount were both highly up-regulated in HMGCS2-rescued CM-HMGCS2+/− mice compared to Ctrl mice 1 day after cI/R injury (Figure 5J and 5K). HMGCS2-rescued CM-HMGCS2+/− mice showed lower infarct area% than Ctrl CM-HMGCS2+/− mice (Figure 5L and 5M). Echocardiography revealed that heart function was also rescued, showing higher EF% at D21 following cI/R surgery in the HMGCS2-rescued mice (Figure 5N). Catheter measurements indicated a trend toward improved systolic and diastolic functions in HMGCS2-rescued mice after cI/R injury for 21 days compared to Ctrl mice (Figure 5O). At the same time, the percentage of fibrotic area was lower in HMGCS2-rescued mice compared to the Ctrl mice (Figure 5P and 5Q).
Figure 5. Loss of CM-specific HMGCS2 expression worsens heart function after cardiac ischemia reperfusion (cI/R) injury and can be rescued by exogenous HMGCS2 expression.
A, Experimental design of cI/R on CM-HMGCS2+/+ or CM-HMGCS2+/− mice. B, Protein expression of HMGCS2 measured by western-blot in CMs isolated from CM-HMGCS2+/+ or CM-HMGCS2+/− mice after cI/R for 1 day. C, OHB level measured by OHB colorimetric assay in isolated CMs from CM-HMGCS2+/+ or CM-HMGCS2+/− mice 1 day after cI/R. D, Infarct area presented by triphenyltetrazolium chloride (TTC) staining and remote area presented by Evans blue staining in heart sections of CM-HMGCS2+/+ or CM-HMGCS2+/− mice 1 day after cI/R. E, Quantification of infarct area% in heart sections of CM-HMGCS2+/+ or CM-HMGCS2+/− mice 1 day after cI/R. IS: infarct size; AAR: area at risk; LV: left ventricle. F, EF% of CM-HMGCS2+/+ or CM-HMGCS2+/− hearts measured by echocardiography at different time points after cI/R. G, Heart function of CM-HMGCS2+/+ or CM-HMGCS2+/− mice determined by catheterization at post-cI/R day 21. H, The fibrotic area in CM-HMGCS2+/+ or CM-HMGCS2+/− hearts shown by Masson Trichrome staining of heart tissue sections at post-cI/R day 21. I, Quantification of fibrotic percentage in CM-HMGCS2+/+ or CM-HMGCS2+/− hearts at post-cI/R day 21 measured by Masson Trichrome staining. J, Protein expression of HMGCS2 measured by western-blot in control or HMGCS2 rescued CM-HMGCS2+/− mice 1 day after cI/R. K, OHB level measured by OHB colorimetric assay in isolated CMs from control or HMGCS2 rescued CM-HMGCS2+/− mice 1 day after cI/R. L, Infarct area presented by triphenyltetrazolium chloride (TTC) staining and remote area presented by Evans blue staining in heart sections of control or HMGCS2-rescued CM-HMGCS2+/− mice 1 day after cI/R. M, Quantification of infarct area% in heart sections of control or HMGCS2-rescued CM-HMGCS2+/− mice 1 day after cI/R. IS: infarct size; AAR: area at risk; LV: left ventricle. N, EF% of control or HMGCS2 rescued CM-HMGCS2+/− hearts measured by echocardiography at different time points after cI/R. O, Heart function of control or HMGCS2 rescued CM-HMGCS2+/− hearts determined by catheterization at post-cI/R day 21. P, The fibrotic area in control or HMGCS2 rescued CM-HMGCS2+/− hearts shown by Masson Trichrome staining of heart tissue sections at post-cI/R day 21. Q, Quantification of fibrotic percentage in control or HMGCS2 rescued CM-HMGCS2+/− hearts at post-cI/R day 21 measured by Masson Trichrome Staining. R, Ratio of 13C-Lactate-to-13C-Succinate in the infarct or remote areas measured by NMR through perfusing 13C-Glucose with isolated hearts from control or HMGCS2-rescued CM-HMGCS2+/− mice at post-cI/R day 3. S, Immunofluorescence staining of heart tissue sections showing morphology of proliferative CMs through H3P and cTnT staining at the border zone of control or HMGCS2 rescued CM-HMGCS2+/− mice at post-cI/R day 3. Arrow heads represented H3P+/cTnT+ proliferative CMs. Scale bars were 50μm. T, Quantification of proliferative CMs (H3P+%) in the heart tissue sections of at the border zone of control or HMGCS2 rescued CM-HMGCS2+/− mice at post-cI/R day 3. U, Schematic diagram of metabolic switch in the infarct and remote areas of control or HMGCS2 rescued CM-HMGCS2+/− hearts at post-cI/R day 3.
According to the results shown in Figure 2 and 3, cardiac ketogenesis should competitively reduce aerobic FA metabolism, leading to a metabolic switch based on oxygen level. This implies that HMGCS2-induced metabolic changes should be different in the hypoxic infarct area versus the normoxic remote area of hearts after cI/R injury. To test this hypothesis, heart tissue from each area was isolated and perfused with 13C-glucose through a working heart system. NMR data revealed that the ratio of 13C-Lactate to 13C-Succinate (Glycolysis/aerobic Glc metabolism) was significantly higher in the infarct area of HMGCS2-rescued hearts (Figure 5R, S14A and S14B). This indicates that a higher rate of adult CM glycolysis in the border zone would induce their dedifferentiation, allowing for proliferation, which we previously found during CM reprogramming (Figure 2N). However, the 13C-Lactate (13C-Lac) to 13C-Succinate (13C-Suc) ratio was lower and both 13C-Lac and 13C-Suc were higher in the remote area of HMGCS2-rescued hearts compared to the infarct area (Figure 5R, S14A and S14B). This indicates that the induction of CM glycolysis and aerobic Glc metabolism in the remote area would help to maintain their OCR and mitochondrial function, as mentioned previously in HMGCS2-overexpressing hearts (Figure 3H). At the same time, more H3P+ CMs was found at the border zone of HMGCS2-rescued hearts compared to the Ctrl (Figure 5S to 5T). In conclusion, cardiac ketogenesis lead to metabolic switching in adaptation to the hypoxic environment, thus supporting heart function and regeneration after injury (Figure 5U).
