Abstract
Background:
Despite the high prevalence of allergic asthma, currently avoidance of the responsible allergens, which is nearly impossible for allergens such as house dust mite (HDM), remains among the most effective treatments. Consequently, determination of the immunogenic epitopes of allergens will aid in developing a better understanding of the condition for diagnostic and therapeutic purposes. Current methods of epitope identification, however, only evaluate IgE-epitope binding interactions, which is not directly related to epitope immunogenicity.
Objective:
To determine and rank the immunogenicity of the epitopes of major HDM allergen, Der p 2.
Methods:
We performed degranulation assays with RBL-SX38 cells primed using patient plasma and challenged with nanoallergens which multivalently displayed epitopes to study the relative immunogenicity of various epitopes of Der p 2. Nanoallergens were used to evaluate epitopes individually or in combinations.
Results:
When evaluated using three patient samples, three epitopes in two distal regions of Der p 2 were identified as highly immunogenic when presented in combination, while no individual epitope triggered significant degranulation. One of the epitopes (69-DPNACHYMKCPLVKGQQY-86) was identified to be cooperatively immunogenic when combined with other epitopes.
Conclusion:
Our study highlights the importance of conformational epitopes in HDM-related allergies. This study also provides further evidence of the versatility of nanoallergens and their value for functional characterization of allergy epitopes, by ranking the Der p 2 epitopes according to immunogenicity. We believe nanoallergens, by aiding in identification and understanding of immunogenic epitopes, will provide a better understanding of the manifestation of the allergic condition and potentially aid in developing new treatments.
Keywords: Der p 2, house dust mite, immunogenicity, allergen epitopes, nanoallergen, allergies, asthma
INTRODUCTION
Asthma, a chronic respiratory disease, affects about 8% of the US population.1 An estimated 60% of asthmatic adults have uncontrolled asthma; a situation which led to 1.6 million emergency room visits and 179,000 people hospitalized in 2018.2,3 Atopic asthma, the most common type of asthma, is triggered or exacerbated by environmental or airborne allergens (e.g. mold, pollen, pet dander, cockroaches, or dust mites) and is characterized by high levels of IgE. 4-6 Approximately 85% of asthma patients are sensitized to house dust mite (HDM) allergens.7-9
HDMs are microscopic organisms, present in >80% of US homes, which consume mammals’ shed skin cells and leave several allergenic proteins in their excrement and exoskeletons.8,10,11 Avoidance of HDMs is nearly impossible, as their allergens are airborne and the mites often live deep in upholstered furniture, bedding, and carpets where vacuuming has little-to-no effect.10,12-14 Clinical effectiveness of avoidance methods, such as frequent cleaning, has been a point of controversy due to an inability to reduce exposure to HDM allergens below sensitization levels.8,10,12,15 The use of impermeable covers has also been reported to improve patient outcomes despite not entirely eliminating dust mites.13,14 While immunotherapy, particularly subcutaneous immunotherapy (SCIT), is shown to be effective in reducing symptoms for select patients, it is not appropriate for all patients.9,16-18 Therefore, improved understanding, diagnosis, and treatment of HDM allergies and related asthma are urgently needed.
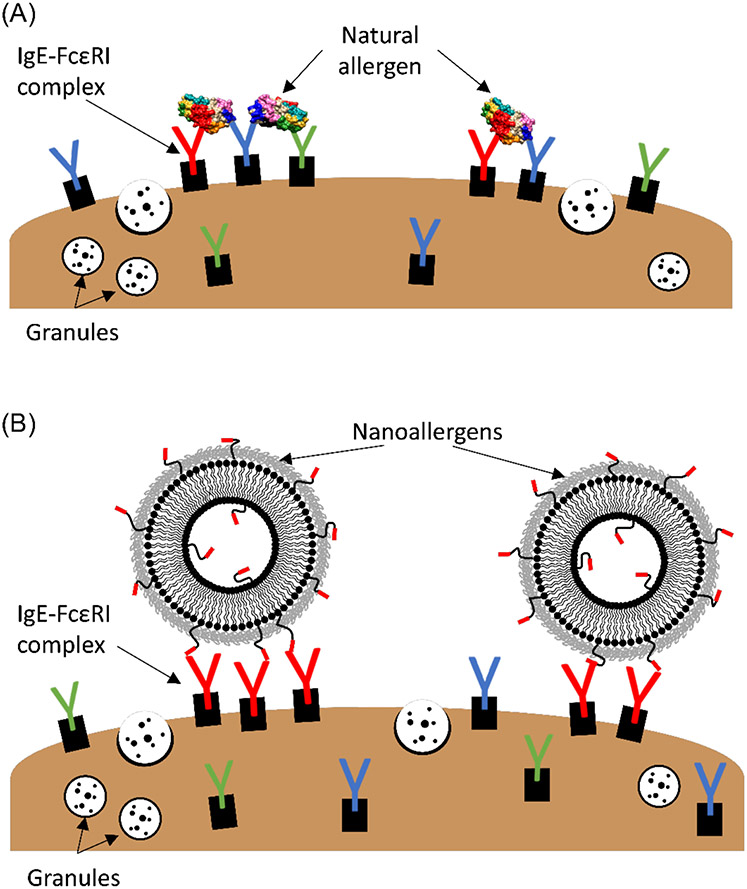
In atopic asthma, the allergic reaction is triggered when specific IgE, presented on the surface of mast cells and basophils, are clustered and crosslinked as a result of multivalent binding to epitopes on the allergenic protein.4,5,19,20 Crosslinking of the IgE results in degranulation, i.e. the release of mediators (e.g. histamine) from the cells, and the body’s subsequent allergic response (Figure 1A).19,20 To induce IgE crosslinking, multivalent binding (i.e., multiple epitopes on an allergen simultaneously binding to multiple IgE molecules) must occur, to cause the IgE to cluster and provide the proximity needed for crosslinking, as shown in Figure 1. The necessity of multivalent presentation of epitopes motivated our development of nanoallergens, a liposomal platform for displaying allergen epitopes (Figure 1B), as previously described.21 Nanoallergens are superior to previous and commonly used allergen models, such as haptenized BSA, due to the ability to precisely control particle size and loading of individual epitopes, or combinations thereof, with high consistency between batches (Figure 1B, 2A, 2B).21-23 The multivalent nature of nanoallergens is also important for the evaluation of short synthetic epitopes. When an epitope is presented as a short peptide, instead of a segment naturally positioned on an intact allergen (globular protein), affinity between epitope and IgE is significantly weaker due largely to the increase in conformational entropy (i.e., increased structural conformations) of the amino acid sequence presented without the surrounding protein framework.24,25 This is particularly important to consider when comparing sequential epitopes (i.e., continuous sequence of amino acids in the protein) versus conformational epitopes (i.e., amino acids in proximity only when the protein is properly folded), as conformational epitopes cannot be mimicked outside of the natural allergen structure without a scaffold for proper structural support. To overcome the weaker monovalent affinity of the short peptide epitopes, here we optimized the valency (i.e., number of epitope copies presented on a nanoallergen) of our nanoallergens by varying the epitope loading and particle size, to increase their avidity (multivalent affinity) for IgE-presenting cells. Increasing the epitope loading could also allow for both FAB arms on a single IgE molecule to interact bivalently with two separate copies of an epitope on the surface of a single nanoallergen, which increases avidity for each IgE-nanoallergen interaction, and in turn increases overall avidity of the nanoallergen, making it more potent. Furthermore, we envision larger-sized particles accommodating a greater number of IgE binding to a single nanoallergen, which results in larger receptor clusters that typically increase the cellular response (i.e., degranulation). Ultimately, synthesis of more potent nanoallergens could allow for the detection of lower affinity immunogenic epitopes that were overlooked in traditional binding assays.
