Abstract
The human transcriptome contains many types of noncoding RNAs, which rival the number of protein-coding species. From long noncoding RNAs (lncRNA) over 200 nucleotides long to piwi-interacting RNAs (piRNA) of only 20 nucleotides, noncoding RNAs play important roles in regulating transcription, epigenetic modifications, translation, and cell signaling. Roles for noncoding RNAs in disease mechanisms are also being uncovered, and several species have been identified as potential drug targets. On May 11 to 14, 2021 the Keystone eSymposium “Noncoding RNAs: Biology and Applications,” brought together researchers working in RNA biology, structure, and technologies to accelerate both the understanding of RNA basic biology and the translation of those findings into clinical applications.
Keywords: circRNA, noncoding RNA, lncRNA, nuclear condensate, phase transition, piRNA, transcription, Xist
Graphical abstract
Noncoding RNA comprise a diverse range of RNAs. Long noncoding RNAs (lncRNA) contain features of mRNAs; the most well-studied localize to the nucleus, help form nuclear condensates, and affect transcription machinery. Short noncoding RNA species play roles in mRNA translation, alternative splicing, and RNA editing and silencing. As new technologies are developed, the prevalence and importance of the diverse array of small RNAs will come to light. On Demand: https://keysym.us/21EK44NYAS.

Introduction
Noncoding RNA are a diverse range of RNA species whose physiological and pathological functions are just beginning to be understood. Long noncoding RNAs (lncRNA) are RNA transcripts of greater than 200 nucleotides that often contain many features of protein-coding RNAs. Approximately 30,000 lncRNAs have been identified in the human genome, putting them on par with the number of protein-coding RNAs. The most well-studied lncRNAs localize to the nucleus, where they can play key roles in formation of nuclear condensates like the nucleolus and affect the transcription machinery to regulate gene expression. Dysregulation of several lncRNAs has been implicated in diseases, including cancer and cardiovascular disease, thus opening the potential for new therapeutic targets.1
Short noncoding RNA species include microRNA (miRNA), small RNA, piwi-interacting RNA (piRNA), and circular RNA (circRNA) as well as more obscure RNAs like tRNA-derived small RNA (tsRNA), rRNA-derived small RNA (rsRNA), and Y RNA-derived small RNA (ysRNA). These RNAs play roles in mRNA translation, alternative splicing events, RNA editing, and RNA silencing.2 Traditional sequencing techniques often miss small RNAs. As new technologies are developed, the prevalence and importance of these small RNA species are beginning to come to light.
On May 11–14, 2021 researchers in noncoding RNA met virtually for the Keystone eSymposium Noncoding RNAs: biology and applications. Speakers highlighted the mechanisms and roles of diverse types of noncoding RNA species in areas like transcription, translation, gene activation and silencing, and protein and cell signaling regulation. In addition, several speakers expounded on the potential for noncoding RNAs to act as drug targets in disease like cancer and cardiovascular disease while others described the structure/function relationships for lncRNAs, long thought to be mostly unstructured. Finally, speakers discussed the emergence of new technologies to systematically and comprehensively identify and map noncoding RNAs
Keynote address: The multiple roles of Xist RNA
Edith Heard from the European Molecular Biology Laboratory presented the keynote address on elucidating the role of Xist RNA in X chromosome inactivation (XCI). In almost all mammals, one of the two X chromosomes is randomly silenced during development. This silencing is epigenetically propagated through cell divisions and enables dosage compensation between the sexes.3
Xist is a long noncoding RNA (lncRNA) of approximately 15,000 nucleotides in mouse, encoded by the gene Xist on the X chromosome. Xist initiates XCI by coating the chromosome from which it is expressed, triggering chromosome-wide gene silencing. Xist contains several conserved regions, including the A-repeat, which is essential for chromosome-wide gene silencing, the B-repeat, which recruits Polycomb repressive complexes to modify epigenetic marks on the X chromosome, and a few other dispersed regions, which collectively help Xist coat and reorganize the X chromosome. The role of Xist is believed to be primarily in initiating X chromosome inactivation, and helping to establish early maintenance.3
Xist initiates gene silencing by associating with sites along the X chromosome that are close in 3D space to Xist. Within a few hours, the entire chromosome is coated with Xist.4 Heard’s group and others have shown that Xist RNA induces massive changes in the 3D organization of the chromosome, characterized by the loss of topologically associated domains and the formation of two poorly structured megadomains.5 As genes are silenced, they relocate into the Xist-coated territory, while a small number of genes which escape inactivation remain located outside.6,7
One possible explanation for how Xist induces gene silencing could be that it physically excludes the transcriptional machinery. This is consistent with observations that soon after Xist coating, the inactive X chromosome is depleted of transcriptional machinery factors like RNA Pol II, splicing factors, and TFs. Heard’s group used single-particle tracking to visualize Xist and RNA Pol II molecules in live embryonic stem cells (ESC) and showed that RNA Pol II was able to freely diffuse in and out of the Xist domain, indicating that the early Xist compartment does not present a physical barrier to the transcriptional machinery.8
To understand the mechanism through which Xist mediates X-chromosome inactivation, Heard’s group conducted a comprehensive analysis of the time- and space-dependent epigenetic changes in the inactive X chromosome upon Xist coating using native ChIP-seq, RNA-seq/TT-seq, and allele-specific analysis. They showed that the two earliest epigenomic events are loss of H3K27ac at promoters and enhancers due to the activity of HDAC3 and accumulation of H2AK119 ubiquitination (catalyzed by PRC1). H2AK119ub1 initially accumulates at intergenic regions and spread into genic regions after gene silencing.9
Studies by Heard and others have identified binding partners of Xist.10–15 One of the most important Xist binding partner that has been identified in multiple screens and pull-down assays is SPEN. SPEN is a large RNA-binding protein (RBP) involved in chromatin regulation. The SPEN SPOC domain associates with nuclear co-repressors (NCoR/SMRT) and recruits HDAC3, which is necessary for the early deacetylation observed upon Xist coating. Heard’s group recently showed that SPEN is essential for initiating X-chromosome inactivation. Depleting SPEN abolished X-chromosome gene silencing while knocking out SPEN in mice phenocopied Xist knock out. Further investigation revealed that the SPOC domain of SPEN is critical for gene silencing and for dampening expression of genes that escape XCI. Live and fixed-cell imaging showed that SPEN accumulates on the X chromosome simultaneously with Xist. While SPEN is not required for Xist to coat the chromosome, it is required for gene silencing. Mass spectrometry analysis revealed that the SPOC domain acts as a platform for multiple protein complexes, including the RNA Pol II transcription machinery.16
Heard put forth a model for the role of SPEN and Xist in gene silencing in which Xist’s A-repeat recruits SPEN to the X chromosome, where it is targeted to transcriptionally active promoters and enhancers, likely via interactions between the SPOC domain and RNA Pol II. There, SPEN triggers gene silencing, following which SPEN disengages from chromatin, even though it continues to be recruited by Xist RNA.
Regulation of long noncoding RNA in nuclear condensates
Most noncoding RNAs retained in the nucleus are retained in membraneless nuclear bodies, like the nucleolus, nuclear speckles, and promyelocytic leukemia (PML) bodies. Recent studies have indicated that these noncoding RNAs can play an important role as regulatory or structural molecules in these nuclear domains.
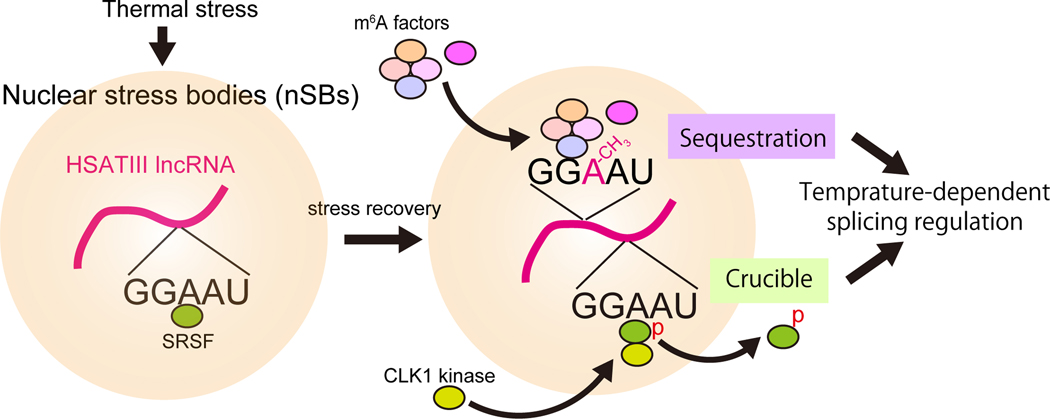
Tetsuro Hirose from Osaka University presented work on delineating the functions of architectural noncoding RNAs (arcRNA), which act as a structural scaffold for membraneless nuclear bodies by sequestering IDR-RBPs and inducing phase separation.17,18 These membraneless bodies may have many functions, including acting as reaction crucibles for biochemical reactions, sequestering proteins, and acting as an organization hub for chromosomes.19 Hirose focused on the role of temperature-induced nuclear stress bodies (nSBs) in regulating RNA splicing. They showed that HSATIII arcRNAs form nSBs upon thermal stress that sequester proteins involved in intron splicing, including SRSF and SAFB.20 Upon recovery from thermal stress, additional factors are recruited to the nSBs, including CLK1 (Fig. 1), which phosphorylates SRSF and allows it to leave the nSB and repress intron splicing.21 In a separate pathway, HSAT III induces intron retention via m6A writer and reader proteins. nSBs sequester the m6A writer complex, which methylates HSATIII, leading to sequestration of the nuclear m6A reader, YTHDC1. Sequestration of the m6A writer and reader represses m6A modification of pre-mRNAs, thereby reducing m6A-dependent splicing during stress recovery.22 Hirose’s work shows that nSBs can control temperature-dependent intron splicing through two distinct RNP complexes.
Figure 1.
HSATIII arcRNAs form nSBs upon thermal stress that sequester proteins involved in intron splicing, including SRSF and SAFB. Upon recovery from thermal stress, additional factors are recruited to the nSBs, including CLK1.
Noncoding RNAs in the nucleolus
Another membraneless nuclear body in which noncoding RNAs play an important role is the nucleolus. The nucleolus is the site of ribosomal RNA (rRNA) gene transcription, processing, and assembly with ribosomal proteins. It is organized around nucleolus organizer regions (NOR), which consist of tandem repeats of ribosomal genes. In humans, each NOR contains ~40 rDNA repeats that form head-to-tail tandem arrays. Noncoding RNAs are central to the nucleolar structure and function. The nucleolus consists of three major compartments: the fibrillar center (FC), which is the site of rRNA transcription, the dense fibrillar component (DFC), the site of early rRNA processing, and the granular component (GC), the site of late rRNA processing.
Ling-Ling Chen from the Chinese Academy of Sciences presented work on RNA regulation in the nucleolus. Chen focused on sno-lncRNA enhances pre-rRNA transcription (SLERT). SLERT is an approximately 700-nucleotide-long snoRNA found in the nucleolus that enhances pre-rRNA transcription.23 Knocking out SLERT reduces pre-rRNA and ribosome synthesis and subsequently reduced cell proliferation.23 SLERT interacts with the DEAD box helicase DDX21, which forms a shell-like structure around RNA Pol I.23 In the nucleolus, fibrillar centers (FCs) and dense fibrillar components (DFCs) are essential morphologically distinct sub-regions of mammalian cell nucleoli for rDNA transcription and pre-rRNA processing.24 DDX21 shells both control FC/DFC size and suppress rDNA transcription. Chen showed that SLERT promotes Pol I transcription by changing the conformation of the DDX21 shells. In brief, DEAD box RNA helicase can adopt an open or closed conformation. In the open conformation, DDX21 can form dense hyper-multimerized clusters that impair the liquidity and size of the FC/DFC units in the nucleolus. In the closed conformation, DDX21 forms loose hypo-multimerized clusters that enable liquidity in the FC/DFC. SLERT keeps DDX21 in the closed state via an RNA chaperone-like mechanism at a substantially low stoichiometric ratio, thereby promoting hypo-multimerization and maintaining the FC/DFC and preventing DDX21 from hijacking rDNA.25
Kannanganattu Prasanth from the University of Illinois presented unpublished data on a family of novel nucleolus-enriched noncoding RNAs. Individual members of this class of ncRNAs associated with specific NOR-containing chromosomes and modulated rDNA gene expression. Several recent studies on the nucleolus-enriched ncRNAs, including the work from Ling-ling Chen and Prasanth labs have highlighted the importance of ncRNAs in rDNA gene expression and nucleolus organization.25–32
Sofia Quinodoz in Clifford Brangwynne’s lab at Princeton University presented work done in Mitchell Guttman’s lab at Cal Tech to comprehensively map the location of all RNAs in the nucleus. Quinodoz developed SPRITE, an iterative split-and-pool tagging method that measures multi-way RNA and DNA interactions in 3D space.33,34 Quinodoz recently developed a new version of SPRITE (SPRITE 2.0), which enables detection of all classes of RNA, including low abundance noncoding RNAs and nascent pre RNAs. They showed that SPRITE 2.0 captures known RNA-DNA interactions as well as known RNA-RNA interactions and global RNA bodies. For example, SPRITE 2.0 identified many RNA hubs via RNA interactions, corresponding to various nuclear bodies, such as the nucleolus, speckles, and Cajal bodies. Quinodoz has used SPRITE to map the 3D organization of RNA and DNA in these bodies. During the talk, Quinodoz focused on the organization of RNA and DNA in nucleolus, though they have also applied the method to nuclear speckles, histone locus body RNAs, and Cajal body-associated RNAs. The results reveal several general principles for the organization of RNA hubs in which nascent RNAs remain near their DNA, multiple DNA loci come together in 3D space, and diffusible regulatory ncRNAs come to the hubs. Quinodoz has also used SPRITE to map the localization of over 600 lncRNAs, showing that the majority associate with chromatin and localize around their loci.33,34
Noncoding RNAs as therapeutic targets
As the functional roles of noncoding RNAs become better appreciated, several have been implicated in diseases such as cancer and cardiovascular disease.35,36 Several talks focused on the potential to targeted misregulated noncoding RNAs to treat disease.