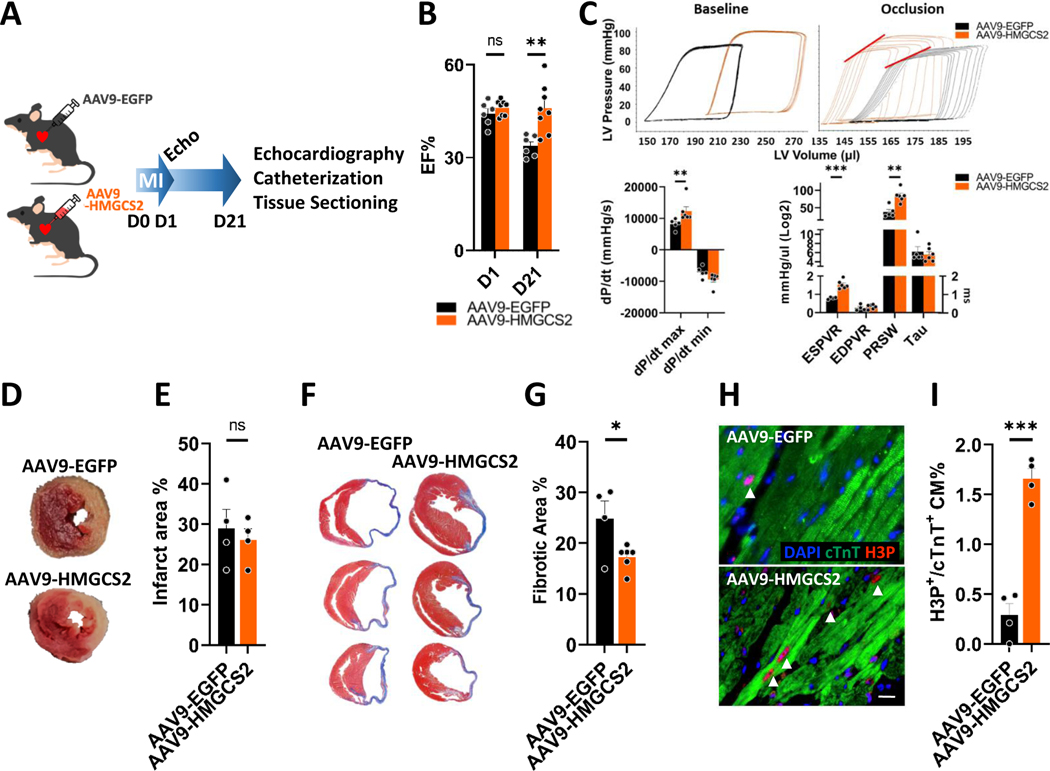
Forced HMGCS2 overexpression increases adult CM dedifferentiation and proliferation for heart function improvement after myocardial infarction (MI) or under hypoxia.
Lastly, we decided to investigate the possible therapeutic role of HMGCS2 on a permanent coronary artery ligation MI model (Figure 6A). After exogenous HMGCS2 induction by intramyocardial AAV9 injection immediately after MI, HMGCS2-overexpressing mice showed a higher EF% at post-MI D21 than Ctrl AAV9-EGFP mice (Figure 6B). Catheter measurements indicated better heart function in HMGCS2-overexpressing mice 21 days after MI injury compared to Ctrl mice (Figure 6C). The infarct area showed no differences in Ctrl or HMGCS2-overexpressing mice 1 day after MI (Figure 6D and 6E), indicating that HMGCS2 overexpression may stimulate regeneration rather than protecting the myocardium. The fibrotic area was also smaller in HMGCS2-overexpressing mice compared to the Ctrl mice (Figure 6F and 6G). More H3P+ CMs were found in HMGCS2-overexpressing hearts 3 days after MI injury compared to controls (Figure 6H and 6I). These proliferative CMs were all confirmed as HMGCS2+ (Figure S15A). Taken together, these findings show that exogenous HMGCS2 expression can support cardiac regeneration and improve heart function after MI.
Figure 6. Forced HMGCS2 overexpression increases adult CM dedifferentiation and proliferation for heart function improvement after myocardial infarction.
A, Experimental design for performing myocardial infarction (MI) in AAV9-EGFP or AAV9-HMGCS2 mice. B, Heart function measured by echocardiography in AAV9-EGFP or AAV9-HMGCS2 mice. C, Heart function measured by catheterization in AAV9-EGFP or AAV9-HMGCS2 mice. D, The infarct area in AAV9-EGFP or AAV9-HMGCS2 hearts presented by TTC staining of heart tissues at post-MI day 21. E, Quantification of infarct percentage in AAV9-EGFP or AAV9-HMGCS2 hearts at post-MI day 21 measured by TTC staining. F, The fibrotic area in AAV9-EGFP or AAV9-HMGCS2 hearts shown by Masson Trichrome staining of heart tissue sections at post-MI day 21. G, Quantification of fibrotic percentage in AAV9-EGFP or AAV9-HMGCS2 hearts at post-MI day 21 measured by Masson Trichrome Staining. H, Immunofluorescence staining of heart tissue sections showing morphology of proliferative CMs through H3P and cTnT staining at the border zone of AAV9-EGFP or AAV9-HMGCS2 mice at post-MI day 3. Arrow heads represented H3P+/cTnT+ proliferative CMs. Scale bars were 50μm. I, Quantification of proliferative CMs (H3P+%) in the heart tissue sections of at the border zone of AAV9-EGFP or AAV9-HMGCS2 mice at post-MI day 3.
