Abstract
Purpose:
The phase III PROfound study (NCT02987543) evaluated olaparib versus abiraterone or enzalutamide (control) in metastatic castration-resistant prostate cancer (mCRPC) with tumor homologous recombination repair (HRR) gene alterations. We present exploratory analyses on the use of circulating tumor DNA (ctDNA) testing as an additional method to identify patients with mCRPC with HRR gene alterations who may be eligible for olaparib treatment.
Patients and Methods:
Plasma samples collected during screening in PROfound were retrospectively sequenced using the FoundationOne®Liquid CDx test for BRCA1, BRCA2 (BRCA), and ATM alterations in ctDNA. Only patients from Cohort A (BRCA/ATM alteration positive by tissue testing) were evaluated. We compared clinical outcomes, including radiographic progression-free survival (rPFS) between the ctDNA subgroup and Cohort A.
Results:
Of the 181 (73.9%) Cohort A patients who gave consent for plasma sample ctDNA testing, 139 (76.8%) yielded a result and BRCA/ATM alterations were identified in 111 (79.9%). Of these, 73 patients received olaparib and 38 received control. Patients’ baseline demographics and characteristics, and the prevalence of HRR alterations were comparable with the Cohort A intention-to-treat (ITT) population. rPFS was longer in the olaparib group versus control [median 7.4 vs. 3.5 months; hazard ratio (HR), 0.33; 95% confidence interval (CI), 0.21–0.53; nominal P < 0.0001], which is consistent with Cohort A ITT population (HR, 0.34; 95% CI, 0.25–0.47).
Conclusions:
When tumor tissue testing is not feasible or has failed, ctDNA testing may be a suitable alternative to identify patients with mCRPC carrying BRCA/ATM alterations who may benefit from olaparib treatment.
Translational Relevance.
Our findings from the phase III PROfound study demonstrate the clinical utility of circulating tumor DNA (ctDNA) next-generation sequencing (NGS) testing as an alternative to tumor-based NGS testing, to identify patients with metastatic castration-resistant prostate cancer (mCRPC) and BRCA1, BCRA2, and ATM (BRCA/ATM) alterations that may benefit from olaparib treatment. In this exploratory analysis, efficacy and safety of olaparib were consistent between patients with mCRPC whose alterations in BRCA/ATM were identified by ctDNA NGS testing and those whose alterations in BRCA/ATM were identified by tissue-based NGS testing in the intention-to-treat Cohort A population of the PROfound study. These results support consideration of ctDNA testing to identify patients with mCRPC BRCA/ATM alterations who may benefit from olaparib treatment when tumor tissue testing is not feasible.
Introduction
Metastatic castration-resistant prostate cancer (mCRPC) is a molecularly heterogeneous disease with a poor prognosis (1–3). Approximately 20%–30% of patients with this disease harbor deleterious alterations in DNA damage repair genes, including those with direct or indirect roles in homologous recombination repair (HRR; refs. 1–3). The PROfound study (NCT02987543) is the first phase III trial to show that the PARP inhibitor olaparib provides a statistically and clinically significant improvement in radiographic progression-free survival (rPFS) and overall survival (OS) versus control (physician's choice of enzalutamide or abiraterone) in patients with mCRPC and tumor HRR gene alterations (4, 5). Patient tumor tissue was tested for detecting alterations in 15 prespecified HRR genes using an investigational use only (IUO) assay based on the FoundationOne®CDx tissue test [Foundation Medicine Inc. (FMI); ref. 6]. Patients with alterations in BRCA1, BRCA2, or ATM (BRCA/ATM) were assigned to Cohort A, while patients with alterations in the other prespecified HRR genes joined Cohort B. On the basis of the findings of the PROfound study, the FDA approved olaparib for patients with germline or somatic 14 HRR gene-altered mCRPC, who have progressed following prior treatment with enzalutamide or abiraterone, while the European Medicines Agency and the Japanese Ministry of Health, Labour and Welfare have approved olaparib for patients with germline or somatic BRCA1 and BRCA2 alterations following new hormonal agent progression (7–9). Identifying eligible patients with mCRPC who may benefit from olaparib treatment using molecular diagnostics is key to improving patients’ survival.
Tissue-based next-generation sequencing (NGS) is currently the gold standard approach for the identification of HRR gene alterations in patients with mCRPC (10–12); however, there may be instances when this may not be an option. For example, the logistical challenges of formalin-fixed paraffin-embedded block retrieval, which can prevent use of existing archival materials (13), suboptimal success rates in patients with nonaccessible visceral lesions (14–16), and technical difficulties in obtaining tissue samples from patients with bone-only metastases (17–20), all can preclude tissue testing. The overall success rate of tissue NGS testing in PROfound was 69% (4), compared with 67% in the IPATential150 trial (21) and 74% in metastatic samples and 56% in primary tumor samples for TRITON2 and TRITON3 (22). The PROfound study highlighted factors which may predict successful generation of NGS results and gives options to optimize the testing approach; practical considerations for the optimization of tissue NGS testing have been published previously (13). Circulating tumor DNA (ctDNA) testing is an evolving technology in the assessment of patients with advanced prostate cancer, that represents an additional approach to tissue testing, that is particularly useful when tumor tissue samples are not available. ctDNA analysis allows for another means of assessing tumor tissue including gene alterations of somatic and germline origin, suggesting that ctDNA analysis may be a suitable surrogate for gene alteration testing in patients with prostate cancer (23). While not the case for all types of prostate cancer, in metastatic disease high concordance between alterations detected by ctDNA testing and tumor tissue testing has been observed, which makes it a potential alternative option when tissue testing is not possible (2, 24–26). The FoundationOne®Liquid CDx test has subsequently been approved by the FDA and Japanese regulatory agency (27) as a companion diagnostic for evaluation of ctDNA samples to enable identification of patients with mCRPC who have alterations in BRCA/ATM in ctDNA who may benefit from PARP inhibitor treatment (28).
The exploratory analyses presented here evaluated the efficacy of olaparib versus control in patients with qualifying alterations in BRCA/ATM in Cohort A of the PROfound study as determined retrospectively by the FoundationOne®Liquid CDx test.
