Abstract
Neisseria meningitidis is a gram-negative bacterium that may cause meningitis, sepsis, or both. The increase in the incidence of meningococcal disease in various countries in the past 2 decades is mainly due the genotypically related lineage III meningococci. The chromosomal DNA differences between lineage III strains and non-lineage III strains were identified using representational difference analysis. Thus, a 1.8-kb locus that is specific for lineage III meningococci was identified. The locus contains three open reading frames encoding the NmeSI restriction-modification system. The methyltransferase gene was cloned and expressed in Escherichia coli. Site AGTACT was found to be modified by the enzyme. In conclusion, lineage III strains differ from endemic strains by the presence of a specific restriction-modification system. This restriction-modification system may contribute to the clonal and hypervirulent character of lineage III strains by influencing horizontal gene transfer and transcription.
Neisseria meningitidis is a gram-negative bacterium that commonly resides in the human nasopharynx. Occasionally, this bacterium causes serious disease, mainly meningitis and sepsis. Since 1980, an increase in the incidence of meningococcal disease has taken place in The Netherlands. Previous studies showed that this increase is due to genotypically related isolates, designated lineage III strains (7). Strains of this cluster have also been isolated in many other western European countries and in Chile and more recently in New Zealand (15). Lineage III strains isolated from patients in various countries over the past 20 years can be recognized as such by both multilocus enzyme electrophoresis (7) and multilocus sequence typing (12). This suggests that the diversifying effect of horizontal gene transfer affecting meningococcal population biology (24) in these lineage III strains is relatively low. The increased incidence of meningococcal disease due to a specific clone may imply that such a clone possesses certain virulence properties that are not present in other isolates (8).
To address the observed differences between lineage III meningococci and other meningococci, we previously used representational difference analysis (RDA) (11) to compare the chromosomal DNA content of lineage III strains with that of two strains that only caused endemic disease. By this method, DNA sequences that are present in one DNA pool (i.e., the lineage III chromosomal DNA) but absent in another DNA pool (i.e., the chromosomal DNA of the endemic strains) are selectively amplified. Parts of the differences and point mutations are expected to go undetected by this method. Recently, we identified three DNA sequences that are present in all lineage III strains tested but absent from a majority of non-lineage III strains (4). Database similarities of the fragments suggested that they formed part of a restriction-modification (RM) locus.
Here we report the identification of the lineage III-specific NmeSI RM system and show that this is an isoschizomer of the ScaI RM system. Sequence analysis indicates that the NmeSI RM system and the ScaI RM system may have evolved separately.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, and reagents.
N. meningitidis strains 800615, 882066, 3532, and 830248 were isolated from patients with meningococcal disease and collected by the Netherlands Reference Laboratory for Bacterial Meningitis (Academic Medical Center, Amsterdam, The Netherlands, and the Rijksinstituut voor Volksgezondheid en Milieuhygiëne, Bilthoven, The Netherlands). Strains 800615 and 882066 belong to the hypervirulent lineage III clone. Strains 3532 and 830248 belong to lineage IV, most closely related to lineage III (7), containing isolates from the period 1958 to 1986 causing endemic disease (23). Meningococci were grown on heated blood (chocolate) agar plates at 37°C in a humidified atmosphere of 5% CO2.
Competent Escherichia coli Top10F′ cells and cloning vector pCR2.1 were obtained from Invitrogen (Groningen, The Netherlands). Plasmid-carrying E. coli strains were routinely grown in Luria-Bertani medium with 100 μg of ampicillin/ml, supplemented with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and IPTG (isopropyl-β-d-thiogalactopyranoside) if necessary for screening purposes, according to the manufacturer's protocol. Expression vector pSE380 was obtained from Invitrogen. Expression was induced by adding IPTG according to the manufacturer's protocol. Oligonucleotides used in this study were synthesized by Perkin-Elmer Nederland B.V., Gouda, The Netherlands.
DNA techniques.
Chromosomal DNA was isolated as described by Akopyanz et al. (1) or using the Puregene kit (Gentra Systems, Minneapolis, Minn.). Plasmid DNA isolations were performed using the QIAGEN kit (Qiagen GmbH, Hilden, Germany) or the Wizard kit (Promega Corp., Madison, Wis.). The concentration of DNA was assessed by measuring the optical density at 260 nm using an Ultraspec 2000 spectrophotometer (Pharmacia, Woerden, The Netherlands). Restriction enzymes and digestion buffers were obtained from Boehringer Mannheim GmbH (Almere, The Netherlands) and used according to the manufacturer's instructions.
