Abstract
Phage tail-like bacteriocins (PTLBs) are large proteomic structures similar to the tail phages. These structures function in bacterial competition by making pores in the membrane of their competitors. The PTLBs identified in Pseudomonas aeruginosa are known as R-type and F-type pyocins, which have a narrow spectrum of action. Their specificity is determined by the tail fiber and is closely related to the lipopolysaccharide type of the target competitor strain. In this study, the genome sequences of 32 clinical of P. aeruginosa clinical isolates were analysed to investigate the presence of R-type and F-type pyocins, and one was detected in all strains tested. The pyocins were classified into 4 groups on the basis of the tail fiber and also the homology, phylogeny and structure of the cluster components. A relationship was established between these groups and the sequence type and serotype of the strain of origin and finally the killing spectrum of the representative pyocins was determined showing a variable range of activity between 0 and 37.5%. The findings showed that these pyocins could potentially be used for typing of P. aeruginosa clinical isolates, on the basis of their genomic sequence and cluster structure, and also as antimicrobial agents.
Subject terms: Molecular medicine, Bacterial genetics
Introduction
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen, responsible for nosocomial infections, including bloodstream infections, pneumonia, urinary tract infections and surgical site infections1. This pathogen represents a serious problem in health care systems because of its capacity to acquire antibiotic resistance and its ability to produce biofilms and persist on surfaces, thereby contributing to its spread and causing outbreaks2. P. aeruginosa contains many intrinsic antibiotic resistance mechanisms that make this species a difficult to treat multidrug resistant bacteria3. In addition, due to the outbreaks, it is necessary to type the causative bacterial pathogen. The traditional typing methods, Pulse Field Gel Electrophoresis (PFGE) and the Multilocus Sequencing Typing (MLST), are the primary election in many clinical laboratories, but although they are very effective it should be necessary to improve the discrimination inside each Sequence Type (ST)4. So, new antimicrobial agents as well as new typing methods are therefore required.
Bacteria utilize phage tail-like bacteriocins (PTLBs) to enable them to compete with other strains of the same species or with different species5 and could therefore be good candidates for use as antimicrobial agents. PTLBs are large, ribosomally synthetized6 protein structures (2 × 106–1 × 107 KDa) encoded in the bacterial genome and structured in genetic clusters similar to the phage tail structure models, always encoding structural and assembly tail proteins, a lysis cassette and preceded by the transcription regulatory proteins5.
P. aeruginosa produces PTLBs called pyocins, traditionally named after the species of origin, in this case P. aeruginosa also known as P. pyocyanea5. The PTBLs from P. aeruginosa are some of the most widely studied PTLBs and are used as PTLBs models5. The P. aeruginosa pyocins are grouped into three types: S-type pyocins, R-type pyocins and F-type pyocins. The S-type pyocins, not considered PTLBs, are colicin-like proteins composed by large multi domain polypeptides with DNase activity7. The R-type pyocins and F-type pyocins are considered PTLBs and differ in their structure. R-type (Rigid-type) pyocins are contractile tail particles similar to the Myoviridae phage family and are composed by a long large tube surrounded by a sheath and ending in a baseplate where the receptor-binding protein (RBP) is located. The F-type (Flexible-type) pyocins are non-contractile particles, similar to the Siphoviridae phage family; they are simpler than the R-type as they do not possess a sheath, but also have an RBP5,8. Despite the great morphological similarity of PTBLs with phages, they cannot be considered degenerate phages but both would have a common cellular ancestry along with the type VI secretion system5,8,9.
The killing mechanism of R-type and F-type pyocins involves the phage tail fiber possessing the RBP, which recognizes the bacterial receptor in the lipopolysaccharide (LPS). The specificity of the RBP leads to a narrow spectrum of activity and the protection of the pyocin producer strain, although the producer cell dies altruistically due to the pyocins release5; once the cell is bound, the R-type pyocin triggers contraction of the sheath by pulling the tail and the core through the envelope, forming a channel or pore that decouples the membrane potential thus inhibiting membrane transport and causing cell death3,5. The killing mechanism of the F-type also involves disruption of the membrane gradient, but as it lacks a tube or core that forms a channel, its mechanism of action is not yet well understood5,10. The S-type pyocins are secreted as binary protein complexes that contain an effector, which is a larger protein with killing activity including DNase activity, the second component is a smaller protein with immunity activity that protects the producer strain from the activity of the released pyocin11,12.
To date, five subtypes of R-type pyocins have been described on the basis of their target specificity: R1, R2, R3, R4 and R58,12,13. Of these, R5 has the broadest spectrum, while R2 has the narrowest spectrum and encompasses the spectra of R3 and R4, which are similar. R1 is a subset of R5 but is considered a different branch. In addition, the amino acid sequence of the tail fiber protein is similar in R2, R3 and R4, but very different in R1 and R55,13,14. The F-type pyocins have been less well studied and are currently classified into three groups (F1, F2 and F3) based on their lytic spectra, although another two groups have also been identified: one in P. aeruginosa PA14 and other in P. aeruginosa M18. The former is similar to F2, identified in the reference strain P. aeruginosa PAO1, but with differences in 210 to 340 residues from the tail fiber protein; the M18 F-type shares 66% homology with the tail fiber protein of the F1 group12.
PTBLs are encoded in gene clusters preceded by a group of regulatory genes and organised in blocks of lytic and structural genes. These clusters can be found encoding individual R-type or F-type pyocins but also in dual R-F pyocin clusters. It has previously been reported that all clusters in P. aeruginosa, i.e. both individual and the dual R-F type, are located in the genome in the intergenic region of the tryptophan operon between the trpE and trpGCD genes5,12. The regulatory system of both individual and dual R-type and F-type pyocins is located upstream of the cluster and is composed by an activator (prtN) and a repressor (prtR); these are related to recA, which is activated by DNA damage, cleaving prtR and producing prtN and thus activating expression of the pyocin cluster5,12,15. The production of both R-type and F-type pyocins in dual R-F pyocins relies on the shared regulatory system and also the shared lysis cassette, suggesting coordinated release of the dual pyocins8,12.
