Abstract
BACKGROUND
Human adenoviruses typically cause self-limited respiratory, gastrointestinal, and conjunctival infections in healthy children. In late 2021 and early 2022, several previously healthy children were identified with acute hepatitis and human adenovirus viremia.
METHODS
We used International Classification of Diseases, 10th Revision, codes to identify all children (<18 years of age) with hepatitis who were admitted to Children’s of Alabama hospital between October 1, 2021, and February 28, 2022; those with acute hepatitis who also tested positive for human adenovirus by whole-blood quantitative polymerase chain reaction (PCR) were included in our case series. Demographic, clinical, laboratory, and treatment data were obtained from medical records. Residual blood specimens were sent for diagnostic confirmation and human adenovirus typing.
RESULTS
A total of 15 children were identified with acute hepatitis — 6 (40%) who had hepatitis with an identified cause and 9 (60%) who had hepatitis without a known cause. Eight (89%) of the patients with hepatitis of unknown cause tested positive for human adenovirus. These 8 patients plus 1 additional patient referred to this facility for follow-up were included in this case series (median age, 2 years 11 months; age range, 1 year 1 month to 6 years 5 months). Liver biopsies indicated mild-to-moderate active hepatitis in 6 children, some with and some without cholestasis, but did not show evidence of human adenovirus on immunohistochemical examination or electron microscopy. PCR testing of liver tissue for human adenovirus was positive in 3 children (50%). Sequencing of specimens from 5 children showed three distinct human adenovirus type 41 hexon variants. Two children underwent liver transplantation; all the others recovered with supportive care.
CONCLUSIONS
Human adenovirus viremia was present in the majority of children with acute hepatitis of unknown cause admitted to Children’s of Alabama from October 1, 2021, to February 28, 2022, but whether human adenovirus was causative remains unclear. Sequencing results suggest that if human adenovirus was causative, this was not an outbreak driven by a single strain. (Funded in part by the Centers for Disease Control and Prevention.)
Human adenoviruses are double-stranded DNA viruses that cause a range of illnesses in humans, including upper respiratory symptoms, fever, pneumonia, gastroenteritis, and conjunctivitis.1 A total of 51 serotypes of human adenoviruses are recognized, and more than 85 genotypes (or “types”) have been described.2 Different types of human adenovirus have different tissue tropism, meaning that a specific type may cause more focal illness by targeting certain organs or organ systems. In immunocompetent patients with human adenovirus infection, liver involvement is typically limited to subclinical acute hepatitis, although severe hepatitis or acute liver failure is sometimes seen in immunocompromised patients.3–6 Published reports of severe hepatitis or acute liver failure associated with human adenovirus in immunocompetent children remain limited.7,8
Clinicians at Children’s of Alabama hospital noted five patients with acute hepatitis who had been admitted in October 2021 and were found to have human adenovirus viremia. On recognition of these patients, in November 2021, clinicians alerted the Alabama Department of Public Health, the Jefferson County Department of Health, and the Centers for Disease Control and Prevention (CDC). This prompted prospective monitoring of additional children who were evaluated for hepatitis and had human adenovirus viremia; four additional patients with acute hepatitis and human adenovirus viremia were identified by February 28, 2022.
To further understand the etiologic and pathophysiological factors contributing to development of hepatitis in these nine patients, we evaluated the demographic, clinical, and virologic characteristics of these patients, including histopathological and molecular findings from liver biopsies and human adenovirus partial hexon genomic sequencing (typing) results from blood specimens.
Methods
Oversight
This investigation was approved by the institutional review board of the University of Alabama, Birmingham, with an approved waiver of informed consent. Nevertheless, informed consent granting permission to publish case information was obtained from the families of the prospective cohort and was documented in the electronic medical records. This investigation was reviewed by the CDC and was conducted in accordance with applicable federal law and CDC policy (see the Supplementary Appendix, available with the full text of this article at NEJM.org).
Patients
From October 1, 2021, to February 28, 2022, clinicians at Children’s of Alabama prospectively identified nine children with acute hepatitis (as defined below) of unknown cause and human adenovirus viremia. To put these patients into epidemiologic context and capture any additional, missed patients, we conducted a systematic, retrospective review of all patients younger than 18 years of age who were admitted with acute hepatitis (defined on the basis of admission International Classification of Diseases, 10th Revision, codes for hepatitis [K75.9], transaminitis [R74.01], elevated liver enzymes [R74.8], liver failure [K72.9], and acute liver failure [K72.00]). All children who were identified as having hepatitis were then assigned to one of two categories: those with an identified cause of their hepatitis and those with an unknown cause. Those with an unknown cause were further categorized on the basis of quantitative polymerase-chain-reaction (qPCR) testing for human adenovirus. Patients with acute hepatitis and human adenovirus viremia (without another cause identified) were included in this case series.
Definitions and Testing
To provide additional epidemiologic context, all children who had whole blood tested for human adenovirus by qPCR at Children’s of Alabama during the period from October 1, 2021, to February 28, 2022 (i.e., the same period), were reviewed by the laboratory, and the percentage of children who tested positive was calculated. In addition, a second retrospective systematic chart review was performed to identify all patients with acute hepatitis who had been admitted to Children’s of Alabama from October 1, 2020, to September 30, 2021 (the year immediately preceding the identification of the cases described in this case series; see the Supplementary Appendix).
Clinicians involved in the medical care of patients included in this case series reviewed electronic medical records and extracted demographic and clinical information. Diagnostic testing for acute hepatitis was not standardized.
Acute hepatitis was defined as levels of alanine aminotransferase (ALT) or aspartate aminotransferase (AST) that were elevated to at least 10 times the upper limit of the normal range (>250 U per liter for ALT and >440 U per liter for AST). Pediatric acute liver failure was defined as an international normalized ratio (INR) of at least 1.5 with documented hepatic encephalopathy or an INR of at least 2.0 regardless of hepatic encephalopathy, no history of liver disease in the previous 8 weeks, and no improvement in the INR after parenteral vitamin K treatment.9
Human adenovirus viremia was defined as a positive human adenovirus viral load detected in whole blood on qPCR testing performed at the University of Alabama at Birmingham Diagnostic Virology Laboratory. The same laboratory also performed human adenovirus PCR on available fresh-frozen liver tissue following a protocol similar to that used for whole blood (additional details are provided in the Supplementary Appendix).