Sequence analysis of parts of the NmeSI locus.
The procedure for inverse PCR (IPCR) was performed as previously described (18, 26). The IPCR template was prepared by self-ligation of digested chromosomal DNA from strain 800615. IPCR with primers ABM3 (5′-ATA CAT TCA ATT TAG ATG CTG TAC G-3′) and ABM4 (5′-GGT GGA GAT GTG ATT GTC ATT TGG A-3′) yielded a single 1.2-kb amplicon for PstI-digested and self-ligated chromosomal DNA. Cloning this amplicon in the pCR2.1 vector yielded recombinant plasmid pMP01. Digestion of pMP01 with SspI and EcoRV yielded two fragments. A 600-bp fragment was subcloned in pUC19, yielding pMP10. Self-ligation of the other fragment yielded 4.5-kb plasmid pMP02. Plasmids pMP01, pMP10, and pMP02 were sequenced with the M13 universal primers.
Similarly, IPCR with primers ABM2 (5′-ATT TAG CAG GAT TTT TCA CAT ACC A-3′) and ABM3 yielded a 1.4-kb fragment for SphI and a 2.5-kb product for ClaI-digested and self-ligated chromosomal DNA. Cloning these products in pCR2.1 did not yield transformants. We suspected that this was due to the lethality of an expressed gene product of this sequence in E. coli; therefore, a second PCR was performed on both ABM2 and -3 amplicons with primers ABM2 and ABM7 (5′-CTC GCC TGC TGG CCT GTC GCT GCA G-3′). By this repeated PCR, we hoped to abolish the lethal effect in E. coli by the introduction of a mutation due to the infidelity of Taq polymerase. Cloning the ABM2 and -7 IPCR products yielded only a few transformants, two of which were sequenced with the M13 dye primers.
The sequence information obtained from the IPCR product (sub)clones was used to design primers ABM5 to ABM14. Sequences were as follows: ABM5, 5′-TTA AAT GGA TGA TTG AAG AAT TGA G-3′; ABM6, 5′-TCT CCA GAG GCT TAT AGA AGT AAA C-3′; ABM8, 5′-GAG ATT GTC CAA CTT TGT TTA GAT A-3′; ABM9, 5′-CTC ATT CAA AGA AGC ATA CGG CGA T-3′; ABM10, 5′-AAG TCG TTT CGA TAA ATC ATA GGA C-3′; ABM11, 5′-TGT AGC CTG CAT CAA ACC GCG TGC A-3′; ABM12, 5′-GCA TCG ACG CGG TTT GAT GCA GGC T-3′; ABM13, 5′-CGG TAT CTA CCT ACC CCA CCT ATT T-3′; ABM14, 5′-ACC CAA TAG TTT TCC AAA CCG CAT A-3′. PCR products amplified with primer pairs ABM5 and -2, ABM5 and -6, ABM5 and -12, ABM1 and -6, ABM1 and -8, ABM3 and -8, ABM3 and -10, ABM7 and -8, ABM7 and -10, ABM7 and -12, ABM9 and -10, ABM9 and -12, ABM9 and -14, ABM11 and -10, ABM11 and -14, ABM13 and -10, and ABM13 and -14 were sequenced directly using dye terminator chemistry.
DNA sequencing.
Automated DNA sequencing was performed with fluorescence dye-labeled universal M13 primers or dye terminators. Analysis was performed on an automatic sequencer (model 373), according to the instructions supplied by Applied Biosystems Incorporated (Foster City, Calif.). Computer analyses of DNA and protein sequences were performed with the programs in the PCGene package and with Genetics Computer Groups programs. Database similarity searches were performed using the BLASTX and BLASTN algorithms (2), and sequence patterns were identified using BEAUTY (BLAST enhanced alignment utility) (29) and using the PROSITE database (3). GC content and codon usage were compared using the CUTG (codon usage tabulated from GenBank) database (17) and the Countcodon program. Pairwise alignments were made using ALIGN (20).
Expression and specificity of the NmeSI methyltransferase in E. coli.
