Abstract
Introduction
The RNA N6-methyladenosine (m6A) regulators play a crucial role in tumorigenesis and could be indicators of prognosis and therapeutic targets in various cancers. However, the expression status and prognostic value of m6A regulators have not been studied in pancreatic neuroendocrine neoplasms (PanNENs). We aimed to investigate the expression patterns and prognostic value of m6A regulators and assess their correlations with immune checkpoints and infiltrates in PanNENs.
Methods
Immunohistochemistry was performed for 15 m6A regulators and immune markers using tissue microarrays obtained from 183 patients with PanNENs. The correlation between m6A protein expression and clinicopathological parameters with recurrence-free survival (RFS) was examined using a random survival forest, Cox regression model, and survival tree analysis.
Results
Among the 15 m6A proteins, high expression of YTHDF2 (p < 0.001) and HNRNPC (p = 0.006) was found to be significantly associated with recurrence and served as independent risk factors in multivariate analysis. High YTHDF2 expression was associated with higher number of CD3+ T cells (p = 0.003), whereas high HNRNPC expression was significantly correlated with the expression of PD-L1 (p = 0.039). A YTHDF2-based signature was determined, including five patterns from survival tree analysis: patients with the LN<sub>neg</sub>YTHDF2<sub>high</sub> signature had a 5-year RFS rate of 92.1%, whereas patients with LN<sub>pos</sub>TumorSize<sub><2.5 cm</sub> signature had the worst 5-year RFS rate of 0% (p < 0.001). The area under receiver operating characteristic curve was 0.870 (95% confidence interval: 0.762–0.915) for the YTHDF2-based signature. The C-index was 0.978, suggesting good discrimination ability; moreover, the risk score of recurrence served as an independent prognostic factor indicating shorter RFS.
Conclusions
YTHDF2 appears to serve as a promising prognostic biomarker and therapeutic target. A YTHDF2-based signature can identify distinct subgroups, which may be helpful to strategize personalized postoperative monitoring.
Keywords: N6-methyladenosine methylation regulators, Pancreatic neuroendocrine neoplasms, Immune markers, Recurrence, Postoperative surveillance
Introduction
Pancreatic neuroendocrine neoplasms (PanNENs) are a rare, heterogeneous group of neoplasms originating from the neuroendocrine cells and classified as functional and nonfunctional depending on whether the tumor overproduces biologically active hormones [1]. However, the incidence of PanNENs has significantly increased in the last decades and has risen to 0.8 per 100,000 individuals per year [2]. This increase may be attributed, at least in part, to the improved and more frequent use of imaging in clinical diagnostic practices [3]. Moreover, both patients with nonfunctional or functional tumors are always diagnosed at a late stage and usually present locally advanced or metastatic disease [4, 5].
Current clinical strategies for most patients with PanNENs primarily involve surgical resection [6]. One of the most significant causes of concern in patients with a PanNEN is cancer recurrence after curative resection. However, clinical outcomes after surgical resection vary widely, with recurrence-free survival (RFS) ranging from 0.96 to 121.9 months [7]. The most frequently used cancer staging system is based on the WHO classification which categorizes tumors into four groups based on proliferative activity and morphology: NET G1, NET G2, NET G3, and NEC [8]. However, patients with PanNEN with the same grade of tumor may demonstrate different clinical courses, whereas patients with low-grade PanNENs show unpredictable disease progression and outcomes [9]. Therefore, a clinically feasible and accurate prediction of the risk of relapse in patients with PanNENs after resection is needed.
N6-methyladenosine (m6A) is the most common epigenetic modification of the mRNA, comprising >60% of all RNA-based modifications [10]. The m6A methylation levels are greatly associated with the immune response, stress response regulation, tumorigenesis, and miRNA processing [11, 12, 13, 14, 15, 16]. The m6A methylation plays a crucial role in disease progression of various cancers [11, 12, 16] as well as in the onset and progression of fetal growth restriction and preeclampsia [17, 18]. The modification of m6A is regulated by methyltransferases (writers) and demethylases (erasers), and m6A performs multiple functions through its interactions with specific binding proteins known as the readers [19]. The writers mainly include KIAA1429, METTL3, WTAP, METTL14, RBM15, and METTL16, which promote m6A RNA methylation. The erasers encompass fat mass and obesity-associated protein (FTO) and alkylation repair homolog protein 5 (ALKBH5). The readers comprise HNRNPC, IGF2BP2, YTHDC1, YTHDC2, YTHDF1, YTHDF2, and YTHDF3, which recognize RNA m6A binding site [19, 20, 21]. Increasing amount of evidence has emerged which shows the vital role of m6A regulators in tumorigenesis and prognosis of various cancers. Chen et al. [22] reported that METTL14 inhibited colorectal cancer (CRC) progression through primary miR-375 processing-based regulation. Cives et al. [23] revealed that METTL3 could promote the proliferation and migration of hepatocellular carcinoma. A previous study found that high YTHDF1 expression predicted a poor clinical outcome in CRC patients [24]. Additionally, the low expression of FTO was associated with a shorter RFS in patients with intrahepatic cholangiocarcinoma [25]. Overexpression of FTO suppressed acute myeloid leukemia cell differentiation [26]. Growing evidence suggests that m6A modification and its regulators play vital roles in human cancers [11, 12, 22, 23, 24, 25, 26]. m6A modification is associated with tumorigenesis, proliferation, invasion, and metastasis [22, 23, 24]. And m6A regulators were indicators of prognosis and therapeutic targets [25, 26]. Moreover, epigenetic modification and especially methylation have been already described as important events during tumorigenesis of PanNEN, and methylation modification is an epigenetic regulatory mechanism for gene expression in PanNENs, including MEN1, which involved in the chromatin remodeling in PanNENs [27]. However, whether m6A regulators could also be specific biomarkers in PanNENs is yet to be determined. And the expression status, functional roles, and clinical significance of m6A methylation regulators also remain largely unknown in PanNENs. In addition, recent studies have shown prognosis of immune infiltrates and immune checkpoints in pancreatic neuroendocrine tumors [23, 28]. However, the correlation between m6A regulators and immune infiltration and immune checkpoints is yet to be explored.
To better identify patients at risk and improve their follow-up management, we explored the expression profiles of m6A proteins in resected PanNENs and investigated the potential prognostic value of m6A proteins. We also analyzed the association among common immune infiltrates, immune checkpoints, and m6A regulators. Furthermore, a YTHDF2-based signature was established, which may help in postoperative monitoring and stratification of risks to guide clinical treatment options.
Materials and Methods
Patient Cohort and Follow-Up
A total of 183 patients with PanNENs who were admitted to the Peking Union Medical College Hospital (PUMCH) and underwent surgeries during the time period 2004–2019 with sufficient tumor tissue for evaluation were included in the current study. The FFPE specimens and matching hematoxylin and eosin slides were retrieved. Patients' clinicopathological data, including age, sex, primary tumor site, tumor size, tumor grading, functional status, and surgical procedures, were collected from the medical records. The American Joint on Cancer Committee (AJCC) 8th edition stages for each patient were defined based on the collected data. Survival and recurrence information were obtained from medical record reviews and telephone interviews. Recurrence and distant metastasis were determined based on biochemical markers, clinical multidisciplinary consultation, radiological, and/or histological examinations. The time between the surgery and tumor recurrence or last follow-up appointment was termed as RFS. Disease-specific survival was calculated from the surgery date to the time of patient death or last follow-up till time point of December 29, 2020. This retrospective study was approved by the Institutional Review Board of PUMCH (approval number: S-K1593) and conducted in accordance with the Declaration of Helsinki. Written consent for this study was not required and formally waived by the hospital's Ethics Committee.