The CDC received formalin-fixed, paraffin-embedded (FFPE) liver-biopsy specimens and completed immunohistochemical analysis for human adenovirus and conventional human adenovirus–specific PCR with an assay targeting the hexon gene, followed by Sanger sequencing of amplicons10,11; this laboratory also completed reverse-transcriptase (RT)–PCR assays for enterovirus and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) on RNA extracted from FFPE liver tissue.12,13
Available residual blood specimens were sent to the Wadsworth Center, New York State Department of Health (Albany), for human adenovirus typing performed by amplifying and sequencing a portion of the hexon gene.14 Sequences generated in this study were deposited in GenBank, and phylogenetic trees were constructed to superimpose sequences from human adenovirus isolated from patients with hepatitis on other background human adenovirus sequences (additional details are provided in the Supplementary Appendix).
Results
Patients Included in Case Series
Between October 1, 2021, and February 28, 2022, a total of 15 patients younger than 18 years of age with acute hepatitis were admitted to Children’s of Alabama. In 6 of these patients (40%), the underlying cause of hepatitis had been identified; the causes included cholangitis associated with complications from biliary atresia (1 patient), newly diagnosed autoimmune hepatitis (2 patients), shocked liver after a code event (1 patient), acetaminophen overdose (1 patient), and complicated urinary tract infection (1 patient). Five of the patients with identified underlying causes of their hepatitis also had whole blood tested by qPCR for human adenovirus, and none had positive results (Fig. 1).
Figure 1. Acute Hepatitis Cases Identified by Retrospective Review.
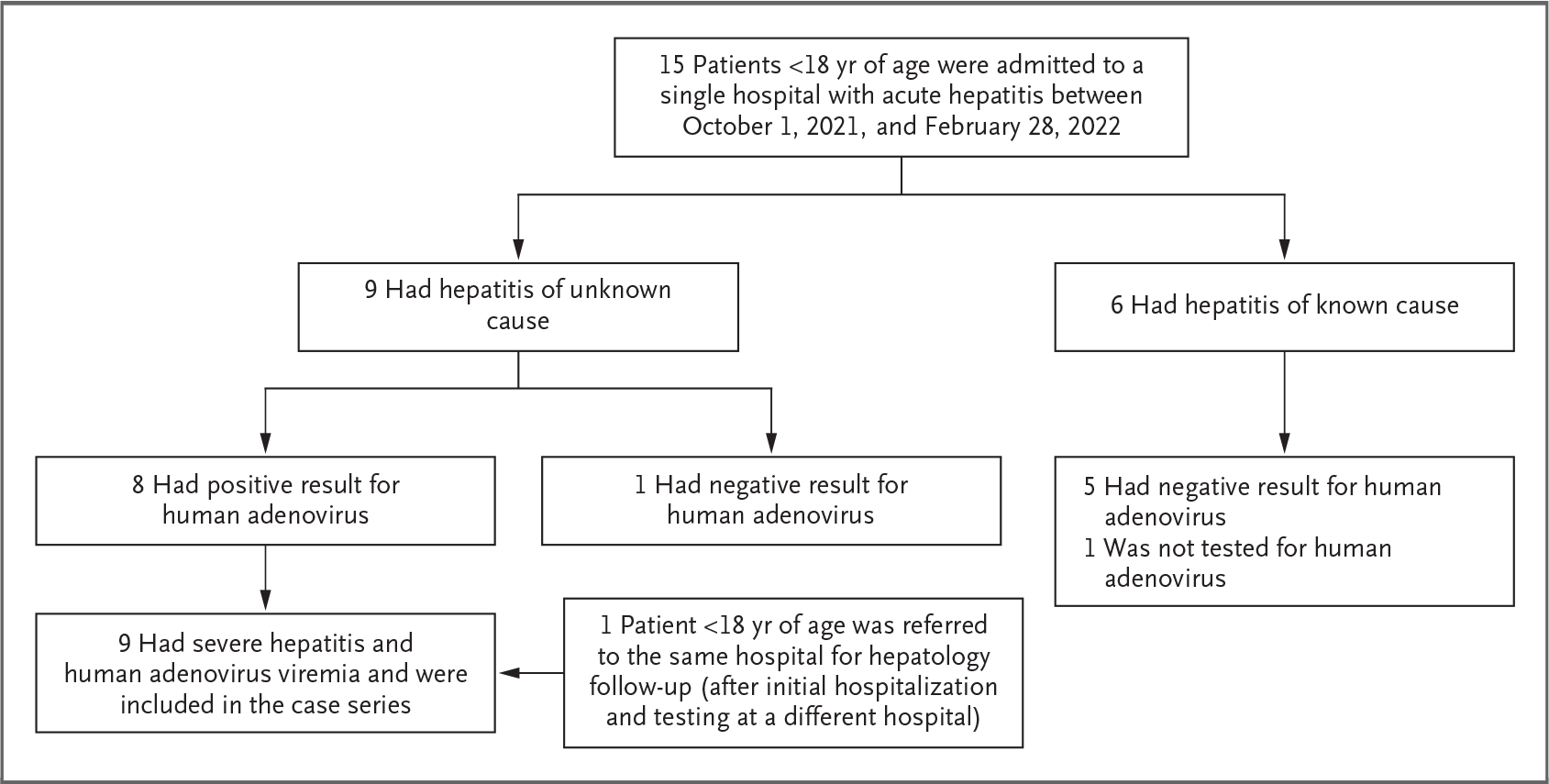
Patients younger than 18 years of age with the following International Classification of Diseases, 10th Revision, codes at admission were included in the case series: hepatitis (K75.9), transaminitis (R74.01), elevated liver enzymes (R74.8), liver failure (K72.9), and acute liver failure (K72.00). Identified causes of hepatitis included cholangitis associated with complications of biliary atresia (1 patient), autoimmune hepatitis (2 patients), liver shock after a code event (1 patient), acetaminophen overdose (1 patient), and complicated urinary tract infection (1 patient). A total of 9 patients with acute hepatitis and human adenovirus viremia were included in this case series.