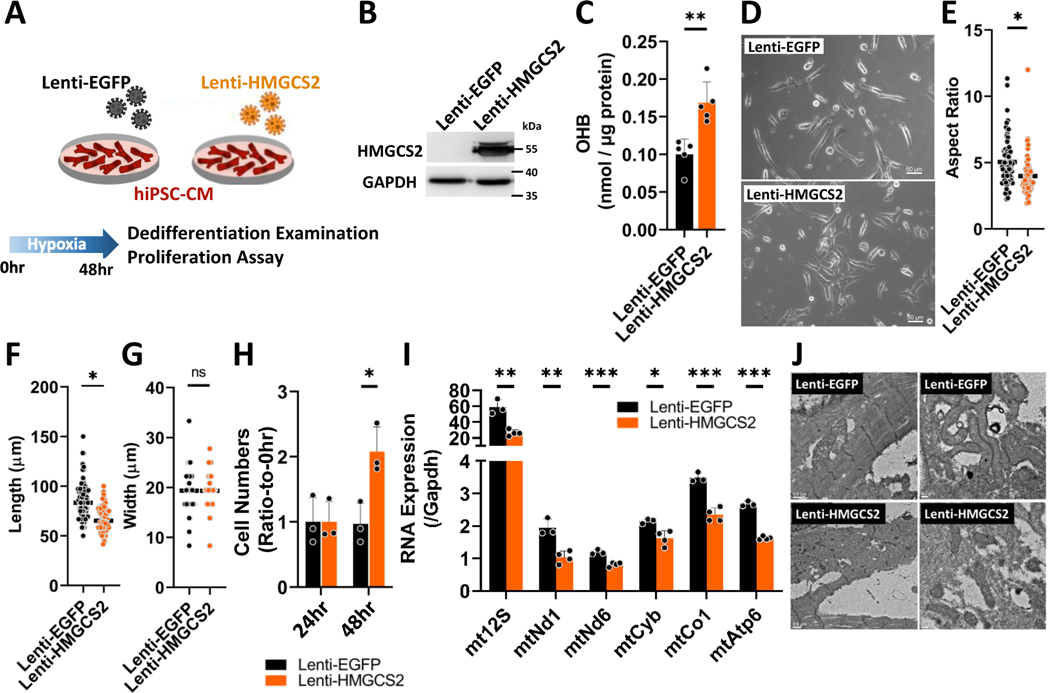
Next, we decided to test these findings in a human system. We utilized an in vitro model using hypoxic (1% O2) human induced pluripotent stem cell-derived CMs (hiPSC-CMs) (Figure 7A). HMGCS2 expression was highly up-regulated in hiPSC-CMs after lentiviral infection (Lenti-HMGCS2) compared to the Ctrl (Lenti-EGFP) (Figure 7B and Figure S15B). HMGCS2 overexpression also induces increased ketone production in hiPSC-CMs (Figure 7C). Furthermore, HMGCS2 overexpressing hiPSC-CMs showed a shorter morphology with a lower length-to-width ratio compared to the Ctrl cells under hypoxia (Figure 7D to 7G). HMGCS2 overexpressing hiPSC-CMs showed a two-fold greater proliferative ability compared to Ctrl cells under hypoxic conditions, while the same proliferation induction could not be detected during normoxia (Figure 7H and Figure S15C). We further examined mitochondrial gene expression and morphology by real-time PCR and TEM respectively. HMGCS2-overexpressing iPSC-CMs showed lower mitochondrial RNA expression compared to controls (Figure 7I). In addition, the same poorly-organized sarcomere structures with smaller, misaligned mitochondria were found in HMGCS2-overexpressing iPSC-CMs, while elongated mitochondria were aligned between sarcomeres in control cells (Figure 7J). These data indicate that forced HMGCS2 overexpression supports CM dedifferentiation and facilitates proliferation under hypoxic condition.
Figure 7. HMGCS2 overexpression increases adult human iPSC-CM dedifferentiation and proliferation under hypoxia.
A, Experimental design for examining effects on forced HMGCS2 expression in hiPSC-CMs after Lenti-EGFP or Lenti-HMGCS2 infection. B, Protein expression of HMGCS2 measured by western-blot in Ctrl or HMGCS2 overexpressed hiPSC-CM under hypoxia. C, OHB level detected by OHB colorimetric assay in Ctrl or HMGCS2 overexpressed hiPSC-CM under hypoxia. D, The morphology of control or HMGCS2 overexpressed hiPSC-CM under hypoxia. E, The length of each control or HMGCS2 overexpressed hiPSC-CM under hypoxia. F, The width of each control or HMGCS2 overexpressed hiPSC-CM under hypoxia. G, Aspect ratio determined by length-to-width ratio of each control or HMGCS2 overexpressed hiPSC-CM under hypoxia. H, The proliferative ability determined by calculation of CM numbers of control or HMGCS2 overexpressed hiPSC-CM after culturing in hypoxia chamber for 48 hours. I, Mitochondrial RNA expression detected by real-time PCR in control or HMGCS2 overexpressed hiPSC-CM after culturing in hypoxia chamber for 48 hours. These RNA expressions were normalized by Gapdh. J, Mitochondrial structure examined by TEM in control or HMGCS2 overexpressed hiPSC-CM after culturing in hypoxia chamber for 48 hours.