PTLBs are characterized by a narrow host spectrum, which in P. aeruginosa is known to be closely related to the LPS type of the target strain that acts as a receptor for the tail fiber proteins14. It has also been reported that each specific type of pyocin recognizes a specific serotype strain determined by the O-polysaccharide repeating portion, which acts as an O-antigen and is linked to the virulence of the strains14,16. The PTLBs, and more specifically the P. aeruginosa pyocins, can be used as antimicrobial agents and owing to their relationship with the O-antigen they were used in typing schemes before the emergence of molecular typing methods17–19. The value of pyocins as antimicrobial agents is currently being considered in research studies. The R-type pyocins were used for the first time in 1969 to rescue chick embryos infected with P. aeruginosa20. In addition, several assays in murine models confirmed the efficacy of R-type pyocins in the treatment of infections with P. aeruginosa13,21,22. Due to the narrow spectrum of the PTLBs, some studies focused on generating recombinant R-type pyocins to retarget them by substituting the tail fiber protein with a phage protein in order to increase the host range in several species such as P. aeruginosa, Clostridioides difficile and Listeria monocytogenes10,23–26.
In the search for new pyocins, 32 genome sequences of clinical strains of P. aeruginosa of different clinical origins were analysed in the present study, and at least one pyocin cluster was identified in each strain. The sequence of these pyocins was analysed and homology and phylogenetic studies were conducted. The pyocins were isolated and purified and their killing spectrum was established, and they were also related to the serotype and ST of the clinical isolates.
Results
Identification and characterization of the phage tail-like bacteriocins
Thirty-two genomic sequences of P. aeruginosa were analysed to search for PTLBs (Table 1). In all of the genomes analysed, at least one cluster corresponding to a pyocin was found between tryptophan operon genes, trpE and trpG (Table 2; Table 1S. Supplementary material). Thus, 21 of the strains contained a cluster that corresponded to a unique pyocin corresponding to an R-type pyocin. Dual clusters were identified in the 11 remaining strains. In the P. aeruginosa PAO1 reference strain5, one of these clusters corresponded to a R-type pyocin, and the contiguous cluster corresponded to an F-type pyocin, both sharing the regulatory and lytic genes, as previously described for P. aeruginosa PAO112.
Table 1.
Characteristics of the 32 clinical isolates of P. aeruginosa which were obtained in previous study30.
| Strain | Genbank | Origin | ST | Serotype |
|---|---|---|---|---|
| 1-13 | SAMN14776823 | IAI | ST235 | O11 |
| 2-29 | SAMN14776826 | UTI | ST235 | O11 |
| 3-49 | SAMN14776829 | IAI | ST235 | O11 |
| 4-17 | SAMN14776829 | UTI | ST235 | O11 |
| 4-71 | SAMN14776839 | IAI | ST235 | O11 |
| 4-79 | SAMN14776840 | UTI | ST235 | O11 |
| 4-86 | SAMN14776841 | IAI | ST235 | O11 |
| 4-92 | SAMN14776842 | UTI | ST235 | O11 |
| 4-93 | SAMN14776843 | UTI | ST235 | O11 |
| 4-94 | SAMN14776844 | IAI | ST235 | O11 |
| 4-120 | SAMN14776833 | UTI | ST235 | O11 |
| 4-121 | SAMN14776834 | UTI | ST235 | O11 |
| 9-41 | SAMN14776860 | LRTI | ST235-1LV | O11 |
| C11 | SAMN14776862 | UTI | ST175 | O4 |
| C58 | SAMN14776863 | UTI | ST175-2LV | O4 |
| G6 | SAMN14776870 | IAI | ST175-1LV | O4 |
| G7 | SAMN14776871 | IAI | ST175-2LV | O4 |
| G26 | SAMN14776868 | IAI | ST175-1LV | O4 |
| G31 | SAMN14776869 | IAI | ST175-1LV | O4 |
| H18 | SAMN14776872 | UTI | ST175-2LV | O4 |
| H52 | SAMN14776874 | UTI | ST309 | O11 |
| 3-5 | SAMN14776830 | UTI | ST348-1LV | O11 |
| 3-38 | SAMN14776827 | LRTI | ST348 | O12 |
| 3-41 | SAMN14776828 | LRTI | ST348-1LV | O12 |
| 3-58 | SAMN14776831 | IAI | ST348 | O11 |
| 9-86 | SAMN14776861 | IAI | ST554 | O5 |
| 5-23 | SAMN14776846 | LRTI | ST244 | O5 |
| 6-25 | SAMN14776848 | LRTI | ST244-1LV | O12 |
| 8-24 | SAMN14776854 | UTI | ST244-1LV | O5 |
| 8-36 | SAMN14776855 | UTI | ST244 | O5 |
| 9-25 | SAMN14776858 | LRTI | ST244-1LV | O12 |
| 10-58 | SAMN14776820 | LRTI | ST244 | O12 |
It is indicated each Genbank code, clinical origin, sequence type (ST) and serotype.
IAI intra-abdominal infection, UTI urinary tract infection, LRTI lower respiratory tract infection.
Table 2.