Figure 1.
Cartoon representation of the multivalent binding and crosslinking of specific IgE-FcεRI complexes on RBL cell surface by natural dust mite allergen Der p 2 (A) and nanoallergens presenting immunogenic epitopes (B), inducing intracellular signaling cascades resulting in degranulation. Drawings are not to scale.
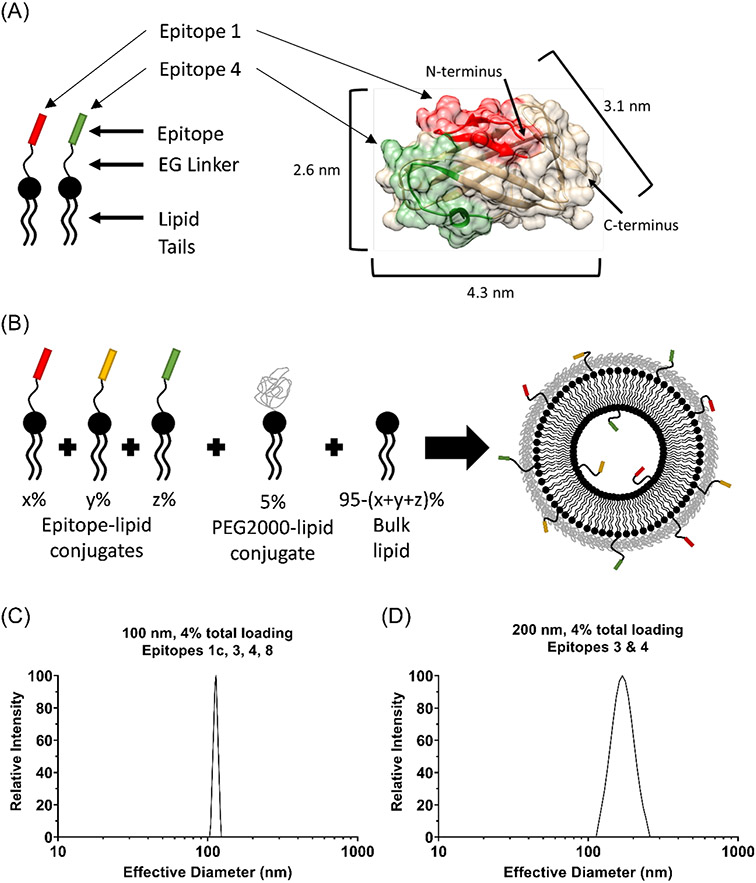
Figure 2.
(A) Epitope-lipid conjugate depiction of Der p 2 epitopes. Der p 2 crystal structure (PDB-ID 1ktj) with epitopes color-coded and allergen dimensions depicted. (B) Cartoon depiction of liposome assembly from components at specific stoichiometries. (C), (D) Representative DLS data shown for nominally 100 nm (C) and 200 nm (D) diameter nanoallergens.
For this study, we focused on a single major allergen from the Dermatophagoides pteronyssinus species: Der p 2, to which 70-80% of HDM patients are reported to be sensitized.26 While HDMs include several species, the most prevalent in the US are D. pteronyssinus (Der p) and D. farinae (Der f), between which there is typically 80-90% sequence homology (e.g. 88% homology between Der p 2 and Der f 2, group 2 allergens).27-30 Literature reports on IgE-binding Der p 2 epitopes vary significantly and disagree on whether sequential epitopes of Der p 2 exist.31-40 Here, we further explore the importance of sequential versus conformational epitopes in HDM allergies.
Literature-reported Der p 2 epitope sequences were determined via studies which tested for epitope binding, typically relying on affinity of the monovalent (one-to-one) epitope-IgE interaction. Such assays fall short of reflecting the complexities of multivalent allergen-IgE-cell interactions in vivo and do not directly correlate to an immunogenic response. Importantly, this strategy may result in low-affinity epitopes being overlooked. Bias towards high-affinity epitopes becomes problematic when analyzing patient samples, where the presence of multivalent interactions (i.e., multiple epitopes interacting with multiple IgE) yields a complex relationship between relative abundance and affinity of each IgE, and the immunogenicity of the IgE. In such cases, lower affinity epitope-IgE pairs can play a crucial role, as previously reported.41,42 Specifically, due to the heterogeneity of IgE on the cell surface, high affinity epitopes alone may not be enough to trigger degranulation, whereas inhibition of lower affinity epitopes may be enough to subdue it.41 Due to these shortcomings, there is currently no consensus on the major HDM epitopes nor any information on their relative immunogenicity.26,31-40 The nanoallergen platform is able to evaluate patient samples in an ex vivo setting and incorporate multivalent interactions to more accurately simulate the immunogenic activities triggered by the patient IgE. The immunogenic activity can then be quantified through an RBL cell degranulation assay. In earlier publications, we reported that the nanoallergen platform overcame the limitations of the traditional methods used to identify IgE-binding epitopes,21,23,43 and here we used a similar approach to identify the relative immunogenicities of Der p 2 epitopes. Furthermore, we have previously demonstrated identification of immunogenic epitopes provides relevant information for the development of new treatments, such as inhibitors.44,45
To determine the immunogenic epitopes of Der p 2, in this study, degranulation assays were performed with plasma from patients sensitized to HDM, using nanoallergens loaded multivalently with copies of one epitope at a time (Figure 1B). Assays with nanoallergens presenting a combination of epitopes were also performed to evaluate the possibility of cooperativity when simultaneously presenting multiple epitopes. By testing the ability of the reported epitopes to induce a cellular response, rather than relying on binding affinity alone, a more robust method for identification of the immunogenic epitopes of major HDM allergen Der p 2 is presented. Ultimately, identification of immunogenic portions of the allergen is an important tool for improving the treatment and diagnostic options for HDM allergies and asthma.
METHODS
Materials
All solvents and peptide synthesis reagents were purchased from Millipore-Sigma (St. Louis, MO): dimethylformamide (DMF), piperidine, dichloromethane (DCM), N,N-diisopropylethylamine (DIEA), 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluroniumhexafluorophosphate (HBTU), hydrazine, Oxyma, N,N′-Diisopropylcarbodiimide (DIC), diethyl ether, trifluoroacetic acid (TFA), triisopropylsilane (TIS), isopropanol (IPA), acetonitrile (ACN), chloroform, methanol, cholesterol, bovine serum albumin (BSA), cytochalasin D, poly-N-acetylglucosamine (PNAG), phosphate buffered saline (PBS), palmitic acid, citric acid, sodium citrate, glycine, magnesium chloride hexahydrate, NovaPEG Rink Amide LL resin, and all Novabiochem fmoc-protected amino acids. Fmoc-N-amido-dPEG8-acid and Fmoc-N-amido-dPEG2-acid were purchased from Quanta BioDesign (Plain City, OH). Sodium hydroxide, sodium chloride, and potassium chloride were purchased from VWR (Radnor, PA). 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-methoxy(polyethylene glycol)-2000 (DSPE-mPEG2000) ammonium salt, membranes, and all mini-extruder components were purchased from Avanti Polar Lipids (Alabaster, AL). Minimum essential media (MEM), fetal bovine serum (FBS), L-glutamine, penicillin/streptomyocin, G418, calcium chloride, glucose monohydrate, and goat anti-human IgE were purchased from Thermo Fisher Scientific (Waltham, MA). Trypsin was purchased from Corning (Corning, NY). Patient plasma was purchased from PlasmaLab International (Seattle, WA). Isotype monoclonal human IgE were kindly provided by Dr. Scott Smith (Vanderbilt University).
