Abstract
The objective of this study was to compare the virulence of 4 porcine circovirus type 2 (PCV-2) genotypes (2a, 2b, 2d, and 2e) in pigs singly infected with 1 of these 4 PCV-2 genotypes and pigs dually infected with a combination of 1 of the 4 PCV-2 genotypes and porcine reproductive and respiratory syndrome virus (PRRSV). Virulence was determined based on levels of PCV-2 loads in the blood and lymph nodes and the severity of lymphoid lesion. Within the singly infected groups, PCV-2a, PCV-2b, and PCV-2d resulted in a similar virulence to each other and all were more virulent than the PCV-2e groups. Within the dually infected groups, the combination of PCV-2d and PRRSV was more virulent than the other 3 PCV-2 genotypes (2a, 2b, and 2e), each in combination with PRRSV. Both PCV-2a+PRRSV and PCV-2b+PRRSV were more virulent than PCV-2e+PRRSV in dually infected pigs. This increased virulence of PCV-2d compared to the other 3 PCV-2 genotypes (2a, 2b, and 2e) may be attributed to an extra amino acid (lysine residue) found within open reading frame 2 (ORF2) of PCV-2d. In contrast, extra amino acids in ORF2 may decrease the virulence of PCV-2e when compared to the other 3 PCV-2 genotypes (2a, 2b, and 2d). The results of this study demonstrated that PCV-2d was the most virulent PCV-2 genotype in pigs co-infected with PRRSV. The results also suggest that genetic differences in the ORF2 of PCV-2 may affect the virulence of PCV-2 genotypes.
Résumé
L’objectif de cette étude était de comparer la virulence de quatre génotypes de circovirus porcin de type 2 (PCV-2) (2a, 2b, 2d et 2e) chez des porcs infectés individuellement par un de ces quatre génotypes de PCV-2 et des porcs doublement infectés par une combinaison d’un des quatre génotypes PCV-2 et du virus du syndrome reproducteur et respiratoire porcin (PRRSV). La virulence a été déterminée en fonction des niveaux de charges de PCV-2 dans le sang et les ganglions lymphatiques et de la gravité des lésions lymphoïdes. Au sein des groupes infectés individuellement, PCV-2a, PCV-2b et PCV-2d ont entraîné une virulence similaire les uns aux autres et tous étaient plus virulents que les groupes PCV-2e. Au sein des groupes doublement infectés, la combinaison du PCV-2d et du PRRSV était plus virulente que les trois autres génotypes du PCV-2 (2a, 2b et 2e), chacun en combinaison avec le PRRSV. Le PCV-2a+PRRSV et le PCV-2b+PRRSV étaient plus virulents que le PCV-2e+PRRSV chez les porcs doublement infectés. Cette virulence accrue du PCV-2d par rapport aux trois autres génotypes du PCV-2 (2a, 2b et 2e) peut être attribuée à un acide aminé supplémentaire (résidu lysine) trouvé dans le cadre de lecture ouvert 2 (ORF2) de PCV-2d. En revanche, des acides aminés supplémentaires dans ORF2 peuvent diminuer la virulence du PCV-2e par rapport aux trois autres génotypes PCV-2 (2a, 2b et 2d). Les résultats de cette étude ont démontré que le PCV-2d était le génotype PCV-2 le plus virulent chez les porcs co-infectés par le PRRSV. Les résultats suggèrent également que des différences génétiques dans l’ORF2 du PCV-2 peuvent affecter la virulence des génotypes du PCV-2.
(Traduit par Docteur Serge Messier)
Introduction
Porcine circovirus type 2 (PCV-2) is a nonenveloped, single-stranded, circular DNA virus belonging to the genus Circovirus in the family Circoviridae (1,2). PCV-2 contains 2 major open reading frames (ORFs): ORF1 encodes 2 replication-associated proteins (rep and rep′) and ORF2 encodes a viral capsid protein (cap), which is the only structural protein (1).
Porcine circovirus type-2 (PCV-2) is the primary etiological agent of several diseases and syndromes collectively referred to as porcine circovirus-associated disease (PCVAD) (3). Porcine reproductive and respiratory syndrome virus (PRRSV) is an enveloped, single-stranded, positive-sense RNA virus belonging to the genus Porarterivirus in the family Arteriviridae with 2 species, PRRSV-1 (formerly known as European genotype) and PRRSV-2 (formerly known as North American genotype) (4).
Infection with PRRSV can cause reproductive failure (abortions, weak and stillborn piglets, and infertility) in sows and respiratory disease in weaned and growing pigs (5). Although PCV-2 is considered the primary causative agent of PCVAD, it requires other pathogens to reach its full expression. Coinfection of pigs with PCV-2 and PRRSV is the most common combination that causes the full spectrum of clinical signs and lesions associated with PCVAD (6,7).
At least 8 distinct PCV-2 genotypes (2a to 2h) have been identified to date (8). Among these, PCV-2a, PCV-2b, and PCV-2d are considered major genotypes, with PCV-2d currently being the most prevalent. Another PCV-2 genotype, PCV-2e, was first detected in the USA and Mexico in 2015, followed by China and Korea in 2017 and 2021, respectively (9–12).
Capsid protein is generally encoded by ORF2, which determines the antigenicity and virulence of PCV-2 (13,14). The capsid proteins of PCV-2a and PCV-2b are considered the traditional forms. However, PCV-2d and PCV-2e are both recently emerged genotypes with different lengths of capsid protein (9,15). The biological function of extra amino acids, particularly in relation to virulence, remains unknown.
The objective of this study was to compare the virulence of 4 PCV-2 genotypes. This was analyzed in both pigs singly infected with 1 of 4 PCV-2 genotypes (PCV-2a, PCV-2b, PCV-2d, PCV-2e), as well as in pigs dually infected with a combination of 1 of the 4 PCV-2 genotypes and PRRSV (PCV-2a/PRRSV, PCV-2b/PRRSV, PCV-2d/PRRSV, PCV-2e/PRRSV).
Materials and methods
Animals
All experimental protocols were approved before the study began by the Seoul National University Institutional Animal Care and Use Committee (SNU-210120-7).
A total of 60 clinically healthy, colostrum-fed conventional pigs from sows not previously vaccinated against PCV-2 were purchased at 38 d of age from a PRRSV-free commercial farm. The farm was also free of Mycoplasma hyopneumoniae based on serological testing and long-term clinical and slaughter history.
