Abstract
Background:
According to several studies, there is an association between human papillomavirus (HPV) and breast cancer. Therefore, detection and genotyping of HPV seem important. The present study aimed to investigate the presence of HPV DNA in breast tissues by analyzing the L1 gene.
Materials and Methods:
This case-control study was conducted on 63 formalin-fixed paraffin-embedded (FFPE) tissues of invasive ductal carcinoma (IDC) as the case group and 32 FFPE tissues of fibroadenoma as the control group. HPV DNA was detected using the polymerase chain reaction assay. Positive samples were then subjected to genotyping. All statistical analyses were performed in SPSS version 22.0.
Results:
The patients’ age ranged from 15 to 92 years, with a mean age of 43.54±16.36 years. HPV DNA was detected in 17/95 (17.89%) samples, including 9/32 (28.12%) fibroadenoma samples and 8/63 (12.69%) IDC samples. No significant difference was observed regarding the presence of HPV DNA between the IDC and fibroadenoma tissues (P=0.08). However, a significant difference was found in the detection of high-risk HPV (HR-HPV) between the case and control groups (P=0.03). In the case group, 87.5% of the detected viruses (7/8 samples) were HR-HPV, while in the control group, 22.22% of positive samples (2/9 samples) were HR-HPV (P=0.03). Based on the results, HR-HPV and low-risk HPV genotypes were detected in 53% (9/17) and 47% (8/17) of positive samples, respectively.
Conclusion:
In this study, 12.69% of IDC samples were positive for HPV genomes, and HR-HPV was detected in 87.5% of these samples. The present results suggest the important role of HR-HPV in the development of breast cancer.
Key Words: Human Papillomavirus, DNA, Invasive Ductal Carcinoma
Introduction
Breast cancer (BC) is caused by an uncontrolled growth of cells in the breast. Although BC may occur in both men and women, women are exposed to a greater risk (Lawson and Heng, 2010b). This cancer is recognized as one of the main global health problems and a leading cause of mortality in women worldwide, with 1.7 million new cases and 522,000 deaths estimated annually (Ferlay et al., 2015). In the United States, it is the most commonly diagnosed cancer and the second leading cause of cancer-related death following lung cancer in women (Al Moustafa et al., 2016).
There are many types of BC, including ductal carcinoma in situ (DCIS), invasive ductal carcinoma (IDC), and invasive lobular carcinoma (ILC). IDC, accounting for 77% of all BC cases, is the most common type of BC in various countries, including Iran (Mousavi et al., 2007). Because of a series of factors, the incidence rate of BC is increasing significantly in South America, Africa, and Asia (Salman et al., 2017). Moreover, according to epidemiological data from Iran, BC is one of the most common malignancies in women, with a frequency of 2.5 per 100,000 people in 2015 (Ghaffari et al., 2018). The risk factors for BC can be genetics, unhealthy lifestyles, age, hormonal problems, and some viral infections (Martin and Weber, 2000; Lawson and Heng, 2010a; Chlebowski, 2013).
Human papillomavirus (HPV), an infectious agent, is one of the most common viral sexually transmitted diseases (Braaten and Laufer, 2008), associated with several cancers, such as cervical, vaginal, vulvar, head and neck, anal, penile, bladder, skin, lung, and breast cancers (Mahmoudvand et al., 2015; Tulay and Serakinci, 2016). HPVs are exclusively intraepithelial pathogens, which can infect the cutaneous and mucosal squamous epithelium and cause both benign and malignant hyperproliferative lesions (Stanley, 2012). The mechanism of carcinogenicity in papillomaviruses depends on the role of E6 and E7 oncoproteins, which degrade two major cellular tumor suppressor proteins, that is, p53 and retinoblastoma tumor suppressor protein (pRb), respectively (Yim and Park, 2005).
Considering the oncogenic characteristics of HPV, its genotypes can be divided into high-risk HPV (HR-HPV) and low-risk HPV (Ahmed et al., 2015). Persistent HR-HPV types, including HPV 16, 18, 31, 33, and 35, are associated with human cancers (Burd, 2003). Evidence suggests that the prevalence of HPV infection ranges from 0% to 86.2% among women with BC (Mou et al., 2011). According to several studies, HPV types 16, 18, and 33 are responsible for 70% of all HPV-related BC cases worldwide (Haghshenas et al., 2016a). The polymerase chain reaction (PCR)-based techniques for the detection of HPV DNA are currently used as the standard diagnostic method in clinical laboratories (Abreu et al., 2012). However, since the PCR assay may not detect all HPV genotypes in samples with a low copy number of viral genomes, the nested PCR technique has been shown to be more sensitive than other methods for the detection of HPV.
