Abstract
Purpose:
Gene fusions are drivers of many pediatric tumors. In fibrolamellar hepatocellular carcinoma (FLC), a fusion of DNAJB1 and PRKACA is the dominant recurrent mutation. Expression of the DNAJB1-PRKACA fusion gene in mice results in a tumor that recapitulates FLC. However, it is not known whether transient expression of DNAJB1-PRKACA is sufficient only to trigger tumor formation or whether ongoing expression is necessary for maintenance and progression.
Experimental Design:
We screened short hairpin RNAs (shRNA) tiled over the fusion junction and identified several potent and specific candidates in vitro and two independent FLC patient-derived xenografts (PDX).
Results:
We show that continued DNAJB1-PRKACA expression is not only required for continued tumor growth, but additionally its inhibition results in cell death. Inhibition of DNAJB1-PRKACA by an inducible shRNA in cells of PDX of FLC resulted in cell death in vitro. Induction of the shRNA inhibits FLC tumors growing in mice with no effect on xenografts from a hepatocellular carcinoma cell line engineered to express DNAJB1-PRKACA.
Conclusions:
Our results validate DNAJB1-PRKACA as the oncogene in FLC and demonstrate both a continued requirement for the oncogene for tumor growth as well as an oncogenic addiction that can be exploited for targeted therapies. We anticipate our approach will be useful for investigations of other fusion genes in pediatric cancers and spur development of precision therapies.
Translational Relevance.
Our previous work demonstrated that DNAJB1-PRKACA is the oncogene in the usually lethal cancer fibrolamellar hepatocellular carcinoma (FLC). It is the only recurrent genomic alteration and expression of this fusion kinase is sufficient to produce FLC in mouse liver. It is unknown whether targeting DNAJB1-PRKACA would be a valid therapeutic target. Simply the presence of a novel fusion gene or overexpression of an oncogene does not mean that intervention would be successful. Here we demonstrate that FLC not only requires the continued activity of a fusion kinase to grow but it is oncogenically dependent on it. Elimination of the oncokinase with an inducible short hairpin RNA in vitro or in vivo results in cell death and tumor clearance. Thus, this article establishes the fusion kinase as a key therapeutic target for intervention by antisense oligonucleotides, siRNA, or PROTACS.
Introduction
Fibrolamellar hepatocellular carcinoma (FLC) is a rare and usually lethal liver cancer affecting adolescents and young adults (1–3). There are no established therapies available for the treatment of FLC. Surgical resection is the only curative treatment for primary tumors but is only palliative for metastatic disease. FLC is classified as a subset of hepatocellular carcinoma (HCC; ref. 4). However, FLC has distinct histopathologic features (3). Unlike HCC, FLC occurs on the background of an otherwise healthy liver and therapeutics that work for HCC are ineffective for FLC (2, 5). FLC also has a molecular pathology distinct from HCC. All patients with FLC have a dysregulation of protein kinase A (PKA). In 3 patients, a mutations has been found in one of the regulatory subunits of PKA (6). The vast majority of patients, however, have a unique pathognomonic molecular driver, a somatic deletion of approximately 400 kb on one copy of chromosome 19. This results in the fusion of exon 1 of DNAJB1, a HSP cofactor, to exons 2–10 of PRKACA, the catalytic subunit of PKA (7, 8). In mice, generation of the DNAJB1-PRKACA fusion, referred to as chimera, by CRISPR/Cas9 drives tumorigenesis and recapitulates molecular and morphologic characteristics of patient tumors (9, 10). The transformation is specifically the result of the expression of the fusion gene, and not loss of genes in the 400 kb as ectopic expression of the Chimera, DNAJB1-PRKACA, with a transposon is sufficient for transformation (10). The catalytic activity of the fusion kinase is essential for transformation (10).