Pigs were confirmed seronegative for PRRSV, PCV-2, and M. hyopneumoniae by use of commercial enzyme-linked immunosorbent assay (ELISA) kits (PRRSV: HerdChek PRRS X3 Ab test; IDEXX Laboratories, Westbrook, Maine, USA; PCV-2: INgezim CIRCO IgG; Ingenasa, Madrid, Spain; M. hyopneumoniae: M. hyo. Ab test; IDEXX Laboratories). Piglets were also confirmed negative for PCV-2 (a, b, d, and e) viremia, PRRSV viremia, and M. hyopneumoniae laryngeal shedding by real-time polymerase chain reaction (PCR) testing upon arrival.
Experimental design
For the study, pigs were allocated to 10 groups (6 pigs per group) using the random number generator function from Excel (Microsoft, Redmond, Washington, USA). Pigs were randomly assigned into 10 separate rooms based on group. At 0 d post-inoculation (dpi), 42 d of age, pigs in the PCV-2a, PCV-2b, PCV-2d, and PCV-2e groups were inoculated intranasally with either 3 mL of PCV-2a (SNUVR100032 strain, GenBank no. KF871067); PCV-2b (SNUVR202155 strain, GenBank no. MZ440696); PCV-2d (SNUVR202003 strain, GenBank no. MZ440695); or PCV-2e (SNUVR199707strain, GenBank no. MN967003) in their respective groups. Each inoculum contained 1.2 ×105 of 50% tissue culture infective dose (TCID50/mL) in the 5th passage in PCV-free PK15 cell lines.
Pigs in the PCV-2a/PRRSV, PCV-2b/PRRSV, PCV-2d/PRRSV, and PCV-2e/PRRSV groups were each inoculated intranasally with a mixture of equal volumes of PCV-2 (PCV-2a, PCV-2b, PCV-2d, and PCV-2e) and PRRSV. Each 3-mL PCV-2 administered challenge contained 1.2 × 105 TCID50/mL per dose, regardless of the PCV-2 genotype. Pigs that received the PRRSV co-challenge were also concurrently administered 3 mL of PRRSV-2 (SNUVR090851 strain, GenBank no. JN315685) inoculum (1.2 × 105 TCID50/mL in the 5th passage in the MARC-145 cells lines).
Pigs in the PRRSV group were inoculated intranasally with 3 mL of PRRSV-2 (SNUVR090851 strain, GenBank no. JN315685) inoculum (1.2 × 105 TCID50/mL in the 5th passage in the MARC-145 cells lines). Pigs in the negative control group were inoculated intranasally with 6 mL (3 mL/nostril) of uninfected cell culture supernatant.
Blood samples were collected from each pig by jugular venipuncture at 0, 7, 14, and 21 dpi. At 21 dpi, pigs were sedated by an intravenous injection of sodium pentobarbital and then euthanized by electrocution as previously described (16). Tissues were collected from each pig at necropsy.
Clinical observation
Pigs were monitored daily for clinical signs and scored weekly using a score-ranking system that ranged from 0 (normal) to 6 (severe dyspnea and abdominal breathing) (17). All observers involved in these processes were blinded to type of challenge virus.
Growth performance
The live weight of each pig was measured at 42 (0 dpi) and 63 (21 dpi) d of age. The average daily weight gain (ADWG; gram/pig/day) was analyzed over the time period between 42 and 63 d of age. The average daily weight gain during the different production stages was calculated as the difference between the starting and final weight divided by the duration of the stage. Data for dead or removed pigs were included as well in the calculation. Body weight and ADWG data are provided in Table I.
Table I.
Body weight and average daily weight gain (ADWG) data (mean ± standard deviation) of pigs in single-infected and dual-infected groups at 42 d of age (0 d postinoculation, 0 dpi) and 63 d of age (21 dpi).
| Groups | Body weight (kg) | ADWG (gram/pig/day) | |
|---|---|---|---|
|
|
|
||
| 42 d old (0 dpi) | 63 d old (21 dpi) | 42 to 63 d old | |
| Single-infection | |||
| PCV-2a | 8.40 ± 0.17 | 16.17 ± 1.05 | 369.84 ± 50.55 |
| PCV-2b | 8.56 ± 0.29 | 16.22 ± 0.61 | 364.84 ± 18.22 |
| PCV-2d | 8.50 ± 0.09 | 16.08 ± 0.76 | 361.11 ± 36.32 |
| PCV-2e | 8.33 ± 0.53 | 16.02 ± 0.88 | 365.87 ± 56.38 |
| Negative control | 8.55 ± 0.30 | 16.37 ± 1.19 | 372.22 ± 48.23 |
| Dual-infection | |||
| PCV-2a/PRRSV | 8.37 ± 0.53 | 13.08 ± 0.86a,b | 224.60 ± 39.40a,b |
| PCV-2b/PRRSV | 8.58 ± 0.56 | 13.38 ± 1.13a,b | 228.57 ± 56.90a,b |
| PCV-2d/PRRSV | 8.47 ± 0.59 | 12.88 ± 1.39a | 210.32 ± 41.01a |
| PCV-2e/PRRSV | 8.18 ± 0.70 | 14.08 ± 0.69a,b | 280.95 ± 9.04a,b |
| PRRSV | 8.45 ± 0.27 | 14.87 ± 0.62b,c | 305.56 ± 31.79b,c |
| Negative control | 8.55 ± 0.30 | 16.37 ± 1.19c | 372.22 ± 48.23c |
Different superscripts (a, b, and c) indicate significant (P < 0.05) difference among either single-infected or dual-infected groups.
Serology
Serum samples were also tested for antibodies against PCV-2 (INgezim CIRCO IgG; Ingenasa) and PRRSV (HerdChek PRRS X3 Ab test; IDEXX Laboratories). Samples were considered positive for PCV-2 antibodies if the optical density (OD) was > 0.3 and for PRRSV antibodies if the sample-to-positive (S/P) ratio was ≥ 0.4, according to the manufacturer’s instructions.
Quantification of PCV-2 DNA
A commercial kit (QIAamp DNA Mini Kit; QIAGEN, Valencia, California, USA) was used to extract DNA from serum samples for PCV-2. The forward and reverse primer (5′-GGGCCAGAATTC AACCTTAA-3′ and 5′-CGCACCTTCGGATATACTATCA-3′) and probe (5′-FAM-GGGGACCAACAAAATCTCTATACCCTTT-TA MRA-3′) were used to detect PCV-2a (18). The forward and reverse primer (5′-GGGCCAGAATTCAACCTTAA-3′ and 5′-CGCACCTTC GGATATACTATCA-3′) and probe (5′-FAM-GGGCTCAAACCCCC GCTCTGTGCCCTTT-TAMRA-3′) were used to detect PCV-2b (18). The forward and reverse primer (5′-GTATTCAAAGGGCACA GTGAGG-3′ and 5′-CACCATCGGTTATACTGTCAAGAAA-3′) and probe (5′-FAM-CATCATGTCCACATTCCAG-TAMRA-3′) were used to detect PCV-2d (19). The forward and reverse primer (5′-CTCTCCC GCTCCTTTGTATATACTG-3′ and 5′-TCCAATATTAAATCTCATC ATGTCCAC-3′) and probe (5′-FAM-TAACCTCCACAGTCACA CCGCCATCAT-TAMRA-3′) were used to detect PCV-2e (20). Genomic DNA copy numbers for PCV-2a, PCV-2b, PCV-2d, and PCV-2e were quantified by real-time PCR (18–20).