David Spector from Cold Spring Harbor Laboratory presented work on identifying and targeting lncRNAs upregulated in breast cancer that might play a role as therapeutic targets. LncRNA misregulation has been associated with various cancer types, both as pro-metastatic factors and tumor suppressors.35 Sarah Diermeier, a former post-doc in Spector’s lab, now at the University of Otago, performed an RNA-seq screen in mouse models of HER2+ and luminal B breast cancer to identify lncRNAs involved in breast cancer progression. They prioritized 30 lncRNAs, dubbed mammary tumor associated RNAs (MaTARs), as promising clinically relevant targets due to location of their genes in the genome and lack of sequence homology with other RNA species.37
Spector focused on MaTAR25, an intergenic lncRNA gene that localizes to the nucleus and is upregulated in several breast cancer models but not expressed in normal tissues.38 Both in vitro and in vivo assays show that loss of MaTAR25 impacts tumor formation, proliferation, invasion, and metastatic ability. Spector showed that MaTAR25 impacts cell migration by upregulating expression of tensin1, a protein found in focal adhesions.38,39 While high tensin1 levels had been implicated in CRC, it had not been previously associated with breast cancer. Knocking out MaTAR25 resulted in disassembly of actin filaments and focal adhesions as well as loss of microvilli, which are critical for cell migration and invasion. Spector showed that MaTAR25 regulates tensin1 expression via association with PURB and the tensin1 promoter. Spector’s group has identified the human ortholog of MaTAR25, named LINC01271.38 LINC01271 is overexpressed in primary breast tumors and lung metastases, with increased expression correlating with poor survival in breast cancer patients.40 Spector’s group is evaluating LINC01271 as a potential therapeutic target that can be manipulated by an antisense-based approach to impact metastatic breast cancer progression.
Diermeier discussed another MaTAR identified in their RNA-seq screen, MaTAR17. In mammary tumor organoids, knockdown of MaTAR17 via antisense oligonucleotides (ASO) developed in collaboration with IONIS reduced organoid branching as a result of reduced cell proliferation and invasiveness.37,41 Previous work has shown that, unlike other lncRNAs, which are typically associated with one tumor type, hMATAR17 is overexpressed in several types of cancer, including head and neck, colorectal, breast and lung cancer.42 Diermeier presented unpublished data elucidating the mechanisms by which MaTAR17 drives breast and colorectal tumor progression and investigating the therapeutic potential of anti-MaTAR17 ASOs for breast cancer. The group is also exploring the utility of MaTAR17 as a predictive or prognostic biomarker for tracking disease progression and treatment response.
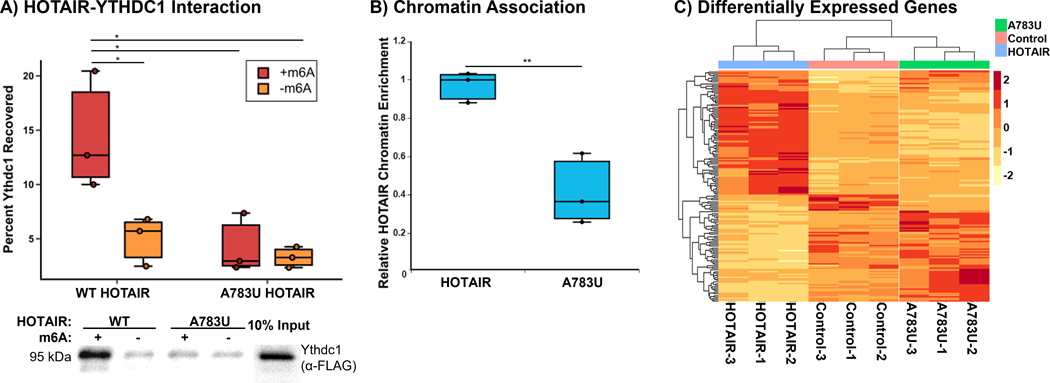
Allison M. Porman (Swain) from Aaron Johnson’s lab at the University of Colorado Anschutz Medical Campus showed how N6-methyladenosine (m6A) modification in the lncRNA HOTAIR regulates its function in breast cancer. HOTAIR promotes metastasis in breast cancer cells by reprogramming the chromatin state. While this was originally believed to occur via interactions with PRC2, subsequent studies found that PRC2 is not required for HOTAIR-mediated transcriptional repression.43,44 Porman identified an m6A modification site in HOTAIR that is required for HOTAIR-mediated cell proliferation and invasion in breast cancer cells. They showed that m6A at this single site enables HOTAIR to interact with the m6A reader YTHDC1 which is required for HOTAIR to associate with chromatin and repress tumor suppressor genes (Fig. 2). Porman’s work demonstrates how a single site of RNA modification within a lncRNA can play a major role in cell phenotype.45
Figure 2.
An m6A modification site in HOTAIR required for HOTAIR-mediated cell proliferation and invasion in breast cancer cells.
Dhiraj Kumar from the University of Texas MD Anderson Cancer Center presented unpublished work on how another lncRNA, Malat1, which has been identified as a mediator of metastatic reactivation46 promotes metastatic reactivation in breast cancer by regulating the immune microenvironment at the metastatic site.
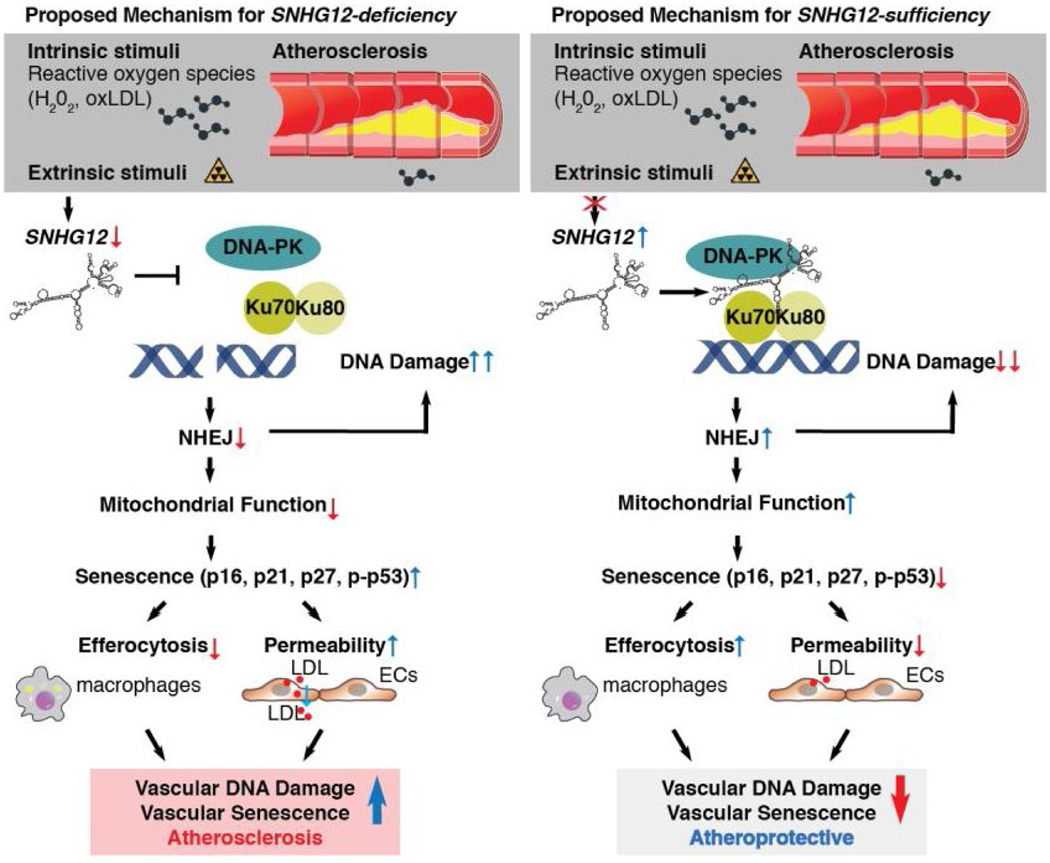
Mark Feinberg from Brigham and Women’s Hospital/Harvard Medical School focused on the role of lncRNAs in atherosclerosis. While roles for lncRNAs have been identified in different aspects of CVD36,47, a direct role in vessels is poorly understood. RNA-seq studies of blood vessels in mice fed a high-cholesterol diet identified SNHG12 as a lncRNA that decreases with atheroprogression and increases with atheroregression (Fig. 3). While knocking down SNHG12 increased plaque burden in mice, the lesions showed no differences in pro-inflammatory signaling pathways compared to wild-type lesions. Feinberg showed that reduced expression of SNGH12 increases atherosclerosis independent of lipid profile or lesional inflammation via DNA damage and vascular senescence. SNHG12 acts to control DNA damage by stabilizing the interaction between DNA-PK and Ku70/Ku80. Knocking down SNGH12 increased DNA damage, impaired NHEJ, and increased cell senescence. These effects can increase endothelial permeability to LDL.48 Feinberg’s work builds upon increasing evidence suggesting that senescence can independently promote atherosclerosis.49,50 Their group has also identified and worked to understand the mechanism for other lncRNAs involved in atherosclerosis.51–54
Figure 3.
Proposed mechanism of lncRNA SNHG12 regulation of atherosclerosis through a DNA-PK-mediated DNA damage response in the vascular endothelium.
Mechanisms of long noncoding RNA function
Transcription by RNA Pol II occurs via a series of sequential events, including promoter binding, transcription initiation, and transcription elongation. Each of these events is quite inefficient such that only 1% of polymerases that bind to a promoter produce an mRNA transcript. Many lncRNAs have been shown to modulate transcription at sites near their genes, though the exact mechanisms are not clear.55
Jonathan Henninger from Richard Young’s lab at the Whitehead institute for Biomedical Research presented a model in which these nascent RNAs regulate Pol II transcription via a dynamic feedback loop. During transcription, short RNAs are produced from enhancer and promoter elements that do not have a clear function.56 Henninger’s model is based on a type of phase separation called complex coacervation, whereby oppositely charged polymers form condensates through electrostatic interactions. In the model, during initial stages of transcription, electrostatic interactions between negatively charged RNA and positively charged proteins balance each other and form condensates. As transcription proceeds, negatively charged RNAs accumulate, the repulsive electrostatic interactions dissolve the condensate, and the system returns to a diffuse phase. The model predicts that nascent RNAs initially stimulate and then arrest transcription, which was validated both in vitro and in vivo. Given that this electrostatic-based model is dependent on RNA size and not sequence, Henninger believes that it may explain the role of many lncRNAs, which often show little sequence conservation but are primarily located near sites of transcription, in regulating gene expression of nearby genes.57
Several lines of evidence suggest that there are key regulatory events that stabilize RNA Pol II binding for transcription elongation57–59, yet the molecular link between RNA and the transcription machinery had not been elucidated.
