Abstract
Autophagy is a process of eliminating damaged or unnecessary proteins and organelles, thereby maintaining intracellular homeostasis. Deregulation of autophagy is associated with several diseases including cancer. Contradictory dual roles of autophagy have been well established in cancer. Cytoprotective mechanism of autophagy has been extensively investigated for overcoming resistance to cancer therapies including radiotherapy, targeted therapy, immunotherapy, and chemotherapy. Selective autophagy inhibitors that directly target autophagic process have been developed for cancer treatment. Efficacies of autophagy inhibitors have been tested in various pre-clinical cancer animal models. Combination therapies of autophagy inhibitors with chemotherapeutics are being evaluated in clinal trials. In this review, we will focus on genetical and pharmacological perturbations of autophagy-related proteins in different steps of autophagic process and their therapeutic benefits. We will also summarize combination therapies of autophagy inhibitors with chemotherapies and their outcomes in pre-clinical and clinical studies. Understanding of current knowledge of development, progress, and application of cytoprotective autophagy inhibitors in combination therapies will open new possibilities for overcoming drug resistance and improving clinical outcomes.
Keywords: Autophagy, Autophagy inhibitor, Anticancer agent, Resistance, Combination therapy
INTRODUCTION
Macroautophagy (hereafter referred to as autophagy) is a highly conserved catabolic process by which damaged or unnecessary proteins or organelles are delivered to lysosomes for degradation, leading to maintenance of intracellular homeostasis (Levy et al., 2017). Autophagic process involves formation of double-membraned vesicles known as autophagosomes that can engulf proteins and organelles prior to delivery to lysosome (Mizushima, 2007; Mizushima et al., 2011). Autophagy occurs at a basal level in all cells. It is induced by various signals and cellular stresses such as hypoxia, starvation, and different cancer therapies as a cytoprotective mechanism. Autophagy has a context-dependent role in cancer. It is closely related to the occurrence and drug resistance of cancer (Eskelinen, 2011; Towers and Thorburn, 2016; Chang and Zou, 2020). Autophagy can limit oxidative stress, chronic tissue damage, and oncogenic signaling by preventing toxic accumulation of damaged proteins and organelles, particularly mitochondria, thereby inhibiting tumorigenesis in the early stage of tumor formation (White et al., 2015). In contrast, some cancers are dependent on autophagy for survival by using autophagy-mediating recycling to maintain mitochondria function and energy homeostasis because of elevated metabolic demand of cancer growth. In established tumors, autophagy can be induced as a response to nutrient deprivation, energy deficits, hypoxia, and chemotherapeutics drugs, finally resulting in acquired resistance in tumors. Some tumor cell types with high basal autophagic flux might show intrinsic drug resistance. Conversely, persistent or excessive autophagy can induce autophagic cell death in cancer therapy (Puissant et al., 2010; Aryal et al., 2014). Clinical interventions to manipulate autophagy in cancer treatment are underway by mainly focusing on inhibiting autophagy, although such interventions are in contradiction with dual roles of autophagy.
In this review, we will focus on application of autophagy inhibitors in cancer treatment. First, we will provide important targets of autophagic process and impact of autophagy-related gene deficiency in genetically engineered mice as basic information. Pharmacological inhibitors developed for targeting autophagic process and their efficacies in preclinical and clinical studies are then reviewed. Specially, we will focus on combination therapies of autophagy inhibitors with chemotherapeutic agents to improve therapeutic benefits of current cancer therapies.
CORE PROCESS OF AUTOPHAGY
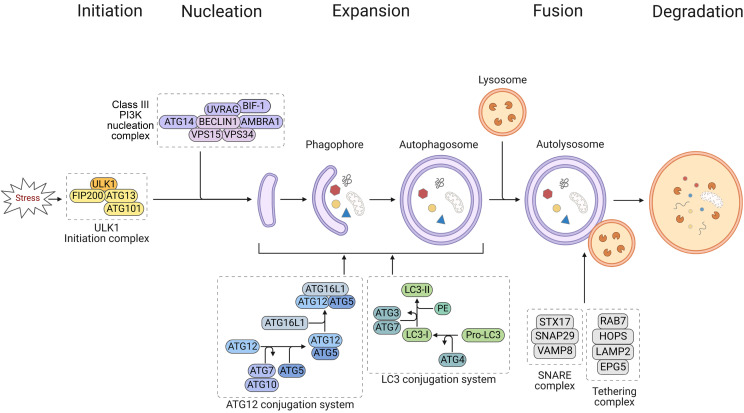
Autophagic process can be divided into distinct stages: initiation, nucleation of the autophagosome, expansion of the autophagosome membrane and maturation, docking and fusion with the lysosome for cargo degradation, and degradation (Fig. 1). Formation and turnover of autophagosome are executed by highly conserved autophagy-related (ATG) proteins (Mizushima et al., 2011). Initiating signals of autophagy to form the autophagosome originate from activated Unc-51-like kinase 1 (ULK1, human homolog of yeast ATG1). ULK1 forms a pre-initiation complex with ATG13, ATG101, and focal adhesion kinase family-interacting protein 200 (FIP200) under stress conditions (Carlsson and Simonsen, 2015). ULK1 initiation complex then recruits class III PI3K nucleation complex composed of BECLIN 1 (human homolog of yeast ATG6), ATG14, type III phosphatidylinositol 3-kinase/vacuolar protein sorting 34 (class III PI3K/VPS34), autophagy and BECLIN 1 regulator-1 (AMBRA1), and UV radiation resistance-associated gene protein (UVRAG) by phosphorylating BECLIN 1 to activate autophagy-specific VPS34 (Russell et al., 2013). BIF-1 is also involved in the formation of nucleation complex by binding to UVRAG (Takahashi et al., 2007). The formed phosphatidylinositol 3-phosphate (PI3P)-binding complex can then direct distribution of the machinery that enable autophagosome formation (Hansen et al., 2018). In the ATG12 conjugation system, ATG12 is attached to ATG5, which is then attached to ATG16L1, followed by dimerization and interaction with the PI3P-binding complex through WD repeat domain phosphoinositide-interacting proteins (WIPIs; ATG18 in yeast) and zinc-finger FYVE domain-containing protein 1 (DFCP1). Under catalysis of E1-like enzyme ATG7 and E2-like enzyme ATG10, the formed ATG5/ATG12/ATG16L1 (E3) complex can facilitate recruitment and conversion of precursor pro-microtubule-associated protein 1 light chain 3 (LC3; ATG8 in yeast) to membrane bound LC3-II form (LC3 conjugation). In the LC3 conjugation system, pro-LC3 is cleaved by protease ATG4 to form cytosolic LC3-I, which is then recognized by E1-like enzyme ATG7 and E2-like enzyme ATG3, leading to conjugation with phosphatidylethanolamine (PE) to form LC3-II (Ichimura et al., 2000; Kabeya et al., 2000; Hanada et al., 2007; Aman et al., 2021). This conjugation is incorporated into pre-autophagosomal and autophagosomal membranes, where LC3 can interact with cargo receptors such as sequestosome 1 (SQSTM1/p62) and neighbor of BRCA1 gene 1 (Nbr1) carrying LIRs (LC3-interacting regions) to target them for autophagic degradation (Birgisdottir et al., 2013; Slobodkin and Elazar, 2013). LC3-II is widely used as a marker for assessing autophagy due to its abundance in autophagosomal membranes (Schaaf et al., 2016). Following expansion and maturation, LC3-I is released from autophagosomes by deconjugation through the action of ATG4 (Tanida et al., 2004). Sealed autophagosome then merges with lysosome through assistance of SNARE complex (STX17, SNAP29, VAMP8) and tethering complex (HOPS complex, RAB7, EPG5, and LAMP2) to form autolysosome (Yu et al., 2018; Xiao et al., 2021). Sequestered autophagic bodies and the inner membrane are then released into the lumen, where they are exposed to acidic hydrolases and lipases for degradation (Mizushima, 2007). Finally, autophagy is completed by allowing the resulting macromolecules to be recycled for reuse in the biosynthesis of essential components required for survival under stress conditions (Yorimitsu and Klionsky, 2005).
Fig. 1.
Schematic overview of core autophagic process.
PHENOTYPES OF AUTOPHAGY-DEFICIENT MICE
Atg gene knockout mice are useful for understanding physiological roles of autophagy in vivo. Among core Atg genes involved in autophagosome formation in mammals, 25 of them have been knocked out in mice (Table 1). Three mortality patterns of Atg knockout mice have been observed. Some die at embryonic period. Some die within 1 d after birth. Some show vitality without obvious abnormalities. Atg gene (such as Atg4a, Atg4b, Atg4c, Lc3b, Ulk1, and Ulk2) knockout mice show no obvious defective phenotypes with close-to-normal development partly due to functional redundancy (Cann et al., 2008; Kundu et al., 2008; Marino et al., 2010; Lee and Tournier, 2011; Groza et al., 2022) (International Mouse Phenotyping Consortium [IMPC], https://www.mousephenotype.org/). However, Ulk1/Ulk2 double knockout mice show neonatal lethality (Cheong et al., 2014). Mice with deletion of nonredundant genes (Atg3, Atg5, Atg7, Atg12, Atg14, and Atg16l1) involved in ATG12 and LC3 conjugation systems after nucleation stage show neonatal lethality (Kuma et al., 2004; Komatsu et al., 2005; Saitoh et al., 2008; Sou et al., 2008; Malhotra et al., 2015) (https://www.mousephenotype.org/), whereas mice with knockout of nonredundant genes (Fip200, Atg13, Beclin 1, Vps15, Vps34, Uvrag, and Ambra1) involved in earlier stages before expansion stage are embryonic lethal (Yue et al., 2003; Gan et al., 2006; Fimia et al., 2007; Zhou et al., 2011; Nemazanyy et al., 2013; Afzal et al., 2015; Kaizuka and Mizushima, 2016). Among genes involved in fusion stage, knockout of Snap29 gene encoding a component of SNARE complex shows neonatal lethality and deletion of Vamp8 gene encoding another component of SNARE complex shows partial lethality and growth retardation with defect in secretion of the pancreas (Wang et al., 2004; Schiller et al., 2016). Deletion of genes (Lamp2 and Epg5) of the tethering complex shows partial lethality, retardation, or reduced survival (Tanaka et al., 2000; Zhao et al., 2013). Loss of Rab7 gene encoding another factor of the tethering complex leads to embryonic lethality (Kawamura et al., 2012). Thus, most autophagy-related genes are very important for embryonic and neonatal development. Reasons for phenotypic differences between whole-body knockout mice of Atg genes are still questionable. No genetic study has been reported for Atg101, Atg10, or Stx17 gene.
Table 1.