Tissue Microarrays and Immunohistochemistry
Representative areas of tumors and adjacent normal tissue were selected from hematoxylin and eosin-stained paraffin blocks and then re-embedded into recipient tissue microarray (TMA) blocks (12 × 8 arrays). The diameter of each core was 2 mm.
Immunohistochemical analysis was performed according to the protocol described by Zong et al. [29]. The 4-μm TMA sections were deparaffinized. Heat-mediated antigen retrieval was performed using sodium citrate buffer (pH 6.0) for 10 min. The endogenous peroxidase activity was quenched using a 3% hydrogen peroxide solution (ZSGB-BIO, Beijing, China) and then the sections were blocked with 5% normal goat serum for 30 min. TMA sections were incubated with primary antibodies against m6A methylation regulators (METTL3, METTL14, METTL16, WTAP, KIAA1429, RBM15, FTO, ALKBH5, IGF2BP2, HNRNPC, YTHDC1, YTHDC2, YTHDF1, YTHDF2, and YTHDF3) and immune markers (PD-L1, B7-H3, CD3, CD4, CD15, CD68) overnight at 4°C; the details and optimal dilutions are summarized in online supplementary Table S1 (see www.karger.com/doi/10.1159/000525228 for all online suppl. material). Stromal cells, normal acinar cells, ductal cells, and islet cells from normal tissues were incubated with primary antibodies and used as internal positive controls, whereas the same tissues without primary antibodies comprised the negative controls.
Evaluation of Immunostaining
The immunostaining performed on the collected patient samples was independently assessed by two pathologists (XL Chen and SW Mo) who were blinded to the patients' clinicopathological features and clinical outcomes. Both pathologists reexamined the slides and reached a consensus in case of any discrepancy. The immunoreactivity of 15 m6A markers, PD-L1, and B7-H3 in tumor specimens was quantified using the method developed by Budwit-Novotny et al. [30]. In brief, the percentage of positively stained tumor cells was multiplied by the relative intensity of specific staining, which was assigned a value of negative (0), weak (1), distinct (2), and strong (3) [30, 31]. The density of stromal CD3, CD4, CD15, and CD68 were quantified in four ×400 high-power fields and the mean counts of four fields were used for statistical analysis as we described earlier [32]. The cutoff values of m6A markers and immune markers were determined using X-tile (Yale University, New Haven, CT, USA). These cutoff values are summarized in the online supplementary Table S2.
Risk Score Models and Random Survival Forest
A random survival forest (RSF) model was constructed based on the minimal depth and variable importance (VIMP). VIMP and minimal depth were used to assess the true effect of the variable on RFS. Given that feature subsets of variables are randomly selected using RSF method, RSF can process high-dimensional (many variables) data. In our current study, we included more than ten variables. Through training of the RSF algorithm, the variables which were the most related to recurrence were selected using minimal depth and VIMP. And in the training process, the interaction between variables can be detected. Moreover, when creating random forest, unbiased estimation is used for generalization error, and the model has strong generalization ability [33]. If a large part of the features is lost, the accuracy can still be maintained [33]. Based on these strengths of RSF, this method was helpful to select relapse-related variables and construct a recurrence signature. Recurrence-specific variables defined by a VIMP of >0 and minimal depth less than the depth threshold were finally entered into a survival tree analysis [33]. The recurrence-specific variables, including nodal status, tumor size, YTHDF2, were selected by using minimal depth and VIMP. By integrating the expression level of YTHDF2, nodal status, and tumor size as well as their corresponding coefficients which were derived from the multivariate Cox model constructed only using these 3 variables, a risk score was generated, as represented below: Risk Score = (1.218 × Nodal Status) + (1.625 × Tumor Size) + (0.745 × expression value of YTHDF2).
Nodal status was divided into negative/positive and scored as 0/1; tumor size was divided into <2.5 cm/≥2.5 cm and scored as 0/1, and expression value of YTHDF2 was divided into low/high and scored as 0/1, and these scores were multiplied by the corresponding coefficients to generate a risk score. The median (median = 1.625) was used as the cutoff value for the risk score.
Statistical Analysis
The continuous and categorical variables were described as median (range) and frequency (percentage), respectively. The Mann-Whitney U test was conducted to compare nonnormally distributed continuous variables, whereas a Student's t test was used to analyze normally distributed continuous variables. The χ2 test or Fisher's exact test was used to evaluate the relationship between the expression of m6A methylation regulators and categorical variables. The survival curves were plotted by Kaplan-Meier method, and the log-rank test was employed to compare the survival curves generated. The Cox proportional hazard regression model was used to estimate the hazard ratio with a 95% confidence interval (CI) for variables associated with RFS. Potential risk factors with a p value of <0.05 in the univariate Cox analysis were entered into the multivariate Cox regression model (backward Wald) after considering collinearity among the variables. We evaluated the discrimination ability of the final model with Harrell concordance index (C-index). The area under the time-dependent receiver operating characteristic (ROC) curve at different cutoff times was measured as predictive performance. Calibration was assessed by visual examination of the calibration plot. p values of <0.05 were considered statistically significant. Statistical analyses were two-sided and accomplished using SPSS software (version 21.0; IBM Corp., Armonk, NY, USA). Statistical analyses of RNA sequencing data from International Cancer Genome Consortium (ICGC) database (https://www.icgc.org/) were conducted with R version 3.5.0 (http://www.r-project.org).
Results
Patients' Characteristics
A total of 96 patients were male (52.4%), whereas 87 patients were female (47.6%). The tumor diameter was >2.5 cm in 53.1% of the patients (97/183). The number of positive lymph nodes ranged from 1 to 29. A total of 50.7% tumors were nonfunctional, and the majority of functional tumors were classified as insulinoma (76/90). Most of the tumors (175/183) were well differentiated, and eight were poorly differentiated (Table 1). The median follow-up time was 39 months (range: 1–197 months), during which there were 43 relapses and 10 deaths, and all of the deaths were owing to disease progression. The 3- and 5-year RFS rates were 83.6% and 77.0%, respectively.
Table 1.
Clinicopathological characteristics of patients (N = 183)
| Variables | N (%)/median (range) |
|---|---|
| Age, years | |
| <50 | 88 (48.1) |
| ≥50 | 95 (51.9) |
| Sex | |
| Male | 96 (52.4) |
| Female | 87 (47.6) |
| Tumor site | |
| Head and neck | 67 (36.6) |
| Body and tail | 116 (63.4) |
| Tumor size, cm | |
| <2.5 | 86 (46.9) |
| ≥2.5 | 97 (53.1) |
| Positive lymph nodes, n | 2.5 (1–29) |
| Ki67 index | 3 (1–15) |
| PNI | |
| No | 171 (93.5) |
| Yes | 12 (6.5) |
| LVI | |
| No | 166 (90.8) |
| Yes | 17 (9.2) |
| AJCC stage | |
| I | 57 (31.6) |
| IIA | 59 (32.2) |
| IIB | 20 (10.9) |
| IIIB | 20 (10.8) |
| IV | 26 (14.5) |
| Functionality | |
| Nonfunctional | 93 (50.7) |
| Functional | 90 (49.3) |
| Insulinoma | 76 (41.5) |
| VIPoma | 6 (3.2) |
| Gastrinoma | 3 (1.6) |
| Glucagonoma | 3 (1.6) |
| ACTH-producing tumor | 2 (1.1) |
| WHO grade | |
| Well differentiated | 175 (95.6) |
| NET G1 | 78 (42.6) |
| NET G2 | 95 (51.9) |
| NET G3 | 2 (0.9) |
| Poorly differentiated | 8 (4.4) |
| NEC | 8 (4.4) |
PNI, perineural invasion; LVI, lymphovascular invasion.