In 9 patients (60%), the primary cause of hepatitis had not been determined. All 9 patients were tested for human adenovirus by qPCR on whole blood, and 8 (89%) tested positive. For context, during this same period, the virology laboratory used by Children’s of Alabama (the University of Alabama at Birmingham Diagnostic Virology Laboratory) tested 106 children for human adenovirus by whole-blood qPCR, and 10 (9%) were positive.
Clinical Presentation and Diagnostic Testing
Eight of the children with acute hepatitis and human adenovirus viremia were originally admitted to Children’s of Alabama; one additional child with acute hepatitis and human adenovirus viremia was initially admitted to a different hospital but later underwent hepatology follow-up at Children’s of Alabama. All nine children had previously been healthy. The median age at admission was 2 years 11 months (range, 1 year 1 month to 6 years 5 months). Additional demographic characteristics are listed in Table 1. The nine children resided in different areas of Alabama, and none of the children were related to one another; all resided in different households, none attended the same day care facility or had known contact with each other, and no shared exposures were identified.
Table 1.
Demographic and Clinical Characteristics of Nine Patients with Acute Hepatitis and Human Adenovirus Viremia, Alabama, October 2021-February 2022.*
| Characteristic | Value (N = 9) |
|---|---|
| Demographic characteristics | |
| Age at admission — no. (%) | |
| 0–2 yr | 5 (56) |
| 3–4 yr | 1 (11) |
| 5–6 yr | 3 (33) |
| Sex — no. (%) | |
| Female | 7 (78) |
| Male | 2 (22) |
| Race — no. (%)† | |
| White | 9 (100) |
| Other | 0 |
| Ethnic group — no. (%)† | |
| Hispanic | 6 (67) |
| Non-Hispanic | 3 (33) |
| Clinical characteristics | |
| Reported symptoms at admission — no. (%) | |
| Emesis | 7 (78) |
| Diarrhea | 6 (67) |
| Fever | 5 (56) |
| Fatigue | 4 (44) |
| Upper respiratory symptoms‡ | 3 (33) |
| Poor appetite | 3 (33) |
| Dark urine | 2 (22) |
| Findings on initial physical examination — no. (%) | |
| Scleral icterus | 8 (89) |
| Hepatomegaly | 7 (78) |
| Jaundice | 6 (67) |
| Hepatic encephalopathy | 1 (11) |
| Splenomegaly | 1 (11) |
| Ascites | 0 |
| Median liver-function measures at admission (range)§ | |
| ALT — U/liter | 1724 (602–4696) |
| AST — U/liter¶ | 1963 (447–4000) |
| Total bilirubin — mg/dl | 7.0 (0.2–13.5) |
| INR | 1.2 (1.0–7.3) |
| Ammonia — μmol/liter | 73 (49–85) |
| Median peak liver-function measures (range)§ | |
| ALT — U/liter | 1724 (602–4696) |
| AST — U/liter | 1963 (447–4000) |
| Total bilirubin — mg/dl | 7.0 (0.2–28.8) |
| INR | 1.3 (1.1–9.1) |
| Ammonia — μmol/liter | 64 (49–210) |
| Pathogen tests | |
| Adenovirus whole-blood qPCR — no. positive/no. tested (%) | 9/9 (100) |
| Median human adenovirus viral load at admission (range) — copies/ml | 11,060 (991–70,680) |
| Median peak human adenovirus viral load (range) — copies/ml | 11,130 (991–156,400) |
| EBV blood qPCR — no. positive/no. tested (%) | 6/9 (67) |
| Median EBV level at admission (range) — IU/ml | 1680 (80–2240) |
| HHV-6 blood qPCR — no. positive/no. tested (%) | 1/9 (11) |
| CMV blood qPCR — no. positive/no. tested | 0/7 |
| HSV blood qPCR — no. positive/no. tested | 0/6 |
| Enterovirus blood PCR — no. positive/no. tested (%) | 2/7 (29) |
| Hepatitis A, B, and C virus testing — no. positive/no. tested∥ | 0/9 |
| Respiratory panel tests — no. positive/no. tested (%)** | |
| Enterovirus or rhinovirus | 4/8 (50) |
| Human metapneumovirus | 1/8 (12) |
| Respiratory syncytial virus | 1/8 (12) |
| Human coronavirus OC43 | 1/8 (12) |
| Human adenovirus | 1/8 (12) |
| SARS-CoV-2†† | 0/9 |
| Liver-biopsy tests — no. positive/no. tested (%) | |
| Enterovirus tissue PCR | 0/6 |
| SARS-CoV-2 tissue PCR | 0/6 |
| Human adenovirus tissue PCR‡‡ | 3/6 (50) |
To convert the values for bilirubin to micromoles per liter, multiply by 17.1. CMV denotes cytomegalovirus, EBV Epstein-Barr virus, HHV-6 human herpesvirus 6, HSV herpes simplex virus, INR international normalized ratio, and qPCR quantitative polymerase chain reaction.
Race and ethnic group were reported by the patients’ parents or guardians.
Upper respiratory symptoms included nasal congestion, nasal discharge, cough, sore throat, wheezing, and dyspnea, among other symptoms.
Normal ranges are as follows: alanine aminotransferase (ALT), 9 to 25 U per liter; aspartate aminotransferase (AST), 21 to 44 U per liter; total bilirubin, 0.1 to 1.0 mg per deciliter; and ammonia, 18 to 72 μmol per liter.
The laboratory has a maximum reported AST level of 4000 U per liter; two of the patients had this maximum value.
Patients were tested for hepatitis A IgM, hepatitis B surface antigen, hepatitis B core IgM, and hepatitis C antibody.
The respiratory viral panels (ePlex Respiratory Pathogen Panel [GenMark] and BioFire Respiratory Panel [Biomérieux]) tested for adenovirus, coronavirus (229E, HKU1, NL63, and OC43), human metapneumovirus, human rhinovirus or enterovirus, influenza A, influenza A/H1, influenza A/H1–2009, influenza A/H3, influenza B, parainfluenza 1, parainfluenza 2, parainfluenza 3, parainfluenza 4, respiratory syncytial virus A, respiratory syncytial virus B, Chlamydia pneumoniae, Mycoplasma pneumoniae, Bordetella parapertussis (BioFire only), and Bordetella pertussis (BioFire only). Specimens were obtained with a nasopharyngeal swab.