In vivo genetic studies of autophagy-related genes
| Autophagic stage | Target | Genetic modification | Phenotypes | Reference |
|---|---|---|---|---|
| Initiation | ULK1 | KO | Viable without overt development defects; delayed mitochondrial clearance in reticulocytes | Kundu et al., 2008 |
| ULK2 | KO | Normal development and fertile | Lee and Tournier, 2011 | |
| ATG13 | KO | Embryonic lethality with growth retardation and myocardial growth defects (E17.5) | Kaizuka and Mizushima, 2016 | |
| ATG101 | N/A | |||
| FIP200 | KO | Embryonic lethality with defective heart and liver development (E14.5) | Gan et al., 2006 | |
| Nucleation | BECLIN 1 | KO | Embryonic lethality (E8.5) High incidence of spontaneous tumors as in heterozygote |
Yue et al., 2003 |
| ATG14 | KO | Neonatal lethality | https://www.mousephenotype.org/ | |
| VPS15 | KO | Embryonic lethality (E7.5) | Nemazanyy et al., 2013 | |
| VPS34 | KO | Embryonic lethality with abnormal embryogenesis (E7.5-E8.5) | Zhou et al., 2011 | |
| UVRAG | KO | Embryonic lethality (E7.5) | Afzal et al., 2015 | |
| BIF-1 | KO | Normal development and high incidence of spontaneous tumorigenesis | Takahashi et al., 2007 | |
| AMBRA1 | KO | Embryonic lethality with neural tube defect (E18.5*) | Fimia et al., 2007 | |
| Expansion | LC3B | KO | Normal development and fertile | Cann et al., 2008 |
| ATG3 | KO | Neonatal lethality (1 d) | Sou et al., 2008 | |
| ATG4A | KO | Close-to-normal development | https://www.mousephenotype.org/ | |
| ATG4B | KO | Close-to-normal development | Marino et al., 2010 | |
| ATG4C | KO | Close-to-normal development | https://www.mousephenotype.org/ | |
| ATG5 | KO | Neonatal lethality (1 d) | Kuma et al., 2004 | |
| ATG7 | KO | Neonatal lethality (1 d) | Komatsu et al., 2005 | |
| ATG10 | N/A | |||
| ATG12 | KO | Neonatal lethality (1 d) | Malhotra et al., 2015 | |
| ATG16L1 | KO | Neonatal lethality (1 d) | Saitoh et al., 2008 | |
| Fusion | STX17 | N/A | ||
| SNAP29 | KO | Neonatal lethality with ichthyotic phenotype (1 d) | Schiller et al., 2016 | |
| VAMP8 | KO | Partial lethality and growth retardation with defect in secretion of the pancreas | Wang et al., 2004 | |
| RAB7 | KO | Embryonic lethality (E7-8) | Kawamura et al., 2012 | |
| LAMP2 | KO | Increased mortality between (20-40 d) with cardio-myopathy | Tanaka et al., 2000 | |
| EPG5 | KO | Growth retardation and reduced survival with selective neuronal vulnerability to degeneration | Zhao et al., 2013 |
N/A, not available; *, available at https://www.mousephenotype.org/.
While biallelic deletion of Beclin 1 gene in mice shows embryonic lethality, monoallelic deletion of Beclin 1 shows normal development with high incidence of spontaneous tumorigenesis and reduced autophagy, indicating that Beclin 1 gene is essential for early embryonic development and a haploinsufficient tumor suppressor (Qu et al., 2003; Yue et al., 2003). In case of Bif-1 gene, biallelic deletion in whole body leads to normal development with high incidence of spontaneous tumorigenesis (Takahashi et al., 2007). Mice with systemic mosaic deletion of Atg5 and mice with liver-specific Atg7 homologous knockout also develop benign liver adenomas, which originate from autophagy-deficient hepatocytes (Takamura et al., 2011). Thus, some Atg genes are necessary for suppression of spontaneous tumorigenesis through a cell-intrinsic protective mechanism. Conversely, autophagy can promote tumor growth by suppressing p53 response, maintaining mitochondrial function, sustaining metabolic homeostasis and survival during stress, and preventing progression of tumor to benign oncocytomas (Kimmelman, 2011; Guo et al., 2013b). Deletion of Atg5 or Atg7 gene in KRAS-transformed cells with proficient autophagy can impair their tumorigenicity by failing to maintain levels of tricarboxylic acid cycle metabolite and mitochondrial respiration under nutrient starvation, which creates an energy crisis that threatens survival (Guo et al., 2011; Yang et al., 2011). Deletion of Atg7 also alters progression of lung cancer cells with KRAS (G12D) and Trp53 mutations by developing into oncocytomas instead of adenomas and carcinomas with suppressed proliferation and reduced tumor burden (Guo et al., 2013a). These observations support that autophagy plays a double-edged sword role in suppressing tumor initiation and in promoting survival and growth of tumors.
PHARMACOLOGICAL INHIBITORS DIRECTLY TARGETING AUTOPHAGY FORMATION
The process of autophagy is divided into four distinct stages (Fig. 1). Each stage has potential targets for inhibiting autophagy. Pharmacological inhibitors that target tumor growth and autophagy formation are summarized in Table 2. At the initiation stage, ULK1 has been mainly studied to develop inhibitors to interfere with growth of various cancer types including lung cancer and leukemia both in vitro and in vivo (Tang et al., 2017; Qiu et al., 2020). SBI-0206965, MRT68921, and ULK101 have been found as ULK1 kinase inhibitors showing cytotoxicity against various cancer cells in vitro (Tang et al., 2017; Martin et al., 2018; Chen et al., 2020; Qiu et al., 2020). In case of MRT68921, its effects on tumor growth inhibition and prolonged survival have been shown in H460 lung cancer and MNK45 gastric cancer xenografted animal models. It has dual targets, NUAK1 and ULK1 (Martin et al., 2018).
Table 2.
Autophagy inhibitors targeting core autophagic process
| Autophagic stage |
Target | Inhibitor | Cancer type (cell lines) | Working concentrations/in vivo dose | Mechanism of autophagy inhibition | Inhibitory effects on cancer | Reference | |||
|---|---|---|---|---|---|---|---|---|---|---|
| In vitro | In vivo | |||||||||
| Cyto- toxicity |
Tumor growth | Survival | ||||||||
| Initiation | ULK1/2 | SBI-0206965 | Non-small cell lung cancer (A549, H460, HCC827) | 0.01-100 μM | Inhibits ULK1 kinase activity (Egan et al., 2015) | + | ND | ND | Tang et al., 2017 | |
| Acute myeloid leukemia (HL60, U937) | 2-15 μM | + | ND | ND | Qiu et al., 2020 | |||||
| MRT68921 | Various cancer (A549, H460, H1299, SW480, SW620, U251, U266, HT-29, HCT-116, Colo320, PC-3, MNK45, 4T1) | 1.76-8.91 μM (IC50)/ 10-40 mg/kg/d, 7 times |
Inhibits ULK1 kinase activity (Petherick et al., 2015) | + | + | + | Chen et al., 2020 | |||
| ULK101 | Various cancer (A549, U2OS, H838, H727, H2030) | 11.3-56.8 μM (IC50) | Inhibits ULK1 kinase activity (Martin et al., 2018) | + | ND | ND | Martin et al., 2018 | |||
| Nucleation | VPS34 | 3-MA | Uterine sarcoma (FU-MMT-1, MES-SA) | 200-300 μM/ 15 mg/kg/d, 7 times |
Inhibits VPS34 kinase activity (Petiot et al., 2000) | + | + | + | Chen and Yao, 2021 | |
| SAR405 | Various cancer (B16-F10, CT26, Renca, YUMM) | 10 mg/kg/d, 10 times | Inhibits VPS34 kinase activity (Ronan et al., 2014) | ND | + | + | Noman et al., 2020 | |||
| SB02024 | Various cancer (B16-F10, CT26, Renca, YUMM) | 20 mg/kg/d, 10 times | Inhibits VPS34 kinase activity (Dyczynski et al., 2018) | ND | + | + | Noman et al., 2020 | |||
| Breast cancer (MCF-7, MDA-MB-231) | 0.1-100 μM/ 20-50 mg/kg/d, 21-30 times |
+ | + | ND | Dyczynski et al., 2018 | |||||
| VPS34-IN1 | Acute myeloid leukemia (HL60, U937, K562, MOLM-14, MV4-11, THP1, KASUMI, OCI-AML2, OCI-AML3) | 1.4-9.6 μM (IC50) | Inhibits VPS34 kinase activity (Bago et al., 2014) | + | ND | ND | Meunier et al., 2020 | |||
| VPS34-IN2 (PIK-III) | Not studied on tumor cell growth and death | Inhibits VPS34 kinase activity (Dowdle et al., 2014) | ||||||||
| Expansion | ATG4B | NSC185058 | Glioblastoma (Patient-derived glioma stem-like cell JK83, 23) | 10-100 μM/ 150 mg/kg/d, 9 times |
Inhibits ATG4B protease activity (Akin et al., 2014) | + | + | + | Huang et al., 2017 | |
| Osteosarcoma (Saos-2) | 3-100 μM/ 100 mg/kg, ~15 times |
+ | + | ND | Akin et al., 2014 | |||||
| UAMC-2526 | Colorectal cancer (HT-29) | 1-100 μM | Inhibits ATG4B protease activity (Kurdi et al., 2017) | + | + | ND | Kurdi et al., 2017 | |||
| FMK-9a | Ovarian cancer (HeLa) | 100 μM | Inhibits ATG4B protease activity (Qiu et al., 2016) | + | ND | ND | Chu et al., 2018b | |||
| S130 | Various cancer (HeLa, HCT116, HL60) | 4.7-16.1 μM (IC50)/ 20 mg/kg/d, 21 times |
Inhibits ATG4B activity (Fu et al., 2019) | + | + | ND | Fu et al., 2019 | |||
| Tioconazole | Various cancer (H4, HCT116, MDA-MB-231) | 40 μM/60 mg/kg/d, ~9 times | Inhibits ATG4B protease activity (Liu et al., 2018) | + | + | ND | Liu et al., 2018 | |||
| Breast cancer (MCF-7) | 14.5 μM (IC50)/ 60 mg/kg/d, ~12 times |
+ | + | + | El-Gowily et al., 2021 | |||||
| Fusion | STX17 | EACC | Not studied on tumor cell growth and death | Inhibits translocation of Stx17 onto autophagosome (Vats and Manjithaya, 2019) | ||||||
| Lysosome | Chloroquine | Hepatocellular carcinoma (HepG2, Huh7) | 5-80 μM/ 160 mg/kg/d, ~15 times |
Deacidifies lysosome and increases lysosomal membrane permeability (Homewood et al., 1972) | + | + | ND | Hu et al., 2016 | ||
| Breast cancer (MCF-7) | 32.5 μM (IC50)/ 50 mg/kg/d, ~12 times |
+ | + | + | El-Gowily et al., 2021 | |||||
| Hydroxy-chloroquine | Glioblastoma (LN18, LN229) | ~15 μM | + | ND | ND | Liu et al., 2019 | ||||
| Hepatocellular carcinoma (HepG2, Huh7) |
12.69-13.60 μM (IC50)/ 30 mg/kg/d, 20 times |
+ | + | ND | Chen et al., 2021 | |||||
| Bafilomycin A1 | Gastric cancer (SGC-7901) | 0.1 μM | Deacidifies lysosome by inhibiting the lysosomal V-ATPase (Yamamoto et al., 1998) | + | ND | ND | Li et al., 2016 | |||
| ROC-325 | Acute myeloid leukemia (MV4-11, HL-60, KG-1, MOLM-13, NOMO-1, PL-21) | 1-10 μM/ 50 mg/kg, ~12 times |
Deacidifies lysosome and increases lysosomal membrane permeability (Carew et al., 2017) | + | + | + | Nawrocki et al., 2019 | |||
| Various cancer (A498, A549, CFPAC-1, COLO-205, DLD-1, IGROV-1, MCF-7, MiaPaCa-2, NC1-H69, PC-3, RL, UACC-62, 786-O, Caki-2, Achn) | 4.6-11 μM (IC50)/ 25-50 mg/kg/d, 30 times |
+ | + | ND | Carew et al., 2017 | |||||
| LS-1-10 | Colon cancer (LoVo, DLD1, HT29, HCT116, SW480) | 0.82-1.31 μM (IC50)/ 40-80 mg/kg/d, 15 times |
Increases lysosomal membrane permeability (Fu et al., 2017) | + | + | ND | Fu et al., 2017 | |||
| BRD1240 | Not studied on tumor cell growth and death | Inhibits lysosomal acidification (Aldrich et al., 2015) | ND | |||||||
| Cytochalasin E | Lung cancer (A549) | 0.25-1 μM | Deacidifies lysosome and increases lysosomal membrane permeability (Takanezawa et al., 2018) | + | ND | ND | Takanezawa et al., 2018 | |||
| Lys05 | Various cancer (LN229, C8161, 1205Lu, HT-29) | 3.6-7.9 μM (IC50)/ 10-80 mg/kg, ~7 times |
Deacidifies lysosome and increases lysosomal membrane permeability (McAfee et al., 2012; Zhou et al., 2020) | + | + | ND | McAfee et al., 2012 | |||
| Glioblastoma (U251, LN229) | 6.0-9.1 μM (IC50) | + | ND | ND | Zhou et al., 2020 | |||||
| DC661 | Hepatocellular carcinoma (Hep3B, Hep1-6) | 0.5-0.6 μM (IC50)/ 3 mg/kg/d, 21 times |
Increases lysosomal membrane permeability by inhibiting PPT1 (Xu et al., 2022) | + | + | + | Xu et al., 2022 | |||
| Various cancer (A375P, WM3918, WM983B, PANC1, HT-29) | 0.1-1 μM/ 3 mg/kg/d, 10 times |
+ | + | ND | Rebecca et al., 2019 | |||||
At the nucleation stage, VPS34/class III PI3K has been extensively studied as a main target for autophagy formation. Several kinase inhibitors including 3-MA, SAR405, SB02024, and VPS34-IN1 have been developed for inhibiting autophagy and tumor growth as shown in Table 2. These compounds show a good relevance to inhibition of autophagy formation and suppression of in vitro tumor cell growth in several cancer types including breast cancer and leukemia. 3-MA, SAR405, and SB02024 show inhibitory effects on tumor growth in vivo with extended survival in xenograft animal models (Dyczynski et al., 2018; Noman et al., 2020; Chen and Yao, 2021).