Patients and Methods
Patients
A detailed description of the methods for the PROfound study (NCT02987543), including patient eligibility criteria, has already been published (4, 29). In brief, the trial included men with confirmed mCRPC whose disease had progressed while receiving enzalutamide or abiraterone. All patients were required to have tumors harboring a qualifying deleterious or suspected deleterious alteration in at least one of 15 prespecified genes selected for their direct or indirect role in HRR. Two cohorts were enrolled: those with alterations in BRCA1, BRCA2, and/or ATM (Cohort A) and those with alterations in ≥1 of 12 other prespecified HRR genes (Cohort B): BRIP1, BARD1, CDK12, CHEK1, CHEK2, FANCL, PALB2, PPP2R2A, RAD51B, RAD51C, RAD51D, and RAD54L. Gene alterations were identified in archival or recently obtained biopsy tissue from the primary or metastatic tumor using an IUO assay based on FMI's FoundationOne®CDx test (30). During screening, matched plasma samples were collected from consenting patients and subsequently assessed using the FoundationOne®Liquid CDx test (31).
Trial design and interventions
Patients were randomized 2:1 to receive olaparib tablets (300 mg twice daily; n = 256) or control [prespecified physician's choice of enzalutamide (160 mg once daily) or abiraterone (1,000 mg once daily) plus prednisone (5 mg twice daily); n = 131]. Patients who were assigned to the control group were eligible to cross over to receive olaparib treatment after independent review confirmed imaging-based progression. All patients provided written informed consent to participate. In the exploratory analysis presented here, only patients from Cohort A who had a positive BRCA/ATM alteration test following tissue testing and who had provided consent for ctDNA testing of their plasma samples were evaluated.
Endpoints
We report results from an exploratory analysis for the subgroup of patients for whom BRCA/ATM alterations were detected by ctDNA testing. The PROfound study was designed so that the primary analysis was conducted on patients in Cohort A only, hence the focus of the article is on Cohort A. Endpoints reported here include rPFS, objective response rate (ORR), and OS. The primary endpoint in PROfound was rPFS—assessed by blinded independent central review (BICR)—in Cohort A for the intention-to-treat (ITT) population [hazard ratio (HR), 0.34; 95% confidence interval (CI), 0.25–0.47)] and has been reported previously (4). Key secondary endpoints, such as rPFS in the overall population (Cohorts A+B), OS, ORR, and time to pain progression in Cohort A, have also been reported previously (4, 5). Safety was also assessed in the overall population (Cohorts A+B) through reporting of adverse events (AE) according to the Common Terminology Criteria Adverse Events, version 4.0 (4, 5). The PROfound trial was approved by an Institutional Review Board and performed in accordance with the principles of the Declaration of Helsinki, the International Conference on Harmonisation Good Clinical Practice guidelines, and the AstraZeneca and Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc. policies on bioethics. All the patients provided written informed consent.
Statistical analysis
For time-to-event endpoints, rPFS and OS, two-sided P values were calculated using log-rank tests stratified by the variables selected in the primary pooling strategy (prior taxane use and measurable disease in Cohort A) and using the Breslow method for handling ties. HRs and 95% CIs were calculated with the use of Cox proportional hazards models, adjusted for the variables selected in the primary pooling strategy. The Efron approach was used for handling ties. The Kaplan–Meier method was used to calculate medians. Objective response was analyzed for patients with measurable disease at baseline. The OR and CI of ORR data were calculated using a logistic regression model adjusted for prior taxane use. Where the number of patients with a response was ≥5, a P value was calculated on the basis of twice the change in log-likelihood resulting from the addition of the treatment factor to the model that contains the specified covariates. Where the number of patients with a response was <5, a two-sided P value was calculated on the basis of the mid P-value modification of Fisher's exact test.
Data availability
Data underlying the findings described in this manuscript may be obtained in accordance with AstraZeneca's data sharing policy described at https://astrazenecagrouptrials.pharmacm.com/ST/Submission/Disclosure. Data can be requested through Vivli at www.vivli.org.
Results
Patients
Of the 245 patients in Cohort A of the ITT population, 181 (73.9%) consented to plasma sample ctDNA testing. Of these, 139 (76.8%) had a ctDNA result reported (either mutation positive or negative) with BRCA/ATM alterations identified in 111 (79.9%) of the 139 patients. In total, 42 patients had a failed ctDNA test; this failure rate was higher than expected because of a technical failure of a single assay plate during testing. Because of the limited DNA yield obtained from ctDNA samples, it was not possible to attempt a repeat analysis from the majority of the samples on the plate which failed.
Overall, 28 patients were reported to have BRCA/ATM alterations by tumor-based NGS testing but were reported negative for BRCA/ATM alterations by the ctDNA NGS test. This includes 4 patients with homozygous deletions or large rearrangements in ATM reported by tissue testing, which were not able to be identified by ctDNA testing as these variant types have not been analytically validated (32). In addition, of the 15 patients with a BRCA2 homozygous deletion reported in the tumor sample, 4 patients reported the presence of a homozygous deletion by the ctDNA NGS test, 2 patients reported other alterations in BRCA/ATM but did not report the homozygous deletion, and 9 patients were reported as negative for BRCA/ATM alterations.
Of the identified 111 ctDNA subgroup patients, 73 received olaparib treatment and 38 received control. Demographics and baseline characteristics of patients with BRCA/ATM alterations evaluated by ctDNA were generally balanced between treatment arms, and were compatible with those of patients in the Cohort A ITT population (Table 1; Supplementary Table S1).
Table 1.