Genomic characteristics and subtype of the 32 pyocins (PTBLs) identified in this study.
| Pyocin name | Pyocin subtype | Genbank | Genomic lenght (pb) | % GC | ORF | Group |
|---|---|---|---|---|---|---|
| 1-13_pyoR5 | R5 | BK062625 | 16,471 | 65.4 | 23 | A |
| 2-29_pyoR5 | R5 | BK062626 | 16,471 | 65.4 | 23 | |
| 3-49_pyoR5 | R5 | BK062627 | 16,471 | 65.4 | 23 | |
| 4-17_pyoR5 | R5 | BK062628 | 16,472 | 65.5 | 23 | |
| 4-71_pyoR5 | R5 | BK062629 | 16,471 | 65.4 | 23 | |
| 4-79_pyoR5 | R5 | BK062630 | 16,471 | 65.4 | 23 | |
| 4-86_pyoR5 | R5 | BK062631 | 16,471 | 65.4 | 23 | |
| 4-92_pyoR5 | R5 | BK062632 | 16,471 | 65.4 | 23 | |
| 4-93_pyoR5 | R5 | BK062633 | 16,471 | 65.4 | 23 | |
| 4-94_pyoR5 | R5 | BK062634 | 16,803 | 65.4 | 23 | |
| 4-120_pyoR5 | R5 | BK062635 | 16,471 | 65.4 | 23 | |
| 4-121_pyoR5 | R5 | BK062636 | 16,803 | 65.4 | 23 | |
| 9-41_pyoR5 | R5 | BK062645 | 16,803 | 65.3 | 23 | |
| C11_pyoR5 | R5 | BK062637 | 16,455 | 65.1 | 23 | B |
| C58_pyoR5 | R5 | BK062638 | 16,455 | 65.1 | 23 | |
| G6_pyoR5 | R5 | BK062639 | 16,312 | 65 | 23 | |
| G7_pyoR5 | R5 | BK062640 | 16,120 | 65.2 | 23 | |
| G26_pyoR5 | R5 | BK062641 | 16,455 | 65.1 | 22 | |
| G31_pyoR5 | R5 | BK062642 | 16,455 | 65.1 | 22 | |
| H18_pyoR5 | R5 | BK062643 | 16,120 | 65.2 | 22 | |
| H52_pyoR5 | R5 | BK062644 | 16,215 | 65.3 | 22 | |
| 3-5_pyoR2-F2 | R2-F2 | BK062615 | 30,282 | 63.9 | 39 | C |
| 3-38_pyoR2-F2 | R2-F2 | BK062614 | 30,282 | 63.9 | 39 | |
| 3-41_pyoR2-F2 | R2-F2 | BK062616 | 30,282 | 63.9 | 39 | |
| 3-58_pyoR2-F2 | R2-F2 | BK062617 | 30,283 | 63.9 | 39 | |
| 9-86_pyoR2-F2 | R2-F2 | BK062622 | 30,510 | 64 | 41 | |
| 5-23_pyoR2-F(PA14) | R2-F(PA14) | BK062618 | 28,526 | 64.2 | 36 | D |
| 6-25_pyoR2-F(PA14) | R2-F(PA14) | BK062619 | 28,805 | 64.2 | 36 | |
| 8-24_pyoR2-F(PA14) | R2-F(PA14) | BK062620 | 28,805 | 64.2 | 36 | |
| 8-36_pyoR2-F(PA14) | R2-F(PA14) | BK062621 | 28,526 | 64.2 | 36 | |
| 9-25_pyoR2-F(PA14) | R2-F(PA14) | BK062623 | 28,805 | 64.2 | 36 | |
| 10-58_pyoR2-F(PA14) | R2-F(PA14) | BK062624 | 28,526 | 64.2 | 36 |
Each pyocin name corresponds to the producer strain. It is indicated the Genbank code, the genome size, % GC, number of ORF and the group to which each was assigned.
In order to identify the PTLBs as R or F subtypes, homology analysis of the tail fiber was conducted. In the R-type pyocins, the tail fiber proteins were compared against the reference sequence for each R subtype (R1, R2, R3, R4 and R5); the results revealed that 21 of the proteins belonged to the R5 subtype, while 11 belonged to the R2 subtype, which corresponded to those that were followed by a F-type pyocin (Fig. 1A,B). For the F-type group, the results showed a group of 5 pyocins belonging to the F2 subtype, comprising a R2-F2 pyocin, similar to the PAO1 R2-F2 pyocin5, and a group of 6 that were similar to the P. aeruginosa PA14 F-type pyocin12, thus giving rise to a pyocin cluster R2-F(PA14) (Fig. 1A,B).
Figure 1.
Identification of the pyocin subgroup. (A) Homology comparison of the tail fiber protein model sequence of R2, R5, F2 and F(PA14) with one representative of each pyocin type identified in this study. (B) Structure of the pyocin cluster of one representative of each group (A, B, C and D), and comparison with the reference pyocin R2-F2 from P. aeruginosa PAO1. Yellow: regulatory genes; red: lysis cassette; blue: R-type pyocin cluster genes, brown: F-type pyocin cluster genes. The representative sequences were selected by numerical order due to the high intragroup similarity.
The genomic analysis of the pyocin clusters identified showed a % GC content very similar, between 63.9 and 65.4 (Table 2). Analysis of the protein sequence of the pyocins revealed some differences in the protein number between the R5-type pyocins (Table 2). Thus, these pyocins were classified in two groups, including a group of 12 pyocins (group A) constituted by an R-type cluster of 14 genes, 4 lytic genes and preceded by 5 regulator genes. The second group (group B) of 8 pyocins differed from group A in the absence of the latter protein belonging to the lytic cassette. In both groups, the cluster was preceded by the regulatory region composed by 5 genes, while in the reference P. aeruginosa PAO1 strain it is composed by 4 genes (Fig. 1B; Table 2)7. The R2-F2 pyocins (group C) were composed by 38 genes, which corresponded to an R-type cluster of 14 proteins and a F-type cluster composed by 16 genes, while in P. aeruginosa PAO1 the F-type cluster is formed by 17 genes. In addition, the two clusters share a regulatory region of five genes and a lytic cassette composed by four proteins (Fig. 1B). The pyocins R2-F(PA14) (group D) comprised two consecutive R and F clusters: the R-type comprised 14 proteins and the F-type comprised 13 proteins, unlike P. aeruginosa PAO1 and the group C pyocins, in which the last three proteins are duplicated (Fig. 1B)7.