At the expansion stage, only ATG4B protease has been targeted for the development of autophagy inhibitors. NSC185058, S130, and tioconazole have shown autophagy formation-inhibiting and tumor-suppressive effects on various cancers including glioblastoma and colorectal cancer both in vitro and in vivo (Akin et al., 2014; Huang et al., 2017; Liu et al., 2018; Fu et al., 2019; El-Gowily et al., 2021). NSC185058 has been found to prolong survival of JK83 primary cancer-xenografted mice (Huang et al., 2017). Tioconazole has a survival benefit in MCF-7 breast cancer-xenografted mice (El-Gowily et al., 2021). UAMC-2526 can dose-dependently inhibit HT-29 cancer cells with potent inhibition of autophagy (Kurdi et al., 2017). FMK-9a has a weak cytotoxicity to HeLa cells although it can potently inhibit autophagy formation (Chu et al., 2018b). Its in vivo efficacy has not been reported yet.
At the fusion stage, EACC can inhibit STX17, resulting in inhibition of autolysosome formation (Vats and Manjithaya, 2019). At present, its inhibitory effect on tumor growth has not been reported yet. Several other inhibitors can also inhibit autolysosome formation. Among them, chloroquine (CQ) and hydroxychloroquine (HCQ) originally developed as anti-malaria drugs have been found to be able to inhibit autolysosome formation by increasing lysosomal pH and lysosomal membrane permeability (Homewood et al., 1972). They can also inhibit autophagic flux by decreasing autophagosome-lysosome fusion presumably by interfering with SNAP29 recruitment (Mauthe et al., 2018). Both compounds have been intensively studied for inhibiting tumor growth of many cancer types. They also have beneficial effects by prolonging survival in preclinical animal models (Hu et al., 2016; Liu et al., 2019; Chen et al., 2021; El-Gowily et al., 2021). Besides these inhibitors, bafilomycin A1, ROC-325, LS-1-10, BRD1240, cytochalasin E, Lys05, and DC661 can also inhibit autolysosome formation during autophagic process. Bafilomycin A1, ROC-325, LS-1-10, BRD1240, cytochalasin E, Lys05, and DC661 compounds can inhibit autolysosome formation by elevating lysosomal pH (Yamamoto et al., 1998; McAfee et al., 2012; Aldrich et al., 2015; Carew et al., 2017; Fu et al., 2017; Takanezawa et al., 2018; Zhou et al., 2020; Xu et al., 2022). DC661 is a dimeric CQ derivative (Xu et al., 2022). ROC-325, LS-1-10, Lys05, and DC661 have been tested as possible cancer therapeutics in pre-clinical animal models. It was found that they could suppress tumor growth in several cancer models (McAfee et al., 2012; Carew et al., 2017; Fu et al., 2017; Nawrocki et al., 2019; Rebecca et al., 2019; Xu et al., 2022). ROC-325 and DC661 have a survival benefit in MV4-11 acute myeloid leukemia or Hep1-6 hepatocellular carcinoma (HCC) xenografted mice (Nawrocki et al., 2019; Xu et al., 2022).
So far, ULK1/2, VPS34, ATG4B, and fusion with lysosome have been mainly targeted to develop inhibitors to block autophagic process. Their inhibitors can suppress tumor growth and prolong survival in various cancers in preclinical setting. Notably, most autophagy inhibitors have been developed to block activities of enzymes such as kinase and protease rather than protein-protein interactions except for inhibitors blocking autophagosome fusion with lysosome.
SYNERGISTIC EFFECTS OF AUTOPHAGY INHIBITORS WITH ANTI-CANCER DRUGS IN PRE-CLINICAL STUDIES
Combination of autophagy inhibitors with various anti-cancer therapeutics have been tested in various cancer cell lines and pre-clinical cancer animal models to increase their efficacies (Table 3). In the initiation stage, SBI-0206965, a ULK1 inhibitor, showed an anti-tumor effect on non-small cell lung cancer (NSCLC) cells (Tang et al., 2017). It has been reported that SBI-0206965 can sensitize these cells to cisplatin (a platinum-based chemotherapeutic agent causing DNA damage) by modulating both autophagy and apoptosis pathways. The sensitivity of acute myeloid leukemia (AML) cell lines to daunorubicin (a DNA alkylating agent) can also be enhanced by SBI-0206965 (Qiu et al., 2020).
Table 3.
Synergistic effects of autophagy inhibitors with anti-cancer drugs in pre-clinical models
| Autophagic stage | Autophagy inhibitor |
Anticancer drug | Cancer type (cell lines) | Synergistic effect | Reference | |
|---|---|---|---|---|---|---|
| In vitro | In vivo | |||||
| Initiation | SBI-0206965 | Cisplatin | Non-small cell lung cancer (A549, H460) | + | ND | Tang et al., 2017 |
| Daunorubicin | Acute myeloid leukemia (HL60, U937) | + | ND | Qiu et al., 2020 | ||
| Nucleation | 3-MA | Apatinib | Osteosarcoma (KHOS) | + | + | Liu et al., 2017a |
| Uterine sarcoma cancer (MES-SA, FU-MMT-1) |
+ | +* | Chen and Yao, 2021 | |||
| Gefitinib | Triple negative breast cancer (MDA-MB-468, MDA-MB-231) | + | + | Liu et al., 2017b | ||
| Sorafenib, Vorinostat | Hepatocellular carcinoma (Hep3B, HepG2, PLC/PRF/5) | + | ND | Yuan et al., 2014 | ||
| Cisplatin | Ovarian cancer (A2780, OVCAR3) | + | ND | Zhang et al., 2012 | ||
| SB02024 | Sunitinib, Erlotinib | Breast cancer (MDA-MB-231, MCF-7) | + | ND | Dyczynski et al., 2018 | |
| Anti-PD-1, Anti-PD-L1 | Various tumor (B16-F10, CT26) | ND | +* | Noman et al., 2020 | ||
| SAR405 | Anti-PD-1, Anti-PD-L1 | Various tumor (B16-F10, CT26) | ND | +* | Noman et al., 2020 | |
| PIK-III | Nilotinib | Chronic myeloid leukemia (Patient-derived CD34+ CML cell) | + | ND | Baquero et al., 2019 | |
| Sunitinib, Erlotinib | Breast cancer (MDA-MB-231, MCF-7) | + | ND | Dyczynski et al., 2018 | ||
| Expansion | UAMC-2526 | Oxaliplatin | Colorectal cancer (HT-29) | + | + | Kurdi et al., 2017 |
| Tioconazole | Doxorubicin | Colorectal cancer (HCT116, H4, MDA-MB-231) | + | + | Liu et al., 2018 | |
| Breast cancer (MCF-7) | + | + | El-Gowily et al., 2021 | |||
| Fusion | Chloroquine | Paclitaxel | Endometrial carcinoma (HEC-1A, JEC) | + | ND | Liu and Li, 2015 |
| Doxorubicin | Various cancer (HCT116, H4, MDA-MB-231) | + | ND | Liu et al., 2018 | ||
| Breast cancer (MCF-7) | + | + | El-Gowily et al., 2021 | |||
| Cisplatin | Bladder cancer (5637, T24) | + | ND | Lin et al., 2017 | ||
| Ovarian cancer (A2780, OVCAR3) | + | ND | Zhang et al., 2012 | |||
| Temozolomide, Mebendazole | Glioblastoma (U87, U373) | + | ND | Jo et al., 2022 | ||
| Apatinib | Anaplastic thyroid cancer (C643, KHM-5M) | + | + | Feng et al., 2018 | ||
| Hydroxy-chloroquine | Sorafenib | Hepatocellular carcinoma (Huh7, HepG2) | + | + | Chen et al., 2021 | |
| Bevacizumab | Glioblastoma (LN18, LN229) | + | ND | Liu et al., 2019 | ||
| Bafilomycin A1 | Cisplatin | Tongue squamous cell carcinoma (Tca8113, Tscca) | + | ND | Chu et al., 2018a | |
| Bladder cancer (5637, T24) | + | ND | Lin et al., 2017 | |||
| Gefitinib | Triple negative breast cancer (MDA-MB-468, MDA-MB-231) | + | + | Liu et al., 2017b | ||
| 5-Fluorouracil | Gastric cancer (SGC-7901) | + | ND | Li et al., 2016 | ||
| ROC-325 | Azacitidine | Acute myeloid leukemia (MV4-11, HL-60, MOLM-13, KG-1) | + | +* | Nawrocki et al., 2019 | |
| Cytochalasin E | Bortezomib | Lung cancer (A549) | + | ND | Takanezawa et al., 2018 | |
| Lys05 | Nilotinib | Chronic myeloid leukemia (Patient-derived CD34+ CML cell) | + | + | Baquero et al., 2019 | |
| DC661 | Sorafenib | Hepatocellular carcinoma (Hep 3B, Hep 1-6) | + | +* | Xu et al., 2022 | |
ND, not determined; Bold letters indicate cell lines used in in vivo experiments; *, increased survival rate in in vivo mouse models.
In the nucleation stage, 3-MA has been tested for combination with several kinase inhibitors such as apatinib, gefitinib, and sorafenib, a HDAC inhibitor (vorinostat), and a platinum-based chemotherapeutic agent (cisplatin). Apatinib, a highly selective inhibitor of vascular endothelial growth factor receptor 2 (VEGFR2) tyrosine kinase, can induce cell cycle arrest, apoptosis, and autophagy in osteosarcoma cells lines. By inhibiting autophagy with 3-MA, apoptosis can be increased in apatinib-treated cells (Liu et al., 2017a). Apatinib also shows combinatorial effect with 3-MA by significantly inhibiting the growth and migration of uterine sarcoma cells (Chen and Yao, 2021). For triple negative breast cancers (TNBCs), effective targeted therapy is lacking. Since epidermal growth factor receptor (EGFR) is over-expressed in about 50% of TNBCs, EGFR inhibitors such as gefitinib treatment have been attempted. However, their effects were disappointing (Nakai et al., 2016). Autophagy was thought to be related to drug resistance. By autophagy inhibition with 3-MA or bafilomycin A1, the sensitivity of gefitinib could be improved (Liu et al., 2017b). Autophagy inhibition by 3-MA can enhance the synergistic effect of a combination of vorinostat with sorafenib in HCC cells (Yuan et al., 2014). Treatment with 3-MA can also enhance cisplatin sensitivity in ovarian cancer cells (Zhang et al., 2012). SB02024, another inhibitor of VPS34, can significantly potentiate cytotoxicities of sunitinib (broad kinase inhibitor) and erlotinib (EGFR kinase inhibitor) to breast cancer cells (Dyczynski et al., 2018). For immunologically cold tumors, antibodies targeting programmed cell death 1 (PD-1) or programmed death-ligand 1 (PD-L1) have limited efficacies (Jiang et al., 2019; Wu et al., 2022). Combination of SB02024 or SAR405 can improve the therapeutic benefit of anti-PD-L1/PD-1 in melanoma and colorectal cancer cells by inhibiting VPS34 (Noman et al., 2020). PIK-III, a recently developed inhibitor of lipid kinase VPS34, can also inhibit tyrosine kinase inhibitor (TKI)-induced autophagy when used in combination with nilotinib (Baquero et al., 2019), showing an enhanced anti-cancer effect when it is combined with sunitinib or erlotinib (Dyczynski et al., 2018).