As BC is the fifth leading cause of cancer-related death in Iranian women (Akbari et al., 2017), the present study aimed to evaluate the presence of HPV DNA in tissues of BC patients in Ahvaz, Iran. Ahvaz is the capital of Khuzestan Province, located in the southwest of Iran, with a population of approximately two million people.
Objectives
This molecular epidemiological study aimed to detect HPV infection in individuals with BC in Ahvaz, Iran.
Materials and Methods
Tissue samples
A total of 95 formalin-fixed paraffin-embedded (FFPE) biopsy specimens, including IDC and fibroadenoma tissues, were collected and examined in this study from March, 2014 to February, 2018. Generally, fibroadenoma is a common benign breast tumor (non-cancerous), which was considered as the control group in the present study. Samples were collected from Imam Khomeini Hospital, a teaching hospital affiliated to Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran.
Ethical approval
This study was approved by the Ethics Committee of Ahvaz Jundishapur University of Medical Sciences (ethics number, IR.AJUMS.REC.1397.239). Informed consent was obtained from each participant.
DNA extraction
Four sections (10 µm) of FFPE tissue blocks were cut and placed in a 1.5-ml eppendorf tube. The first step of deparaffinization was performed by adding 1.2 mL of xylene to the 1.5-ml tubes, containing the tissue sections. After tube vortexing and incubation for five minutes at room temperature, the tubes underwent centrifugation at 14,000 rpm for five minutes. Next, the supernatant was removed, and 1 mL of 100% ethanol was added to each tube and incubated for five minutes at room temperature. Finally, the tubes underwent centrifugation at 14,000 rpm for five minutes, and the supernatant was removed; both steps were performed once. In the final step, the tubes were incubated at 37°C on a heating block until ethanol totally evaporated. DNA was then extracted using a High-Pure Viral Nucleic Acid Kit (Roche, Germany), according to the manufacturer’s instructions. The extracted DNA was stored at -20°C until further use.
PCR assay
To examine the quality of extracted DNA, β-globin gene was used as an internal control. All DNA samples were initially subjected to PCR with specific primers, including PCO3 and PCO4 (Table 1). The PCR reactions were performed in a total volume of 25 μL, containing 2 μL of extracted DNA, 0.25 µL of each primer (25 pmol), 12 µL of amplification premix (PCRBIO Taq Mix Red 2x), and 10.5 µL of distilled water. The PCR assays were carried out under the following conditions: five minutes of initial denaturation at 95°C, 40 cycles of denaturation at 95°C for one minute, annealing at 55°C for one minute, extension at 72°C for one minute, and a final extension at 72°C for 10 minutes. Next, using MY09/11 and GP5+/6+ primers, HPV-L1 amplification was performed on samples that were positive for β-globin gene (Table 1).
Table1.
The Sequences and other Characteristics of Primers Used in this Study
| Locus | Primers | ORF | Oligonucleotide sequence | |
|---|---|---|---|---|
| β-globin | PCO3 | β-globin | 5′-ACACAACTGTGTTCACTAGC-3′ | Fragment size |
| PCO4 | 5′-CAACTTCATCCACGTTCACC-3′ | 110 | ||
| HPV | MY09 | L1 | 5′-CGTCCMARRGGAWACTGATC-3′ | 450 |
| MY11 | 5′-GCMCAGGGWCATAAYAATGG-3′ | |||
| HPV | GP5+ | L1 | 5′-TTTGTTACTGTGGTAGATACTAC-3′ | 150 |
| GP6+ | 5′-GAAAAATAAACTGTAAATCATATTC-3′ |
ORF, pen Reading Frame; HPV, Human papillomavirus
The HPV-L1 gene was detected using the PCR assay in two consecutive amplification reactions. The first round of PCR was performed in a total volume of 25 μL, containing 2 μL of extracted DNA, 0.25 µL of each primer, 10 µL of amplification premix (PCRBIO Taq Mix Red 2x), and 12.5 µL of distilled water. The thermal conditions of the first PCR assay were as follows: five minutes of initial denaturation at 95°C, 40 cycles of denaturation at 95°C for one minute, annealing at 56°C for one minute, extension at 72°C for one minute, and a final extension at 72°C for 10 minutes.