All patients were tested for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with the use of nucleic acid amplification tests.
Two of six patients (33%) were positive for human adenovirus in formalin-fixed, paraffin-embedded (FFPE) liver tissue, and two of three patients (66%) were positive for human adenovirus in fresh-frozen liver tissue. One patient was negative in FFPE tissue but positive in fresh-frozen tissue. This resulted in a total of three patients who were positive for human adenovirus by polymerase-chain-reaction (PCR) testing of either FFPE or fresh-frozen liver tissue.
Symptoms were reported to have begun days to weeks before admission and are described in Table 1. At admission, all nine children had elevated aminotransferase levels (median ALT level, 1724 U per liter; median AST level, 1963 U per liter) and eight had hyperbilirubinemia (median total bilirubin level, 7.0 mg per deciliter [120 μmol per liter]; median direct bilirubin level, 5.5 mg per deciliter [94 μmol per liter]). The initial INRs were variable (median, 1.2; range, 1.0 to 7.3). Three of the patients met criteria for pediatric acute liver failure.
Human adenovirus whole-blood qPCR was performed with specimens obtained from all nine children on the first day of hospitalization, and the median initial human adenovirus viral load was 11,060 copies per milliliter (range, 991 to 70,680). Respiratory specimens tested positive for human adenovirus in one of eight patients tested, and stool specimens tested positive in one of three patients tested. All the children tested negative for hepatitis A, B, and C viruses. Six of the nine children tested positive for Epstein–Barr virus (EBV) on whole-blood qPCR (viral load range, 80 to 2240 IU per liter). EBV IgM titers and EBV nuclear antigen (EBNA) levels were available for five patients in this subcohort; all five patients were negative for EBV IgM, and four (80%) were positive for EBNA, which suggested previous infection. Seven of the nine children (78%) had respiratory viral coinfections diagnosed by multipathogen PCR respiratory panel testing; four of eight children were found to be infected with rhinovirus or enterovirus, and one of the children was infected with human metapneumovirus, respiratory syncytial virus, and human coronavirus OC43.
All nine children tested negative for SARS-CoV-2 by nucleic acid amplification testing, and none of the children had received a SARS-CoV-2 vaccine before admission. No patients underwent SARS-CoV-2 antibody testing. None of these children were reported to have had past SARS-CoV-2 infections or met criteria for multisystem inflammatory syndrome in children (MIS-C).15
Five children had elevated total IgG levels (median total IgG level, 1468 mg per deciliter; range, 875 to 2198), and four had autoantibodies (antinuclear antibodies or smooth-muscle antibody; titer range, 1:40 to 1:80). None of the children met scoring criteria for a probable or definite diagnosis according to the autoimmune hepatitis simplified scoring system.16 Further details regarding other diagnostic testing are provided in the Supplementary Appendix.
Liver biopsies were performed in six of the children — two of whom had progression to acute liver failure (biopsy performed on mean hospital day 1.5 [one on hospital day 1 and the other on hospital day 2]) and four of whom did not have progression to acute liver failure (biopsy performed on median hospital day 4.5; range, 2 to 8). The liver biopsies showed variable degrees of chronic and acute portal and lobular hepatitis (Fig. S1 in the Supplementary Appendix) characterized by mixed inflammation, consisting of lymphocytes, histiocytes, and neutrophils with interface activity in the majority of cases. Inflammation in the lobules was associated with extensive hepatocyte damage and foci of apoptosis. No viral cytopathic effect or characteristic viral inclusions were identified. Immunohistochemical testing for human adenovirus was negative in all six patients, and none of the six patients had viral particles identified on electron microscopy. However, FFPE liver-tissue specimens from two of the six children (33%) were found to be positive for human adenovirus (species F) by human adenovirus PCR and Sanger sequencing. Two of three children (66%) were found to be positive for human adenovirus by PCR performed on fresh-frozen liver tissue. Combined, three of the six patients (50%) were positive for human adenovirus on PCR testing of FFPE or fresh-frozen liver-tissue. In contrast, FFPE biopsy specimens from all six children were found to be negative for enterovirus and SARS-CoV-2 by RT-PCR. Liver biopsies in all six children were negative for EBV-encoded small RNAs (EBERs).
Patients with Acute Liver Failure
Three children had acute liver failure, as defined above. All three children tested positive for enterovirus or rhinovirus on the viral respiratory panel, two were positive for EBV on whole-blood qPCR, and none were positive for autoantibodies.
One child had been brought to an outside hospital in acute liver failure (INR, >2.0) and was subsequently transferred to another quaternary children’s hospital because of concerns about hepatic encephalopathy. After transfer, within 3 days this patient recovered spontaneously through supportive measures only. Testing indicated human adenovirus viremia and positivity for human adenovirus in PCR analysis of stool. This patient has since been followed up in an outpatient liver clinic and, at the time of this report, was doing well.
Two children who presented with acute hepatitis and an INR of less than 2 had progression to acute liver failure, with a mean peak INR of 8.8 and a mean peak ammonia level of 169.6 μmol per liter, and both patients eventually underwent liver transplantation. The median human adenovirus viral load at admission in these two children who subsequently underwent liver transplantation was substantially higher than that in the children who did not undergo transplantation (63,010 vs. 7465 copies per milliliter).
One of the two children received high-dose glucocorticoid treatment for possible immune dysregulation on hospital days 3 to 6; however, this treatment was stopped when the human adenovirus viral load was not decreasing. Cidofovir was given on hospital days 1 (5 mg per kilogram of body weight), 7 (1 mg per kilogram), and 9 (5 mg per kilogram), and intravenous immune globulin (500 mg per kilogram) was given on hospital days 3 to 5; despite treatment, the human adenovirus viral load remained persistently elevated. Ascites and hepatic encephalopathy developed, leading to the initiation of a continuous infusion of furosemide.