For the expansion stage, ATG4B inhibiting compound UAMC-2526 is a benzotropolone derivative with fair plasma stability (Kurdi et al., 2017). It has been demonstrated that UAMC-2526 can improve inhibition of tumor growth with oxaliplatin (a platinum-based chemotherapeutic agent) in colorectal cancer cells. It has been shown that tioconazole, a clinical anti-fungal drug, can inhibit activities of ATG4A and ATG4B in a drug repurposing study (Liu et al., 2018). In HCT116 colorectal cancer cells, tioconazole combined with doxorubicin (a DNA alkylating agent) resulted in significantly enhanced chemotherapeutic efficacy in spheroid cell culture and xenografted tumors. In MCF breast cancer cells, combination of tioconazole with doxorubicin significantly inhibited PI3K/AKT/mTOR and ATG4B pathways, resulting in tumor growth inhibition with various antioxidant effects (El-Gowily et al., 2021).
In the late autophagy stage, multiple inhibitors can affect the fusion process. CQ and HCQ are clinically approved anti-malarial agents. They have been tested for combination with various anti-cancer drugs. Anti-microtubule drug [paclitaxel, mebendazole (MBZ)], DNA alkylating agents [doxorubicin, daunorubicin, temozolomide (TMZ)], platinum-based chemotherapeutic agent (cisplatin), a Raf kinase inhibitor (sorafenib), and an anti-VEGF antibody (bevacizumab) have been successfully used for combination with CQ or HCQ to sensitize cancer cells to anti-cancer drugs. In endometrial carcinoma cell lines, paclitaxel-mediated cell death is further potentiated by pretreatment with CQ (Liu and Li, 2015). Combination of CQ with doxorubicin can also significantly sensitize various cancer cells to doxorubicin treatment in vitro (Liu et al., 2018; El-Gowily et al., 2021) and in vivo (El-Gowily et al., 2021). Cisplatin-based chemotherapy is the first line treatment for bladder cancer. Cisplatin-induced autophagy is considered to be responsible for cisplatin resistance. Autophagy inhibitors bafilomycin A1 and CQ can significantly enhance cytotoxicity of cisplatin toward bladder cancer cells (Lin et al., 2017). CQ treatment can also sensitize ovarian cancer cells to cisplatin in vitro (Zhang et al., 2012). TMZ is the first line chemotherapeutic drug of choice in glioblastoma. It can induce autophagy (Singh et al., 2021). However, glioblastoma with a grim prognosis (median overall survival (OS) of 14.6 months) demands further therapeutic modalities. MBZ, a widely used anthelmintic drug, has shown cytotoxic effects on several cancer cells including melanoma, gastric cancer, lung cancer, and glioblastomas (Guerini et al., 2019). Addition of CQ can also enhance anti-proliferative effect of TMZ or MBZ (Kanzawa et al., 2004; Lee et al., 2015). Such effect is further potentiated by triple combination with TMZ (Jo et al., 2022). Combination of CQ with apatinib (VEGFR2 inhibitor) can also effectively inhibit in vivo growth of thyroid cancer cells (KHM-5M) xenografted in mice (Feng et al., 2018).
HCQ has also been studied for combination with sorafenib or bevacizumab to inhibit cancer cell growth. While sorafenib is an effective chemotherapeutic agent in advanced HCC, sorafenib resistance can lead to treatment failure. A combination therapy of sorafenib with HCQ provides better therapeutic outcomes even for sorafenib-resistant HCC cells partly by modulating autophagy (Chen et al., 2021). For recurrent glioblastomas, bevacizumab (BEV) is widely used for disease control. However, BEV treatment only shows extended progression free survival (PFS). OS benefit could not be gained for patients (Wick et al., 2017). Recent evidence has demonstrated that BEV-induced cytoprotective autophagy is a cause of treatment failure (Huang et al., 2018). By combining HCQ with BEV for glioblastoma cell lines, the anti-cancer effect of BEV can be enhanced by blocking the autophagic process (Liu et al., 2019). Bafilomycin A1 can also increase cisplatin cytotoxicity in tongue squamous cell carcinoma (TSCC) and bladder cancer cells by inhibiting lysosomal uptake of platinum and enhancing intracellular platinum ion binding to DNA (Lin et al., 2017; Chu et al., 2018a). Combination of bafilomycin A1 with gefitinib (EGFR kinase inhibitor) can also enhance anti-tumor effects in vitro and in vivo (Liu et al., 2017b). When gastric cancer cell line was treated with 5-fluorouracil, chemotherapy-induced autophagy was recognizable. Bafilomycin A1 decreased the viability and clone formation, inhibited the invasive and migratory ability, and increased apoptosis (Li et al., 2016).
ROC-325, a novel autophagy inhibitor, can effectively inhibit autophagy in AML cells. Azacitidine (AZA), a hypomethylating agent, is frequently used in the management of myelodysplastic syndromes and AML. AZA treatment can trigger autophagy in AML cells. AZA in combination with ROC-325 can significantly increase the benefit in both in vitro and in vivo studies (Nawrocki et al., 2019). Cytochalasin E in combination with bortezomib, an inhibitor of the 26S proteasome, has also been used to treat human lung cancer cells (Takanezawa et al., 2018). In chronic myeloid leukemia (CML) patients, TKI treatment could induce autophagy that leads to treatment failure. To overcome such resistance, the effect of Lys05, a highly potent lysosomotropic agent, has been studied (Baquero et al., 2019). Lys05-mediated autophagy inhibition can reduce numbers of leukemic stem cells both in vivo and in vitro. Furthermore, Lys05 can sensitize patient-derived CMLs to TKI treatment. Palmitoyl-protein thioesterase 1 (PPT1) plays a critical role in various cancers (Rebecca et al., 2019; Sharma et al., 2020; Luo et al., 2021). It is significantly upregulated in HCC tissues compared with that in normal tissues (Xu et al., 2022). Increased PPT1 levels are also associated with poor prognosis. DC661, a selective and potent small-molecule PPT1-inhibitor, can inhibit autophagy and enhance sensitivity of HCC cells to sorafenib by inducing lysosomal membrane permeabilization, leading to lysosomal deacidification.
Synergy in anti-tumor effects has been observed by combining chemotherapeutics with all autophagy inhibitors to block each stage of autophagic process. In addition, all cytotoxic drugs ranging from platinum-based chemotherapeutic agents and DNA alkylating drugs to anti-angiogenic agent (bevacizumab) and immune modulating drug (anti-PD-1/anti-PD-L1) have been effectively combined with various autophagy inhibitors. In the future, it is necessary to evaluate the best combinations by examining which chemotherapeutics can be more effectively combined with which type of autophagy inhibitors.
CLINICAL TRIALS OF AUTOPHAGY INHIBITORS WITH OR WITHOUT CHEMOTHERAPEUTICS FOR CANCER TREATMENT
Various phases of clinical trials have been performed regarding autophagy inhibitors in combination with or without several chemotherapeutic drugs (Table 4). Although autophagy inhibitors showed potential benefits from pre-clinical studies, only those affecting the late autophagy stage are studied as potential candidates for clinical trials. Since most trials were designed as phase 1 or 2 studies, majority of trials were single arm trials without masking. At present, several solid tumors have been subjected to ongoing or completed clinical trials of CQ or HCQ by single treatment or combination treatment with various anti-cancer agents (https://clinicaltrials.gov/). Only published results or recognizable results from completed clinical trials will be reviewed.
Table 4.
Clinical trials of autophagy inhibitors with or without anti-cancer drugs for cancer treatment
| Autophagic stage | Autophagy inhibitor | Anti-cancer drugs | Cancer type | Phase | Study design | Masking | Enrollment | Trial no. | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Fusion | Chloroquine | NA | Glioblastoma | 3 | Randomized | Double | 30 | NCT00224978* | Sotelo et al., 2006 |
| NA | Breast cancer | 1/2 | Single arm | None | 12 | NCT01023477 | https://clinicalTrials.gov/ | ||
| Docetaxel, paclitaxel, nab-paclitaxel, ixabepilone | Breast cancer | 2 | Single arm | None | 38 | NCT01446016* | Anand et al., 2021 | ||
| Metformin | IDH1/2-mutated solid tumors | 1/2 | Single arm | None | 15 | NCT02496741 | Molenaar et al., 2017 | ||
| Gemcitabine | Pancreatic cancer | 1 | Single arm | None | 9 | NCT01777477 | https://clinicalTrials.gov/ | ||
| DT01 | Melanoma | 1 | Single arm | None | 27 | NCT01469455 | https://clinicalTrials.gov/ | ||
| Temozolomide | Glioblastoma | 1 | Single arm | None | 13 | NCT02378532 | https://clinicalTrials.gov/ | ||
| Temozolomide | Glioblastoma | 2 | Single arm | None | 10 | NCT04397679 | https://clinicalTrials.gov/ | ||
| Bortezomib, cyclophosphamide | Multiple myeloma | 2 | Single arm | None | 11 | NCT01438177 | https://clinicalTrials.gov/ | ||
| Hydroxy-chloroquine | NA | Solid tumor | 1 | Single arm | None | 10 | NCT02232243* | Wang et al., 2018 | |
| NA | Prostate cancer | 2 | Single arm | None | 64 | NCT00726596 | https://clinicalTrials.gov/ | ||
| NA | Colorectal cancer | 2 | Single arm | None | 38 | NCT01006369 | https://clinicalTrials.gov/ | ||
| NA | Pancreatic cancer | 2 | Single arm | None | 20 | NCT01273805* | Wolpin et al., 2014 | ||
| Carboplatin, gemcitabine | Advanced solid tumors | 1 | Single arm | None | 22 | NCT02071537* | Karim et al., 2022 | ||
| Gemcitabine, nab-paclitaxel | Pancreatic cancer | 2 | Randomized | None | 104 | NCT01978184 | https://clinicalTrials.gov/ | ||
| Paclitaxel, carboplatin, bevacizumab | Lung cancer | 2 | Single arm | None | 32 | NCT01649947* | Malhotra et al., 2019 | ||
| Gemcitabine, nab-paclitaxel | Pancreatic cancer | 1/2 | Randomized | None | 119 | NCT01506973* | Karasic et al., 2019 | ||
| Temozolomide | Brain and CNS tumors | 1/2 | Single arm | None | 92 | NCT00486603* | Rosenfeld et al., 2014 | ||
| Gemcitabine | Pancreatic cancer | 1/2 | Single arm | None | 35 | NCT01128296 | https://clinicalTrials.gov/ | ||
| IL-2 | Renal cell cancer | 1/2 | Single arm | None | 30 | NCT01550367* | https://clinicalTrials.gov/ | ||
| NA | Prostate cancer | 2 | Single arm | None | 20 | NCT04011410 | https://clinicalTrials.gov/ | ||
| Ulixertinib | Gastrointestinal cancer | 2 | Single arm | None | 215 | NCT05221320 | https://clinicalTrials.gov/ | ||
| Sorafenib | Hepatocellular carcinoma | 2 | Single arm | None | 68 | NCT03037437 | https://clinicalTrials.gov/ | ||
| Abemaciclib | Breast cancer | 2 | Randomized | None | 66 | NCT04523857 | https://clinicalTrials.gov/ | ||
| Binimetinib | Lung cancer | 2 | Single arm | None | 29 | NCT04735068 | https://clinicalTrials.gov/ | ||
| Chlorphensin carbamate, mFOLFIRINOX | Pancreatic cancer | 1 | Single arm | None | 40 | NCT05083780 | https://clinicalTrials.gov/ | ||
| Trametinib | Pancreatic cancer | 2 | Single arm | None | 22 | NCT05518110 | https://clinicalTrials.gov/ |
NA, not applicable; *, Clinical trials with published results.