The second reaction was also carried out in a volume of 25 μL, containing 3 μL of extracted DNA, 0.25 µL of each primer, 12 µL of amplification premix (PCRBIO Taq Mix Red 2x), and 9.5 µL of distilled water. The thermal conditions of the second PCR assay were as follows: five minutes of initial denaturation at 95°C, 40 cycles of denaturation at 95°C for one minute, annealing at 52°C for one minute, extension at 72°C for one minute, and a final extension at 72°C for four minutes. Finally, the PCR products were subjected to electrophoresis on 1.7% agarose gel, supplemented with DNA Safe Stain (CinnaGen, Iran) and visualized under a UV transilluminator. To determine HPV genotypes, all positive specimens were sequenced in an ABI 3130xl DNA sequencer (Applied Biosystems).
Statistical analysis
Data analysis was performed using SPSS version 22.0 (SPSS Inc., Chicago, IL, USA). Chi-square test and Fisher’s exact test were used to evaluate the data. A P-value less than 0.05 was considered significant.
Results
In the present study, a total of 95 breast samples, including 63 IDC (66.3%) and 32 fibroadenoma (33.7%) samples, were examined to identify the presence of HPV. The study population consisted of women, aged 15-92 years (mean age, 43.54±16.36 years). All samples were positive for β-globin, indicating the good quality of DNA, and underwent HPV genome detection. The HPV DNA was found in 17.89% (17/95) of the specimens (Table 2), including nine out of 32 fibroadenoma samples (28.12%) and eight out of 63 IDC samples (12.69%).
Table 2.
Distribution of Human Papillomaviruses (HPVs) in Invasive Ductal Carcinoma (IDC) Tissues, Fibroadenoma Tissues, and Different Age Groups
| Parameter | Total samples N (%) |
Human Papilloma Virus Positive Samples N (%) |
Genotype |
|---|---|---|---|
| Invasive Ductal Carcinoma (IDC) | 63/95 | 8/63 | |
| 66.30 | 12.69 | 16 (2), 31 (2), 33 (3), 11 (1) | |
| Fibroadenoma | 32/95 | 9/32 | |
| 33.70 | 28.12 | 55 (2), 30 (2), 51 (2) 11, 6, 50 | |
| Age (year) (15–92) | |||
| <20 | 7/95 (7.4) | 2/7 (28.57) | |
| 20-40 | 35/95 (36.9) | 7/35 (20) | |
| 41-60 | 37/95 (38.9) | 5/37 (13.51) | |
| >60 | 16/95 (16.8) | 3/16 (18.75) |
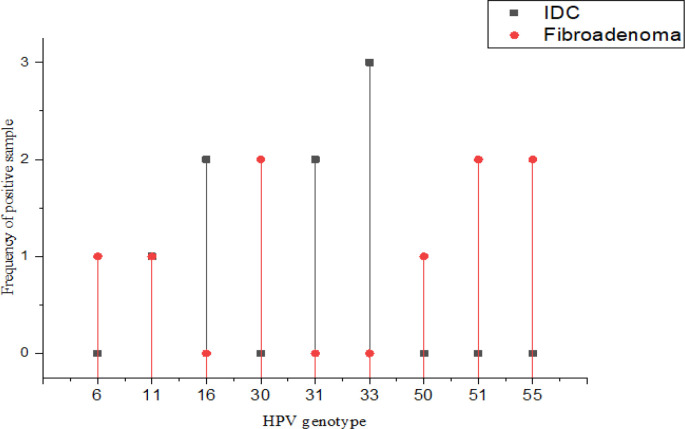
No significant difference was found regarding the presence of HPV DNA between the IDC and fibroadenoma tissues (P=0.08), while a significant difference was found in HR-HPV between the case and control groups (P=0.03). In the case group, 87.5% (7/8) of detected viruses were HR-HPV, while 22.22% (2/9) of positive samples were HR-HPV in the control group (P=0.03) (Figure 1). Regarding the distribution of HPV genotypes (Figure 2), genotypes 31, 33, 16, and 11 were detected in the case group, while genotypes 50, 51, 55, 6, 11, and 30 were identified in the control group.
Figure 1.