Quantification of PRRSV
RNA was extracted from serum samples using the QIAamp Viral RNA Mini Kit (QIAGEN). The forward and reverse primer (5′-GTGGTGAATGGCACTGATTG-3′ and 5′-CCCCACA CGGTCGCC-3′) and probe (5′-FAM-TCCTCTAAGTCACCTAT TCAATTAGGGCGA-TAMRA-3′) were used to detect PRRSV-2. Real-time PCR for PRRSV was carried out to quantify PRRSV genomic cDNA copy (21).
Histopathology
For the morphometric analysis of histopathological changes in lung and superficial inguinal lymph nodes, 3 sections of lung and lymph node were stained with hematoxylin and eosin and examined blindly. Lung sections were selected based on lesional areas and were evaluated for the severity of the interstitial pneumonia and given a score ranging from 0 to 4 (0 = normal; 1 = mild interstitial pneumonia; 2 = moderate multifocal interstitial pneumonia; 3 = moderate diffuse interstitial pneumonia; and 4 = severe interstitial pneumonia) (17).
Lymph node sections were evaluated for the presence of lymphoid depletion and inflammation and given a score ranging from 0 to 5 (0 = normal; 1 = mild lymphoid depletion; 2 = mild-to-moderate lymphoid depletion and histiocytic replacement; 3 = moderate diffuse lymphoid depletion and histiocytic replacement; 4 = moderateto-severe lymphoid depletion and histiocytic replacement; and 5 = severe lymphoid depletion and histiocytic replacement) (22).
Immunohistochemistry
Immunohistochemistry (IHC) using a polyclonal PCV-2a antibody (Veterinary Diagnostic Laboratory, Iowa State University, Ames, Iowa, USA) and morphometric analysis of IHC was carried out as described in a previous study (23). Positive signal was quantified using the NIH Image J 1.45s Program, available from http://imagej.nih.gov/ij/download.html For each slide of lymph node tissue, 10 fields were randomly selected and the number of positive cells per unit area (0.25 mm2) was counted. The mean values were also calculated (23).
Statistical analysis
Prior to statistical analysis, real-time PCR data were log-transformed to reduce variance and positive skewness. Data were tested for normal distribution using the Shapiro-Wilk test. The linear mixed model (LMM) was employed to examine differences in genomic copies of viral DNA and total antibodies against the virus at each time point among groups. The LMM consisted of both fixed and random effects to adjust non-independence in the data. The fixed effects of the model included treatment and time point. The random effects of the model included room and animal. The Kruskal-Wallis test was used for variables without a normal distribution, e.g., microscopic lymphoid and lung lesion scores. The test results that showed a statistical significance were further evaluated with a Dunn’s nonparametric comparison for post-hoc test.
Results
Clinical signs
Pigs inoculated with 1 of 4 PCV-2 genotypes exhibited mild tachypnea and sneezing, whereas pigs inoculated with PRRSV showed moderate tachypnea and abdominal breathing. Pigs dually inoculated with 1 of 4 PCV-2 genotypes and PRRSV exhibited moderate-to-severe respiratory disease that was characterized mainly by dyspnea, pronounced abdominal breathing, lethargy, coughing, and occasional sneezing.
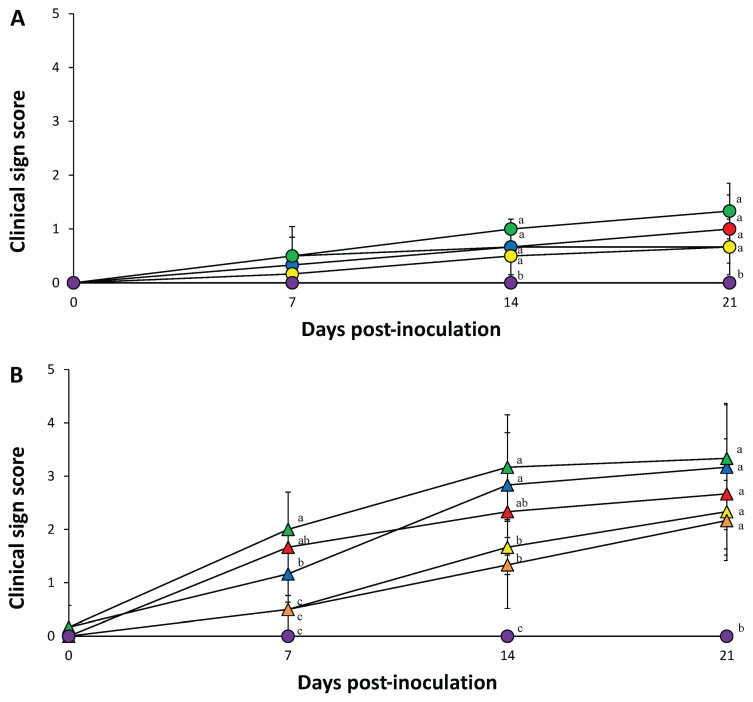
Respiratory sign scores in each of the 4 dually inoculated (PCV-2a, -2b, -2d, and -2e/PRRSV) groups and PRRSV-inoculated group were significantly greater (P < 0.05) than any of those from groups inoculated with 1 PCV-2 genotype or the control groups at 14 and 21 dpi. Respiratory sign scores in the 4 single-inoculated groups (PCV-2a, -2b, -2d, and -2e) were significantly greater (P < 0.05) than those of the control group at 14 and 21 dpi (Figure 1). No respiratory signs were observed in the control pigs throughout the entire experiment.
Figure 1.