Homology and phylogenetic analysis of pyocins
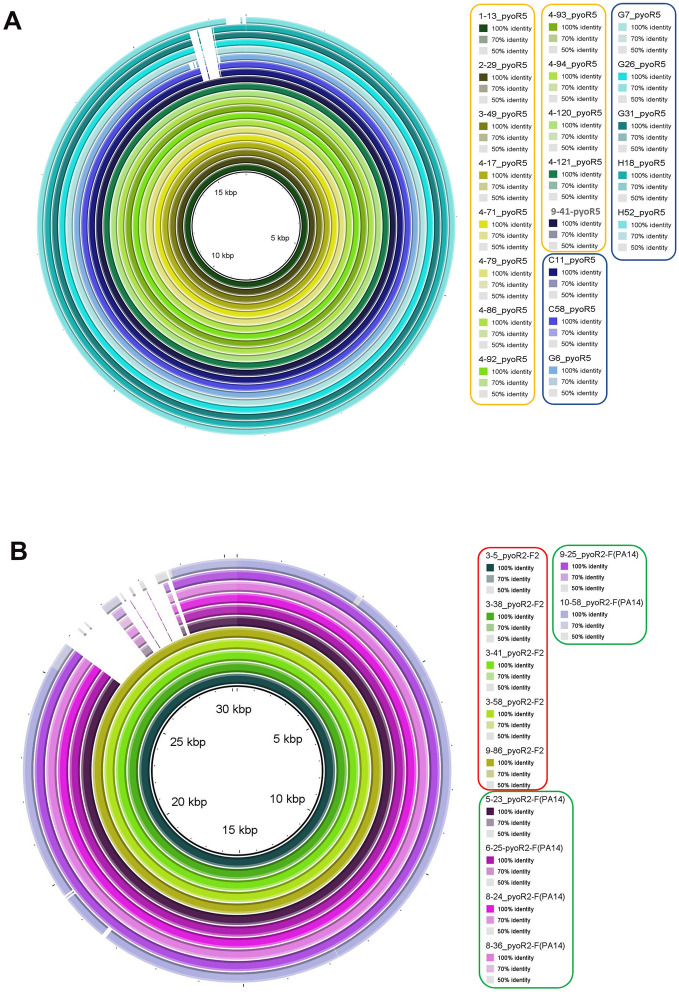
The results obtained by the homology studies of the tail fiber specificity genes were confirmed by the homology and phylogenetic analysis of the complete pyocin genomes. The homology analysis showed that the R5-type pyocins were very similar and can be grouped in two blocks corresponding to the established groups A and B, sharing a query cover value of 97–98% and an identity value of 99.35%. The pyocin H52-R5 homology BRIG differed slightly from the two blocks but had similar homology values (Fig. 2). In the case of the R-F pyocin clusters, the homology results showed two blocks of homology, one corresponding to the R2-F2 pyocins (group C) and another corresponding to the R2-F(PA14) pyocin (group D) (Fig. 2). Despite the presence of two groups, the pyocin clusters represented by them were also similar, with a query cover value between 88 and 89% and an identity value of 98%.
Figure 2.
Homology of the pyocin clusters determined by BRIG 0.95. (A) R-type pyocin clusters. The clusters corresponding to group A are rounded in yellow and to group B in blue. (B) R-F type clusters. The cluster corresponding to group C are rounded in red and to group D in green.
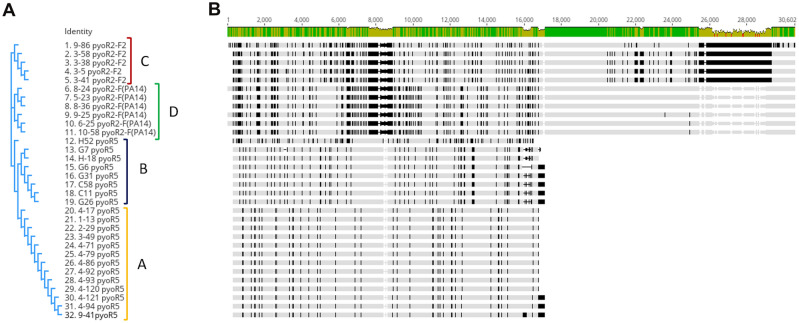
The phylogenetic study of all the pyocin clusters revealed, as previously observed, that the pyocins identified are divided into four phylogenetic groups. Two closely related clades of the phylogenetic tree were represented by two blocks, one corresponding to the R5-type pyocins included in group A and another also corresponding to an R5-type pyocin grouped in B. Another two closely related clades included one corresponding to group C, which was constituted by the R2-F2 pyocin and group D, constituted by the R2-F(PA14) (Fig. 3).
Figure 3.
Similarity study of the 32 pyocin clusters identified and its relation with the established groups. (A) Phylogenetic tree. (B) Alignment of the 32 pyocins clusters.
Identification of pyocins by transmission electron microscopy (TEM)
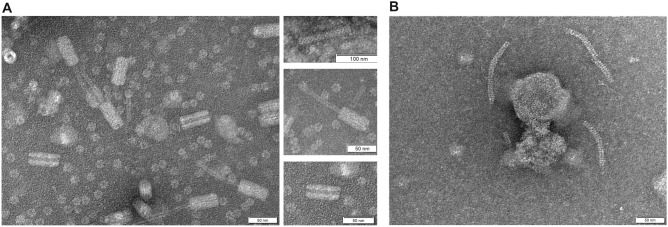
The purified pyocins were examined by TEM, and the images obtained (Fig. 4) revealed the presence of two different type of pyocins. One type had a structure similar to a tail of the viral family Myoviridae, corresponding to the R-type pyocins, observed in three conformations: a complete form, a contracted form and an empty sheath27. The other type was the F-type, observed as a flexible structure similar to a phage tail of the viral family Syphoviridae.
Figure 4.
TEM images of pyocins. (A) R-type pyocin with a similar structure to a myovirus, in three conformations. Top: complete form; middle: contracted form; bottom: empty sheath of the pyocin. (B) F-type pyocin with a similar structure to a syphovirus.