Demographics and baseline characteristics of patients in Cohort A ctDNA subgroup and Cohort A ITT population with BRCA/ATM alterations.
| Cohort A ctDNA subgroupa | Cohort A ITT populationb | |||
|---|---|---|---|---|
| Olaparib | Olaparib | |||
| 300 mg bid | Control | 300 mg bid | Control | |
| (n = 73) | (n = 38) | (n = 162) | (n = 83) | |
| Age, median (years) | 65.0 | 68.5 | 68.0 | 67.0 |
| Site of metastases | ||||
| Lymph nodes | 46 (63.0) | 26 (68.4) | 94 (58.0) | 52 (62.6) |
| Bone | 64 (87.7) | 34 (89.5) | 140 (86.4) | 73 (88.0) |
| Respiratory | 18 (24.7) | 6 (15.8) | 30 (18.5) | 11 (13.3) |
| Liver | 8 (11.0) | 8 (21.1) | 18 (11.1) | 13 (15.7) |
| Other distant sites | 17 (23.3) | 9 (23.7) | 34 (21.0) | 15 (8.1) |
| Measurable disease at baseline | 43 (58.9) | 21 (55.3) | 84 (51.9) | 43 (51.8) |
| PSA (μg/L) | ||||
| n | 72 | 38 | 160 | 81 |
| Median | 51.65 | 193.80 | 62.18 | 112.92 |
| Range | 0.22–2,980.75 | 1.85–7,115.00 | 0.20–7,240.74 | 1.85–7,115.00 |
| ECOG performance status | ||||
| 0–1 | 67 (91.8) | 36 (94.7) | 151 (93.2) | 80 (96.4) |
| 2 | 6 (8.2) | 2 (5.3) | 11 (6.8) | 3 (3.6) |
| Prior taxane use | ||||
| Yes | 54 (74.0) | 25 (65.8) | 106 (65.4) | 52 (62.7) |
| No | 19 (26.0) | 13 (34.2) | 56 (34.6) | 31 (37.3) |
Note: Data presented as n (%) unless stated otherwise.
Abbreviations: bid, twice daily; ECOG, Eastern Cooperative Oncology Group; PSA, prostate-specific antigen.
aPatients with BRCA/ATM alterations identified by ctDNA testing.
bPatients with BRCA/ATM alterations identified by tissue testing.
Efficacy analyses
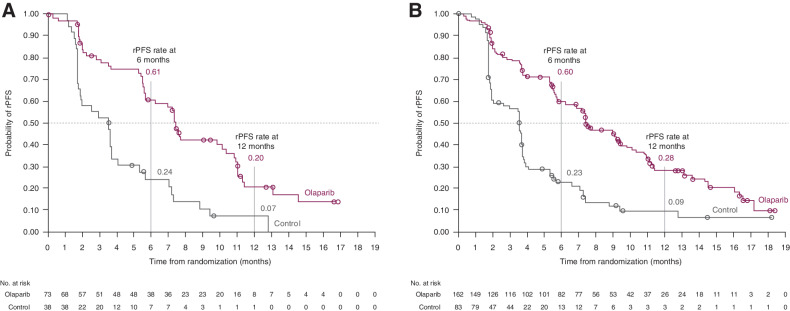
In the subgroup of patients with alterations in BRCA/ATM determined by ctDNA testing, rPFS was longer in the olaparib group than in the control group (median 7.4 vs. 3.5 months; HR, 0.33; 95% CI, 0.21–0.53; nominal P < 0.0001; Fig. 1A; Table 2). In addition, rPFS percentage rates at 6 and 12 months were 61% and 20%, respectively, in the olaparib group and 24% and 7%, respectively, in the control group. These findings are comparable with the overall Cohort A ITT population in which rPFS was statistically significantly longer in the olaparib group than the control group (HR, 0.34; 95% CI, 0.25–0.47; P < 0.001; Fig. 1B), with similar rPFS rates at 6 and 12 months for olaparib and control treatment (Table 2; ref. 4). Within the 28 patients who were reported to have BRCA/ATM alterations by tumor-based NGS testing but were reported negative for BRCA/ATM alterations by the ctDNA NGS test, rPFS was also longer in the olaparib group compared with the control group (median 10.9 vs. 3.5 months; HR, 0.11; 95% CI, 0.03–0.35).
Figure 1.
Kaplan–Meier plot of rPFS in Cohort A ctDNA subgroup* (A), and Cohort A ITT population† (B). *Patients with BRCA/ATM alterations identified by ctDNA testing; †Patients with BRCA/ATM alterations identified by tissue testing. (Figure 1B from N Engl J Med, de Bono J, et al., Olaparib for Metastatic Castration-Resistant Prostate Cancer, Vol. 382, pp. 2091–102, Copyright 2020 Massachusetts Medical Society. Reprinted with permission from Massachusetts Medical Society.)
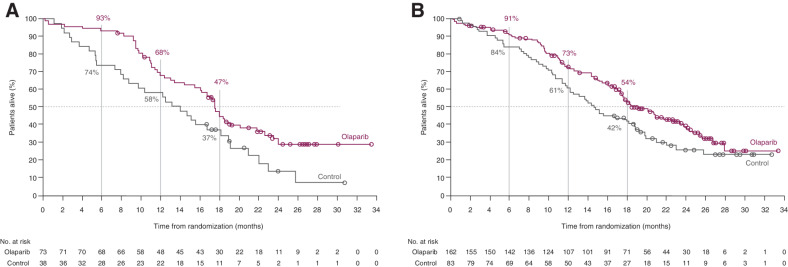
Figure 2.
Kaplan–Meier plot of OS in Cohort A ctDNA subgroup* (A), and Cohort A ITT population (B)†. *Patients with BRCA/ATM alterations identified by ctDNA testing; †Patients with BRCA/ATM alterations identified by tissue testing. (Figure 2B from N Engl J Med, Hussain M, et al., Survival with Olaparib in Metastatic Castration-Resistant Prostate Cancer, Vol. 383, pp. 2345–57, Copyright 2020 Massachusetts Medical Society. Reprinted with permission from Massachusetts Medical Society.)