Furthermore, the mechanism how HMGCS2 increases adult CM proliferation was examined. SRC has long been defined as an oncogene with the ability to increase cell proliferation, with its activation controlled by peroxisome proliferator-activated receptors (PPARs).23 HMGCS2 has also been reported to regulate transcriptional expression through association with PPARα24. Thus, we hypothesized that HMGCS2 may activate Src expression to increase adult CM proliferation via associating with PPARα. We found that both Pparα and Src were significantly up-regulated in HMGCS2-overexpressing iPSC-CMs only under hypoxic conditions (Figure S16A). Down-regulation of Src or Pparα expression was performed to test adult CM proliferation after HMGCS2 overexpression during hypoxia. Src down-regulation by shRNA reduced expression to approximately 10% (Figure S16B), and adult CMs showed 2 times lower proliferative ability than the control cells after culturing in hypoxia chamber for 48 hours (Figure S16C). Down-regulation of Pparα by shRNA was then performed in human iPSC-CMs during HMGCS2 overexpression, successfully reducing Pparα expression to 20% (Figure S16D). Src expression was also found to be down-regulated to approximately 50% compared to control cells (Figure S16D). The proliferative ability was 1.6 times lower in Pparα knockdown cells than in the control cells (Figure S16E). Pparα knockdown by specific inhibitor GW6471 was also performed and showed the similar effects on reducing human iPSC-CM proliferation (Figure S16F and S16G). Furthermore, a mutant HMGCS2, unable to interact with PPARα, was generated. Palmitoylation of HMGCS2 has been previously reported to play an important role for its interaction with PPARα, and HMGCS2-C305S (mutant) could be generated to block this interaction without affecting its active site.25 Following the same strategy, HMGCS2-WT (wild type) or HMGCS2-C305S protein expression was confirmed to be upregulated in 293 cells after inducing overexpression by lentivirus (Figure S16H). Immunoprecipitation results presented that strong interaction with PPARα was detected in HMGCS2-WT overexpressing human iPSC-CMs, while the interaction was decreased in HMGCS2-C305S cells (Figure S16I). These data indicate that the position 305 is essential for HMGCS2 interaction with PPARα. Furthermore, Src expression was downregulated in HMGCS2-C305S overexpressing human iPSC-CMs compared to the wild type cells (Figure S16J). Finally, the proliferative ability was significantly decreased in mutant HMGCS2-overexpressing cells compared to the wild type (Figure S16K). We further examined this mechanism in HMGCS2-overexpressing CM-OSKM hearts. Src expression was only significantly up-regulated in reprogramming hearts compared to the control hearts while simultaneously inducing HMGCS2 overexpression (Figure S16L). We also investigated expression in the infarct border zone and remote area of hearts after HMGCS2 overexpression. Although Pparα expression was high in both areas of the MI-performed hearts and even higher in the remote area, Src expression was only significantly up-regulated in the border zone area (Figure S16M). Taken together, these data indicate that HMGCS2 supports adult CM proliferation through association with PPARα for inducing SRC expression.
Discussion
In a previous study our group showed that in vitro reprogramming of neonatal mouse CMs up-regulated expression of proliferation-related genes. We then delivered these genes to successfully drive CM proliferation via activation of the cell cycle.18 However, neonatal CMs and adult CMs differ significantly in their structure, function, metabolism and response to injury.26 As a result, the gene cocktail described in our previous study was unable to efficiently induce proliferation in adult CMs. Through specific induction of adult CM reprogramming in vivo, we can not only investigate the transformation of CMs during the process, but the effects on whole mouse can be also detected. The data in this manuscript suggests that inducing a metabolic switch in adult CMs, rather than directly inducing cell cycle-related activators, may be a more efficient way to regain proliferative ability (Figure 1 and 2). A similar approach was published recently showing that short-term in vivo expression of OSKM in adult CMs induced dedifferentiation, enabling reentry into the cell cycle and facilitating heart regeneration.19 Our two studies therefore clearly demonstrate that partial reprogramming is a potential tool in the regeneration toolkit. One notable difference is that our study used intraperitoneal doxycycline administration rather than oral (drinking water) administration. This evidently led to a different progression of CM reprogramming. In our study, CM proliferation peaked 2 days after doxycycline injection, and 6 days was lethal, with mice showing apoptotic CMs. However, the study by Chen and colleagues showed a slower timeline of reprogramming, with dedifferentiation occurring at day 6. Extended administration of doxycycline resulted in cardiac tumors. Thus, it is clear that the reprogramming-based approach has limitations. Therefore, we were motivated to examine the adult CM reprogramming process and identify any potentially modifiable downstream targets which may allow induction of temporary adult CM dedifferentiation. Our microarray analysis of CMs during early reprogramming revealed changes in genes associated with metabolism and we also note that Chen and colleagues’ study found metabolism-related gene expression changes. In this current study we have comprehensively profiled the metabolism of early-reprogramming CMs using labelled substrates and identified a switch to ketogenesis.