HPV Genotypes are Illustrated by Types of Specimens. Detected HR-HPVs were significantly higher in the IDC group (p=0.03). In addition, HPV-11 genotype found in both IDC and fibroadenoma
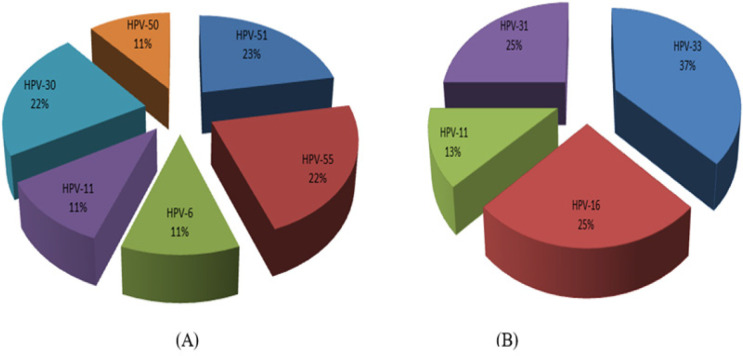
Figure 2.
Distribution of HPV Genotypes in Fibroadenoma (A) and in IDC Samples (B), which Revealed HPV16 was the only Highly Oncogenic Type of HPV Present in IDC Group (2/8, 25%).
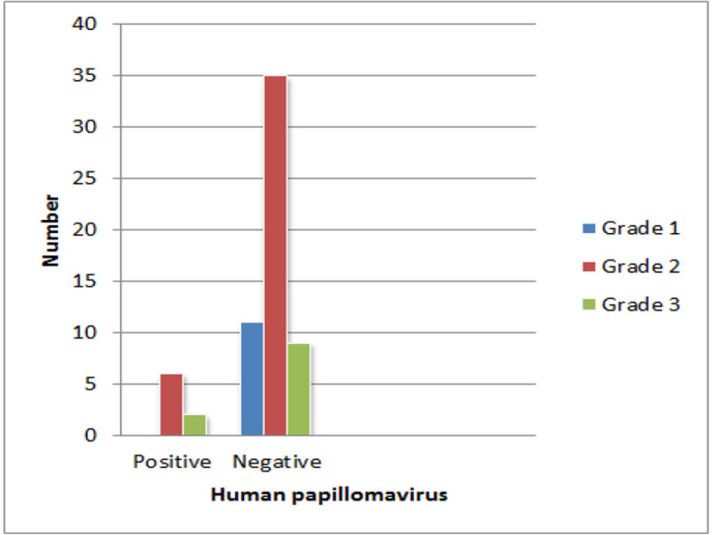
In the IDC specimens, the most frequent genotype was HR-HPV33 (3/8, 37%), and the high-risk type, HPV16, was only found in the case group (2/8, 25%). HPV11 was the only low-risk genotype in the case group (1/8, 11%). The relationship between the HPV status and cancer grade is presented in Figure 3.
Figure 3.
Data Demonstrated most of HPV Positive Samples (6 samples) in IDC Patients were Related to Grade 2 and others (2 samples) were Related to Grade 3 Malignancy
Discussion
The burden of BC has markedly increased around the world. BC is responsible for women’s death around the world (Azubuike et al., 2018). Therefore, identifying the risk factors that contribute to the development of this cancer is very important. Viruses are one of the main suspected contributors to cancer development, as they are involved in approximately 15-20% of human cancers (Sarvari et al., 2018). HPV is one of the most commonly known agents associated with various types of human cancers. The association between HR-HPV and cervical cancer and other types of cancer is well established (Bansal et al., 2016). The findings of previous studies demonstrated that HPV is associated with 60% of oropharyngeal cancers, 35% of penile cancers, and 90% of anal cancers (Zandberg et al., 2013). However, the role of HPV in BC development remains controversial.
For the first time, in 1992, Di Lonardo et al. showed that HPV might be involved in BC pathogenesis; they detected the presence of HPV in 29.4% of BC samples (Di Lonardo et al., 1992). There is an association between BC and HPV according to studies conducted in different areas around the world, including the United States, Australia, Italy, Japan, Norway, Greece, Korea, Mexico, and Taiwan (Lawson and Heng, 2010c; Haghshenas et al., 2016c); nevertheless, there are still many controversies about this association. A meta-analysis by Haghshenas et al. in Iran on 1,119 BC patients demonstrated that the prevalence of HPV was 23.6% in the case group (Haghshenas et al., 2016c). In agreement with the study by Haghshenas et al.,(2016c) another meta-analysis by Simoes et al. on 2,211 European, North American, and Australian women with BC showed that the prevalence of HPV was 23%, ranging from 13.4% in Europe to 42.9% in North America and Australia; in the control group, the prevalence of HPV was 12.9% (Simões et al., 2012).