PCR products were generated with primer combination ABM6 and MNCO1 (5′-TAG CAC CAT GGG TTT AGA AAA TTT TCA AT-3′) and with primer combination ABM6 and MNCO2 (5′-AAA TTT CCA TGG ATA CTA TAA GTA GC-3′) and cloned into vector pCR2.1, introducing NcoI sites (underlined in the primer sequences). Resulting plasmids pCRT1 and pCRT2 were checked by sequence analysis, after which the NcoI-SpeI fragments containing the methyltransferase genes of both plasmids were cloned into expression vector pSE380 and transformed to E. coli Top10F′, yielding plasmids pT1 and pT2, respectively. To detect expression of M.NmeSI, cells containing pSE380, pT1, and pT2 grown with or without IPTG were collected and lysed by boiling at 100°C for 5 min. The lysates were used for sodium dodecyl sulfate–polyacrylamide gel electrophoresis (11% separating gels), and the gels were stained with Coomassie brilliant blue. To assess the effects of M.NmeSI expression, plasmid DNA was isolated from these cells and the sensitivity to digestion with restriction endonucleases with recognition sites of interest was examined. Sequence analysis of the modified site was performed as described above using primers AB1249F (5′-GTG AAA GTA AAA GAT GCT GA-3′) and AB1658R (5′-TGT CAC GCT CGT CGT TTG GT-3′), which target vector sequences present in pSE380.
Nucleotide sequence accession numbers.
The nucleotide sequence data are available in the EMBL/GenBank/DDBJ nucleotide sequence databases under accession no. AF123569.
RESULTS AND DISCUSSION
Sequencing of the flanking sequences of the RDA amplicons.
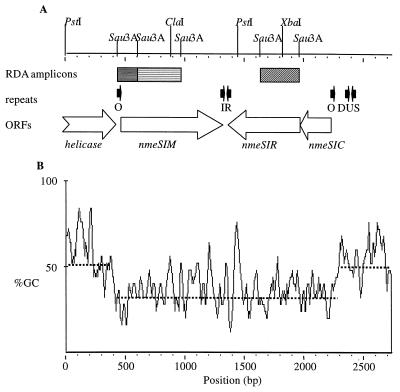
Previously, three DNA fragments that contained sequences specific for the hyperendemic lineage III cluster were identified by RDA using lineage III strain 800615 as the tester and strains 3532 and 830248 as the drivers (4). The sequence of the 3-kb locus containing these sequences was obtained as described in Materials and Methods. The region contains three complete open reading frames (ORFs) as indicated in Fig. 1A.
FIG. 1.
(A) Schematic representation of the NmeSI locus. Relevant restriction sites are indicated. Hatched bars, RDA fragments; open arrows, ORFs; solid arrows, repeats; O, 12-bp direct repeat, putative NmeSIC binding site (5); IR, 10-bp inverted repeat. (B) GC content of the NmeSI locus. Sliding windows with a size of 25 bp were used. Dotted lines, mean values.
The predicted protein sequences encoded by two of the ORFs are 53 and 33% identical to the predicted sequences of the ScaI methyltransferase and restriction endonuclease (30), respectively. The genes were named nmeSIM and nmeSIR. The putative target recognition domains (21) of the predicted M.NmeSI sequence and the M.ScaI methyltransferase are highly conserved between the two proteins. Both sequences contain conserved motif IV sequence TSPP, which is shared by N-4-cytosine-specific methyltransferases. Based on the order of the nine conserved motifs of methyltransferases, M.NmeSI belongs to the β group of methyltransferases (13).
The third ORF is 40% identical to the control element of the SmaI RM system. An alignment, given in Fig. 2, shows that the recognition helices of the helix-turn-helix regions of the two control element proteins are identical. The NmeSI locus is flanked by two 12-bp direct repeats (Fig. 1A), which have been postulated to contain the operator sequences for the control gene product (5). Notably, the ScaI RM system does not contain such a regulatory control element.
FIG. 2.
Alignment of NmeSIC and SmaIC. The putative helix-turn-helix regions of these short proteins are underlined. The second helices, the recognition helices, are identical and indicated by double underlining (5). Double dots, identical amino acids; single dots, similar amino acids.
