Abstract
Spinocerebellar ataxia type 14 (SCA14) is a neurodegenerative disease caused by germline variants in the diacylglycerol (DAG)/Ca2+-regulated protein kinase C gamma (PKCγ), leading to Purkinje cell degeneration and progressive cerebellar dysfunction. Most of the identified mutations cluster in the DAG-sensing C1 domains. Here, we found with a FRET-based activity reporter that SCA14-associated PKCγ mutations, including a previously undescribed variant, D115Y, enhanced the basal activity of the kinase by compromising its autoinhibition. Unlike other mutations in PKC that impair its autoinhibition but lead to its degradation, the C1 domain mutations protected PKCγ from such down-regulation. This enhanced basal signaling rewired the brain phosphoproteome, as revealed by phosphoproteomic analysis of cerebella from mice expressing a human SCA14-associated H101Y mutant PKCγ transgene. Mutations that induced a high basal activity in vitro were associated with earlier average age of onset in patients. Furthermore, the extent of disrupted autoinhibition, but not agonist-stimulated activity, correlated with disease severity. Molecular modeling indicated that almost all SCA14 variants not within the C1 domain were located at interfaces with the C1B domain, suggesting that mutations in, and proximal to, the C1B domain are a susceptibility for SCA14 because they uniquely enhance PKCγ basal activity while protecting the enzyme from downregulation. These results provide insight into how PKCγ activation is modulated and how deregulation of the cerebellar phosphoproteome by SCA14-associated mutations affects disease progression.
Introduction
Conventional protein kinase C (PKC) isozymes play key roles in normal brain physiology, where they regulate neuronal functions such as synapse morphology, receptor turnover, and cytoskeletal integrity (1). These isozymes are transiently and reversibly activated by Ca2+ and diacylglycerol (DG), the two second messenger products of receptor-mediated hydrolysis of phosphatidylinositol-4,5-bisphosphate (PIP2) (2). Tight control of not only activity, but also steady-state protein levels, is necessary for cellular homeostasis, with deregulation of either resulting in pathophysiology. For conventional PKC isozymes, loss-of-function somatic mutations, or reduced protein levels, are associated with cancer (3); in contrast, gain-of-function variants have been identified in neurodegenerative diseases (4–6). Thus, whereas reduced protein levels and activity of conventional PKC isozymes are associated with poorer patient survival in cancers such as colon and pancreatic cancer, enhanced activity of the conventional PKCα is associated with Alzheimer’s disease (4, 6–8).
Spinocerebellar ataxias (SCAs) are a group of over 40 autosomal dominant neurodegenerative diseases characterized by Purkinje cell degeneration and cerebellar dysfunction, resulting in progressive ataxia and loss of motor coordination and control (9). Each subtype of SCA is caused by germline variants in a distinct gene. A majority of these genes encode proteins that regulate Ca2+ homeostasis, including the IP3 receptor, IP3R1 (SCA 15, 16 and 29), ataxins 2 and 3, which regulate IP3R1 function (SCA2 and 3, respectively) (10, 11), the cation channel TRPC3 (SCA41) (12), and mGluR1 which couples to phospholipase C (SCA44) (13). Spinocerebellar ataxia type 14 (SCA14) is caused by missense variants in PKCγ (14), a conventional PKC isozyme whose expression is restricted to neurons, particularly Purkinje cells (15–17). Given that Ca2+ is an important activator of PKC, one intriguing theory is that enhanced PKCγ activity is not only central to SCA14 pathology but is also at the epicenter of many other types of SCA. Thus, understanding how SCA14-associated variants deregulate the function of PKCγ has strong potential clinical relevance.
Exquisite regulation of the spatiotemporal dynamics of conventional PKC signaling ensures that these enzymes are only activated for a specific time, at defined locations, and in response to appropriate stimuli. In the absence of specific stimuli, these enzymes are maintained in an autoinhibited conformation by an N-terminal regulatory moiety that constrains the catalytic activity of the C-terminal kinase domain (18). Specifically, an autoinhibitory pseudosubstrate segment occupies the substrate-binding cavity to maintain the enzyme in an inactive conformation. Additionally, multiple interactions of the kinase domain with modules in the regulatory moiety secure the pseudosubstrate in place to prevent aberrant basal signaling. These modules are the DG-sensing C1A and C1B domains and Ca2+-sensing C2 domain, which pack against the kinase domain to maintain it in an autoinhibited conformation until the relevant second messengers are generated (19). Release of the pseudosubstrate occurs upon generation of the appropriate second messengers. Specifically, following phospholipase C-catalyzed hydrolysis of PIP2, Ca2+ binds to the C2 domain causing it to translocate to the plasma membrane where it is anchored by interaction of a basic surface with PIP2 (20). At the membrane, the C1B domain engages its membrane-embedded allosteric activator, DG, resulting in release of the pseudosubstrate from the active site, allowing PKC to phosphorylate its substrates (21). This process is readily reversible upon decay of the second messengers, and thus normal PKC activity is transient. Before PKC can adopt an autoinhibited but signaling-competent conformation, newly synthesized enzyme must be processed by a series of ordered phosphorylations in the kinase domain. In particular, phosphorylation at a residue termed the hydrophobic motif is required for PKC to adopt the autoinhibited conformation (22). Aberrant PKC that is not properly autoinhibited is dephosphorylated by the phosphatase PHLPP, ubiquitinated, and degraded by a proteasomal pathway (22). This quality control mechanism ensures that only properly autoinhibited PKC accumulates in the cell. For example, cancer-associated variants that prevent autoinhibition of PKC are paradoxically loss-of-function because the mutant protein is degraded by this quality control pathway (22). Thus, autoinhibited PKC is stable and prolonged activation renders PKC sensitive to dephosphorylation and degradation. In this regard, phorbol esters which bind PKC with high affinity and are not readily metabolized cause the acute activation but long-term downregulation of PKC.
Since the original discovery of germline variants in PKCγ by Raskind and colleagues that defined SCA14 (14, 23, 24), approximately 50 variants across all domains of PKCγ have now been identified in SCA14 (25–28). Mouse model studies by Kapfhammer and colleagues have established that a single SCA14-associated point mutation in PKCγ is sufficient to drive pathophysiology characteristic of the human disease, including Purkinje cell degeneration and motor deficits (29, 30). Cellular studies by several groups have addressed the mechanism by which cerebellar degeneration in SCA14 may precipitate. Schrenk et al. have shown that stimulation of PKC in mouse cerebellar slices by treatment with phorbol esters leads to a decrease in Purkinje cell dendrites, whereas inhibition of PKC leads to hyper-arborization, suggesting a causative role for enhanced PKC activity in Purkinje cell degeneration (31). Others have observed that PKC inhibition prevents Purkinje cell death (32). Verbeek and colleagues have also identified a role for altered PKCγ activity in SCA14, showing in some cases that SCA14-associated PKCγ mutations lead to unmasking of the C1 domains to enhance ‘openness’ and thus membrane accessibility of PKCγ, but concluded that these mutants have lower kinase activity (33, 34). A sizable body of work has also focused on the role of PKCγ aggregation in SCA14. Notably, Saito and colleagues have shown that in both overexpression and in vitro systems, wild-type and mutant PKCγ form amyloid-like fibrils and aggregates that lead to cell death, which can be decreased by pharmacological induction of heat shock proteins (35, 36). Other studies have also demonstrated the presence of such aggregates in iPSCs from SCA14 patients or primary culture mouse Purkinje cells (26, 37). However, the precise biochemical mechanisms in which SCA14 mutations alter PKCγ function to ultimately drive neurodegeneration in SCA14 is still unknown.
Here, we addressed the mechanism by which SCA14 mutations affect PKCγ function using our previously developed, genetically encoded biosensor for PKC activity coupled with biochemical, molecular modeling, and bioinformatics approaches. Our studies revealed that SCA14-associated mutations in every segment or domain of PKCγ (pseudosubstrate, C1A, C1B, C2, kinase) produced the same defect: impaired autoinhibition leading to increased basal activity. Furthermore, we found that SCA14-associated PKCγ mutations in the C1A and C1B domains, mutational hotspots for the disease, rendered PKCγ insensitive to phorbol ester-mediated downregulation, an effect also observed upon deletion of either domain. Specifically, mutating (or deleting) the C1A domain prevented dephosphorylation, the first step in downregulation, and mutating (or deleting) the C1B domain permitted dephosphorylation but prevented the next step, protein degradation. Thus, C1A and C1B domain mutations provide unique mechanisms to deregulate PKC without subjecting it to degradation. Focusing on one mutation in the C1A domain, ΔF48, we found that deletion of this single residue (or the entire C1A domain) not only reduced autoinhibition resulting in high basal activity but also uncoupled the communication between the pseudosubstrate and the kinase domain, thus trapping this PKC variant in an unresponsive but slightly “open” state. Structural analyses revealed that most SCA14 mutations are either in the C1 domains or at common interfaces with the C1 domains. Furthermore, bioinformatics analyses revealed that mutations in the C1 domains are relatively under-represented in cancer, a disease where conventional PKC function is generally lost. This is consistent with our findings that mutations in these domains will enhance, not suppress, PKC activity. Validating altered signaling in a physiological context, phosphoproteomic analysis of cerebella from mice expressing a human bacterial artificial chromosome (BAC) WT or H101Y PKCγ transgene revealed significant alteration in the phosphorylation of components related to cytoskeletal organization and neuronal development. Lastly, compilation of the age of SCA14 onset for patients with C1 domain mutations revealed that the magnitude of the biochemical defect (reduced autoinhibition) inversely correlated with age of SCA14 onset. Together, our results suggest that sustained “leaky” activity of PKCγ, by mechanisms that protect it from degradation, alters the cerebellar phosphoproteome to drive SCA14 pathology.
Results
Previously undescribed PKCγ D115Y is a pathogenic variant for SCA14
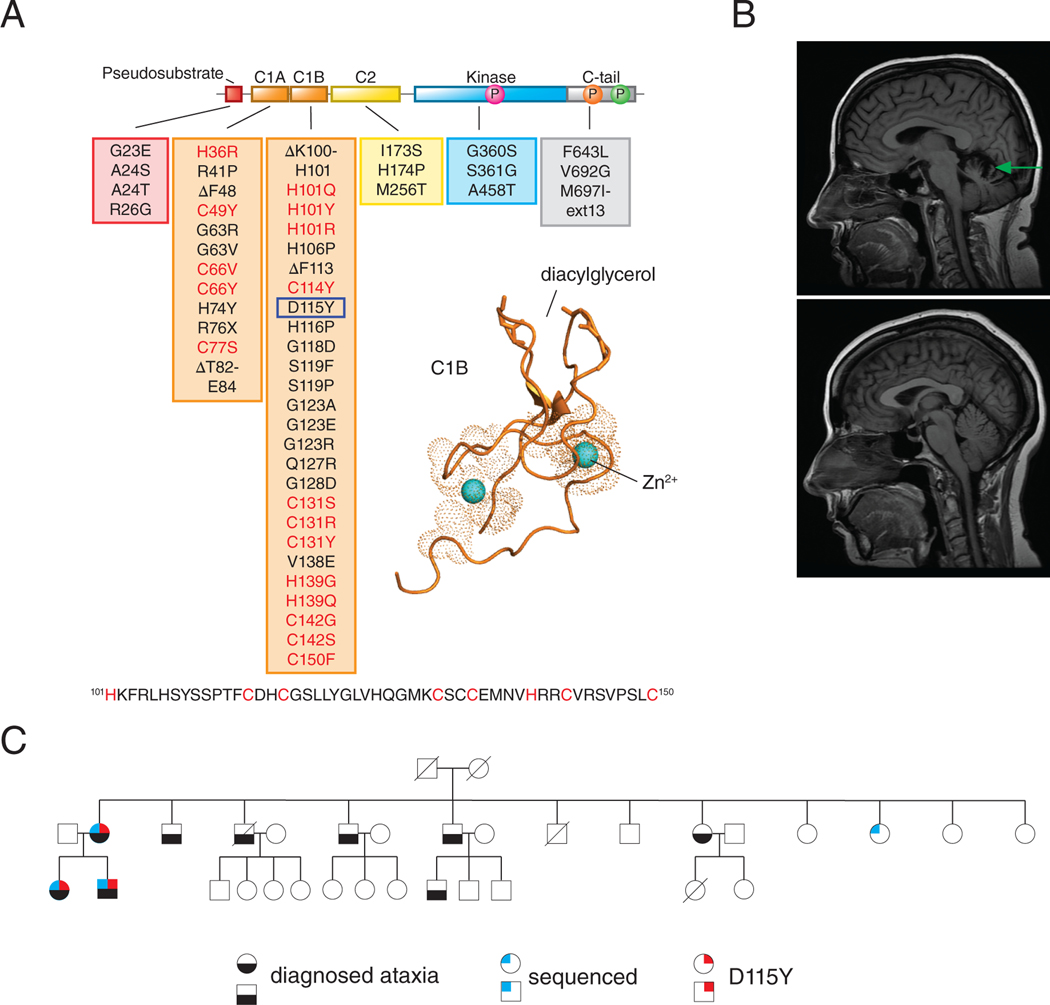
SCA14 is caused by germline variants in PKCγ, of which over 50 unique variants have been identified (Fig. 1A) (25–27). Although these variants occur in every domain of the kinase, the majority cluster to the C1 domains, particularly the C1B domain. This small globular DG-binding domain coordinates two Zn2+ ions through invariant histidine and cysteine residues (Fig. 1A, residues of motif in red). Mutation of any of the Zn2+-coordinating residues abolishes or severely impairs phorbol ester binding (38). The SCA14 C1B variants occur with the highest frequency at residues within the zinc finger motif, suggesting that these mutants may affect ligand binding to C1B, and thus, proper regulation of kinase activity. Here, we also report on a previously undescribed variant, D115Y, identified by whole-genome sequencing of a patient who was diagnosed with ataxia. Magnetic resonance imaging (MRI) on the patient harboring the novel D115Y variant revealed significant cerebellar degeneration when compared with a healthy, age-matched individual (Fig. 1B), a hallmark of SCA. This patient’s mother came from a large family with 6 out of 12 siblings diagnosed with ataxia (Fig. 1C; bottom, black fill), consistent with the autosomal dominant nature of the disease. Of the subset of this patient’s family who underwent whole-genome sequencing (Fig. 1C; top left, blue fill), three individuals diagnosed with ataxia harbored the D115Y variant (Fig. 1C; top right, red fill), whereas the one healthy individual that was sequenced did not harbor this variant (Fig. 1C; no fill), indicating segregation of the variant with the disease.