The second child received cidofovir (5 mg per kilogram) on hospital days 2 and 8 and intravenous immune globulin (2 g per kilogram) on day 4; similarly, the human adenovirus viral load remained persistently elevated. Ascites and hepatic encephalopathy developed. Continuous renal replacement therapy was initiated for hyperammonemia, and total plasma exchange was performed because of worsening coagulopathy and active bleeding. This patient received etoposide and dexamethasone on hospital day 9, after meeting criteria for secondary hemophagocytic lymphohistiocytosis.
Both children had persistent viremia and deteriorating clinical status and were transferred to a quaternary care center for potential initiation of extracorporeal liver support as a bridge to liver transplantation. On transfer, both children tested negative for human adenovirus by plasma qPCR. Repeat testing of the same specimens by whole-blood qPCR was positive. Because of further clinical deterioration, both children underwent liver transplantation. At the time of this report, both were doing well without recurrence of human adenovirus viremia.
Patients without Acute Liver Failure
The six children without acute liver failure recovered with supportive therapy. As compared with the three patients who had liver failure, these six patients had a lower median ALT level (912 vs. 3854 U per liter) and AST level (1626 vs. 4000 U per liter); the median INR and total bilirubin level did not differ significantly from those of the children who had acute liver failure (median INR, 1.2 among children without liver failure and 1.6 among children with liver failure; median total bilirubin level, 6.9 and 13.2 mg per deciliter, respectively [118 and 226 μmol per liter, respectively]). The median human adenovirus viral load at admission was lower among patients without acute liver failure than among those with acute liver failure (9262.5 vs. 55,340 copies per milliliter). The median length of hospitalization (counted as the total days before either discharge or transfer to a quaternary care center) among the six children who did not have acute liver failure was 3 days, as compared with 10 days for the three children with acute liver failure. At clinic follow-up, human adenovirus was detected in three of five patients (4200, 2970, and 3140 copies per milliliter at 47, 42, and 25 days after discharge, respectively). One patient did not have human adenovirus quantified at follow-up. At the time of this report, all six children were doing well, with four patients having complete normalization of liver-enzyme levels at first follow-up at a median of 27 days after discharge (range, 14 to 47 days).
Human Adenovirus Hexon Gene Molecular Typing
Residual blood specimens obtained from eight patients were typed by human adenovirus hexon gene sequencing. Five specimens generated sufficient-quality sequence for typing, and the sequences in all five were identified as human adenovirus 41; these specimens had been obtained from two patients with acute liver failure and three patients with acute hepatitis without acute liver failure. Low viral loads in specimens obtained from three patients did not produce quality sequence, and residual specimens were unavailable from one patient.
Phylogenetic analysis of the sequences obtained showed that human adenoviruses in the five patients consisted of three distinct hexon variants (Fig. 2). Sequences from the two patients with acute liver failure (GenBank accession numbers, ON565007 and ON565008) and from one of the patients without acute liver failure (GenBank accession number, ON565009) were identical to each other and to a strain isolated from the Netherlands in 1981. A fourth sequence (GenBank accession number, ON565010), from one of the patients without acute liver failure, was identical to previously identified background strains in circulation, including the human adenovirus 41 prototype strain (GenBank accession number, DQ315364), which was isolated in the 1970s. The last sequence (GenBank accession number, ON565011), from one of the patients without acute liver failure, had one amino acid change (G697R [amino acid numbering from sequence DQ315364]) relative to the sequences from other countries within the same cluster.
Figure 2. Phylogenetic Analysis.
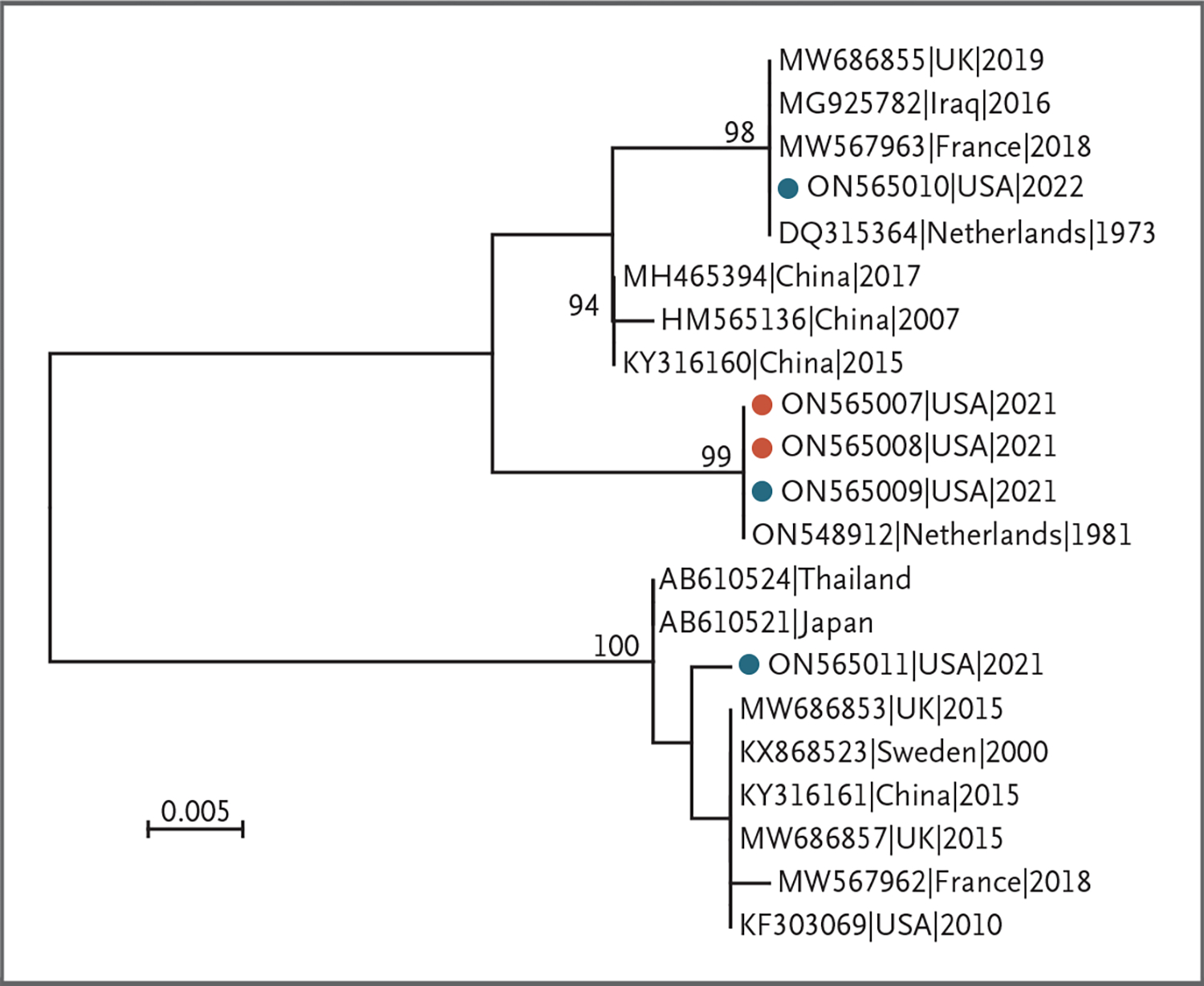
Results of the analysis of human adenovirus 41 hexon hypervariable regions 1 through 6 in specimens obtained from five patients in this study and representative sequences available in GenBank are shown. Sequences obtained in this study are marked with solid circles; red circles indicate specimens obtained from two patients with acute liver failure who underwent liver transplantation, and blue circles indicate specimens obtained from three patients with acute hepatitis who did not have acute liver failure. The tree was constructed by the maximum-likelihood method with a Kimura two-parameter model of nucleotide substitution and five rate categories of gamma distribution, and bootstrap resampling values (1000 replicates) of at least 70% are indicated above the respective nodes. The scale bar indicates genetic distance in nucleotide substitutions per site.