Moreover, the results of two meta-analyses by Li et al., (2011) and Zhou et al., (2015) showed that 24.49% and 30.30% of BC tissues were positive for HPV DNA, respectively. In the current study, to increase the sensitivity of HPV genome identification in the samples (Ahangar-Oskouee et al., 2014), a nested PCR method was used to confirm the presence of viral genomes. Also, to better understand the relationship between HPV and the risk of BC, fibroadenoma (a non-cancerous tumor) samples were used as the control group. In the present study, HPV DNA was detected in 17.89% of samples (17/95), including 28.12% of fibroadenoma samples (9/32) and 12.69% of IDC samples (8/63). However, the results did not indicate a significant difference in the presence of HPV DNA between cancerous and non-cancerous samples (P=0.08). The results of statistical analysis indicated a difference in the identified HR-HPV between the case and control groups. The current study also reported a higher percentage of HR-HPV in IDC (87.5%) compared to fibroadenoma specimens (22.22%).
Different rates of HPV have been reported among BC patients in different parts of Iran. In agreement with our findings, a study by Malekpour Afshar et al., (2018), detected HPV DNA in 8.2% (8/98) of BC patients. Overall, 62.5% and 37.5% of positive samples were related to oncogenic genotypes 16/18 and 31/33, respectively. Another study on paraffin-embedded BC specimens in Iran in 2014 detected HPV DNA in 33.8% of samples in the case group (22/65), with only 4.5% of positive samples associated with HR-HPV (Ahangar-Oskouee et al., 2014). In this study, no virus was found in benign breast lesions as the control group.
Moreover, Sigaroodi et al., (2012) reported the presence of HPV DNA in 25.95% of tumor specimens versus 2.4% of specimens in the control group in north of Iran. They suggested HR-HPV 16/18 as the predominant HPV type (53.34%) in patients with BC. Some studies conducted in Iran have reported equal prevalence rates for HR-HPV and low-risk HPV. For example, Doosti et al., (2016) detected HPV in 22.9% (20/87) of BC tissue samples, including HPV18 (15%), HPV16 (35%), HPV6 (45%), and HPV11 (5%) in Yazd, Iran. In contrast to the present study, a study by Eslamifar et al., (2015) in Tehran, Iran found no HPV in 100 samples of IDC in the breasts. Overall, the results of studies conducted in Iran confirm the findings of other investigations from other geographic regions, including Australia, Brazil, Spain, and Venezuela, which identified HPV DNA in 48%, 24.75%, 51.8%, and 41.67% of BC samples, respectively (Damin et al., 2004; Kan et al., 2005; Fernandes et al., 2015; Delgado-García et al., 2017).
Some studies have indicated the higher frequency of HPV in BC tissues compared to benign breast tumor or normal breast tissues (Tsai et al., 2005; Gumus et al., 2006). In a study by Heng et al., HPV DNA was found in 14.28% (3/21) of IDC samples, whereas 18% (3 of 17) of normal breast tissue samples were positive for HPV. The results of this study are consistent with the present findings, which showed that the prevalence of HPV was higher in non-cancer tissues compared to cancerous tissues (Heng et al., 2009). Nevertheless, in some studies, HPV DNA was not detected in BC tissues (Hedau et al., 2011; Kwong et al., 2013; Vernet-Tomas et al., 2015).
As described above, several studies have found an association between HPV and BC, while some studies did not approve this association. This discrepancy between the results of previous studies may be affected by the geographic region, detection method, genetic background, cultural differences (e.g., different sexual behaviors), and sample type (e.g., paraffin-embedded tissue vs. fresh frozen tissue). As mentioned earlier, in the present study, we identified HPV genome in 17.89% of the samples. This rate of virus detection may be explained by the use of low-quality DNA extracted from paraffin-embedded tissues (Bae and Kim, 2016). Also, the age of the selected population could affect the detection rate of HPV, because it is assumed that the prevalence of HPV decreases with advancing age (de Cremoux et al., 2008).