Relationship between the pyocin type and the source P. aeruginosa strain serotype, sequence type and clinical origin
In this analysis, the relationship between the serotype, the sequence type (ST) and the clinical origin of the source strains and the pyocin type was determined (Table 1). The R5-type pyocin in group A was almost always present in ST235 and serotype O11 clinical isolates. However, pyocin H52_pyoR5 was quite different in the homology and phylogenetic analysis, as it was present in the H52 clinical isolate belonging to the ST309 but like the rest of group A had the O11 serotype. The R5-type pyocins from group B were associated with clinical isolates belonging to the ST175 with serotype O4. On the other hand, the group C of R2-F2 pyocins was the most variable, with isolates belonging to the ST348 and ST554 and serotype O5, O11 and O12. Finally, group D, constituted by R2-F(PA14) pyocins, was represented by isolates belonging to ST244 and serotypes O5 and O12. Finally, no relationship between the clinical origin of the isolate and each pyocin was observed.
Killing spectrum of the pyocins
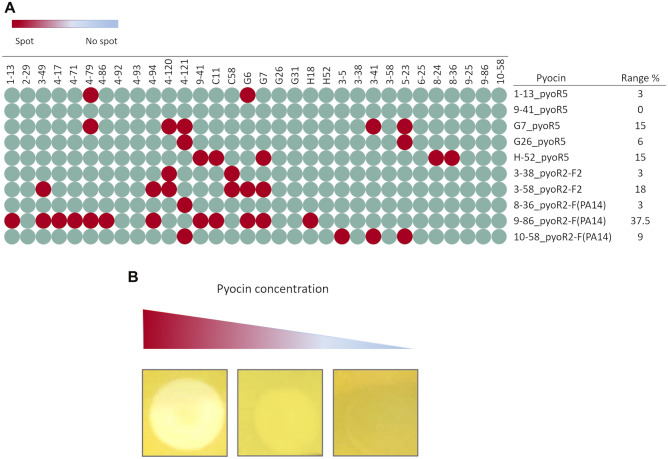
The target range of the pyocins was studied by the spot test technique (Fig. 5). The pyocins included in this analysis were selected by according to ST and serotype of the strain from which they were isolated. The results revealed a great variability in the susceptibility of the strains to the pyocins of the same subtype. In addition, no spots occurred when the target strain belonged to the same ST and serotype as the source strain of the pyocin, except for pyocin 10-58_R2-F(PA14), which produced a spot in strain 3–5, which belongs to a different ST but has the same serotype (O11). Furthermore, a variable percentage of target range was observed for all of the pyocins tested, with 9-86_pyoR2F2 showing the highest value and being able to lyse 37.5% of the strains tested.
Figure 5.
Target range of the pyocins. (A) Spot test of the pyocins originating from strains with different ST and serotype. (B) Spot production dependent of the pyocin concentration.
Discussion
Pyocins are PTLBs produced by P. aeruginosa. Like all PTBLs they are protein complexes with the same structure as phage tails. Like other phage tail particles, such as the type VI secretion systems (T6SS), they also play a role in defence and in interbacterial competition28. The genetic and structural similarities between the phage tails and PTBLs initially suggested that the PTBLs evolved as defective phages; however, structural comparison between the T6SS, R-type pyocins and the contractile tail phages suggests evolution from a common ancestor5,28.
The presence of pyocins in clinical strains of P. aeruginosa seems to be variable. Thus, in a study conducted by Mei et al.3, from an analysis of 852 clinical isolates of P. aeruginosa they found that 448 belonged to the R-type pyocin and 300 contained genes of the F-type pyocins. From those included in the R-type pyocins 144 belonged to the R1 subtype, 76 to the R2 subtype, 43 to the R5 subtype and the remaining strains were untypeables. Köhler et al.14 analysed the tracheal aspirates of 61 patients, in search of R-type pyocins, detecting different R subtype pyocins in 77% of the isolates. In another study of 24 isolates from the lungs of CF patients, all isolates were found to contain R-type pyocins, also mainly of the R1 subtype, and it was concluded that this subtype confers a competitive advantage in biofilms, explaining why certain strains displace others in the CF lungs6. In the present study, the genome of 32 clinical isolates of P. aeruginosa from several origins, including urinary tract infections, lower respiratory tract infections and intra-abdominal infections, were analysed in search of pyocins, and at least one cluster was found in all isolates. In contrast to the previously mentioned studies, both R-type clusters and F-type clusters were found, resulting in R pyocins and dual R-F pyocins. Analysis of the tail fibers, which determines the specificity (Fig. 1), showed that only R2, R5, F2 and F(PA14) were present in these isolates, with an equal representation of the R5 and R2 subtypes, and a lower representation of F2 than F(PA14) (Fig. 2). In other studies of CF isolates, the R1 subtype was the most commonly identified and in contrast to the results of the present study, R5 was the least well represented subtype3,6. In a study conducted by Köhler et al.14, pyocins from R1, R2 and R5 were found in the same proportions.
Homology modelling and phylogenetic studies of the pyocins identified in these clinical isolates confirmed the grouping of the pyocins and also revealed the great similarity between them. The main difference observed between the R2-type and the R5-type pyocins was in the genomic region corresponding to the tail fiber, and the same was observed between the F2-type and the F(PA14)-type pyocins (Fig. 3). The tail fiber region has been described as being responsible for the specificity of the pyocins, as it acts as an RBP recognizing the target in the LPS of the target bacteria5,8.