Acute Hepatitis Cases during the Previous Year
From October 1, 2020, to September 30, 2021, eight patients younger than 18 years of age were admitted to Children’s of Alabama with acute hepatitis. Five of the patients (62%) had an underlying cause of hepatitis identified, and three (38%) had acute hepatitis of unknown cause. Seven of the children were tested for human adenovirus by whole-blood qPCR, and all were negative. Additional details about this cohort are provided in the Supplementary Appendix.
Discussion
We describe nine previously healthy children who, during a 5-month period, received care at a single hospital in Alabama for acute hepatitis and were also found to have human adenovirus viremia. Although we do not know whether human adenovirus infection was the cause of their hepatitis, the remainder of the diagnostic investigations did not identify other common possible causes. Although six of the children were found to be positive for EBV by qPCR, EBV IgM negativity and EBNA positivity suggest that these patients had past EBV infection (i.e., not acute infection). This was further validated by negativity for EBER in the liver biopsies, providing further evidence against EBV-driven hepatitis. Although several patients had other viral infections detected in a respiratory panel, no clear pattern was found because of the diversity of the other viruses identified in patients. Other common causes of acute hepatitis in children, including autoimmune hepatitis, were ruled out. The median human adenovirus viral load was substantially higher in patients with acute liver failure than in those without acute liver failure. Although we cannot determine whether human adenovirus caused acute hepatitis, the diagnostic results and the clinical courses of these nine children suggest a possible association.
In our patients, liver biopsies showed different degrees of inflammation, which could be seen in the context of an infection; however, the liver-biopsy specimens were negative for human adenovirus on immunohistochemical testing, no viral inclusions were observed, and human adenovirus was also not observed by electron microscopy. Similar biopsy findings of inflammation and negative results on human adenovirus immunohistochemical testing have been described in immunocompetent children with acute hepatitis and human adenovirus viremia.7,8 It is unclear whether these biopsies merely missed evidence of a direct viral cytopathic effect because of sampling error or whether a human adenovirus infection could have played an indirect role (e.g., by triggering a dysregulated immune response that resulted in liver injury). It is also possible that human adenovirus was not the cause of hepatitis in these patients but instead was only an incidental finding. PCR testing for human adenovirus in FFPE tissue-biopsy specimens or fresh-frozen liver tissue was positive in three of our patients. However, this finding does not differentiate between the presence of human adenovirus DNA within liver cells and contamination with viremic blood during the biopsy procedure. In addition, no substantial histopathological differences were noted between the liver-biopsy specimens obtained from two of the patients with acute liver failure and those obtained from four of the patients with acute hepatitis without liver failure.
The differences observed in the hexon hypervariable region sequences in the five patients with human adenovirus 41 suggest that at least three known and distinct human adenovirus 41 hexon variants infected these patients. This observation is important because it suggests that if human adenovirus was causative in this group of patients with hepatitis, it was not an outbreak driven by a single new strain of human adenovirus 41. In addition, the partial genomic sequencing results are consistent with background circulating strains of adenovirus that were available in GenBank, at least preliminarily suggesting the viruses associated with these hepatitis cases are probably not new strains.
The time frame in which hepatitis developed in these patients warrants consideration of whether SARS-CoV-2 may be playing a role. SARS-CoV-2 is known to cause elevation of liver-enzyme levels17 and has also been associated with case reports of acute liver failure.18 MIS-C after SARS-CoV-2 has also been associated with liver involvement.19 However, the children in this study were found to be negative for SARS-CoV-2 by nucleic acid amplification testing, hence ruling out acute infection, and they also did not meet criteria for MIS-C. These children did not have antibody testing performed, and therefore it remains unclear whether past SARS-CoV-2 infection played a pathophysiologic role in their hepatitis.
A limitation of this study was the lack of standardization of diagnostic testing. Diagnostic studies completed for each patient differed on the basis of their clinical presentations, treating clinicians, and the different times in which they received medical care. Although genotyping was completed, direct whole-genome sequencing was not possible because of the low viral loads. As a result, genetic variation across the full genome is unknown. Although the hexon protein contains the majority of the antigen determinants, it is unknown whether recombination in other portions of the genome, such as the fiber gene, could have resulted in a change in the tissue tropism of human adenovirus 41. Finally, the small sample of nine patients prevented a larger epidemiologic interrogation of other possible exposures or cofactors that may have caused these cases of hepatitis.