Expression Patterns of the 15 m6A Regulators
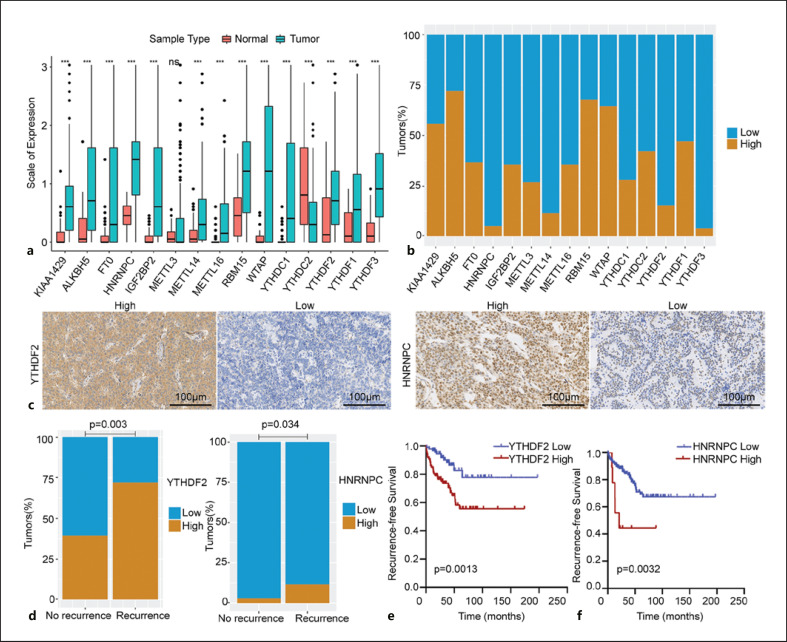
We found that the expression levels of m6A proteins in PanNENs and normal tissues were evident (Fig. 1a). The expression levels of writers (i.e., KIAA1429, METTL14, RBM15, WTAP, and METTL16), readers (i.e., IGF2BP2, HNRNPC, YTHDC1, YTHDC2, YTHDF1, YTHDF2, and YTHDF3), and erasers (ALKBH5 and FTO) were higher in PanNEN tissues than in normal tissues (p < 0.001). No statistically significant difference was evident between the normal and PanNENs tissues in the context of METTL3 expression level. Regarding METTL3, METTL14, METTL16, WTAP, RBM15, and KIAA1429 expression, a high level was observed in 26.7%, 11.5%, 35.5%, 64.4%, 67.7%, and 55.7% of tumors, respectively. Among the 183 tumors, 36.6% were deemed to have high FTO expression and 72.1% had high ALKBH5 expression. Regarding reader molecules (i.e., IGF2BP2, HNRNPC, YTHDC1, YTHDC2, YTHDF1, YTHDF2, and YTHDF3), immunostaining revealed low expression in 64.5%, 95.1%, 72.1%, 57.9%, 84.7%, 50.3%, and 96.2%, respectively, of these samples and high expression in the remaining samples (Fig. 1b). The immunohistochemical patterns of the 15 m6A proteins are shown in Figure 1c and online supplementary Figure S1. All 183 PanNENs showed nuclear immunoreactivity for METTL3, METTL14, METTL16, WTAP, RBM15, KIAA1429, FTO, ALKBH5, HNRNPC, and YTHDC1. The positive sites of IGF2BP2, YTHDC2, YTHDF1, YTHDF2, and YTHDF3 were located in the cytoplasm.
Fig. 1.
a Expression level of 15 m6A RNA regulators in tumor and adjacent normal tissues. *p < 0.05, **p < 0.01, and ***p < 0.001. b Hundred percent stacked bar chart showing the immunohistochemical expression pattern of each m6A protein. c Representative micrographs of IHC stained tissue sections for YTHDF2 and HNRNPC (×200 magnification). d Positive correlation of YTHDF2 and HNRNPC expression with PanNEN recurrence (p < 0.001 and p = 0.034, respectively). e Kaplan-Meier curves and log-rank tests showing the association between YTHDF2 and RFS. f Kaplan-Meier curves and log-rank tests showing the association between HNRNPC and RFS.
Associations among m6A Regulators, Immune Markers, and Clinicopathological Parameters in PanNENs
Among the 15 proteins studied, only YTHDF2 and HNRNPC expressions had significant prognostic value. Of the 43 patients with recurrence, 12 (27.9%) showed low YTHDF2 expression, which was dramatically lower than the proportion of patients without recurrence having low YTHDF2 expression (85 of 140, 60.7%; p = 0.003; Fig. 1d). Similarly, 88.4% of the patients (38 of 43 patients) with recurrence showed low tumoral HNRNPC expression; in contrast, 97.1% of the patients without recurrence (136 of 140 patients) showed low tumoral expression of HNRNPC (p = 0.034; Fig. 1d). The log-rank test revealed that both high HNRNPC and YTHDF2 expressions predicted a significantly shortened RFS (p = 0.003 and p = 0.001, respectively; Fig. 1e, f). In the correlation analysis between YTHDF2 and HNRNPC expression and clinicopathological variables, we found that YTHDF2 expression was positively associated with tumor size (p = 0.021), nodal status (p = 0.007), functional status (p = 0.010), Ki67 index (p = 0.023), and liver metastasis (p < 0.001). There were no significant associations between YTHDF2 expression and the other clinicopathological variables, including age, sex, tumor site, lymphovascular invasion (LVI), and perineural invasion (PNI) (online suppl. Table S3).
High YTHDF2 expression was associated with higher number of CD3+ T cells (p = 0.003). There were no significant associations between YTHDF2 expression and other immune markers (PD-L1, B7-H3, CD3, CD4, and CD68). However, high HNRNPC expression was significantly correlated with positive PD-L1 expression (p = 0.039) and high densities of CD3+ T cells. Moreover, there were no significant associations between HNRNPC expression and other immune markers (Table 2). Representative images of PD-L1, B7-H3 CD3, CD4, CD15, and CD68 are presented in online supplementary Figure S2.
Table 2.
Associations between immune markers and YTHDF2 and HNRNPC expression in patients with PanNENs
| Variables | YTHDF2 expression |
HNRNPC expression |
||||
|---|---|---|---|---|---|---|
| low | high | p value | low | high | p value | |
| PD-L1 | ||||||
| Negative | 49 | 36 | 0.241 | 84 | 1 | 0.039 |
| Positive | 48 | 50 | 90 | 8 | ||
| B7-H3 | ||||||
| Negative | 79 | 71 | 0.845 | 142 | 8 | 0.601 |
| Positive | 18 | 15 | 32 | 1 | ||
| CD3 | ||||||
| Low | 88 | 64 | 0.003 | 147 | 5 | 0.046 |
| High | 9 | 22 | 27 | 4 | ||
| CD4 | ||||||
| Low | 33 | 26 | 0.652 | 55 | 4 | 0.469 |
| High | 66 | 60 | 121 | 5 | ||
| CD15 | ||||||
| Low | 76 | 75 | 0.115 | 144 | 7 | 0.701 |
| High | 21 | 11 | 30 | 2 | ||
| CD68 | ||||||
| Low | 59 | 40 | 0.052 | 114 | 7 | 0.449 |
| High | 38 | 46 | 60 | 2 | ||
PNI, perineural invasion; LVI, lymphovascular invasion.