In a single-center, randomized, double blinded, placebo-controlled trial of single treatment of CQ for glioblastoma patient, median OS after surgery was extended to 24 months for CQ-treated patients compared to 11 months for controls, although that trial failed to show statistical significance probably due to a small sample size (n=30) (Sotelo et al., 2006). Although this result warrants further a larger scale study, it provides implications that CQ, in conjunction with other treatments, might prolong survival of patients with glioblastoma.
Combination therapy of CQ with taxane or taxane-like chemotherapeutic agents (Docetaxel, paclitaxel, nab-paclitaxel, ixabepilone) against advanced or metastatic breast cancer which is refractory to anthracycline-based therapy has demonstrated a higher objective response rate (ORR) of 45 % than the expected ORR of 30% (Anand et al., 2021). In addition, the combination was well-tolerated without showing significant toxicity. A phase 1B/2 clinical trial of metformin and CQ has been registered for a dose-finding study in patients with IDH1-mutated or IDH2-mutated solid tumors (Molenaar et al., 2017).
In a phase 1 trial for surgically removable early-stage solid tumors, oral HCQ of 200 or 400 mg twice daily for 14 days as a neoadjuvant regimen showed no serious adverse events (Wang et al., 2018). It elevated plasma prostate apoptosis response- 4 (Par-4) levels over basal levels. Four patients had prostate adenocarcinomas. Two patients had NSCLC. Others had papillary thyroid carcinoma, squamous cell carcinoma of larynx, or carcinoid tumor of the lung. All nine HCQ-treated patients showed p62 induction indicative of autophagy inhibition by HCQ. Resected tumors from eight patients with elevated plasma Par-4 levels all exhibited TUNEL-positivity indicative of apoptosis. A single administration of HCQ was also performed for previously treated metastatic pancreatic cancer patients as a phase 2 trial (Wolpin et al., 2014). The primary endpoint was 2-month PFS. Among 20 patients enrolled, only 2 (10%) had no disease progression. Median PFS and OS were 46.5 days and 69.0 days, respectively. The HCQ monotherapy failed to show therapeutic efficacy. Thus, further combinatorial treatment strategies are needed.
In a phase 1 dose-escalation study of HCQ in combination with carboplatin and gemcitabine, HCQ 100 mg daily was found to be the maximum tolerated dose (MTD) (Karim et al., 2022). Dose-limiting toxicity was thrombocytopenia and/or neutropenia. This MTD was lower than that from previously reported outcomes with concomitant use of chemotherapeutics probably due to the myelosuppressive nature of these agents and previous treatment history of patients. When response rate was assessed in that study, one patient showed partial response (PR), 15 patients showed stable disease (SD), and six patients had progressive disease (PD). The disease control rate (DCR) was 48% for more than 6 months duration, 21% for more than 12 months, and 14% for more than 18 months. Combinatorial effects of HCQ on pancreatic cancer, gastrointestinal cancer, HCC, breast cancer, prostate cancer, and lung cancer are also under investigation in various phase 1-2 clinical trials (Table 4). A randomized phase 2 trial of pancreatic cancer to examine the ability of HCQ combined with a pre-operative regimen of gemcitabine and nab-paclitaxel (GA) was completed. The exact clinical impact of this study is yet to be determined. Further reports with proper analysis are warranted.
In a phase 2 trial, untreated metastatic NSCLC patients underwent a single arm designed study of HCQ in combination with carboplatin, paclitaxel, or bevacizumab (Malhotra et al., 2019). The ORR was 33% in 30 patients evaluable for response. It was found that 20% of patients demonstrated SD. The medium PFS was 3.3 months. In nine patients with KRAS positive tumors, the ORR was 44 % with median PFS higher than 6.4 months. Addition of HCQ provided a beneficial effect on clinical response. The benefit seemed to be higher for a certain subgroup of molecularly targeted patients.
Another phase 2 trial of GA regimen with or without HCQ on patients with advanced pancreatic cancer has been performed (Karasic et al., 2019). Primary end point of OS at 12 months was not improved by HCQ treatment. However, ORR was 38.2% (n=21) in the HCQ group and 21.1% (n=12) in the non-HCQ group. Treatment-related grade 3 or 4 adverse events that were higher in the HCQ group were neutropenia, fatigue, nausea, peripheral neuropathy, visual changes, and neuropsychiatric symptoms. Although HCQ combination failed to provide enhanced OS, higher response rate indicated that HCQ combination could be beneficial under certain clinical circumstances such as locally advanced tumor that might be potentially resectable upon treatment response.
A phase 1/2 trial of HCQ in combination with the standard of care for newly diagnosed glioblastoma has also been performed (Rosenfeld et al., 2014). Regarding phase 1 trial results, 3/3 subjects experienced Grade 3 or 4 neutropenia and thrombocytopenia. The MTD for HCQ was found to be 600 mg/d in this combination. Phase 2 trial results revealed that the median survival was 15.6 months with survival rates of 70%, 36%, and 25% at 12, 18, and 24 months, respectively. Pharmacokinetics analysis demonstrated a dose-proportional exposure for HCQ. However, since the MTD for HCQ was 600 mg/d, autophagy inhibition was not constantly achieved. Further development of compounds with lower toxicities and/or more inhibitory potential for autophagy is mandatory.
Another phase 1/2 trial of HCQ in combination with IL-2, a standard treatment for metastatic renal cell cancer, has been performed (https://clinicaltrials.gov/). Among 30 enrolled participants, initial 13 patients were administered with 1200 mg/d of HCQ. However, due to severe unexpected adverse events, the dose of HCQ was reduced to 600 mg/d. Overall, 29 patients were analyzed. Control rate (CR), PR, and SD were achieved in 3 (10.3%), 3 (10.3%), and 14 (48.3%) patients, respectively. Interestingly, 3/3 CR patients and 2/3 PR patients belonged to the 600 mg/d HCQ cohort. Proper interpretation of these results by relevant authorities has not been reported yet. The combinatorial effect of HCQ should be discussed in further details.
Up to date, CQ and its derivative HCQ are the only autophagy inhibitors that have been investigated in clinical trials for cancer treatment. However, most studies are still in phase 1 or 2. Clinical benefits of single and combinatorial treatments are not clearly demonstrated yet. Several dozens of clinical trials are still on-going or planned. We hope that more positive results would follow to provide cancer patients better treatment options.
FUTURE PERSPECTIVES
Despite various autophagy inhibitors targeting each autophagy stage have been tested in pre-clinical in vitro and in vivo studies, only CQ and HCQ have been translated into clinical trials. While CQ and HCQ are clinically approved anti-malarial agents, other autophagy inhibitors do not have clinical implications yet. Since anti-cancer effect of CQ and HCQ might also come from other activities besides inhibition of autophagy, more specific autophagy inhibitors with safety that allow use in clinics should be developed. At present, clinically available inhibitors that can act in early stages of autophagic process are very limited. Such inhibitors also should be developed as effective adjuvant therapeutics for blocking cytoprotective autophagy to cure cancers. Thus, the development of more clinically effective and more selective autophagy inhibitors with various modes of action and acceptable toxicities is mandatory.
In the future, the positive impact of autophagy inhibition on cancer treatment needs further clarification. In order to determine whether the combinatorial strategy is beneficial or not, measures to identify autophagy inhibition needs to be standardized. Different MTDs from various clinical trials with different measures for autophagy inhibition can lead to confused interpretation for the presence of autophagy inhibition and their effects on clinical outcomes.
Utilizing an autophagy inhibitor in combination with chemotherapeutics may show potential benefits in cancer treatment because treatment resistance is in part correlated with increased autophagy reaction to cancer treatment. However, limited pool of autophagy inhibitors applicable to real world practice cripples our capability of validating autophagy inhibition in oncology practice. Further development of clinically available autophagy inhibitors with sufficient efficacy and safety is mandatory. By properly assessing autophagy inhibition in cancer treatment, more sophisticated clinical trials can be designed, leading to more informative results regarding combinatorial effects of autophagy inhibitors.
ACKNOWLEDGMENTS
This research was supported by a grant (NRF-2020R1A2C2006189) from the National Research Foundation of Korea grants funded by the Korean government.
Footnotes
CONFLICT OF INTEREST
The authors have no competing financial interests relevant to this study to disclose.