A − Clinical signs of pigs from PCV-2a (
 ), PCV-2b (
), PCV-2b (
 ), PCV-2d (
), PCV-2d (
 ), PCV-2e (
), PCV-2e (
 ), and negative control (
), and negative control (
 ) groups. B − Clinical signs of pigs from PCV-2a/PRRSV (
) groups. B − Clinical signs of pigs from PCV-2a/PRRSV (
 ), PCV-2b/PRRSV (
), PCV-2b/PRRSV (
 ), PCV-2d/PRRSV (
), PCV-2d/PRRSV (
 ), PCV-2e/PRRSV (
), PCV-2e/PRRSV (
 ), PRRSV (
), PRRSV (
 ), and negative control (
), and negative control (
 ) groups. Variation is expressed as the standard deviation. Different superscripts (a and b) indicate significant (P < 0.05) difference among either single-infected or dual-infected groups.
) groups. Variation is expressed as the standard deviation. Different superscripts (a and b) indicate significant (P < 0.05) difference among either single-infected or dual-infected groups.
Growth performance
There was no statistical difference in average body weight among the 10 groups at the start of the experiment (42-day-old pigs). At 21 dpi, pigs in the PCV-2d/PRRSV groups had significantly lower (P < 0.05) average body weights than pigs in the PRRSV-inoculated and control groups. At 21 dpi, pigs in the PCV-2a/PRRSV, PCV-2b/PRRSV, and PCV-2e/PRRSV groups had significantly lower (P < 0.05) average body weights than pigs in the control groups.
Pigs in the PCV-2d/PRRSV groups had significantly lower (P < 0.05) average daily weight gain (ADWG) from 42 to 63 d of age than pigs in the PRRSV-inoculated and control groups. Pigs in the PCV-2a/PRRSV, PCV-2b/PRRSV, and PCV-2e/PRRSV groups had significantly lower (P < 0.05) ADWG from 42 to 63 d of age than pigs in the control groups (Table I).
PCV-2 serology
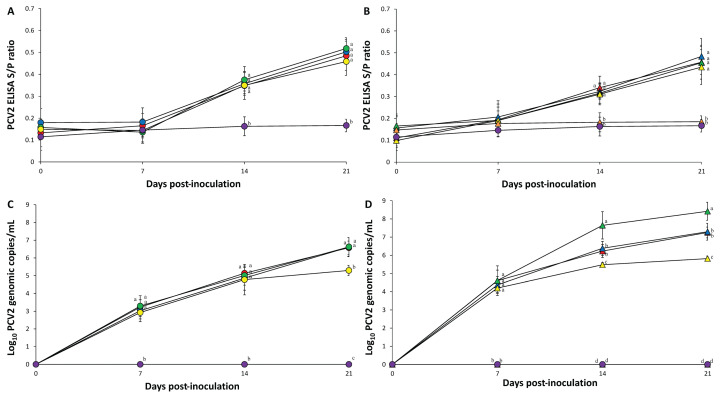
Prior to inoculation, all serum samples collected from the 10 groups were seronegative against PCV-2 and PRRSV. There was no statistical difference in PCV-2 S/P ratios at 0, 7, 14, and 21 dpi in pigs from the 4 single-inoculated groups (PCV-2a, -2b, -2d, and -2e) (Figure 2 A) and 4 dually inoculated groups (PCV-2a, -2b, -2d, and -2e/PRRSV) (Figure 2 B). PCV-2 antibodies were not detected in pigs from the PRRSV and control groups at any time.
Figure 2.
A − Porcine circovirus type 2 (PCV-2)-specific ELISA antibody levels in serum of pigs from PCV-2a (
 ), PCV-2b (
), PCV-2b (
 ), PCV-2d (
), PCV-2d (
 ), PCV-2e (
), PCV-2e (
 ), and negative control (
), and negative control (
 ) groups. B − PCV-2-specific ELISA antibody levels in serum of pigs from PCV-2a/PRRSV (
) groups. B − PCV-2-specific ELISA antibody levels in serum of pigs from PCV-2a/PRRSV (
 ), PCV-2b/PRRSV (
), PCV-2b/PRRSV (
 ), PCV-2d/PRRSV (
), PCV-2d/PRRSV (
 ), PCV-2e/PRRSV (
), PCV-2e/PRRSV (
 ), PRRSV (
), PRRSV (
 ;), and negative control (
;), and negative control (
 ) groups. C − Mean values of the genomic copy number of porcine circovirus type 2 (PCV-2) DNA in serum of pigs from PCV-2a (
) groups. C − Mean values of the genomic copy number of porcine circovirus type 2 (PCV-2) DNA in serum of pigs from PCV-2a (
 ), PCV-2b (
), PCV-2b (
 ), PCV-2d (
), PCV-2d (
 ), PCV-2e (
), PCV-2e (
 ), and negative control (
), and negative control (
 ) groups. D − Mean values of the genomic copy number of porcine circovirus type 2 (PCV-2) DNA in serum of pigs from PCV-2a/PRRSV (
) groups. D − Mean values of the genomic copy number of porcine circovirus type 2 (PCV-2) DNA in serum of pigs from PCV-2a/PRRSV (
 ), PCV-2b/PRRSV (
), PCV-2b/PRRSV (
 ), PCV-2d/PRRSV (
), PCV-2d/PRRSV (
 ), PCV-2e/PRRSV (
), PCV-2e/PRRSV (
 ), PRRSV (
), PRRSV (
 ), and negative control (
), and negative control (
 ) groups. Variation is expressed as the standard deviation. Different superscripts (a, b, c, and d) indicate significant (P < 0.05) difference among either single-infected or dual-infected groups.
) groups. Variation is expressed as the standard deviation. Different superscripts (a, b, c, and d) indicate significant (P < 0.05) difference among either single-infected or dual-infected groups.
Quantification of PCV-2 DNA in blood
Prior to inoculation, all serum samples collected from the 10 groups were negative for PCV-2. At 21 dpi, pigs in the PCV-2a, PCV-2b, and PCV-2d groups had a significantly higher (P < 0.05) number of PCV-2 genomic copies than pigs in the PCV-2e group (Figure 2 C). Pigs in the PCV-2d/PRRSV groups had a significantly higher (P < 0.05) number of PCV-2 genomic copies than pigs in the PCV-2a/PRRSV, PCV-2b/PRRSV, and PCV-2e/PRRSV groups at 14 and 21 dpi. Pigs in the PCV-2a/PRRSV and PCV-2b/PRRSV groups had a significantly higher (P < 0.05) number of PCV-2 genomic copies than pigs in the PCV-2e/PRRSV group at 14 and 21 dpi (Figure 2 D). PCV-2 genomic copies were not detected in pigs from the PRRSV and control groups at any time.