Ketogenesis is mainly carried out in the liver where ketones can be easily transferred to other tissues for utilization.27 Ketone utilization is common as an alternative energy source while fasting or exercising,28 and ketones are used as a metabolic substrate in the heart after injury.29–31 Under normal conditions with sufficient oxygen, the adult myocardium primarily utilizes fatty acid and glucose oxidation, with ketone bodies used minimally for energy production.32 However, if extra ketones are supplied, they can be utilized by the heart, subsequently reducing fatty acid and glucose metabolism.33–35 Failing hearts gradually lose their ability to oxidize fatty acids or glucose, and alternatively use ketone oxidation to bypass the dysregulation of β-oxidation pathway and pyruvate dehydrogenase complex. Despite mitochondrial damage during heart failure, ketones still can be completely oxidized with less steps.29, 36 Increased ketone use is thought to be adaptive since overexpression of ketolysis enzyme BDH1 has been shown to attenuate cardiac remodeling in a pressure-overloaded model.30, 37 However, there are few studies clearly defining the role of ketone synthesis in the heart tissue itself. Here, we provide evidence that HMGCS2-induced ketogenesis in adult CMs competitively reduces FA metabolism leading to a metabolic switch and mitochondrial changes (Figure 2 and 3).
Metabolic flexibility allows cells to adapt changing conditions, and primarily occurs due to the antagonism between glucose and FA for providing energy production.38 Several studies have demonstrated the importance of metabolic reprogramming in the heart during periods of stress. Decreased aerobic respiration has also been shown to provide a suitable environment for adult CM proliferation by reduction of fatty acid or glucose oxidation.39–41 In a previous study, ectopic expression of cell cycle genes in human iPSC-CMs resulted in reduced oxidative phosphorylation and increased anabolic metabolism, including gluconeogenesis.40 Our study corroborates this, since ketogenesis is also a form of anabolic metabolism, and thus may create a favorable environment for increasing CM proliferation. In addition, ketogenesis regulates FA metabolism, Glc metabolism, and the TCA cycle for maintaining hepatic metabolic homeostasis.28, 42, 43 The same scenario is presented in our current study, showing that an increase of HMGCS2-induced ketogenesis in adult CMs decreases FA metabolism. Glucose is then used via anaerobic or aerobic respiration, depending on the available oxygen. As such, ketogenesis-induced adult CM reprogramming can be specifically induced in the border zone but not the remote area of injury hearts (Figure 5).
HMGCS2 is a rate-limiting enzyme for catalyzing the first reaction of ketogenesis.44 3-hydroxy-3-methylglutaryl-CoA lyase deficiency (HMGCLD) is a rare autosomal recessive error of ketone body synthesis and leucine degradation. There are more than 20 different HMGCS2 mutations reported and these patients suffer from hepatomegaly and hypoglycemia during acute infection and prolonged fasting.45–49 HMGCS2 is up-regulated in the mouse ventricle within one week after birth, and its expression is diminished at postnatal day 12.50 However, the role of HMGCS2 in maintenance of heart function during development, or after injury, had not been previously investigated. In our study, we generated AAV9-HMGCS2 and CM-specific HMGCS2 knockout mice to thoroughly demonstrate the role of HMGCS2 in initiating adult CM dedifferentiation and proliferation, thus offering a degree of cardiac protection and regeneration after injury. Our study also provides other possible targets identified by microarray and metabolic profiling experiments including up-regulation of immune response and apoptosis-related genes, and down-regulation of nucleotide synthesis-related genes during early reprogramming. Each of these provides new research directions for future studies hoping to induce adult CM proliferation.
Overall, we have demonstrated the importance of HMGCS2-induced ketogenesis as a means to regulate metabolic response to CM injury, thus allowing cell dedifferentiation and proliferation as a regenerative response. Overlaps between OSKM-induced CM reprogramming, heart development and maturation, and the response to heart injury become readily apparent. Since myocardial infarction remains the greatest cause of death in developed countries, we hope this study provides a foundation for future research, exploiting metabolism as a mechanism to drive myocardial regeneration following injury.
Supplementary Material
Clinical Perspective.
What Is New?
Temporary reprogramming of adult cardiomyocytes (CMs) in vivo results in dedifferentiation with changes in cell morphology, proliferation, and metabolism.
Upregulation of HMGCS2-mediated ketogenesis leads to a metabolic switch in the adult CMs during early reprogramming.
Loss of CM-specific HMGCS2 expression worsens heart function after cardiac ischemia reperfusion (cI/R) injury, which can be rescued by exogenous HMGCS2 expression.
What Are the Clinical Implications?
Exploiting CM dedifferentiation and manipulating CM metabolism are potential mechanisms for cardiac regeneration after injury.
HMGCS2 overexpression improves heart function following myocardial infarction, and increases human iPSC-CM dedifferentiation and proliferation under hypoxic conditions.
Acknowledgement
The authors are grateful to the Genomic Technology Core Laboratory at the Institute of Plant and Microbial Biology, Academia Sinica for microarray analysis; We thank the Metabolomics Core Laboratory at the Center of Genomic Medicine of National Taiwan University for Metabolomic experiments. We thank Academia Sinica High-Field NMR Center (HFNMRC) for technical support; HFNMRC is funded by Academia Sinica Core Facility and Innovative Instrument Project (AS-CFII-108-112). FACS, high-content imaging, and animal studies were supported by Core Facility located at the Institute of Biomedical Sciences and TEM imaging was supported by Core Facility located at the Institute of Cellular and Organismic Biology, Academia Sinica. We thank the Taiwan Mouse Clinic, Academia Sinica and Taiwan Animal Consortium for the technical support in Seahorse analysis. This work was supported by the Ministry of Science and Technology, Taiwan (109-2321-B-001-012, 109-2327-B-001-001, 109-2740-B-001-002), the National Health Research Institutes (EX109-10907SI), the Academia Sinica Thematic Research Program (AS-107-TPL12), Healthy Longevity Grand Challenge (AS-HLGC-109-05) and Translational Medical Research Program (AS-KPQ-110-BioMed) and the University of Wisconsin Institute for Clinical and Translational Research (UL1TR002373 from NIH/NCATS).