It is unknown whether FLC tumors are dependent on the continuous presence of the fusion protein. Acquisition of other mutations, rewiring of gene expression and metabolism are well established hallmarks of cancer, that can allow disease progression subsequent to the action of the inciting oncogene (11). For instance, KRAS-mutated cell lines have been found to fall into either KRAS-dependent or KRAS-independent groups. The dependency on KRAS or other oncogenes can be changed by various mechanisms, genetic, and nongenetic. For example, autocrine IGF1 signaling has been found to confer resistance to suppression of oncogenic drivers (12). In other cases, overexpression of YAP1 was driving KRAS independence in initially addicted cells (13). In addition, metabolic remodeling and oxidative phosphorylation can protect KRAS-driven cells from apoptosis (14). Furthermore, in prostate cancer, overexpression of FGFR1 was found to be indispensable in prostatic neoplasia, while prostate cancer lesions became independent of FGFR1 expression (15).
Here, we test whether FLC tumors are addicted to the DNAJB1-PRKACA fusion protein and can be a valid therapeutic target. We identified a short hairpin RNA (shRNA) inhibiting the chimeric transcript and protein, but with minimal effect on the wildtype DNAJB1 or PRKACA. Knockdown of the chimera resulted in cell death of FLC cells in vitro and in vivo, but not in a model of HCC artificially created to express the chimera. These results demonstrate that DNAJB1-PRKACA, unlike some other oncogenes, is required as a continuous driver for FLC. Furthermore, elimination of the DNAJB1-PRKACA results in cell death, rather than a return to wildtype phenotype. Thus, DNAJB1-PRKACA is a promising therapeutic target for precision medicines.
Materials and Methods
Cloning
shRNA plasmids were constructed as previously described and cloned into the pLKO.1 vector (16). Short oligonucleotides of shRNA sequences were ordered from IDT (Fig. 1A). Control (Ctr) shRNA and shRNA P were cloned into the inducible plasmid as described previously (17).
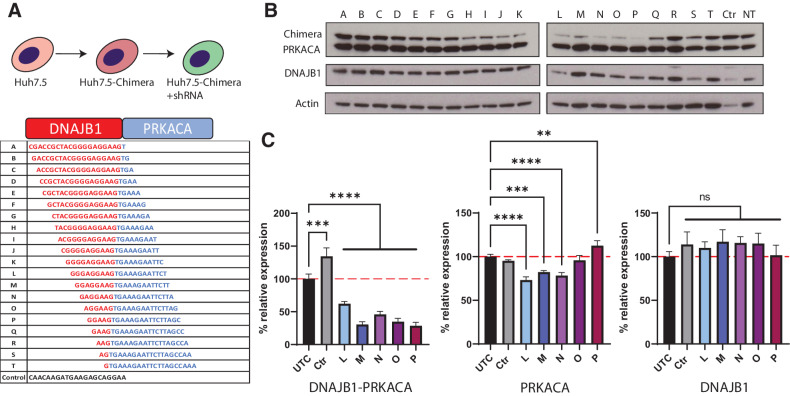
Figure 1.
Potent and specific shRNA-mediated knockdown of DNAJB1-PRKACA. A, Generation of stable Huh7.5 cells expressing the chimeric transcript and shRNAs by serial lentiviral transduction. Sequences of shRNAs tiling over the fusion junction, red and blue letters denote DNAJB1 and PRKACA sequence, respectively. A Ctrl shRNA was used for all experiments. B, Western blot analysis of Huh7-Chimera cells stably transduced with shRNAs A-T, Ctrl shRNA or nontransduced (NT). C, qPCR results of Huh7-Chimera cells stably expressing shRNAs L-P, Ctrl shRNA or untreated cells (UTC). Left graph measures Chimera knockdown, middle graph measures PRKACA knockdown, and right graph measures DNAJB1 knockdown. N = 3 for all conditions, one-way ANOVA with Dunnett multiple comparison test. **, P = 0.0021; ***, P = 0.0002; ****, P < 0.0001.