PRRSV serology
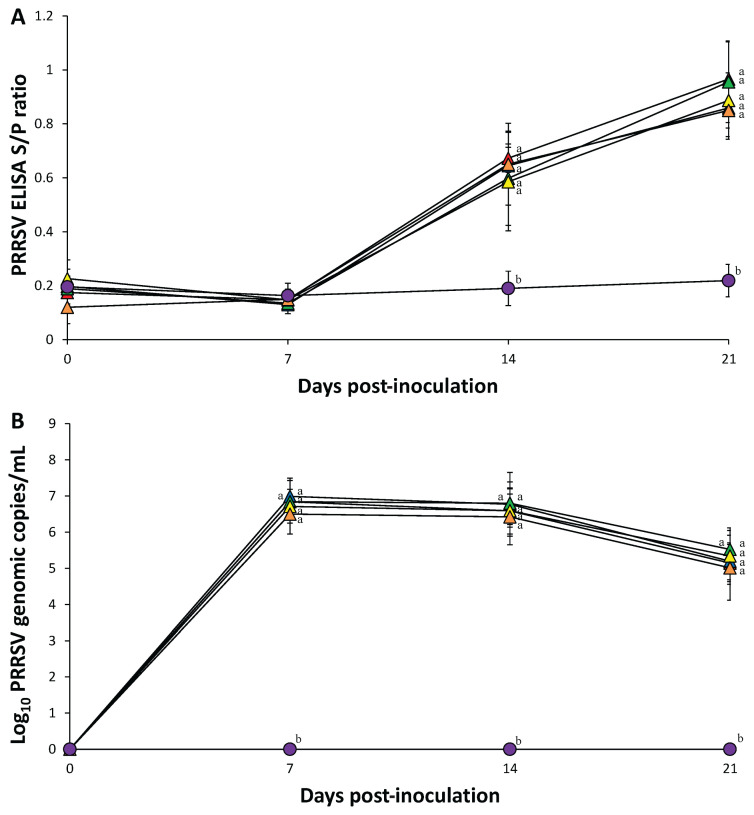
There was no statistical difference in PRRSV S/P ratios at 7, 14, and 21 dpi in pigs in the 4 dually inoculated (PCV-2a, -2b, -2d, and -2e/PRRSV) and PRRSV-inoculated groups. PRRSV antibodies were not detected in pigs from the control groups at any time (Figure 3 A).
Figure. 3.
A − Porcine reproductive and respiratory syndrome virus (PRRSV)-specific ELISA antibody levels. B − Mean values of the genomic copy number of porcine reproductive and respiratory syndrome virus (PRRSV) cDNA in serum of pigs from PCV-2a/PRRSV (
 ), PCV-2b/PRRSV (
), PCV-2b/PRRSV (
 ), PCV-2d/PRRSV (
), PCV-2d/PRRSV (
 ), PCV-2e/PRRSV (
), PCV-2e/PRRSV (
 ), PRRSV (
), PRRSV (
 ), and negative control (
), and negative control (
 ) groups. Variation is expressed as the standard deviation. Different superscripts (a and b) indicate significant (P < 0.05) difference among 6 groups.
) groups. Variation is expressed as the standard deviation. Different superscripts (a and b) indicate significant (P < 0.05) difference among 6 groups.
Quantification of PRRSV cDNA in blood
Prior to inoculation, all serum samples collected from the 10 groups were negative for PRRSV. There were no statistical differences in the number of PRRSV genomic copies in pigs from the 4 dually inoculated (PCV-2a, -2b, -2d, and -2e/PRRSV) and PRRSV-inoculated groups at any time. PRRSV genomic copies were not detected in pigs from the control groups at any time (Figure 3 B).
Pathology
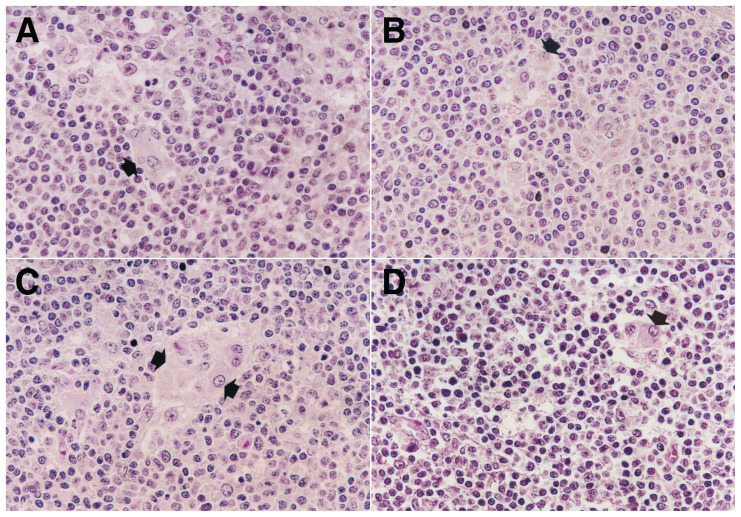
Pigs in the 4 single inoculated (PCV-2a, -2b, -2d, and -2e) groups had minimal-to-mild lymphoid depletion in their lymph nodes. Pigs in the dually inoculated (PCV-2a, -2b, -2d, and -2e/PRRSV) groups had typical PCV-2-associated lymphoid lesions with moderate-to-severe lymphoid depletion, moderate-to-severe granulomatous inflammation, and prominent multinucleated giant cells (Figure 4).
Figure 4.
Lymph node histopathology. A − Moderate granulomatous inflammation characterized by infiltration with reactive histiocytes (arrow) from pigs inoculated with porcine circovirus type 2a (PCV-2a) and porcine reproductive and respiratory syndrome virus (PRRSV). B − Moderate granulomatous inflammation characterized by infiltration with reactive histiocytes (arrow) from pigs inoculated with PCV-2b and PRRSV. C − Severe granulomatous inflammation characterized by infiltration with reactive histiocytes and multinucleated giant cells (arrows) from pigs inoculated with PCV-2d and PRRSV. D − Mild granulomatous inflammation characterized by infiltration with reactive histiocytes from pigs inoculated with PCV-2e and PRRSV. Hematoxylin and eosin. ×200.