Figure 1. PKCγ in Spinocerebellar Ataxia Type 14.
(A) Primary structure of PKCγ with all known SCA14 variants indicated in boxes beneath each domain (24–27). Newly identified patient variant D115Y is indicated by the blue box. Previously published crystal structure (61) of PKCβII C1B domain is shown with Zn2+ (cyan spheres) and diacylglycerol binding sites labeled (PDB: 3PFQ). Conserved His and Cys residues of Zn2+ finger motif are shown in red in PKCγ primary sequence. (B) MRI of patient at age 46 with D115Y variant (top) compared to age-matched healthy control (bottom); green arrow indicates cerebellar atrophy. (C) Pedigree of family with PKCγ D115Y variant; black shape-fill indicates family members diagnosed with ataxia, blue shape-fill indicate family members that have been sequenced, red shape-fill indicates family members with D115Y variant.
SCA14-associated PKCγ mutants display decreased autoinhibition
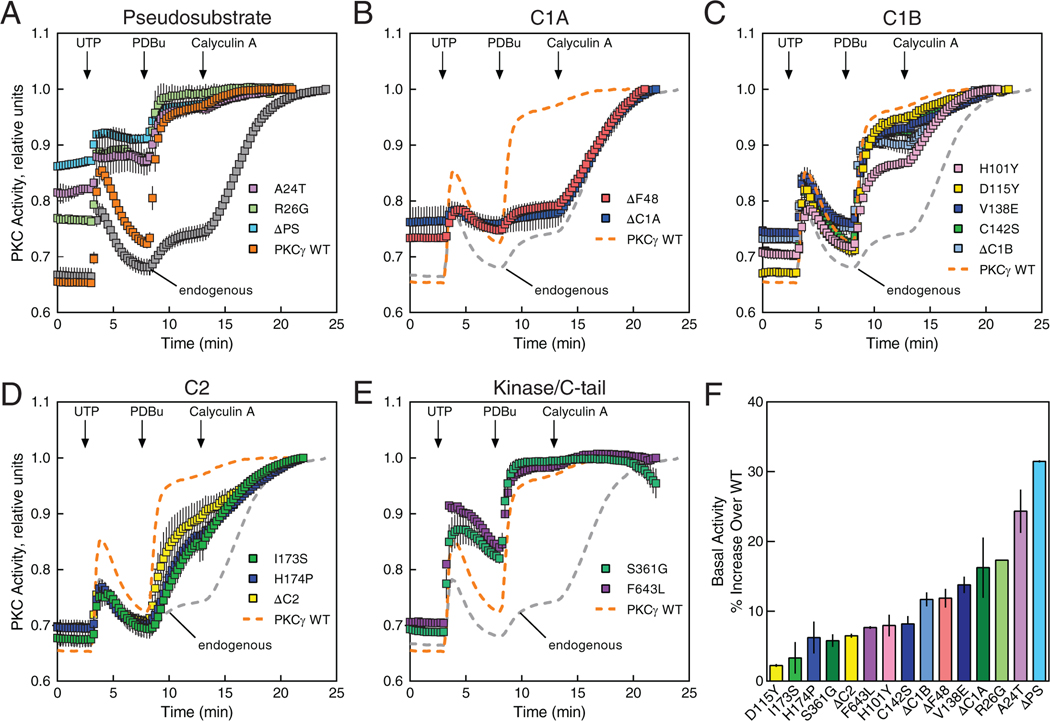
To assess how SCA14 mutations affect PKCγ function, we first addressed their effect on the basal and agonist-evoked activity of PKCγ in cells using the genetically-encoded FRET-based biosensor, C Kinase Activity Reporter 2 (CKAR2) (39). Mutations in each domain were selected for analysis, including the new D115Y mutation in the C1B domain. Additionally, constructs lacking the pseudosubstrate segment (ΔPS) or regulatory domain (ΔC1A, ΔC1B, or ΔC2) were analyzed. COS7 cells co-expressing mCherry-tagged PKCγ constructs and the reporter were sequentially treated with 1] uridine-5′-triphosphate (UTP), which activates purinergic receptors to elevate diacylglycerol (DG) and Ca2+, to transiently activate PKC and observe differences in activation and re-autoinhibition after stimulus, 2] phorbol 12,13-dibutyrate (PDBu) to maximally activate PKC, and 3] the phosphatase inhibitor Calyculin A to assess maximal phosphorylation of the reporter; traces were normalized to this endpoint. UTP stimulation of cells caused a transient activation of endogenous (grey) and overexpressed wild-type (WT) PKCγ (orange) that was reversed as the enzyme regained the autoinhibited conformation following second messenger decay, as previously reported (40) (Fig. 2A). Treatment with phorbol ester stimulated nearly maximal phosphorylation of the reporter in cells overexpressing WT PKCγ; endogenous PKC required phosphatase suppression with calyculin A to observe maximal reporter phosphorylation (Fig. 2A). These kinetics are characteristic of properly autoinhibited PKC (22). In contrast, the two SCA14 pseudosubstrate mutants (A24T and R26G) had high basal activity resulting in only modest additional activation by UTP and phorbol esters, approaching the level of deregulated autoinhibition observed upon deletion of the entire pseudosubstrate segment (ΔPS) (Fig. 2A). The C1A SCA14 mutation ΔF48, in which a single residue is deleted (no frameshift), also had high basal activity but was relatively unresponsive to stimulation with UTP or PDBu (Fig. 2B). This signature of high basal activity and lack of response to agonists was also observed upon deletion of the entire C1A domain (ΔC1A). Mutations in the C1B domain, including the new D115Y, all caused an increase in basal activity but, in contrast to the C1A mutations, did not uncouple responsiveness to UTP and PDBu, similar to the effect observed with C1B domain deletion (ΔC1B) (Fig. 2C). Mutations in the C2 domain, as well as deletion of the entire C2 domain, resulted in slightly enhanced basal activity but reduced response to agonist (Fig. 2D). Lastly, mutations in the kinase domain (S361G) and C-tail (F643L) resulted in both an increase in basal activity and an increase in agonist-evoked activity compared with WT PKCγ (Fig. 2E). Note that experiments using the previously characterized CKAR1 (41), under similar experimental conditions, produced the same qualitative results as CKAR2, although CKAR2 displayed a larger dynamic range (fig. S1). Every mutant tested exhibited higher basal activity compared to WT, but with varying degrees of deregulation, as revealed by quantitation of the initial FRET ratio of each trace, normalized to that of WT enzyme (Fig. 2F). The higher basal activity observed in these assays was not due to higher expression of the ataxia mutants in cells, because quantitation of mCherry fluorescence revealed similar protein levels of WT and ataxia mutants (fig. S2). Thus, SCA14 mutations in every domain of PKCγ consistently display impaired autoinhibition.
Figure 2. SCA14 mutants exhibit impaired autoinhibition compared to WT PKCγ.
(A to E) COS7 cells were transfected with CKAR2 alone (endogenous; gray) or co-transfected with CKAR2 and the indicated mCherry-tagged PKCγ construct: WT PKCγ (orange), a PKCγ lacking a pseudosubstrate (ΔPS), or the indicated pseudosubstrate SCA14 mutants in (A); the ΔC1A or ΔF48 (C1A domain) in (B); the ΔC1B or C1B SCA14 mutants in (C); the ΔC2 or C2 SCA14 mutants in (D); or the kinase-domain and C-tail SCA14 mutants in (E). PKC activity was monitored by measuring FRET/CFP ratio changes after sequential addition of 100 μM UTP, 200 nM PDBu, and 50 nM Calyculin A at the indicated times. Data were normalized to the endpoint (1.0) and show mean ± S.E.M. from at least two independent experiments, N ≥ 17 (A), ≥ 16 (B), ≥ 19 (C), ≥ 11 (D), and ≥ 33 (E) cells per condition. In (B) through (E), the PKCγ WT and endogenous data (dashed lines) are reproduced from (A) for comparison. (F) Quantification of percent increase in basal activity in (A to E) over WT PKCγ.
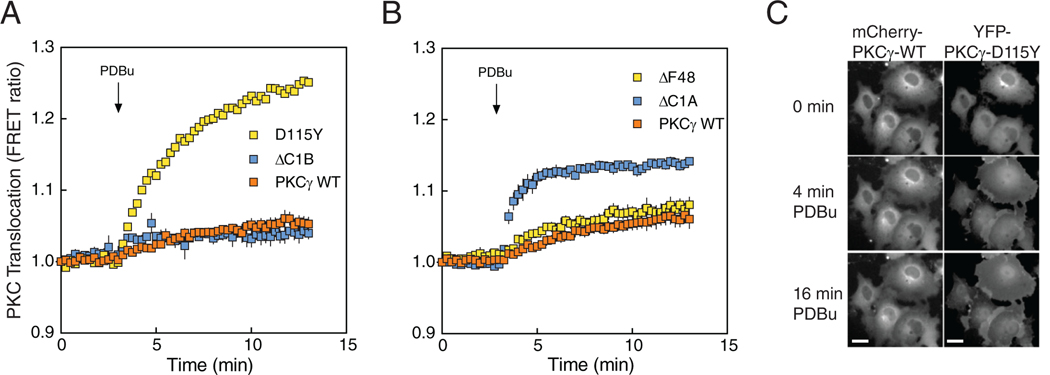
PKC with impaired autoinhibition is in a more “open” conformation with its membrane-targeting modules unmasked, resulting in enhanced membrane affinity and faster kinetics of agonist-dependent membrane translocation (21). To further characterize how SCA14-associated mutations in the C1 domains affect the “openness” of PKCγ, we examined the translocation of the SCA14 mutants D115Y and ΔF48 compared to WT using a FRET-based translocation assay. Plasma membrane-targeted CFP (MyrPalm-CFP) was co-transfected with YFP-tagged WT, D115Y, ΔC1B, ΔF48, or ΔC1A PKCγ in COS7 cells and the increase in FRET following stimulation of cells with PDBu, a measure of membrane association, was determined (Fig. 3). In response to PDBu, the D115Y mutant associated much more robustly with plasma membrane compared to WT, consistent with unmasking of membrane-targeting modules (Fig. 3A). Furthermore, deletion of the C1B domain (ΔC1B) prevented translocation above WT levels, suggesting that the C1B domain is the predominant binder of plasma membrane-embedded PDBu. On the other hand, deletion of the C1A domain (ΔC1A) enhanced plasma membrane binding, suggesting that the loss of the C1A unmasked the C1B domain to facilitate PDBu binding. ΔF48 translocated with comparable kinetics and magnitude as WT, which could be accounted for by proper masking of its C1B domain (with normal accessibility to ligand) (Fig. 3B). To further assess enhanced membrane association of the D115Y mutant, mCherry-tagged PKCγ WT and YFP-tagged PKCγ D115Y were co-expressed in COS7 cells, and phorbol ester-stimulated translocation was monitored within the same cells (Fig. 3C). Both WT and D115Y displayed diffuse localization in the cytosol before PDBu treatment. Whereas there was little detectable difference in translocation of the WT PKCγ 4 min following addition of PDBu, D115Y PKCγ displayed enhanced plasma membrane association, which was sustained at 16 minutes post-PDBu addition. Thus, these results are consistent with the D115Y being in a more basally “open” conformation resulting in enhanced association with plasma membrane following phorbol ester treatment.
Figure 3. SCA14 mutations affect translocation of PKCγ.
(A) COS7 cells were co-transfected with MyrPalm-CFP and YFP-tagged WT PKCγ (orange), PKCγ D115Y (yellow), or PKCγ ΔC1B (blue). Rate of translocation to plasma membrane was monitored by measuring FRET/CFP ratio changes after addition of 200 nM PDBu. Data were normalized to the starting point (1.0) and are representative of two independent experiments, N ≥ 22 cells per condition. (B) COS7 cells were co-transfected with MyrPalm-CFP and YFP-tagged ΔF48 or ΔC1A. Data are mean ± S.E.M. from at least three independent experiments, N ≥ 23 cells per condition. (C) COS7 cells were co-transfected with mCherry-tagged WT PKCγ and YFP-tagged PKCγ D115Y. Localization of mCherry-PKCγ (WT; left) and YFP-PKCγ-D115Y (right) in the same cells under basal conditions and after addition of 200 nM PDBu was observed by fluorescence microscopy. Images are representative of three independent experiments. Scale bar = 20 μm.