Xiaohua Shen from Tsinghua University presented a phase-separation mechanism by which noncoding RNA act with RNA-binding proteins (RBPs) to aid in the rate-limiting step of polymerase condensate formation to promote polymerase engagement and transcription. Shen showed that RBPs are prevalent among chromatin-bound proteins and are susceptible to liquid-liquid phase condensation on chromatin. Many RBPs co-localize with RNA Pol II at sites of transcription, and the degree of RBP binding to a promoter positively correlated with the level of mRNA expression, suggesting that RBPs might act collaboratively to regulate transcription. She showed the paraspeckle protein PSPC1 inhibits the RNA-induced premature release of Pol II and makes use of RNA as multivalent molecules to enhance the formation of transcription condensates and subsequent phosphorylation and release of Pol II. This synergistic interplay enhances polymerase engagement and activity via the RNA-binding and phase-separation activities of PSPC1. In embryonic stem cells, auxin-induced acute degradation of PSPC1 leads to genome-wide defects in Pol II binding and nascent transcription. Shen put forth a model in which RBPs stabilize the interaction between RNA Pol II and the transcription site via phase separation. During transcription, RNA Pol II produces nascent transcripts which can evict it from the gene promoter before phosphorylation of the C-terminal domain, which is required for transcription elongation. The nascent transcripts can also recruit RBPs, which once reaching a critical threshold, create an RBP-rich condensate that occlude RNA Pol II eviction, allow it to be phosphorylated, and proceed to the elongation phase.60 The interplay between noncoding transcripts and RBPs around the transcription sites regulates dynamic assembly of polymerase clusters in cells, expanding the horizon in understanding transcription regulation beyond classic transcription and epigenetic factors.
One of the barriers to understanding the role of lncRNAs on gene transcription is the lack of explicit definitions of the types of regulatory lncRNAs. Some lncRNAs regulate gene expression in cis, meaning that their gene targets are located nearby on the same chromosome, while some regulated gene expression in trans, meaning that their targets are on a different chromosome or distally on the same chromosome. Rory Johnson from University College Dublin presented a new tool to identify cis-regulatory lncRNAs. Johnson’s group is developing tools to classify lncRNAs as either cis or trans acting to determine the prevalence of each modality and to define general rules for how cis-acting lncRNAs work.
Joshua Mendell from Howard Hughes Medical Institute UT Southwestern discussed how NORAD regulates Pumilio (PUM) activity. The lncRNA NORAD is a ubiquitously expressed cytoplasmic noncoding RNA that is activated by DNA damage.61–66 In contrast to most lncRNAs, it contains several repeat units, some of which form conserved secondary structures while some consist of Pumilio recognition elements (PRE) and bind PUM proteins. PUM represses mRNA translation by binding to PREs in mRNA transcripts and accelerating their degradation.
Given that the number of PUM target mRNAs vastly outnumbers the number of PUM binding sites on NORAD, Mendell’s group is interested in how NORAD can outcompete PUM target mRNA binding sites to effectively inhibit PUM activity. Mendell showed that NORAD and PUM co-localize in punctate foci in the cytoplasm, which they termed NORAD-Pumilio (NP) bodies. These foci disperse in the absence of NORAD. PUM contains several features associated with the ability to form condensates, including large IDRs and the ability to bind RNA. They proposed that NORAD sequesters PUM in NP bodies, which are formed through NORAD-induced PUM phase separation. In vitro studies showed that at physiologic concentrations, NORAD drives phase separation of PUM. Mendell showed that NORAD can recruit PUM into the droplet phase not only via direct binding to PREs but also via PUM-PUM IDR interactions. This allows NORAD to outcompete other PRE-containing RNAs as it can sequester approximately three times as many PUM proteins as it would be able to via PRE binding alone. Moreover, each PRE in NORAD can recruit PUM ~40 times more efficiently than an isolated PRE in a target mRNA. Therefore, in the cell, the effective number of NORAD PREs is comparable to the total number of PUM binding sites on target mRNAs. Mendell also showed that sequestration of PUM in phase-separated condensates is important for regulating PUM activity and maintaining genomic stability in mammalian cells.63
Juan Pablo Unfried from Puri Fortes’s lab at the University of Navarra discussed their work on lncRNAs associated with hepatocellular carcinoma (HCC). Unfried previously published a list of lncRNAs that are highly, preferentially expressed in cancer, including HCC, that may serve as attractive drug targets. Several of these are associated with clinical outcomes, including survival, suggesting that they may play a role in disease pathology.67 Unfried presented data on one such lncRNA, NIHCOLE.68 Depletion of NIHCOLE from HCC cells led to impaired proliferation, increased apoptosis and DNA damage accumulation due to a specific decrease in the activity of the non-homologous end-joining (NHEJ) pathway of DNA double-stranded break (DSB) repair. DNA damage induction further decreased the growth of NIHCOLE-depleted cells. Authors found that NIHCOLE associates with DSB markers and recruits several molecules of the Ku70/80 heterodimer. Band-shift analyses showed that structural domains within NIHCOLE support multimeric repair complexes including the ligation complex formed by XRCC4 and DNA Ligase IV. This allows NIHCOLE to promote the ligation efficiency in NHEJ reconstitution assays by serving as a scaffold and facilitator of the NHEJ pathway. Collectively they showed that NIHCOLE’s role in DSB repair confers an advantage to HCC cells uncovering a novel cancer vulnerability that could be targeted with therapeutic purposes.
Enhancer RNAs, circular RNAs, and small RNA
Several talks focused on the regulatory roles of small RNA species, such as small nuclear RNAs (snRNA), piRNA, microRNA (miRNA), and circular RNA (circRNA).
Jeremy E. Wilusz from the University of Pennsylvania showed how Integrator (Int), a multi-subunit complex that associates with RNA Pol II is regulated.69 Integrator contains an endonuclease, IntS11, that cleaves nascent snRNA transcripts, which mature into small nuclear ribonucleoproteins (snRNP) and catalyze splicing.70 Wilusz’s lab has shown that Int can also regulate protein-coding genes by cleaving nascent mRNA transcripts, triggering premature transcription termination that results in short, nonfunctional transcripts that are degraded.71,72 While these two roles for Int have similar characteristics molecularly—they both involve recruitment of Int by RNA Pol II, endonuclease-mediated cleavage, and subsequent release of an RNA transcript—they seem to have highly distinct regulatory needs. When Int cleaves at an snRNA gene, it results in a functional transcript; therefore, constitutive Int activity is required for the production of functional snRNPs. Conversely, Int activity at a protein-coding gene abolishes the production of functional mRNA, indicating that Int activity must be regulated so that protein-coding genes can be expressed under the appropriate cellular conditions. Wilusz presented unpublished data elucidating how Int is differentially regulated at snRNA genes versus at protein-coding genes.
The piRNA pathway is the largest and most diverse class of small RNAs. Its most well-understood function is as a defense mechanism against transposons to protect germ cells against the threat of mobile genetic elements. piRNAs bind to and cleave mRNAs, thus negatively impacting translation. In the nucleus, this can also lead to epigenetic changes and transposon silencing. While many piRNAs match to transposons, many do not.73–75 Lamia Wahba from Andrew Fire’s lab at Stanford University presented work on understanding the function of non-transposon piRNA. Many possible mechanisms for non-transposon piRNAs have been proposed, including transposon defense through imperfect complementarity, massive RNA elimination, and translational activation or repression. Wahba presented unpublished data in C. elegans, in which over 99% of the 16,000 piRNAs do not map to transposons, to elucidate the mechanism of how loss of the piRNA pathway results in gradual sterility across generations.
Adelheid Lempradl from the Van Andel Institute discussed how the piRNA pathway can play a role on inheritance of metabolic phenotype. Several studies indicate that effects of a parent’s environment can be inherited in their offspring. For example, Lempradl had previously established a model for diet-induced intergenerational metabolic reprogramming in Drosophila in which fathers fed a high-sugar diet produced offspring with a higher susceptibility of diet-induced obesity.76 Other studies have observed similar phenotypic inheritance patterns with metabolic disorders and trauma.77,78 Lempradl has investigated the role of the piRNA pathway in mediating this intergenerational heritability. Most research on the piRNA pathway in Drosophila has been in females. Lempradl showed that the piRNA pathway is also present in mature sperm and is necessary for intergenerational inheritance of paternal metabolic state. They identified piwi-bound small RNAs in sperm and provided evidence that these RNAs repress target gene expression in offspring.79
Sean McGeary from David Bartel’s group at the Whitehead Institute presented work on identifying the rules that govern miRNA binding to target mRNAs. miRNAs mediate repression of targets by binding to sites within the 3′ UTRs of target mRNAs. Binding occurs via pairing between two key regions: the seed region and the 3′-pairing region.80 Previous studies to identify the rules governing pairing to the miRNA 3′ end were based on transcriptome-wide analyses of 11 miRNA transfections, which may have caused miRNA-specific differences in 3′-pairing preferences to go undetected. Several studies have shown there might be more miRNA-specific differences that govern 3′-pairing rules.81,82 McGeary used RNA bind-n-seq (RBNS) to more precisely measure the binding affinity of thousands of 3′-pairing architectures by varying the length of the 3′ pairing, the starting position of the 3′ pairing, the offset of the 3′ pairing with respect to the seed pairing, and the nature of the seed pairing. Analyses of these measurements revealed two distinct binding modes shared by multiple miRNAs and allowed them to identify optimal 3′-pairing architectures for different miRNAs. McGeary has also developed a mathematical model–based framework to understand the relationship between 3′-pairing length, position, and offset.83
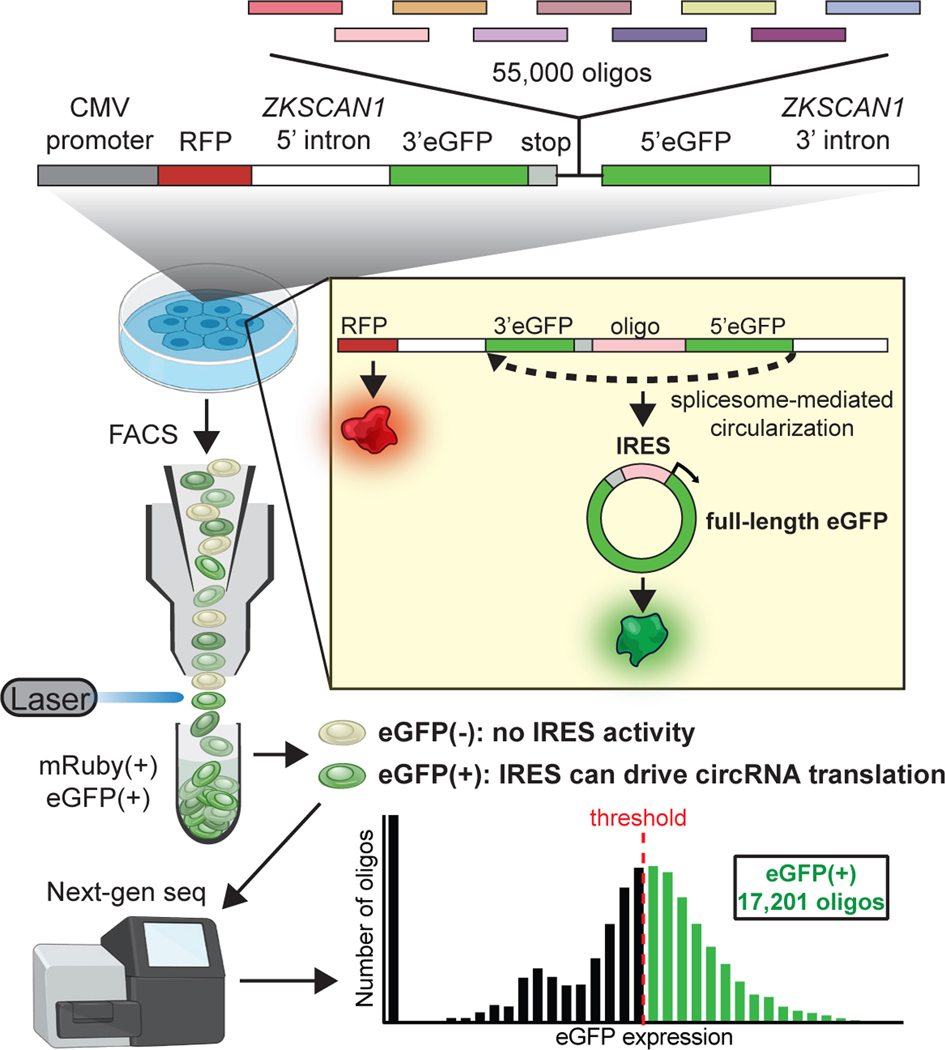
Chun-Kan Chen from Howard Chang’s lab at Stanford University discussed the elements that drive translation of circRNA. Circular RNA is generated by spliceosome-mediated back splicing forming a head-to-tail closed loop.84–86 Previous work has shown that circRNAs can act as sponges for miRNA and RBPs.87–92 However, circRNAs can also serve as templates for translation.93–99 Translation of linear mRNA is initiated by ribosomal recognition of the 5′ cap. Circular RNA has no 5′ cap. Instead, the ribosome recognizes internal ribosomal entry sites (IRES) that mimic the structure of the 5′ cap.96,98,100–106 Given that most of the research on translation has focused on linear RNA, much less is known about the RNA elements that are required for circRNA translation. In unpublished work, Chen has systematically identified IRES sequences in circRNAs (Fig. 4). Using these, they were able to identify potential protein-coding circRNAs and showed how the resulting peptides can regulate cell function.