Non-standard Abbreviations and Acronyms
- CM
Cardiomyocyte
- MI
Myocardial Infarction
- FA
Fatty Acid
- Glc
Glucose
- cI/R
Cardiac Ischemia Reperfusion
- Dox
Doxycycline
Footnotes
Disclosures
None.
References
- 1.Kubin T, Poling J, Kostin S, Gajawada P, Hein S, Rees W, Wietelmann A, Tanaka M, Lorchner H, Schimanski S, Szibor M, Warnecke H and Braun T. Oncostatin M is a major mediator of cardiomyocyte dedifferentiation and remodeling. Cell Stem Cell. 2011;9:420–32. [DOI] [PubMed] [Google Scholar]
- 2.Jopling C, Sleep E, Raya M, Marti M, Raya A and Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta V and Poss KD. Clonally dominant cardiomyocytes direct heart morphogenesis. Nature. 2012;484:479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sturzu AC, Rajarajan K, Passer D, Plonowska K, Riley A, Tan TC, Sharma A, Xu AF, Engels MC, Feistritzer R, Li G, Selig MK, Geissler R, Robertson KD, Scherrer-Crosbie M, Domian IJ and Wu SM. Fetal Mammalian Heart Generates a Robust Compensatory Response to Cell Loss. Circulation. 2015;132:109–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sereti KI, Nguyen NB, Kamran P, Zhao P, Ranjbarvaziri S, Park S, Sabri S, Engel JL, Sung K, Kulkarni RP, Ding Y, Hsiai TK, Plath K, Ernst J, Sahoo D, Mikkola HKA, Iruela-Arispe ML and Ardehali R. Analysis of cardiomyocyte clonal expansion during mouse heart development and injury. Nat Commun. 2018;9:754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bersell K, Arab S, Haring B and Kuhn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138:257–70. [DOI] [PubMed] [Google Scholar]
- 7.Hsieh PCH and Kamp TJ. To Be Young at Heart. Cell Stem Cell. 2018;22:475–476. [DOI] [PubMed] [Google Scholar]
- 8.Mohamed TMA, Ang YS, Radzinsky E, Zhou P, Huang Y, Elfenbein A, Foley A, Magnitsky S and Srivastava D. Regulation of Cell Cycle to Stimulate Adult Cardiomyocyte Proliferation and Cardiac Regeneration. Cell. 2018;173:104–116 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu S, Li K, Wagner Florencio L, Tang L, Heallen TR, Leach JP, Wang Y, Grisanti F, Willerson JT, Perin EC, Zhang S and Martin JF. Gene therapy knockdown of Hippo signaling induces cardiomyocyte renewal in pigs after myocardial infarction. Sci Transl Med. 2021;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahuja P, Perriard E, Perriard JC and Ehler E. Sequential myofibrillar breakdown accompanies mitotic division of mammalian cardiomyocytes. J Cell Sci. 2004;117:3295–306. [DOI] [PubMed] [Google Scholar]
- 11.Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG and Poss KD. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–19. [DOI] [PubMed] [Google Scholar]
- 12.Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY and Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karwi QG, Uddin GM, Ho KL and Lopaschuk GD. Loss of Metabolic Flexibility in the Failing Heart. Front Cardiovasc Med. 2018;5:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergmann O, Zdunek S, Felker A, Salehpour M, Alkass K, Bernard S, Sjostrom SL, Szewczykowska M, Jackowska T, Dos Remedios C, Malm T, Andra M, Jashari R, Nyengaard JR, Possnert G, Jovinge S, Druid H and Frisen J. Dynamics of Cell Generation and Turnover in the Human Heart. Cell. 2015;161:1566–75. [DOI] [PubMed] [Google Scholar]
- 15.Neubauer S. The failing heart--an engine out of fuel. N Engl J Med. 2007;356:1140–51. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi K and Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. [DOI] [PubMed] [Google Scholar]
- 17.Wu J, Ocampo A and Belmonte JCI. Cellular Metabolism and Induced Pluripotency. Cell. 2016;166:1371–1385. [DOI] [PubMed] [Google Scholar]
- 18.Cheng YY, Yan YT, Lundy DJ, Lo AH, Wang YP, Ruan SC, Lin PJ and Hsieh PC. Reprogramming-derived gene cocktail increases cardiomyocyte proliferation for heart regeneration. EMBO Mol Med. 2017;9:251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Luttmann FF, Schoger E, Scholer HR, Zelarayan LC, Kim KP, Haigh JJ, Kim J and Braun T. Reversible reprogramming of cardiomyocytes to a fetal state drives heart regeneration in mice. Science. 2021;373:1537–1540. [DOI] [PubMed] [Google Scholar]
- 20.Li T, Zhang Z, Kolwicz SC Jr, Abell L, Roe ND, Kim M, Zhou B, Cao Y, Ritterhoff J, Gu H, Raftery D, Sun H and Tian R. Defective Branched-Chain Amino Acid Catabolism Disrupts Glucose Metabolism and Sensitizes the Heart to Ischemia-Reperfusion Injury. Cell Metab. 2017;25:374–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsboom G, Toth PT, Ryan JJ, Hong Z, Wu X, Fang YH, Thenappan T, Piao L, Zhang HJ, Pogoriler J, Chen Y, Morrow E, Weir EK, Rehman J and Archer SL. Dynamin-related protein 1-mediated mitochondrial mitotic fission permits hyperproliferation of vascular smooth muscle cells and offers a novel therapeutic target in pulmonary hypertension. Circ Res. 2012;110:1484–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Camarero N, Mascaro C, Mayordomo C, Vilardell F, Haro D and Marrero PF. Ketogenic HMGCS2 Is a c-Myc target gene expressed in differentiated cells of human colonic epithelium and down-regulated in colon cancer. Mol Cancer Res. 2006;4:645–53. [DOI] [PubMed] [Google Scholar]