SCA14 mutants evade phorbol ester-mediated degradation, yet display higher turnover
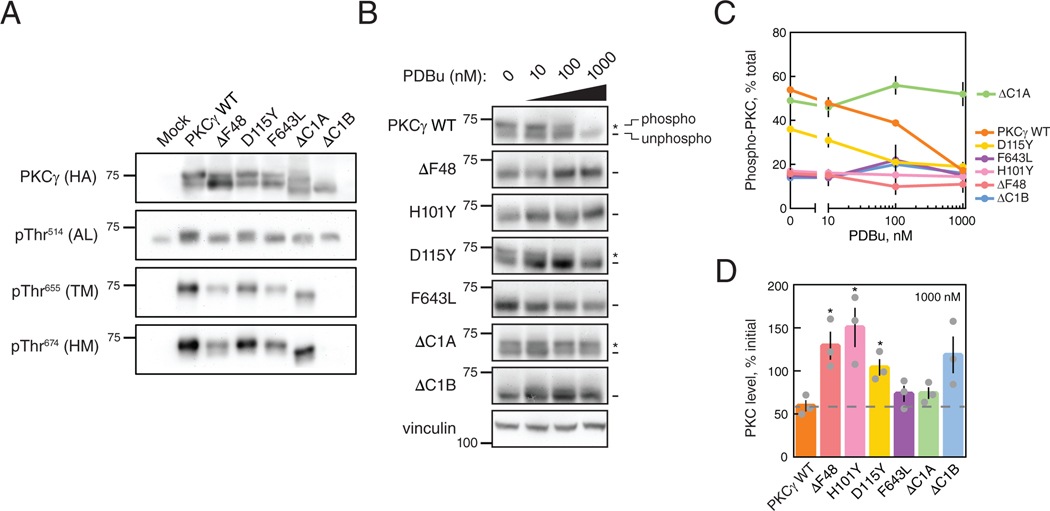
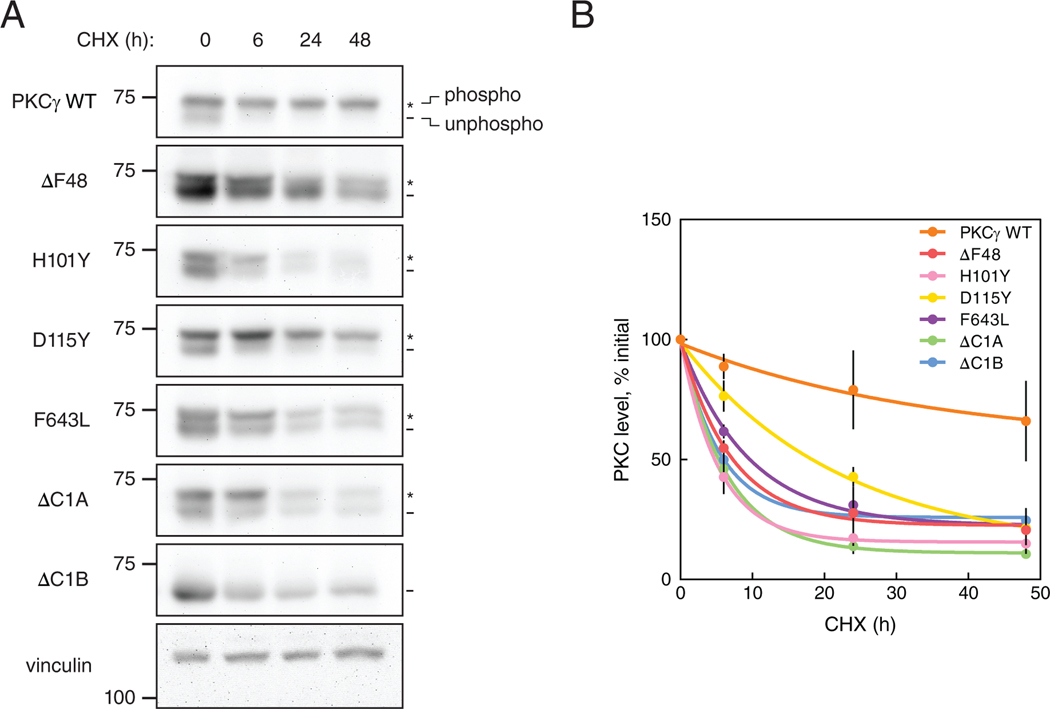
Because reduced autoinhibition of PKC renders the constitutive phosphorylation sites within the kinase domain and C-tail highly phosphatase labile, we examined the phosphorylation state of the basally active SCA14 mutants in the Triton-soluble lysate fraction. Phosphorylation of HA-tagged PKCγ WT, the indicated SCA14 mutants, ΔC1A, or ΔC1B overexpressed in COS7 cells was assessed by monitoring the previously-characterized phosphorylation-induced mobility shift that accompanies phosphorylation of the two C-terminal sites (42) or using phospho-specific antibodies to the activation loop (pThr514), the turn motif (pThr655), and the hydrophobic motif (pThr674) by Western blot (Fig. 4A). WT PKCγ migrated predominantly as a slower mobility species (phosphorylated); this slower mobility species was detected with each of the phospho-specific antibodies. In contrast, the ΔC1B migrated as a single species and was not phosphorylated at any of the processing sites (note that for the activation loop (pThr514) blot, the band present represents endogenous PKC). Each SCA14 mutant had reduced phosphorylation compared to WT as assessed by the ratio of upper (phosphorylated) to lower (unphosphorylated) bands, with D115Y having the smallest defect and the ΔF48 having the largest defect. The accumulation of dephosphorylated mutant PKC is consistent with increased PHLPP-mediated dephosphorylation of defectively autoinhibited PKC at the hydrophobic motif (22).
Figure 4. SCA14 mutants are resistant to phorbol ester-mediated downregulation.
(A) Western blot of Triton-soluble lysates from COS7 transfected with HA-tagged WT PKCγ, PKCγ lacking a C1A domain (ΔC1A), PKCγ lacking a C1B domain (ΔC1B), the indicated SCA14 mutants, or with empty vector (Mock). Membranes were probed with anti-HA (PKCγ) or phospho- specific antibodies. N = three independent experiments. (B) Western blot of whole-cell lysates from COS7 cells transfected with HA-tagged WT PKCγ, PKCγ lacking a C1B domain (ΔC1B), PKCγ lacking a C1A domain (ΔC1A), or the indicated SCA14 mutants. COS7 cells were treated with the indicated concentrations of PDBu for 24 hours prior to lysis. Endogenous expression of vinculin was also probed as a loading control. N = three independent experiments. *, phosphorylated species; -, unphosphorylated species. (C) Quantification of percent phosphorylation of total PKC as a function of PDBu concentration. Data are mean + S.E.M. (D) Quantification of total levels of PKC with 1000 nM PDBu shown as a percentage of initial levels of PKC (0 nM) and represents mean ± S.E.M. WT levels after 24 hours with 1000 nM PDBu indicated (grey dashed line). *P < 0.05 by Welch’s t-test.
Given the increase in dephosphorylated species of SCA14 mutants, we next addressed whether these mutants were more susceptible to downregulation (loss of total protein) than WT PKCγ. COS7 cells overexpressing HA-tagged PKCγ WT, the indicated SCA14 mutants, ΔC1A, or ΔC1B were treated with increasing concentrations of PDBu for 24 hours (Fig. 4B) and PKC levels were probed by Western blot analysis of whole-cell lysates. Dephosphorylation of WT PKCγ was observed at the lowest concentration of PDBu (10 nM) as assessed by the increase in the ratio of unphosphorylated PKC (faster mobility species) over phosphorylated species (slower mobility species) (Fig. 4C), and this dephosphorylated species was degraded at the highest concentration of PDBu (1000 nM). Surprisingly, every C1 domain SCA14 mutant tested (ΔF48, H101Y, D115Y) was significantly more resistant to PDBu-mediated downregulation than WT PKCγ (Fig. 4D). The catalytic domain mutant F643L was also moderately less sensitive to PDBu downregulation than WT enzyme. Furthermore, ΔC1B, ΔF48, and H101Y levels increased with increasing concentrations of PDBu compared to levels in untreated cells. Although the C1B mutant D115Y was effectively dephosphorylated (Fig. 4, B and C), the dephosphorylated species was resistant to degradation (Fig. 4, B and D). In contrast, deletion of the C1A prevented dephosphorylation of the upper mobility, phosphorylated species (Fig. 4, B and C), but allowed degradation of the faster mobility, dephosphorylated species (Fig. 4, B and D). This demonstrates an uncoupling within the degradative pathway of PKC, such that a PKC that lacks a C1A domain is less susceptible to dephosphorylation, whereas a PKC without a functional C1B domain loses the ability to be degraded in a phorbol ester-dependent manner. These results indicate that C1 domain mutants render PKC resistant to phorbol ester-mediated downregulation by impairing dephosphorylation (as observed upon deletion of C1A) or impairing degradation (as observed upon deletion of C1B). The kinase domain mutant F643L mirrored C1B domain mutations in resistance to phorbol ester-mediated degradation.
We next addressed whether SCA14-associated mutations altered the steady-state turnover of PKC in unstimulated cells. COS7 cells overexpressing HA-tagged PKCγ WT, the indicated SCA14 mutants, ΔC1A, or ΔC1B were treated with cycloheximide to prevent protein synthesis for increasing time and lysates were analyzed for PKC levels (Fig. 5A). PKCγ WT was remarkably stable, with a half-life of over 48 hours, as previously reported for other conventional PKC isozymes (22). In marked contrast, the ataxia mutants were considerably less stable, with half-lives of approximately 10 hours for mutations that had strong effects on autoinhibition (ΔF48, H101Y, F643L) and 20 hours for the D115Y mutation, which had a modest effect on autoinhibition (Fig. 5B). Deletion of the C1A or C1B domains (ΔC1A, ΔC1B) also had a strong effect on stability, consistent with decreased autoinhibition due to the loss of a regulatory domain. Thus, whereas SCA14 mutations render activated PKC resistant to phorbol ester-induced downregulation, they increase the steady-state turnover of unstimulated PKC.
Figure 5. SCA14 mutants are more rapidly turned over in the presence of cycloheximide.
(A and B) Western blot analysis of lysates from COS7 cells transfected with HA-tagged WT PKCγ, PKCγ lacking the C1B domain (ΔC1B), PKCγ lacking the C1A domain (ΔC1A), or the indicated SCA14 mutants. COS7 cells were treated with CHX (355 μM) for 0, 6, 24, or 48 hours prior to lysis. Membranes were probed for HA (PKCγ) as well as endogenous expression of vinculin as a loading control. *, phosphorylated species; -, unphosphorylated species. Blot is representative (A) with quantification of total levels of PKCγ at each time point from three independent experiments shown in (B) as a percentage of initial level of PKC (0 hours) and represents mean ± S.E.M. Points were curve fit by non-linear regression.
PKCγ C1A residue Phe48 is critical for proper autoinhibition and activation
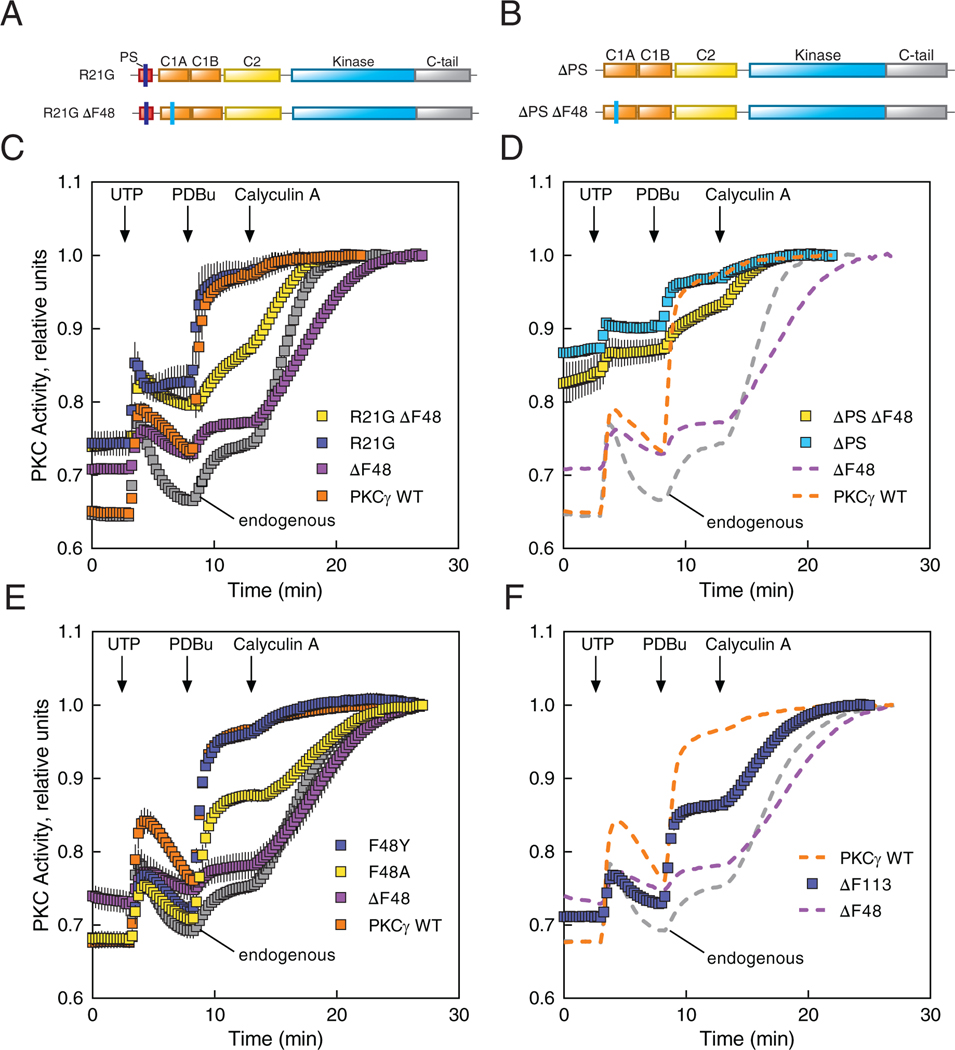
The characterized SCA14 mutants displayed an increase in basal activity, and all but one retained the ability to have this increased basal activity further enhanced in response to agonist stimulation (Fig. 2, A to E). To gain insight into this uncoupling from agonist stimulated activity, we further characterized the deletion mutation in the C1A domain (deletion of Phe48: ΔF48) whose activity was unresponsive to stimulation by UTP or PDBu, an uncoupling also observed upon deletion of the entire C1A domain (Fig. 2B). We first asked whether reducing the affinity of the pseudosubstrate for the active site pocket (Fig. 6A) or deleting the pseudosubstrate (Fig. 6B) would promote agonist-responsiveness of ΔF48. Mutation of arginine at the P-3 position to a glycine in WT (R21G) or ΔF48 (R21G ΔF48) PKCγ enhanced basal activity for both WT PKCγ and ΔF48 (Fig. 6C). However, UTP and PDBu caused additional activation of only the WT PKCγ with the pseudosubstrate mutation. Whereas the pseudosubstrate mutation caused an even greater increase in basal activity of the ΔF48 mutant, this still did not permit activation by UTP and PDBu (note that the small responses seen are those of the endogenous PKC). Similarly, deletion of the entire pseudosubstrate elevated basal activity even more for both WT and ΔF48, but further activation by PDBu was only observed for the PKCγ without the mutation in the C1A (Fig. 6D). Lastly, we addressed whether substitution (rather than deletion) of Phe48 restored agonist responsiveness. Mutation to either alanine (F48A) or the structurally more similar tyrosine (F48Y) restored autoinhibition to that observed for WT enzyme (Fig. 6E). However, whereas F48Y responded similarly to PDBu as WT PKCγ, F48A only partially rescued the WT response to PDBu. These data reveal that it is the loss of F48 that uncouples the pseudosubstrate from ligand engagement; substitution with Ala or Tyr may reduce activation kinetics and response to UTP, but still allows response to phorbol esters. We next examined a SCA14 deletion mutation at the corresponding position in the C1B domain (ΔF113) (Fig. 6F). Similar to ΔF48, ΔF113 had higher basal activity indicating impaired autoinhibition. However, the ΔF113 retained some responsiveness to phorbol esters, as evidenced by the increase in activity following PDBu stimulation. Thus, deletion of Phe48 in the C1A domain impairs autoinhibition but locks PKC in a conformation that prevents communication between the pseudosubstrate and membrane binding modules, whereas deletion of the corresponding Phe113 in the C1B impairs autoinhibition but allows more communication between the pseudosubstrate and membrane engagement.