Figure 4.
IRES sequences identify potential protein-coding circRNAs.
RNA structure and function
Anna Marie Pyle from Yale University discussed the importance of structure in lncRNAs. Many lncRNAs do not have specific structural elements. Given that one of their key roles is to increase the effective RNA Pol II concentration at a locus, many believe that their sequence and structure is largely irrelevant. Pyle’s group was the first to solve a high-resolution structure of a group II self-splicing intron107, which were largely believed to lack any tertiary structure due to their lack of sequence conservation. Similar assumptions have been made about lncRNAs. Pyle’s group has conducted secondary structure analyses on the lncRNA HOTAIR. As noted by Swain previously during the meeting, HOTAIR is differentially expressed along the body axis and overexpressed in cancer.108 Pyle showed that HOTAIR contains four distinct, modular secondary structural domains. Covariance analysis identified patterns of phylogenetic covariance and conservation in some of the stem regions.109 While lncRNAs are often noted for their lack of sequence conservation, Pyle stressed that experimentally determined secondary structures can be instrumental in identifying regions of sequence conservation and covariance by providing a framework for organizing elements of primary sequence in space. Pyle’s group had adapted the tool R-Scape110 to make it more robust at identifying conservation and co-variation in longer RNAs. Their tool was released as R-Scape APC-RAFS.111 Pyle also discussed unpublished structural studies on another lncRNA, Pnky, as well as a new high-throughput approach their lab is developing to rapidly detect tightly folded tertiary motifs.
Amanda Hargrove from Duke University discussed work on targeting lncRNAs with small molecules. One challenge to developing RNA-targeting small molecules is the relative paucity of high-resolution structures. Hargrove’s group has been working to optimize selective RNA-binding small molecules in the absence of such structures. Toward this goal, they have developed a database, R-BIND, that contains approximately 150 bioactive ligands that target non-rRNA.112,113 Hargrove focused on their group’s work in scaffold-based library design, which has revealed insights on the factors that drive RNA binding affinity and specificity. Based on data showing that dimethyl amiloride is a weak, selective binder of HIV-1 TAR RNA114, Hargrove’s group synthesized and screened a library of amiloride (AMR)-based small molecules and identified a molecule with 100-fold increased binding affinity. They developed a model whereby simple molecular properties, such as molecular weight and charges, can be used to predict which ligands selectively bind to HIV-1 TAR RNA.115,116 Hargrove’s group has expanded upon this model to make it more generalizable for a range of RNAs and scaffolds. They hope that general rules can guide prediction of ligand structure-activity relationships in the absence of high-resolution structures. In a similar vein, screening AMR compounds for binding to an enterovirus RNA identified a small molecule that selectively inhibited viral translation and replication.117 Finally, Hargrove discussed work on a separate scaffold, DPF, to target the triple helix of MALAT-1, the stability of which is important to prevent degradation.118 DPF-based compounds bind to the triple helix with a range of affinities and selectivities. The data revealed the importance of shape, with rod-like compounds displaying higher affinity and selectivity.119 Interestingly, the degree of destabilization did not correlate with binding affinity.120 Hargrove’s group is screening additional small molecules to identify stabilizing compounds and tease out which features are important for de/stabilization, selectivity, and affinity.
Matthew D. Simon, from Yale University presented work on understanding the role of promoter proximal pausing in gene regulation. During the initial stages of transcription, RNA Pol II pauses after transcribing approximately 40 nucleotides. This pause allows cells to either release the paused RNA Pol II and continue transcription or to terminate transcription. Simon presented data on determining the rates of transcription termination and RNA Pol II release at promoter proximal pause sites throughout the genome and demonstrated how various perturbations can affect these rates to regulate transcription.121
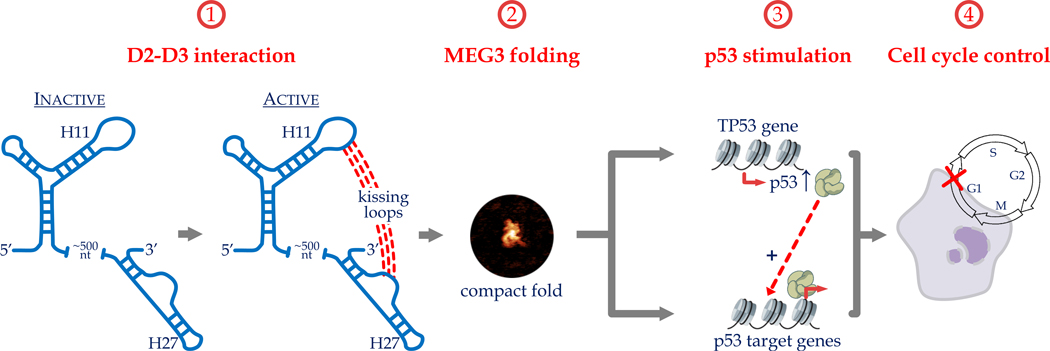
Marco Marcia from EMBL Grenoble presented structural-functional studies for the lncRNA MEG3. Marcia was involved in developing a representation of group II intron splicing using nearly 20 high-resolution structures and molecular dynamics to capture catalytic RNA in the act of splicing.122–124 They plan to similarly visualize the activity of lncRNAs using structural studies. MEG3 is involved in tumor progression via regulating expression of p53.125–127 In vitro and in vivo probing analyses of MEG3 revealed five secondary structure domains. Using a gene reporter assay of p53 activation, Marcia’s group demonstrated that two of these domains, domain 2 (D2) and domain 3 (D3), are essential for MEG3-mediated p53 activation. They also identified elements within these domains that contribute to p53 stimulation. Some of these regions are highly conserved, although conservation was not picked up via other structure-free methods. Marcia focused on an invariant loop within D2 that is required for p53 activation. Regions within D3 are complementary to this loop, suggesting the potential for a long-range tertiary interaction (Fig. 5). Indeed, introducing compensatory mutations between these two regions rescued the activity of the single point mutations, suggesting that a long-range tertiary interaction between these sites does control MEG3-mediated p53 expression. Consistent with this, atomic force microscopy studies showed that MEG3 forms a distinct, compact 3D structure that functionally inactive mutants cannot adopt.128,129
Figure 5.
An invariant loop within domain 2 (D2) of MEG3 required for p53 activation mediated by MEG3.
Róża K. Przanowska from Anindya Dutta’s lab at the University of Virginia presented work on the structure-function relationship in the lncRNA MUNC. MUNC has two isoforms, MUNC spliced and MUNC genomic, both of which are involved in muscle satellite cell differentiation.130 RNA-seq analysis of myoblasts overexpressing either isoform revealed that, while both isoforms are myogenic, they regulate different sets of genes, and MUNC spliced is a stronger pro-myogenic factor. SHAPE-MaP revealed six secondary structural elements present in both isoforms that were not predicted by folding algorithms.131 The data also revealed that MUNC spliced is more stable structurally, whereas MUNC genomic exists in a more dynamic unfolded state. Przanowska used the SHAPE-MaP-predicted secondary structure to delete and mutate various structural regions and determine which regions were responsible for MUNC function. They identified distinct structural domains responsible for regulating different genes and for binding to different genomic sites and cohesin complex.132
RNA technologies
Howard Y. Chang from Stanford University discussed work in understanding the role of the X chromosome and Xist in sex-biased immunity. Several lines of evidence support a sex bias in immunity, including particular the preponderance of autoimmune diseases among women133 and the improved outcomes in women to COVID-19.134 As Heard discussed in the keynote address, Xist is a lncRNA that coats and silences the X chromosome. While long thought to be primarily involved in establishing X-chromosome inactivation, Chang showed that a subset of X-linked genes requires ongoing Xist silencing in adult female B cells, including TLR7, a gene involved in innate immunity, in adult female B cells. Genes that re-activate upon Xist silencing have low levels of methylation, which is known to maintain X-chromosome inactivation. Chang showed that Xist binds to many factors in a cell type-dependent fashion. In B cells, interactions between Xist and TRIM28, which enforces RNA Pol II pausing, is key to maintaining X-chromosome inactivation. Chang put forth a model in which TRIM28 binds to target genes and interacts with Xist. This interaction enhances the ability of TRIM28 to pause RNA Pol II, thereby blocking gene expression. Chang also showed single-cell RNA-seq data of B cells from female COVID-19 patients that demonstrates a population of atypical B memory cells (ABC) that have high levels of Xist escape. Chang proposed that expansion of these ABCs increases reactivity to self-antigens but can also lead to a more effective immune response against some pathogens, such as SARS-CoV-2.135
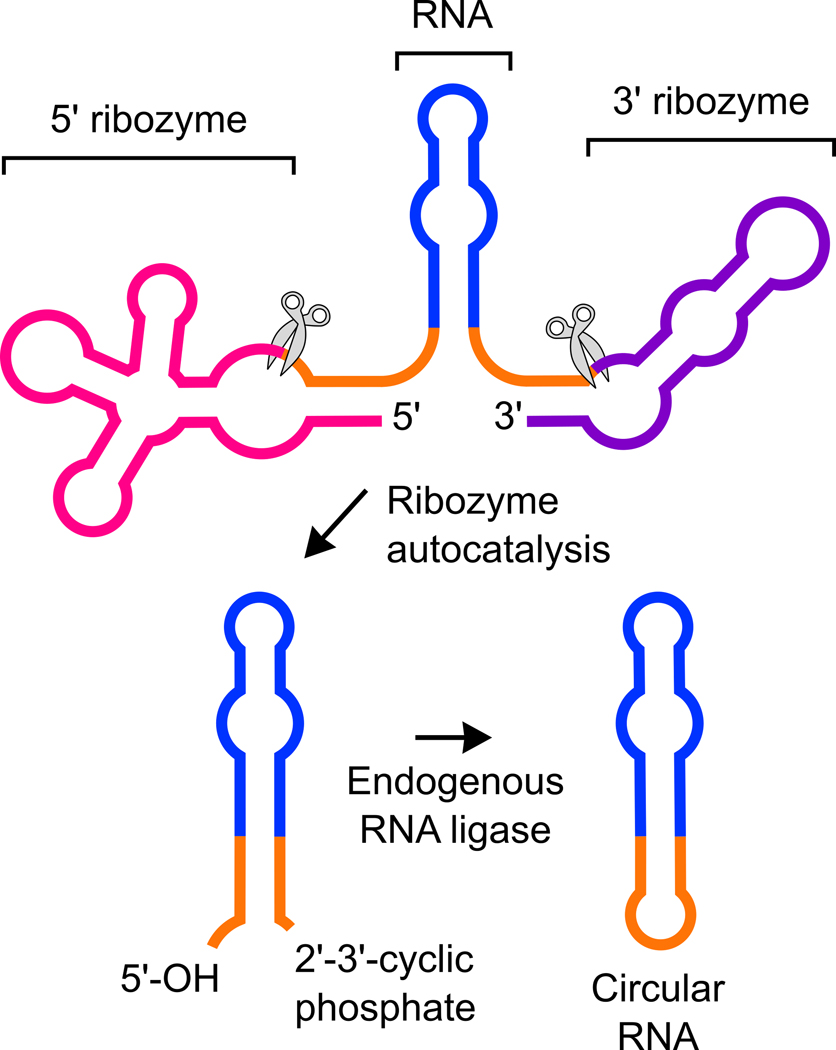
Samie Jaffrey from Weill Medical College of Cornell University discussed work on developing a system to express circRNA in mammalian cells. Expressing RNA at high levels in mammalian cells is a major challenge due to their high susceptibility to exonucleases. Jaffrey’s group had previously leveraged an endogenous pathway that generates circRNAs, which are resistant to exonucleases, from a tRNA intron.136 The resulting circRNA was stable in mammalian cells but only present at low levels. To optimize the production of circRNA, Jaffrey’s group insert RNA sequences into a self-splicing bacterial Twister ribozyme that generates 3′ and 5′ ends that are recognized by the tRNA ligase that creates circRNA (Fig. 6). The resulting system, dubbed Tornado, generates circRNA species that are highly stable and expressed at high levels in mammalian cells.137 Jaffrey showed how the Tornado system can be used to express engineered RNA-based aptamers that can at as metabolite sensors, interact with endogenous proteins to affect signaling pathways, act as decoys to block the activity of RBPs, or be translated into protein. Jaffrey hopes that the Tornado expression system will expand the use of RNA as tools to control mammalian cell function and potentially as a new therapeutic modality.