RNA extraction and gene expression analysis
Cultured cells were directly lysed in RLT buffer (QIAGEN) supplemented with 1% 2-mercaptoethanol. Total mRNA was extracted with the RNeasy Kit (QIAGEN) according to the manufacturer's instructions. Gene expression was measured with the Luna Universal Probe OneStep qRT-PCR kit (NEB) normalizing to beta-actin. Sequences of primer–probe sets are:
Chimera:
Probe: 56-FAM/AAGCGCGAG/ZEN/ATCTTCGACCGCTA/3IABkFQ
Forward Primer: GAAGTTCAAGGAGATCGCTGAG
Reverse Primer: GAGCGGGACTTTCCCATTT
DNAJB1:
Forward Primer: CAAGCGCGAGATCTTCGAC
Reverse Primer: GAACTCAGCAAACATGGCAT
Probe: 56-FAM/CCACTCCCC/ZEN/TTTAGGCCTTCCTC/3IABkFQ
PRKACA:
Forward Primer: CAAGAAGGGCAGCGAGCA
Reverse Primer: CTGTGTTCTGAGCGGGACTT
Probe: 56-FAM/AGAGCGTGA/ZEN/AAGAATTCTTAGCCAAAGCC/3IABkFQ
Actin:
Forward Primer: CCGACTATGACTTAGTTGCGTTACA
Reverse Primer: GCCATGCCAATCTCATCTTGT
Probe: 5HEX/CCTTTCTTG/ZEN/ACAAAACCTAACTTGCGCAGA/3IABkFQ
TetR:
Forward Primer: GAGGTCGGAATCGAAGGTTTA
Reverse Primer: GAGCAAAGCCCGCTTATTT
Probe: 56-FAM/ACAACCCGT/ZEN/AAACTCGCCCAGAAG/3IABkFQ
Cell culture and viral transduction
Huh7-chimera cells were cultured in DMEM supplemented with 10% FBS. Dissociated patient-derived xenograft (PDX) cells were cultured in RPMI, supplemented with 10% FBS and 2% penicillin/streptomycin. Cells were transduced with lentivirus expressing shRNA with 10 μg/mL polybrene, incubated for 48 hours for protein expression, before selection with 2–3 μg/mL puromycin for a minimum of 72 hours.
Cell viability assays
Cultured cells were assayed for viability using PrestoBlue HS (Thermo Fisher Scientific) based on manufacturer's instructions. Briefly, FLC cells were cultured in 96-well plates in RPMI medium supplemented with 10% FBS and 2% penicillin/streptomycin. Huh7-Chimera cells were cultured in 96-well plates with DMEM supplemented with 10% FBS and 2% penicillin/streptomycin. Huh7-Chimera cells were seeded at 5,000 cells/well and growth measured daily. For experiments with doxycycline, 100 ng/mL of doxycycline was added for the indicated time periods before PrestoBlue readout. PrestoBlue was added at 1:10 and cells were incubated for 1–2 hours before plate reader fluorescence measurement at 560 nm excitation and 590 nm emission in a Spark Tecan instrument.
For experiments with Huh7-Chimera cells, raw absorbance values were used and normalized by wells not containing cells. For experiments with FLC cells, viability was calculated compared with cells treated with 20 μmol/L chaetocin (Positive, SelleckChem S8068) and untreated (Negative) as percent survival = (Positive − Treated)/(Positive − Negative) * 100.
Tumor dissociation
Tumor-bearing mice were euthanized in accordance with Institutional Animal Care and Use Committee approval (#20027-H). PDXs were harvested and cut into 2-mm pieces, connective tissue, blood vessels and necrotic tissue was discarded and then transferred into 50-mL Falcon tubes with RPMI, 2% penicillin/streptomycin, collagenase 5 (Worthington 1 mg/mL), neutral protease (Worthington 0.5 U/mL) and DNAse (Roche, 1 μg/mL), and digested while rotating at 37°C until digestion was complete (Benchmark scientific Roto-therm). All following steps were done on ice or at 4°C. The digested tissue was passed through a 200-μm (Pluriselect) strainer using a syringe plunger for remaining pieces, and then through a 100 μm strainer (Thermo Fisher Scientific). The cells were spun down at 321 × g for 5 minutes at 4°C and the pellet depleted of red blood cells by a 10-second exposure to 1 mL of water followed by the addition of 49 mL of PBS. The cells were counted and plated on collagen-coated plates for in vitro experiments. For in vitro experiments, the cells from PDX tumors were subjected to mouse cell depletion according to the manufacturer's instructions (Miltenyi Biotec).