Figure 6. SCA14 mutant ΔF48 displays an abrogated response to agonists.
(A and B) Domain structures of PKCγ constructs containing the mutated pseudosubstrate (R21G) alone or combined with Phe48-deleted (R21G ΔF48) (A), and the pseudosubstrate-deleted (ΔPS) alone or combined with Phe48-deleted (ΔPS ΔF48) (B). (C and D) COS7 cells were transfected with CKAR2 alone (endogenous; gray) or co-transfected with CKAR2 and the indicated mCherry-tagged PKCγ constructs: WT (orange), ΔF48 (purple), or the mutants shown in (A) and (B), respectively. PKC activity was monitored by measuring FRET/CFP ratio changes after addition of 100 μM UTP, 200 nM PDBu, and 50 nM calyculin A. Data were normalized to the endpoint (1.0) and represent mean ± S.E.M. from at least two independent experiments, N ≥ 20 cells per condition. In (D), the PKCγ WT, ΔF48, and endogenous data (dashed lines) are reproduced from (C) for direct comparison purposes. (E and F) COS7 cells were transfected with CKAR2 alone (endogenous) or co-transfected with CKAR2 and the indicated mCherry-tagged PKCγ constructs (E) or the SCA14 mutant ΔF113 (F). PKC activity was monitored, analyzed, and shown as described in (C and D), from at least three independent experiments of N ≥ 49 (E) or 31 (F) cells per condition. In (F), the PKCγ WT, ΔF48, and endogenous data (dashed lines) are reproduced from (E) for direct comparison purposes.
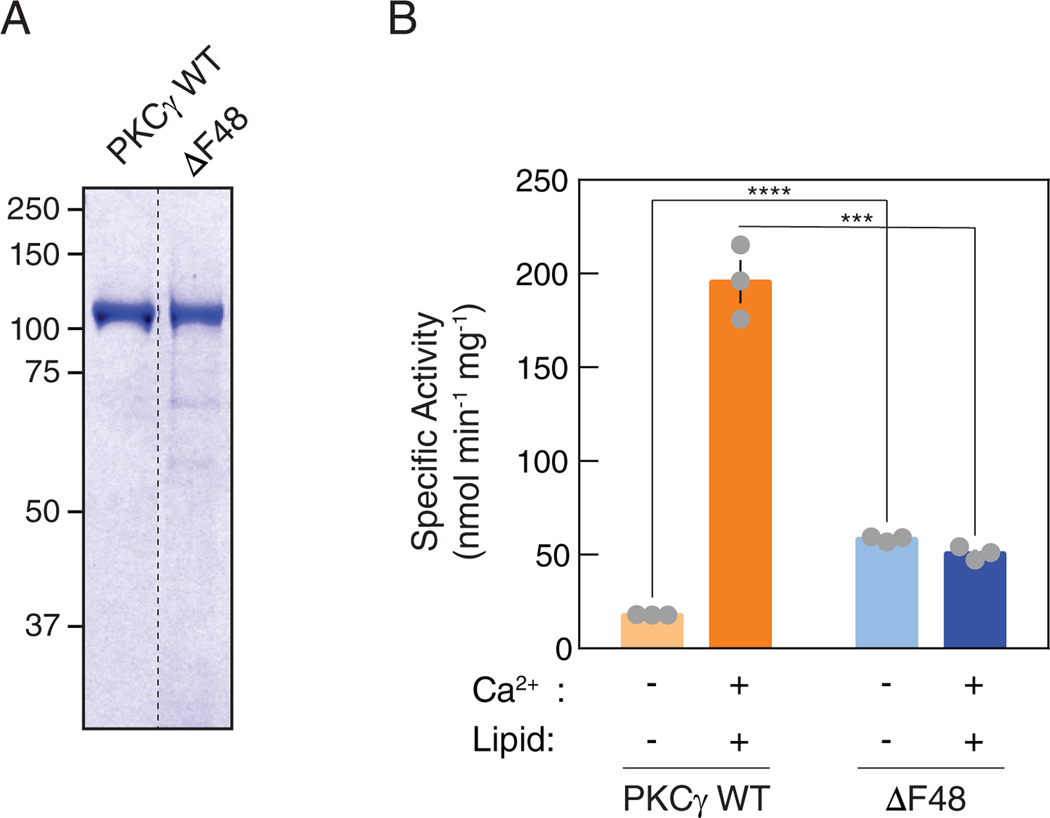
To validate whether the ΔF48 protein has lost the ability to be allosterically activated, we examined the activity of pure protein in vitro in the absence and presence of Ca2+ and lipid. GST-tagged PKCγ WT or ΔF48 produced in insect cells using a baculovirus expression system was purified to homogeneity (Fig. 7A) and activity was measured in the absence (non-activating conditions) or presence (activating conditions) of Ca2+ and multilamellar lipid structures (Fig. 7B). The activity of WT PKCγ was stimulated approximately 10-fold by Ca2+ and lipid, as reported previously (43), reflecting effective autoinhibition. In contrast, the specific activity of ΔF48 was approximately 3-fold higher than that of WT enzyme in the absence of cofactors, indicating impaired autoinhibition. Furthermore, addition of Ca2+/lipid had no effect on the activity of the ΔF48 mutant. Together with the activity data in live cells, these results establish that the ΔF48 C1A domain has reduced autoinhibition and is locked in a conformation that prevents communication between the pseudosubstrate and the membrane-binding regulatory domains.
Figure 7. Purified ΔF48 exhibits increased activity compared to WT PKCγ under non- activating conditions.
(A) Coomassie Blue-stained SDS-PAGE gel of purified GST-PKCγ WT or ΔF48. (B) In vitro kinase assays of purified GST-PKCγ WT or ΔF48 (6.1 nM per reaction). PKC activity was measured under non-activating conditions (EGTA, absence of Ca2+ or lipids) or activating conditions (presence of Ca2+ and lipids). Data are graphed in nanomoles phosphate per minute per milligram GST-PKC. Data are mean ± S.E.M. from three independent experiments, N = 9 reactions per condition. ***P<0.001, ****P<0.0001 by Holm-Sidak multiple comparison t-tests.
Altered phosphoproteome in cerebellum of mice harboring SCA14-associated PKCγ mutation
Every SCA14 C1 domain mutant tested displayed increased basal activity (Fig. 2A) and resistance to phorbol ester downregulation in cell-based studies (Fig. 4B). To address whether this leaky activity altered downstream signaling in a physiological setting, we took advantage of an ataxic transgenic mouse expressing human PKCγ H101Y and compared the cerebellar phosphoproteome to that of mice expressing PKCγ WT in a C57BL/6 background. The H101Y mice displayed an ataxic phenotype based on cerebellar morphology (fig. S3A, wherein calbindin staining revealed that Purkinje cells in H101Y-expressing mice displayed less fine development of dendritic arbor compared to WT-expressing mice) and behavior using the rotarod test for motor coordination (fig. S3B, wherein H101Y-expressing mice exhibited modestly decreased fall latency at 1 and 3 months old, and significantly decreased fall latency at 9 months of age compared to WT-expressing mice). Thus, the H101Y-expressing mice displayed progressive motor impairment consistent with an ataxic phenotype.
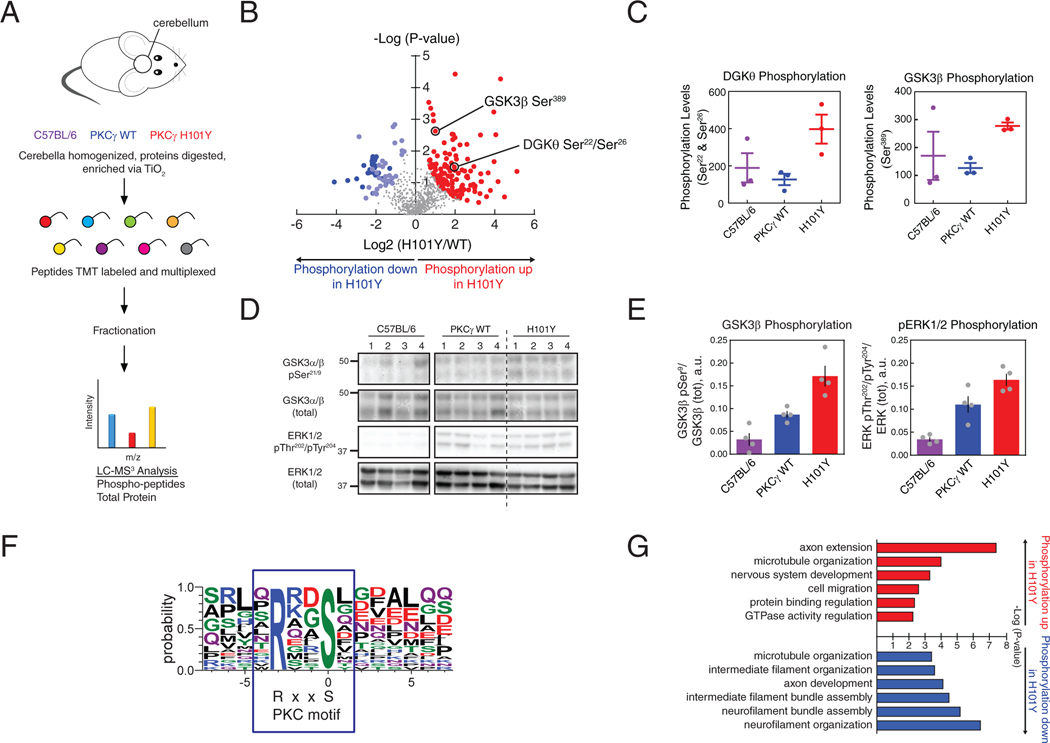
We next undertook a phosphoproteomic analysis of the cerebella of the PKCγ WT, H101Y, and control C57BL/6 background mice at 6 months of age (Fig. 8A). We quantified nearly 7000 unique proteins, from which 914 contained quantifiable phosphopeptide results across all samples (data file S1). After correction for protein amount, a total of 195 phosphopeptides on 166 unique proteins were identified, with 135 phosphopeptides significantly increasing in abundance and 60 phosphopeptides significantly decreasing in abundance in H101Y mice (Fig. 8B). Changes in phosphopeptide abundance were corrected by dividing phosphopeptide relative abundance by the corresponding protein abundance. Statistical significance was determined using a ranking method that simultaneously considers fold change and p-value (44) setting the α-value less than or greater than .05. Of the phosphopeptides whose phosphorylation decreased in the H101Y cerebella, a striking 30 of them were contained within neurofilament proteins (Fig. 8B, light blue circles), consistent with a general reduction in neurofilament phosphorylation in H101Y mouse cerebellum. Of those that increased, we noted an increase in phosphorylation at two sites (Ser22 and Ser26) on a single phosphopeptide of diacylglycerol kinase θ (DGKθ), which catalyzes the phosphorylation of DG to phosphatidic acid, in the H101Y mice, consistent with either direct or indirect regulation of DGKθ by PKCγ (Fig. 8C, left). Furthermore, the phosphorylation of one of the major kinases of neurofilaments, glycogen synthase kinase 3 beta (GSK3β) (45, 46), was increased on an inhibitory site, Ser389 (47) (Fig. 8C, right). This site is in an -SP- motif that is not a direct PKC phosphorylation site, rather, it is phosphorylated by MAPK (47), a kinase that is activated following PKC activation (48). To validate that enhanced basal signaling by PKC inhibits GSK3β, we examined whether phosphorylation on Ser9, a bonafide PKC consensus RxxS site (7), was altered. Western blot analysis revealed an approximately 70% increase in the phosphorylation of Ser9 in the H101Y cerebella compared to WT. Additionally, phosphorylation of ERK itself on the activating sites Thr202/Tyr204 was elevated in the cerebella of H101Y mice compared to WT (Fig. 8D-E). We also performed a motif analysis on the phosphopeptides that were significantly increased in H101Y-expressing mice to determine the fraction of these peptides that contain the PKC substrate motif, RxxS (7) (Fig. 8F). Out of 77 significantly increased phosphopeptides analyzed, 24 contained an RxxpS motif, representing an over 5-fold increase in phosphorylation of PKC consensus site substrates in H101Y-expressing mice compared to PKCγ WT-expressing mice. Given that many of the other changes in phosphorylation detected in H101Y mice are likely targets that are farther downstream of PKCγ, this motif analysis is consistent with enhanced PKCγ activity in SCA14 mutant-expressing mice driving the rewiring of the H101Y mouse phosphoproteome. Lastly, gene ontology analysis by DAVID GO (Fig. 8G) (49, 50) revealed that phosphopeptides with increased abundance in H101Y-expressing mice were primarily involved in processes related to axon extension, neural development, and cytoskeletal organization, and, similarly, phosphopeptides with decreased abundance in H101Y mice were mainly involved in neurofilament organization and axon development. This analysis suggests H101Y-expressing mice display dysregulation of signaling pathways involved in developing and maintaining neuron cytoskeletal structure and function, which may be regulated upstream by PKCγ.