Prognostic Predictors of RFS in PanNENs
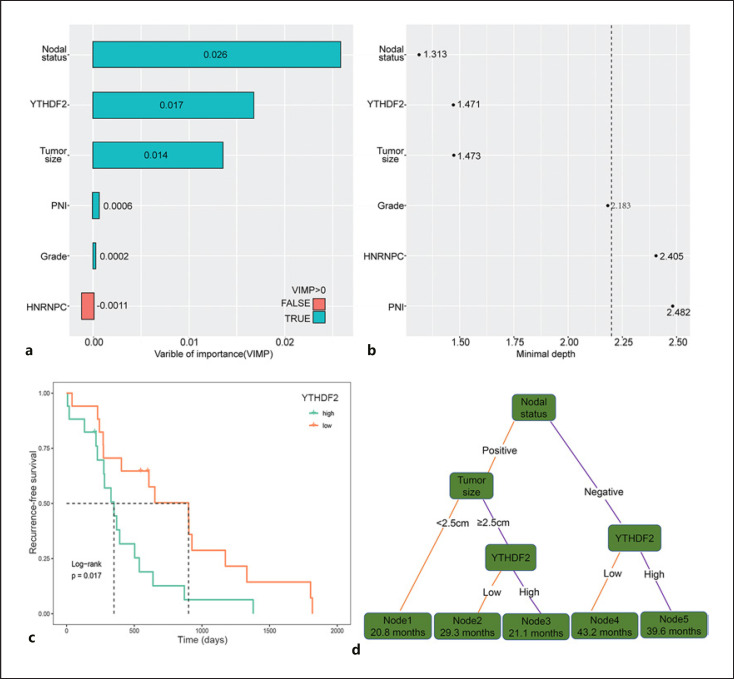
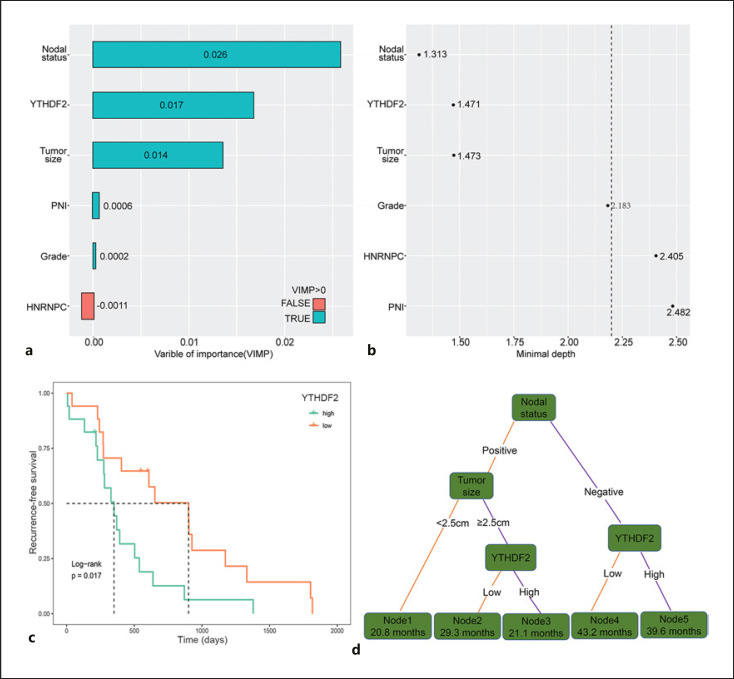
A univariate Cox analysis for both the clinicopathological parameters and m6A proteins was conducted. Nine clinicopathological factors (tumor size, lymph node status, PNI, LVI, grade, functional status, liver metastasis, and Ki67 index) and five m6A proteins (METTL14, METTL16, ALKBH5, HNRNPC, YTHDF1, YTHDF2, and YTHDF3) were identified to be associated with RFS, with p values of <0.05 (Table 3). Considering analysis of collinear factors, tumor grade was found to be associated with Ki67 index and liver metastasis, and LVI was associated with lymph node status. Therefore, PNI, tumor size, lymph node status, grade, functional status, METTL14, METTL16, ALKBH5, HNRNPC, YTHDF1, YTHDF2, and YTHDF3 were included in the multivariate Cox regression model. Multivariate Cox regression analysis identified tumor size, lymph node status, tumor grade, PNI, HNRNPC expression, and YTHDF2 expression as independent risk factors for RFS after excluding collinear factors. Next, we applied an RSF model to evaluate the importance of these independent risk factors. In the analysis of variable of importance, we found that VIMP of HNRNPC was −0.0011 (Fig. 2a). In the minimal depth analysis, HNRNPC, PNI, and grade had values of 2.405, 2.482, and 2.183, respectively, which were very close to or more than the depth threshold (Fig. 2b). Therefore, they were all excluded from the RSF model. We also performed survival analyses to determine the prognostic value of YTHDF2 mRNA expression in patients with PanNET in ICGC cohort. The Kaplan-Meier plot revealed that high YTHDF2 mRNA expression was significantly associated with poor RFS (p = 0.017, Fig. 2c). Furthermore, three variables (tumor size, lymph node status, and YTHDF2 expression) significantly affected RFS. Nodal status had the largest effect on RFS, followed by YTHDF2 expression and tumor size.
Table 3.
Univariate and multivariate analysis of factors associated with RFS
| Variables | Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | |
| Age, years | ||||
| <50 versus ≥50 | 0.69 (0.377–1.266) | 0.232 | − | − |
| Sex | ||||
| Male versus female | 1.61 (0.766–3.102) | 0.125 | − | − |
| PNI | ||||
| No versus yes | 3.49 (1.461–8.333) | 0.005 | 3.97 (1.361–11.557) | 0.012 |
| LVI | ||||
| No versus yes | 5.84 (2.974–11.458) | 0.001 | − | − |
| Tumor size, cm | ||||
| <2.5 versus ≥2.5 | 8.05 (3.168–20.473) | <0.001 | 4.62 (1.531–13.923) | 0.007 |
| Nodal status | ||||
| Negative versus positive | 6.25 (1.734–7.527) | <0.001 | 3.58 (1.799–7.120) | <0.001 |
| Grade | ||||
| Low versus high | 6.28 (2.589–15.213) | <0.001 | 6.19 (2.040–18.800) | 0.001 |
| Functional | ||||
| Yes versus no | 3.26 (1.642–6.471) | 0.027 | 0.98 (0.439–2.165) | 0.950 |
| Liver metastasis | ||||
| No versus yes | 14.39 (7.729–26.799) | <0.001 | − | − |
| Ki67 index | ||||
| <5% versus ≥5% | 5.75 (3.050–10.827) | <0.001 | − | − |
| METTL3 | ||||
| Low versus high | 0.84 (0.399–1.751) | 0.634 | − | − |
| METTL14 | ||||
| Low versus high | 2.22 (1.093–4.504) | 0.027 | 0.56 (0.245–1.012) | 0.530 |
| METTL16 | ||||
| Low versus high | 1.98 (1.087–3.617) | 0.026 | 1.34 (0.663–2.727) | 0.414 |
| WTAP | ||||
| Low versus high | 1.22 (0.637–2.342) | 0.547 | − | − |
| KIAA1429 | ||||
| Low versus high | 0.71 (0.392–1.301) | 0.271 | − | − |
| RBM15 | ||||
| Low versus high | 0.69 (0.377–1.282) | 0.245 | − | − |
| FTO | ||||
| Low versus high | 0.65 (0.338–1.245) | 0.193 | − | − |
| ALKBH5 | ||||
| Low versus high | 2.37 (1.052–5.331) | 0.037 | 1.57 (0.625–3.927) | 0.338 |
| IGF2BP2 | ||||
| Low versus high | 0.76 (0.399–1.440) | 0.397 | − | − |
| HNRNPC | ||||
| Low versus high | 3.38 (1.460–9.561) | 0.006 | 2.99 (1.005–8.849) | 0.041 |
| YTHDC1 | ||||
| Low versus high | 1.14 (0.583–2.218) | 0.700 | − | − |
| YTHDC2 | ||||
| Low versus high | 1.69 (0.920–3.103) | 0.091 | − | − |
| YTHDF1 | ||||
| Low versus high | 2.56 (1.331–4.903) | 0.005 | 1.52 (0.696–3.332) | 0.293 |
| YTHDF2 | ||||
| Low versus high | 5.30 (2.358–11.931) | <0.001 | 4.81 (1.879–12.306) | 0.001 |
| YTHDF3 | ||||
| Low versus high | 3.94 (1.546–10.026) | 0.004 | 1.24 (0.412–3.731) | 0.702 |
Analyses were performed using the former category of each variable as the reference. CI, confidence interval; HR, hazard ratio; PNI, perineural invasion; LVI, lymphovascular invasion.