Table 2.
rPFS, OS, and ORR in patients from the Cohort A ctDNA subgroup and Cohort A ITT population.
| Cohort A ctDNA subgroupa | Cohort A ITT populationb | |||
|---|---|---|---|---|
| Olaparib | Olaparib | |||
| 300 mg bid | Control | 300 mg bid | Control | |
| (n = 73) | (n = 38) | (n = 162) | (n = 83) | |
| rPFS | ||||
| Events, n (%) | 49 (67.1) | 34 (89.5) | 106 (65.4) | 68 (81.9) |
| Median (95% CI), months | 7.39 (5.65–10.38) | 3.53 (1.77–3.71) | 7.39 (6.24–9.33) | 3.55 (1.91–3.71) |
| HR (95% CI) | 0.33 (0.21–0.53) | 0.34 (0.25–0.47) | ||
| P | <0.0001c | <0.0001 | ||
| OS d | ||||
| Events, n (%) | 47 (64.4) | 31 (81.6) | 91 (56.2) | 57 (68.7) |
| Median (95% CI), months | 17.51 (15.47–20.11) | 13.59 (8.21–18.10) | 19.1 (17.4–23.4) | 14.7 (11.9–18.8) |
| HR (95% CI) | 0.58 (0.37–0.92) | 0.69 (0.50–0.97) | ||
| P | 0.0100a | 0.0200 | ||
| Confirmed ORR by BICR | ||||
| Objective responders/total number of patients with measurable disease at baseline, n (%) | 17/43 (39.5) | 1/21 (4.8) | 28/84 (33.3) | 1/43 (2.3) |
| OR (95% CI) | 13.17 (2.37–247.69) | 20.86 (4.18–379.18) | ||
| P | 0.0013a | <0.0001 | ||
aPatients with BRCA/ATM alterations identified by ctDNA testing.
bPatients with BRCA/ATM alterations, identified by tissue testing.
c P values are two-sided nominals for Cohort A ctDNA.
dNot adjusted for crossover.
Patients in the olaparib group also had a longer duration of OS compared with control in patients with alterations in BRCA1, BRCA2, or ATM detected by ctDNA testing (HR, 0.58; 95% CI, 0.37–0.92; nominal P = 0.01; Fig. 2A; Table 2). Survival rates at 6, 12, and 18 months were 93%, 68%, and 47%, respectively, in the olaparib group and 74%, 58%, and 37%, respectively, in the control group for the Cohort A ctDNA subgroup. These findings are also comparable with the overall Cohort A ITT population in which OS was statistically significantly longer in the olaparib group than the control group (HR, 0.69; 95% CI, 0.50–0.97; P = 0.02; Fig. 2B). These results have not been adjusted for patients crossing over from control therapy to olaparib following disease progression.
The ORR was higher in the olaparib group (39.5%) compared with control group (4.8%) in patients with alterations in BRCA/ATM as detected by ctDNA testing (Table 2). These response rates are comparable to those reported for the overall Cohort A ITT population.
Discussion
The findings presented here from the phase III PROfound study demonstrate the clinical utility of ctDNA testing as an alternative to tumor-based NGS testing to identify patients with mCRPC with HRR alterations that may benefit from olaparib treatment when tumor testing cannot be undertaken due to insufficient tissue or when a tumor test has failed. In the PROfound study, a NGS result generation rate of 69% was reported for tumor tissue-based testing; there is a need to further optimize tumor-based NGS testing approaches and the study recognized areas for improvement and ways to increase success rates (4, 13, 33). In Cohort A, for BRCA/ATM alterations, ctDNA testing was concordant with tumor tissue testing in approximately 80% of patients (34). This concordance rate is similar to that reported for the rucaparib TRITON-2 study (74%) evaluating BRCA alterations in tumor tissue and matched ctDNA samples in patients with mCRPC (35). Other studies comparing ctDNA and tumor-based NGS testing in samples from patients with prostate cancer, including de novo metastatic disease, have shown concordance rates of 80%–90% for a wider range of gene alterations beyond HRR (24, 36).
Our exploratory analyses of the subgroup of 111 patients from Cohort A of the PROfound study who had BRCA/ATM alterations by retrospective ctDNA testing showed that olaparib resulted in improved efficacy compared with control treatment. Nominally significant improvements in rPFS, ORR, and OS, were reported for the olaparib group compared with the control treatment group for the Cohort A ctDNA subgroup; these improvements were comparable with those that have previously been reported for the 245 patients from the overall Cohort A ITT population with BRCA/ATM alterations identified by prospective tumor-based NGS testing (4, 5). The benefit of olaparib on OS in this study may be underestimated as the results reported here were not adjusted for patient crossover from the control arm to the olaparib arm following disease progression. Approximately 60% of patients in the control arm switched to olaparib treatment, and previous analyses showed that when adjusted for switching, the benefit of olaparib treatment for patients with mCRPC could be even greater for patients in Cohort A, as well as the overall population (crossover-adjusted analysis of OS in Cohort A: HR, 0.42; 95% CI, 0.19–0.91; refs. 5, 37). The safety profile of olaparib in the Cohort A ctDNA subgroup was also consistent with that in the overall PROfound study population (Cohorts A + B) with similar proportions of patients reporting AEs, grade ≥3 AEs and serious AEs, as well as requiring dose modifications in the two cohorts, which for the overall population includes patients with HRR gene alterations beyond BRCA/ATM (Supplementary Table S2).
The TRITON2 and TRITON3 studies evaluating rucaparib have also assessed the feasibility of using ctDNA testing (FoundationACT™ and FoundationOne®Liquid assays) to identify specific gene alterations in patients with mCRPC (35). In an evaluation of 620 plasma ctDNA samples and 888 tissue samples from screened patients, the incidence of BRCA/ATM alterations was marginally higher in ctDNA samples than tumor tissue for both studies. Homozygous BRCA alterations could not be detected in these ctDNA samples as the FoundationOne®Liquid CDx test is not validated to detect them. A subsequent analysis of >3,000 samples, including those from patients screened for TRITON2 and TRITON3, reported a high level of agreement in detection of BRCA alterations by ctDNA testing compared with tissue testing (38). Of the 94% of samples that had detectable ctDNA, the median tumor fraction was 7.5%. Another analysis of data from the TRITON2 study reported that the ORR was 46.3% (95% CI, 30.7–62.6) in 41/62 (66%) patients from the primary efficacy population with BRCA alterations retrospectively confirmed by ctDNA testing, which is comparable with the ORR of 43.5% (95% CI, 31.0–56.7) reported in patients with BRCA alterations identified by tumor testing (32).