Mice
NSG mice were purchased from Jackson Laboratories and bred at The Rockefeller University animal facility specific pathogen-free immune core. Mice were kept in 12 hours light/dark cycle, fed an amoxicillin diet, and had ad libitum access to food and water. Both male and female mice were used for PDX passaging and in vivo studies. Mice used for in vivo studies were between 1 and 2 months at the time of implantation. Mice were inspected at least twice a week for health and tumor growth. All experiments were conducted under animal use protocols approved by The Rockefeller University (New York, NY).
In vivo tumor studies
In accordance with Institutional Animal Care and Use Committee approval (#20027-H), mice were anesthetized using isoflurane. Cells were injected subcutaneous with 200k to 500k cells in a 1:1 ratio mix or cells in RPMI and Matrigel (Corning). Tumors were allowed to engraft until formation of sizeable tumors and then randomly assigned to receive doxycycline. Mice in the doxycycline group of the in vivo studies, received 1,000 ppm doxycycline feed. Tumor size was measured by caliper measure once to three times per week, depending on tumor aggressiveness.
Protein isolation and immunoblotting
Total protein from the cells in culture was extracted using RIPA buffer (Sigma) supplemented with protease and phosphatase inhibitors [complete ethylenediaminetetraacetic acid (EDTA)-free and PhosSTOP, Roche]. Samples were incubated on ice for 20 minutes and then centrifuged at 21,000 × g for 10 minutes. Supernatants were collected, and protein concentrations were measured by a modified Lowry assay (DC protein assay, Bio-Rad). Protein (20–30 μg) per sample was diluted with 4× Nupage LDX sample buffer (Life Technologies) containing 10% β-mercaptoethanol. Samples were heated at 67°C for 10 minutes and then loaded on 4%–12% Bis–Tris gels (Nupage, Invitrogen) and run in MOPS buffer for 50 minutes at 200 V. Transfer was performed using the iBlot (Life Technologies). Membranes were blocked for 1 hour in 2.5%–5% milk (Carnation powdered milk) in TBS with Tween (TBST), washed in TBST, and then probed with primary antibodies against PRKACA [PKA C-α (D38C6) Rabbit mAb, Cell Signaling Technology, 1:1,000 dilution], actin [Actin (A5316) mouse mAb, Sigma, 1:1,000] and DNAJB1 [Hsp40, (4868S), Rabbit polyclonal, Cell Signaling Technology, 1:1,000] in 5% milk and incubated overnight shaking at 4°C. After washing in TBST, membranes were incubated with horseradish peroxidase–conjugated appropriate secondary antibodies (Sigma, A0545 goat anti-rabbit, A9917, goat anti-mouse, 1:10,000) in 5% milk in TBST for 1 hour. Membranes were washed in TBST and then incubated with Amersham ECL prime Western blotting detection reagent (GE Healthcare), exposed to film in a dark room or imaged on a LI-COR Blot Scanner (C-DiGit). Blots were stripped with Restore Plus Western Blot Stripping buffer (46430, Thermo Fisher Scientific) if multiple proteins were probed for.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 9.0.1. To compare the means of two groups, we used a two-sided unpaired t test. All in vivo studies were performed with n = 3 per group. For doxycycline studies in PDX, mice with shRNA P or Ctr were randomly assigned to receive doxycycline.
ANOVA with Dunnett test for post hoc comparisons was used for Fig. 1. Linear mixed-effects regression model with compound symmetry covariance structure was used to compare the growth trajectories between the different experimental groups (18). The model included group, day (categorical), and group*day interaction as fixed effects. A significant group*day interaction indicated a difference in tumor growth over time between groups. Prior to analysis, due to a skewed tumor volume distribution, a log transformation was used for tumor volume of PDXA. Because PDXB had some tumors whose volume was undetectable, for that model we used a cube root transformation. Residual plots were examined to evaluate the validity of model assumptions. Data analysis was performed using Proc Mixed in SAS Studio v3.8.
Data availability
Data were generated by the authors and are available on request from the corresponding author.