REFERENCES
- Afzal S., Hao Z., Itsumi M., Abouelkheer Y., Brenner D., Gao Y., Wakeham A., Hong C., Li W. Y., Sylvester J., Gilani S. O., Brustle A., Haight J., You-Ten A. J., Lin G. H., Inoue S., Mak T. W. Autophagy-independent functions of UVRAG are essential for peripheral naive T-cell homeostasis. Proc. Natl. Acad. Sci. U. S. A. 2015;112:1119–1124. doi: 10.1073/pnas.1423588112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akin D., Wang S. K., Habibzadegah-Tari P., Law B., Ostrov D., Li M., Yin X. M., Kim J. S., Horenstein N., Dunn W. A., Jr. A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors. Autophagy. 2014;10:2021–2035. doi: 10.4161/auto.32229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldrich L. N., Kuo S. Y., Castoreno A. B., Goel G., Kuballa P., Rees M. G., Seashore-Ludlow B. A., Cheah J. H., Latorre I. J., Schreiber S. L., Shamji A. F., Xavier R. J. Discovery of a small-molecule probe for V-ATPase function. J. Am. Chem. Soc. 2015;137:5563–5568. doi: 10.1021/jacs.5b02150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman Y., Schmauck-Medina T., Hansen M., Morimoto R. I., Simon A. K., Bjedov I., Palikaras K., Simonsen A., Johansen T., Tavernarakis N., Rubinsztein D. C., Partridge L., Kroemer G., Labbadia J., Fang E. F. Autophagy in healthy aging and disease. Nat. Aging. 2021;1:634–650. doi: 10.1038/s43587-021-00098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Niravath P., Patel T., Ensor J., Rodriguez A., Boone T., Wong S. T., Chang J. C. A phase II study of the efficacy and safety of chloroquine in combination with taxanes in the treatment of patients with advanced or metastatic anthracycline-refractory breast cancer. Clin. Breast Cancer. 2021;21:199–204. doi: 10.1016/j.clbc.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryal P., Kim K., Park P. H., Ham S., Cho J., Song K. Baicalein induces autophagic cell death through AMPK/ULK1 activation and downregulation of mTORC1 complex components in human cancer cells. FEBS J. 2014;281:4644–4658. doi: 10.1111/febs.12969. [DOI] [PubMed] [Google Scholar]
- Bago R., Malik N., Munson M. J., Prescott A. R., Davies P., Sommer E., Shpiro N., Ward R., Cross D., Ganley I. G., Alessi D. R. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem. J. 2014;463:413–427. doi: 10.1042/BJ20140889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero P., Dawson A., Mukhopadhyay A., Kuntz E. M., Mitchell R., Olivares O., Ianniciello A., Scott M. T., Dunn K., Nicastri M. C., Winkler J. D., Michie A. M., Ryan K. M., Halsey C., Gottlieb E., Keaney E. P., Murphy L. O., Amaravadi R. K., Holyoake T. L., Helgason G. V. Targeting quiescent leukemic stem cells using second generation autophagy inhibitors. Leukemia. 2019;33:981–994. doi: 10.1038/s41375-018-0252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgisdottir A. B., Lamark T., Johansen T. The LIR motif - crucial for selective autophagy. J. Cell Sci. 2013;126:3237–3247. doi: 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
- Cann G. M., Guignabert C., Ying L., Deshpande N., Bekker J. M., Wang L., Zhou B., Rabinovitch M. Developmental expression of LC3alpha and beta: absence of fibronectin or autophagy phenotype in LC3beta knockout mice. Dev. Dyn. 2008;237:187–195. doi: 10.1002/dvdy.21392. [DOI] [PubMed] [Google Scholar]
- Carew J. S., Espitia C. M., Zhao W., Han Y., Visconte V., Phillips J., Nawrocki S. T. Disruption of autophagic degradation with ROC-325 antagonizes renal cell carcinoma pathogenesis. Clin. Cancer Res. 2017;23:2869–2879. doi: 10.1158/1078-0432.CCR-16-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson S. R., Simonsen A. Membrane dynamics in autophagosome biogenesis. J. Cell Sci. 2015;128:193–205. doi: 10.1242/jcs.141036. [DOI] [PubMed] [Google Scholar]
- Chang H., Zou Z. Targeting autophagy to overcome drug resistance: further developments. J. Hematol. Oncol. 2020;13:159. doi: 10.1186/s13045-020-01000-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. Y., Yadav V. K., Chu Y. C., Ong J. R., Huang T. Y., Lee K. F., Lee K. H., Yeh C. T., Lee W. H. Hydroxychloroquine (HCQ) modulates autophagy and oxidative DNA damage stress in hepatocellular carcinoma to overcome sorafenib resistance via TLR9/SOD1/hsa-miR-30a-5p/Beclin-1 axis. Cancers (Basel) 2021;13:3227. doi: 10.3390/cancers13133227.73d596a59b6e40499a8e37f31b19113a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Yao L. Autophagy inhibitor potentiates the antitumor efficacy of apatinib in uterine sarcoma by stimulating PI3K/Akt/mTOR pathway. Cancer Chemother. Pharmacol. 2021;88:323–334. doi: 10.1007/s00280-021-04291-5. [DOI] [PubMed] [Google Scholar]
- Chen Y., Xie X., Wang C., Hu Y., Zhang H., Zhang L., Tu S., He Y., Li Y. Dual targeting of NUAK1 and ULK1 using the multitargeted inhibitor MRT68921 exerts potent antitumor activities. Cell Death Dis. 2020;11:712. doi: 10.1038/s41419-020-02885-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Wu J., Gonzales L. K., Guttentag S. H., Thompson C. B., Lindsten T. Analysis of a lung defect in autophagy-deficient mouse strains. Autophagy. 2014;10:45–56. doi: 10.4161/auto.26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H. Y., Wang W., Chen X., Jiang Y. E., Cheng R., Qi X., Zhong Z. M., Zeng M. S., Zhu X. F., Sun C. Z. Bafilomycin A1 increases the sensitivity of tongue squamous cell carcinoma cells to cisplatin by inhibiting the lysosomal uptake of platinum ions but not autophagy. Cancer Lett. 2018a;423:105–112. doi: 10.1016/j.canlet.2018.03.003. [DOI] [PubMed] [Google Scholar]
- Chu J., Fu Y., Xu J., Zheng X., Gu Q., Luo X., Dai Q., Zhang S., Liu P., Hong L., Li M. ATG4B inhibitor FMK-9a induces autophagy independent on its enzyme inhibition. Arch. Biochem. Biophys. 2018b;644:29–36. doi: 10.1016/j.abb.2018.03.001. [DOI] [PubMed] [Google Scholar]
- Dowdle W. E., Nyfeler B., Nagel J., Elling R. A., Liu S., Triantafellow E., Menon S., Wang Z., Honda A., Pardee G., Cantwell J., Luu C., Cornella-Taracido I., Harrington E., Fekkes P., Lei H., Fang Q., Digan M. E., Burdick D., Powers A. F., Helliwell S. B., D'Aquin S., Bastien J., Wang H., Wiederschain D., Kuerth J., Bergman P., Schwalb D., Thomas J., Ugwonali S., Harbinski F., Tallarico J., Wilson C. J., Myer V. E., Porter J. A., Bussiere D. E., Finan P. M., Labow M. A., Mao X., Hamann L. G., Manning B. D., Valdez R. A., Nicholson T., Schirle M., Knapp M. S., Keaney E. P., Murphy L. O. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat. Cell Biol. 2014;16:1069–1079. doi: 10.1038/ncb3053. [DOI] [PubMed] [Google Scholar]
- Dyczynski M., Yu Y., Otrocka M., Parpal S., Braga T., Henley A. B., Zazzi H., Lerner M., Wennerberg K., Viklund J., Martinsson J., Grander D., De Milito A., Pokrovskaja Tamm K. Targeting autophagy by small molecule inhibitors of vacuolar protein sorting 34 (Vps34) improves the sensitivity of breast cancer cells to Sunitinib. Cancer Lett. 2018;435:32–43. doi: 10.1016/j.canlet.2018.07.028. [DOI] [PubMed] [Google Scholar]
- Egan D. F., Chun M. G., Vamos M., Zou H., Rong J., Miller C. J., Lou H. J., Raveendra-Panickar D., Yang C. C., Sheffler D. J., Teriete P., Asara J. M., Turk B. E., Cosford N. D., Shaw R. J. Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol. Cell. 2015;59:285–297. doi: 10.1016/j.molcel.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gowily A. H., Loutfy S. A., Ali E. M. M., Mohamed T. M., Mansour M. A. Tioconazole and chloroquine act synergistically to combat doxorubicin-induced toxicity via inactivation of PI3K/AKT/mTOR signaling mediated ROS-dependent apoptosis and autophagic flux inhibition in MCF-7 breast cancer cells. Pharmaceuticals (Basel) 2021;14:254. doi: 10.3390/ph14030254.f5d3fd4a8f1945c194b8ddbd224d5d0c [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Eskelinen E. L. The dual role of autophagy in cancer. Curr. Opin. Pharmacol. 2011;11:294–300. doi: 10.1016/j.coph.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Feng H., Cheng X., Kuang J., Chen L., Yuen S., Shi M., Liang J., Shen B., Jin Z., Yan J., Qiu W. Apatinib-induced protective autophagy and apoptosis through the AKT-mTOR pathway in anaplastic thyroid cancer. Cell Death Dis. 2018;9:1030. doi: 10.1038/s41419-018-1054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia G. M., Stoykova A., Romagnoli A., Giunta L., Di Bartolomeo S., Nardacci R., Corazzari M., Fuoco C., Ucar A., Schwartz P., Gruss P., Piacentini M., Chowdhury K., Cecconi F. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- Fu W., Li X., Lu X., Zhang L., Li R., Zhang N., Liu S., Yang X., Wang Y., Zhao Y., Meng X., Zhu W. G. A novel acridine derivative, LS-1-10 inhibits autophagic degradation and triggers apoptosis in colon cancer cells. Cell Death Dis. 2017;8:e3086. doi: 10.1038/cddis.2017.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Hong L., Xu J., Zhong G., Gu Q., Gu Q., Guan Y., Zheng X., Dai Q., Luo X., Liu C., Huang Z., Yin X. M., Liu P., Li M. Discovery of a small molecule targeting autophagy via ATG4B inhibition and cell death of colorectal cancer cells in vitro and in vivo. Autophagy. 2019;15:295–311. doi: 10.1080/15548627.2018.1517073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B., Peng X., Nagy T., Alcaraz A., Gu H., Guan J. L. Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J. Cell Biol. 2006;175:121–133. doi: 10.1083/jcb.200604129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groza T., Gomez F. L., Mashhadi H. H., Munoz-Fuentes V., Gunes O., Wilson R., Cacheiro P., Frost A., Keskivali-Bond P., Vardal B., McCoy A., Cheng T. K., Santos L., Wells S., Smedley D., Mallon A. M., Parkinson H. The International Mouse Phenotyping Consortium: comprehensive knockout phenotyping underpinning the study of human disease. Nucleic Acids Res. 2022:gkac972. doi: 10.1093/nar/gkac972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerini A. E., Triggiani L., Maddalo M., Bonu M. L., Frassine F., Baiguini A., Alghisi A., Tomasini D., Borghetti P., Pasinetti N., Bresciani R., Magrini S. M., Buglione M. Mebendazole as a candidate for drug repurposing in oncology: an extensive review of current literature. Cancers (Basel) 2019;11:1284. doi: 10.3390/cancers11091284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. Y., Chen H. Y., Mathew R., Fan J., Strohecker A. M., Karsli-Uzunbas G., Kamphorst J. J., Chen G., Lemons J. M., Karantza V., Coller H. A., Dipaola R. S., Gelinas C., Rabinowitz J. D., White E. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. Y., Karsli-Uzunbas G., Mathew R., Aisner S. C., Kamphorst J. J., Strohecker A. M., Chen G., Price S., Lu W., Teng X., Snyder E., Santanam U., Dipaola R. S., Jacks T., Rabinowitz J. D., White E. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013a;27:1447–1461. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. Y., Xia B., White E. Autophagy-mediated tumor promotion. Cell. 2013b;155:1216–1219. doi: 10.1016/j.cell.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T., Noda N. N., Satomi Y., Ichimura Y., Fujioka Y., Takao T., Inagaki F., Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- Hansen M., Rubinsztein D. C., Walker D. W. Autophagy as a promoter of longevity: insights from model organisms. Nat. Rev. Mol. Cell Biol. 2018;19:579–593. doi: 10.1038/s41580-018-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homewood C. A., Warhurst D. C., Peters W., Baggaley V. C. Lysosomes, pH and the anti-malarial action of chloroquine. Nature. 1972;235:50–52. doi: 10.1038/235050a0. [DOI] [PubMed] [Google Scholar]
