Abstract
Objectives
This study assessed the clinical performance of the cobas Liat SARS‑CoV‑2 & Influenza A/B assay (LiatCOVID/flu) for the detection of both severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and influenza viruses during the SARS-CoV-2 Omicron outbreak.
Methods
Residual nasopharyngeal swab samples (NPS) previously tested with cobas SARS-CoV-2 & Influenza A/B for SARS-CoV-2 and with the Allplex Respiratory Panel 1 for influenza viruses were collected. All samples were submitted to the LiatCOVID/flu assay.
Results
A total of 1147 samples were collected comprising 167 SARS-CoV-2-positive, 556 SARS-CoV-2-negative, 224 influenza-positive, and 200 influenza-negative cases. The positive percent agreement (PPA)/negative percent agreement (NPA) of LiatCOVID/flu for SARS-CoV-2 and influenza viruses compared to the previously tested methods were 100% of 100% and 99.6% of 100%, respectively.
Conclusions
The LiatCOVID/flu assay shows an acceptable performance in the detection of SARS-CoV-2 and influenza viruses using NPS samples.
Keywords: SARS-CoV-2, COVID-19, POCT, rapid diagnostic test, influenza, nasopharyngeal swab
1. Introduction
Data from the World Health Organization (WHO) indicate that approximately 520 million people worldwide have now been infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), of which 1.2% have died from coronavirus disease 2019 (COVID-19) caused by this virus [1]. Since February 2022, the Omicron variant of SARS-CoV-2 has represented 90% of the newly identified worldwide SARS-CoV-2 genome, according to the Global Initiative on Sharing All Influenza Data (GISAID) dataset [2]. In Korea, the SARS-CoV-2 Omicron variant was first identified on November 25, 2021, and 99.9% of the newly identified SARS-CoV-2 genomes were replaced by this variant by March 2022 [3,4]. With the emergence of Omicron variants, worldwide weekly confirmed case numbers for SARS-CoV-2 were dramatically increased by up to 23 million in mid-January 2022, but the fatality rate from COVID-19 had declined [1]. Notably however, the Omicron variants of SARS-CoV-2 remain a big threat for high-risk patients. Furthermore, rapid antigen testing has a role in rapid SARS-CoV-2 detection and subsequent rapid isolation of COVID-19 patients [5], but the low sensitivities of this testing method remain problematic. Further to this, the majority of the currently-available rapid diagnostic tools for SARS-CoV-2, with the exception of rapid real-time reverse transcription polymerase chain reaction (rRT-PCR) tests, are not as sensitive as rRT-PCR tests, which are still considered to be the gold standard [6,7]. Thus, there is a critical need for highly sensitive, rapid testing to screen inpatients for SARS-CoV-2, to improve patient flow.
Several reports have indicated that well-known respiratory viruses such as parainfluenza virus and respiratory syncytial virus (RSV) also caused outbreaks during the SARS-CoV-2 pandemic period [8,9]. The rapid screening of these viruses in addition to SARS-CoV-2 is essential for preventing the concurrent outbreak of SARS-CoV-2 and respiratory viruses. In this regard, the cobas Liat SARS‑CoV‑2 & Influenza A/B assay (LiatCOVID/flu; Roche Molecular Systems; Pleasanton, CA) is an automated rRT-PCR assay. The LiatCOVID/flu assay tests 1 patient sample at a time with short turn-around time of 20 minutes, and testing of influenza viruses and SARS-CoV-2 occurs simultaneously. LiatCOVID/flu has shown a reliable performance with clinical specimens in several studies [10], [11], [12], [13], but false-positive SARS-CoV-2 rates with this assay of up to 9.6% were notably described in 2 studies, and more problematic in asymptomatic groups [10,14]. In addition, this test has demonstrated an acceptable performance for nasal samples and posterior oropharyngeal saliva in addition to nasopharyngeal swab samples (NPS) [15,16]. However, to the best of our knowledge, LiatCOVID/flu has rarely been evaluated for influenza testing. We here assessed the clinical performance of LiatCOVID/flu for the detection of both SARS-CoV-2 and influenza viruses in the context of the emerging SARS-CoV-2 Omicron variant.