Figure 6.
Tornado generates circRNA species that are highly stable and expressed at high levels in mammalian cells.
Given that high-resolution structures of RNA species are difficult to obtain, other technologies are needed that can characterize RNA structural elements.
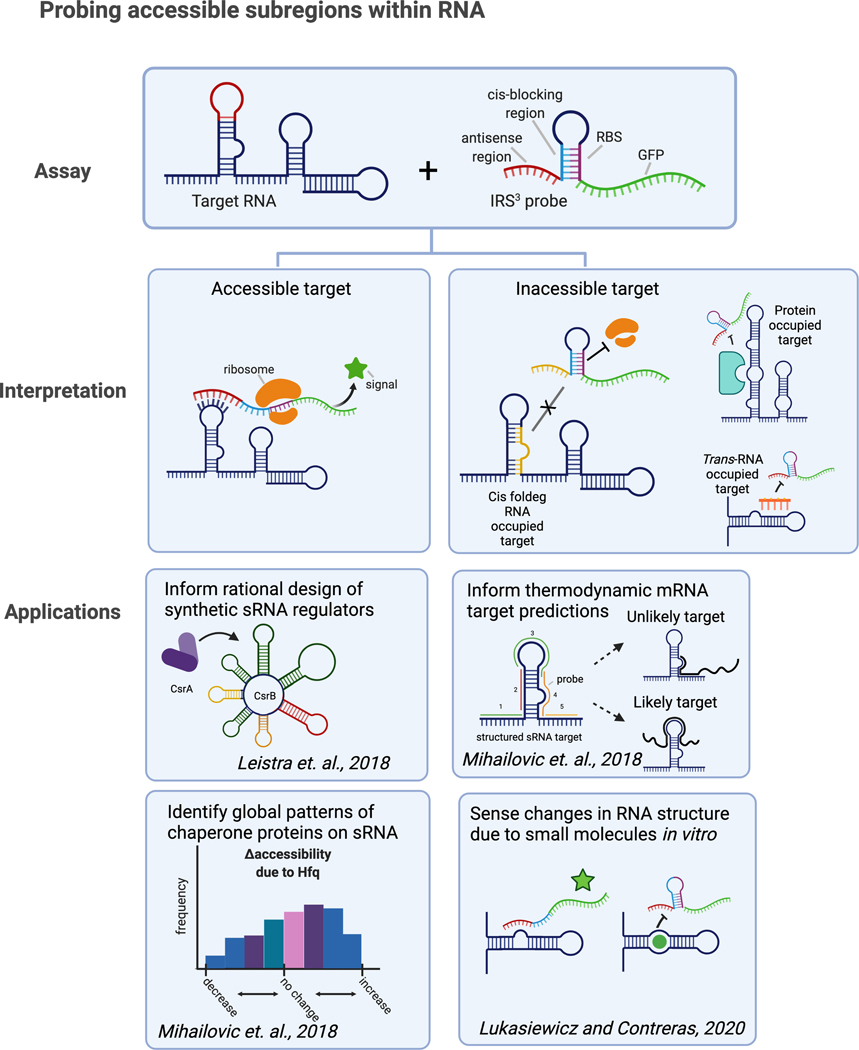
Lydia Contreras from the University of Texas at Austin presented work on quantifying accessibility within RNA species to potentially characterize functionally important areas. Contreras noted that accessibility can give insights into which areas are more likely to mediate interactions with binding partners (Fig. 7). Their group has developed a high-throughput method that measures regional RNA accessibility in vivo (and, more recently, in vitro138) to understand interfaces available for binding interactions that may play a role in regulatory processes.139,140 In this system, an RNA probed designed to target a region (9–16 nucleotides) of a target RNA is incorporated into a riboregulator, which leads to GFP expression upon RNA binding.139–141 A high-throughput version for use in E. coli was also developed that uses RNA transcript length as the output and can probe hundreds of molecules at a time.140 Contreras showed how RNA accessibility can be assessed under various conditions within cells, to pinpoint regions important for regulation and/or to understand how intracellular factors affect RNA accessibility.140,142 For example, by using accessibility as a proxy for CsrA occupancy (low accessibility = high occupancy), the CsrB sRNA was rationally reengineered for modular activity.142 Accessibility insights for other sRNAs, namely extreme accessibility found indicative of target binding, have supported the rational selection of predicted mRNA targets for experimental follow-up.143 They are currently modifying the method for detection of long-range tertiary interactions within RNA.
Figure 7.
Models of understanding regional RNA accessibility and interfaces available for binding interactions that may play roles in regulatory processes.
Small RNAs species are often underestimated in RNA-seq studies, this is in part, because RNA modifications can block reverse transcription and/or unique termini features can occlude detection.
Qi Chen from the University of California Riverside presented a new RNA-seq method that detects small RNAs often missed by traditional sequencing, dubbed Panoramic RNA Display by Overcoming RNA Modification Aborted Sequencing (PANDORA-seq). In PANDORA-seq, samples are treated with enzymes that eliminates reverse transcriptase-blocking modification and converts non-canonical termini so that they can be sequenced.144 Conducting PANDORA-seq on multiple human and mouse tissues/cells revealed a RNA landscape containing enriched tsRNA and rsRNA species that had not previously been identified. Additional data suggests that these small RNAs may be functionally relevant. Chen showed that tsRNA and rsRNA expression is tissue specific and that they can regulate mouse embryonic stem cell (mESC) differentiation process. To annotate and analyze the different types of small ncRNAs, Junchao Shi, from Chen’s group developed the software SPORTS.145 Shi showed that SPORTS performs admirable both in precision and sensitivity compared with other annotating software at identifying miRNA.146 Moreover, SPORTS can also annotate other RNA species, including piRNA, tsRNA, and rsRNA. SPORTS showed that tsRNAs and rsRNAs are derived from different regions within tRNAs and rRNAs, respectively. As an example of translational application, SPORTS was used to identify a small RNA-based signature, TRY-RNA, that consists of tsRNA, rsRNA, and ysRNA and can distinguish between control, lung cancer, and tuberculosis subjects, demonstrating the potential for these types of tools in lung cancer screening and potentially other diseases.147
Noncoding RNA in development and disease
Lin He from the University of California at Berkeley discussed how retrotransposons can affect cell differentiation during preimplantation development. The transcriptome is highly enriched with retrotransposons during pre-implantation development148,149 and thus these mobile elements may play an important developmental role during this stage. For example, MERVL, a family of LTR retrotransposons, is an important marker of totipotency.150–153 RNA-seq studies of various mammals showed that retrotransposons exhibit dynamic expression during preimplantation development. He showed that retrotransposon derived promoters can affect host gene regulation, generating gene isoforms with an altered ORF. One such example is the MT2B2 promoter for Cdk2ap1, which is strongly induced in morulae. Activation of MT2B2 shifts the transcription start site of Cdk2ap1, resulting in an N-terminally truncated protein isoform. This alternative promoter is essential for preimplantation development; deleting it in mice resulted in partial embryonic lethality. Interestingly, while MT2B2 retrotransposons are mosue specific, the resulted, MT2B2 induced Cdk2ap1 isoform is evolutionarily conserved, albeit with species-specific expression regulation. The level of the N-terminally truncated Cdk2ap1 inversely correlated with development timing in mammalian preimplantation embryos. The high level of MT2B2 dependent Cdk2ap1 isoform in mice is correlated with a rapid progression of its preimplantation development. He put forth a model in which specific retrotransposon promoters can yield novel gene isoforms during preimplantation embryos that are essential for proper, species-specific development.154
Erwei Song from Sun Yat-sen University discussed how the lncRNA IRENA affects macrophage phenotype in response to chemotherapy. Tumor-associated macrophages (TAM) are the most abundant infiltrating inflammatory cells in various cancers.155 In treatment-naive breast cancer patients, TAMs exhibit an immunosuppressive phenotype, which may drive out T cells and promote tumor progression.156 Song showed that chemotherapy treatment changes the phenotype of TAMs toward an IFN-activated, proinflammatory phenotype, which enhances antitumor immunity but also promotes resistance to chemotherapy. TAMs isolated from patients after receiving neoadjuvant chemotherapy were able to enhance chemotaxis and tumor-specific cytotoxicity of CD8+ T cells whereas TAMs isolated before chemotherapy were not. In mice, macrophage deficiency reduced chemotherapy-induced infiltration of CD8+ T cells but enhanced response to chemotherapy. These two effects of post-chemotherapy TAMs—increased immune response and chemoresistance—were mediated by distinct mechanisms. Song showed that chemoresistance was mediated by NF-κB activation while anti-tumor immunity was mediated by JAK1–STAT1 signaling. Song showed that the lncRNA IRENA can activate the NF-κB pathway following chemotherapy via its interaction with PKR. Knocking down IRENA abrogated the chemoresistance effect of macrophages in co-culture with tumor cells without affecting the anti-tumor immunity effects on T cells. Song showed that IRENA activates NF-κB by interacting with PKR. In a mice breast cancer model, IRENA knock out improves response to chemotherapy and abrogates NF-κB but not JAK1-STAT1 signaling. In human breast cancer tissue, IRENA expression in TAMs correlates with chemoresistance and poorer survival.157
John Rinn from the University of Colorado Boulder discussed the importance of splicing in lncRNA function. Rinn focused on the lncRNA Tug1 and TERT mRNA. Both RNA species often retain some of their introns. Rinn showed that splicing is important for subcellular localization. Specifically, transcripts that maintain their introns localize to the nucleus whereas spliced transcripts localize to the cytoplasm. Rinn showed that the localization pattern of Tug1 is important for its function. Blocking Tug1 splicing with a nuclear localized modified ASO, thiomorpholino (TMO), depleted the cytoplasmic Tug1 pool and shifted localization to the nucleus, which had a negative impact on cell viability. In the case of TERT, the mRNA normally localizes to the nucleus, which would preclude its translation. Rinn showed that retention of two introns (intron 11 and 14) maintains TERT mRNA nuclear localization. During mitosis, the nuclear membrane breaks down, TERT mRNA enters the cytoplasm, and intron 11 is spliced. Blocking intron 11 splicing with TMOs also reduced cell viability. Rinn’s work suggests that splicing is important for lncRNA localization and function.158
Mukesh Lalwani from Andrew Baker’s lab at the University of Edinburgh presented unpublished work on the role of the lncRNA CARMN in heart regeneration. After injury, the heart suffers from loss of cardiomyocytes, which are replaced by noncontractile fibroblasts, which can lead to contractile dysfunction and heart failure. While adult mammals cannot induce cardiomyocyte proliferation, neonates and lower vertebrate species can regenerate and proliferate cardiomyocytes after injury.159 Several lncRNAs are induced upon injury in cardiomyocytes.160,161 Lalwani focused on the impact of CARMN. Baker’s group previously showed that loss of CARMN can enhance vascular smooth muscle cell proliferation, migration, inflammation, and plasticity and accelerate atherosclerosis in mice.162 In addition, differential expression of CARMN isoforms in cardiac progenitor cells affects differentiation at different stages of development. In the fetal heart, CARMN isoforms promote differentiation into cardiomyocytes while in the adult heart, CARMN isoforms promote differentiation to smooth muscle cells.163,164 Lalwani is investigating the role of CARMN in cardiomyocyte proliferation and cardiac regeneration in zebrafish.
Murat Can Kalem from John Panepinto’s lab at the State University of New York, University at Buffalo presented unpublished work on protein post-translational modification can regulate lncRNA function in C. neoformans infection. C. neoformans is an environmental fungal pathogen that can cause Cryptococcal meningitis in immunocompromised people. RNA-seq data has identified approximately 3000 differentially expressed genes in C. neoformans at the site of human meningitis, of which approximately 500 of which are lncRNAs.165 Gene regulation is important for C. neoformans to adapt to different microenvironments, such as elevated temperature. One of the key factors that enables C. neoformans to grow at elevated temperature is the arginine methyltransferase Rmt5. Arginine methylation can potentially impact protein-protein and protein-RNA interactions. Given the importance of Rmt5 in C. neoformans survival and of lncRNAs at sites of infection, Kalem is investigating whether there is a link between arginine Rmt5 target proteins and lncRNA function.