We have described a cohort of nine children with acute hepatitis in the context of human adenovirus viremia. Although these findings do not prove causality, they mirror what has recently also been identified in Europe20 and is now being reported across the United States.21 The World Health Organization has reported 920 probable cases from 33 countries,22 as of the time of this report. As of June 13, 2022, the U.K. Health Security Agency has reported 260 cases of non-A–E hepatitis in children younger than 16 years of age (since January 1, 2022)23; 156 of 241 patients (64.7%) tested for human adenovirus were positive, and 27 of 35 patients (77%) with available typing by partial hexon gene sequencing have had results consistent with human adenovirus 41.24 In addition, 44 children with acute hepatitis of unknown cause were reported in a single-center study from the United Kingdom, and 27 of 30 patients (90%) were positive for human adenovirus.25 Moreover, numbers of cases of acute hepatitis of unknown cause were higher during that study period than in previous years, which parallels our experience at Children’s of Alabama.
It remains unclear whether human adenovirus infection — alone or in combination with what remains an unidentified additional cofactor — may result in development of hepatitis in children. It is difficult to know whether hepatitis associated with human adenovirus is new or may have previously been below the threshold of recognition, or whether what we are observing simply represents two unrelated findings. It is also possible that immunologic changes in a large cohort of young children may have occurred because of a lack of exposure to adenovirus and other common childhood viral illnesses during the coronavirus disease 2019 pandemic and, as a result, some children may have a dysregulated response to their first human adenovirus infection.
Supplementary Material
Acknowledgments
Supported in part by the CDC (cooperative agreement 5NU600E000104) through the Association of Public Health Laboratories.
We thank the patients and their parents for providing informed consent for the publication of the case details; Paige A. Armstrong, Neil Gupta, Senad Handanagic, Megan Hofmeister, and Philip Spradling (CDC) for their guidance in interpreting and contextualizing pediatric hepatitis data; Luciana Flannery (CDC) for performing adenovirus immunohistochemical testing; Jana M. Ritter (CDC) for assistance with preparing histology images; James J. Dunn (Texas Children’s Hospital) for running previously negative adenovirus plasma samples using whole blood; the Wadsworth Center Advanced Genomics Technology Core for performing the sequencing reactions; and Simon Ogbamikael for performing the hexon gene-sequencing assays.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention (CDC), the Department of Health and Human Services, the Association of Public Health Laboratories, the Jefferson County Department of Health, the Alabama Department of Public Health, Children’s of Alabama, the University of Alabama at Birmingham, Cincinnati Children’s Hospital Medical Center, the University of Cincinnati, Texas Children’s Hospital, Baylor College of Medicine, the Wadsworth Center, and the University of Albany.
Contributor Information
L. Helena Gutierrez Sanchez, Division of Pediatric Gastroenterology, Hepatology, and Nutrition, University of Alabama at Birmingham, Alabama Children’s of Alabama, Birmingham, Alabama.
Henry Shiau, Division of Pediatric Gastroenterology, Hepatology, and Nutrition, University of Alabama at Birmingham, Alabama Children’s of Alabama, Birmingham, Alabama.
Julia M. Baker, Division of Viral Diseases, Centers for Disease Control and Prevention, Atlanta, Georgia Epidemic Intelligence Service, Centers for Disease Control and Prevention, Atlanta, Georgia.
Stephanie Saaybi, Division of Pediatric Gastroenterology, Hepatology, and Nutrition, University of Alabama at Birmingham, Alabama Children’s of Alabama, Birmingham, Alabama.
Markus Buchfellner, Division of Pediatric Infectious Diseases, University of Alabama at Birmingham, Alabama Children’s of Alabama, Birmingham, Alabama.
William Britt, Division of Pediatric Infectious Diseases, University of Alabama at Birmingham, Alabama
Veronica Sanchez, Division of Pediatric Infectious Diseases, University of Alabama at Birmingham, Alabama
Jennifer L. Potter, Division of Pediatric Infectious Diseases, University of Alabama at Birmingham, Alabama
L. Amanda Ingram, Alabama Department of Public Health, Montgomery, Alabama
David Kelly, Department of Pediatrics, University of Alabama at Birmingham, Alabama Department of Pathology, University of Alabama at Birmingham, Alabama; Children’s of Alabama, Birmingham, Alabama.
Xiaoyan Lu, Division of Viral Diseases, Centers for Disease Control and Prevention, Atlanta, Georgia
Stephanie Ayers-Millsap, Jefferson County Department of Health, Birmingham, Alabama
Wesley G. Willeford, Jefferson County Department of Health, Birmingham, Alabama
Negar Rassaei, Division of High-Consequence Pathogens and Pathology, Centers for Disease Control and Prevention, Atlanta, Georgia
Julu Bhatnagar, Division of High-Consequence Pathogens and Pathology, Centers for Disease Control and Prevention, Atlanta, Georgia
Hannah Bullock, Division of High-Consequence Pathogens and Pathology, Centers for Disease Control and Prevention, Atlanta, Georgia Synergy America, Duluth, Georgia.
Sarah Reagan-Steiner, Division of High-Consequence Pathogens and Pathology, Centers for Disease Control and Prevention, Atlanta, Georgia
Ali Martin, Alabama Department of Public Health, Montgomery, Alabama
Michael E. Rogers, Department of Pediatric Gastroenterology, Hepatology and Nutrition, Cincinnati Children’s Hospital Medical Center, Cincinnati Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati.
Anna M. Banc-Husu, Division of Pediatric Gastroenterology, Hepatology and Nutrition, Texas Children’s Hospital, Houston Department of Pediatrics, Baylor College of Medicine, Houston.
Sanjiv Harpavat, Division of Pediatric Gastroenterology, Hepatology and Nutrition, Texas Children’s Hospital, Houston Department of Pediatrics, Baylor College of Medicine, Houston.
Daniel H. Leung, Division of Pediatric Gastroenterology, Hepatology and Nutrition, Texas Children’s Hospital, Houston Department of Pediatrics, Baylor College of Medicine, Houston.
Elizabeth A. Moulton, Division of Pediatric Infectious Diseases, Texas Children’s Hospital, Houston Department of Pediatrics, Baylor College of Medicine, Houston.
Daryl M. Lamson, Wadsworth Center, New York State Department of Health, University at Albany, Albany
Kirsten St. George, Wadsworth Center, New York State Department of Health, University at Albany, Albany Department of Biomedical Sciences, University at Albany, Albany.