In the patients who had BRCA/ATM alterations reported by tumor tissue testing, but who were BRCA/ATM negative by ctDNA testing, the observed efficacy in olaparib-treated patients demonstrated a longer median PFS (10.9 months) than in olaparib-treated patients who were positive in both tissue and ctDNA testing (7.4 months). The reasons for this difference are unclear. However, it should be noted that only 28 patients contributed to this analysis. Furthermore, ctDNA shedding, and hence ability to detect alterations in ctDNA have been linked with increased disease burden in other tumor types (39). ctDNA testing has several advantages over tumor testing, including being less invasive. Although the majority of HRR gene alterations appear truncal and so present in primary tumor tissue with changes over time being rarely observed (40–42), it has been shown that BRCA2/RB1/RNASEH2B loss can develop post–next-generation endocrine agent therapy (43). In these instances, where multiple plasma samples can be collected at different timepoints, ctDNA NGS testing could be a useful tool to provide an overview of the patient's mutational status over disease progression and to monitor the emergence of new acquired alterations and resistance mechanisms that cannot be identified by single-site biopsies or archival sample analyses. Timelines for obtaining ctDNA test results may be shorter, as there is no requirement for retrieval of archived samples, and as a consequence costs for ctDNA testing may be lower than for tumor testing.
There are also several limitations of ctDNA testing. The amount of detectable ctDNA can vary greatly depending on the timing of testing in relation to treatment, as well as the histologic type and cancer clinical stage (44). Higher ctDNA fractions are associated with poor prognosis and are correlated with overall tumor burden in patients with mCRPC, suggesting that those with lower ctDNA fractions, who might be more likely to have a non-HRRm ctDNA test result, might have a more favorable prognosis (45). However, in PROfound Cohort A, the ctDNA subgroups positive and negative for HRRm and the ITT cohort all showed comparable baseline characteristics, although patient numbers for the non-HRRm ctDNA subgroup were small (olaparib, n = 20; control, n = 8; Supplementary Table S1). A comparable rPFS for the ctDNA subgroup positive for HRRm and the ITT cohort was also observed. These similarities in baseline characteristics across the different subgroups mean the detectability of ctDNA is not affected by demographic factors or by the disease characteristics assessed in PROfound.
In PROfound, in approximately 20% of patients, BRCA/ATM alterations detected by tumor testing were not detected by ctDNA NGS testing. This is to be expected for two main reasons. First, some patients may not shed, or shed very low quantities of ctDNA into the blood stream. For these patients, detection of BRCA/ATM gene mutations using ctDNA NGS may not be possible due to insufficient target ctDNA in the collected samples. Assessing the ctDNA content in samples is therefore important. Second, in contrast to tumor tissue testing, ctDNA test assays are not validated to accurately detect homozygous loss and zygosity of mutations. For example, ctDNA testing can detect homozygous deletions of BRCA2 but only with 30%–40% positive percentage agreement with tissue testing due to low tumor content in at least half of ctDNA samples with this being critically important because these patients are the ones that appear to benefit most from PARP inhibition (46). As a consequence, the FDA-approved label for the FoundationOne®Liquid CDx test reports that a negative result does not rule out the presence of an actionable key alteration, such as BRCA2 homozygous deletion, and that patients who are negative for companion diagnostic mutations should undergo tumor tissue testing and alteration status confirmed using an FDA-approved tumor tissue test, if feasible (28). Finally, a known confounder of ctDNA testing is clonal hematopoiesis of indeterminate potential (CHIP), which can lead to false-positive results during ctDNA analysis. CHIP is an age-related phenomenon, which may be particularly relevant in patients with metastatic prostate cancer who tend to be older at diagnosis (38). A recent study found that around 10% of patients with advanced prostate cancer showed CHIP interference in plasma ctDNA in HRR genes, such as ATM and CHEK2, to mitigate the risk of misdiagnoses; ctDNA results should therefore be compared with results from a paired whole-blood control or tumor tissue (47).
While our study has demonstrated that the efficacy and safety of olaparib in the Cohort A subgroup who had BRCA/ATM alterations by ctDNA testing is similar to that for the Cohort A ITT population, there are some limitations of these exploratory analyses. All patients in Cohort A had already been identified as having BRCA/ATM alterations by a tissue test, and this is important to consider when interpreting these findings. Further studies are needed to investigate the efficacy of olaparib in patients whose BRCA/ATM alterations were detected by ctDNA testing in the absence of a tumor test result. Furthermore, data from ctDNA testing were not performed to validate alterations in the 12 prespecified genes with a direct or indirect role in HRR, other than BRCA/ATM, that were identified by tumor tissue testing in patients from Cohort B. As such, the impact of these alterations on olaparib efficacy was not assessed. In addition, the efficacy of olaparib in patients identified solely using ctDNA testing remains to be determined. The exploratory analyses reported here were not part of the prespecified statistical testing hierarchy for the PROfound study and so were not alpha controlled, and the sample size of the Cohort A ctDNA subgroup included in this exploratory analysis was relatively small, with only 111 patients being eligible for evaluation. Furthermore, although baseline characteristics appeared balanced overall between the olaparib group and the control groups, analyses of this subgroup were not adjusted for confounding factors (e.g., baseline prognostic factors, differences in disease burden and treatment history at baseline, and crossover from the control arm to the olaparib arm following disease progression).
In conclusion, the efficacy of olaparib was consistent between patients with mCRPC whose alterations in BRCA/ATM were identified by ctDNA testing and those whose alterations in BRCA/ATM were identified by tissue testing in the ITT Cohort A population of the PROfound study. The overall safety profile of olaparib of Cohort A ctDNA subgroup was similar to that reported for the overall ITT (Cohorts A+B) population. These results support consideration of ctDNA testing to identify patients with mCRPC and BRCA/ATM alterations who may benefit from olaparib treatment when tumor tissue testing is not feasible (43).