Figure 8. Phosphoproteomics analysis from cerebella of mice expressing human WT or H101Y PKCγ transgene.
(A) Experimental design for processing of mouse tissue and proteins. Cerebella from B6 background (purple), PKCγ WT transgenic (blue), and PKCγ H101Y transgenic (red) mice at 6 months of age were subjected to phosphoprotemics analysis. 6893 total proteins were quantified in the standard proteomics and 914 quantifiable phosphopeptides were detected in the phosphoproteomics. After correction for protein expression, 195 phosphopeptides on 166 unique proteins were identified in H101Y-expressing mice. N = 3 mice of C57BL/6, PKCγ WT, PKCγ H101Y. (B) Volcano plot of phosphopeptide replicates of cerebella from WT and H101Y transgenic mice. Graph represents the log-transformed p-values (Student’s t-test) linked to individual phosphopeptides versus the log-transformed fold change in phosphopeptide abundance between WT and H101Y cerebella. Color represents phosphopeptides with significant changes in P-value and fold change; red, increased phosphorylation in H101Y mice; blue, lower phosphorylation in H101Y mice (dark blue indicates significantly decreased neurofilament phosphopeptides, light blue indicates all other significantly decreased phosphopeptides). (C) Graphs representing quantification of either a DGKθ phosphopeptide (left) or a GSK3β phosphopeptide (right) from the volcano plot in (B) in cerebella from C57BL/6 mice (purple), WT mice (blue), and H101Y mice (red). (D and E) Western blotting analysis of Triton-soluble cerebellar tissue lysates from C57BL/6, PKCγ WT, or H101Y mice. Membranes were probed with antibodies against GSK3α/β pSer21/9, GSK3α/β (total), ERK1/2 pThr202/pTyr204, or ERK1/2 (total). Quantification of blots from N = 4 mice per genotype, for GSK3β phosphorylation (pSer9; left) and ERK1/2 phosphorylation (pThr202/pTyr204; right), are shown in (E). Data are mean + S.E.M. (F) Motif analysis of RxxpS PKC consensus substrate sequence in significantly increased phosphopeptides. RxxpS was detected in 24 of 77 sequences of length 15 after removing background. Fold increase of RxxpS phosphopeptide abundance in H101Y:WT cerebellum = 5.3. (G) Gene ontology analysis of significantly increased (red) or decreased (blue) phosphopeptides representing significantly changed biological processes.
Conventional PKC C1 domains are protected from mutation in cancer
We have previously shown that cancer-associated mutations in conventional PKC isozymes are generally loss-of-function (3), with mutations that impair autoinhibition triggering degradation by a PHLPP-mediated quality control mechanisms (22). However, SCA14 mutations, which occur with high frequency in the C1 domains, impair autoinhibition without triggering downregulation. None of the identified SCA14 mutations are currently annotated in cancer data bases such as cBioPortal (51, 52). Thus, we assessed whether the frequency of cancer-associated mutations in conventional PKC isozymes is lower in the C1 domains compared to the C2 domain. The number of missense mutations at each aligned residue position of PKCα, β, and γ was obtained from GDC Data Portal (fig. S4A) (53) and the total mutation frequency within each domain (number of mutations per residues in the domain) was analyzed (fig. S4B, left, and table S1). The mutational frequency of the C1 domains was approximately half that of the C2 domain when all three conventional isozymes were analyzed together. Furthermore, we compared mutation frequencies of the C1B domain to all other domains and found that the C1B has significantly lower missense mutation frequency than other domains in PKC (fig. S4B, right). Notably, analysis of the individual isozymes revealed that the C1A domain of PKCα was more protected from mutation than the C1B domain (table S1). Notably, our analysis suggests that the C1B domain—a mutational hotspot in SCA14—is more protected overall from mutation in cancer compared to other domains.
Age of SCA14 onset inversely correlates with the degree of impaired PKCγ autoinhibition
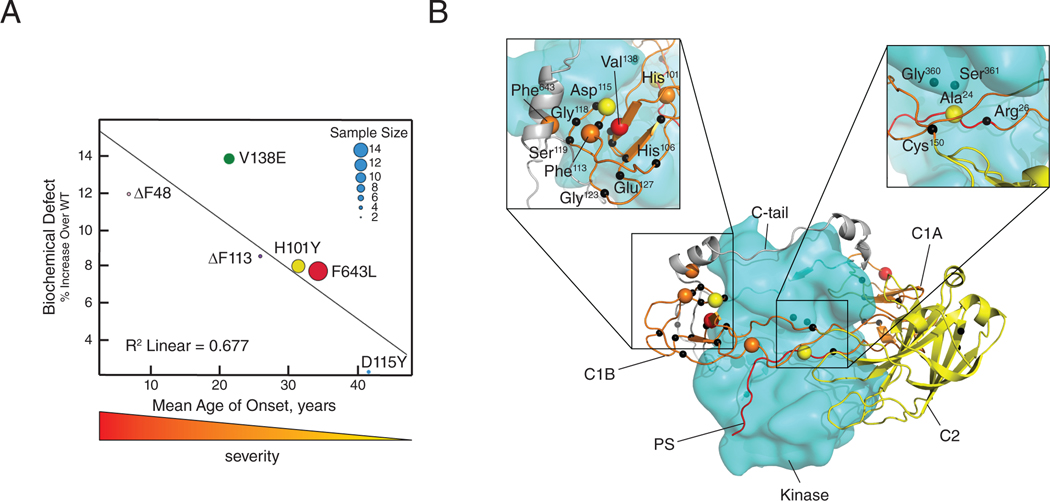
To understand the degree to which the enhanced basal activity of the SCA14 mutants may contribute to disease, we plotted the level of biochemical defect (basal activity) the average age of onset of disease in the patients with the respective variants (14, 54–59) (Fig. 9A). For this analysis, we focused on mutations that do not impair the stability of PKC (C1 domain mutations and F643L) because mutations that impair stability (for instance, pseudosubstrate mutations) would reduce steady-state levels and thus reduce the impact of enhanced basal activity (22, 60). These data reveal a trend between the degree of biochemical defect and disease severity: C1 domain mutants with high basal activity, such as V138E and ΔF48, were associated with an age of disease onset in early childhood (high disease severity), whereas those with lower levels of autoinhibitory defect, such as D115Y, were associated with an older age of onset (lower disease severity). Taking into account the varying patient sample sizes for each mutation, we calculated an R2 value of approximately 0.67, supporting the idea that enhanced PKCγ basal activity may be a key contributor to the development of SCA14.
Figure 9. Degree of ataxia mutant biochemical defect correlates with SCA14 severity.
(A) Graph of the indicated SCA14 mutant basal activities from Fig. 2, B, C, and E, and Fig. 6F plotted against average age of disease onset in patients (14, 53–58). Sample size, between 2 and 14 patients, is indicated by dot size. (B) PKCγ model based on the previously published model of PKCβII (18). Indicated SCA14 mutations are represented as black spheres; the five mutations presenting in (A) are color coded by disease severity.
Lastly, we used a homology model for the architecture of conventional PKC isozymes (19) to predict where the 54 known SCA14-associated mutations (Fig. 1A) would occur within the 3-dimensional structure of PKCγ (Fig. 9B). In the autoinhibited conformation, the kinase adopts a compact conformation with the regulatory modules packed against the kinase domain and C-tail, and the pseudosubstrate segment (red) in the substrate binding cavity. Notably, many of the SCA14 mutations are predicted to exist either at an interface between the C1B and kinase domain or between the C1B domain and the C-tail. Notably, Asp115 is predicted to interface with the kinase domain, providing an explanation for why the D115Y mutation, which does not alter the affinity for phorbol ester binding in the isolated C1B domain (38), reduces autoinhibition in the context of the full-length protein: the bulkier side chain of tyrosine compared to aspartate, and the loss of negative charge, may break interdomain interactions to favor the ‘open’ conformation. Similarly, Phe643 is predicted to interface with the C-tail. This residue is part of the conserved NFD motif, a key regulatory determinant of AGC kinases (61), which anchors the C1B in place (Fig. 9B, left inset) (62), and its mutation of Leu is likely to reduce the affinity of the tail for the C1B. Additionally, two mutations (A24T and R26G) are located in the pseudosubstrate, both of which are predicted to disrupt autoinhibition. The first, A24T, occurs at the phospho-acceptor site, which likely introduces a phosphorylation site, whereas R26G may disrupt a possible H-bond to Gly500 of the conserved DFG motif in the kinase domain (Fig. 9B, right inset). Only the two mutations in the C2 domain (I173S and H174P) were not at an interface with the kinase domain or regulatory domains. Thus, our model indicates that almost all SCA14 mutations target the C1 domains and their interfaces with the rest of the protein.
Discussion
An abundance of germline variants in PKCγ are causal in SCA14, yet establishing whether a unifying mechanism accounts for the defect in these aberrant enzymes has remained elusive (26, 31–37). Here we show that SCA14 PKCγ mutations in every domain of PKC display a shared autoinhibitory defect that leads to enhanced basal activity. Furthermore, by analyzing a mutant that uncouples pseudosubstrate regulation from phorbol ester binding (ΔF48), we show that increased basal signaling, rather than changes in agonist-evoked signaling, is the determinant associated with the ataxic phenotype. Remarkably, the degree of biochemical defect (impaired autoinhibition) of the C1 domain mutants correlated inversely with average age of disease onset in patients. Our data support a model in which aberrant basal signaling by PKCγ is the driver behind SCA14.
A key finding from our study is that C1 mutations represent a susceptibility that allows for deregulated PKC activity without the paradoxical down-regulation accompanying the “open” conformation of PKC (60). Namely, whereas disruption of autoinhibition of conventional PKC isozymes generally results in unstable enzyme that is dephosphorylated and degraded (63), mutations in, or deletion of, the C1A or C1B domains renders PKCγ insensitive to phorbol ester-mediated downregulation, suggesting why these domains harbor the highest number of SCA14 mutations. Despite the the resistance of SCA14 C1 domain mutants to phorbol ester-mediated downregulation, the steady-state turnover of the mutants was enhanced compared to WT PKCγ. This uncoupling of agonist-dependent turnover and basal turnover has been reported previously. For example, the E3 ligase RINCK was shown to promote PKC ubiquitination and degradation under non-activating conditions but its deletion did not affect phorbol ester-mediated downregulation (64). Similarly, Leontieva and Black have identified two distinct pathways that mediate PKCα downregulation, one that is proteosome-dependent and one that is not (65). How the increased basal turnover of SCA14 mutants affects the steady-state levels of PKCγ in the disease awaits further studies.A particularly informative SCA14 mutation was the recurrent deletion of a Phe (ΔF48) on the ligand binding loop of the C1A domain. This mutation (or deletion of the entire C1A) destroys communication between the pseudosubstrate and the C1B-C2 membrane-targeting modules such that phorbol ester-induced membrane engagement does not allosterically activate the enzyme. Thus, the mutant is ‘frozen’ in a partially active conformation, uncoupling it from DG and Ca2+ signaling. Yet patients with this mutation have early childhood onset of the disease. A major ramification is that enhanced basal signaling, and not agonist-evoked signaling, drives the pathology of SCA14.