2. Materials and methods
2.1. Sample collection
This study included residual NPS obtained using SG Medical swabs (Seegene; Seoul, Korea) and both in- and outpatient samples previously tested for influenza viruses and SARS-CoV-2, which had been randomly collected from patients in our tertiary-care hospital in Korea between January and March 2022. In alignment with the Ministry of Food and Drug Safety of Korea regulations, more than 10% of samples for evaluation of SARS-COV-2 tests had a cycle threshold value >30. Samples of influenza-positive cases during the time period of elevated influenza activity from January 2020, to March 2020, which had been randomly collected, were used, owing to the low level of influenza occurrence between May 2020 and March 2022. All samples were assigned a random number to facilitate blinding of the technicians performing the tests to the patient identifiers.
2.2. Sample size calculation
Sample sizes were calculated according to the approval guidelines for SARS-CoV-2 diagnostics issued by the Korean government [17]. Based on an estimated clinical sensitivity of 95%, with a lower confidence interval limit of 90%, an estimated clinical specificity of 97% with a lower confidence interval of 95%, and an estimated failure rate of 10%, the minimum sample sizes of SARS-CoV-2-positive and -negative samples were determined to be 167 and 556, respectively. For influenza, based on an estimated clinical sensitivity of 96% with a lower confidence interval of 90%, an estimated clinical sensitivity of 95% with a lower confidence interval of 90%, and a significance level (type I error) of 0.05, a power (type II error) of 80% and an estimated failure rate of 10%, the minimum sample sizes for influenza A-positive, influenza B-positive and influenza-negative samples were determined to be 94, 94 and 167, respectively.
2.3. Study design and laboratory testing
Results from the cobas SARS-CoV-2 & Influenza A/B (cobas 6800) assay obtained using a cobas 6800 instrument (Roche Molecular Systems) were used as the reference results for samples for SARS-CoV-2 testing. Results from the Allplex Respiratory Panel 1 (AllplexRP1; Seegene), which is a diagnostic assay for influenza A and B viruses, RSV A and -B, and influenza A virus subtyping, obtained using the STARlet system (Seegene) for extraction and a Bio-Rad CFX96 instrument (Bio-Rad; Hercules, CA) for rRT-PCR were used as the reference results for samples for influenza testing. All residual samples were stored at -70℃. Fig. 1 depicts study design. This study was approved by the Institutional Review Board of Asan Medical Center (#2022-0190).
Fig. 1.
Flow chart of the study design. (Left) For SARS-CoV-2 testing, samples were previously tested with the cobas SARS-CoV-2 & Influenza A/B as a reference assay. All samples were submitted to a test assay, the Liat SARS-CoV-2 Influenza A/B assay, and the Allplex SARS-CoV-2 assay was used as a comparison assay. (Right) For influenza testing, samples were previously tested with the Allplex Respiratory Panel 1 as a reference assay. All samples were submitted to a test assay, Liat SARS-CoV-2 Influenza A/B assay, and Liat Influenza A/B & RSV assay was used as a comparison assay.
Samples for SARS-CoV-2 testing were simultaneously submitted to the LiatCOVID/flu assay using the cobas Liat Analyzer and the Allplex SARS-CoV-2 assay (AllplexCOVID). Samples for influenza testing were simultaneously submitted to the LiatCOVID/flu assay and the cobas Liat Influenza A/B & RSV assay (LiatFlu/RSV) using the cobas Liat Analyzer. AllplexCOVID for SARS-CoV-2 testing and LiatFlu/RSV for influenza testing were performed concurrently as the comparison method.
2.4. Performance evaluation and comparison analysis
The positive percent agreement (PPA), negative percent agreement (NPA), and Cohen's kappa value for the LiatCOVID/flu test were calculated in comparison to the reference methods. The overall agreement rates and Cohen's kappa value were also calculated between LiatCOVID/flu and the comparison methods. An “invalid” result was defined as a negative finding for all target genes and the internal control. Invalid results were excluded from subsequent analysis. In the AllplexCOVID assay, an “inconclusive” result was defined by positivity for 1 or 2 only of the 3 target genes and such cases were also excluded from the evaluation owing to the uncertainty behind these test data, including prolonged shedding, genomic variations, contamination, and nonspecific amplification [18], [19], [20].