Supplementary Material
Supplementary data presenting additional demographic data and safety analysis.
Acknowledgments
This study was funded by AstraZeneca and is part of an alliance between AstraZeneca and Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc. Medical writing assistance was provided by Kristin Almond, PhD, from Mudskipper Business Ltd, funded by AstraZeneca and Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
N. Matsubara reports grants from AstraZeneca during the conduct of the study; N. Matsubara also reports personal fees from Sanofi, as well as grants from Janssen, AstraZeneca, Bayer, Roche, MSD, Taiho, Astellas, Amgen, Eisai, Eli Lilly, PRA Health Sciences, Takeda, Pfizer, Seagen, Chugai, AbbVie, and Novartis outside the submitted work. J. de Bono reports personal fees from AstraZeneca during the conduct of the study. J. de Bono also reports personal fees and non-financial support from Astellas Pharma and Sanofi; grants, personal fees, and non-financial support from AstraZeneca; personal fees from Bayer, Boehringer Ingelheim, Genentech/Roche, Merck Serono, Merck Sharp & Dohme, Janssen, and Pfizer; non-financial support from Genmab, Orion Pharma GmbH, Qiagen, Taiho Pharmaceuticals, and Vertex; personal fees, non-financial support, and other support from GlaxoSmithKline; and personal fees and other support from Cellcentric, Daiichi, Menarini/Silicon Biosystems, and Sierra Oncology outside the submitted work. D. Olmos reports grants from AstraZeneca during the conduct of the study. D. Olmos also reports grants, personal fees, and other support from AstraZeneca, Bayer, and Janssen; personal fees from Clovis, Daiichi-Sankyo, and Merck Sharp & Dohme; personal fees and other support from F. Hoffman-La Roche and Pfizer; and other support from Ipsen outside the submitted work. S. Kawakami reports personal fees from AstraZeneca during the conduct of the study, as well as personal fees from Astellas, Bayer, Janssen, and Sanofi outside the submitted work. Y. Ürün reports other support from AstraZeneca during the conduct of the study, as well as other support from Astellas, AstraZeneca, Bristol Myers Squibb, Janssen Oncology, MSD, Pfizer, Roche, and Amgen outside the submitted work. A. Flechon reports personal fees and other support from AstraZeneca, Janssen, Astellas, AAA, Novartis, and Bayer, as well as personal fees from Lilly outside the submitted work. M.A. Carducci reports personal fees from Pfizer outside the submitted work. S.J. Hotte reports personal fees from Merck Canada and AAA/Novartis; grants from AstraZeneca; and grants and personal fees from Bayer, Janssen, and Astellas outside the submitted work. G. Kramer reports other support from Sanofi, Amgen, AstraZeneca, Bayer, Astellas, Janssen, Ferring, MSD, and Novartis during the conduct of the study, as well as other support from Sanofi, Amgen, AstraZeneca, Bayer, Astellas, Janssen, Ferring, MSD, and Novartis outside the submitted work. N. Agarwal reports consultancy to Astellas, AstraZeneca, Aveo, Bayer, Bristol Myers Squibb, Calithera, Clovis, Eisai, Eli Lilly, EMD Serono, Exelixis, Foundation Medicine, Genentech, Gilead, Janssen, Merck, MEI Pharma, Nektar, Novartis, Pfizer, Pharmacyclics, and Seattle Genetics, as well as research funding to institution from Astellas, AstraZeneca, Bavarian Nordic, Bayer, Bristol Myers Squibb, Calithera, Celldex, Clovis, Eisai, Eli Lilly, EMD Serono, Exelixis, Genentech, Gilead, GlaxoSmithKline, Immunomedics, Janssen, Medivation, Merck, Nektar, New Link Genetics, Novartis, Pfizer, Prometheus, Rexahn, Roche, Sanofi, Seattle Genetics, Takeda, and Tracon. K.N. Chi reports grants and personal fees from AstraZeneca during the conduct of the study; K.N. Chi also reports personal fees from Astellas and Sanofi, as well as grants and personal fees from Janssen, Merck, Novartis, Pfizer, Point Biopharma, and Roche outside the submitted work. S. Dearden reports other support from AstraZeneca during the conduct of the study, as well as other support from AstraZeneca outside the submitted work. C. Gresty reports personal fees from AstraZeneca during the conduct of the study. J. Kang is an employee of AstraZeneca, the sponsor of this study. C. Poehlein reports other support from Merck & Co during the conduct of the study, as well as other support from Merck & Company outside the submitted work. E.A. Harrington reports other support from AstraZeneca during the conduct of the study; other support from AstraZeneca outside the submitted work; and is an employee and shareholder for AstraZeneca. M. Hussain reports grants from AstraZeneca during the conduct of the study, as well as personal fees from AstraZeneca outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
N. Matsubara: Conceptualization, data curation, formal analysis, writing–review and editing. J. de Bono: Resources, data curation, formal analysis, methodology, writing–review and editing. D. Olmos: Writing–review and editing. G. Procopio: Conceptualization, data curation, writing–review and editing. S. Kawakami: Writing–review and editing. Y. Ürün: Conceptualization, data curation, methodology, writing–review and editing. R. van Alphen: Writing–review and editing. A. Flechon: Writing–review and editing. M.A. Carducci: Writing–review and editing. Y.D. Choi: Writing–review and editing. S.J. Hotte: Writing–review and editing. E. Korbenfeld: Investigation, writing–review and editing. G. Kramer: Writing–review and editing. N. Agarwal: Writing–review and editing. K.N. Chi: Writing–review and editing. S. Dearden: Conceptualization, resources, data curation, methodology, writing–review and editing. C. Gresty: Writing–review and editing. J. Kang: Writing–review and editing. C. Poehlein: Resources, funding acquisition. E.A. Harrington: Resources, funding acquisition. M. Hussain: Conceptualization, formal analysis, investigation, methodology, writing–review and editing.