Fig. 2.
Variables of importance and construction of the recurrence signatures. a RSF using the minimal depth and VIMP of recurrence-specific variables in predicting RFS. Smaller minimal depth and longer VIMP bars are more related to recurrence. VIMP of variables with negative properties were excluded. b The most important variables were at the top in the minimal depth. The dotted line suggested the depth threshold for recurrence-related variables. c Kaplan-Meier curve and log-rank test showing the association between high YTHDF2 mRNA expression and poor RFS in the International Cancer Genome Consortium cohort. d Decision tree was established using variables from the RSF model. Each variable has two nodes per branch based on relapse. Corresponding median RFS (months) is shown in each node box.
Construction and Evaluation of the Prognostic Model for Predicting RFS in PanNENs
To construct a recurrence signature that can classify patients into subpopulations according to RFS, we further performed a recursive partitioning analysis. After pruning the decision trees using the post-pruning method, five terminal nodes representing a recurrence signature were identified (Fig. 2d). LNposTumorSize<2.5 cm (node 1; 20.8 months) and LNposTumorSize≥2.5 cmYTHDF2high (node 3; 21.1 months) patients had worse median RFS than LNnegYTHDF2high patients (node 5; 39.6 months, node 1 vs. node 5, p < 0.001), LNnegYTHDF2low patients (node 4; 43.2 months), and LNposTumorSize≥2.5 cmYTHDF2low patients (node 2; 29.3 months). Patients with the LNnegYTHDF2high (node 5) signature had a 5-year RFS rate of 92.1%, whereas patients with the LNposTumorSize<2.5 cm (node 1) signature had the worst 5-year RFS rate of 0% (Fig. 3a). To better identify risk groups for the recurrence signatures, we performed pair-wise comparisons for each recurrence signature subpopulation (on RFS) and the corresponding risk score of recurrence. The results showed that patients within the five terminal nodes can be categorized into three risk groups: low (node 4), intermediate (node 2 and node 5), and high (node 1 and node 3) with well-separated RFS curves (p < 0.001; Fig. 3b). Among the risk groups, the 5-year RFS rates were 6.0%, 70.4%, and 90.1% for the high-, intermediate-, and low-risk groups, respectively. To further test our proposed recurrence signature, we performed a subanalysis on a specific group of patients with nonsyndromic and nonfunctional PanNENs. A total of 93 PanNENs from the full cohort were eligible for this analysis (online suppl. Table S4). The Kaplan-Meier RFS curves showed clear separation among the risk groups (p = 0.018; Fig. 3c).
Fig. 3.
Evaluation and validation of recurrence signature. a RFS curves of five terminal nodes. b Kaplan-Meier curves of the three relapse risk groups (low, intermediate, and high) categorized from the five terminal nodes (recurrence signatures) in the decision tree. c Kaplan-Meier curves showing RFS in patients with nonfunctional, nonsyndromic PanNENs in the high, intermediate, and low groups. d Time-dependent ROC curves indicating the predictive accuracy of the signature after 1, 3, and 5 years. e Calibration curves estimating the 1-, 3- and 5-year RFS.
Furthermore, a risk score was generated to evaluate the risk of recurrence based on variables selected from the survival tree analysis: Risk Score = (1.218 × Nodal Status) + (1.625 × Tumor Size) + (0.745 × expression value of YTHDF2) (Table 4). Multivariate analysis using a Cox proportional hazards model was performed which included PNI, tumor size, nodal status, grade, functional status, METTL14, METTL16, ALKBH5, HNRNPC, YTHDF1, YTHDF2, YTHDF3, and the risk score. The risk score was determined to be an independent prognostic factor for patients with resected PanNEN in our study based on the multivariate Cox regression, and higher risk scores were found to be associated with shorter survival (hazard ratio: 33.04, 95% CI: 4.341–251.434; p = 0.001) (Table 5). Specificity and sensitivity comparisons were performed via time-dependent ROC curve analysis of risk score. The predictive accuracy of the recurrence signature was relatively high and the area under ROC curve was 0.858 (95% CI: 0.747–0.913) at 1 year, 0.824 (95% CI: 0.754–0.912) at 3 years, and 0.870 (95% CI: 0.762–0.915) at 5 years (Fig. 3d). The C-index was 0.978 (95% CI: 0.936–1), suggesting good discrimination ability. The calibration curves also showed good agreement between the predicted and observed RFS (Fig. 3e).
Table 4.
Multivariate analysis of factors associated with RFS in the risk score selected by using minimal depth and VIMP
| Variables | Multivariate analysis |
|
|---|---|---|
| HR (95% CI) | p value | |
| Tumor size, cm | ||
| <2.5 versus ≥2.5 | 5.08 (1.928–13.371) | 0.001 |
| Nodal status | ||
| Negative versus positive | 3.38 (1.804–6.337) | <0.001 |
| YTHDF2 | ||
| Low versus high | 2.11 (1.074–4.132) | 0.030 |
HR, hazard ratio.
Table 5.
Factors associated with RFS in the final multivariable Cox model
| Variables | Multivariate analysis |
|
|---|---|---|
| HR (95% CI) | p value | |
| PNI | ||
| No versus yes | 3.94 (1.077–8.027) | 0.035 |
| Tumor size, cm | ||
| <2.5 versus ≥2.5 | 3.70 (1.016–13.443) | 0.047 |
| Nodal status | ||
| Negative versus positive | 3.33 (1.479–7.747) | 0.004 |
| Grade | ||
| Low versus high | 4.87 (1.606–14.247) | 0.004 |
| Functional | ||
| Yes versus no | 1.05 (0.468–2.335) | 0.913 |
| METTL14 | ||
| Low versus high | 0.38 (0.154–0.917) | 0.032 |
| METTL16 | ||
| Low versus high | 1.51 (0.747–3.033) | 0.253 |
| ALKBH5 | ||
| Low versus high | 1.57 (0.638–3.870) | 0.326 |
| HNRNPC | ||
| Low versus high | 3.21 (1.063–9.096) | 0.039 |
| YTHDF1 | ||
| Low versus high | 1.58 (0.741–3.347) | 0.238 |
| YTHDF2 | ||
| Low versus high | 1.72 (1.034–4.653) | 0.028 |
| YTHDF3 | ||
| Low versus high | 1.53 (0.594–3.943) | 0.379 |
| Risk score | ||
| Low versus high | 33.04 (4.341–251.434) | 0.001 |
Analyses were performed using the former category of each variable as the reference. CI, confidence interval; HR, hazard ratio; PNI, perineural invasion.