- 23.Montagner A, Delgado MB, Tallichet-Blanc C, Chan JS, Sng MK, Mottaz H, Degueurce G, Lippi Y, Moret C, Baruchet M, Antsiferova M, Werner S, Hohl D, Saati TA, Farmer PJ, Tan NS, Michalik L and Wahli W. Src is activated by the nuclear receptor peroxisome proliferator-activated receptor beta/delta in ultraviolet radiation-induced skin cancer. EMBO Mol Med. 2014;6:80–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meertens LM, Miyata KS, Cechetto JD, Rachubinski RA and Capone JP. A mitochondrial ketogenic enzyme regulates its gene expression by association with the nuclear hormone receptor PPARalpha. EMBO J. 1998;17:6972–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kostiuk MA, Keller BO and Berthiaume LG. Palmitoylation of ketogenic enzyme HMGCS2 enhances its interaction with PPARalpha and transcription at the Hmgcs2 PPRE. FASEB J. 2010;24:1914–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szibor M, Poling J, Warnecke H, Kubin T and Braun T. Remodeling and dedifferentiation of adult cardiomyocytes during disease and regeneration. Cell Mol Life Sci. 2014;71:1907–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grabacka M, Pierzchalska M, Dean M and Reiss K. Regulation of Ketone Body Metabolism and the Role of PPARalpha. Int J Mol Sci. 2016;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puchalska P and Crawford PA. Multi-dimensional Roles of Ketone Bodies in Fuel Metabolism, Signaling, and Therapeutics. Cell Metab. 2017;25:262–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aubert G, Martin OJ, Horton JL, Lai L, Vega RB, Leone TC, Koves T, Gardell SJ, Kruger M, Hoppel CL, Lewandowski ED, Crawford PA, Muoio DM and Kelly DP. The Failing Heart Relies on Ketone Bodies as a Fuel. Circulation. 2016;133:698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horton JL, Davidson MT, Kurishima C, Vega RB, Powers JC, Matsuura TR, Petucci C, Lewandowski ED, Crawford PA, Muoio DM, Recchia FA and Kelly DP. The failing heart utilizes 3-hydroxybutyrate as a metabolic stress defense. JCI Insight. 2019;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen R, Moller N, Gormsen LC, Tolbod LP, Hansson NH, Sorensen J, Harms HJ, Frokiaer J, Eiskjaer H, Jespersen NR, Mellemkjaer S, Lassen TR, Pryds K, Botker HE and Wiggers H. Cardiovascular Effects of Treatment With the Ketone Body 3-Hydroxybutyrate in Chronic Heart Failure Patients. Circulation. 2019;139:2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho KL, Zhang L, Wagg C, Al Batran R, Gopal K, Levasseur J, Leone T, Dyck JRB, Ussher JR, Muoio DM, Kelly DP and Lopaschuk GD. Increased ketone body oxidation provides additional energy for the failing heart without improving cardiac efficiency. Cardiovasc Res. 2019;115:1606–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bassenge E, Wendt VE, Schollmeyer P, Bluemchen G, Gudbjarnason S and Bing RJ. Effect of Ketone Bodies on Cardiac Metabolism. Am J Physiol. 1965;208:162–8. [DOI] [PubMed] [Google Scholar]
- 34.Stanley WC, Meadows SR, Kivilo KM, Roth BA and Lopaschuk GD. beta-Hydroxybutyrate inhibits myocardial fatty acid oxidation in vivo independent of changes in malonyl-CoA content. Am J Physiol Heart Circ Physiol. 2003;285:H1626–31. [DOI] [PubMed] [Google Scholar]
- 35.Ziegler A, Zaugg CE, Buser PT, Seelig J and Kunnecke B. Non-invasive measurements of myocardial carbon metabolism using in vivo 13C NMR spectroscopy. NMR Biomed. 2002;15:222–34. [DOI] [PubMed] [Google Scholar]
- 36.Bedi KC Jr., Snyder NW, Brandimarto J, Aziz M, Mesaros C, Worth AJ, Wang LL, Javaheri A, Blair IA, Margulies KB and Rame JE Evidence for Intramyocardial Disruption of Lipid Metabolism and Increased Myocardial Ketone Utilization in Advanced Human Heart Failure. Circulation. 2016;133:706–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uchihashi M, Hoshino A, Okawa Y, Ariyoshi M, Kaimoto S, Tateishi S, Ono K, Yamanaka R, Hato D, Fushimura Y, Honda S, Fukai K, Higuchi Y, Ogata T, Iwai-Kanai E and Matoba S. Cardiac-Specific Bdh1 Overexpression Ameliorates Oxidative Stress and Cardiac Remodeling in Pressure Overload-Induced Heart Failure. Circ Heart Fail. 2017;10. [DOI] [PubMed] [Google Scholar]
- 38.Goodpaster BH and Sparks LM. Metabolic Flexibility in Health and Disease. Cell Metab. 2017;25:1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cardoso AC, Lam NT, Savla JJ, Nakada Y, Pereira AHM, Elnwasany A, Menendez-Montes I, Ensley EL, Petric UB, Sharma G, Sherry AD, Malloy CR, Khemtong C, Kinter MT, Tan WLW, Anene-Nzelu CG, Foo RS, Nguyen NUN, Li S, Ahmed MS, Elhelaly WM, Abdisalaam S, Asaithamby A, Xing C, Kanchwala M, Vale G, Eckert KM, Mitsche MA, McDonald JG, Hill JA, Huang L, Shaul PW, Szweda LI and Sadek HA. Mitochondrial Substrate Utilization Regulates Cardiomyocyte Cell Cycle Progression. Nat Metab. 2020;2:167–178. [PMC free article] [PubMed] [Google Scholar]