Results
To identify shRNAs that specifically reduce DNAJB1-PRKACA fusion transcripts (referred to as chimera), we created a panel of shRNA spanning the fusion junction, each shifted by a single nucleotide (Fig. 1A). The efficacy of these shRNAs was first tested in vitro on a human HCC cell line, Huh7 (19), stably transduced with the FLC oncogenic chimeric transcript under the control of the DNAJB1 promoter (Huh7-Chimera). Huh7 cells do not show an FLC phenotype and do not depend on expression of the chimera for proliferation or survival. We designed primer probe pairs to specifically measure knockdown of the transcripts for chimera, DNAJB1 and PRKACA. The shRNAs L to P were the most effective at reducing protein, (Fig. 1B) and RNA, (Fig. 1C) of the chimera, with minimal effect on the native DNAJB1 or PRKACA fusion partners. Continuous expression of the shRNA did not alter the growth of Huh7-Chimera cells (Fig. 2A). Thus, in a cell ectopically expressing the chimera, shRNA to the chimeric junction had no effect.
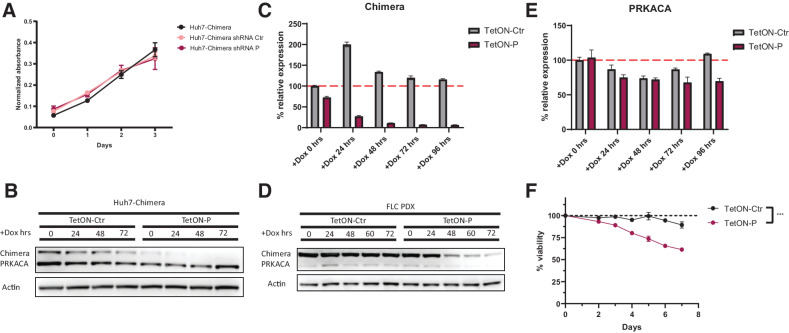
Figure 2.
Cytotoxic effect of shRNA-mediated knockdown of DNAJB1-PRKACA fusion in FLC. A, Cell viability measurements of Huh7-Chimera cells expressing Ctrl or targeting shRNA P. B, Western blot time course of doxycycline induction of shRNA expression and knockdown of the Chimera protein in Huh7-Chimera cells. C, qPCR time course of doxycycline mediated induction of shRNA expression and knockdown of the chimera transcript in PDX cells. Expression was normalized to actin. D, Time course of doxycycline-mediated induction of shRNA expression and knockdown of the chimera protein in PDX cells as assessed by Western blot. E, Time course of doxycycline-mediated induction of shRNA expression and effect on PRKACA transcript as assessed by qPCR. Expression was normalized to actin. F, Time course of doxycycline-mediated shRNA induction and effect on cell viability of PDX cells. Unpaired t test, P = 0.0002 at the last timepoint.
We selected shRNA P for further studies in FLC due to its high efficacy of knockdown and specificity for chimera over DNAJB1 or PRKACA. To test whether the shRNA affected cellular fitness, we cloned the shRNA P into a doxycycline-inducible plasmid (17). This allowed us to transduce and select a population of cells with the ability to control expression of the shRNA for cell viability studies. We first confirmed efficient knockdown with the inducible system in Huh7-Chimera cells before transferring to FLC cells (Fig. 2B). Upon induction with doxycycline the protein levels of the chimera decreased in the Huh7-Chimera cells within the first 24 hours, with no detected effect on expression of PRKACA. The chimera protein in the cells was reduced even at time 0, likely due both to the incomplete suppression of promoter activity in the absence of doxycycline and the high potency of the shRNA.
The inducible shRNA was next tested in cells dissociated from an FLC PDX, denoted as PDXA. PDXA was derived from a patient who had an unusually rapidly growing FLC tumor and is our most rapidly growing FLC PDX. Cells were transduced and selected before induction with doxycycline. We observed a strong decrease in chimera RNA within 24 hours. The protein level showed a delayed decrease, as compared with the rapid reduction in protein seen in Huh7-Chimera cells (Fig. 2C and D). The delayed effect on chimera protein in FLC cells may be a consequence of reduced overall metabolic activity in these cells, which may affect protein turnover. We have previously observed that cells dissociated from FLC PDXs do not actively grow or divide after dissociation and plating in vitro. Alternatively, there may be specific stabilization of the chimera in the FLC cells. We also observed a slight reduction in PRKACA levels both in Ctr and P shRNA groups. This could be a doxycycline-mediated or cell death–dependent effect in FLC cells, as we did not see effects on PRKACA in Huh7-cells (ref. 20; Fig. 2D–F).