2.5. Subsequent analysis of Ct values
The cycle threshold (Ct) values from the LiatCOVID/flu assay for SARS-CoV-2 were compared to those from E gene target of the cobas 6800 and AllplexCOVID using linear regression and scatter plots. For influenza viruses, a comparison of Ct values from LiatCOVID/flu to those obtained with the AllplexRP1 and LiatFlu/RSV test systems was also made using the same analysis. In addition, the statistical significance of any Ct differences between 2 tests was evaluated using a paired samples t test. MedCalc 20.015 (MedCalc Software Ltd; Ostend, Belgium) was used for all statistical analyses including linear regression, Cohen's Kappa test and paired samples t test.
3. Results
3.1. Sample collection
A total of 1147 samples were collected. For SARS-CoV-2 evaluations, 167 SARS-CoV-2-positive and 556 SARS-CoV-2-negative samples were included. In the influenza virus evaluations, 224 influenza-positive and 200 influenza-negative samples were included. The 224 influenza-positive cases comprised 112 A and 112 B subtypes.
3.2. Clinical performance
The PPAs/NPAs of the LiatCOVID/flu assay when compared to the cobas 6800 for SARS-CoV-2 in 723 samples and to the AllplexRP1 for influenza viruses in 424 samples were 100%/100% and 99.6%/100%, respectively (Table 1 ). One Liat false-negative case in the influenza virus testing was found to be influenza A-positive on AllplexRP1 2 years prior but was also invalid on the LiatFlu/RSV test. Five influenza B-positive cases were also positive for the influenza A target of LiatCOVID/flu but all had Ct values from this target of more than 30. From the LiatCOVID/flu testing, all Ct values from the influenza A target in the influenza A-positive cases were less than 30. In addition, 48 (24%) influenza-negative samples, of which 47 were confirmed COVID-19 patients, were SARS-CoV-2-positive on LiatCOVID/flu. The remaining Liat SARS-CoV-2 positive among the influenza-negative samples was positioned between 2 SARS-CoV-2 positive samples during testing. One influenza A-positive sample showed SARS-CoV-2-positivity, but was tested between 2 SARS-CoV-2-positive samples and was taken when the SARS-CoV-2 prevalence was extremely low in Korea.
Table 1.
Clinical performance of the cobas Liat SARS‑CoV‑2 & Influenza A/B assay (LiatCOVID/flu) compared with Allplex Respiratory Panel 1 (AllplexRP1), cobas SARS-CoV-2 and Influenza A/B (cobas 6800) in the detection of influenza viruses and SARS-CoV-2.
| cobas 6800 (SARS-CoV-2) |
AllplexRP1 (Influenza) |
|||||
|---|---|---|---|---|---|---|
| LiatCOVID/flu | Positive | Negative | Total | Positive | Negative | Total |
| Positive | 167 | 0 | 167 | 223a | 0 | 223 |
| Negative | 0 | 556 | 556 | 1b | 200c | 201 |
| Total | 167 | 556 | 723 | 224 | 200 | 424 |
| Positive percent agreement% | 100.0% | 99.6% | ||||
| Negative percent agreement% | 100.0% | 100.0% | ||||
| Cohen's kappa (95% confidence interval) | 0.995 (0.986–1.000) | 1.000 (1.000–1.000) | ||||
Five influenza B-positives were also influenza A-positive with cycle threshold value of >30 on the LiatCOVID/flu.
this sample was positive 2 years ago.
Forty-eight samples were SARS-CoV-2 positive by LiatCOVID/flu.