- 40.Bae J, Salamon RJ, Brandt EB, Paltzer WG, Zhang Z, Britt EC, Hacker TA, Fan J and Mahmoud AI. Malonate Promotes Adult Cardiomyocyte Proliferation and Heart Regeneration. Circulation. 2021;143:1973–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abouleisa RRE, McNally L, Salama ABM, Hammad SK, Ou Q, Wells C, Lorkiewicz PK, Bolli R, Mohamed TMA and Hill BG. Cell cycle induction in human cardiomyocytes is dependent on biosynthetic pathway activation. Redox Biol. 2021;46:102094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cotter DG, Ercal B, Huang X, Leid JM, d'Avignon DA, Graham MJ, Dietzen DJ, Brunt EM, Patti GJ and Crawford PA. Ketogenesis prevents diet-induced fatty liver injury and hyperglycemia. J Clin Invest. 2014;124:5175–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arima Y, Nakagawa Y, Takeo T, Ishida T, Yamada T, Hino S, Nakao M, Hanada S, Umemoto T, Suda T, Sakuma T, Yamamoto T, Watanabe T, Nagaoka K, Tanaka Y, Kawamura YK, Tonami K, Kurihara H, Sato Y, Yamagata K, Nakamura T, Araki S, Yamamoto E, Izumiya Y, Sakamoto K, Kaikita K, Matsushita K, Nishiyama K, Nakagata N and Tsujita K. Murine neonatal ketogenesis preserves mitochondrial energetics by preventing protein hyperacetylation. Nat Metab. 2021;3:196–210. [DOI] [PubMed] [Google Scholar]
- 44.Hegardt FG. Mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase: a control enzyme in ketogenesis. Biochem J. 1999;338 ( Pt 3):569–82. [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson GN, Hsu BY, Pitt JJ, Treacy E and Stanley CA. Fasting hypoketotic coma in a child with deficiency of mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase. N Engl J Med. 1997;337:1203–7. [DOI] [PubMed] [Google Scholar]
- 46.Wolf NI, Rahman S, Clayton PT and Zschocke J. Mitochondrial HMG-CoA synthase deficiency: identification of two further patients carrying two novel mutations. Eur J Pediatr. 2003;162:279–80. [DOI] [PubMed] [Google Scholar]
- 47.Pitt JJ, Peters H, Boneh A, Yaplito-Lee J, Wieser S, Hinderhofer K, Johnson D and Zschocke J. Mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase deficiency: urinary organic acid profiles and expanded spectrum of mutations. J Inherit Metab Dis. 2015;38:459–66. [DOI] [PubMed] [Google Scholar]
- 48.Puisac B, Marcos-Alcalde I, Hernandez-Marcos M, Tobajas Morlana P, Levtova A, Schwahn BC, DeLaet C, Lace B, Gomez-Puertas P and Pie J. Human Mitochondrial HMG-CoA Synthase Deficiency: Role of Enzyme Dimerization Surface and Characterization of Three New Patients. Int J Mol Sci. 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramos M, Menao S, Arnedo M, Puisac B, Gil-Rodriguez MC, Teresa-Rodrigo ME, Hernandez-Marcos M, Pierre G, Ramaswami U, Baquero-Montoya C, Bueno G, Casale C, Hegardt FG, Gomez-Puertas P and Pie J. New case of mitochondrial HMG-CoA synthase deficiency. Functional analysis of eight mutations. Eur J Med Genet. 2013;56:411–5. [DOI] [PubMed] [Google Scholar]
- 50.Talman V, Teppo J, Poho P, Movahedi P, Vaikkinen A, Karhu ST, Trost K, Suvitaival T, Heikkonen J, Pahikkala T, Kotiaho T, Kostiainen R, Varjosalo M and Ruskoaho H. Molecular Atlas of Postnatal Mouse Heart Development. J Am Heart Assoc. 2018;7:e010378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stadtfeld M, Maherali N, Borkent M and Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat Methods. 2010;7:53–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vinegoni C, Aguirre AD, Lee S and Weissleder R. Imaging the beating heart in the mouse using intravital microscopy techniques. Nat Protoc. 2015;10:1802–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang SY, Kuo CH and Tseng YJ. Ion trace detection algorithm to extract pure ion chromatograms to improve untargeted peak detection quality for liquid chromatography/time-of-flight mass spectrometry-based metabolomics data. Anal Chem. 2015;87:3048–55. [DOI] [PubMed] [Google Scholar]
- 54.van der Heijden R, de Boer-Hlupá V, Verpoorte R and Duine JA. Enzymes involved in the metabolism of 3-hydroxy-3-methylglutaryl-coenzyme A in Catharanthus roseus. Plant Cell, Tissue and Organ Culture. 1994;38:345–349. [Google Scholar]
- 55.Karamanlidis G, Lee CF, Garcia-Menendez L, Kolwicz SC Jr., Suthammarak W, Gong G, Sedensky MM, Morgan PG, Wang W and Tian R. Mitochondrial complex I deficiency increases protein acetylation and accelerates heart failure. Cell Metab. 2013;18:239–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bohl S, Medway DJ, Schulz-Menger J, Schneider JE, Neubauer S and Lygate CA. Refined approach for quantification of in vivo ischemia-reperfusion injury in the mouse heart. Am J Physiol Heart Circ Physiol. 2009;297:H2054–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets are deposited and publicly available in the GEO database and the accession number is GSE169035.