Statistical analysis
All error bars represent standard deviations of triplicates in a single experiment, unless otherwise noted. Degranulation assay data are representative of several experiments. Means, standard deviation, and p-values were calculated using GraphPad Prism software (San Diego, CA). P-values were calculated with 2-way ANOVA.
Protein analysis
Analysis and study of protein crystal structures for the structural and spatial identification of potential epitopes were performed using UCSF-Chimera, and Der p 2 crystal structure was obtained from the protein data bank (www.rscb.org).
Peptide synthesis
Peptides were synthesized either using a Liberty Blue microwave-assisted peptide synthesizer or manually on a wrist-action shaker following conventional Fmoc chemistry on a NovaPEG Rink Amide LL resin. Fmoc deprotection reactions were performed using 20% piperidine in DMF and ivdde deprotections with 2% hydrazine in DMF. Fmoc-protected amino acids were activated using either DIC/Oxyma on the synthesizer or HBTU/DIEA during manual synthesis. Palmitic acid conjugations were performed manually using HBTU/DIEA for activation. Peptides were cleaved using cocktails of TFA/H2O/TIS (95/2.5/2.5%) or, if the epitopes contained cysteine residues, TFA/H2O/EDT/TIS (94/2.5/2.5/1%). Post cleaving, TFA cocktails were removed via a Buchi rotavapor.
Peptide cyclization was performed after initial purification of the epitope-lipid conjugates. Epitope-lipid conjugate powder was dissolved in 10 mL DMF and 100 μL DIEA (per ≤ 10 mg of product), then the solution was allowed to react overnight at RT with gentle stirring. After reacting, the solvent was evaporated using a rotavapor, and the cyclized epitope-lipid conjugates were re-purified, as described below.
Peptide purification and characterization
Peptides were purified using RP-HPLC on an Agilent 1200 Series. Peptide-lipid conjugates were purified with a Zorbax semi-prep C3 column and IPA/ACN/H2O gradient. All products were purified to >90% purity. Following purification, product identity was confirmed by Q-TOF mass spectrometry.
Cell culture
RBL-SX38 cells were cultured in Minimum Essential Media with 10% fetal bovine serum, 1% L-glutamine, 1% penicillin/streptomyocin, and 1.2 mg/mL of G418, a selective agent for the human FcεRI-expressing RBL cells, as previously described.21 Cells were split every two to three days. For the purposes of this experiment no cells were kept in culture longer than 4 weeks. Cells were kindly provided by Dr Jean-Pierre Kinet.
Nanoallergen preparation and characterization
Nanoallergens were prepared by the dry film hydration method, as described previously.46 In vials, the purified epitope-lipid conjugates, at desired molar stoichiometric ratios, were combined with bulk lipid and polyethylene glycol (PEG2000) conjugated lipids, all dissolved in volatile solvent. The PEG2000-lipids were added to limit aggregation of nanoallergens and nonspecific interactions between nanoallergens and cells. For the nanoallergens that displayed multiple copies of one or more epitopes simultaneously, a lipid mixture was used following the formula 95-(X+Y+Z):5:X:Y:Z Bulk-Lipid:PEG2000:EpitopeX:EpitopeY:EpitopeZ, where X, Y, and Z were varied from 0-8 mol% (Figure 2B). An additional 5 mol% of cholesterol was included to further stabilize the liposomes. Volatile solvents were evaporated with a gentle flow of nitrogen, and then vials were kept in a desiccator overnight to form a thin dry lipid film. Films were rehydrated in PBS above TG (55°C) and extruded through polycarbonate membrane filters, with pores of the desired size (i.e. 100 or 200 nm). Nanoallergen size was characterized with dynamic light scattering (DLS) analysis (Figure 2C & D), using 658 nm light observed at a fixed angle of 90°, on a Brookhaven NanoBrook Omni. DLS analyses were performed at the Center for Environmental Science and Technology at the University of Notre Dame.
Liposome extruder purification (LEP)
To determine epitope incorporation, nanoallergens were first purified via liposome extruder purification (LEP), as described previously.46 Briefly, nanoallergens solutions were extruded through 30 nm polycarbonate membranes at room temperature, to remove any unincorporated lipids. Then, pre-LEP (before purification) and post-LEP (purified) samples of each nanoallergen were evaluated via RP-HPLC, and area under the curve (AUC) from the chromatogram of each was determined. Pre-LEP AUC was 100% incorporation; therefore, percent incorporation was calculated as: [post-LEP AUC]/[pre-LEP AUC] x 100%.
Degranulation assays
RBL degranulation assays are performed as previously described.21,43 Briefly, RBL cells are plated at 50,000 cells/mL in a 96-well culture plate (100 μL per well) and allowed to attach overnight until cells are confluent on plate. The media is then removed, and cells are incubated overnight in 10% (v/v) human plasma in full growth media. The next morning, the cells are washed twice with sterile PBS and media is replaced to remove impurities and stabilize the cells before the assay. After a 3-hour incubation, cells are washed twice with Tyrodes buffer, and allergens are introduced to elicit degranulation responses. Positive and negative control wells are included on each plate. Negative control wells contain cells only. Either TritonX or anti-IgE were added to positive control wells. After incubating for 1.5 h, the PNAG substrate is introduced to initiate β-hexosaminidase enzymatic cleavage reactions. The reaction is stopped with glycine buffer (pH 10.7) after 1 hour at 37°C. Enzymatic activity is monitored and analyzed with a SpectraMax M2e plate reader (λ = 405 nm). All degranulation assays are run in triplicate. Degranulation is shown as a percent of the positive control.
A control group was used to confirm the epitopes caused degranulation due to interaction with specific IgE-FcERI complexes. For the “No IgE” group, no IgE or plasma was used to prime the RBL-SX38 cells, instead only fresh media was added during incubation. For the “Isotype Control” group, the RBL-SX38 cells were primed with monoclonal human IgE specific to peanuts. We confirmed that RBL-SX38 cells primed with this antibody do not react to the dust mite allergen (Figure 3B & C). Isotype control was used for all assays, unless otherwise noted.