Upstream (5′ of position 413) and downstream (3′ of position 2281) of the nmeSI locus are sequences that are present in both completely sequenced N. meningitidis strains (19, 25). Flanking the 5′ end of the region containing the RDA fragments is the 3′ end of an ORF that probably encodes a helicase, which is also present in the sequences from the three Neisseria genome projects (19, 25; B. A. Roe, L. Song, S. P. Lin, X. Yuan, S. Clifton, T. Ducey, L. Lewis, and D. W. Dyer, http://www.genome.ou.edu/gono. or http://www.ncbi.nlm.nih.gov/BLAST/ouacgtbl.html). Directly flanking the 3′ end (position 2375) of the NmeSI locus are two degenerate Neisseria DNA uptake sequences (DUSs), which form a 15-bp inverted repeat (Fig. 1A). DUSs are typical features of Neisseria DNA sequences. Sequence analysis suggests that the 3′ flanking region contains repetitive IS1106 DNA sequences typical for Neisseria DNA. This implies that only the NmeSI locus is lineage III specific, in contrast to the flanking sequences.
Strikingly, the GC content of the sequence containing the NmeSI locus is much lower (33% for both the full sequence and only the coding sequence) than those of the flanking sequences (54% for the partial helicase coding sequence and 50% for the 3′ region) and the expected value for N. meningitidis coding sequences (51%), as illustrated in Fig. 1B. The low GC content is not the result of overrepresentation of certain amino acids, as the locus encoding the ScaI RM system has a 58% GC content. The lower GC content suggests that the NmeSI locus was recently acquired by a lineage III N. meningitidis ancestor from an organism with a low GC content. Strikingly, sequences in the region downstream of the helicase in the serogroup A strain Z2491 encode a different RM system (19) and those of serogroup B strain MC58 encode a putative regulatory protein (25). This suggests that the NmeSI locus was not inserted, but rather replaced genes. In addition, this demonstrates the plasticity of the neisserial genome at this position.
Specificity of the methyltransferase.
Apart from protecting the bacterial chromosome from the action of its own restriction enzymes, DNA methylation also plays an important role in chromosome repair and replication (14). The methylation state of DNA has been shown to influence gene expression, the paradigm of this mechanism being the regulation of uropathogenic E. coli adhesin expression by DNA methylation (10, 27). Through the action of the methylase, a lineage III-specific RM system could interfere with expression of Neisseria virulence factors and thus be involved in the observed hypervirulent phenotype of these strains. Therefore, we determined the specificity of the methylase gene product as well as its expression in lineage III meningococcal strains.
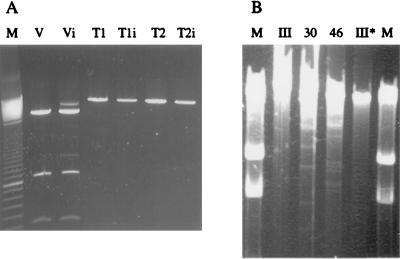
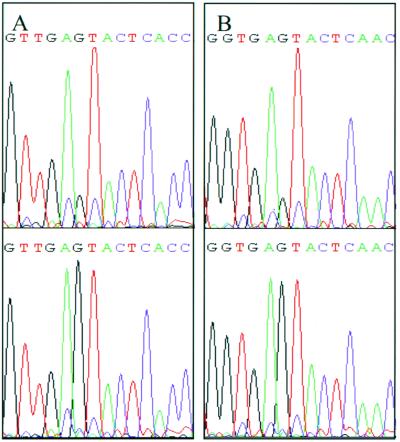
Since the M.NmeSI sequence was highly similar to the M.ScaI sequence in the target recognition domain, we expected the specificities of both methylases to be similar as well. To test this assumption nmeSIM was cloned in E. coli and plasmid DNA of the transformants was digested with ScaI. Since nmeSIM contains two possible ATG start codons, two primers (MNCO1 and MNCO2) were developed to clone nmeSIM in expression vector pSE380. E. coli cells containing recombinant expression systems pT1 and pT2 were grown with IPTG (induced) or without IPTG (uninduced) for 4 h. Expression of the methyltransferase was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis SDS-PAGE. Both induced Top10F′ (pT1) and induced Top10F′ (pT2) produced a protein of approximately the expected size (approximately 32 kDa), which was not detectable in induced Top10F′(pSE380) (data not shown). From noninduced and induced cultures, plasmids were isolated. The plasmids were digested with NcoI, resulting in a linear plasmid for pSE380, pT1, and pT2. This linearization was performed since the ScaI restriction endonuclease is known to cut a supercoiled plasmid poorly. As shown in Fig. 3A, pT1 and pT2 were not digested by ScaI, in contrast to linearized pSE380. This shows that ScaI site 5′-AGTACT-3′ in both pT1 and pT2 was modified by the methyltransferase so that it was no longer a substrate for ScaI. Apparently, induction of expression is not required to protect the plasmid DNA from digestion, since plasmids from both noninduced and induced cultures were resistant to digestion with ScaI. Also, the six-amino-acid difference at the N termini of the pT1 and pT2 products does not abolish methylation by one of the recombinant proteins. To certify that the observed inhibition of digestion was due to an N-4-cytosine modification, the regions containing the ScaI sites of pSE380 and pT1 were sequenced using dye terminator chemistry. It was previously shown that template methylation influences the incorporation of fluorescently labeled dideoxynucleoside triphosphates by AmpliTaq FS polymerase (22). As shown in Fig. 4, unmethylated template pSE380 yields lower G signals in the AGTACT sequence than methylated template pT1. This shows that the cytosine of the template is modified and rules out N5 methylation, as this results in lower G signals (22). Next, we confirmed that the methyltransferase is expressed and active in lineage III meningococci by incubating chromosomal DNA of two lineage III strains (800615 and 882066) and of two non-lineage III strains (3532 and 830248) with restriction enzymes ScaI (recognizing 5′-AGTACT-3′) and SphI (recognizing 5′-GCATGC-3′), respectively. The chromosomal DNA of the four strains was digested by SphI (results not shown), confirming that the DNA was susceptible to restriction endonucleases. Incubation with ScaI showed that the DNA from the non-lineage III strains is digested (Fig. 3B). However, the DNA of the lineage III strains is protected from digestion with ScaI, indicating that the methylation pattern of lineage III strains differs from that of non-lineage III strains. Possibly, this differential methylation pattern affects transcriptional regulation. Differential methylation of DNA has also been implicated in the frequency of phase variation in meningococci (6). Whether the methylation by M.NmeSI contributes to the hypervirulent character of lineage III isolates in either way awaits further investigations.
FIG. 3.
Digestion of methylated and nonmethylated plasmid DNA. (A) ScaI digestion of NcoI-linearized plasmid DNA. M, marker (100-bp marker; the lower intense band equals 800 bp); V, vector pSE380; T1, plasmid T1; T2, plasmid T2. The subscript i denotes induction of expression in the E. coli culture with IPTG. (B) ScaI digestion of N. meningitidis chromosomal DNA. M, marker (23.1-, 9.4-, and 6.6-kbp bands); III, lineage III strain 800615; 30, non-lineage III strain 3532; 46, non-lineage III strain 830246, III∗, lineage III strain 882066. To visualize the large resulting fragments, a high input of chromosomal DNA was used.
FIG. 4.
Dye terminator sequences of methylated and nonmethylated template. (A) Forward sequence obtained using primer AB1249F on nonmethylated template DNA pSE380 (top) and M.NmeSI methylated template DNA pT1 (bottom). The G signal in sequence AGTACT is different. (B) Reverse sequence obtained using primer AB1658R on nonmethylated template DNA pSE380 (top) and M.NmeSI methylated template DNA pT1 (bottom). The G signal in the sequence AGTACT is different.
The population biology of N. meningitidis is supposed to be influenced by horizontal gene transfer to a large extent (24). Horizontal gene transfer can be affected by RM systems (16), and this phenomenon has been implicated in maintenance of clonality of hypervirulent N. meningitidis clones (9). Therefore, the clonality of lineage III may in part be the consequence of the presence of the NmeSI RM system. However, the digestion of chromosomal DNA of the non-lineage III strains with ScaI yields large restriction fragments, indicating that sequence AGTACT is underrepresented in these meningococcal genomes (Fig. 3B). This is in accordance with the low number of AGTACT sites (only 65) in the genome sequence of strain MC58 (26). This suggests that NmeSI RM may have a limited effect on the clonal character of lineage III via its influence on horizontal gene transfer.
Evolutionary implications.