Upon induction of the shRNA in FLC PDXA cells, there was a progressive decrease in viability compared with Ctrl shRNA, starting after 3 days. The time course of the decrease of viability paralleled the time course of the decrease of protein. These results suggest a dependence of FLC on DNAJB1-PRKACA protein in vitro (Fig. 2F).
To test whether continuous expression of the chimera is necessary for FLC cells that are actively growing, we implanted PDX cells transduced with inducible shRNA P or control into immune compromised mice. Cells were allowed to engraft for 2–3 weeks until tumors were visible, before induction with doxycycline (Fig. 3A). We monitored body weight to identify potential confounding effects from the prolonged doxycycline diet. We observed that over long time periods continuous doxycycline feed resulted in weight reduction possibly due to an interplay of reduced food intake, physiologic burden by the tumor growth and the tumor suppressive effect of doxycycline (ref. 21; Fig. 3B). However, tumor growth was similar to groups not receiving doxycycline (Fig. 3C).
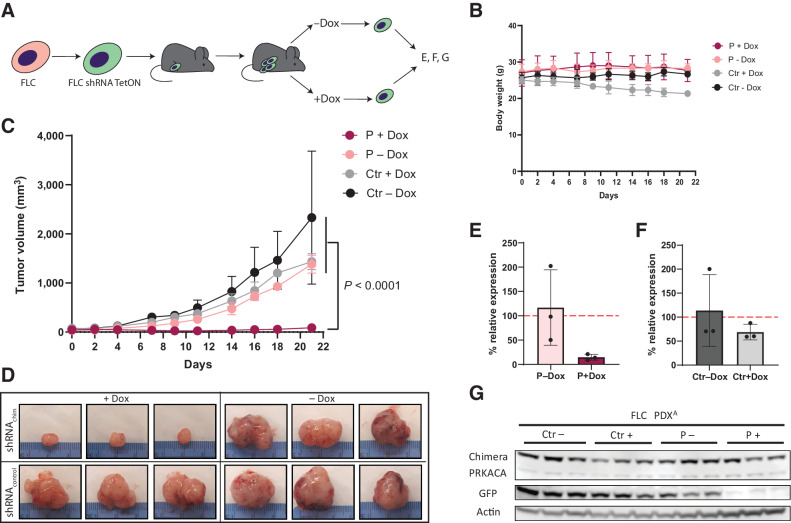
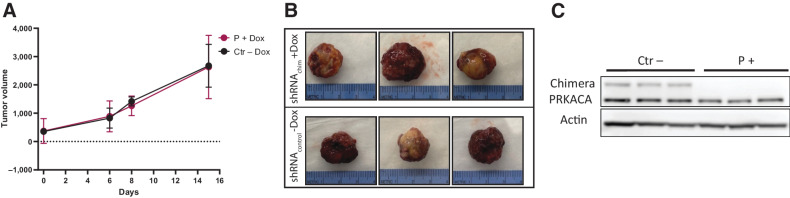
Figure 3.
DNAJB1-PRKACA is necessary for FLC growth in vivo in PDXA, an aggressive model. A, Schematic of the experimental design. B, Weight of tumor-bearing mice over time. C, Tumor volume over time of PDXA transduced with inducible Ctrl shRNA or Chimera targeting shRNA, ± addition of doxycycline (Dox) feed. N = 3 for all groups. Linear mixed-effects model with log tumor volume was used to analyze data across the study duration. Group: < 0.0001; Day: < 0.0001; Group*Day: < 0.0001. Significant post hoc pairwise comparisons of growth trajectories at Bonferroni-adjusted alpha level < 0.0083 (0.05/6) are indicated. Inset is a higher resolution graph of shRNA P+Dox. D, Images of extracted PDX transduced with Ctrl shRNA or Chimera targeting shRNA, ± Dox feed. E, qPCR of the TetR gene normalized to actin in tumors of the Ctr− and + Dox group at the end of the study. F, qPCR of the TetR gene normalized to actin in tumors of P− and P+ doxycycline at the end of the study. G, Western blot analysis of PDX from A and C, showing loss of Chimera knockdown in the P+Dox group together with loss of GFP signal.