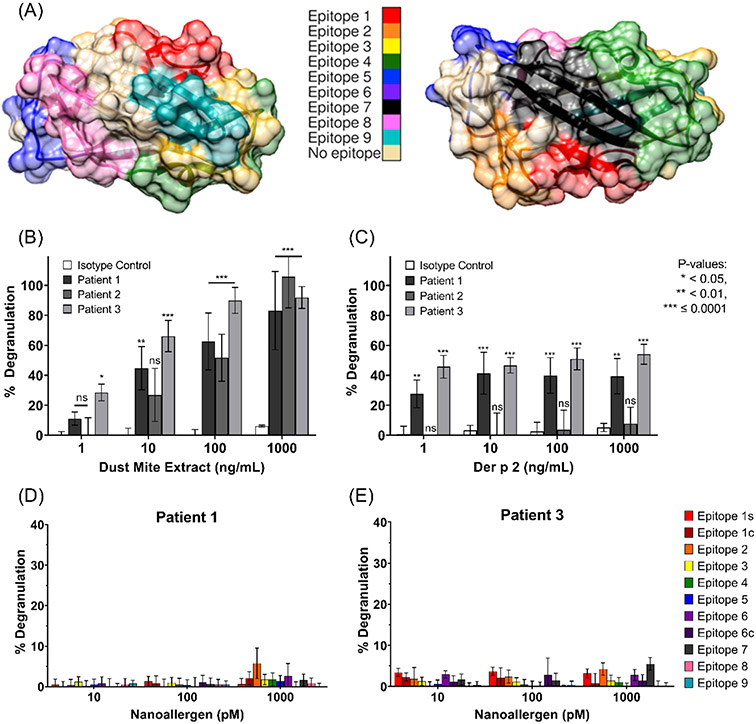
Figure 3.
(A) Der p 2 crystal structure (PDB-ID 1ktj) with epitopes color-coded. Evaluating sensitivity of patient plasma or isotype IgE primed RBL-cells to (B) dust mite extract or (C) Der p 2 via degranulation assays. Cells primed with patient-1 (D) or patient-3 (E) plasma were challenged with nanoallergens presenting copies of a single epitope.
RESULTS
Design, synthesis, and characterization of nanoallergens
The nanoallergen platform provides a means to evaluate epitope immunogenicity with precise control over epitope loading and particle size with low variability between batches (Figure 2).21 The precision in the stoichiometry of loaded epitope per nanoallergen is achieved by assembling the liposome from pure building blocks, including the epitope-presenting peptide-lipid conjugates (Figure 2A & B, eFigure 1A). The epitope-lipid conjugates were produced with solid phase peptide synthesis, using Fmoc chemistry as previously described.43,45,47 The conjugates were purified via reverse phase HPLC (RP-HPLC) and characterized with Q-TOF mass spectrometry (eFigure 1B, eTable 1). A typical concern, when displaying peptides or small molecules on a surface, is that the peptide may hinder itself by becoming buried in the hydrophobic region of the lipid bilayer. To overcome this concern and achieve maximum efficiency in displaying the epitopes on the nanoallergen surface, a linker composed of ethylene glycol (EG) and three lysines was used (eFigure 1A), which enhances the presentation of the epitope to the IgE by improving partitioning of the epitope into the aqueous phase, as previously reported.43
The nanoallergens were prepared from pure components (Figure 2B) via the dry film hydration method, as previously described.21,22,46 Nanoallergen size was characterized with dynamic light scattering (DLS) analysis (Figure 2C & D). For the 200 nm nanoallergens, effective particle diameter ranged 150-175 nm with low polydispersity (<0.17); the 100 nm effective particle diameters ranged 95-120 nm with polydispersity <0.15. Percent epitope loading of >80% incorporation was confirmed using RP-HPLC analysis on nanoallergen samples purified via liposome extruder purification (LEP), as described previously (eFigure 1C & D).46
Cross-analysis of literature-reports to identify allergen epitopes for further immunogenicity evaluation
For Der p 2, literature studies report both continuous (sequential amino acids) and conformational (nonsequential) IgE-binding epitopes. Overlapping portions of both sequential and conformational epitopes reported in multiple literature sources were identified for immunogenic evaluation in this study. The evaluated epitopes are highlighted in the crystal structure of Der p 2 (Figure 3A) and listed in Table 1. Several epitopes were designed and synthesized in multiple configurations (i.e., linear or cyclic; full or truncated), to evaluate whether differences in 3D conformational structure of said epitopes, when presented outside of the protein scaffold, had any meaningful causative effect on their ability to bind sIgE and induce degranulation.
Table 1.
Sequences of the Der p 2 epitopes identified from literature
| # | Sequence | Residues | Notes | References |
|---|---|---|---|---|
| 1s | KDSANHE | 6-12 | Loop without β-sheets | 22,23,30 |
| 1c | CEVDVKDSANHEIKKVLC | 1-18 | N-terminus; cyclized | 22,23 |
| 2 | GCHGSEPCI | 20-28 | cyclized | 15,23-25 |
| 3 | EANQNTKTAK | 42-51 | 15,23,26,27 | |
| 4c | DPNACHYMKCPLVKGQQY | 69-86 | α-helix; cyclized | 15,22,23,25,27,28 |
| 5 | VPKIAPKSEN | 94-103 | 23,27,28 | |
| 6 | KASIDGLEVD | 55-64 | 26,27 | |
| 6c | CSIDGLEC | 56-63 | cyclized | 26,27 |
| 7 | CPFQLEAVGGGDIKYTWNC | 34-40,87-93 | 2 β-sheets, attached via 3 Gly; cyclized | 15,23,26,27 |
| 8 | ASIDGLEVDVPGID | 56-69 | extension of Epitope 6 | 26,27 |
| 9 | CKVMGDDGVLAC | 108-119 | cyclized | 25,27,29 |
The relative immunogenicity of each of the eleven synthesized epitopes was determined using the nanoallergens to display the epitopes and perform degranulation assays. Nanoallergens were loaded with a precisely controlled mole percent (typically 1-8%) of each epitope-lipid conjugate, either presenting copies of a single epitope or a combination of multiple epitopes. Degranulation assays with patient plasma-primed RBL-SX38 cells were performed to compare the intensity of degranulation induced by the nanoallergens.
Verification of patient sample Der p 2 sensitivity
Three samples of plasma were obtained from patients reported to be allergic to dust mites. Since the epitopes of Der p 2 were under closer analysis, it was imperative to verify that the patient samples would respond to dust mite proteins as well as to Der p 2, specifically. Degranulation assays were performed on cells primed with each of the three patient samples and challenged with varying concentrations of either dust mite extract (Figure 3B) or purified Der p 2 (Figure 3C). All three samples demonstrated degranulation to the extract, but only patients 1 and 3 proved to have IgE specific to Der p 2. Therefore, all future assays reported here were performed with patients 1 and 3 only.
Single-epitope immunogenicity analysis with nanoallergen assay
To determine the immunogenicity of the epitopes identified from literature reports, each epitope was loaded at a previously optimized43 valency of 2 mol% on 100 nm nanoallergens, such that nanoallergens presented multivalent copies of a certain epitope. Multivalent presentation of a single epitope or a combination of epitopes on the same platform is necessary for IgE-crosslinking and to trigger degranulation. Patient plasma-primed RBL-SX38 cells were challenged with each of the single-epitope presenting nanoallergens (Figure 3D & E; TritonX positive control, no IgE negative control). Even at the maximum nanoallergen concentrations (1 nM) tested, no significant triggering of degranulation was observed from any of the nanoallergens presenting single epitopes. The nanoallergen concentration was limited to a maximum of 1 nM due to concerns relating to specificity, where controls indicated that some nanoallergens, at concentrations above 1nM, nonspecifically interacted with the cells and yielded potentially unreliable degranulation measurements (data not shown).