Analysis of the pyocins identified in the genomic sequence of the P. aeruginosa clinical isolates and the clinical ST, serotype and clinical origin established a relationship between the pyocin type and the ST and serotype, but not the clinical origin of the isolate. Although a relationship with both ST and serotype was identified, it seems that there is more variability between the serotype and the presence of pyocins than with the ST. Thus, the association between the pyocin subtype and the ST and serotype can be extended to the groups established in this work beyond the pyocin subtype, as the A and B groups, which both correspond to an R5-type pyocin, were associated with a different ST and serotype. Although the association between serotype and pyocin subtype has long been recognised14, to our knowledge the present is the first report of the relationship with ST. The relationship between the serotype and the pyocin subtype14 has always been established with the R-type cluster, but in the present study a relationship between the serotype and the F-type pyocin was demonstrated, as the group C and D pyocins only differ in the F-type cluster. Both groups were related to two different serotypes, but both share O12 and differ in O5 and O11, possibly as a consequence of the presence of the R-pyocin and the F-pyocin, which in this case would confer a competitive advantage to the strain as they are protected by different pyocins28. The observed relationship between the serotype, ST and pyocin from the clinical isolates of P. aeruginosa tested in this study suggests that the pyocins could potentially be used via analysis of the cluster genomic sequences. The pyocin-serotype association has been used for bacterial typing, but unlike in the present study, the typing was based on the killing activity of a pyocin from an unknown isolate over a collection of indicator strains; this method was abandoned, as it is slow and laborious, and was substituted by molecular methods17–19. Currently, thanks to the genome analysis, the study of the PTBLs can complement the traditional typing methods employed in the clinical laboratories.
The serotype in P. aeruginosa is determined by the O-antigen, which is a B-band repeating unit of LPS considered a virulence factor14. It is known that the pyocin tail fiber proteins recognize the LPS of the competitor strains but do not recognize their own LPS as a target, so they cannot lyse those strains with the same serotype. In a study carried out in 2010, Köhler et al.14 deleted different genes responsible for the synthesis of the O-antigen and determined which LPS residues act as receptors for R1-type, R2-type and R5-type pyocins. When the 32 clinical isolates were tested against the 32 isolated pyocins, none were able to lyse the source strain or those strains with the same serotype or ST. As previously described, variability in the susceptibility to the pyocins was observed between those isolates that shared ST and/or serotype and the same type of pyocin, probably due to the frequently observed mutations in the LPS genes in CF, affecting recognition by the RBP3,29,30. Pyocins have been considered potential alternatives to antibiotics, and several studies have demonstrated antimicrobial activity of pyocins alone or in combination with other antimicrobials and those sharing the LPS as target13,22,23,31,32.
In this study, 32 pyocins belonging to the R5-type, R2-F2 type and R2-F(PA14) type were identified. Homology and phylogenetic analysis established four groups of pyocins (A, B, C and D), each of which was found to be related to the serotype and ST of the source strain. Pyocins are therefore good candidate markers for typing strains of P. aeruginosa by analysis of the tail fiber protein of the pyocin. We also observed that they could be used to type R5-type pyocins, by the number and distribution of the genes comprising the pyocin cluster. These pyocins also displayed potential antimicrobial activity as they were able to lyse some of the clinical isolates tested, particularly the 9-86_pyoR2-F(PA14) pyocin, which exhibited the highest range of activity.
Material and methods
Strains and culture conditions
Thirty-two clinical strains of P. aeruginosa isolated in Portuguese and Spanish hospitals within the framework of two multicentre studies, STEP in Portugal and SUPERIOR in Spain33, were used in the study. The software mlst (v2.16.1) (https://github.com/tseemann/mlst) was used for the in silico MLST assignment33. Serogroups based on the O-specific antigen (OSA) gene cluster sequences were determined using Blastn tool (v2.9.0+) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and the OSA database33. The origin, ST and serotype of the isolates are shown in Table 1. All strains were grown in Luria–Bertani broth (LB) medium (0.5% NaCl, 0.5% yeast extract, 1% tryptone) at 37 °C and 180 rpm. LB was supplemented with agar (1.5%) when necessary.
Identification of pyocin clusters in silico
The genomes of 32 clinical isolates P. aeruginosa (NCBI BioProject: PRJNA629475) were analysed to search for pyocins. For this purpose, the anthranilate flanking genes, trpD and trpE, were identified in the search for the genes that typically compose the pyocin clusters, which have been reported to be included between these genes5.
When the pyocin cluster sequences were distributed in several contigs, they were then compared by homology and assembled using BLASTn and Vector NTI Advance™ 11 (Invitrogen) programs. The complete sequences of the pyocin clusters were annotated by RAST34, HMMER (http://hmmer.org) and HHPRED35. The pyocin cluster R2-F2 of P. aeruginosa PAO1 was used as a reference sequence, which corresponds to the region between the genes PAO610-PAO648 (Genbank: AE004091.2-AE004091.2) from the Pseudomonas aeruginosa database36.
All the nucleotide sequence data reported are available in the Third Party Annotation Section of the DDBJ/ENA/GenBank databases under the accession numbers TPA: BK062614–BK062645 (Table 2).
Pyocin cluster type identification
The pyocin cluster types were assigned by homology with the tail fiber corresponding to the protein of the reference strain P. aeruginosa PAO1: PAO620 for the R-type pyocins and by PAO646 for the F-type pyocins12.
These strains were compared by BLASTp against the corresponding protein of the different pyocin types: R1 (ARI05994.1), R2 (AAG04009.1), R3 (ABP93392.1), R4 (ABP93394.1), R5 (ABP93396.1), F2 (AAG04035.1) and F(PA14) (ABJ15607.1). The pyocin clusters were considered to belong to a subtype when the homology value was higher than 90%.
Homology and phylogenetic analysis
The sequences obtained for the different pyocin clusters were analysed in order to study their homology. The studies were carried out using the Easyfig 2.2.537 and BRIG 0.9538 tools with the tBLASTx option. The Query Cover and Identity values were analysed with BLASTn.
The sequences were aligned and a phylogenetic tree was constructed with the bioinformatic software Geneious Prime (Dotmatics).
Extraction and concentration of phage tail-like bacteriocins
The selected strains used as sources of pyocins were cultured overnight in LB broth at 37 °C. The culture was then diluted 1:100 in LB and incubated at 37 °C and 180 rpm. Once the optical density measured at a wavelength of 600 nm (OD600nm) of 0.4 was reached, 10 µg/ml of mitomycin C (Sigma-Aldrich) was added and the culture was incubated until it turned clear. The lysed cultures were centrifuged at 4000 rpm for 10 min, and the supernatant with the pyocins was recovered and incubated with 1% chloroform for 30 min. Finally, the supernatant with the pyocins was filtered through a 45 µm filter (FILTER-LAB®PES Syringe filter).