Discussion/Conclusion
Identification of predictors of relapse in patients with PanNEN is important. Previous studies that investigated risk predictors identified that tumor size, lymph node status, WHO grade, Ki67 index, tertiary lymphoid structures, and tumor-infiltrating neutrophils may predict recurrence [34, 35, 36, 37]. However, a practical and effective model is urgently needed. In our study, we constructed a risk model for recurrence with a corresponding risk score to stratify patients on the basis of RFS. The recurrence signature classifies patients into three risk groups independent of grade, with each group representing a distinct RFS outcome.
This is the first large-scale study exploring 15 m6A proteins as potential relapse biomarkers for PanNENs, to the best of our knowledge. To date, all of these 15 proteins have not been described in PanNENs. High expression of METTL14 was observed in 11.5% of the entire cohort, and high METTL14 expression predicted poor RFS outcomes (p = 0.024), similar to pancreatic cancer [38], whereas there are opposite results in other types of cancers, such as CRC [39] and bladder cancer [40]. METTL16, another m6A methyltransferase, was also a predictor of relapse and associated with poor RFS in this cohort (p = 0.023); however, recent studies suggested that METTL16 deletion is correlated with poor disease-specific survival or RFS in hepatocellular carcinoma [41], and no difference was found between patients with METTL16-high and METTL16-low CRC [42]. We found that patients with low ALKBH5 expression had a better clinical outcome. Recent studies have also suggested that YTHDF1 and YTHDF3 play a vital role in various types of cancers [43, 44]. Liu et al. [44] showed that YTHDF1 promotes ovarian cancer progression by augmenting EIF3C translation, and its high expression indicated a poor clinical outcome. The expression of YTHDF1 and YTHDF3 was reported to be associated with poor prognosis in breast cancer patients [43]. We attempted to examine the prognostic value of this factor in patients with resected PanNENs. We found that the high expression of both YTHDF1 and YTHDF3 predicted extremely low RFS (p = 0.003 and p = 0.002).
YTHDF2 is the most effective m6A reader that can selectively bind to the m6A site. Recent reports have described the ability of YTHDF2 to degrade both tumor promoter as well as suppressor gene mRNAs and adversely affect inflammatory reaction, vascular abnormalization, and self-renewal of leukemic stem cells [45, 46, 47, 48, 49]. Compared with normal tissues, YTHDF2 expression was upregulated in prostate cancer, pancreatic cancer, and lung adenocarcinoma; moreover, high YTHDF2 expression was associated with unfavorable clinicopathological parameters and poor survival, which was consistent with our study [50, 51, 52]. However, Shen et al. [53] found that the expression level of YTHDF2 was lower in gastric cancer than in normal gastric tissues, and low YTHDF2 expression was correlated with poor prognosis. In our PanNENs, high YTHDF2 expression was observed in 49.7% of the tumors; high YTHDF2 expression was strongly associated with poorer RFS (p = 0.0013), and in an independent ICGC cohort, the same correlation was observed (p = 0.017), which validated this finding. We also identified associations between YTHDF2 expression and clinicopathological features, including liver metastasis (p < 0.001), although we did not determine YTHDF2 expression in metastatic lesions. After adjusting for other covariables, high YTHDF2 expression was identified as an independent risk factor for recurrence in patients with PanNENs, suggesting the role of YTHDF2 in metastasis. Recent studies revealed complex biological functions of YTHDF2 in different types of cancers. YTHDF2 could mediate mRNA degradation of tumor suppressors (LHPP and NKX3-1) to promote the proliferation and migration of prostate cancer cells [50]. In pancreatic cancer, YTHDF2 knockdown resulted in epithelial-mesenchymal transition through YAP pathway to promote the invasion and migration of cancer cells [51]. Another study showed that upregulated YTHDF2 decreased FOXC2 expression level to inhibit the proliferation, invasion, and migration of gastric cancer cells [53]. However, the regulatory mechanisms of YTHDF2 require further investigations in PanNENs.
Another finding is the discovery that HNRNPC has prognostic significance in PanNEN recurrence. High HNRNPC expression was observed more frequently in patients with recurrence than in those without recurrence and was associated with a significantly shortened RFS. HNRNPC contributes to tumorigenesis, which may partially explain the association between high HNRNPC expression and the high recurrence rate in PanNENs. Additionally, we observed a positive correlation between HNRNPC expression and tumoral PD-L1 expression (p < 0.001). No existing literature describes the influence of HNRNPC on PD-L1 expression of PanNEN cells. Therefore, detailed mechanistic investigations are required to understand the functional link between HNRNPC and PD-L1 in PanNENs.
The current clinical practice of regular follow-up is the main method for postoperative monitoring of patients and allows early identification of recurrence. However, an important concern with postoperative monitoring of patients is the balance of effectiveness against cost because the radiation exposure over prolonged follow-up periods can be harmful and detrimental to the patient [54]. Current international guidelines of the European Society for Medical Oncology provide follow-up recommendations after PanNEN resection based on grade [55]. In our study, PanNENs were classified into three risk groups: low-, intermediate-, and high-risk groups. Interestingly, grade 2 tumors were distributed across different recurrence risk groups. These results imply that owing to significant heterogeneity in disease behavior, particularly with grade 2 tumors, such distinctions are insufficient and postoperative surveillance should not merely be based on grade. Considering the high accuracy of recurrence signature and its independence from grade, postoperative follow-up regimens could be customized on the basis of these alternative recurrence signatures and risk groups. In addition, adjuvant therapy for high-risk patients may be useful in improving clinical outcomes, although this strategy will need to be validated prospectively in future studies.
The high YTHDF2 expression (47.1%) in patients with PanNEN and its correlation with recurrence may have important therapeutic implications with potential for development and clinical use of YTHDF2 inhibitors for the treatment of high YTHDF2-expressing tumors. High HNRNPC expression was correlated with recurrence and PD-L1 expression, which may allow the development of a combinatorial therapeutic regimen using HNRNPC inhibitor and PD-L1 monoclonal antibody. Recent studies have shown that genomic mutations in PanNENs can be relevant to classify patients beyond their tumor grade and identify novel prognostic markers and therapeutic targets that could be relevant in the future for adjuvant therapy [56]. It will be interesting to investigate whether such a clinical approach is feasible in PanNENs with high HNRNPC expression in prospective clinical trials.
Some limitations exist in the current study, despite several valuable findings. First, its retrospective nature has inherent limitations. Second, the study was performed in patients from a single institution. Although we conducted a subgroup analysis to validate the recurrence signature in a selected group, additional external validation is required to investigate whether these signatures are useful markers in other cohorts. However, the low prevalence of PanNEN in the population is an obstacle to conducting the study in a larger sample size. Therefore, clinical data from multiple centers and prospective evidence are required to validate these results.
In conclusion, this is the first study to explore the expression profiles of m6A proteins and association between m6A regulators and immune microenvironment in PanNENs. We identified high YTHDF2 and HNRNPC expression as independent risk factors for disease relapse after surgery. We further established a recurrence signature to identify patients with PanNEN at a higher risk of recurrence. A comprehensive understanding of m6A regulators in PanNEN may help in developing novel treatment strategies.