Cooperative epitope immunogenicity analysis with nanoallergen degranulation assay
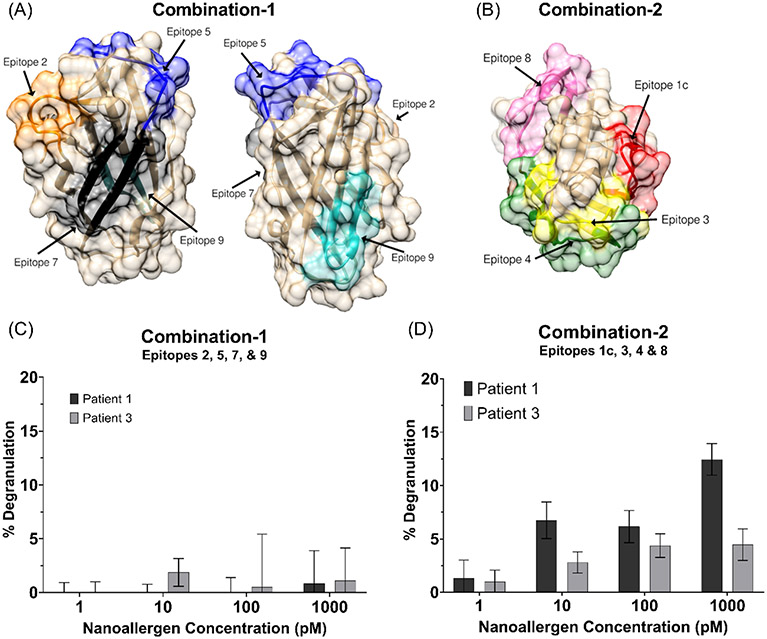
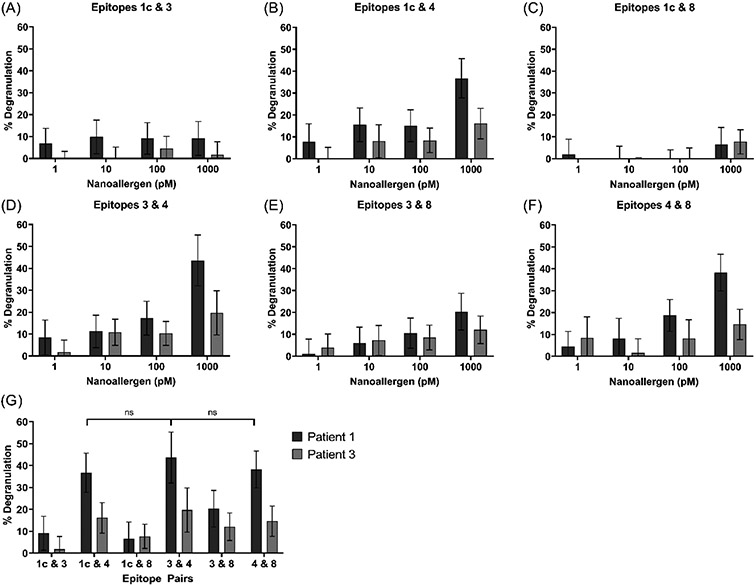
Since nanoallergens presenting single epitopes did not demonstrate any detectable immunogenicity, cooperativity between epitopes was investigated next by presenting the multiple epitopes simultaneously, in combinations, on the same nanoallergen. Using epitopes which are spatially distant on the natural allergen (Figure 4A & 4B), two nanoallergen batches presenting two separate combinations of epitopes were prepared. Nanoallergens presenting combination-1, epitopes 2, 5, 7, and 9, showed no statistically significant degranulation compared to the isotype control (Figure 4C; no IgE negative control). Nanoallergens presenting combination-2, epitopes 1c, 3, 4, and 8, induced degranulation at nanoallergen concentrations of 10-1000 pM for both patient samples (Figure 4D; no IgE negative control). These results indicated that two or more of the epitopes in combination-2 were immunogenic and that the combination of epitopes was having a cooperative effect.
Figure 4.
(A) Location of epitopes 2, 5, 7, and 9 (combination-1) or (B) epitopes 1c, 3, 4, and 8 (combination-2) are shown on Der p 2 crystal structure (PDB-ID 1ktj). Degranulation assays with nanoallergens presenting (C) combination-1 or (D) combination-2 epitopes loaded at 4% total on 100 nm liposomes.
To improve the signal-to-noise ratio of the assay, combination-2 nanoallergens were used to optimize nanoallergen diameter and percent epitope loading for maximum degranulation sensitivity with minimum nonspecificity (eFigure 2). For subsequent experiments described below, 200nm nanoallergens and 4% total loading were used.
Nanoallergens (200 nm, 4% total peptide) loaded with pairs of epitopes from combination-2, (i.e. epitopes 1c&3, 1c&4, 1c&8, 3&4, 3&8, 4&8), were prepared and used in degranulation assays. Analysis of the results with the epitope pair nanoallergens indicated that epitope 4 had the strongest observable immunogenicity among combination-2 epitopes, as it produced degranulation when paired with any of the other three epitopes (Figure 5B, D, F). Epitope 1c was the least immunogenic of the four, inducing no significant degranulation when paired with either epitope 3 or 8 (Figure 5A & 5C). Degranulation induced by epitope pair 3&8 indicated these epitopes have slightly higher immunogenicity than epitope 1c, as the epitope pair 3&8 induced 2-fold higher degranulation on average than either 1c&3 or 1c&8 (Figure 5E & 5G).
Figure 5.
Degranulation triggered by 200 nm nanoallergens presenting 4% total loading of each epitope pair: (A) 1c & 3, (B) 1c & 4, (C) 1c & 8, (D) 3 & 4, (E) 3 & 8, and (F) 4 & 8. (G) Comparison of degranulation due to pairs of epitopes, as observed at 1000 pM nanoallergen concentration.
For patient 1, at 1 nM nanoallergen concentration, epitope pairs 1c&4, 3&4, and 4&8 all achieved 36-44% degranulation in comparison to only 20% achieved with epitope pair 3&8 (Figure 5G). While the degranulation response was 2-to-3-fold lower for patient 3 than for patient 1, patient 3 showed similar trends in immunogenicity.
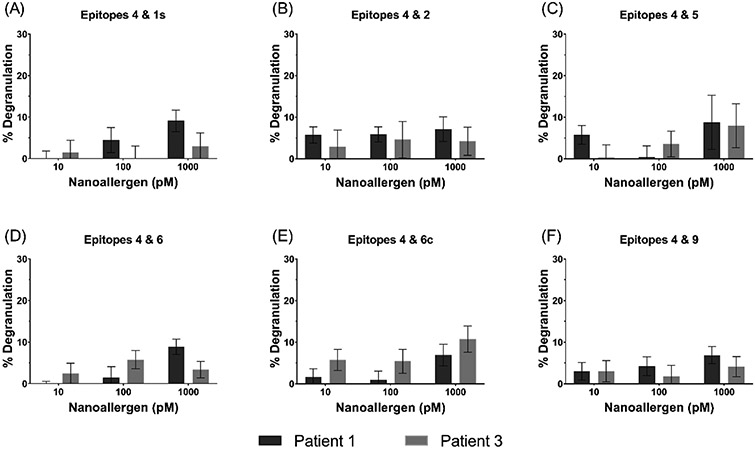
Epitopes 1s, 2, 5, 6, 6c, and 9 also triggered degranulation, to varying degrees, when presented in combination with epitope 4. These results can be seen in Figure 6.