3.3. Comparison with other rRT-PCR methods
The AllplexCOVID assay yielded 4 inconclusive results, all of which were positive on both the LiatCOVID/flu and cobas 6800 tests. These 4 cases were excluded from any subsequent comparative analysis. The overall percentage agreement (OPA) between the LiatCOVID/flu and AllplexCOVID test findings was 99.9% (718/719). The only mismatch between these 2 assays was a SARS-CoV-2-positive sample on LiatCOVID/flu and cobas 6800 that was negative on AllplexCOVID (Table 2 ). LiatFlu/RSV yielded 2 invalid results in 1 influenza A-positive sample and 1 influenza B-positive sample. These cases were excluded from subsequent comparisons between the LiatCOVID/flu and LiatFlu/RSV data. The OPA between LiatCOVID/flu and LiatFlu/RSV was 100% (422/422; Table 3 ). Five positive cases for both influenza A and B targets in the LiatCOVID/flu assay system were positive for only the influenza B target of LiatFlu/RSV. The LiatFlu/RSV test detected RSV on 7 influenza A-positive, 3 influenza B-positive, and 1 influenza-negative samples. Of these specimens, 2 influenza A-positive samples with Ct values >35 for the RSV target were not positive for RSV on AllplexRP1. The other samples were positive for RSV subtype B on AllplexRP1.
Table 2.
Comparison of cobas Liat SARS‑CoV‑2 & Influenza A/B assay (LiatCOVID/flu) and the Allplex SARS-CoV-2 assay (AllplexCOVID) in the detection of SARS-CoV-2.
| AllplexCOVID | |||
|---|---|---|---|
| LiatCOVID/flu | Positive | Negative | Total |
| Positive | 162 | 1 | 163 |
| Negative | 0 | 556 | 556 |
| Total | 162 | 557 | 719 |
| Overall percent agreement | 99.9% | ||
| Cohen's kappa (95% confidence interval) | 1.000 (1.000–1.000) | ||
Four cases showing positivity for 1 or 2 of the 3 target genes from AllplexCOVID. Subsequently, these samples were excluded from the analysis.
Table 3.
Comparison of cobas Liat SARS‑CoV‑2 & Influenza A/B assay (LiatCOVID/flu) and cobas Liat Influenza A/B & RSV assay (LiatFlu/RSV) in the detection of influenza viruses.
| LiatFlu/RSV | |||
|---|---|---|---|
| LiatCOVID/flu | Positive | Negative | Total |
| Positive | 222 | 0 | 222 |
| Negative | 0 | 200 | 200 |
| Total | 222 | 200 | 422 |
| Overall percent agreement | 100.0% | ||
| Cohen's kappa (95% confidence interval) | 0.996 (0.998–1.000) | ||
Two cases showing invalid results on the LiatFlu/RSV. Subsequently, these samples were excluded from the analysis.
3.4. Correlations between the Ct values
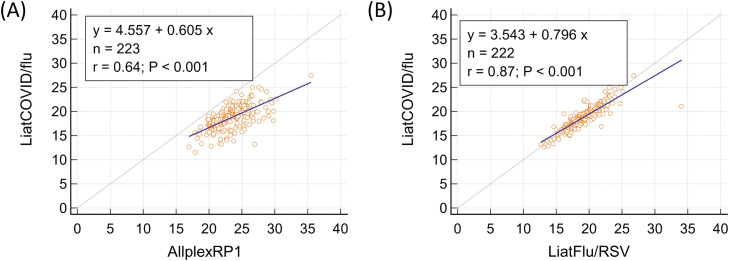
In relation to SARS-CoV-2 testing, correlations between the Ct values obtained from the 3 assay panels are depicted in Fig. 2 . The Ct value for the SARS-CoV-2 target of LiatCOVID/flu was significantly lower than that for the E gene target in both the cobas 6800 (Difference: 3.63±0.12; P < 0.0001) and AllplexCOVID systems (Difference: 3.94±0.16; P < 0.0001). Correlations between the Ct values of the 3 panels with regard to influenza testing are depicted in Fig. 3 . This value for LiatCOVID/flu was lower than for AllplexRP1 (Difference: 4.70±0.15; P < 0.0001) and marginally, but still statistically significantly, lower than that for LiatFlu/RSV (Difference: 0.36±0.09; P < 0.0001).
Fig. 2.
Correlation between the Ct values for the cobas Liat SARS‑CoV‑2 & Influenza A/B assay (LiatCOVID/flu) and E gene target of the cobas SARS-CoV-2 & Influenza A/B (cobas 6800) tests (A), and for SARS-CoV-2 testing, those for the LiatCOVID/flu assay and E gene target of the Allplex SARS-CoV-2 assay (AllplexCOVID) (B).
Fig. 3.