Repetitive RNAs in gene regulation
Transposable elements (TE) are highly repetitive DNA regions that represent over half of the genome. While most TEs are not protein coding, the have been shown to be an abundant source of various kinds of noncoding regulatory sequences, such as promoters, enhancers, noncoding RNAs, and short RNAs.166
Edward B. Chuong from the University of Colorado Boulder discussed how retrotransposons can regulate innate immunity. They focused on the endogenous retrovirus (ERV) MER41, of which approximately 9000 copies are present in primate genomes. MER41 contains a pair of STAT1 binding motifs that can interact with STAT1 and impact the expression of nearby genes. In particular, Chuong showed that MER41 elements exhibit IFN-inducible regulatory activity in macrophages. Upon IFN stimulation, STAT1 can bind to MER41.AIM2, which is located near the inflammasome component absent in melanoma 2 (AIM2) and induce AIM2 expression. Chuong also showed that MER41 elements can impact expression of several other genes, indicating that ERVs have been co-opted to act as IFN-stimulated enhancers for a number of anti-viral and inflammatory genes.167 Their group is continuing to study how transposable elements influence the evolution of immune regulation and variation and how they impact regulatory elements in disease. Toward that goal, Chuong presented unpublished work on a separate EVR, LTR10, and its role in cancer. Transposable element activation is common in cancer cells.168–172 Chuong’s groups is working to understand how the elements may cause gene dysregulation that can contribute to disease.
Lynne E. Maquat from the University of Rochester Medical Center described how loss of the fragile X retardation protein (FMRP) upregulates nonsense-mediated mRNA decay (NMD), potentially contributing to the developmental and cognitive defects observed in individuals with fragile X syndrome. Approximately 5 to 10% of mRNAs are degraded by NMD, which allows the cell to regulate mRNA concentration and adapt to environmental changes. For example, Maquat’s group has shown that NMD competes with a separate mRNA decay pathway, Staufen-mediated mRNA decay (SMD), due to the fact that both pathways require the protein UPF1.173 This competition contributes to myogenesis as decreased levels of the NMD factor UPF2 enables UPF1 to interact with the SMD factor Staufen and increase SMD efficiency. Downregulation of SMD targets, such as PAX3, and upregulation of NMD targets, such as myogenin, ultimately promote myogenesis.142 More recently, Maquat’s lab has shown that UPF1 interacts directly with FMRP. This interaction promotes and/or stabilizes FMRP binding to NMD targets and thus suppresses NMD activity. In the brain, many NMD targets encode proteins important for axon guidance and synaptic transmission. Therefore, in the absence of FMRP, such as in fragile X syndrome and depending on the stage of neural differentiation, NMD is hyperactivated, and NMD targets are destabilized, which negatively affects neurogenesis. Maquat showed that fibroblasts and iPSCs from subjects with fragile X syndrome manifest hyperactivated NMD. The differentiation of these iPSCs to neurons manifests deficits in neurogenesis that are partially normalized upon administration of an NMD inhibitor.143
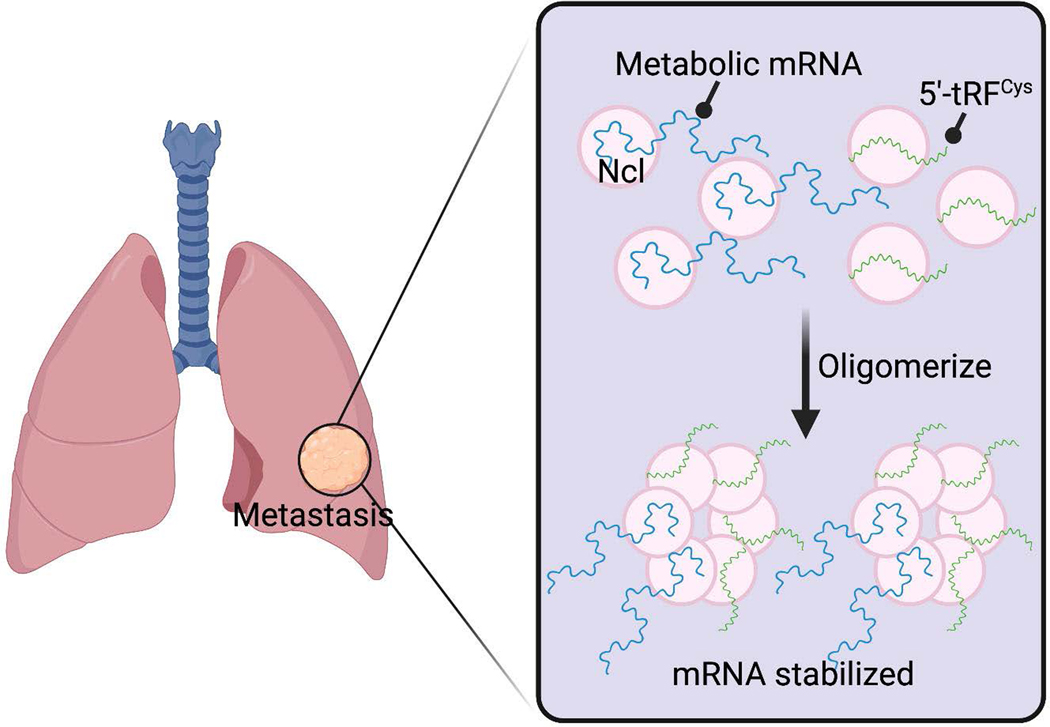
Xuhang Liu from Sohail Tavazoie’s lab at The Rockefeller University showed how a tRNA fragment can impact metastasis. Small RNA-seq analysis of mouse breast cancer cells with differing metastatic capacity identified 5′-tRFCys as one of the most upregulated tRFs in highly metastatic cells. High expression of 5′-tRFCys also correlates with poor clinical outcomes in breast cancer patients and is increased in breast tumors compared to normal breast tissues. Liu showed that interactions between nucleolin and 5′-tRFCys enhances nucleolin binding to a subset of transcripts, including several metabolic enzymes, and enhances their expression (Fig. 8). Liu put forth a model in which breast cancer cells contain a subset of pro-metastatic transcripts that bind to nucleolin and form nucleolin monomers. In the absence of 5′-tRFCys, these monomers are not able to efficiently oligomerize, the complexes disassemble, and cells have limited metastatic potential. However, upregulation of 5′-tRFCys promotes efficient oligomerization of nucleolin-based complexes, which stabilizes pro-metastatic transcripts and increases the metastatic potential of cells.174
Figure 8.
Interactions between nucleolin and 5′-tRFCys enhances nucleolin binding to a subset of transcripts, including several metabolic enzymes, and enhances their expression.
Acknowledgements
R.K.P. is supported by a Predoctoral Fellowship from the American Heart Association (18PRE33990261); a Wagner Fellowship from the University of Virginia; and F99/K00 NCI Predoctoral to Postdoctoral Fellow Transition Award (F99CA253732).
J.E.W. is supported by NIH grant R35-GM119735.
X.L. was supported by a Bristol Myers Squibb postdoctoral fellowship.
The research of S.F.T. was supported in part by a Faculty Scholar grant from the HHMI, by the DOD Collaborative Scholars and Innovators Award (W81XWH-12-1-0301), Pershing Square Sohn Cancer Research Alliance award, Breast Cancer Research Foundation award, Emerald Foundation, NIH grant 5R01CA215491-05, and the Black Family Metastasis Center.
Footnotes
Competing Interests
J. Mendell is a scientific advisor for Ribometrix and Circ Bio.
Sohail Tavazoie is a co-founder and shareholder of Rgenix and member of its scientific advisory board.
References
- 1.Chowdhary A, Satagopam V. & Schneider R. 2021. Long Non-coding RNAs: Mechanisms, Experimental, and Computational Approaches in Identification, Characterization, and Their Biomarker Potential in Cancer. Front. Genet 12: 649619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mattick JS & Makunin IV 2006. Non-coding RNA. Hum. Mol. Genet 15 Spec No 1: R17–29. [DOI] [PubMed] [Google Scholar]
- 3.Dossin F. & Heard E. 2021. The Molecular and Nuclear Dynamics of X-Chromosome Inactivation. Cold Spring Harb. Perspect. Biol a040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engreitz JM, Pandya-Jones A, McDonel P, et al. 2013. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341: 1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giorgetti L, Lajoie BR, Carter AC, et al. 2016. Structural organization of the inactive X chromosome in the mouse. Nature 535: 575–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaumeil J, Le Baccon P, Wutz A, et al. 2006. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 20: 2223–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chow JC, Ciaudo C, Fazzari MJ, et al. 2010. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell 141: 956–969. [DOI] [PubMed] [Google Scholar]
- 8.Collombet S, Rall I, Dugast-Darzacq C, et al. 2021. RNA polymerase II depletion from the inactive X chromosome territory is not mediated by physical compartmentalization. bioRxiv 2021.03.26.437188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Żylicz JJ, Bousard A, Žumer K, et al. 2019. The Implication of Early Chromatin Changes in X Chromosome Inactivation. Cell 176: 182–197.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu C, Zhang QC, da Rocha ST, et al. 2015. Systematic discovery of Xist RNA binding proteins. Cell 161: 404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McHugh CA, Chen C-K, Chow A, et al. 2015. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521: 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C-K, Blanco M, Jackson C, et al. 2016. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354: 468–472. [DOI] [PubMed] [Google Scholar]
- 13.Minajigi A, Froberg J, Wei C, et al. 2015. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moindrot B, Cerase A, Coker H, et al. 2015. A Pooled shRNA Screen Identifies Rbm15, Spen, and Wtap as Factors Required for Xist RNA-Mediated Silencing. Cell Rep. 12: 562–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monfort A, Di Minin G, Postlmayr A, et al. 2015. Identification of Spen as a Crucial Factor for Xist Function through Forward Genetic Screening in Haploid Embryonic Stem Cells. Cell Rep. 12: 554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dossin F, Pinheiro I, Żylicz JJ, et al. 2020. SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature 578: 455–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamazaki T, Souquere S, Chujo T, et al. 2018. Functional Domains of NEAT1 Architectural lncRNA Induce Paraspeckle Assembly through Phase Separation. Mol. Cell 70: 1038–1053.e7. [DOI] [PubMed] [Google Scholar]
- 18.Yamazaki T, Nakagawa S. & Hirose T. 2019. Architectural RNAs for Membraneless Nuclear Body Formation. Cold Spring Harb. Symp. Quant. Biol 84: 227–237. [DOI] [PubMed] [Google Scholar]
- 19.Shin Y. & Brangwynne CP 2017. Liquid phase condensation in cell physiology and disease. Science 357: eaaf4382. [DOI] [PubMed] [Google Scholar]
- 20.Biamonti G. 2004. Nuclear stress bodies: a heterochromatin affair? Nat. Rev. Mol. Cell Biol 5: 493–498. [DOI] [PubMed] [Google Scholar]
- 21.Ninomiya K, Adachi S, Natsume T, et al. 2020. LncRNA-dependent nuclear stress bodies promote intron retention through SR protein phosphorylation. EMBO J. 39: e102729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ninomiya K, Iwakiri J, Aly MK, et al. 2021. m6 A modification of HSATIII lncRNAs regulates temperature-dependent splicing. EMBO J. e107976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xing Y-H, Yao R-W, Zhang Y, et al. 2017. SLERT Regulates DDX21 Rings Associated with Pol I Transcription. Cell 169: 664–678.e16. [DOI] [PubMed] [Google Scholar]
- 24.Yao R-W, Xu G, Wang Y, et al. 2019. Nascent Pre-rRNA Sorting via Phase Separation Drives the Assembly of Dense Fibrillar Components in the Human Nucleolus. Mol. Cell 76: 767–783.e11. [DOI] [PubMed] [Google Scholar]
- 25.Wu M, Xu G, Han C, et al. 2021. lncRNA SLERT controls phase separation of FC/DFCs to facilitate Pol I transcription. Science 373: 547–555. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Hu X, Song W, et al. 2021. Mutual dependency between lncRNA LETN and protein NPM1 in controlling the nucleolar structure and functions sustaining cell proliferation. Cell Res. 31: 664–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caudron-Herger M, Pankert T, Seiler J, et al. 2015. Alu element-containing RNAs maintain nucleolar structure and function. EMBO J. 34: 2758–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li D, Zhang J, Wang M, et al. 2018. Activity dependent LoNA regulates translation by coordinating rRNA transcription and methylation. Nat. Commun 9: 1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yap K, Mukhina S, Zhang G, et al. 2018. A Short Tandem Repeat-Enriched RNA Assembles a Nuclear Compartment to Control Alternative Splicing and Promote Cell Survival. Mol. Cell 72: 525–540.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Audas TE, Jacob MD & Lee S. 2012. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol. Cell 45: 147–157. [DOI] [PubMed] [Google Scholar]
- 31.Bierhoff H, Schmitz K, Maass F, et al. 2010. Noncoding transcripts in sense and antisense orientation regulate the epigenetic state of ribosomal RNA genes. Cold Spring Harb. Symp. Quant. Biol 75: 357–364. [DOI] [PubMed] [Google Scholar]