Figure 6.
Cooperativity analysis of epitopes 1s, 2, 5, 6, 6c, or 9 paired with epitope 4.
DISCUSSION
Here, we described how nanoallergens were used to identify the relative immunogenicity of allergen epitopes of dust mite protein Der p 2. The results of this study, upon evaluation of linear and cyclic versions, demonstrated that sequential epitopes of Der p 2 were not highly immunogenic on their own. Instead, presenting combinations of Der p 2 epitopes was necessary to produce significant cellular response, suggesting that immunodominant Der p 2 epitopes had to be conformational. The well-formulated nanoallergens made it possible to identify cooperative immunogenicity among epitopes 1c, 3, 4, and 8 and to rank the relative levels of immunogenicity between these epitopes. In the end, epitope 4 proved to be the most immunogenic, followed by epitopes 3 and 8, and with epitope 1c the least immunogenic. The remainder of the literature-reported epitopes did not display any significant immunogenicity in our analysis. It is noteworthy that the epitopes identified throughout this report align with results from a recently published study that determined two regions on Der p 2 to which certain human monoclonal IgE bind.48 Specifically, one of these regions supports our identification of epitopes 3 and 4 as immunogenic epitopes, while the other region partially overlaps with epitope 8. While powerful, previous studies required complicated NMR-based evaluations, which necessitate large financial input, time-consuming processes, and difficult to obtain resources, such as large amounts of purified human monoclonal IgE. Our methodology, on the other hand, achieved similar results with the heterogeneous pool of IgE in patient samples, at much lower cost and without the need for synthesis of human monoclonal IgE.
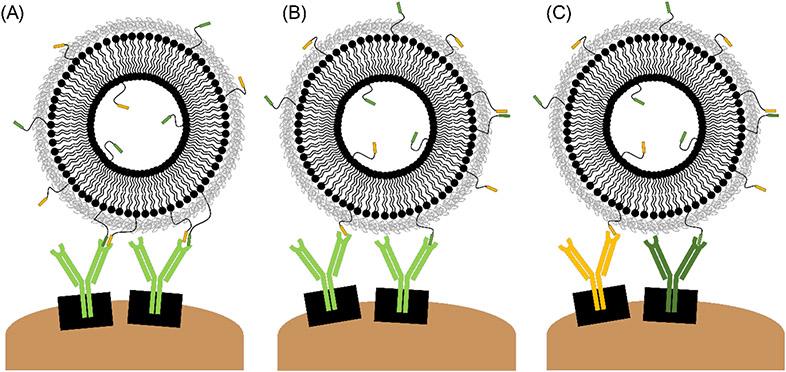
Given the proximity of epitopes 3 and 4 on the natural allergen (Figure 4B), they are unlikely to be simultaneously binding to two separate sIgE, as steric hindrances would prevent two IgE from binding to two very closely positioned sites. We hypothesize that the mechanism by which these epitopes induce degranulation, when presented on a nanoallergen, is related to one or more of the following binding scenarios: i) portions of Epitopes 3 and 4 may simultaneously bind to the antigen binding site of a single IgE clone, analogous to the interaction of an IgE with a conformational epitope (Figure 7A), ii) alternatively, it is possible epitopes 3 and 4 are recognized individually by the same IgE clone, albeit with different affinity for each epitope (Figure 7B), or iii) two distinct IgE clones may exist, one that binds epitope 3 and one that binds epitope 4. (Figure 7C). If the first case was true, it is possible that epitope 3 provided structural support to keep epitope 4 in its suitable conformation for binding, or vice versa. Conversely, epitopes 1c and 8 are located more spatially distant from epitope 4 on the natural allergen, therefore we would expect epitopes 1c, 8 and 4 to be binding to different monoclonal sIgE (Figure 7C).
Figure 7.
Hypothesized binding scenarios for cooperative epitopes may occur individually or in combination: (A) Two epitopes bind to the same antigen binding site. (B) Two epitopes bind to separate copies of the same IgE. (C) Epitopes bind to separate IgE specific for different regions of the allergen. Not to scale.
In the case of epitope pairs 1c&4 and 4&8, we believe that epitope 4, possibly a higher affinity epitope, may be acting as an anchor (Figure 7C). Association rate constants for biomolecular interactions are generally independent of overall binding affinity (i.e., the on-rate constant (kon) should be similar for most epitopes). Instead, variations in dissociation rate constants (koff), as well as relative abundance of each sIgE in the patient sample, are expected to correlate to the residence time of the nanoallergen staying bound to the cell-surface IgE. Epitope 4 could have served as an anchor by providing sufficient residence time for cellular receptor-IgE complexes to rearrange and bind to the other epitopes on a nanoallergen, thereby promoting crosslinking and initiating degranulation. We predict that the pairing of epitopes 3&8 acts in a similar manner, as these epitopes are also positioned on opposite ends of the natural allergen.
As reported in previous publications, identification of immunodominant epitopes has led to the development of inhibitors, specific for the immunogenic sIgE, which successfully blocked or reduced the intensity of allergic responses to peanuts and drug allergens.44,45,49 Moreover, identification of immunogenic sequential epitopes (which may be a portion of, or mimic, conformational epitopes) provides a more cost-effective and technically feasible roadmap to the development of allergen-specific inhibitors, compared to conformational epitopes. Thus, we predict that understanding and ranking HDM epitope immunogenicity will aid in the development of a potential preventative treatment for HDM-related asthma. The extension of such technology to the clinic could translate to personalized medicine, as diagnostic identification of immunogenic epitopes in patients can allow for use of personalized therapeutics for inhibition of specific IgE relevant to the patient’s allergy. Using a platform such as nanoallergens could determine which epitopes a patient is allergic to, via an ex vivo diagnostic similar to those described in this study or as a skin prick test. Then epitope-specific therapeutics would allow for more accurate and specific treatment of allergic condition. For example, identification of immunodominant epitopes can allow for the development of IgE-specific inhibitors, which could treat HDM allergies and asthma by selectively blocking IgE from binding to HDM allergens. Another potential approach would be developing sIgG against the immunogenic HDM epitopes, which could be used as a monoclonal antibody therapy to capture allergens before allergen-IgE-FcεRI binding can occur.50 IgG that is randomly generated from the parent allergen will not be guaranteed to block or outcompete IgE binding. Therefore, if a patient is administered only the IgG specific to the relevant epitopes, instead of a mixture of IgG of varying efficacy, the cost of the IgG therapy should decrease.
In conclusion, this report described a novel method to investigate HDM allergic asthma at the molecular level, and a potential new approach to inhibitor development for this condition. Inarguably, the more alternative approaches clinicians have in their arsenal with which to treat asthma and allergies, the better their chances are to effectively treat the condition and improve patient outcomes.