In the present study, high-risk genotypes 16, 31, 33, and 51 were detected in 11.76% (2/17), 11.76% (2/17), 17.64% (3/17), and 11.76% (2/17) of positive samples, respectively. Genotypes 16, 31, and 33 were also found in 11.11% (7/63) of IDC samples. Besides, low-risk genotypes 6, 11, 30, 50, and 55 were detected in 47% (8/17) of positive samples. The present results indicated that 53% of the identified HPVs belonged to the high-risk group. This finding is consistent with previous studies by Ngamkham et al., (2017); Malekpour Afshar et al., (2018) and Sigaroodi et al., (2012) indicating high-risk genotypes as the predominant type in breast tissue samples.
In conclusion, the present results indicated the presence of HPV genomes in 12.69% of breast tumor tissues of women in Ahvaz, Iran. Overall, 53% of specimens were infected with high-risk genotypes of HPV. The current findings also revealed that 87.5% of the detected viruses in the case group were HR-HPV, while 22.22% of positive samples were HR-HPV in the control group (P=0.03). Therefore, HPV may be an important contributor to the process of BC carcinogenesis; however, further research with a larger sample size is needed to better understand the carcinogenic role of HPV in BC.
Author Contribution Statement
Study concept and design: Gholam abbas Kaydani, Manoochehr Makvandi; analysis and interpretation of data: Manoochehr Makvandi, GA Kaydani, Seyed Nematollah Jazayeri, Javad Charostad, and Abdolhassan Talaiezadeh; drafting of the manuscript: Javad Charostad; and statistical analysis: Kambiz ahmadi Angali.
Acknowledgements
We would like to thank Dr. Mahdi Parsa for his contribution to data analysis.
Ethical Approval
The study protocol was approved by the Ethics Committee of Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran (Code: IR.AJUMS.REC.1397.239).
Informed Consent
Informed consent was obtained from all the participants included in this study.
Funding/Support
This study was supported by Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran (grant number: CRC-9706).
Conflict of Interest
The authors declare no conflicts of interest.
References
- Abreu ALP, Souza RP, Gimenes F, et al. A review of methods for detect human Papillomavirus infection. Virol J . 2012;9:262. doi: 10.1186/1743-422X-9-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahangar-Oskouee M, Shahmahmoodi S, Jalilvand S, et al. No detection of ‘high-risk’human papillomaviruses in a group of Iranian women with breast cancer. Asian Pac J Cancer Prev. 2014;15:4061–5. doi: 10.7314/apjcp.2014.15.9.4061. [DOI] [PubMed] [Google Scholar]
- Ahmed HG, Bensumaidea SH, Ashankyty IM. Frequency of Human Papilloma Virus (HPV) subtypes 31, 33, 35, 39 and 45 among Yemeni women with cervical cancer. Infect Agents Cancer. 2015;10:29. doi: 10.1186/s13027-015-0026-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari ME, Sayad S, Sayad S, et al. Breast cancer status in Iran: Statistical Analysis of 3010 Cases between 1998 and 2014. Int J Breast Cancer. 2017:2017. doi: 10.1155/2017/2481021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Moustafa A-E, Al-Antary N, Aboulkassim T, et al. Co-prevalence of Epstein–Barr virus and high-risk human papillomaviruses in Syrian women with breast cancer. Hum Vaccines Immunotherapeutics. 2016;12:1936–9. doi: 10.1080/21645515.2016.1139255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azubuike SO, Muirhead C, Hayes L, et al. Rising global burden of breast cancer: the case of sub-Saharan Africa (with emphasis on Nigeria) and implications for regional development: a review. World J Surg Oncol. 2018;16:63. doi: 10.1186/s12957-018-1345-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae J-M, Kim EH. Human papillomavirus infection and risk of breast cancer: a meta-analysis of case-control studies. Infect Agents Cancer. 2016;11:14. doi: 10.1186/s13027-016-0058-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal A, Singh MP, Rai B. Human papillomavirus-associated cancers: A growing global problem. Int J Appl Basic Med Res. 2016;6:84–9. doi: 10.4103/2229-516X.179027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braaten KP, Laufer MR. Human Papillomavirus (HPV), HPV-Related Disease, and the HPV Vaccine. Rev Obstet Gynecol. 2008;1:2–10. [PMC free article] [PubMed] [Google Scholar]
- Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev. 2003;16:1–17. doi: 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowski RT. Nutrition and physical activity influence on breast cancer incidence and outcome. Breast J. 2013;22:30–7. doi: 10.1016/j.breast.2013.07.006. [DOI] [PubMed] [Google Scholar]
- Damin AP, Karam R, Zettler CG, et al. Evidence for an association of human papillomavirus and breast carcinomas. Breast Cancer Res Treat. 2004;84:131–7. doi: 10.1023/B:BREA.0000018411.89667.0d. [DOI] [PubMed] [Google Scholar]
- de Cremoux P, Thioux M, Lebigot I, et al. No evidence of human papillomavirus DNA sequences in invasive breast carcinoma. Breast Cancer Res Treat. 2008;109:55–8. doi: 10.1007/s10549-007-9626-4. [DOI] [PubMed] [Google Scholar]
- Delgado-García S, Martínez-Escoriza J-C, Alba A, et al. Presence of human papillomavirus DNA in breast cancer: a Spanish case-control study. BMC Cancer. 2017;17:320. doi: 10.1186/s12885-017-3308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lonardo A, Venuti A, Marcante ML. Human papillomavirus in breast cancer. Breast Cancer Res Treat. 1992;21:95–100. doi: 10.1007/BF01836955. [DOI] [PubMed] [Google Scholar]
- Doosti M, Bakhshesh M, Zahir ST, et al. Lack of evidence for a relationship between high risk human papillomaviruses and breast cancer in Iranian patients. Asian Pac J Cancer Prev. 2016;17:4357–61. [PubMed] [Google Scholar]
- Eslamifar A, Ramezani A, Azadmanesh K, et al. Assessment of the association between human papillomavirus infection and breast carcinoma. Iran J Pathol. 2015;10:41–6. [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- Fernandes A, Bianchi G, Feltri AP, et al. Presence of human papillomavirus in breast cancer and its association with prognostic factors. Ecancermedicalscience. 2015:9. doi: 10.3332/ecancer.2015.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaffari H, Nafissi N, Hashemi-Bahremani M, et al. Molecular prevalence of human papillomavirus infection among Iranian women with breast cancer. Breast Dis. 2018;37:207–13. doi: 10.3233/BD-180333. [DOI] [PubMed] [Google Scholar]
- Gumus M, Yumuk P, Salepci T, et al. HPV DNA frequency and subset analysis in human breast cancer patients’ normal and tumoral tissue samples. J Exp Clin Cancer Res. 2006;25:515–21. [PubMed] [Google Scholar]
- Haghshenas MR, Mousavi T, Moosazadeh M, et al. Human papillomavirus and breast cancer in Iran: a meta- analysis. Iran J Basic Med Sci. 2016a;19:231–7. [PMC free article] [PubMed] [Google Scholar]
- Haghshenas MR, Mousavi T, Moosazadeh M, et al. Human papillomavirus and breast cancer in Iran: a meta- analysis. Iran J Basic Med Sci. 2016b;19:231–7. [PMC free article] [PubMed] [Google Scholar]
- Haghshenas MR, Mousavi T, Moosazadeh M, et al. Human papillomavirus and breast cancer in Iran: a meta-analysis. Iran J Basic Med Sci. 2016c;19:231. [PMC free article] [PubMed] [Google Scholar]
- Hedau S, Kumar U, Hussain S, et al. Breast cancer and human papillomavirus infection: no evidence of HPV etiology of breast cancer in Indian women. BMC Cancer. 2011;11:27. doi: 10.1186/1471-2407-11-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng B, Glenn W, Ye Y, et al. Human papilloma virus is associated with breast cancer. Br J Cancer. 2009;101:1345. doi: 10.1038/sj.bjc.6605282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan CY, Iacopetta BJ, Lawson JS, et al. Identification of human papillomavirus DNA gene sequences in human breast cancer. Br J Cancer. 2005;93:946–8. doi: 10.1038/sj.bjc.6602778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong A, Leung CP, Shin VY, et al. No evidence of human papillomavirus in patients with breast cancer in Hong Kong, Southern China. ISRN Virology. 2013:2013. [Google Scholar]