Correlation between the Ct values for the cobas Liat SARS‑CoV‑2 & Influenza A/B assay (LiatCOVID/flu) and Allplex Respiratory Panel 1 (AllplexRP1) (A), and LiatCOVID/flu and cobas Liat Influenza A/B & RSV assay (LiatFlu/RSV) (B) for influenza testing.
4. Discussion
We here present our clinical evaluation of the LiatCOVID/flu testing system for the detection and subtyping of influenza viruses and SARS-CoV-2 using a cohort of more than 1000 clinical NPS specimens. Because some previous studies have described meaningful false-positive rates for SARS-CoV-2 detection [10,14], we have also here reevaluated more than 500 previously documented SARS-CoV-2-negative samples. Nevertheless, LiatCOVID/flu showed a reliable performance delivering a PPA and NPA of nearly 100% for both viruses. Moreover, an OPA of nearly 100% was found for both viruses.
One of the SARS-CoV-2-positive cases in our sample population was positive on the cobas 6800 and LiatCOVID/flu, but negative on AllplexCOVID. Considering a limit of detection (LoD) of 46.4 copies/mL for the cobas 6800 and 214.5 copies/mL for AllplexCOVID (unpublished data) and a manufacturer stated LoD of 24 copies/mL for LiatCOVID/flu, this case could possibly be explained by differences in the analytical sensitivities of these tests. SARS-CoV-2 positivity in the influenza virus evaluation was mostly due to the high prevalence of COVID-19 during the Omicron variant outbreak period in Korea [4]. Notably however, 2 of our cases, which were tested in positions adjacent to SARS-CoV-2 positive samples, were possibly affected by cross-contamination. Although the false-positivity rates with the LiatCOVID/flu assay is a topic of some debate [10,[13], [14], [15]], caution should be exercised in this regard.
One false-negative case emerged from our influenza detection analyses on LiatFlu/RSV, but may have been caused by degeneration of the virus during the 2-year storage period for our samples. However, 5 influenza B-positive cases in our current cohort were also found to be positive for influenza A with a Ct value > 30 for the influenza A target. Because these samples were not positive for the influenza A targets on LiatFlu/RSV and AllplexCOVID, this finding is possibly related to cross-reactivity of influenza B to the influenza A target on LiatCOVID/flu, in addition to possible cross-contamination. Hence, influenza A-positivity with a Ct value >30 in an influenza B-positive sample for the LiatCOVID/flu assay should be assessed as a possible false-positive result.
The LiatCOVID/flu assay had a significantly lower Ct value for the SARS-CoV-2 target than either the AllplexCOVID or cobas 6800 tests. On the other hand, LiatFlu/RSV and LiatCOVID/flu had similar Ct values for their influenza target, for which AllplexRP1 showed a significantly delayed Ct value in comparison. Although the reporting and clinical utilization of Ct values is not recommended by various professional bodies [21,22], numerous laboratories have been doing this for SARS-CoV-2 testing. In line with this, findings from this study also discourage the reporting and clinical utilization of Ct value.
We found in our present analyses LiatFlu/RSV detected 11 RSV-positive cases that were not detected using a conventional single target approach. Among these 11 samples, 8 cases were positive for both the RSV subtype B and influenza viruses. Because the cocirculation of several respiratory viruses has occasionally been observed [23,24], a dual target approach using LiatFlu/RSV and LiatCOVID/flu would likely detect patients infected by both the SARS-CoV-2 and influenza viruses. Notably however, false positivity should be considered when the Ct value for the RSV target is greater than 35.
This study had some limitations of note. First, not all of our study samples were subtyped. However, all of the SARS-CoV-2-positive samples were regarded as delta or Omicron variants due to the regional epidemiology during the study period [4]. Second, samples with both influenza virus and SARS-CoV-2 could not be analyzed. There were no positive cases for the influenza target among 20,668 respiratory cases tested by multiplex rRT-PCR between May 2020 and March 2022 in this institution. Third, many influenza-negative samples were SARS-CoV-2-positive, and this was not considered during the study design. Hence, the influenza-negative samples showing SARS-CoV-2 positivity were not included in our SARS-CoV-2 detection analyses.