For concentration, the pyocins were precipitated with polyethylene glycol (PEG). The pyocin solution was precipitated overnight at 4 °C with 10% PEG and 0.5 M NaCl. The pyocins were collected by centrifugation for 15 min at 11,000 rpm and 4 °C. The supernatant was discarded and the pellet suspended in SM buffer (0.1 M NaCl, 1 mM MgSO4, 0.2 M Tris–HCl, pH 7.5), to obtain a tenfold concentration. Finally, a 1:1 volume of chloroform was added and incubated with gentle shaking for 20 min, and the phases were then separated by centrifugation for 10 min at 4000 rpm. The aqueous phase with the pyocin suspension was recovered and stored at 4 °C until use.
Pyocin transmission electron microscopy (TEM)
The pyocin solutions were fixed in a grid and negatively stained in 1% aqueous uranil acetate for 5 min and examined in a transmission electron microscope JEOL JEM-1011.
Pyocin killing spectrum
In order to determine the target range of each isolated pyocin, each was tested against the 32 strains from which they were isolated (Table 1). The killing activity was assayed by spot test39. Briefly, double agar layer plates were prepared with the putative host strain mixed with the soft upper agar layer (0.4% agar). Once solidified, a drop (10 μl) of the pyocin solution was deposited on top of the agar layer, and the plates were incubated at 37 °C for 20 h.
To differentiate the R-type and F-type pyocins from the S-type pyocins, proteinase K was added to the plates, as R and F-type pyocins are protease resistant and S-type is protease sensitive12. In order to differentiate the pyocins from prophages induced with mitomycin C, serial dilutions were spotted on agar plates. When no individual plaques were observed at the higher dilutions, the presence of a spot was considered to be the result of the killing activity of the pyocin.
Supplementary Information
Acknowledgements
This study was financed by grant PI19/00878 and PI22/00323 awarded to M. Tomás within the State Plan for R+D+I 2013-2016 (National Plan for Scientific Research, Technological Development and Innovation 2008-2011) and co-financed by the ISCIII-Deputy General Directorate for Evaluation and Promotion of Research—European Regional Development Fund "A way of Making Europe" and Instituto de Salud Carlos III FEDER, Spanish Network for the Research in Infectious Diseases (REIPI, RD16/0016/0006 and CIBER CB21/13/00084), and by the Study Group on Mechanisms of Action and Resistance to Antimicrobials, GEMARA (SEIMC, http://www.seimc.org/). M. Tomás was financially supported by the Miguel Servet Research Programme (SERGAS and ISCIII). I. Bleriot was financially supported by the pFIS program (ISCIII, FI20/00302). O. Pacios and M. López were financially supported by grant IN606A-2020/035 and IN606C-2022/002, respectively (GAIN, Xunta de Galicia).
Author contributions
L.B., developed the experiments, analysed the results and wrote the original manuscript. M.G.A, C.O-C., I.B, O.P., M.L., L.F.-G., A.B.-P., M.H.G., visualization the results and manuscript. R.C.., M.T., re-written the manuscript, financed and directed the experiments and supervised the writing of the original manuscript.
Data availability
All data generated or analysed during this study are included in this published article [and its supplementary information files].
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lucía Blasco, Manuel González de Aledo, Rafael Cantón and María Tomás.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-27341-1.
References
- 1.Pires DP, Vilas Boas D, Sillankorva S, Azeredo J. Phage therapy: A step forward in the treatment of Pseudomonas aeruginosa infections. J. Virol. 2015;89:7449–7456. doi: 10.1128/JVI.00385-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Sales RO, Migliorini LB, Puga R, Kocsis B, Severino P. A core genome multilocus sequence typing scheme for Pseudomonas aeruginosa. Front. Microbiol. 2020;11:1049. doi: 10.3389/fmicb.2020.01049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mei M, Thomas J, Diggle SP. Heterogenous susceptibility to R-Pyocins in populations of Pseudomonas aeruginosa sourced from cystic fibrosis lungs. MBio. 2021 doi: 10.1128/mBio.00458-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simar SR, Hanson BM, Arias CA. Techniques in bacterial strain typing: Past, present, and future. Curr. Opin. Infect. Dis. 2021;34:339–345. doi: 10.1097/QCO.0000000000000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scholl D. Phage tail-like bacteriocins. Annu. Rev. Virol. 2017;4:453–467. doi: 10.1146/annurev-virology-101416-041632. [DOI] [PubMed] [Google Scholar]
- 6.Oluyombo O, Penfold CN, Diggle SP. Competition in biofilms between cystic fibrosis isolates of Pseudomonas aeruginosa is shaped by R-Pyocins. MBio. 2019 doi: 10.1128/mBio.01828-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakayama K, et al. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol. 2000;38:213–231. doi: 10.1046/j.1365-2958.2000.02135.x. [DOI] [PubMed] [Google Scholar]
- 8.Ghequire MGK, De Mot R. The tailocin tale: Peeling off phage tails. Trends Microbiol. 2015;23:587–590. doi: 10.1016/j.tim.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 9.Vacheron J, Heiman CM, Keel C. Live cell dynamics of production, explosive release and killing activity of phage tail-like weapons for Pseudomonas kin exclusion. Commun. Biol. 2021;4:87. doi: 10.1038/s42003-020-01581-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee G, et al. F-type bacteriocins of Listeria monocytogenes: A new class of phage tail-like structures reveals broad parallel coevolution between tailed bacteriophages and high-molecular-weight bacteriocins. J. Bacteriol. 2016;198:2784–2793. doi: 10.1128/JB.00489-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ling H, Saeidi N, Rasouliha BH, Chang MW. A predicted S-type pyocin shows a bactericidal activity against clinical Pseudomonas aeruginosa isolates through membrane damage. FEBS Lett. 2010;584:3354–3358. doi: 10.1016/j.febslet.2010.06.021. [DOI] [PubMed] [Google Scholar]