The serine residue in motif IV (SPPY) is conserved in N-4-cytosine methyltransferases and is part of the active site of the protein (28). This amino acid is encoded by the codon TCT in nmeSIM, whereas it is encoded by AGT in scaIM. Mutation of one of these codons to the other requires two point mutations, resulting in a threonine (ACT) or cysteine (TGT) residue after the first mutation. Neither of these residues has been found in any aminomethyltransferase. Possibly either of these residues results in an inactive protein. Therefore, it is unlikely that the two methyltransferases have a recent common ancestor. Moreover, the similarity between the predicted protein sequences of M.ScaI and M.NmeSI is more concentrated in the putative target recognition domain (TRD) than in conserved domains I to X of methyltransferases. In addition, the genes of the respective systems mentioned above have a large difference in GC content, and the NmeSI RM system seems to contain a regulatory protein, in contrast to the ScaI system. In conclusion, the similarity of the respective TRDs of the two methyltransferases is likely to be the consequence of convergent evolution.
ACKNOWLEDGMENTS
We acknowledge the Gonococcal Genome Sequencing Project supported by USPHS/NIH grant AI38399.
We thank Wendy Keijzers for expert technical assistance. We acknowledge B. A. Roe, L. Song, S. P. Lin, X. Yuan, S. Clifton, T. Ducey, L. Lewis and D. W. Dyer at the University of Oklahoma for sequence data prior to publication.
REFERENCES
- 1.Akopyanz N, Bukanov N O, Westblom T U, Kresovich S, Berg D E. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bairoch A. The PROSITE dictionary of sites and patterns in proteins, its current status. Nucleic Acids Res. 1993;21:3097–3103. doi: 10.1093/nar/21.13.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bart A, Dankert J, van der Ende A. Representational difference analysis of Neisseria meningitidis identifies sequences that are specific for the hyper-virulent lineage III clone. FEMS Microbiol Lett. 2000;188:111–114. doi: 10.1111/j.1574-6968.2000.tb09179.x. [DOI] [PubMed] [Google Scholar]
- 5.Bart A, Dankert J, van der Ende A. Operator sequences for the regulatory proteins of restriction modification systems. Mol Microbiol. 1999;31:1277–1278. doi: 10.1046/j.1365-2958.1999.01253.x. [DOI] [PubMed] [Google Scholar]
- 6.Bucci C, Lavitola A, Salvatore P, Del Giudice L, Massardo D R, Bruni C B, Alifano P. Hypermutation in pathogenic bacteria: frequent phase variation in meningococci is a phenotypic trait of a specialized mutator biotype. Mol Cell. 1999;3:435–445. doi: 10.1016/s1097-2765(00)80471-2. [DOI] [PubMed] [Google Scholar]
- 7.Caugant D A, Bol P, Høiby E A, Zanen H C, Frøholm L O. Clones of serogroup B Neisseria meningitidis causing systemic disease in the Netherlands, 1958-1986. J Infect Dis. 1990;162:867–874. doi: 10.1093/infdis/162.4.867. [DOI] [PubMed] [Google Scholar]
- 8.Caugant D A. Population genetics and molecular epidemiology of Neisseria meningitidis. APMIS. 1998;106:505–525. [PubMed] [Google Scholar]
- 9.Claus H, Friedrich A, Frosch M, Vogel U. Differential distribution of novel restriction-modification systems in clonal lineages of Neisseria meningitidis. J Bacteriol. 2000;182:1296–1303. doi: 10.1128/jb.182.5.1296-1303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hale W B, van der Woude M W, Braaten B A, Low D A. Regulation of uropathogenic Escherichia coli adhesin expression by DNA methylation. Mol Genet Metab. 1998;65:191–196. doi: 10.1006/mgme.1998.2744. [DOI] [PubMed] [Google Scholar]
- 11.Lisitsyn N, Lisitsyn N, Wigler M. Cloning the differences between two complex genomes. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 12.Maiden M C J, Bygraves J A, Feil E, Morelli G, Russell J E, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D A, Feavers I M, Achtman M, Spratt B G. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malone T, Blumenthal R M, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J Mol Biol. 1995;253:618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 14.Marinus M G. Methylation of DNA, P. 782–791. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. [Google Scholar]