5. Conclusions
In conclusion, LiatCOVID/flu shows satisfactory performance in the detection of influenza viruses and SARS-CoV-2 from an NPS. Considering that rapid detection and isolation are critical for combating outbreaks [25], a rapid rRT-PCR test is a valuable tool. Concurrent testing of SARS-CoV-2 and the influenza viruses within 20 minutes may be helpful in combating a possible future dual pandemic of these 2 viruses.
Author's contributions
KP was responsible for data curation, writing – original draft. HS contributed to the supervision, conceptualization, methodology, funding acquisition. MNK completed supervision, writing – review & editing.
Funding
This work was funded by Roche Molecular Systems, Inc. (Pleasanton, CA). This funding source had no role in study design, data collection, data analysis, data interpretation, or writing of the report. This work was supported by the Korean Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health and Welfare, Korea (grant no. HI18C2383).
Declaration of competing interest
The authors report no conflicts of interest relevant to this article.
Acknowledgments
We would like to thank Boston BioEdit and Editage for English language editing.
References
- 1.World Health Organization. WHO coronavirus (COVID-19) dashboard, Available from: https://covid19.who.int/; 2022 [17th May 2022].
- 2.Elbe S, Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall. 2017;1(1):33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JJ, Choe YJ, Jeong H, Kim M, Kim S, Yoo H, et al. Importation and transmission of SARS-CoV-2 B.1.1.529 (Omicron) variant of concern in Korea, November 2021. J Korean Med Sci. 2021;36(50):e346. doi: 10.3346/jkms.2021.36.e346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ministry of Health and Welfare. Coronavirus (COVID-19), Republic of Korea, Available from: http://ncov.mohw.go.kr/en; 2022 [ 17th May 2022].
- 5.Walensky RP, Del Rio C. From mitigation to containment of the COVID-19 pandemic: putting the SARS-CoV-2 genie back in the bottle. JAMA. 2020;323(19):1889–1890. doi: 10.1001/jama.2020.6572. [DOI] [PubMed] [Google Scholar]
- 6.Corman VM, Haage VC, Bleicker T, Schmidt ML, Mühlemann B, Zuchowski M, et al. Comparison of seven commercial SARS-CoV-2 rapid point-of-care antigen tests: a single-centre laboratory evaluation study. Lancet Microbe. 2021;2(7):e311–e319. doi: 10.1016/S2666-5247(21)00056-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramachandran A, Noble J, Deucher A, Miller S, Tang PW, Wang RC. Performance of Abbott ID-Now rapid nucleic amplification test for laboratory identification of COVID-19 in asymptomatic emergency department patients. J Am Coll Emerg Physicians Open. 2021;2(6):e12592. doi: 10.1002/emp2.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ujiie M, Tsuzuki S, Nakamoto T, Iwamoto N. Resurgence of respiratory syncytial virus infections during COVID-19 pandemic, Tokyo, Japan. Emerg Infect Dis. 2021;27(11):2969–2970. doi: 10.3201/eid2711.211565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim KR, Park H, Kim DR, Kim YJ. Changes in epidemiology of parainfluenza virus and respiratory syncytial virus infection during coronavirus disease 2019 pandemic in Korea. Clin Exp Pediatr. 2022;65(6):320–321. doi: 10.3345/cep.2021.01823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blackall D, Moreno R, Jin J, Plotinsky R, Dworkin R, Oethinger M. Performance characteristics of the Roche Diagnostics cobas Liat PCR system as a COVID-19 screening tool for hospital admissions in a regional health care delivery system. J Clin Microbiol. 2021;59(10) doi: 10.1128/JCM.01278-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Er TK, Chou YC, Chen SY, Huang JW. Rapid Cobas Liat SARS-CoV-2 assay in comparison with the laboratory-developed real-time RT-PCR test. Clin Lab. 2021;67(11) doi: 10.7754/Clin.Lab.2021.210316. [DOI] [PubMed] [Google Scholar]