References
- 1. Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med 2016;375:443–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Robinson D, Van Allen EM, Wu Y-M, Schultz N, Lonigro RJ, Mosquera J-M, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;162:454. [DOI] [PubMed] [Google Scholar]
- 3. Abida W, Armenia J, Gopalan A, Brennan R, Walsh M, Barron D, et al. Prospective genomic profiling of prostate cancer across disease states reveals germline and somatic alterations that may affect clinical decision making. JCO Precis Oncol 2017;2017:PO.17.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Bono J, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med 2020;382:2091–102. [DOI] [PubMed] [Google Scholar]
- 5. Hussain M, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. Survival with olaparib in metastatic castration-resistant prostate cancer. N Engl J Med 2020;383:2345–57. [DOI] [PubMed] [Google Scholar]
- 6. Food and Drug Administration Agency. FoundationOne CDx - P170019; 2020. Available from: https://www.fda.gov/medical-devices/recently-approved-devices/foundationoner-cdx-p170019s017.
- 7. Lynparza (olaparib) prescribing information; 2020. Available from: https://www.ema.europa.eu/en/documents/product-information/lynparza-epar-product-information_en.pdf.
- 8. European Medicines Agency. European Medicines Agency - Lynparza; 2020. Available from: https://www.ema.europa.eu/en/medicines/human/EPAR/lynparza.
- 9. AstraZeneca. Lynparza approved in Japan for the treatment of advanced ovarian, prostate and pancreatic cancers; 2020. Available from: https://www.astrazeneca.com/media-centre/press-releases/2020/lynparza-approved-in-japan-for-three-cancers.html#:~:text=Lynparza%20is%20the%20first%20and,for%20over%20100%2C000%20new%20cases.&text=With%20limited%20treatment%20options%2C%20the,is%20only%209%2D13%20months.
- 10. Parker C, Castro E, Fizazi K, Heidenreich A, Ost P, Procopio G, et al. Cancer of the prostate: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol 2020;31:1119–34. [DOI] [PubMed] [Google Scholar]
- 11. Schaeffer E, Srinivas S, Antonarakis ES, Armstrong AJ, Bekelman JE, Cheng H, et al. NCCN guidelines insights: prostate cancer, version 1.2021. J Natl Compr Canc Netw 2021;19:134–43. [DOI] [PubMed] [Google Scholar]
- 12. Lowrance WT, Breau RH, Chou R, Chapin BF, Crispino T, Dreicer R, et al. Advanced prostate cancer: AUA/ASTRO/SUO guideline part II. J Urol 2021;205:22–9. [DOI] [PubMed] [Google Scholar]
- 13. Gonzalez D, Mateo J, Stenzinger A, Rojo F, Shiller M, Wyatt AW, et al. Practical considerations for optimising homologous recombination repair mutation testing in patients with metastatic prostate cancer. J Pathol Clin Res 2021;7:311–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Efstathiou E, Titus M, Wen S, Hoang A, Karlou M, Ashe R, et al. Molecular characterization of enzalutamide-treated bone metastatic castration-resistant prostate cancer. Eur Urol 2015;67:53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ross RW, Halabi S, Ou S-S, Rajeshkumar BR, Woda BA, Vogelzang NJ, et al. Predictors of prostate cancer tissue acquisition by an undirected core bone marrow biopsy in metastatic castration-resistant prostate cancer–a cancer and leukemia group B study. Clin Cancer Res 2005;11:8109–13. [DOI] [PubMed] [Google Scholar]
- 16. Spritzer CE, Afonso PD, Vinson EN, Turnbull JD, Morris KK, Foye A, et al. Bone marrow biopsy: RNA isolation with expression profiling in men with metastatic castration-resistant prostate cancer–factors affecting diagnostic success. Radiology 2013;269:816–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jimenez RE, Atwell TD, Sicotte H, Eckloff B, Wang L, Barman P, et al. A prospective correlation of tissue histopathology with nucleic acid yield in metastatic castration-resistant prostate cancer biopsy specimens. Mayo Clin Proc Innov Qual Outcomes 2019;3:14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zheng G, Lin M-T, Lokhandwala PM, Beierl K, Netto GJ, Gocke CD, et al. Clinical mutational profiling of bone metastases of lung and colon carcinoma and malignant melanoma using next-generation sequencing. Cancer Cytopathol 2016;124:744–53. [DOI] [PubMed] [Google Scholar]
- 19. Bashir MN. Epidemiology of prostate cancer. Asian Pac J Cancer Prev 2015;16:5137–41. [DOI] [PubMed] [Google Scholar]
- 20. Friedlander TW, Pritchard CC, Beltran H. Personalizing therapy for metastatic prostate cancer: the role of solid and liquid tumor biopsies. Am Soc Clin Oncol Educ Book 2017;37:358–69. [DOI] [PubMed] [Google Scholar]
- 21. De Bono JS, Sweeney C, Bracarda S, Sternberg CN, Chi KN, Olmos D, et al. PI3K/AKT pathway biomarkers analysis from the phase III IPATential150 trial of ipatasertib plus abiraterone in metastatic castration-resistant prostate cancer. J Clin Oncol 39: 6s, 2021. (suppl; abstr 13). [Google Scholar]
- 22. Abida W, Campbell D, Patnaik A, Shapiro JD, Sautois B, Vogelzang NJ, et al. Non-BRCA DNA damage repair gene alterations and response to the PARP inhibitor rucaparib in metastatic castration-resistant prostate cancer: analysis from the Phase II TRITON2 study. Clin Cancer Res 2020;26:2487–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vandekerkhove G, Struss WJ, Annala M, Kallio HML, Khalaf D, Warner EW, et al. Circulating tumor DNA abundance and potential utility in de novo metastatic prostate cancer. Eur Urol 2019;75:667–75. [DOI] [PubMed] [Google Scholar]
- 24. Grasso CS, Wu Y-M, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012;487:239–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hong MKH, Macintyre G, Wedge DC, Van Loo P, Patel K, Lunke S, et al. Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer. Nat Commun 2015;6:6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ulz P, Belic J, Graf R, Auer M, Lafer I, Fischereder K, et al. Whole-genome plasma sequencing reveals focal amplifications as a driving force in metastatic prostate cancer. Nat Commun 2016;7:12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Japanese regulatory approval of FoundationOne® Liquid CDx test; 2021. Available from: https://www.chugai-pharm.co.jp/english/news/detail/20210323170004_807.html.