Supplementary Material
FUNDING SOURCE:
NIH (award # R01 AI108884)
Footnotes
CONFLICT OF INTEREST: none
REFERENCES
- 1.National Center for Health Statistics. 2019 National Health Interview Survey (NHIS) Data.; 2020. Accessed January 6, 2022. https://www.cdc.gov/asthma/nhis/2019/table3-1.htm
- 2.Center for Disease Control and Prevention. AsthmaStats.; 2019. Accessed January 5, 2022. https://www.cdc.gov/asthma/asthma_stats/default.htm
- 3.National Hospital Ambulatory Medical Care Survey. Asthma-Related Healthcare Use Data.; 2018. Accessed January 6, 2022. https://www.cdc.gov/asthma/healthcare-use/2018/table_a.html
- 4.Comberiati P, di Cicco ME, D’Elios S, Peroni DG. How much asthma is atopic in children? Frontiers in Pediatrics. 2017;5. doi: 10.3389/fped.2017.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ali FR. Does this patient have atopic asthma? Clinical Medicine, Journal of the Royal College of Physicians of London. 2011;11(4):376–380. doi: 10.7861/clinmedicine.11-4-376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akar-Ghibril N, Casale T, Custovic A, Phipatanakul W. Allergic Endotypes and Phenotypes of Asthma. Journal of Allergy and Clinical Immunology: In Practice. 2020;8(2):429–440. doi: 10.1016/j.jaip.2019.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prabhala P, Wright DB, Robbe P, et al. Laminin α4 contributes to airway remodeling and inflammation in asthma. American Journal of Physiology-Lung Cellular and Molecular Physiology. Published online September 25, 2019:ajplung.00222.2019. doi: 10.1152/ajplung.00222.2019 [DOI] [PubMed] [Google Scholar]
- 8.Calderón MA, Linneberg A, Kleine-Tebbe J, et al. Respiratory allergy caused by house dust mites: What do we really know? Journal of Allergy and Clinical Immunology. 2015;136(1):38–48. doi: 10.1016/j.jaci.2014.10.012 [DOI] [PubMed] [Google Scholar]
- 9.Eifan AO, Calderon MA, Durham SR. Allergen immunotherapy for house dust mite: clinical efficacy and immunological mechanisms in allergic rhinitis and asthma. Expert Opinion on Biological Therapy. 2013;13(11):1543–1556. doi: 10.1517/14712598.2013.844226 [DOI] [PubMed] [Google Scholar]
- 10.Arbes SJ, Cohn RD, Yin M, et al. House dust mite allergen in US beds: Results from the first National Survey of Lead and Allergens in Housing. Journal of Allergy and Clinical Immunology. 2003;111(2):408–414. doi: 10.1067/mai.2003.16 [DOI] [PubMed] [Google Scholar]
- 11.Portnoy J, Miller JD, Williams PB, et al. Environmental assessment and exposure control of dust mites: a practice parameter. Annals of allergy, asthma & immunology : official publication of the American College of Allergy, Asthma, & Immunology. 2013;111(6):465–507. doi: 10.1016/J.ANAI.2013.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arlian LG, Neal JS, Morgan MS, Vyszenski-Moher DL, Rapp CM, Alexander AK. Reducing relative humidity is a practical way to control dust mites and their allergens in homes in temperate climates. Journal of Allergy and Clinical Immunology. 2001;107(1):99–104. doi: 10.1067/MAI.2001.112119 [DOI] [PubMed] [Google Scholar]
- 13.Wilson JM, Platts-Mills TAE. Home Environmental Interventions for House Dust Mite. The Journal of Allergy and Clinical Immunology: In Practice. 2018;6(1):1–7. doi: 10.1016/JJAIP.2017.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calderón MA, Kleine-Tebbe J, Linneberg A, et al. House Dust Mite Respiratory Allergy: An Overview of Current Therapeutic Strategies. The Journal of Allergy and Clinical Immunology: In Practice. 2015;3(6):843–855. doi: 10.1016/j.jaip.2015.06.019 [DOI] [PubMed] [Google Scholar]
- 15.Tovey ER, Marks GB. It’s time to rethink mite allergen avoidance. Journal of Allergy and Clinical Immunology. 2011;128(4):723–727. doi: 10.1016/j.jaci.2011.07.009 [DOI] [PubMed] [Google Scholar]
- 16.Karakoc-Aydiner E, Eifan AO, Baris S, et al. Long-term effect of sublingual and subcutaneous immunotherapy in dust mite–allergic children with asthma/rhinitis: A 3-year prospective randomized controlled trial. Journal of Investigational Allergology and Clinical Immunology. 2015;25(5):334–342. [PubMed] [Google Scholar]
- 17.Tortajada-Girbés M, Mesa del Castillo M, Larramona H, et al. Decision-making for pediatric allergy immunotherapy for aeroallergens: a narrative review. European Journal of Pediatrics. 2019;178(12):1801–1812. doi: 10.1007/s00431-019-03444-2 [DOI] [PubMed] [Google Scholar]
- 18.Calderon MA, Casale TB, Nelson HS, Demoly P. An evidence-based analysis of house dust mite allergen immunotherapy: a call for more rigorous clinical studies. The Journal of allergy and clinical immunology. 2013;132(6):1322–1336. doi: 10.1016/J.JACI.2013.09.004 [DOI] [PubMed] [Google Scholar]
- 19.Gould HJ, Sutton BJ. IgE in allergy and asthma today. Nature Reviews Immunology. 2008;8(3):205–217. doi: 10.1038/nri2273 [DOI] [PubMed] [Google Scholar]
- 20.Shamji MH, Valenta R, Jardetzky T, et al. The role of allergen-specific IgE, IgG and IgA in allergic disease. Allergy. 2021;00:1–15. doi: 10.1111/ALL.14908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deak PE, Vrabel MR, Pizzuti VJ, Kiziltepe T, Bilgicer B. Nanoallergens: A multivalent platform for studying and evaluating potency of allergen epitopes in cellular degranulation. Experimental Biology and Medicine. 2016;241(9):996–1006. doi: 10.1177/1535370216644533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noble GT, Stefanick JF, Ashley JD, Kiziltepe T, Bilgicer B. Ligand-targeted liposome design: Challenges and fundamental considerations. Trends in Biotechnology. 2014;32(1):32–45. doi: 10.1016/j.tibtech.2013.09.007 [DOI] [PubMed] [Google Scholar]
- 23.Deak PE, Kim B, Adnan A, et al. Nanoallergen platform for detection of platin drug allergies. Journal of Allergy and Clinical Immunology. 2019;143(5):1957–1960.e12. doi: 10.1016/j.jaci.2019.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caro JA, Harpole KW, Kasinath V, et al. Entropy in molecular recognition by proteins. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(25):6563–6568. doi: 10.1073/pnas.1621154114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verteramo ML, Stenström O, Ignjatović MM, et al. Interplay between Conformational Entropy and Solvation Entropy in Protein-Ligand Binding. Journal of the American Chemical Society. 2019;141(5):2012–2026. doi: 10.1021/JACS.8B11099/SUPPL_FILE/JA8B11099_SI_001.PDF [DOI] [PubMed] [Google Scholar]
- 26.Glesner J, Kapingidza AB, Godzwon M, et al. A Human IgE Antibody Binding Site on Der p 2 for the Design of a Recombinant Allergen for Immunotherapy. The Journal of Immunology. 2019;9(24):ji1900580. doi: 10.4049/jimmunol.1900580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chapter 5 : Indoor Air Pollutants and Toxic Materials. In: Healthy Housing Reference Manual. US Department of Health and Human Services; 2006:1–26. Accessed February 17, 2020. https://www.cdc.gov/nceh/publications/books/housing/cha05.htm [Google Scholar]