Statement of Ethics
The study was approved by the Institutional Review Board of Peking Union Medical College Hospital (approval number S-K1593) and conducted in accordance with the Declaration of Helsinki. Written consent for this study was not required and formally waived by the hospital's Ethics Committee.
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
Funding Sources
This work was supported by grants from the Chinese Academy of Medical Sciences Initiative for Innovative Medicine (CAMS-2016-I2M-1–001), the National Scientific Data Sharing Platform for Population and Health (NCMI–YF01N–201906), and the National Natural Science Foundation of China (Nos. 81672648).
Author Contributions
Jie Chen made substantial contributions to the conception, design, and critical revision of the manuscript. Shengwei Mo, Liju Zong, Shuangni Yu, and Zhaohui Lu made substantial contributions to tissue microarray construction. Shengwei Mo made substantial contributions to data acquisition. Xianlong Chen and Shengwei Mo performed analysis of the data and drafting of the manuscript. All the authors read and approved the final manuscript.
Data Availability Statement
The data used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgments
We thank Ruiyu Li and Yong Wang for technical help.
Funding Statement
This work was supported by grants from the Chinese Academy of Medical Sciences Initiative for Innovative Medicine (CAMS-2016-I2M-1–001), the National Scientific Data Sharing Platform for Population and Health (NCMI–YF01N–201906), and the National Natural Science Foundation of China (Nos. 81672648).
References
- 1.Yao JC, Eisner MP, Leary C, Dagohoy C, Phan A, Rashid A, et al. Population-based study of islet cell carcinoma. Ann Surg Oncol. 2007;14((12)):3492–500. doi: 10.1245/s10434-007-9566-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muscogiuri G, Altieri B, Albertelli M, Dotto A, Modica R, Barrea L, et al. Epidemiology of pancreatic neuroendocrine neoplasms: a gender perspective. Endocrine. 2020;69((2)):441–450. doi: 10.1007/s12020-020-02331-3. [DOI] [PubMed] [Google Scholar]
- 3.Dromain C, Déandréis D, Scoazec JY, Goere D, Ducreux M, Baudin E, et al. Imaging of neuroendocrine tumors of the pancreas. Diagn Interv Imaging. 2016;97((12)):1241–1257. doi: 10.1016/j.diii.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 4.Halfdanarson TR, Rubin J, Farnell MB, Grant CS, Petersen GM. Pancreatic endocrine neoplasms: epidemiology and prognosis of pancreatic endocrine tumors. Endocr Relat Cancer. 2008;15((2)):409–427. doi: 10.1677/ERC-07-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capodanno Y, Altieri B, Elders R, Colao A, Faggiano A, Schrader J. Canine insulinoma as a model for human malignant insulinoma research: novel perspectives for translational clinical studies. Transl Oncol. 2022;15((1)):101269. doi: 10.1016/j.tranon.2021.101269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunz PL, Reidy-Lagunes D, Anthony LB, Bertino EM, Brendtro K, Chan JA, et al. Consensus guidelines for the management and treatment of neuroendocrine tumors. Pancreas. 2013;42((4)):557–577. doi: 10.1097/MPA.0b013e31828e34a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Genç CG, Falconi M, Partelli S, Muffatti F, van Eeden S, Doglioni C, et al. Recurrence of pancreatic neuroendocrine tumors and survival predicted by Ki67. Ann Surg Oncol. 2018;25((8)):2467–2474. doi: 10.1245/s10434-018-6518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scoazec JY, Couvelard A. Classification of pancreatic neuroendocrine tumours: changes made in the 2017 WHO classification of tumours of endocrine organs and perspectives for the future. Ann Pathol. 2017;37((6)):444–456. doi: 10.1016/j.annpat.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Hain E, Sindayigaya R, Fawaz J, Gharios J, Bouteloup G, Soyer P, et al. Surgical management of pancreatic neuroendocrine tumors: an introduction. Expert Rev Anticancer Ther. 2019;19((12)):1089–100. doi: 10.1080/14737140.2019.1703677. [DOI] [PubMed] [Google Scholar]
- 10.Du K, Zhang L, Lee T, Sun T. m6A RNA methylation controls neural development and is involved in human diseases. Mol Neurobiol. 2019;56((3)):1596–606. doi: 10.1007/s12035-018-1138-1. [DOI] [PubMed] [Google Scholar]
- 11.Wang S, Sun C, Li J, Zhang E, Ma Z, Xu W, et al. Roles of RNA methylation by means of N(6)-methyladenosine (m(6)A) in human cancers. Cancer Lett. 2017;408:112–120. doi: 10.1016/j.canlet.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 12.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347((6225)):1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 13.Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548((7667)):338–342. doi: 10.1038/nature23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han W, et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16((3)):289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 15.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m(6A Demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31((4)):591–606.e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, et al. m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18((11)):2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laganà AS, Vitale SG, Sapia F, Valenti G, Corrado F, Padula F, et al. miRNA expression for early diagnosis of preeclampsia onset: hope or hype? J Matern Fetal Neonatal Med. 2018;31((6)):817–821. doi: 10.1080/14767058.2017.1296426. [DOI] [PubMed] [Google Scholar]
- 18.Chiofalo B, Laganà AS, Vaiarelli A, La Rosa VL, Rossetti D, Palmara V, et al. Do miRNAs play a role in fetal growth restriction? A fresh look to a busy corner. Biomed Res Int. 2017;2017:6073167. doi: 10.1155/2017/6073167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27((3)):444–447. doi: 10.1038/cr.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28((6)):616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schöller E, Weichmann F, Treiber T, Ringle S, Treiber N, Flatley A, et al. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA. 2018;24((4)):499–512. doi: 10.1261/rna.064063.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Xu M, Xu X, Zeng K, Liu X, Sun L, et al. METTL14 suppresses CRC progression via regulating N6-methyladenosine-dependent primary miR-375 processing. Mol Ther. 2020;28((2)):599–612. doi: 10.1016/j.ymthe.2019.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Cives M, Pelle' E, Quaresmini D, Rizzo FM, Tucci M, Silvestris F. The tumor microenvironment in neuroendocrine tumors: biology and therapeutic implications. Neuroendocrinology. 2019;109((2)):83–99. doi: 10.1159/000497355. [DOI] [PubMed] [Google Scholar]
- 24.Nishizawa Y, Konno M, Asai A, Koseki J, Kawamoto K, Miyoshi N, et al. Oncogene c-Myc promotes epitranscriptome m(6)A reader YTHDF1 expression in colorectal cancer. Oncotarget. 2018;9((7)):7476–7486. doi: 10.18632/oncotarget.23554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rong ZX, Li Z, He JJ, Liu LY, Ren XX, Gao J, et al. Downregulation of fat mass and obesity associated (FTO) promotes the progression of intrahepatic cholangiocarcinoma. Front Oncol. 2019;9:369. doi: 10.3389/fonc.2019.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell. 2017;31((1)):127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mafficini A, Scarpa A. Genomic landscape of pancreatic neuroendocrine tumours: the International Cancer Genome Consortium. J Endocrinol. 2018 Mar;236((3)):R161–7. doi: 10.1530/JOE-17-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi D, Kojima M, Suzuki T, Sugimoto M, Kobayashi S, Takahashi S, et al. Profiling the tumour immune microenvironment in pancreatic neuroendocrine neoplasms with multispectral imaging indicates distinct subpopulation characteristics concordant with WHO 2017 classification. Sci Rep. 2018;8((1)):13166. doi: 10.1038/s41598-018-31383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zong L, Zhang M, Wang W, Wan X, Yang J, Xiang Y. PD-L1, B7-H3 and VISTA are highly expressed in gestational trophoblastic neoplasia. Histopathology. 2019;75((3)):421–430. doi: 10.1111/his.13882. [DOI] [PubMed] [Google Scholar]