- 28. Food and Drug Administration Agency. FoundationOne Liquid CDx – P190032; 2020. Available from: https://www.fda.gov/medical-devices/recently-approved-devices/foundationone-liquid-cdx-p190032.
- 29. Matsubara N, Nishimura K, Kawakami S, Joung JY, Uemura H, Goto T, et al. Olaparib in patients with mCRPC with homologous recombination repair gene alterations: PROfound Asian subset analysis. Jpn J Clin Oncol 2022;52:441–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Foundation Medicine Inc. FoundationOne®CDx Technical Information; 2020. Available from: https://assets.ctfassets.net/w98cd481qyp0/41rJj28gFwtxCwHQxopaEb/2725881bbc67d6f323ab893851344c4a/FoundationOne_CDx_Label_Technical_Info.pdf.
- 31. Foundation Medicine Inc. FoundationOne® Liquid CDx Technical Information; 2020. Available from: https://assets.ctfassets.net/w98cd481qyp0/41rJj28gFwtxCwHQxopaEb/2725881bbc67d6f323ab893851344c4a/FoundationOne_CDx_Label_Technical_Info.pdf.
- 32. FoundationOne Liquid CDx Technical Information. Available from: http://info.foundationmedicine.com/hubfs/FMI%20Labels/FoundationOne_Liquid_CDx_Label_Technical_Info.pdf.
- 33. Hussain M, Mateo J, Sandhu SK, Fizazi K, Saad F, Shore ND, et al. Next-generation sequencing (NGS) of tumor tissue from >4000 men with metastatic castration-resistant prostate cancer (mCRPC): the PROfound phase III study experience. J Clin Oncol 38: 6s, 2020. (suppl; abstr 195). [Google Scholar]
- 34. Chi KN, Barnicle A, Sibilla C, Lai Z, Corcoran C, Williams JA, et al. Concordance of BRCA1, BRCA2 (BRCA), and ATM mutations identified in matched tumor tissue and circulating tumor DNA (ctDNA) in men with metastatic castration-resistant prostate cancer (mCRPC) screened in the PROfound study. J Clin Oncol 39: 6s, 2021. (suppl; abstr 26). [Google Scholar]
- 35. Chowdhury S, McDermott R, Piulats JM, Shapiro JD, Mejlholm I, Morris D, et al. Genomic profiling of circulating tumour DNA (ctDNA) and tumour tissue for the evaluation of rucaparib in metastatic castration-resistant prostate cancer (mCRPC). Ann Oncol 2018;29:viii273. [Google Scholar]
- 36. Wyatt AW, Annala M, Aggarwal R, Beja K, Feng F, Youngren J, et al. Concordance of circulating tumor DNA and matched metastatic tissue biopsy in prostate cancer. J Natl Cancer Inst 2017;109:djx118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Agarwal N, Evans R, Abrams K, Dequen-O'Byrne P, McCrea C, Muston D, et al. Exploring the impact of treatment switching on the interim overall survival (OS) results of the PROfound study. Ann Oncol 2020;31:S514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tukachinsky H, Madison RW, Chung JH, Gjoerup OV, Severson EA, Dennis L, et al. Genomic analysis of circulating tumor DNA in 3,334 patients with advanced prostate cancer identifies targetable BRCA alterations and AR resistance mechanisms. Clin Cancer Res 2021;27:3094–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lam VK, Zhang J, Wu CC, Tran HT, Li L, Diao L, et al. Genotype-specific differences in circulating tumor DNA levels in advanced NSCLC. J Thorac Oncol 2021;16:601–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Warner E, Herberts C, Fu S, Yip S, Wong A, Wang G, et al. BRCA2, ATM, and CDK12 defects differentially shape prostate tumor driver genomics and clinical aggression. Clin Cancer Res 2021;27:1650–62. [DOI] [PubMed] [Google Scholar]
- 41. Mateo J, Seed G, Bertan C, Rescigno P, Dolling D, Figueiredo I, et al. Genomics of lethal prostate cancer at diagnosis and castration resistance. J Clin Invest 2020;130:1743–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schweizer MT, Sivakumar S, Tukachinsky H, Coleman I, De Sarkar N, Yu EY, et al. Concordance of DNA repair gene mutations in paired primary prostate cancer samples and metastatic tissue or cell-free DNA. JAMA Oncol 2021;7:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sumanasuriya S, Seed G, Parr H, Christova R, Pope L, Bertan C, et al. Elucidating prostate cancer behaviour during treatment via low-pass whole-genome sequencing of circulating tumour DNA. Eur Urol 2021;80:243–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wyatt AW, Annala M, Beja K, Parimi S, Vandekerkhove G, Warner E, et al. Genomic alterations in circulating tumor DNA (ctDNA) are associated with clinical outcomes in treatment-naive metastatic castration-resistant prostate cancer (mCRPC) patients commencing androgen receptor (AR)-targeted therapy. Ann Oncol 2016;27:vi17. [Google Scholar]
- 46. Carreira S, Porta N, Arce-Gallego S, Seed G, Llop-Guevara A, Bianchini D, et al. Biomarkers associating with PARP inhibitor benefit in prostate cancer in the TOPARP-B trial. Cancer Discov 2021;11:2812–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jensen K, Konnick EQ, Schweizer MT, Sokolova AO, Grivas P, Cheng HH, et al. Association of clonal hematopoiesis in DNA repair genes with prostate cancer plasma cell-free DNA testing interference. JAMA Oncol 2021;7:107–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data presenting additional demographic data and safety analysis.
Data Availability Statement
Data underlying the findings described in this manuscript may be obtained in accordance with AstraZeneca's data sharing policy described at https://astrazenecagrouptrials.pharmacm.com/ST/Submission/Disclosure. Data can be requested through Vivli at www.vivli.org.