- 32.Hao Q. & Prasanth KV 2021. Regulatory roles of nucleolus organizer region-derived long non-coding RNAs. Mamm. Genome Off. J. Int. Mamm. Genome Soc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quinodoz SA, Ollikainen N, Tabak B, et al. 2018. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell 174: 744–757.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinodoz SA, Bhat P, Ollikainen N, et al. 2020. RNA promotes the formation of spatial compartments in the nucleus. bioRxiv 2020.08.25.267435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balas MM & Johnson AM 2018. Exploring the mechanisms behind long noncoding RNAs and cancer. Non-Coding RNA Res. 3: 108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haemmig S. & Feinberg MW 2017. Targeting LncRNAs in Cardiovascular Disease: Options and Expeditions. Circ. Res 120: 620–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diermeier SD, Chang K-C, Freier SM, et al. 2016. Mammary Tumor-Associated RNAs Impact Tumor Cell Proliferation, Invasion, and Migration. Cell Rep. 17: 261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang K-C, Diermeier SD, Yu AT, et al. 2020. MaTAR25 lncRNA regulates the Tensin1 gene to impact breast cancer progression. Nat. Commun 11: 6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen H. & Lo SH 2003. Regulation of tensin-promoted cell migration by its focal adhesion binding and Src homology domain 2. Biochem. J 370: 1039–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, Han L, Roebuck P, et al. 2015. TANRIC: An Interactive Open Platform to Explore the Function of lncRNAs in Cancer. Cancer Res. 75: 3728–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diermeier SD & Spector DL 2017. Antisense Oligonucleotide-mediated Knockdown in Mammary Tumor Organoids. Bio-Protoc. 7: e2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan X, Hu Z, Feng Y, et al. 2015. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell 28: 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta RA, Shah N, Wang KC, et al. 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Portoso M, Ragazzini R, Brenčič Ž, et al. 2017. PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J. 36: 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porman AM, Roberts JT, Duncan ED, et al. 2021. A single N6-methyladenosine site in lncRNA HOTAIR regulates its function in breast cancer cells. bioRxiv 2020.06.08.140954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao H, Chakraborty G, Lee-Lim AP, et al. 2014. Forward genetic screens in mice uncover mediators and suppressors of metastatic reactivation. Proc. Natl. Acad. Sci. U. S. A 111: 16532–16537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haemmig S, Simion V. & Feinberg MW 2018. Long Non-Coding RNAs in Vascular Inflammation. Front. Cardiovasc. Med 5: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haemmig S, Yang D, Sun X, et al. 2020. Long noncoding RNA SNHG12 integrates a DNA-PK-mediated DNA damage response and vascular senescence. Sci. Transl. Med 12: eaaw1868. [DOI] [PubMed] [Google Scholar]
- 49.Childs BG, Baker DJ, Wijshake T, et al. 2016. Senescent intimal foam cells are deleterious at all stages of atherosclerosis. Science 354: 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang JC & Bennett M. 2012. Aging and atherosclerosis: mechanisms, functional consequences, and potential therapeutics for cellular senescence. Circ. Res 111: 245–259. [DOI] [PubMed] [Google Scholar]
- 51.Simion V, Zhou H, Haemmig S, et al. 2020. A macrophage-specific lncRNA regulates apoptosis and atherosclerosis by tethering HuR in the nucleus. Nat. Commun 11: 6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simion V, Zhou H, Pierce JB, et al. 2020. LncRNA VINAS regulates atherosclerosis by modulating NF-κB and MAPK signaling. JCI Insight 5: 140627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou H, Simion V, Pierce JB, et al. 2021. LncRNA-MAP3K4 regulates vascular inflammation through the p38 MAPK signaling pathway and cis-modulation of MAP3K4. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol 35: e21133. [DOI] [PubMed] [Google Scholar]
- 54.Ni H, Haemmig S, Deng Y, et al. 2021. A Smooth Muscle Cell-Enriched Long Noncoding RNA Regulates Cell Plasticity and Atherosclerosis by Interacting With Serum Response Factor. Arterioscler. Thromb. Vasc. Biol 41: 2399–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin Y, Lu JY, Zhang X, et al. 2020. U1 snRNP regulates chromatin retention of noncoding RNAs. Nature 580: 147–150. [DOI] [PubMed] [Google Scholar]
- 56.Seila AC, Calabrese JM, Levine SS, et al. 2008. Divergent transcription from active promoters. Science 322: 1849–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Henninger JE, Oksuz O, Shrinivas K, et al. 2021. RNA-Mediated Feedback Control of Transcriptional Condensates. Cell 184: 207–225.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cisse II, Izeddin I, Causse SZ, et al. 2013. Real-time dynamics of RNA polymerase II clustering in live human cells. Science 341: 664–667. [DOI] [PubMed] [Google Scholar]
- 59.Hnisz D, Shrinivas K, Young RA, et al. 2017. A Phase Separation Model for Transcriptional Control. Cell 169: 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shao W, Bi X, Gao B, et al. 2021. Phase separation of RNA-binding protein promotes polymerase engagement and transcription. bioRxiv 2021.03.26.436939. [DOI] [PubMed] [Google Scholar]
- 61.Lee S, Kopp F, Chang T-C, et al. 2016. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell 164: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kopp F, Elguindy MM, Yalvac ME, et al. 2019. PUMILIO hyperactivity drives premature aging of Norad-deficient mice. eLife 8: e42650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elguindy MM, Kopp F, Goodarzi M, et al. 2019. PUMILIO, but not RBMX, binding is required for regulation of genomic stability by noncoding RNA NORAD. eLife 8: e48625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tichon A, Gil N, Lubelsky Y, et al. 2016. A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat. Commun 7: 12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tichon A, Perry RB-T, Stojic L, et al. 2018. SAM68 is required for regulation of Pumilio by the NORAD long noncoding RNA. Genes Dev. 32: 70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Munschauer M, Nguyen CT, Sirokman K, et al. 2018. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature 561: 132–136. [DOI] [PubMed] [Google Scholar]
- 67.Unfried JP, Serrano G, Suárez B, et al. 2019. Identification of Coding and Long Noncoding RNAs Differentially Expressed in Tumors and Preferentially Expressed in Healthy Tissues. Cancer Res. 79: 5167–5180. [DOI] [PubMed] [Google Scholar]
- 68.Unfried JP, Marín-Baquero M, Rivera-Calzada Á, et al. 2021. Long noncoding RNA NIHCOLE promotes ligation efficiency of DNA double-strand breaks in hepatocellular carcinoma. Cancer Res. canresCAN-21–0463-A.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mendoza-Figueroa MS, Tatomer DC & Wilusz JE 2020. The Integrator Complex in Transcription and Development. Trends Biochem. Sci 45: 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baillat D, Hakimi M-A, Näär AM, et al. 2005. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123: 265–276. [DOI] [PubMed] [Google Scholar]
- 71.Tatomer DC, Elrod ND, Liang D, et al. 2019. The Integrator complex cleaves nascent mRNAs to attenuate transcription. Genes Dev. 33: 1525–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elrod ND, Henriques T, Huang K-L, et al. 2019. The Integrator Complex Attenuates Promoter-Proximal Transcription at Protein-Coding Genes. Mol. Cell 76: 738–752.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mani SR & Juliano CE 2013. Untangling the web: the diverse functions of the PIWI/piRNA pathway. Mol. Reprod. Dev 80: 632–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Williams Z, Morozov P, Mihailovic A, et al. 2015. Discovery and Characterization of piRNAs in the Human Fetal Ovary. Cell Rep. 13: 854–863. [DOI] [PubMed] [Google Scholar]
- 75.Özata DM, Yu T, Mou H, et al. 2020. Evolutionarily conserved pachytene piRNA loci are highly divergent among modern humans. Nat. Ecol. Evol 4: 156–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Öst A, Lempradl A, Casas E, et al. 2014. Paternal diet defines offspring chromatin state and intergenerational obesity. Cell 159: 1352–1364. [DOI] [PubMed] [Google Scholar]
- 77.Carone BR, Fauquier L, Habib N, et al. 2010. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell 143: 1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gapp K, Jawaid A, Sarkies P, et al. 2014. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat. Neurosci 17: 667–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lempradl A, Kugelberg U, Iconomou M, et al. 2021. Intergenerational metabolic priming by sperm piRNAs. bioRxiv 2021.03.29.436592. [Google Scholar]
- 80.Grimson A, Farh KK-H, Johnston WK, et al. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sheu-Gruttadauria J, Pawlica P, Klum SM, et al. 2019. Structural Basis for Target-Directed MicroRNA Degradation. Mol. Cell 75: 1243–1255.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sheu-Gruttadauria J, Xiao Y, Gebert LF, et al. 2019. Beyond the seed: structural basis for supplementary microRNA targeting by human Argonaute2. EMBO J. 38: e101153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McGeary SE, Bisaria N. & Bartel DP 2021. Pairing to the microRNA 3′ region occurs through two alternative binding modes, with affinity shaped by nucleotide identity as well as pairing position. bioRxiv 2021.04.13.439700. [Google Scholar]
- 84.Vicens Q. & Westhof E. 2014. Biogenesis of Circular RNAs. Cell 159: 13–14. [DOI] [PubMed] [Google Scholar]
- 85.Chen L. & Shan G. 2015. Circular RNAs remain peculiarly unclear in biogenesis and function. Sci. China Life Sci 58: 616–618. [DOI] [PubMed] [Google Scholar]
- 86.Chen I, Chen C-Y & Chuang T-J 2015. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip. Rev. RNA 6: 563–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hansen TB, Jensen TI, Clausen BH, et al. 2013. Natural RNA circles function as efficient microRNA sponges. Nature 495: 384–388. [DOI] [PubMed] [Google Scholar]
- 88.Memczak S, Jens M, Elefsinioti A, et al. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495: 333–338. [DOI] [PubMed] [Google Scholar]
- 89.Xu H, Guo S, Li W, et al. 2015. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci. Rep 5: 12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. 2014. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56: 55–66. [DOI] [PubMed] [Google Scholar]
- 91.Kulcheski FR, Christoff AP & Margis R. 2016. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J. Biotechnol 238: 42–51. [DOI] [PubMed] [Google Scholar]
- 92.Zheng Q, Bao C, Guo W, et al. 2016. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun 7: 11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang Y, Gao X, Zhang M, et al. 2018. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J. Natl. Cancer Inst 110:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang M, Zhao K, Xu X, et al. 2018. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun 9: 4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang M, Huang N, Yang X, et al. 2018. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 37: 1805–1814. [DOI] [PubMed] [Google Scholar]
- 96.Legnini I, Di Timoteo G, Rossi F, et al. 2017. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 66: 22–37.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zheng X, Chen L, Zhou Y, et al. 2019. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol. Cancer 18: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pamudurti NR, Bartok O, Jens M, et al. 2017. Translation of CircRNAs. Mol. Cell 66: 9–21.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liang W-C, Wong C-W, Liang P-P, et al. 2019. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 20: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Granados-Riveron JT & Aquino-Jarquin G. 2016. The complexity of the translation ability of circRNAs. Biochim. Biophys. Acta 1859: 1245–1251. [DOI] [PubMed] [Google Scholar]
- 101.Schneider T, Hung L-H, Schreiner S, et al. 2016. CircRNA-protein complexes: IMP3 protein component defines subfamily of circRNPs. Sci. Rep 6: 31313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li L-J, Leng R-X, Fan Y-G, et al. 2017. Translation of noncoding RNAs: Focus on lncRNAs, pri-miRNAs, and circRNAs. Exp. Cell Res 361: 1–8. [DOI] [PubMed] [Google Scholar]
- 103.Wang Y. & Wang Z. 2015. Efficient backsplicing produces translatable circular mRNAs. RNA N. Y. N 21: 172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen X, Han P, Zhou T, et al. 2016. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci. Rep 6: 34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jang SK, Kräusslich HG, Nicklin MJ, et al. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol 62: 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pelletier J. & Sonenberg N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334: 320–325. [DOI] [PubMed] [Google Scholar]