- 30.Budwit-Novotny DA, McCarty KS, Cox EB, Soper JT, Mutch DG, Creasman WT, et al. Immunohistochemical analyses of estrogen receptor in endometrial adenocarcinoma using a monoclonal antibody. Cancer Res. 1986;46((10)):5419–5425. [PubMed] [Google Scholar]
- 31.Zhang Y, Xu J, Hua J, Liu J, Liang C, Meng Q, et al. A PD-L2-based immune marker signature helps to predict survival in resected pancreatic ductal adenocarcinoma. J Immunother Cancer. 2019;7((1)):233. doi: 10.1186/s40425-019-0703-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mo S, Zong L, Chen X, Chang X, Lu Z, Yu S, et al. High mast cell density predicts a favorable prognosis in patients with pancreatic neuroendocrine neoplasms. Neuroendocrinology. 2021 Dec 28; doi: 10.1159/000521651. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 33.Zhu R, Kosorok MR. Recursively imputed survival trees. J Am Stat Assoc. 2012;107((497)):331–340. doi: 10.1080/01621459.2011.637468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grillo F, Albertelli M, Brisigotti MP, Borra T, Boschetti M, Fiocca R, et al. Grade increases in gastroenteropancreatic neuroendocrine tumor metastases compared to the primary tumor. Neuroendocrinology. 2016;103((5)):452–459. doi: 10.1159/000439434. [DOI] [PubMed] [Google Scholar]
- 35.Nanno Y, Toyama H, Otani K, Asari S, Goto T, Terai S, et al. Microscopic venous invasion in patients with pancreatic neuroendocrine tumor as a potential predictor of postoperative recurrence. Pancreatology. 2016;16((5)):882–887. doi: 10.1016/j.pan.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 36.Zhang WH, Wang WQ, Gao HL, Xu SS, Li S, Li TJ, et al. Tumor-infiltrating neutrophils predict poor survival of non-functional pancreatic neuroendocrine tumor. J Clin Endocrinol Metab. 2020;105((7)) doi: 10.1210/clinem/dgaa196. [DOI] [PubMed] [Google Scholar]
- 37.Zhang WH, Wang WQ, Han X, Gao HL, Xu SS, Li S, et al. Infiltrating pattern and prognostic value of tertiary lymphoid structures in resected non-functional pancreatic neuroendocrine tumors. J Immunother Cancer. 2020;8((2)):e001188. doi: 10.1136/jitc-2020-001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang M, Liu J, Zhao Y, He R, Xu X, Guo X, et al. Upregulation of METTL14 mediates the elevation of PERP mRNA N(6) adenosine methylation promoting the growth and metastasis of pancreatic cancer. Mol Cancer. 2020;19((1)):130. doi: 10.1186/s12943-020-01249-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B, et al. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol Cancer. 2020;19((1)):106. doi: 10.1186/s12943-020-01220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu C, Wang Z, Zhou N, Li G, Kou Y, Luo Y, et al. Mettl14 inhibits bladder TIC self-renewal and bladder tumorigenesis through N(6)-methyladenosine of Notch1. Mol Cancer. 2019;18((1)):168. doi: 10.1186/s12943-019-1084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang P, Wang X, Zheng L, Zhuang C. Gene signatures and prognostic values of m6a regulators in hepatocellular carcinoma. Front Genet. 2020;11:540186. doi: 10.3389/fgene.2020.540186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X, Liu L, Dong Z, Li J, Yu Y, Chen X, et al. Expression patterns and prognostic value of m(6)A-related genes in colorectal cancer. Am J Transl Res. 2019;11((7)):3972–3991. [PMC free article] [PubMed] [Google Scholar]
- 43.Anita R, Paramasivam A, Priyadharsini JV, Chitra S. The m6A readers YTHDF1 and YTHDF3 aberrations associated with metastasis and predict poor prognosis in breast cancer patients. Am J Cancer Res. 2020;10((8)):2546–2554. [PMC free article] [PubMed] [Google Scholar]
- 44.Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020;48((7)):3816–3831. doi: 10.1093/nar/gkaa048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu R, Li Q, Feng Z, Cai L, Xu Q. m6A reader YTHDF2 regulates LPS-induced inflammatory response. Int J Mol Sci. 2019;20((6)) doi: 10.3390/ijms20061323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hou J, Zhang H, Liu J, Zhao Z, Wang J, Lu Z, et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol Cancer. 2019;18((1)):163. doi: 10.1186/s12943-019-1082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67((6)):2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 48.Zhong L, Liao D, Zhang M, Zeng C, Li X, Zhang R, et al. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252–261. doi: 10.1016/j.canlet.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 49.Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, et al. Targeting the RNA m(6)A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell. 2019;25((1)):137–48.e6. doi: 10.1016/j.stem.2019.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li J, Xie H, Ying Y, Chen H, Yan H, He L, et al. YTHDF2 mediates the mRNA degradation of the tumor suppressors to induce AKT phosphorylation in N6-methyladenosine-dependent way in prostate cancer. Mol Cancer. 2020;19((1)):152. doi: 10.1186/s12943-020-01267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen J, Sun Y, Xu X, Wang D, He J, Zhou H, et al. YTH domain family 2 orchestrates epithelial-mesenchymal transition/proliferation dichotomy in pancreatic cancer cells. Cell Cycle. 2017;16((23)):2259–2271. doi: 10.1080/15384101.2017.1380125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu J, Wang M, Hu D. Deciphering N6-methyladenosine-related genes signature to predict survival in lung adenocarcinoma. Biomed Res Int. 2020;2020:2514230. doi: 10.1155/2020/2514230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen X, Zhao K, Xu L, Cheng G, Zhu J, Gan L, et al. YTHDF2 inhibits gastric cancer cell growth by regulating FOXC2 signaling pathway. Front Genet. 2021;11:592042. doi: 10.3389/fgene.2020.592042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh S, Moody L, Chan DL, Metz DC, Strosberg J, Asmis T, et al. Follow-up recommendations for completely resected gastroenteropancreatic neuroendocrine tumors. JAMA Oncol. 2018;4((11)):1597–604. doi: 10.1001/jamaoncol.2018.2428. [DOI] [PubMed] [Google Scholar]
- 55.Falconi M, Eriksson B, Kaltsas G, Bartsch DK, Capdevila J, Caplin M, et al. ENETS Consensus guidelines update for the management of patients with functional pancreatic neuroendocrine tumors and non-functional pancreatic neuroendocrine tumors. Neuroendocrinology. 2016;103((2)):153–171. doi: 10.1159/000443171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Capodanno Y, Chen Y, Schrader J, Tomosugi M, Sumi S, Yokoyama A, et al. Cross-talk among MEN1, p53 and notch regulates the proliferation of pancreatic neuroendocrine tumor cells by modulating INSM1 expression and subcellular localization. Neoplasia. 2021;23((9)):979–992. doi: 10.1016/j.neo.2021.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Data Availability Statement
The data used and/or analyzed during the current study are available from the corresponding author on reasonable request.