Aron J. Hall, Division of Viral Diseases, Centers for Disease Control and Prevention, Atlanta, Georgia
Umesh Parashar, Division of Viral Diseases, Centers for Disease Control and Prevention, Atlanta, Georgia
Adam MacNeil, Division of Viral Diseases, Centers for Disease Control and Prevention, Atlanta, Georgia
Jacqueline E. Tate, Division of Viral Diseases, Centers for Disease Control and Prevention, Atlanta, Georgia
Hannah L. Kirking, Division of Viral Diseases, Centers for Disease Control and Prevention, Atlanta, Georgia
References
- 1.Lynch JP III, Kajon AE. Adenovirus: epidemiology, global spread of novel serotypes, and advances in treatment and prevention. Semin Respir Crit Care Med 2016;37:586–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kajon AE, Weinberg JB, Spindler KR. Adenoviruses. In: Mitchell R, Bradshaw R, McManus L, eds. Reference module in biomedical sciences. New York: Elsevier, 2019. [Google Scholar]
- 3.Schaberg KB, Kambham N, Sibley RK, Higgins JPT. Adenovirus hepatitis: clinicopathologic analysis of 12 consecutive cases from a single institution. Am J Surg Pathol 2017;41:810–9. [DOI] [PubMed] [Google Scholar]
- 4.Ronan BA, Agrwal N, Carey EJ, et al. Fulminant hepatitis due to human adenovirus. Infection 2014;42:105–11. [DOI] [PubMed] [Google Scholar]
- 5.Cames B, Rahier J, Burtomboy G, et al. Acute adenovirus hepatitis in liver transplant recipients. J Pediatr 1992;120:33–7. [DOI] [PubMed] [Google Scholar]
- 6.Onda Y, Kanda J, Sakamoto S, et al. Detection of adenovirus hepatitis and acute liver failure in allogeneic hematopoietic stem cell transplant patients. Transpl Infect Dis 2021;23(2):e13496. [DOI] [PubMed] [Google Scholar]
- 7.Ozbay Hoşnut F, Canan O, Ozçay F, Bilezikçi B. Adenovirus infection as possible cause of acute liver failure in a healthy child: a case report. Turk J Gastroenterol 2008;19:281–3. [PubMed] [Google Scholar]
- 8.Vayngortin T, Pai A, Thomas D. Not just the stomach flu: a case of acute liver failure after adenovirus infection. Ann Pediatr Child Health 2014;2:1014. [Google Scholar]
- 9.Squires RH Jr, Shneider BL, Bucuvalas J, et al. Acute liver failure in children: the first 348 patients in the pediatric acute liver failure study group. J Pediatr 2006;148:652–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matoq A, Salahuddin A. Acute hepatitis and pancytopenia in healthy infant with adenovirus. Case Rep Pediatr 2016;2016:8648190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heim A, Ebnet C, Harste G, Pring-Akerblom P. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J Med Virol 2003;70:228–39. [DOI] [PubMed] [Google Scholar]
- 12.Bhatnagar J, Gary J, Reagan-Steiner S, et al. Evidence of severe acute respiratory syndrome coronavirus 2 replication and tropism in the lungs, airways, and vascular endothelium of patients with fatal coronavirus disease 2019: an autopsy case series. J Infect Dis 2021;223:752–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guarner J, Bhatnagar J, Shieh WJ, et al. Histopathologic, immunohistochemical, and polymerase chain reaction assays in the study of cases with fatal sporadic myocarditis. Hum Pathol 2007;38:1412–9. [DOI] [PubMed] [Google Scholar]
- 14.Okada M, Ogawa T, Kubonoya H, Yoshizumi H, Shinozaki K. Detection and sequence-based typing of human adenoviruses using sensitive universal primer sets for the hexon gene. Arch Virol 2007;152:1–9. [DOI] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. Information for healthcare providers about multisystem inflammatory syndrome in children (MIS-C). May 20, 2021 (https://www.cdc.gov/mis/mis-c/hcp/index.html).
- 16.Hennes EM, Zeniya M, Czaja AJ, et al. Simplified criteria for the diagnosis of autoimmune hepatitis. Hepatology 2008;48:169–76. [DOI] [PubMed] [Google Scholar]
- 17.Hundt MA, Deng Y, Ciarleglio MM, Nathanson MH, Lim JK. Abnormal liver tests in COVID-19: a retrospective observational cohort study of 1,827 patients in a major U.S. hospital network. Hepatology 2020;72:1169–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orandi BJ, Li G, Dhall D, et al. Acute liver failure in a healthy young female with COVID-19. JPGN Reports 2021;2(3):e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez A, Cantor A, Rudolph B, et al. Liver involvement in children with SARS-COV-2 infection: two distinct clinical phenotypes caused by the same virus. Liver Int 2021;41:2068–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.World Health Organization. Multi-country — acute, severe hepatitis of unknown origin in children. April 23, 2022 (https://www.who.int/emergencies/disease-outbreak-news/item/multi-country-acute-severe-hepatitis-of-unknown-origin-in-children).
- 21.Centers for Disease Control and Prevention. Persons under investigation: children with hepatitis of unknown etiology. June 22, 2022 (https://www.cdc.gov/ncird/investigation/hepatitis-unknown-cause/updates.html).
- 22.World Health Organization. Severe acute hepatitis of unknown aetiology in children — multi-country. June 24, 2022 (https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON394).
- 23.UK Health Security Agency. Investigation into acute hepatitis of unknown aetiology in children in England: case update. June 17, 2022 (https://www.gov.uk/government/publications/acute-hepatitis-technical-briefing/investigation-into-acute-hepatitis-of-unknown-aetiology-in-children-in-england-case-update).
- 24.UK Health Security Agency. Investigation into acute hepatitis of unknown aetiology in children in England — technical briefing 3. May 19, 2022 (https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1077027/acute-hepatitis-technical-briefering_3.pdf).
- 25.Kelgeri C, Couper M, Gupte GL, et al. Clinical spectrum of children with acute hepatitis of unknown cause. N Engl J Med. DOI: 10.1056/NEJMoa2206704. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