Mutations that reduce the affinity of the pseudosubstrate for the active site destabilize PKC, promoting dephosphorylation and degradation (22); yet, four SCA14 mutations have been identified in the pseudosubstrate. Shimobayashi and Kapfhammer have provided key insight to this paradox by their analysis of a transgenic mouse harboring a mutation in the pseudosubstrate, A24E (29). This mutation, which caused an ataxic phenotype in mice and impaired Purkinje cell maturation, greatly reduced the stability of the enzyme and decreased steady-state levels approximately 10-fold compared with levels in WT mice. However, the unrestrained activity of the aberrant PKC that was present was sufficient to cause an increase in substrate phosphorylation in the cerebellum of these mice. Thus, although this PKC is unstable and steady-state levels are reduced, the basal activity is sufficiently increased to drive the ataxic phenotype.
Supporting a unifying model of enhanced basal activity driving the pathology of SCA14, transgenic mouse models harboring PKCγ mutations display an ataxic phenotype regardless of where in the structure the mutation occurs: in the C1 domain (such as H101Y in this study), in the pseudosubstrate (such as A24E), or in the kinase domain (S361G and S360S) (66)(25)(67). Interestingly, a variant that introduces a premature stop codon in PRKCG at Arg76 (R76X) has been proposed to produce a fragment of PKCγ that may activate remaining PKC isozymes through RACKs (28).
Our analysis of the H101Y SCA14 mouse model revealed that ‘leaky’ PKC activity causes significant changes in the phosphorylation state of components of numerous processes in the cerebellum. Most strikingly, the phosphorylation of neurofilament proteins, which play key roles in growth of axons, with aberrations associated with neurodegeneration (68), decreased. This could arise from increased phosphatase output by phosphatases such as PP2A B56δ, whose phosphorylation at a proposed PKC activating site, Ser566 (69, 70), increased two-fold in the H101Y cerebellum compared to WT. Decreased neurofilament phosphorylation could also arise from inhibition of GSK3β, a major kinase regulating neurofilaments (45, 46), whose phosphorylation on two inhibitory PKC sites (47)(71) increased two-fold in the H101Y cerebellum. Future focus on the role of basal PKCγ activity in neurofilament organization and axon development may provide insight into the mechanisms of Purkinje cell degeneration.
The possibility that “leaky” PKC signaling may be a driver in cerebellar ataxia, in general, has been suggested by both unbiased network analyses and by specific mechanisms. First, a network analysis by Verbeek and colleagues identified alterations in synaptic transmission as one of the main shared mechanisms underlying genetically diverse SCAs (72), similar to the alterations we observed in the H101Y mouse model. Second, several other genetic causes of SCA directly impact PKCγ regulation, including genes that regulate intracellular Ca2+ and DG (10–13, 17, 73). Indeed, increased DG is sufficient to produce an ataxic phenotype: Shirai and colleagues found that mice deficient in diacylglycerol kinase γ (DGKγ), which converts DG into phosphatidic acid, display an ataxic phenotype (74). Whether the majority of SCAs converge on deregulated PKCγ signaling is an intriguing possibility that would provide a tangible therapeutic target for the disease.
Small changes in the intrinsic activity of PKCα similar to those observed in PKCγ in the present study are associated with Alzheimer’s Disease (6) (5) (75). In contrast to the SCA14 variants, characterized germline mutations in PKCα in Alzheimer’s Disease do not affect autoinhibition, rather, they enhance the catalytic rate of the kinase domain such that a stronger response is evoked in response to agonist (5, 6). Thus, these two neurodegenerative diseases both have subtle gain-of-function mutations in a conventional PKC, that lead to changes in activity (either basal or agonist-evoked), suggesting that small changes over a lifetime accumulate neuronal damage that eventually manifests in the disease.
In summary, our study reveals that SCA14 mutations are uniformly associated with enhanced basal signaling of PKCγ, indicating that therapies that inhibit this enzyme may have therapeutic potential. In addition to identifying PKCγ as an actionable target in this neurodegenerative disease, our studies also provide a framework to predict disease severity in SCA14. Specifically, the direct correlation between the degree of impaired autoinhibition and disease severity allows prediction of patient prognosis of new mutations, such as the D115Y reported here. Lastly, given the direct regulation of PKCγ by intracellular Ca2+, and that many of the proteins mutated in other SCAs regulate Ca2+ homeostasis, one intriguing possibility is that enhanced PKCγ activity is not only central to SCA14 pathology, but is also at the epicenter of many other types of ataxia. This raises exciting possibilities for therapeutically targeting PKCγ in not just SCA14, but in many other subtypes of spinocerebellar ataxia.
Materials and Methods
Chemicals and antibodies
Uridine-5-triphosphate (UTP; cat #6701) and phorbol 12,13-dibutyrate (PDBu; cat #524390) were purchased from Calbiochem. Calyculin A (cat #9902) was purchased from Cell Signaling Technology. The anti-HA antibody (cat #11867423001) was purchased from Roche. The anti-phospho-PKCα/βII turn motif (pThr638/641; cat #9375) was from Cell Signaling Technology. Lipids used in kinase assays (DG, cat #800811C and PS, cat #840034C) were purchased from Avanti Polar Lipids. The anti-phospho-PKCγ hydrophobic motif (pThr674; cat# ab5797) antibody was from Abcam. The anti-phospho-PKCα/β/γ activation loop (pThr497/500/514) antibody was previously described (44). Ladder (cat #161-0394), bis/acrylamide solution (cat # 161–0156), and PVDF (cat# 162–0177) used for SDS-PAGE and Western blotting were purchased from Bio-Rad. Luminol (cat #A-8511) and p-coumaric acid (cat # C-9008) used to make chemiluminescence solution for western blotting were purchased from Sigma-Aldrich. [γ−32P]-ATP (cat # BLU002Z500UC) was purchased from Perkin Elmer. Phosphatidylserine (cat # 840034C) and diacylglycerol (cat # 800811C) were from Avanti Polar Lipids. The PKC substrate peptide (Ac-FKKSFKL-NH2) was synthesized by GenScript.
Magnetic resonance imaging of ataxia patient brains
MRI Imaging was performed on a 1.5 Tesla Siemens MRI Scanner. Sagittal T1 Flair images were taken. Patients have consented to their anonymized scans being used in this publication.
Plasmid constructs and mutagenesis
All mutants were generated using QuikChange site-directed mutagenesis (Agilent). PKC pseudosubstrate-deleted constructs were created by deletion of residues 19–36 by QuikChange mutagenesis (Agilent) using WT PKCγ, R21G, or ΔF48-containing mCherry-pcDNA3 plasmid. PKC C1A-, C1B-, and C2-deleted constructs were created by deletion of residues 36–75 (ΔC1A), 100–150 (ΔC1B), or 179–257 (ΔC2) by QuikChange mutagenesis (Agilent) using WT PKCγ mCherry- or HA-pcDNA3 plasmid. The C Kinase Activity Reporter 1 (CKAR1) (27) and C Kinase Activity Reporter 2 (CKAR2) (26) were previously described.
Cell culture and transfection
COS7 cells were maintained in DMEM (Corning) containing 10% FBS (Atlanta Biologicals) and 1% penicillin/streptomycin (Gibco) at 37 °C in 5% CO2. Transient transfection was carried out using the Lipofectamine 3000 kit (ThermoFisher) per the manufacturer’s instructions, and constructs were allowed to express for 24 hours for imaging and CHX assays, or 48 hours for PDBu downregulation assays and phosphorylation site Western blotting.
FRET imaging and analysis
COS7 cells were seeded into individual dishes, and imaging was performed under conditions and parameters previously described (76). Images were acquired on a Zeiss Axiovert microscope (Carl Zeiss Micro-Imaging, Inc.) using a MicroMax digital camera (Roper-Princeton Instruments) controlled by MetaFluor software (Universal Imaging Corp.). For CKAR activity assays, COS7 cells were co-transfected with the indicated mCherry-tagged PKCγ constructs and CKAR2 for 24 h before imaging; cells were sequentially treated with 100 μM UTP, 200 nM PDBu, and 50 nM calyculin A at the times indicated in the associated figures. For translocation assays, COS7 cells were co-transfected with the indicated YFP-tagged constructs and MyrPalm-mCFP (plasma-membrane targeted) (41) for 24 h before imaging, and cells were treated with 200 nM PDBu. For co-translocation assays, COS7 cells were co-transfected with mCherry-tagged PKCγ and YFP-tagged D115Y for 24 hours before imaging, and cells were treated with 200 nM PDBu. Baseline images were acquired every 15 s for 3 min prior to treatment with agonists. For CKAR activity assays, all FRET ratios were normalized to the endpoint of the assay. Translocation assays are normalized to the starting point of the assay.
Phorbol ester downregulation assay
COS7 cells were seeded in 6-well plates at 1.5×105 cells per well. After 24 hours, cells were transfected with indicated HA-tagged PKCγ constructs (100ng DNA per well) for 48 hours before PDBu treatment. Cells were treated with 10 to 1000 nM PDBu or DMSO for 24 hours. Cells were then washed with DPBS (Corning) and lysed in buffer containing 50 mM NaPO4 (pH 7.5), 1% Triton X-100, 20 mM NaF, 1 mM Na4P2O7, 100 mM NaCl, 2 mM EDTA, 2 mM EGTA, 1 mM Na3VO4, 1 mM PMSF, 40 μg/mL leupeptin, 1 mM DTT, and 1 μM microcystin. For PDBu downregulation assays, whole-cell lysate was loaded on gels. Benzonase was added to whole-cell lysates at 1:100 to digest nucleic acids.
Cycloheximide assay
COS7 cells were seeded in 6-well plates at 1.5×105 cells per well. After 24 hours, cells were transfected with indicated HA-tagged PKCγ constructs (100ng DNA per well) for 24 h before CHX treatment. Cells were treated with 355 μM or DMSO for 0, 6, 24, or 48 hours. Cells were then washed with DPBS (Corning) and lysed in buffer containing 50 mM NaPO4 (pH 7.5), 1% Triton X-100, 20 mM NaF, 1 mM Na4P2O7, 100 mM NaCl, 2 mM EDTA, 2 mM EGTA, 1 mM Na3VO4, 1 mM PMSF, 40 μg/mL leupeptin, 1 mM DTT, and 1 μM microcystin. Whole-cell lysate was loaded on gels. Benzonase was added to whole-cell lysates at 1:100 to digest nucleic acids.
Western blots
All cell lysates were analyzed by SDS-PAGE on 6.5% big gels, run overnight at 9 mA per gel, to observe phosphorylation-induced mobility shift. Gels were transferred to PVDF membrane (Bio-Rad) by a wet transfer method at 4 °C for 2 hours at 80 V. Membranes were blocked in 5% BSA in phosphate buffered saline containing 0.05% Tween-20 (PBST) for 1 hour at room temperature, then incubated in primary antibodies overnight at 4 °C. Membranes were washed for 5 min three times in PBST, incubated in appropriate secondary antibodies for 1 hour at room temperature, washed for 5 min three times in PBST, then imaged by chemiluminescence (100 mM Tris pH 8.5, 1.25 mM Luminol, 198 μM coumaric acid, 1% H2O2) on a FluorChem Q imaging system (ProteinSimple). In the displayed Western blots, the asterisk (*) indicates phosphorylated PKC species, while a dash (−) indicates unphosphorylated species.
Purification of GST-PKC from Sf9 insect cells
PKCγ wild-type and ΔF48 were cloned into the pFastBac vector (Invitrogen) containing an N-terminal GST tag. Using the Bac-to-Bac Baculovirus Expression System (Invitrogen), the pFastBac plasmids were transformed into DH10Bac cells, and the resulting bacmid DNA was transfected into Sf9 insect cells by CellFECTIN (ThermoFisher). Sf9 cells were grown in Sf-900 II SFM media (Gibco) in shaking cultures at 27 °C. The recombinant baculoviruses were harvested and amplified three times. Sf9 cells were seeded in 125mL spinner flasks at 1×106 cells per mL and infected with baculovirus. After 2 days of infection, Sf9 cells were lysed in buffer containing 50 mM HEPES (pH 7.5), 1 mM EDTA, 100 mM NaCl, 1% Triton X-100, 100 μM PMSF, 1 mM DTT, 2 mM benzamidine, 50 μg/ml leupeptin, and 1 μM microcystin. Lysates were centrifuged at 10,000 x g for 15 min at 4 °C and the supernatants, containing the detergent-soluble fraction, were incubated with GST-Bind resin (EMD Millipore) for 30 min at 4 °C, washed three times, then GST-PKCγ was eluted off the beads with buffer containing 50 mM HEPES (pH 7.5), 1 mM EDTA, 100 mM NaCl, 1 mM DTT, and 10 mM glutathione. Purified protein was concentrated with Amicon Ultra-0.5 mL centrifugal filters (50kDa cutoff; EMD Millipore) to 100 μL. Protein purity and concentration were determined using BSA standards on an 8% SDS-PAGE mini-gel stained with Coomassie Brilliant Blue stain.
In vitro kinase activity assays
The activity of purified GST-PKCγ (6 nM) was assayed as previously described (77) except the substrate was the MARCKS peptide substrate (Ac-FKKSFKL-NH2) and with the specific assay conditions as follows. Reactions contained 20 mM HEPES (pH 7.4), 0.06 mg/mL BSA, 1.2 mM DTT, 100 μM [γ−32P]-ATP, 100 μM peptide substrate, and 5 mM MgCl2. Assays were performed in the presence of Ca2+ (100 μM) and PS (140 μM):DG (3.8 μM) multilamellar vesicles (activating conditions) or 50 μM EGTA (non-activating conditions). Reactions were allowed to proceed at 30 °C for 10 min before quenched as described (78).