- 28.Arlian LG, Bernstein D, Bernstein IL, et al. Prevalence of dust mites in the homes of people with asthma living in eight different geographic areas of the United States. The Journal of Allergy and Clinical Immunology. 1992;90(3 PART 1):292–300. doi: 10.1016/S0091-6749(05)80006-5 [DOI] [PubMed] [Google Scholar]
- 29.Thomas WR, Hales BJ, Smith WA. House dust mite allergens in asthma and allergy. Trends in Molecular Medicine. 2010;16(7):321–328. doi: 10.1016/J.M0LMED.2010.04.008 [DOI] [PubMed] [Google Scholar]
- 30.Reginald K, Chew FT. The major allergen Der p 2 is a cholesterol binding protein. Published online 2019:1–8. doi: 10.1038/s41598-018-38313-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hof WV t., Driedijk PC, Van Den Berg M, Beck-sickinger AG, Jung G, Aalberse RC. Epitope mapping of the Dermatophagoides pteronyssinus house dust mite major allergen Der p II using overlapping synthetic peptides. Molecular Immunology. 1991;28(11):1225–1232. doi: 10.1016/0161-5890(91)90009-9 [DOI] [PubMed] [Google Scholar]
- 32.Hakkaart GAJ, Chapman MD, Aalberse RC, Van Ree R. Immune-reactivity of recombinant isoforms of the major house dust mite allergen Der p 2. Clinical and Experimental Allergy. 1998;28(2):169–174. doi: 10.1046/j.1365-2222.1998.00205.x [DOI] [PubMed] [Google Scholar]
- 33.Hakkaart GAJ, Aalberse RC, Van Ree R. Epitope mapping of the house-dust-mite allergen Der p 2 by means of site-directed mutagenesis. Allergy: European Journal of Allergy and Clinical Immunology. 1998;53(2):165–172. doi: 10.1111/j.1398-9995.1998.tb03865.x [DOI] [PubMed] [Google Scholar]
- 34.Smith AM, Chapman MD. Reduction in IgE binding to allergen variants generated by site-directed mutagenesis: Contribution of disulfide bonds to the antigenic structure of the major house dust mite allergen Der p 2. Molecular Immunology. 1996;33(4-5):399–405. doi: 10.1016/0161-5890(95)00150-6 [DOI] [PubMed] [Google Scholar]
- 35.Yong TS, Lee SM, Park GM, et al. Monoclonal antibodies to recombinant Der p 2, a major house dust mite allergen: specificity, epitope analysis and development of two-site capture ELISA. The Korean journal of parasitology. 1999;37(3):163–169. doi: 10.3347/kjp.1999.37.3.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobayashi I, Sakiyama Y, Tame A, Kobayashi K, Matsumoto S. IgE and IgG4 antibodies from patients with mite allergy recognize different epitopes of Dermatophagoides pteronyssinus group II antigen (Der p 2). Journal of Allergy and Clinical Immunology. 1996;97(2):638–645. doi: 10.1016/S0091-6749(96)70309-3 [DOI] [PubMed] [Google Scholar]
- 37.Szalai K, Fuhrmann J, Pavkov T, et al. Mimotopes identify conformational B-cell epitopes on the two major house dust mite allergens Der p 1 and Der p 2. 2008;45:1308–1317. doi: 10.1016/j.molimm.2007.09.012 [DOI] [PubMed] [Google Scholar]
- 38.Smith AM, Chapman MD. Localization of antigenic sites on Der p 2 using oligonucleotide-directed mutagenesis targeted to predicted surface residues. Clinical and Experimental Allergy. 1997;27(5):593–599. doi: 10.1111/j.1365-2222.1997.tb00750.x [DOI] [PubMed] [Google Scholar]
- 39.Derewenda U, Li J, Derewenda Z, et al. The crystal structure of a major dust mite allergen Der p 2, and its biological implications. Journal of Molecular Biology. 2002;318(1):189–197. doi: 10.1016/S0022-2836(02)00027-X [DOI] [PubMed] [Google Scholar]
- 40.Chen KW, Focke-Tejkl M, Blatt K, et al. Carrier-bound nonallergenic Der p 2 peptides induce IgG antibodies blocking allergen-induced basophil activation in allergic patients. Allergy: European Journal of Allergy and Clinical Immunology. 2012;67(5):609–621. doi: 10.1111/j.1398-9995.2012.02794.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Handlogten MW, Kiziltepe T, Serezani AP, Kaplan MH, Bilgicer B. Inhibition of weak-affinity epitope-IgE interactions prevents mast cell degranulation. Nature Chemical Biology. 2013;9(12):789–795. doi: 10.1038/nchembio.1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Handlogten MW, Kiziltepe T, Alves NJ, Bilgicer B. Synthetic allergen design reveals the significance of moderate affinity epitopes in mast cell degranulation. ACS Chemical Biology. 2012;7(11):1796–1801. doi: 10.1021/cb300193f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deak PE, Vrabel MR, Kiziltepe T, Bilgicer B. Determination of Crucial Immunogenic Epitopes in Major Peanut Allergy Protein, Ara h2, via Novel Nanoallergen Platform. Scientific Reports. 2017;7(1):1–13. doi: 10.1038/s41598-017-04268-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deak PE, Kim B, Koh B, et al. Covalent Heterobivalent Inhibitor Design for Inhibition of IgE-Dependent Penicillin Allergy in a Murine Model. The Journal of Immunology. 2019;203(1):21–30. doi: 10.4049/jimmunol.1900225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deak PE, Kim B, Abdul Qayum A, et al. Designer covalent heterobivalent inhibitors prevent IgE-dependent responses to peanut allergen. Proceedings of the National Academy of Sciences. 2019;116(18):8966–8974. doi: 10.1073/pnas.1820417116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alves NJ, Cusick W, Stefanick JF, Ashley JD, Handlogten MW, Bilgicer B. Functionalized liposome purification via Liposome Extruder Purification (LEP). Analyst. 2013;138(17):4746–4751. doi: 10.1039/c3an00680h [DOI] [PubMed] [Google Scholar]
- 47.Handlogten MW, Kiziltepe T, Bilgicer B. Design of a heterotetravalent synthetic allergen that reflects epitope heterogeneity and IgE antibody variability to study mast cell degranulation. Biochemical Journal. 2013;449(1):91–99. doi: 10.1042/BJ20121088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.GA M, J G, JL D, et al. Mapping Human Monoclonal IgE Epitopes on the Major Dust Mite Allergen Der p 2. Journal of immunology (Baltimore, Md: 1950). 2020;205(8):1999–2007. doi: 10.4049/JIMMUNOL.2000295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Handlogten MW, Serezani AP, Sinn AL, Pollok KE, Kaplan MH, Bilgicer B. A Heterobivalent Ligand Inhibits Mast Cell Degranulation via Selective Inhibition of Allergen–IgE Interactions In Vivo. The Journal of Immunology. 2014;192(5):2035–2041. doi: 10.4049/jimmunol.1301371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orengo JM, Radin AR, Kamat V, et al. Treating cat allergy with monoclonal IgG antibodies that bind allergen and prevent IgE engagement. Nature Communications 2018 9:1. 2018;9(1):1–15. doi: 10.1038/s41467-018-03636-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.