- Hu T., Li P., Luo Z., Chen X., Zhang J., Wang C., Chen P., Dong Z. Chloroquine inhibits hepatocellular carcinoma cell growth in vitro and in vivo. Oncol. Rep. 2016;35:43–49. doi: 10.3892/or.2015.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Song J., Liu Z., Pan L., Xu G. Autophagy activation promotes bevacizumab resistance in glioblastoma by suppressing Akt/mTOR signaling pathway. Oncol. Lett. 2018;15:1487–1494. doi: 10.3892/ol.2017.7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T., Kim C. K., Alvarez A. A., Pangeni R. P., Wan X., Song X., Shi T., Yang Y., Sastry N., Horbinski C. M., Lu S., Stupp R., Kessler J. A., Nishikawa R., Nakano I., Sulman E. P., Lu X., James C. D., Yin X. M., Hu B., Cheng S. Y. MST4 phosphorylation of ATG4B regulates autophagic activity, tumorigenicity, and radioresistance in glioblastoma. Cancer Cell. 2017;32:840–855.e8. doi: 10.1016/j.ccell.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Chen M., Nie H., Yuan Y. PD-1 and PD-L1 in cancer immunotherapy: clinical implications and future considerations. Hum. Vaccin. Immunother. 2019;15:1111–1122. doi: 10.1080/21645515.2019.1571892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S. B., Sung S. J., Choi H. S., Park J. S., Hong Y. K., Joe Y. A. Modulation of autophagy is a potential strategy for enhancing the anti-tumor effect of mebendazole in glioblastoma cells. Biomol. Ther. (Seoul) 2022;30:616–624. doi: 10.4062/biomolther.2022.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaizuka T., Mizushima N. Atg13 is essential for autophagy and cardiac development in mice. Mol. Cell. Biol. 2016;36:585–595. doi: 10.1128/MCB.01005-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzawa T., Germano I. M., Komata T., Ito H., Kondo Y., Kondo S. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004;11:448–457. doi: 10.1038/sj.cdd.4401359. [DOI] [PubMed] [Google Scholar]
- Karasic T. B., O'Hara M. H., Loaiza-Bonilla A., Reiss K. A., Teitelbaum U. R., Borazanci E., De Jesus-Acosta A., Redlinger C., Burrell J. A., Laheru D. A., Von Hoff D. D., Amaravadi R. K., Drebin J. A., O'Dwyer P. J. Effect of gemcitabine and nab-paclitaxel with or without hydroxychloroquine on patients with advanced pancreatic cancer: a phase 2 randomized clinical trial. JAMA Oncol. 2019;5:993–998. doi: 10.1001/jamaoncol.2019.0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim N. A., Ullah A., Ahmad I., Bahassi E., Olowokure O., Khaled A., Davis H., Morris J. C. A phase I trial to determine the safety and tolerability of autophagy inhibition using chloroquine or hydroxychloroquine in combination with carboplatin and gemcitabine in patients with advanced solid tumors. Front. Oncol. 2022;12:811411. doi: 10.3389/fonc.2022.811411.7ab53619d7e34bc9a80e18c1663cfa50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura N., Sun-Wada G. H., Aoyama M., Harada A., Takasuga S., Sasaki T., Wada Y. Delivery of endosomes to lysosomes via microautophagy in the visceral endoderm of mouse embryos. Nat. Commun. 2012;3:1071. doi: 10.1038/ncomms2069. [DOI] [PubMed] [Google Scholar]
- Kimmelman A. C. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Kominami E., Tanaka K., Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Kundu M., Lindsten T., Yang C. Y., Wu J., Zhao F., Zhang J., Selak M. A., Ney P. A., Thompson C. B. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdi A., Cleenewerck M., Vangestel C., Lyssens S., Declercq W., Timmermans J. P., Stroobants S., Augustyns K., De Meyer G. R. Y., Van Der Veken P., Martinet W. ATG4B inhibitors with a benzotropolone core structure block autophagy and augment efficiency of chemotherapy in mice. Biochem. Pharmacol. 2017;138:150–162. doi: 10.1016/j.bcp.2017.06.119. [DOI] [PubMed] [Google Scholar]
- Lee E. J., Tournier C. The requirement of uncoordinated 51-like kinase 1 (ULK1) and ULK2 in the regulation of autophagy. Autophagy. 2011;7:689–695. doi: 10.4161/auto.7.7.15450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. W., Kim H. K., Lee N. H., Yi H. Y., Kim H. S., Hong S. H., Hong Y. K., Joe Y. A. The synergistic effect of combination temozolomide and chloroquine treatment is dependent on autophagy formation and p53 status in glioma cells. Cancer Lett. 2015;360:195–204. doi: 10.1016/j.canlet.2015.02.012. [DOI] [PubMed] [Google Scholar]
- Levy J. M. M., Towers C. G., Thorburn A. Targeting autophagy in cancer. Nat. Rev. Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. Q., Xie W. J., Pan D., Chen H., Zhang L. Inhibition of autophagy by bafilomycin A1 promotes chemosensitivity of gastric cancer cells. Tumour Biol. 2016;37:653–659. doi: 10.1007/s13277-015-3842-z. [DOI] [PubMed] [Google Scholar]
- Lin J. F., Lin Y. C., Tsai T. F., Chen H. E., Chou K. Y., Hwang T. I. Cisplatin induces protective autophagy through activation of BECN1 in human bladder cancer cells. Drug Des. Devel. Ther. 2017;11:1517–1533. doi: 10.2147/DDDT.S126464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Ren T., Huang Y., Sun K., Bao X., Wang S., Zheng B., Guo W. Apatinib promotes autophagy and apoptosis through VEGFR2/STAT3/BCL-2 signaling in osteosarcoma. Cell Death Dis. 2017a;8:e3015. doi: 10.1038/cddis.2017.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. Q., Wang S. B., Shao Y. F., Shi J. N., Wang W., Chen W. Y., Ye Z. Q., Jiang J. Y., Fang Q. X., Zhang G. B., Xuan Z. X. Hydroxychloroquine potentiates the anti-cancer effect of bevacizumab on glioblastoma via the inhibition of autophagy. Biomed. Pharmacother. 2019;118:109339. doi: 10.1016/j.biopha.2019.109339. [DOI] [PubMed] [Google Scholar]
- Liu P. F., Tsai K. L., Hsu C. J., Tsai W. L., Cheng J. S., Chang H. W., Shiau C. W., Goan Y. G., Tseng H. H., Wu C. H., Reed J. C., Yang L. W., Shu C. W. Drug repurposing screening identifies tioconazole as an ATG4 inhibitor that suppresses autophagy and sensitizes cancer cells to chemotherapy. Theranostics. 2018;8:830–845. doi: 10.7150/thno.22012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Li X. Autophagy inhibition enhances sensitivity of endometrial carcinoma cells to paclitaxel. Int. J. Oncol. 2015;46:2399–2408. doi: 10.3892/ijo.2015.2937. [DOI] [PubMed] [Google Scholar]
- Liu Z., He K., Ma Q., Yu Q., Liu C., Ndege I., Wang X., Yu Z. Autophagy inhibitor facilitates gefitinib sensitivity in vitro and in vivo by activating mitochondrial apoptosis in triple negative breast cancer. PLoS One. 2017b;12:e0177694. doi: 10.1371/journal.pone.0177694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q., Li X., Gan G., Yang M., Chen X., Chen F. PPT1 reduction contributes to erianin-induced growth inhibition in oral squamous carcinoma cells. Front. Cell Dev. Biol. 2021;9:764263. doi: 10.3389/fcell.2021.764263.f892e77656b7487da654516330515984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra J., Jabbour S., Orlick M., Riedlinger G., Guo Y., White E., Aisner J. Phase Ib/II study of hydroxychloroquine in combination with chemotherapy in patients with metastatic non-small cell lung cancer (NSCLC) Cancer Treat. Res. Commun. 2019;21:100158. doi: 10.1016/j.ctarc.2019.100158. [DOI] [PubMed] [Google Scholar]
- Malhotra R., Warne J. P., Salas E., Xu A. W., Debnath J. Loss of Atg12, but not Atg5, in pro-opiomelanocortin neurons exacerbates diet-induced obesity. Autophagy. 2015;11:145–154. doi: 10.1080/15548627.2014.998917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G., Fernandez A. F., Cabrera S., Lundberg Y. W., Cabanillas R., Rodriguez F., Salvador-Montoliu N., Vega J. A., Germana A., Fueyo A., Freije J. M., Lopez-Otin C. Autophagy is essential for mouse sense of balance. J. Clin. Invest. 2010;120:2331–2344. doi: 10.1172/JCI42601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K. R., Celano S. L., Solitro A. R., Gunaydin H., Scott M., O'Hagan R. C., Shumway S. D., Fuller P., MacKeigan J. P. A potent and selective ULK1 inhibitor suppresses autophagy and sensitizes cancer cells to nutrient stress. iScience. 2018;8:74–84. doi: 10.1016/j.isci.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauthe M., Orhon I., Rocchi C., Zhou X., Luhr M., Hijlkema K. J., Coppes R. P., Engedal N., Mari M., Reggiori F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14:1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAfee Q., Zhang Z., Samanta A., Levi S. M., Ma X. H., Piao S., Lynch J. P., Uehara T., Sepulveda A. R., Davis L. E., Winkler J. D., Amaravadi R. K. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc. Natl. Acad. Sci. U. S. A. 2012;109:8253–8258. doi: 10.1073/pnas.1118193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier G., Birsen R., Cazelles C., Belhadj M., Cantero-Aguilar L., Kosmider O., Fontenay M., Azar N., Mayeux P., Chapuis N., Tamburini J., Bouscary D. Antileukemic activity of the VPS34-IN1 inhibitor in acute myeloid leukemia. Oncogenesis. 2020;9:94. doi: 10.1038/s41389-020-00278-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Molenaar R. J., Coelen R. J. S., Khurshed M., Roos E., Caan M. W. A., van Linde M. E., Kouwenhoven M., Bramer J. A. M., Bovée J., Mathôt R. A., Klümpen H. J., van Laarhoven H. W. M., van Noorden C. J. F., Vandertop W. P., Gelderblom H., van Gulik T. M., Wilmink J. W. Study protocol of a phase IB/II clinical trial of metformin and chloroquine in patients with IDH1-mutated or IDH2-mutated solid tumours. BMJ Open. 2017;7:e014961. doi: 10.1136/bmjopen-2016-014961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K., Hung M. C., Yamaguchi H. A perspective on anti-EGFR therapies targeting triple-negative breast cancer. Am. J. Cancer Res. 2016;6:1609–1623. [PMC free article] [PubMed] [Google Scholar]
- Nawrocki S. T., Han Y., Visconte V., Przychodzen B., Espitia C. M., Phillips J., Anwer F., Advani A., Carraway H. E., Kelly K. R., Sekeres M. A., Maciejewski J. P., Carew J. S. The novel autophagy inhibitor ROC-325 augments the antileukemic activity of azacitidine. Leukemia. 2019;33:2971–2974. doi: 10.1038/s41375-019-0529-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemazanyy I., Blaauw B., Paolini C., Caillaud C., Protasi F., Mueller A., Proikas-Cezanne T., Russell R. C., Guan K. L., Nishino I., Sandri M., Pende M., Panasyuk G. Defects of Vps15 in skeletal muscles lead to autophagic vacuolar myopathy and lysosomal disease. EMBO Mol. Med. 2013;5:870–890. doi: 10.1002/emmm.201202057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noman M. Z., Parpal S., Van Moer K., Xiao M., Yu Y., Viklund J., De Milito A., Hasmim M., Andersson M., Amaravadi R. K., Martinsson J., Berchem G., Janji B. Inhibition of Vps34 reprograms cold into hot inflamed tumors and improves anti-PD-1/PD-L1 immunotherapy. Sci. Adv. 2020;6:eaax7881. doi: 10.1126/sciadv.aax7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petherick K. J., Conway O. J., Mpamhanga C., Osborne S. A., Kamal A., Saxty B., Ganley I. G. Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J. Biol. Chem. 2015;290:11376–11383. doi: 10.1074/jbc.C114.627778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petiot A., Ogier-Denis E., Blommaart E. F., Meijer A. J., Codogno P. Distinct classes of phosphatidylinositol 3'-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- Puissant A., Robert G., Fenouille N., Luciano F., Cassuto J. P., Raynaud S., Auberger P. Resveratrol promotes autophagic cell death in chronic myelogenous leukemia cells via JNK-mediated p62/SQSTM1 expression and AMPK activation. Cancer Res. 2010;70:1042–1052. doi: 10.1158/0008-5472.CAN-09-3537. [DOI] [PubMed] [Google Scholar]