Mouse model and harvest of cerebella
Under an approved University of Washington Institutional Animal Care and Use Committee (IACUC) protocol (IACUC 4039–01), SCA14 mutant (H101Y) and wild-type (WT) PKCγ transgenic (Tg) mice were generated using modified human-BAC constructs, where expression of human PKCγ is regulated by the endogenous human PRKCG promoter. Flanking neuronal-expressed genes were removed and an eGFP-tag was introduced to enable detection and visualization of the transgene. Purified PRKCG fragments were microinjected into C3H/C57BL6 (inbred mouse background strain, RRID:IMSR_JAX:000664) hybrid oocyte pronuclei. RNA, western blot and immunohistochemical analyses all confirmed expression of the transgene. We interbred the lines to homozygosity for the transgenes (WT- and H101Y-PRKCG++). The level of expression of the transgene was lower than that of the endogenous PRKCG as detected by fused eGFP-PKCγ. Rotarod tests of transgenic PKCγ WT-expressing or PKCγ H101Y-expressing mice were performed at 1, 3, and 9 months of age. Mice were tested over the course of three trial days and latency to fall off the rotarod was measured. The single technician who performed the rotarod testing was blinded to the genotype of the mice. At 6 months of age, three mice of each homozygous-Tg genotype and three C576BL/6 mice were sacrificed by cervical dislocation. Animal numbers were justified in the neurologic testing teams by a power analysis (http://www.stat.uiowa.edu/~rlenth/Power/index.html) using a standard two-tailed t-test. We also accounted for the natural attrition rate of mice with neurologic impairments. For each genotype, the team consisted of 20 mice to ensure that 15 mice completed the test battery. Cerebella were dissected, snap-frozen in liquid nitrogen and kept at −80°C until shipment on dry ice to UCSD for protein extraction and proteomics analysis.
Mass spectrometry-based proteomics
Sample processing, phosphopeptide enrichment and mass spectrometry analysis followed methods described previously (79), but are described here briefly to highlight modifications. Snap frozen cerebellum (approximately 45 mg) were homogenized by bead beating at 37 °C in lysis buffer (1 mL) composed of 3% SDS, 75 mM NaCl, 1 mM NaF, 1 mM β-glycerophosphate, 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, 1 mM PMSF and 1X Roche Complete mini EDTA free protease inhibitors in 50 mM HEPES, pH 8.5. Rough homogenates were then further subjected to probe sonication (Q500 QSonica sonicator with 1.6 mm microtip). To the protein mixture was added an equal volume of Urea (8 M in 50 mM HEPES). Samples were reduced and alkylated using dithiothreitol (5 mM) and iodoacetamide (15 mM) respectively. Proteins were precipitated using chloroform/methanol, dried, and resuspended in 1 M urea in 50 mM HEPES (600 μL). Proteins were then digested using LysC, followed by trypsin before purification by SepPak cartridges. Protein aliquots (50 μg) from each sample were lyophilized and stored at −80 °C for labeling and proteomic analysis, along with 7 μg per sample pooled to generate two bridge channels. From each sample 1 mg peptide was subjected to phospho-enrichment using 6 mg Titanium dioxide beads. Enriched peptides were desalted using solid-phase extraction columns, lyophilized and stored at −80 °C until labeling.
For both the phospho-enriched peptides and reserved peptides for proteomics, peptides were labeled with tandem mass tag (TMT) reagents, reserving the 126 and 131 mass labels for the two bridge channels. Labeled samples were then pooled into multiplex experiments and desalted by solid phase extraction. Sample fractionation was performed using spin columns to generate eight fractions per multiplex experiment. Fractions were lyophilized, re-suspended in 5% formic acid/5% acetonitrile for LC-MS2/MS3 identification and quantification. LC-MS2/MS3 analysis were performed on an Orbitrap Fusion mass spectrometer and data processing was carried out using the ProteomeDiscoverer 2.1.0.81 software package as described previously (79).
Motif enrichment analysis was done using motifx (R package rmotifx version 1.0). Foreground sequences were set to sequences of length 15 flanking unique phospho-sites either significantly increased or decreased in H101Y:WT. Background sequences were extracted from the mouse proteome (Uniprot UP000000589, downloaded 1/28/2022) using parseDB and extractBackground (R package PTMphinder, version 0.1.0). Central residue was set to “S” or “T” as appropriate, minimum sequence cut-off to 5, and p-value cut-off to 1e-5. Logos for significantly enriched motifs were generated using WebLogo (version 3.7.4).
PKCγ structural model
The PKCγ model was built in UCSF Chimera 1.13.1 (80) with integrated Modeller 9.21 (81). The kinase domain was modelled using the structure of PKCβII (PDB: 2I0E) as a template. The structure of the C1B domain was modelled using the structure of the C1A of PKCγ (PDB: 2E73). The C1 domains were docked to the kinase domain according to the previously published model of PKCβ (62). The structure of the PKCγ C2 domain (PDB: 2UZP) was docked to the kinase domain and C1 domains complex using the PKCβII model as a starting point using ClusPro web server (82).
Quantification and data analysis
FRET ratios for CKAR assays were acquired with MetaFluor software (Molecular Devices). Ratios were normalized to starting point or endpoint (1.0) as indicated in figure legends. Western blots were quantified by densitometry using the AlphaView software (ProteinSimple). Gene ontology was performed by DAVID GO (49, 50) and was background adjusted using the Mus musculus species background. Statistical significance was determined by unequal variances (Welch’s) t-test or multiple t-tests (with the Holm-Sidak method of determining significance) performed in GraphPad Prism 8 (GraphPad Software).
Supplementary Material
Acknowledgments:
We thank Dr. Natasha Fullerton for help with image analysis and providing the image control of the MRI scans. We thank the laboratory of J. Zhang (UCSD) for generously providing the CKAR2. We thank Dr. James T. Yurkovich for advice on initial analysis of frequency of cancer-associated mutations. We also thank all members of our laboratories for their technical work and helpful comments on this manuscript.
Funding:
This work was supported by NIH R35 GM122523 (A.C.N), NIH NINDS 1 R01 NS069719, the Zionic Ataxia Fund, and the National Ataxia Foundation (W.H.R.), and NIH NIGMS R35 GM139656 (N.K.). C.A.P. was supported in part by the UCSD Graduate Training Program in Cellular and Molecular Pharmacology (T32 GM007752).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and Materials Availability:
The phosphoproteomics data have been depostied on UCSD CCMS MassIVE with the accession number: MSV000088896. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Callender JA, Newton AC, Conventional protein kinase C in the brain: 40 years later. Neuronal Signaling. 1, 20160005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishizuka Y, Protein kinase C and lipid signaling for sustained cellular responses. The FASEB Journal. 9, 484–496 (1995). [PubMed] [Google Scholar]
- 3.Antal CE, Hudson AM, Kang E, Zanca C, Wirth C, Stephenson NL, Trotter EW, Gallegos LL, Miller CJ, Furnari FB, Hunter T, Brognard J, Newton AC, Cancer-associated protein kinase C mutations reveal kinase’s role as tumor suppressor. Cell. 160, 489–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newton AC, Brognard J, Reversing the Paradigm: Protein Kinase C as a Tumor Suppressor. Trends in Pharmacological Sciences. 38 (2017), pp. 438–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callender JA, Yang Y, Lordén G, Stephenson NL, Jones AC, Brognard J, Newton AC, Protein kinase Cα gain-of-function variant in Alzheimer’s disease displays enhanced catalysis by a mechanism that evades down-regulation. Proceedings of the National Academy of Sciences of the United States of America. 115, E5497–E5505 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alfonso SI, Callender JA, Hooli B, Antal CE, Mullin K, Sherman MA, Lesné SE, Leitges M, Newton AC, Tanzi RE, Malinow R, Gain-of-function mutations in protein kinase Cα (PKCα) may promote synaptic defects in Alzheimer’s disease. Science Signaling. 9 (2016), doi: 10.1126/scisignal.aaf6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tovell H, Newton AC, PHLPPing the balance: restoration of protein kinase C in cancer. Biochemical Journal. 478 (2021), pp. 341–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lordén G, Newton AC, Conventional protein kinase C in the brain: repurposing cancer drugs for neurodegenerative treatment? Neuronal Signaling. 5, 20210036 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun YM, Lu C, Wu ZY, Spinocerebellar ataxia: relationship between phenotype and genotype – a review. Clinical Genetics. 90, 305–314 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Tada M, Nishizawa M, Onodera O, Roles of inositol 1,4,5-trisphosphate receptors in spinocerebellar ataxias. Neurochemistry International. 94, 1–8 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Tang TS, Tu H, Nelson O, Herndon E, Huynh DP, Pulst SM, Bezprozvanny I, Deranged calcium signaling and neurodegeneration in spinocerebellar ataxia type 2. Journal of Neuroscience. 29, 9148–9162 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fogel BL, Hanson SM, Becker EBE, Do mutations in the murine ataxia gene TRPC3 cause cerebellar ataxia in humans? Movement Disorders. 30 (2015), pp. 284–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watson LM, Bamber E, Schnekenberg RP, Williams J, Bettencourt C, Lickiss J, Fawcett K, Clokie S, Wallis Y, Clouston P, Sims D, Houlden H, Becker EBE, Németh AH, Dominant Mutations in GRM1 Cause Spinocerebellar Ataxia Type 44. American Journal of Human Genetics. 101, 451–458 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen DH, Brkanac Z, Verlinde CLMJ, Tan XJ, Bylenok L, Nochlin D, Matsushita M, Lipe H, Wolff J, Fernandez M, Cimino PJ, Bird TD, Raskind WH, Missense mutations in the regulatory domain of PKCγ: A new mechanism for dominant nonepisodic cerebellar ataxia. American Journal of Human Genetics. 72, 839–849 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saito N, Shirai Y, Protein kinase Cγ (PKCγ): Function of neuron specific isotype. Journal of Biochemistry. 132 (2002), pp. 683–687. [DOI] [PubMed] [Google Scholar]
- 16.Metzger F, Kapfhammer JP, Protein-kinase C: Its role in activity-dependent Purkinje cell dendritic development and plasticity. Cerebellum. 2 (2003), pp. 206–214. [DOI] [PubMed] [Google Scholar]
- 17.Pilo CA, Newton AC, Two Sides of the Same Coin: Protein Kinase C γ in Cancer and Neurodegeneration. Frontiers in Cell and Developmental Biology. 10 (2022), doi: 10.3389/FCELL.2022.929510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newton AC, Protein kinase C: perfectly balanced. Critical Reviews in Biochemistry and Molecular Biology. 53, 208–230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones AC, Taylor SS, Newton AC, Kornev AP, Hypothesis: Unifying model of domain architecture for conventional and novel protein kinase C isozymes. IUBMB Life. 72, 2584–2590 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans JH, Murray D, Leslie CC, Falke JJ, Specific translocation of protein kinase Ca to the plasma membrane requires both Ca2+ and PIP2 recognition by its C2 domain. Molecular Biology of the Cell. 17, 56–66 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antal CE, Violin JD, Kunkel MT, Skovsø S, Newton AC, Intramolecular conformational changes optimize protein kinase C signaling. Chemistry and Biology. 21, 459–469 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baffi TR, Van AAN, Zhao W, Mills GB, Newton AC, Protein Kinase C Quality Control by Phosphatase PHLPP1 Unveils Loss-of-Function Mechanism in Cancer. Molecular Cell. 74, 378–392.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yabe I, Sasaki H, Chen DH, Raskind WH, Bird TD, Yamashita I, Tsuji S, Kikuchi S, Tashiro K, Spinocerebellar Ataxia Type 14 Caused by a Mutation in Protein Kinase C γ. Archives of Neurology. 60, 1749–1751 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Yamashita I, Sasaki H, Yabe I, Fukazawa T, Nogoshi S, Komeichi K, Takada A, Shiraishi K, Takiyama Y, Nishizawa M, Kaneko J, Tanaka H, Tsuji S, Tashiro K, A novel locus for dominant cerebellar ataxia (SCA14) maps to a 10.2-cM interval flanked by D19S206 and D19S605 on chromosome 19q13.4-qter. Annals of Neurology. 48, 156–163 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Adachi N, Kobayashi T, Takahashi H, Kawasaki T, Shirai Y, Ueyama T, Matsuda T, Seki T, Sakai N, Saito N, Enzymological analysis of mutant protein kinase Cγ causing spinocerebellar ataxia type 14 and dysfunction in Ca2+ homeostasis. Journal of Biological Chemistry. 283, 19854–19863 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Wong MMK, Hoekstra SD, Vowles J, Watson LM, Fuller G, Németh AH, Cowley SA, Ansorge O, Talbot K, Becker EBE, Neurodegeneration in SCA14 is associated with increased PKCγ kinase activity, mislocalization and aggregation. Acta neuropathologica communications. 6, 99 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitz-Hübsch T, Lux S, Bauer P, Brandt AU, Schlapakow E, Greschus S, Scheel M, Gärtner H, Kirlangic ME, Gras V, Timmann D, Synofzik M, Giorgetti A, Carloni P, Shah JN, Schöls L, Kopp U, Bußenius L, Oberwahrenbrock T, Zimmermann H, Pfueller C, Kadas EM, Rönnefarth M, Grosch AS, Endres M, Amunts K, Paul F, Doss S, Minnerop M, Spinocerebellar ataxia type 14: refining clinicogenetic diagnosis in a rare adult-onset disorder. Annals of Clinical and Translational Neurology. 8, 774–789 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shirafuji T, Shimazaki H, Miyagi T, Ueyama T, Adachi N, Tanaka S, Hide I, Saito N, Sakai N, Spinocerebellar ataxia type 14 caused by a nonsense mutation in the PRKCG gene. Molecular and Cellular Neuroscience. 