- 15.Martin D R, Walker S J, Baker M G, Lennon D R. New Zealand epidemic of meningococcal disease identified by a strain with phenotype B:4:P1.4. J Infect Dis. 1998;177:497–500. doi: 10.1086/517385. [DOI] [PubMed] [Google Scholar]
- 16.Milkman R. Recombination and population structure in Escherichia coli. Genetics. 1997;146:745–750. doi: 10.1093/genetics/146.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ochman H, Gerber A S, Hartl D L. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parkhill J, Achtman M, James K D, Bentley K S D, Churcher C, Klee S R, Morelli G, Basham D, Brown D, Chillingworth T, Davies R M, Davis P, Devlin K, Feltwell T, Hamlin N, Holroyd S, Jagels K, Leather S, Moule S, Mungall K, Quail M A, Rajandream M A, Rutherford K M, Simmonds M, Skelton J, Whitehead S, Spratt B G, Barrell B G. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 20.Pearson W R, Lipman D J. Improved tools for biological sequence analysis. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Posfai J, Bhagwat A S, Posfai G, Roberts R J. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989;17:2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao B S, Buckler-White A. Direct visualization of site-specific and strand-specific DNA methylation patterns in automated DNA sequencing data. Nucleic Acids Res. 1998;26:2505–2507. doi: 10.1093/nar/26.10.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scholten R J P M, Bijlmer H A, Poolman J T, Kuipers B, Caugant D A, van Alphen L, Dankert J, Valkenburg H A. Meningococcal disease in the Netherlands, 1958-1990: a steady increase in the incidence since 1982 partially caused by new serotypes and subtypes of Neisseria meningitidis. Clin Infect Dis. 1993;16:237–246. doi: 10.1093/clind/16.2.237. [DOI] [PubMed] [Google Scholar]
- 24.Spratt B G, Smith N H, Zhou J, O'Rourke M, Feil E. The population genetics of the pathogenic Neisseriap. 143–160. In: Baumberg S, Young J P W, Wellington E M H, Saunders J R, editors. Population genetics in bacteria. Cambridge, United Kingdom: Cambridge University Press; 1995. [Google Scholar]
- 25.Tettelin H, Saunders N J, Heidelberg J, Jeffries A C, Nelson K E, Eisen J A, Ketchum K A, Hood D W, Peden J F, Dodson R J, Nelson W C, Gwinn M L, DeBoy R, Peterson J D, Hickey E K, Haft D H, Salzberg S L, White O, Fleischmann R D, Dougherty B A, Mason T, Ciecko A, Parksey D S, Blair E, Cittone H, Clark E B, Cotton M D, Utterback T R, Khouri H, Qin H, Vamathevan J, Gill J, Scarlato V, Masignani V, Pizza M, Grandi G, Sun L, Smith H O, Fraser C M, Moxon E R, Rappuoli R, Venter J C. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 26.Triglia T, Peterson M G, Kemp D J. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988;16:8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Woude M W, Braaten B A, Low D A. Evidence for global regulatory control of pilus expression in Escherichia coli by Lrp and DNA methylation: model building based on analysis of pap. Mol Microbiol. 1992;6:2429–2435. doi: 10.1111/j.1365-2958.1992.tb01418.x. [DOI] [PubMed] [Google Scholar]
- 28.Willcock D F, Dryden D T, Murray N E. A mutational analysis of the two motifs common to adenine methyltransferases. EMBO J. 1994;13:3902–3908. doi: 10.1002/j.1460-2075.1994.tb06701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Worley K C, Wiese B A, Smith R F. BEAUTY: an enhanced BLAST-based search tool that integrates multiple biological information resources into sequence similarity search results. Genome Res. 1995;5:173–184. doi: 10.1101/gr.5.2.173. [DOI] [PubMed] [Google Scholar]
- 30.Xu S-Y, Xiao J-P, Ettwiller L, Holden M, Aliotta J, Poh C L, Dalton M, Robinson D P, Petronzio T R, Moran L, Ganatra M, Ware J, Slatko B, Benner J. Cloning and expression of the Apa LI, Nsp HI, Sac I, Sca I, and Sap I restriction-modification systems in Escherichia coli. Mol Gen Genet. 1998;260:226–231. doi: 10.1007/s004380050890. [DOI] [PubMed] [Google Scholar]