- 12.Ghequire MG, De Mot R. Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol. Rev. 2014;38:523–568. doi: 10.1111/1574-6976.12079. [DOI] [PubMed] [Google Scholar]
- 13.Scholl D, Martin DW. Antibacterial efficacy of R-type pyocins towards Pseudomonas aeruginosa in a murine peritonitis model. Antimicrob. Agents Chemother. 2008;52:1647–1652. doi: 10.1128/AAC.01479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohler T, Donner V, van Delden C. Lipopolysaccharide as shield and receptor for R-pyocin-mediated killing in Pseudomonas aeruginosa. J. Bacteriol. 2010;192:1921–1928. doi: 10.1128/JB.01459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsui H, Sano Y, Ishihara H, Shinomiya T. Regulation of pyocin genes in Pseudomonas aeruginosa by positive (prtN) and negative (prtR) regulatory genes. J. Bacteriol. 1993;175:1257–1263. doi: 10.1128/jb.175.5.1257-1263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Q, et al. Pseudomonas aeruginosa serotypes in nosocomial pneumonia: Prevalence and clinical outcomes. Crit. Care. 2014;18:R17. doi: 10.1186/cc13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farmer JJ, 3rd, Herman LG. Epidemiological fingerprinting of Pseudomonas aeruginosa by the production of and sensitivity of pyocin and bacteriophage. Appl. Microbiol. 1969;18:760–765. doi: 10.1128/am.18.5.760-765.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rampling A, Whitby JL, Wildy P. Pyocin-sensitivity testing as a method of typing Pseudomonas aeruginosa: Use of "phage-free" preparations of pyocin. J. Med. Microbiol. 1975;8:531–541. doi: 10.1099/00222615-8-4-531. [DOI] [PubMed] [Google Scholar]
- 19.Fyfe JA, Harris G, Govan JR. Revised pyocin typing method for Pseudomonas aeruginosa. J. Clin. Microbiol. 1984;20:47–50. doi: 10.1128/jcm.20.1.47-50.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bird TJ, Grieble HG. Pyocin antibiosis in chick embryos. Antimicrob. Agents Chemother. (Bethesda) 1969;9:495–498. [PubMed] [Google Scholar]
- 21.Haas H, Sacks T, Saltz N. Protective effect of pyocin against lethal Pseudomonas aeruginosa infections in mice. J. Infect. Dis. 1974;129:470–472. doi: 10.1093/infdis/129.4.470. [DOI] [PubMed] [Google Scholar]
- 22.Six A, et al. Pyocin efficacy in a murine model of Pseudomonas aeruginosa sepsis. J. Antimicrob. Chemother. 2021;76:2317–2324. doi: 10.1093/jac/dkab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams SR, Gebhart D, Martin DW, Scholl D. Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl. Environ. Microbiol. 2008;74:3868–3876. doi: 10.1128/AEM.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gebhart D, et al. A modified R-type bacteriocin specifically targeting Clostridium difficile prevents colonization of mice without affecting gut microbiota diversity. MBio. 2015 doi: 10.1128/mBio.02368-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gebhart D, et al. Novel high-molecular-weight, R-type bacteriocins of Clostridium difficile. J. Bacteriol. 2012;194:6240–6247. doi: 10.1128/JB.01272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alqahtani A, et al. Recombinant R2-pyocin cream is effective in treating Pseudomonas aeruginosa-infected wounds. Can. J. Microbiol. 2021;67:919–932. doi: 10.1139/cjm-2021-0207. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, Chen P, Zheng C, Huang YP. Characterization of maltocin P28, a novel phage tail-like bacteriocin from Stenotrophomonas maltophilia. Appl. Environ. Microbiol. 2013;79:5593–5600. doi: 10.1128/AEM.01648-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patz S, et al. Phage tail-like particles are versatile bacterial nanomachines—A mini-review. J. Adv. Res. 2019;19:75–84. doi: 10.1016/j.jare.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penterman J, et al. Rapid evolution of culture-impaired bacteria during adaptation to biofilm growth. Cell Rep. 2014;6:293–300. doi: 10.1016/j.celrep.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis MR, Jr, et al. Identification of the mutation responsible for the temperature-sensitive lipopolysaccharide O-antigen defect in the Pseudomonas aeruginosa cystic fibrosis isolate 2192. J. Bacteriol. 2013;195:1504–1514. doi: 10.1128/JB.01999-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Redero M, Aznar J, Prieto AI. Antibacterial efficacy of R-type pyocins against Pseudomonas aeruginosa on biofilms and in a murine model of acute lung infection. J. Antimicrob. Chemother. 2020 doi: 10.1093/jac/dkaa121. [DOI] [PubMed] [Google Scholar]
- 32.Redero M, et al. Susceptibility to R-pyocins of Pseudomonas aeruginosa clinical isolates from cystic fibrosis patients. J. Antimicrob. Chemother. 2018;73:2770–2776. doi: 10.1093/jac/dky261. [DOI] [PubMed] [Google Scholar]
- 33.Hernandez-Garcia M, et al. Distinct epidemiology and resistance mechanisms affecting ceftolozane/tazobactam in Pseudomonas aeruginosa isolates recovered from ICU patients in Spain and Portugal depicted by WGS. J. Antimicrob. Chemother. 2021;76:370–379. doi: 10.1093/jac/dkaa430. [DOI] [PubMed] [Google Scholar]
- 34.Overbeek R, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:D206–214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gabler F, et al. Protein sequence analysis using the MPI bioinformatics toolkit. Curr. Protoc. Bioinform. 2020;72:e108. doi: 10.1002/cpbi.108. [DOI] [PubMed] [Google Scholar]
- 36.Winsor GL, et al. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44:D646–653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sullivan MJ, Petty NK, Beatson SA. Easyfig: A genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST ring image generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raya RR, H’Bert EM. Isolation of phage via induction of lysogens. Methods Mol. Biol. 2009;501:23–32. doi: 10.1007/978-1-60327-164-6_3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article [and its supplementary information files].