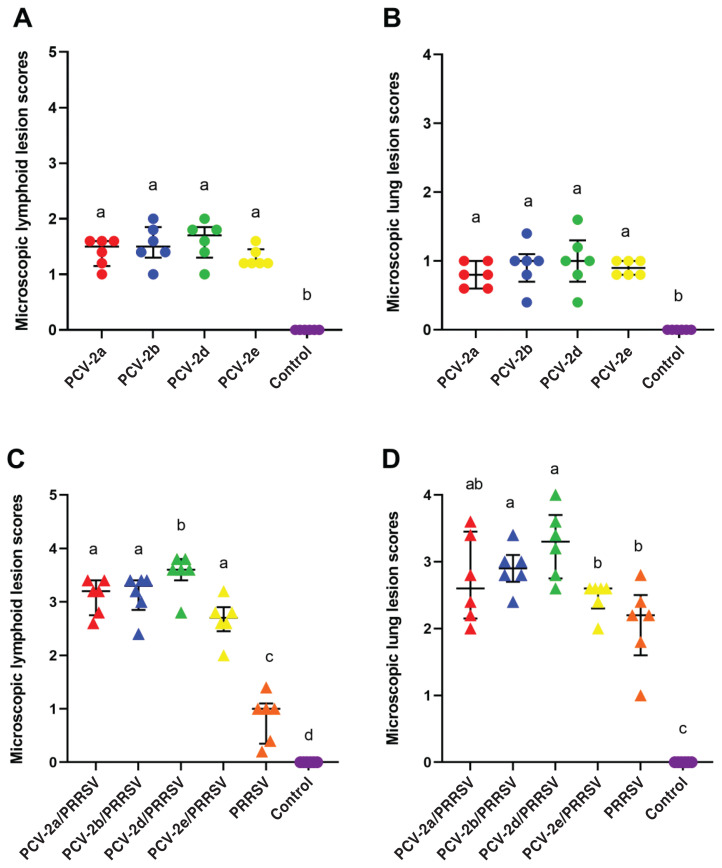
At 21 dpi, pigs in the PCV-2d/PRRSV groups had significantly higher (P < 0.05) microscopic lymphoid lesion scores than pigs in the other 3 dually inoculated (PCV-2a, -2b, and -2e/PRRSV) groups (Figure 5) (Table II).
Figure 5.
A − Median values of the microscopic lymphoid lesion scores in pigs from PCV-2a (
 ), PCV-2b (
), PCV-2b (
 ), PCV-2d (
), PCV-2d (
 ), PCV-2e (
), PCV-2e (
 ), and negative control (
), and negative control (
 ) groups. B − Median values of the microscopic lung lesion scores in pigs from PCV-2a (
) groups. B − Median values of the microscopic lung lesion scores in pigs from PCV-2a (
 ), PCV-2b (
), PCV-2b (
 ), PCV-2d (
), PCV-2d (
 ), PCV-2e (
), PCV-2e (
 ), and negative control (
), and negative control (
 ) groups. C − Median values of the microscopic lymphoid lesion scores in pigs from PCV-2a/PRRSV (
) groups. C − Median values of the microscopic lymphoid lesion scores in pigs from PCV-2a/PRRSV (
 ), PCV-2b/PRRSV (
), PCV-2b/PRRSV (
 ), PCV-2d/PRRSV (
), PCV-2d/PRRSV (
 ), PCV-2e/PRRSV (
), PCV-2e/PRRSV (
 ), PRRSV (
), PRRSV (
 ), and negative control (
), and negative control (
 ) groups. D − Median values of the microscopic lung lesion scores in pigs from PCV-2a/PRRSV (
) groups. D − Median values of the microscopic lung lesion scores in pigs from PCV-2a/PRRSV (
 ), PCV-2b/PRRSV (
), PCV-2b/PRRSV (
 ), PCV-2d/PRRSV (
), PCV-2d/PRRSV (
 ), PCV-2e/PRRSV (
), PCV-2e/PRRSV (
 ), PRRSV (
), PRRSV (
 ), and negative control (
), and negative control (
 ) groups. Variation is expressed as the interquartile range (IQR). Different superscripts (a, b, c, and d) indicate significant (P < 0.05) difference among either single-infected or dual-infected groups.
) groups. Variation is expressed as the interquartile range (IQR). Different superscripts (a, b, c, and d) indicate significant (P < 0.05) difference among either single-infected or dual-infected groups.
Table II.
Pathology data of 6 pigs in each of 10 groups at 21 d post-inoculation (dpi).
| Groups | Microscopic lymphoid lzesion scores | Microscopic lung lesion scores | Number of PCV-2 antigen- positive cells |
|---|---|---|---|
| Single-infection | |||
| PCV-2a | 1.50 ± 0.35a | 0.80 ± 0.30a | 16.67 ± 2.00a |
| PCV-2b | 1.50 ± 0.35a | 1.00 ± 0.15a | 18.44 ± 3.53a |
| PCV-2d | 1.70 ± 0.35a | 1.00 ± 0.30a | 17.89 ± 4.17a |
| PCV-2e | 1.20 ± 0.15a | 0.90 ± 0.20a | 12.39 ± 1.41b |
| Negative control | 0.00 ± 0.00b | 0.00 ± 0.00b | 0.00 ± 0.00c |
| Dual-infection | |||
| PCV-2a/PRRSV | 3.20 ± 0.45a | 2.60 ± 1.00a,b | 31.17 ± 3.42a |
| PCV-2b/PRRSV | 3.30 ± 0.35a | 2.90 ± 0.20a | 30.17 ± 7.43a |
| PCV-2d/PRRSV | 3.60 ± 0.15b | 3.30 ± 0.65a | 41.72 ± 5.20b |
| PCV-2e/PRRSV | 2.70 ± 0.20a | 2.60 ± 0.15b | 14.17 ± 4.04c |
| PRRSV | 1.00 ± 0.45c | 2.20 ± 0.45b | 0.00 ± 0.00d |
| Negative control | 0.00 ± 0.00d | 0.00 ± 0.00c | 0.00 ± 0.00d |
Different superscripts (a, b, c, and d) indicate significant (P < 0.05) difference among either single-infected or dual-infected groups. Microscopic lymphoid and lung lesion scores: median ± interquartile range (IQR) and number of PCV-2 antigen-positive cells: mean ± standard deviation (SD).
Pigs in the dually inoculated (PCV-2a, -2b, -2d, and -2e/PRRSV) groups developed typical interstitial pneumonia characterized by septal thickening with mainly infiltrates of macrophages and accumulation of macrophages and necrotic debris in alveolar space. Many alveolar septa were entirely lined with hypertrophied type-2 pneumocytes. At 21 dpi, pigs in the PCV-2b/PRRSV and PCV-2d/PRRSV groups had significantly higher (P < 0.05) interstitial pneumonia lesion scores than pigs in the PCV-2e/PRRSV or PRRSV inoculated groups (Figure 5) (Table II).
Immunohistochemistry
All pigs inoculated with PCV-2, whether alone or in combination with PRRSV, were immunolabelled for PCV-2 antigen in their lymph nodes. PCV-2 antigens were mainly detected in follicular macrophages in the lymphoid-depleted germinal center. At 21 dpi, pigs in the PCV-2a, PCV-2b, and PCV-2d groups had a significantly higher (P < 0.05) number of PCV-2 antigen-positive cells per unit area (0.25 mm2) in their lymph nodes than those of pigs in the PCV-2e groups.