- Lawson JS, Heng B. Viruses and breast cancer. Cancers. 2010a;2:752–72. doi: 10.3390/cancers2020752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson JS, Heng B. Viruses and breast cancer. Cancers. 2010b;2:752–72. doi: 10.3390/cancers2020752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson JS, Heng B. Viruses and breast cancer. Cancers. 2010c;2:752–72. doi: 10.3390/cancers2020752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Bi X, Zhang Y, et al. Human papillomavirus infection and sporadic breast carcinoma risk: a meta-analysis. Breast Cancer Res Treat. 2011;126:515–20. doi: 10.1007/s10549-010-1128-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudvand S, Safaei A, Erfani N, et al. Presence of human papillomavirus DNA in colorectal cancer tissues in Shiraz, Southwest Iran. Asian Pac J Cancer Prev. 2015;16:7883–7. doi: 10.7314/apjcp.2015.16.17.7883. [DOI] [PubMed] [Google Scholar]
- Malekpour Afshar R, Balar N, Mollaei HR, et al. Low Prevalence of Human Papilloma Virus in Patients with Breast Cancer, Kerman; Iran. Asian Pac J Cancer Prev. 2018;19:3039–44. doi: 10.31557/APJCP.2018.19.11.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A-M, Weber BL. Genetic and hormonal risk factors in breast cancer. J Nat Cancer Instit. 2000;92:1126–35. doi: 10.1093/jnci/92.14.1126. [DOI] [PubMed] [Google Scholar]
- Mou X, Chen L, Liu F, et al. Low prevalence of human papillomavirus (HPV) in Chinese patients with breast cancer. J Int Med Res. 2011;39:1636–44. doi: 10.1177/147323001103900506. [DOI] [PubMed] [Google Scholar]
- Mousavi SM, Montazeri A, Mohagheghi MA, et al. Breast cancer in Iran: an epidemiological review. Breast J. 2007;13:383–91. doi: 10.1111/j.1524-4741.2007.00446.x. [DOI] [PubMed] [Google Scholar]
- Ngamkham J, Karalak A, Chaiwerawattana A, et al. Prevalence of human papillomavirus infection in breast cancer cells from Thai women. Asian Pac J Cancer Prev. 2017;18:1839. doi: 10.22034/APJCP.2017.18.7.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salman NA, Davies G, Majidy F, et al. Association of high risk human papillomavirus and breast cancer: A UK based Study. Sci Rep. 2017;7:43591. doi: 10.1038/srep43591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvari J, Mahmoudvand S, Pirbonyeh N, et al. The very low frequency of Epstein-Barr JC and BK viruses DNA in colorectal cancer tissues in Shiraz, Southwest Iran. Polish J Microbiol. 2018;67:73–9. doi: 10.5604/01.3001.0011.6146. [DOI] [PubMed] [Google Scholar]
- Sigaroodi A, Nadji SA, Naghshvar F, et al. Human papillomavirus is associated with breast cancer in the north part of Iran. Sci World J. 2012:2012. doi: 10.1100/2012/837191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões PW, Medeiros LR, Pires PDS, et al. Prevalence of human papillomavirus in breast cancer: a systematic review. Int J Gynecol Cancer. 2012;22:343–7. doi: 10.1097/IGC.0b013e31823c712e. [DOI] [PubMed] [Google Scholar]
- Stanley MA. Epithelial cell responses to infection with human papillomavirus. Clin Microbiol Rev. 2012;25:215–22. doi: 10.1128/CMR.05028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JH, Tsai CH, Cheng MH, et al. Association of viral factors with non-familial breast cancer in Taiwan by comparison with non-cancerous, fibroadenoma, and thyroid tumor tissues. J Med Virol. 2005;75:276–81. doi: 10.1002/jmv.20267. [DOI] [PubMed] [Google Scholar]
- Tulay P, Serakinci N. The role of human papillomaviruses in cancer progression. J Cancer Metastasis Treat. 2016;2 [Google Scholar]
- Vernet-Tomas M, Mena M, Alemany L, et al. Human papillomavirus and breast cancer: no evidence of association in a Spanish set of cases. Anticancer Res. 2015;35:851–6. [PubMed] [Google Scholar]
- Yim E-K, Park J-S. The role of HPV E6 and E7 oncoproteins in HPV-associated cervical carcinogenesis. Cancer Res Treat. 2005;37:319–24. doi: 10.4143/crt.2005.37.6.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandberg DP, Bhargava R, Badin S, et al. The role of human papillomavirus in nongenital cancers. CA Cancer J Clin. 2013;63:57–81. doi: 10.3322/caac.21167. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Li J, Ji Y, et al. Inconclusive role of human papillomavirus infection in breast cancer. Infect Agent Cancer. 2015;10:36. doi: 10.1186/s13027-015-0029-6. [DOI] [PMC free article] [PubMed] [Google Scholar]