- 107.Toor N, Keating KS, Taylor SD, et al. 2008. Crystal structure of a self-spliced group II intron. Science 320: 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rinn JL, Kertesz M, Wang JK, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Somarowthu S, Legiewicz M, Chillón I, et al. 2015. HOTAIR forms an intricate and modular secondary structure. Mol. Cell 58: 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rivas E, Clements J. & Eddy SR 2017. A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat. Methods 14: 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tavares RCA, Pyle AM & Somarowthu S. 2019. Phylogenetic Analysis with Improved Parameters Reveals Conservation in lncRNA Structures. J. Mol. Biol 431: 1592–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Morgan BS, Sanaba BG, Donlic A, et al. 2019. R-BIND: An Interactive Database for Exploring and Developing RNA-Targeted Chemical Probes. ACS Chem. Biol 14: 2691–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Morgan BS, Forte JE, Culver RN, et al. 2017. Discovery of Key Physicochemical, Structural, and Spatial Properties of RNA-Targeted Bioactive Ligands. Angew. Chem. Int. Ed Engl 56: 13498–13502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stelzer AC, Frank AT, Kratz JD, et al. 2011. Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat. Chem. Biol 7: 553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Patwardhan NN, Ganser LR, Kapral GJ, et al. 2017. Amiloride as a new RNA-binding scaffold with activity against HIV-1 TAR. MedChemComm 8: 1022–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Patwardhan NN, Cai Z, Umuhire Juru A, et al. 2019. Driving factors in amiloride recognition of HIV RNA targets. Org. Biomol. Chem 17: 9313–9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Davila-Calderon J, Patwardhan NN, Chiu L-Y, et al. 2020. IRES-targeting small molecule inhibits enterovirus 71 replication via allosteric stabilization of a ternary complex. Nat. Commun 11: 4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brown JA, Bulkley D, Wang J, et al. 2014. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol 21: 633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Donlic A, Morgan BS, Xu JL, et al. 2018. Discovery of Small Molecule Ligands for MALAT1 by Tuning an RNA-Binding Scaffold. Angew. Chem. Int. Ed Engl 57: 13242–13247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Donlic A, Zafferani M, Padroni G, et al. 2020. Regulation of MALAT1 triple helix stability and in vitro degradation by diphenylfurans. Nucleic Acids Res. 48: 7653–7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zimmer JT, Rosa-Mercado NA, Canzio D, et al. 2021. STL-seq reveals pause-release and termination kinetics for promoter-proximal paused RNA polymerase II transcripts. Mol. Cell S1097–2765(21)00686–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Marcia M. & Pyle AM 2012. Visualizing group II intron catalysis through the stages of splicing. Cell 151: 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhao C, Rajashankar KR, Marcia M, et al. 2015. Crystal structure of group II intron domain 1 reveals a template for RNA assembly. Nat. Chem. Biol 11: 967–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Manigrasso J, Chillón I, Genna V, et al. 2020. Visualizing group II intron dynamics between the first and second steps of splicing. Nat. Commun 11: 2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chunharojrith P, Nakayama Y, Jiang X, et al. 2015. Tumor suppression by MEG3 lncRNA in a human pituitary tumor derived cell line. Mol. Cell. Endocrinol 416: 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang X, Rice K, Wang Y, et al. 2010. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology 151: 939–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhu J, Liu S, Ye F, et al. 2015. Long Noncoding RNA MEG3 Interacts with p53 Protein and Regulates Partial p53 Target Genes in Hepatoma Cells. PloS One 10: e0139790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Uroda T, Anastasakou E, Rossi A, et al. 2019. Conserved Pseudoknots in lncRNA MEG3 Are Essential for Stimulation of the p53 Pathway. Mol. Cell 75: 982–995.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Uroda T, Chillón I, Annibale P, et al. 2020. Visualizing the functional 3D shape and topography of long noncoding RNAs by single-particle atomic force microscopy and in-solution hydrodynamic techniques. Nat. Protoc 15: 2107–2139. [DOI] [PubMed] [Google Scholar]
- 130.Mueller AC, Cichewicz MA, Dey BK, et al. 2015. MUNC, a long noncoding RNA that facilitates the function of MyoD in skeletal myogenesis. Mol. Cell. Biol 35: 498–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cichewicz MA, Kiran M, Przanowska RK, et al. 2018. MUNC, an Enhancer RNA Upstream from the MYOD Gene, Induces a Subgroup of Myogenic Transcripts in trans Independently of MyoD. Mol. Cell. Biol 38: e00655–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Przanowska RK, Weidmann CA, Saha S, et al. 2021. Distinct MUNC lncRNA structural domains regulate transcription of different promyogenic factors. bioRxiv 2021.06.22.449443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Invernizzi P. & Gershwin ME 2009. The genetics of human autoimmune disease. J. Autoimmun 33: 290–299. [DOI] [PubMed] [Google Scholar]
- 134.Peckham H, de Gruijter NM, Raine C, et al. 2020. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat. Commun 11: 6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yu B, Qi Y, Li R, et al. 2021. B cell-specific XIST complex enforces X-inactivation and restrains atypical B cells. Cell 184: 1790–1803.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lu Z, Filonov GS, Noto JJ, et al. 2015. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA N. Y. N 21: 1554–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Litke JL & Jaffrey SR 2021. Trans ligation of RNAs to generate hybrid circular RNAs using highly efficient autocatalytic transcripts. Methods San Diego Calif S1046–2023(21)00135–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lukasiewicz AJ & Contreras LM 2020. Antisense probing of dynamic RNA structures. Methods San Diego Calif 183: 76–83. [DOI] [PubMed] [Google Scholar]
- 139.Sowa SW, Vazquez-Anderson J, Clark CA, et al. 2015. Exploiting post-transcriptional regulation to probe RNA structures in vivo via fluorescence. Nucleic Acids Res. 43: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mihailovic MK, Vazquez-Anderson J, Li Y, et al. 2018. High-throughput in vivo mapping of RNA accessible interfaces to identify functional sRNA binding sites. Nat. Commun 9: 4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Isaacs FJ, Dwyer DJ, Ding C, et al. 2004. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol 22: 841–847. [DOI] [PubMed] [Google Scholar]
- 142.Leistra AN, Amador P, Buvanendiran A, et al. 2017. Rational Modular RNA Engineering Based on In Vivo Profiling of Structural Accessibility. ACS Synth. Biol 6: 2228–2240. [DOI] [PubMed] [Google Scholar]
- 143.Mihailovic MK, Ekdahl AM, Chen A, et al. 2021. Uncovering Transcriptional Regulators and Targets of sRNAs Using an Integrative Data-Mining Approach: H-NS-Regulated RseX as a Case Study. Front. Cell. Infect. Microbiol 11: 696533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shi J, Zhang Y, Tan D, et al. 2021. PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat. Cell Biol 23: 424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shi J, Ko E-A, Sanders KM, et al. 2018. SPORTS1.0: A Tool for Annotating and Profiling Non-coding RNAs Optimized for rRNA- and tRNA-derived Small RNAs. Genomics Proteomics Bioinformatics 16: 144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Di Bella S, La Ferlita A, Carapezza G, et al. 2020. A benchmarking of pipelines for detecting ncRNAs from RNA-Seq data. Brief. Bioinform 21: 1987–1998. [DOI] [PubMed] [Google Scholar]
- 147.Gu W, Shi J, Liu H, et al. 2020. Peripheral blood non-canonical small non-coding RNAs as novel biomarkers in lung cancer. Mol. Cancer 19: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Peaston AE, Evsikov AV, Graber JH, et al. 2004. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell 7: 597–606. [DOI] [PubMed] [Google Scholar]
- 149.Kigami D, Minami N, Takayama H, et al. 2003. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol. Reprod 68: 651–654. [DOI] [PubMed] [Google Scholar]
- 150.Macfarlan TS, Gifford WD, Driscoll S, et al. 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Macfarlan TS, Gifford WD, Agarwal S, et al. 2011. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 25: 594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ishiuchi T, Enriquez-Gasca R, Mizutani E, et al. 2015. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat. Struct. Mol. Biol 22: 662–671. [DOI] [PubMed] [Google Scholar]
- 153.Choi YJ, Lin C-P, Risso D, et al. 2017. Deficiency of microRNA miR-34a expands cell fate potential in pluripotent stem cells. Science 355: eaag1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Modzelewski A, Shao W, Chen J, et al. 2021. A species-specific retrotransposon drives a conserved Cdk2ap1 isoform essential for preimplantation development. bioRxiv 2021.03.24.436683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gentles AJ, Newman AM, Liu CL, et al. 2015. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med 21: 938–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sica A, Larghi P, Mancino A, et al. 2008. Macrophage polarization in tumour progression. Semin. Cancer Biol 18: 349–355. [DOI] [PubMed] [Google Scholar]
- 157.Liu J, Lao L, Chen J, et al. 2021. The IRENA lncRNA converts chemotherapy-polarized tumor-suppressing macrophages to tumor-promoting phenotypes in breast cancer. Nat. Cancer 2: 457–473. [DOI] [PubMed] [Google Scholar]
- 158.Dumbović G, Braunschweig U, Langner HK, et al. 2021. Nuclear compartmentalization of TERT mRNA and TUG1 lncRNA is driven by intron retention. Nat. Commun 12: 3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xin M, Olson EN & Bassel-Duby R. 2013. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat. Rev. Mol. Cell Biol 14: 529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chen J, Hu Q, Zhang B-F, et al. 2019. Long noncoding RNA UCA1 inhibits ischaemia/reperfusion injury induced cardiomyocytes apoptosis via suppression of endoplasmic reticulum stress. Genes Genomics 41: 803–810. [DOI] [PubMed] [Google Scholar]
- 161.Ponnusamy M, Liu F, Zhang Y-H, et al. 2019. Long Noncoding RNA CPR (Cardiomyocyte Proliferation Regulator) Regulates Cardiomyocyte Proliferation and Cardiac Repair. Circulation 139: 2668–2684. [DOI] [PubMed] [Google Scholar]
- 162.Vacante F, Rodor J, Lalwani MK, et al. 2021. CARMN Loss Regulates Smooth Muscle Cells and Accelerates Atherosclerosis in Mice. Circ. Res 128: 1258–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ounzain S, Micheletti R, Arnan C, et al. 2015. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J. Mol. Cell. Cardiol 89: 98–112. [DOI] [PubMed] [Google Scholar]
- 164.Plaisance I, Perruchoud S, Fernandez-Tenorio M, et al. 2016. Cardiomyocyte Lineage Specification in Adult Human Cardiac Precursor Cells Via Modulation of Enhancer-Associated Long Noncoding RNA Expression. JACC Basic Transl. Sci 1: 472–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chen Y, Toffaletti DL, Tenor JL, et al. 2014. The Cryptococcus neoformans transcriptome at the site of human meningitis. mBio 5: e01087–01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Misiak B, Ricceri L. & Sąsiadek MM 2019. Transposable Elements and Their Epigenetic Regulation in Mental Disorders: Current Evidence in the Field. Front. Genet 0: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chuong EB, Elde NC & Feschotte C. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351: 1083–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Deniz Ö, Ahmed M, Todd CD, et al. 2020. Endogenous retroviruses are a source of enhancers with oncogenic potential in acute myeloid leukaemia. Nat. Commun 11: 3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Jang HS, Shah NM, Du AY, et al. 2019. Transposable elements drive widespread expression of oncogenes in human cancers. Nat. Genet 51: 611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Edginton-White B, Cauchy P, Assi SA, et al. 2019. Global long terminal repeat activation participates in establishing the unique gene expression programme of classical Hodgkin lymphoma. Leukemia 33: 1463–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Babaian A. & Mager DL 2016. Endogenous retroviral promoter exaptation in human cancer. Mob. DNA 7: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lamprecht B, Walter K, Kreher S, et al. 2010. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat. Med 16: 571–579, 1p following 579. [DOI] [PubMed] [Google Scholar]
- 173.Kim YK, Furic L, Desgroseillers L, et al. 2005. Mammalian Staufen1 recruits Upf1 to specific mRNA 3’UTRs so as to elicit mRNA decay. Cell 120: 195–208. [DOI] [PubMed] [Google Scholar]
- 174.Liu X, Alwaseem H, Molina H, et al. 2021. A pro-metastatic tRNA fragment drives Nucleolin oligomerization and stabilization of bound metabolic mRNAs. bioRxiv 2021.04.26.441477. [DOI] [PMC free article] [PubMed] [Google Scholar]