Pigs in the PCV-2d/PRRSV group had a significantly higher (P < 0.05) number of PCV-2 antigen-positive cells per unit area (0.25 mm2) in their lymph nodes at 21 dpi than those of pigs in the other 3 dually inoculated (PCV-2a/PRRSV, -2b/PRRSV, and -2e/PRRSV) groups. Pigs in the PCV-2a/PRRSV and PCV-2b/PRRSV groups had a significantly higher (P < 0.05) number of PCV-2 antigen-positive cells per unit area (0.25 mm2) in their lymph nodes at 21 dpi than those of pigs in the PCV-2e/PRRSV group. At 21 dpi, PCV-2 antigen was not detected in the lymph nodes of pigs from PRRSV-inoculated and control groups (Table II).
Discussion
The present study compared the virulence of 4 different PCV-2 genotypes that are currently circulating in Korean pig herds. In general, virulence was associated with the level of PCV-2 loads in blood and in lymph nodes, along with severity of lymphoid lesions. The results of the present study demonstrated that PCV-2d infection was the most virulent of the PCV-2 genotypes, whereas PCV-2e was the least virulent.
Although PCV-2a and PCV-2b typically fell between these 2 extremities, there were subtle differences in virulence based on the single- and dual-infection models. Within the single-infected groups, pigs singly infected with PCV-2a, PCV-2b, and PCV-2d were more virulent than PCV-2e pigs, based on the level of PCV-2 loads in the blood and lymph nodes, along with severity of lymphoid lesions. These results were consistent with previous studies in which no significant differences in the virulence were observed among PCV-2 genotypes 2a, 2b, and 2d (24,25).
In contrast to these findings, the Chinese PCV-2d strain produced more serious symptoms of disease in pigs than the PCV-2a and PCV-2b strains when pigs were singly infected with 1 of 3 PCV-2 genotypes (15). Since PCV-2 alone is not enough to induce PCVAD, it is necessary to compare the virulence of the 4 main PCV-2 genotypes with the dual-infection model. PCV-2d dually infected with PRRSV was more virulent in pigs than the other 3 dually infected PCV-2/PRRSV combinations. Pigs dually infected with PCV-2a/PRRSV and PCV-2b/PRRSV were more virulent that those dually infected with PCV-2e/PRRSV.
The capsid protein encoded by ORF2 determines the antigenicity and virulence of PCV-2 (13,14). The ORF2 of PCV-2d has 234 amino acids (aa) and resulted in a mutation of the stop codon (from UAA to AAG) in ORF2, which is only 1 lysine residue more than that of the PCV-2a and PCV-2b genotypes, which have 233 aa (15). Mutation of amino acids in the capsid protein may affect the virulence and pathogenicity of PCV-2 (26,27).
Interestingly, PCV-2d produced similar virulence to that of PCV-2a and PCV-2b in the single-infected pigs, whereas PCV-2d produced a greater virulence than that of PCV-2a/PRRSV and PCV-2b/PRRSV in dually infected pigs. The increased virulence is tentatively attributed to an extra lysine residue in the capsid protein encoded by ORF2. Our data suggest, however, that the increased virulence is also dependent on the potentiation of PRRSV on PCV-2d. Even though the potentiating effect of PRRSV on PCV-2 is well established (7), the mechanism of this effect is not well-known. Since monocyte/macrophage lineage is the same target cells for PCV-2 and PRRSV, lymphoid and pulmonary environments induced by PRRSV infection appear to enhance the replication of PCV-2, particularly PCV-2d. In contrast to PCV-2d, extra amino acids in ORF2 of PCV-2e may decrease the virulence compared to the other 3 PCV-2 genotypes (2a, 2b, and 2d). The ORF2 of PCV-2e is 5 amino acids longer than that of PCV-2a or PCV-2b and 4 amino acids longer than that of PCV-2d (9). The functional role of extra amino acids in relation to virulence is unknown. As PCV-2e was the least virulent compared with the remaining 3 PCV-2 genotypes evaluated in both the single-and dually infected groups, these extra amino acids may be related to the decreased virulence of PCV-2e when compared to the other 3 PCV-2 genotypes (2a, 2b, and 2d).
The potentiating effect of PRRSV on PCV-2 was observed, but the potentiation effect of PCV-2 on PRRSV was not observed in the present study. Regardless of PCV-2 genotypes, PRRSV potentiated the level of PCV-2 loads in blood and the severity of PCV-2-associated lymphoid lesions in all dually infected groups compared to their respective single-infected groups. By contrast, no significant differences in levels of PRRSV loads in blood and severity of PRRSV-induced lung lesions were measured in any dually infected groups when compared to their counterpart (respective) single-infected group.
To the authors’ knowledge this was the first virulence comparison of the currently circulating, dominant 4 PCV-2 genotypes. Despite the fact that PCV-2d was the most virulent and PCV-2e was the least virulent among the 4 PCV-2 genotypes, these data do not necessarily mean that the specific genotype is more virulent or less virulent than the other PCV-2 genotypes, as identifying virulence determinants from genotypic comparison is complex. Alternatively, virulence may almost certainly be strain-specific. Additional studies are needed to elucidate the virulent determinants among PCV-2 genotypes.
Acknowledgments
This work was supported by the Research Institute for Veterinary Science (RIVS) and the BK 21 FOUR Future Veterinary Medicine Leading Education and Research Center (Grant no. A0449-20200100) at Seoul National University in Korea.