98, 46–53 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Shimobayashi E, Kapfhammer JP, A New Mouse Model Related to SCA14 Carrying a Pseudosubstrate Domain Mutation in PKCγ Shows Perturbed Purkinje Cell Maturation and Ataxic Motor Behavior. Journal of Neuroscience. 41, 2053–2068 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trzesniewski J, Altmann S, Jäger L, Kapfhammer JP, Reduced Purkinje cell size is compatible with near normal morphology and function of the cerebellar cortex in a mouse model of spinocerebellar ataxia. Experimental Neurology. 311, 205–212 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Schrenk K, Kapfhammer JP, Metzger F, Altered dendritic development of cerebellar Purkinje cells in slice cultures from protein kinase Cγ-deficient mice. Neuroscience. 110, 675–689 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Ghoumari AM, Wehrlé R, De Zeeuw CI, Sotelo C, Dusart I, Inhibition of Protein Kinase C Prevents Purkinje Cell Death but Does Not Affect Axonal Regeneration. Journal of Neuroscience. 22, 3531–3542 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verbeek DS, Goedhart J, Bruinsma L, Sinke RJ, Reits EA, PKCγ mutations in spinocerebellar ataxia type 14 affect C1 domain accessibility and kinase activity leading to aberrant MAPK signaling. Journal of Cell Science. 121, 2339–2349 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Verbeek DS, Knight MA, Harmison GG, Fischbeck KH, Howell BW, Protein kinase C gamma mutations in spinocerebellar ataxia 14 increase kinase activity and alter membrane targeting. Brain. 128, 436–442 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Takahashi H, Adachi N, Shirafuji T, Danno S, Ueyama T, Vendruscolo M, Shuvaev AN, Sugimoto T, Seki T, Hamada D, Irie K, Hirai H, Sakai N, Saito N, Identification and characterization of PKCγ, a kinase associated with SCA14, as an amyloidogenic protein. Human Molecular Genetics. 24, 525–539 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Nakazono A, Adachi N, Takahashi H, Seki T, Hamada D, Ueyama T, Sakai N, Saito N, Pharmacological induction of heat shock proteins ameliorates toxicity of mutant PKCɣ in spinocerebellar ataxia type 14. Journal of Biological Chemistry. 293, 14758–14774 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seki T, Shimahara T, Yamamoto K, Abe N, Amano T, Adachi N, Takahashi H, Kashiwagi K, Saito N, Sakai N, Mutant γPKC found in spinocerebellar ataxia type 14 induces aggregate-independent maldevelopment of dendrites in primary cultured Purkinje cells. Neurobiology of Disease. 33, 260–273 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Kazanietz MG, Lewin NE, Bruns JD, Blumberg PM, Characterization of the cysteine-rich region of the Caenorhabditis elegans protein Unc-13 as a high affinity phorbol ester receptor. Analysis of ligand- binding interactions, lipid cofactor requirements, and inhibitor sensitivity. Journal of Biological Chemistry. 270, 10777–10783 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Ross BL, Tenner B, Markwardt ML, Zviman A, Shi G, Kerr JP, Snell NE, McFarland JJ, Mauban JR, Ward CW, Rizzo MA, Zhang J, Single-color, ratiometric biosensors for detecting signaling activities in live cells. eLife. 7 (2018), doi: 10.7554/elife.35458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallegos LL, Kunkel MT, Newton AC, Targeting protein kinase C activity reporter to discrete intracellular regions reveals spatiotemporal differences in agonist-dependent signaling. Journal of Biological Chemistry. 281, 30947–30956 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Violin JD, Zhang J, Tsien RY, Newton AC, A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. Journal of Cell Biology. 161, 899–909 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keranen LM, Dutil EM, Newton AC, Protein kinase C is regulated in vivo by three functionally distinct phosphorylations. Current Biology. 5, 1394–1403 (1995). [DOI] [PubMed] [Google Scholar]
- 43.Burns DJ, Bell RM, Protein kinase C contains two phorbol ester binding domains. Journal of Biological Chemistry. 266, 18330–18338 (1991). [PubMed] [Google Scholar]
- 44.Xiao Y, Hsiao TH, Suresh U, Chen HIH, Wu X, Wolf SE, Chen Y, A novel significance score for gene selection and ranking. Bioinformatics. 30, 801–807 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guidato S, Tsai LH, Woodgett J, Miller CCJ, Differential cellular phosphorylation of neurofilament heavy side-arms by glycogen synthase kinase-3 and cydin-dependent kinase-5. Journal of Neurochemistry. 66, 1698–1706 (1996). [DOI] [PubMed] [Google Scholar]
- 46.Lee S, Pant HC, Shea TB, Divergent and convergent roles for kinases and phosphatases in neurofilament dynamics. Journal of Cell Science. 127, 4064–4077 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thornton TM, Pedraza-Alva G, Deng B, Wood CD, Aronshtam A, Clements JL, Sabio G, Davis RJ, Matthews DE, Doble B, Rincon M, Phosphorylation by p38 MAPK as an alternative pathway for GSK3β inactivation. Science. 320, 667–670 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schönwasser DC, Marais RM, Marshall CJ, Parker PJ, Activation of the Mitogen-Activated Protein Kinase/Extracellular Signal-Regulated Kinase Pathway by Conventional, Novel, and Atypical Protein Kinase C Isotypes. Molecular and Cellular Biology. 18, 790–798 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang DW, Sherman BT, Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Huang DW, Sherman BT, Lempicki RA, Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N, The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discovery. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N, Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling. 6, 1–1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, Staudt LM, Toward a Shared Vision for Cancer Genomic Data. New England Journal of Medicine. 375, 1109–1112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vlak MHM, Sinke RJ, Rabelink GM, Kremer BPH, van de Warrenburg BPC, Novel PRKCG/SCA14 mutation in a Dutch spinocerebellar ataxia family: Expanding the phenotype. Movement Disorders. 21, 1025–1028 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Ganos C, Zittel S, Minnerop M, Schunke O, Heinbokel C, Gerloff C, Zühlke C, Bauer P, Klockgether T, Münchau A, Bäumer T, Clinical and neurophysiological profile of four German families with spinocerebellar ataxia type 14. Cerebellum. 13, 89–96 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Chelban V, Wiethoff S, Fabian-Jessing BK, Haridy NA, Khan A, Efthymiou S, Becker EBE, O’Connor E, Hersheson J, Newland K, Hojland AT, Gregersen PA, Lindquist SG, Petersen MB, Nielsen JE, Nielsen M, Wood NW, Giunti P, Houlden H, Genotype-phenotype correlations, dystonia and disease progression in spinocerebellar ataxia type 14. Movement Disorders. 33, 1119–1129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stevanin G, Hahn V, Lohmann E, Bouslam N, Gouttard M, Soumphonphakdy C, Welter ML, Ollagnon-Roman E, Lemainque A, Ruberg M, Brice A, Durr A, Mutation in the catalytic domain of protein kinase C γ and extension of the phenotype associated with spinocerebellar ataxia type 14. Archives of Neurology. 61, 1242–1248 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Klebe S, Durr A, Rentschler A, Hahn-Barma V, Abele M, Bouslam N, Schöls L, Jedynak P, Forlani S, Denis E, Dussert C, Agid Y, Bauer P, Globas C, Wüllner U, Brice A, Riess O, Stevanin G, New mutations in protein kinase Cγ associated with spinocerebellar ataxia type 14. Annals of Neurology. 58, 720–729 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Chen D, Cimino P, Ranum L, Zoghbi H, Yabe I, Schut L, Margolis R, Lipe H, Feleke A, Matsushita M, Wolff J, Morgan C, Lau D, Fernandez M, Sasaki H, Raskind W, Bird T, The clinical and genetic spectrum of spinocerebellar ataxia 14. Neurology. 64, 1258–60 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Van AAN, Kunkel MT, Baffi TR, Lordén G, Antal CE, Banerjee S, Newton AC, Protein kinase C fusion proteins are paradoxically loss of function in cancer. Journal of Biological Chemistry. 296, 1–18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kannan N, Haste N, Taylor SS, Neuwald AF, The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proceedings of the National Academy of Sciences of the United States of America. 105 (2008), p. 9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leonard TA, Róycki B, Saidi LF, Hummer G, Hurley JH, Crystal structure and allosteric activation of protein kinase C βiI. Cell. 144, 55–66 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hansra G, Garcia-Paramio P, Prevostel C, Whelan RDH, Bornancin F, Parker PJ, Multisite dephosphorylation and desensitization of conventional protein kinase C isotypes. Biochemical Journal. 342, 337–344 (1999). [PMC free article] [PubMed] [Google Scholar]
- 64.Chen D, Gould C, Garza R, Gao T, Hampton RY, Newton AC, Amplitude control of protein kinase C by RINCK, a novel E3 ubiquitin ligase. Journal of Biological Chemistry. 282, 33776–33787 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Leontieva OV, Black JD, Identification of Two Distinct Pathways of Protein Kinase Cα Down-regulation in Intestinal Epithelial Cells. Journal of Biological Chemistry. 279, 5788–5801 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Ji J, Hassler ML, Shimobayashi E, Paka N, Streit R, Kapfhammer JP, Increased protein kinase C gamma activity induces Purkinje cell pathology in a mouse model of spinocerebellar ataxia 14. Neurobiology of Disease. 70, 1–11 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Asai H, Hirano M, Shimada K, Kiriyama T, Furiya Y, Ikeda M, Iwamoto T, Mori T, Nishinaka K, Konishi N, Udaka F, Ueno S, Protein kinase Cγ, a protein causative for dominant ataxia, negatively regulates nuclear import of recessive-ataxia-related aprataxin. Human Molecular Genetics. 18, 3533–3543 (2009). [DOI] [PubMed] [Google Scholar]
- 68.Yuan A, Rao MV, Veeranna RA Nixon, Neurofilaments at a glance. Journal of Cell Science. 125, 3257–3263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahn JH, McAvoy T, V Rakhilin S, Nishi A, Greengard P, Nairn AC, Protein kinase A activates protein phosphatase 2A by phosphorylation of the B56δ subunit. Proceedings of the National Academy of Sciences of the United States of America. 104, 2979–2984 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ahn JH, Kim Y, Kim HS, Greengard P, Nairn AC, Protein kinase C-dependent dephosphorylation of tyrosine hydroxylase requires the B56δ heterotrimeric form of protein phosphatase 2A. PLoS ONE. 6 (2011), doi: 10.1371/journal.pone.0026292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goode N, Hughes K, Woodgett JR, Parker PJ, Differential regulation of glycogen synthase kinase-3β by protein kinase C isotypes. Journal of Biological Chemistry. 267, 16878–16882 (1992). [PubMed] [Google Scholar]
- 72.Nibbeling EAR, Duarri A, Verschuuren-Bemelmans CC, Fokkens MR, Karjalainen JM, Smeets CJLM, De Boer-Bergsma JJ, Van Der Vries G, Dooijes D, Bampi GB, Van Diemen C, Brunt E, Ippel E, Kremer B, Vlak M, Adir N, Wijmenga C, Van De Warrenburg BPC, Franke L, Sinke RJ, Verbeek DS, Exome sequencing and network analysis identifies shared mechanisms underlying spinocerebellar ataxia. Brain. 140, 2860–2878 (2017). [DOI] [PubMed] [Google Scholar]
- 73.Prestori F, Moccia F, D’angelo E, Disrupted calcium signaling in animal models of human spinocerebellar ataxia (SCA). International Journal of Molecular Sciences. 21 (2020), doi: 10.3390/ijms21010216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsumagari R, Kakizawa S, Kikunaga S, Fujihara Y, Ueda S, Yamanoue M, Saito N, Ikawa M, Shirai Y, DGKγ Knock-Out Mice Show Impairments in Cerebellar Motor Coordination, LTD, and the Dendritic Development of Purkinje Cells through the Activation of PKCγ. eNeuro. 7 (2020), doi: 10.1523/ENEURO.0319-19.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lorden G, Wozniak J, Dozier L, Cates-Gatto C, Patrick G, Roberts A, in press, doi: 10.21203/rs.3.rs-894083/v1. [DOI]
- 76.Gallegos LL, Newton AC, Genetically encoded fluorescent reporters to visualize protein kinase C activation in live cells. Methods in Molecular Biology. 756, 295–310 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Orr JW, Keranen LM, Newton AC, Reversible exposure of the pseudosubstrate domain of protein kinase C by phosphatidylserine and diacylglycerol. Journal of Biological Chemistry. 267, 15263–15266 (1992). [PubMed] [Google Scholar]
- 78.Keranen LM, Newton AC, Ca2+ differentially regulates conventional protein kinase Cs’ membrane interaction and activation. Journal of Biological Chemistry. 272, 25959–25967 (1997). [DOI] [PubMed] [Google Scholar]
- 79.Lapek JD, Lewinski MK, Wozniak JM, Guatelli J, Gonzalez DJ, Quantitative temporal viromics of an inducible HIV-1 model yields insight to global host targets and phospho-dynamics associated with protein Vpr. Molecular and Cellular Proteomics. 16, 1447–1461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, UCSF Chimera - A visualization system for exploratory research and analysis. Journal of Computational Chemistry. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 81.Šali A, Blundell TL, Comparative Protein Modelling by Satisfaction of Spatial Restraints. Journal of Molecular Biology. 234 (1993), pp. 779–815. [DOI] [PubMed] [Google Scholar]
- 82.Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, Beglov D, Vajda S, The ClusPro web server for protein-protein docking. Nature Protocols. 12, 255–278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The phosphoproteomics data have been depostied on UCSD CCMS MassIVE with the accession number: MSV000088896. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.