- 12.Hansen G, Marino J, Wang ZX, Beavis KG, Rodrigo J, Labog K, et al. Clinical performance of the point-of-care cobas Liat for detection of SARS-CoV-2 in 20 minutes: a multicenter study. J Clin Microbiol. 2021;59(2):e02811–e02820. doi: 10.1128/JCM.02811-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jian MJ, Chung HY, Chang CK, Lin JC, Yeh KM, Chen CW, et al. Clinical comparison of three sample-to-answer systems for detecting SARS-CoV-2 in B.1.1.7 lineage emergence. Infect Drug Resist. 2021;14:3255–3261. doi: 10.2147/IDR.S328327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahmoud SA, Ganesan S, Ibrahim E, Thakre B, Teddy JG, Raheja P, et al. Evaluation of six different rapid methods for nucleic acid detection of SARS-COV-2 virus. J Med Virol. 2021;93(9):5538–5543. doi: 10.1002/jmv.27090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsang HF, Leung WMS, Chan LWC, Cho WCS, Wong SCC. Performance comparison of the Cobas® Liat® and Cepheid® GeneXpert® systems on SARS-CoV-2 detection in nasopharyngeal swab and posterior oropharyngeal saliva. Expert Rev Mol Diagn. 2021;21(5):515–518. doi: 10.1080/14737159.2021.1919513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akashi Y, Horie M, Kiyotaki J, Takeuchi Y, Togashi K, Adachi Y, et al. Clinical performance of the cobas Liat SARS-CoV-2 & Influenza A/B Assay in nasal samples. Mol Diagn Ther. 2022;26(3):323–331. doi: 10.1007/s40291-022-00580-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ministry of Food and Drug Safety. Guideline on the review and approval of in vitro diagnostic devices for COVID-19, Available from: https://www.mfds.go.kr/eng/brd/m_40/list.do; 2022 [24th May 2022].
- 18.Peñarrubia L, Ruiz M, Porco R, Rao SN, Juanola-Falgarona M, Manissero D, et al. Multiple assays in a real-time RT-PCR SARS-CoV-2 panel can mitigate the risk of loss of sensitivity by new genomic variants during the COVID-19 outbreak. Int J Infect Dis. 2020;97:225–229. doi: 10.1016/j.ijid.2020.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sung H, Roh KH, Hong KH, Seong MW, Ryoo N, Kim HS, et al. COVID-19 molecular testing in Korea: practical essentials and answers from experts based on experiences of emergency use authorization assays. Ann Lab Med. 2020;40(6):439–447. doi: 10.3343/alm.2020.40.6.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alsaud AE, Nair AP, Matarneh AS, Sasi S, El Hassan R, Khan F, et al. Case report: prolonged viral shedding in six COVID-19 patients. Am J Trop Med Hyg. 2021;104(4):1472–1475. doi: 10.4269/ajtmh.20-0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.American Association for Clinical Chemistry. AACC Recommendation for reporting SARS-CoV-2 cycle threshold (CT) values, Available from: https://www.aacc.org/science-and-research/covid-19-resources/statements-on-covid-19-testing/aacc-recommendation-for-reporting-sars-cov-2-cycle-threshold-ct-values; 2021 [17th May 2022].
- 22.Infectious Disease Society of America and Association for Molecular Pathology. IDSA and AMP joint statement on the use of SARS-CoV-2 PCR cycle threshold (Ct) values for clinical decision-making, Available from: https://www.idsociety.org/globalassets/idsa/public-health/covid-19/idsa-amp-statement.pdf; 2021 [17th May 2022].
- 23.Pascalis H, Temmam S, Turpin M, Rollot O, Flahault A, Carrat F, et al. Intense co-circulation of non-influenza respiratory viruses during the first wave of pandemic influenza pH1N1/2009: a cohort study in Reunion Island. PLoS One. 2012;7(9):e44755. doi: 10.1371/journal.pone.0044755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackay IM, Arden KE, Speicher DJ, O'Neil NT, McErlean PK, Greer RM, et al. Co-circulation of four human coronaviruses (HCoVs) in Queensland children with acute respiratory tract illnesses in 2004. Viruses. 2012;4(4):637–653. doi: 10.3390/v4040637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luciani M, Di Febo T, Zilli K, Di Giannatale E, Armillotta G, Manna L, et al. Rapid Detection and Isolation of Escherichia coli O104:H4 from Milk Using Monoclonal Antibody-coated Magnetic Beads. Front Microbiol. 2016;7:942. doi: 10.3389/fmicb.2016.00942. [DOI] [PMC free article] [PubMed] [Google Scholar]