References
- 1.Lv Q-Z, Guo K-K, Zhang Y-M. Current understanding of genomic DNA of porcine circovirus type 2. Virus Genes. 2014;49:1–10. doi: 10.1007/s11262-014-1099-z. [DOI] [PubMed] [Google Scholar]
- 2.Tischer I, Gelderblom H, Vettermann W, Koch MA. A very small porcine virus with circular single-stranded DNA. Nature. 1982;295:64–66. doi: 10.1038/295064a0. [DOI] [PubMed] [Google Scholar]
- 3.Chae C. A review of porcine circovirus 2-associated syndromes and diseases. Vet J. 2005;169:326–336. doi: 10.1016/j.tvjl.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Adams MJ, Lefkowitz EJ, King AMQ, et al. Ratification vote on taxonomic proposals to the international committee on taxonomy of viruses (2016) Arch Virol. 2016;161:2921–2949. doi: 10.1007/s00705-016-2977-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zimmerman JJ, Dee SA, Holtkamp DJ, et al. Porcine reproductive and respiratory syndrome viruses (porcine arteriviruses) In: Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW, Zhang J, editors. Diseases of Swine. 11th ed. Ames, Iowa: Wiley-Blackwell; 2019. pp. 685–708. [Google Scholar]
- 6.Park C, Seo HW, Park S-J, Han K, Chae C. Comparison of porcine circovirus type 2 (PCV-2)-associated lesions produced by co- infection between two genotypes of PCV-2 and two genotypes of porcine reproductive and respiratory syndrome virus. J Gen Virol. 2014;95:2486–2494. doi: 10.1099/vir.0.066290-0. [DOI] [PubMed] [Google Scholar]
- 7.Harms PA, Sorden SD, Halbur PG, et al. Experimental reproduction of severe disease in CD/CD pigs concurrently infected with type 2 porcine circovirus and porcine reproductive and respiratory syndrome virus. Vet Pathol. 2001;38:528–539. doi: 10.1354/vp.38-5-528. [DOI] [PubMed] [Google Scholar]
- 8.Franzo G, Segalés J. Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology. PLoS One. 2018;13:e0208585. doi: 10.1371/journal.pone.0208585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies B, Wang X, Dvorak CMT, Marthaler D, Murtaugh MP. Diagnostic phylogenetics reveals a new porcine circovirus 2 cluster. Virus Res. 2016;217:32–37. doi: 10.1016/j.virusres.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Harmon KM, Gauger PC, Zhang J, Piñeyro PE, Dunn DD, Chriswell AJ. Whole-genome sequences of novel porcine circovirus type 2 viruses detected in swine from Mexico and the United States. Genome Announc. 2015;3:e01315–15. doi: 10.1128/genomeA.01315-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Wei C, Dai A, et al. Detection of PCV-2e strains in southeast China. Peer J. 2018;6:e4476. doi: 10.7717/peerj.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park KH, Oh T, Cho H, Yang S, Chae C. The first isolation of porcine circovirus type 2e from a Korean pig. Arch Virol. 2020;165:2927–2930. doi: 10.1007/s00705-020-04827-9. [DOI] [PubMed] [Google Scholar]
- 13.Lefebvre DJ, Costers S, Van Doorsselaere J, Misinzo G, Delpute PL, Nauwynck HJ. Antigenic differences among porcine circovirus type 2 strains, as demonstrated by the use of monoclonal antibodies. J Gen Virol. 2008;89:177–187. doi: 10.1099/vir.0.83280-0. [DOI] [PubMed] [Google Scholar]
- 14.Saha D, Huang L, Bussalleu E, et al. Antigenic subtyping and epitopes’ competition analysis of porcine circovirus type 2 using monoclonal antibodies. Vet Microbiol. 2012;157:13–22. doi: 10.1016/j.vetmic.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 15.Guo L, Fu Y, Wang Y, et al. A porcine circovirus type 2 (PCV-2) mutant with 234 amino acids in capsid protein showed more virulence in vivo, compared with classical PCV-2a/b strain. PLoS One. 2012;7:e41463. doi: 10.1371/journal.pone.0041463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beaver BV, Reed W, Leary S, et al. Report of the AVMA panel on euthanasia. J Am Vet Med Assoc. 2001;218:669–696. doi: 10.2460/javma.2001.218.669. [DOI] [PubMed] [Google Scholar]
- 17.Halbur PG, Paul PS, Frey ML, et al. Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet Pathol. 1995;32:648–660. doi: 10.1177/030098589503200606. [DOI] [PubMed] [Google Scholar]
- 18.Gagnon CA, del Castillo JRE, Music N, Fontaine G, Harel J, Tremblay D. Development and use of a multiplex real-time quantitative polymerase chain reaction assay for detection and differentiation of Porcine circovirus-2 genotypes 2a and 2b in an epidemiological survey. J Vet Diagn Invest. 2008;20:545–558. doi: 10.1177/104063870802000503. [DOI] [PubMed] [Google Scholar]
- 19.Opriessnig T, Xiao C-T, Gerber PF, Halbur PG. Emergence of a novel mutant PCV-2b variant associated with clinical PCVAD in two vaccinated pig farms in the U.S. concurrently infected with PPV2. Vet. Microbiol. 2013;163:177–183. doi: 10.1016/j.vetmic.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 20.Xiao CT, Harmon KM, Halbur PG, Opriessnig T. PCV-2d-2 is the predominant type of PCV-2 DNA in pig samples collected in the U.S. during 2014–2016. Vet Microbiol. 2016;197:72–77. doi: 10.1016/j.vetmic.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 21.Wasilk A, Callahan JD, Christopher-Hennings J, et al. Detection of U.S., Lelystad, and European-like porcine reproductive and respiratory syndrome viruses and relative quantitation in boar semen and serum samples by real-time PCR. J Clin Microbiol. 2004;42:4453–4461. doi: 10.1128/JCM.42.10.4453-4461.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J, Chae C. Expression of monocyte chemoattractant protein-1 and macrophage inflammatory protein-1 in porcine circovirus 2-induced granulomatous inflammation. J Comp Pathol. 2004;131:121–126. doi: 10.1016/j.jcpa.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Kim D, Kim CH, Han K, et al. Comparative efficacy of commercial Mycoplasma hyopneumoniae and porcine circovirus 2 (PCV-2) vaccines in pigs experimentally infected with M. hyopneumoniae and PCV-2. Vaccine. 2011;29:3206–3212. doi: 10.1016/j.vaccine.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 24.Opriessnig T, Xiao C-T, Gerber PF, Halbur PG, Matzinger SR, Meng X-J. Mutant USA strain of porcine circovirus type 2 (mPCV-2) exhibits similar virulence to the classical PCV-2a and PCV-2b strains in caesarean-derived, colostrum-deprived pigs. J Gen Virol. 2014;95:2495–2503. doi: 10.1099/vir.0.066423-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho H, Kang I, Oh T, et al. Comparative study of the virulence of 3 major Korean porcine circovirus type 2 genotypes (a, b, and d) Can J Vet Res. 2020;84:235–240. [PMC free article] [PubMed] [Google Scholar]
- 26.Huang LP, Lu YH, Wei YW, Guo LJ, Liu CM. Identification of one critical amino acid that determines a conformational neutralizing epitope in the capsid protein of porcine circovirus type 2. BMC Microbiol. 2011;11:188. doi: 10.1186/1471-2180-11-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saha D, Lefebvre DJ, Ooms K, et al. Single amino acid mutations in the capsid switch the neutralization phenotype of porcine circovirus 2. J Gen Virol. 2012;93:1548–1555. doi: 10.1099/vir.0.042085-0. [DOI] [PubMed] [Google Scholar]