- Qiu L., Zhou G., Cao S. Targeted inhibition of ULK1 enhances daunorubicin sensitivity in acute myeloid leukemia. Life Sci. 2020;243:117234. doi: 10.1016/j.lfs.2019.117234. [DOI] [PubMed] [Google Scholar]
- Qiu Z., Kuhn B., Aebi J., Lin X., Ding H., Zhou Z., Xu Z., Xu D., Han L., Liu C., Qiu H., Zhang Y., Haap W., Riemer C., Stahl M., Qin N., Shen H. C., Tang G. Discovery of fluoromethylketone-based peptidomimetics as covalent ATG4B (autophagin-1) inhibitors. ACS Med. Chem. Lett. 2016;7:802–806. doi: 10.1021/acsmedchemlett.6b00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E. L., Mizushima N., Ohsumi Y., Cattoretti G., Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebecca V. W., Nicastri M. C., Fennelly C., Chude C. I., Barber-Rotenberg J. S., Ronghe A., McAfee Q., McLaughlin N. P., Zhang G., Goldman A. R., Ojha R., Piao S., Noguera-Ortega E., Martorella A., Alicea G. M., Lee J. J., Schuchter L. M., Xu X., Herlyn M., Marmorstein R., Gimotty P. A., Speicher D. W., Winkler J. D., Amaravadi R. K. PPT1 promotes tumor growth and is the molecular target of chloroquine derivatives in cancer. Cancer Discov. 2019;9:220–229. doi: 10.1158/2159-8290.CD-18-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronan B., Flamand O., Vescovi L., Dureuil C., Durand L., Fassy F., Bachelot M. F., Lamberton A., Mathieu M., Bertrand T., Marquette J. P., El-Ahmad Y., Filoche-Romme B., Schio L., Garcia-Echeverria C., Goulaouic H., Pasquier B. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat. Chem. Biol. 2014;10:1013–1019. doi: 10.1038/nchembio.1681. [DOI] [PubMed] [Google Scholar]
- Rosenfeld M. R., Ye X., Supko J. G., Desideri S., Grossman S. A., Brem S., Mikkelson T., Wang D., Chang Y. C., Hu J., McAfee Q., Fisher J., Troxel A. B., Piao S., Heitjan D. F., Tan K. S., Pontiggia L., O'Dwyer P. J., Davis L. E., Amaravadi R. K. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy. 2014;10:1359–1368. doi: 10.4161/auto.28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R. C., Tian Y., Yuan H., Park H. W., Chang Y. Y., Kim J., Kim H., Neufeld T. P., Dillin A., Guan K. L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T., Fujita N., Jang M. H., Uematsu S., Yang B. G., Satoh T., Omori H., Noda T., Yamamoto N., Komatsu M., Tanaka K., Kawai T., Tsujimura T., Takeuchi O., Yoshimori T., Akira S. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- Schaaf M. B., Keulers T. G., Vooijs M. A., Rouschop K. M. LC3/GABARAP family proteins: autophagy-(un)related functions. FASEB J. 2016;30:3961–3978. doi: 10.1096/fj.201600698R. [DOI] [PubMed] [Google Scholar]
- Schiller S. A., Seebode C., Wieser G. L., Goebbels S., Mobius W., Horowitz M., Sarig O., Sprecher E., Emmert S. Establishment of two mouse models for CEDNIK syndrome reveals the pivotal role of SNAP29 in epidermal differentiation. J. Invest. Dermatol. 2016;136:672–679. doi: 10.1016/j.jid.2015.12.020. [DOI] [PubMed] [Google Scholar]
- Sharma G., Ojha R., Noguera-Ortega E., Rebecca V. W., Attanasio J., Liu S., Piao S., Lee J. J., Nicastri M. C., Harper S. L., Ronghe A., Jain V., Winkler J. D., Speicher D. W., Mastio J., Gimotty P. A., Xu X., Wherry E. J., Gabrilovich D. I., Amaravadi R. K. PPT1 inhibition enhances the antitumor activity of anti-PD-1 antibody in melanoma. JCI Insight. 2020;5:e133225. doi: 10.1172/jci.insight.133225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Miner A., Hennis L., Mittal S. Mechanisms of temozolomide resistance in glioblastoma - a comprehensive review. Cancer Drug Resist. 2021;4:17–43. doi: 10.20517/cdr.2020.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slobodkin M. R., Elazar Z. The Atg8 family: multifunctional ubiquitin-like key regulators of autophagy. Essays Biochem. 2013;55:51–64. doi: 10.1042/bse0550051. [DOI] [PubMed] [Google Scholar]
- Sotelo J., Briceño E., López-González M. A. Adding chloroquine to conventional treatment for glioblastoma multiforme: a randomized, double-blind, placebo-controlled trial. Ann. Intern. Med. 2006;144:337–343. doi: 10.7326/0003-4819-144-5-200603070-00008. [DOI] [PubMed] [Google Scholar]
- Sou Y. S., Waguri S., Iwata J., Ueno T., Fujimura T., Hara T., Sawada N., Yamada A., Mizushima N., Uchiyama Y., Kominami E., Tanaka K., Komatsu M. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell. 2008;19:4762–4775. doi: 10.1091/mbc.e08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Coppola D., Matsushita N., Cualing H. D., Sun M., Sato Y., Liang C., Jung J. U., Cheng J. Q., Mule J. J., Pledger W. J., Wang H. G. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat. Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura A., Komatsu M., Hara T., Sakamoto A., Kishi C., Waguri S., Eishi Y., Hino O., Tanaka K., Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanezawa Y., Nakamura R., Kojima Y., Sone Y., Uraguchi S., Kiyono M. Cytochalasin E increased the sensitivity of human lung cancer A549cells to bortezomib via inhibition of autophagy. Biochem. Biophys. Res. Commun. 2018;498:603–608. doi: 10.1016/j.bbrc.2018.03.029. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Guhde G., Suter A., Eskelinen E. L., Hartmann D., Lullmann-Rauch R., Janssen P. M., Blanz J., von Figura K., Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- Tang F., Hu P., Yang Z., Xue C., Gong J., Sun S., Shi L., Zhang S., Li Z., Yang C., Zhang J., Xie C. SBI0206965, a novel inhibitor of Ulk1, suppresses non-small cell lung cancer cell growth by modulating both autophagy and apoptosis pathways. Oncol. Rep. 2017;37:3449–3458. doi: 10.3892/or.2017.5635. [DOI] [PubMed] [Google Scholar]
- Tanida I., Sou Y. S., Ezaki J., Minematsu-Ikeguchi N., Ueno T., Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J. Biol. Chem. 2004;279:36268–36276. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- Towers C. G., Thorburn A. Therapeutic targeting of autophagy. EBioMedicine. 2016;14:15–23. doi: 10.1016/j.ebiom.2016.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vats S., Manjithaya R. A reversible autophagy inhibitor blocks autophagosome-lysosome fusion by preventing Stx17 loading onto autophagosomes. Mol. Biol. Cell. 2019;30:2283–2295. doi: 10.1091/mbc.E18-08-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. C., Ng C. P., Lu L., Atlashkin V., Zhang W., Seet L. F., Hong W. A role of VAMP8/endobrevin in regulated exocytosis of pancreatic acinar cells. Dev. Cell. 2004;7:359–371. doi: 10.1016/j.devcel.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Wang P., Burikhanov R., Jayswal R., Weiss H. L., Arnold S. M., Villano J. L., Rangnekar V. M. Neoadjuvant administration of hydroxychloroquine in a phase 1 clinical trial induced plasma Par-4 levels and apoptosis in diverse tumors. Genes Cancer. 2018;9:190–197. doi: 10.18632/genesandcancer.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E., Mehnert J. M., Chan C. S. Autophagy, metabolism, and cancer. Clin. Cancer Res. 2015;21:5037–5046. doi: 10.1158/1078-0432.CCR-15-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick W., Gorlia T., Bendszus M., Taphoorn M., Sahm F., Harting I., Brandes A. A., Taal W., Domont J., Idbaih A., Campone M., Clement P. M., Stupp R., Fabbro M., Le Rhun E., Dubois F., Weller M., von Deimling A., Golfinopoulos V., Bromberg J. C., Platten M., Klein M., van den Bent M. J. Lomustine and bevacizumab in progressive glioblastoma. N. Engl. J. Med. 2017;377:1954–1963. doi: 10.1056/NEJMoa1707358. [DOI] [PubMed] [Google Scholar]
- Wolpin B. M., Rubinson D. A., Wang X., Chan J. A., Cleary J. M., Enzinger P. C., Fuchs C. S., McCleary N. J., Meyerhardt J. A., Ng K., Schrag D., Sikora A. L., Spicer B. A., Killion L., Mamon H., Kimmelman A. C. Phase II and pharmacodynamic study of autophagy inhibition using hydroxychloroquine in patients with metastatic pancreatic adenocarcinoma. Oncologist. 2014;19:637–638. doi: 10.1634/theoncologist.2014-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Huang Q., Xie Y., Wu X., Ma H., Zhang Y., Xia Y. Improvement of the anticancer efficacy of PD-1/PD-L1 blockade via combination therapy and PD-L1 regulation. J. Hematol. Oncol. 2022;15:24. doi: 10.1186/s13045-022-01242-2.48e1f8aff6144c91ab4080684079709a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M., Benoit A., Hasmim M., Duhem C., Vogin G., Berchem G., Noman M. Z., Janji B. Targeting cytoprotective autophagy to enhance anticancer therapies. Front. Oncol. 2021;11:626309. doi: 10.3389/fonc.2021.626309.e4db7c7526394c549c53f4abda3f7663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Su Z., Cheng X., Hu S., Wang W., Zou T., Zhou X., Song Z., Xia Y., Gao Y., Zheng Q. High PPT1 expression predicts poor clinical outcome and PPT1 inhibitor DC661 enhances sorafenib sensitivity in hepatocellular carcinoma. Cancer Cell Int. 2022;22:115. doi: 10.1186/s12935-021-02433-6.efb235ba546041178e50b8e0feb382f7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Tagawa Y., Yoshimori T., Moriyama Y., Masaki R., Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- Yang S., Wang X., Contino G., Liesa M., Sahin E., Ying H., Bause A., Li Y., Stommel J. M., Dell'antonio G., Mautner J., Tonon G., Haigis M., Shirihai O. S., Doglioni C., Bardeesy N., Kimmelman A. C. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D. J. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12 Suppl 2:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Chen Y., Tooze S. A. Autophagy pathway: cellular and molecular mechanisms. Autophagy. 2018;14:207–215. doi: 10.1080/15548627.2017.1378838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Li A. J., Ma S. L., Cui L. J., Wu B., Yin L., Wu M. C. Inhibition of autophagy signi fi cantly enhances combination therapy with sorafenib and HDAC inhibitors for human hepatoma cells. World J. Gastroenterol. 2014;20:4953–4962. doi: 10.3748/wjg.v20.i17.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z., Jin S., Yang C., Levine A. J., Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Cheng Y., Ren X., Zhang L., Yap K. L., Wu H., Patel R., Liu D., Qin Z. H., Shih I. M., Yang J. M. NAC1 modulates sensitivity of ovarian cancer cells to cisplatin by altering the HMGB1-mediated autophagic response. Oncogene. 2012;31:1055–1064. doi: 10.1038/onc.2011.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Zhao Y. G., Wang X., Xu L., Miao L., Feng D., Chen Q., Kovacs A. L., Fan D., Zhang H. Mice deficient in Epg5 exhibit selective neuronal vulnerability to degeneration. J. Cell Biol. 2013;200:731–741. doi: 10.1083/jcb.201211014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Guo Y., Zhang X., Jiang Z. Lys05 induces lysosomal membrane permeabilization and increases radiosensitivity in glioblastoma. J. Cell. Biochem. 2020;121:2027–2037. doi: 10.1002/jcb.29437. [DOI] [PubMed] [Google Scholar]
- Zhou X., Takatoh J., Wang F. The mammalian class 3 PI3K (PIK3C3) is required for early embryogenesis and cell proliferation. PLoS One. 2011;6:e16358. doi: 10.1371/journal.pone.0016358.6ec84d9096f7418685c3001390af8c3f [DOI] [PMC free article] [PubMed] [Google Scholar]