We used a linear mixed-effects model to analyze tumor volume over time to capture differences in growth trajectories (18). In PDXA, induction of shRNA P strongly inhibited tumor growth as compared with Ctrl shRNA or noninduced shRNAs (Fig. 3C and D). The PDXA of FLC grew for 2 days after induction of shRNA P, decreased in volume for the next 9 days, and then started to regrow (Fig. 3C inset, higher resolution of shRNA P+Dox group). At the end of the study, residual tumor was probed by qPCR for the TetR gene as a proxy for the levels of transduced cells expressing the shRNA. The results demonstrate the level of plasmid encoding the shRNA was reduced compared to tumors that were not induced with doxycycline (Fig. 3E and F). Consistent with this, the residual tumor did not have a reduction of DNAJB1-PRKACA. In addition, we found reduced levels of GFP, coexpressed from the shRNA plasmid (Fig. 3G). These results indicate that residual tumor is largely composed of a small subset of cells that survived puromycin selection but were not transduced. Cells that did express the shRNA and had reduced levels of the chimera died, demonstrating a strong dependence of FLC cells on the oncogene.
This study was repeated on PDXB which was independently derived from another patient. The original tumor for PDXB grew more slowly in the patient, typical of most FLC. Accordingly, PDXB grows slowly in the mouse, similar to most of our FLC PDX, but unlike PDXA. Thus, while the slow growth of PDXB makes it a more difficult model for studies, it is likely a more representative model of FLC. Consistent with the results in PDXA, upon induction of the shRNA in PDXB, there was remission and clearance of the tumor (Fig. 4A–C), demonstrating a dependence on continuous expression of the chimera.
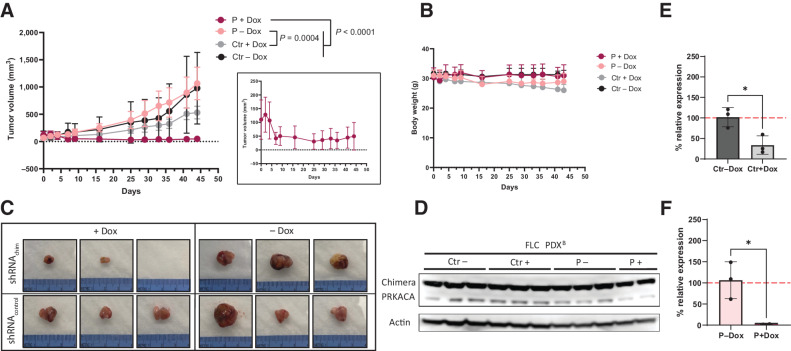
Figure 4.
DNAJB1-PRKACA is necessary for FLC growth in vivo in a slow growing PDX model. A, Tumor volume over time of a slow growing PDX (PDXB) transduced with inducible Ctrl shRNA or Chimera targeting shRNA, ± addition of doxycycline (Dox) feed. N = 3 for all groups. Linear mixed-effects model with cube root tumor volume was used to analyze data across the study duration. Group: P = 0.0423; Day: < 0.0001; Group*Day: < 0.0001. Only significant post hoc pairwise comparisons of growth trajectories at Bonferroni-adjusted alpha level < 0.0083 (0.05/6) are indicated. Inset is higher resolution graph of P+Dox group. B, Weight of tumor-bearing mice over time. C, Images of extracted PDX transduced with Ctrl shRNA or Chimera targeting shRNA, ± Dox feed. D, Western blot analysis of PDX from A and C, showing loss of Chimera knockdown in the P+Dox group together. E, qPCR of the TetR gene normalized to actin in tumors of Ctr− and + Dox at the end of the study. Two-tailed t test P = 0.0225. F, qPCR of the TetR gene normalized to actin in tumors of P− and P+ Dox at the end of the study. Two-tailed t test P = 0.0491.
Similar to what was observed with PDXA, the small residual tumors of the P+Dox group had reduced levels of the shRNA plasmid and there was no decrease of the chimera protein in the two remaining tumors (Fig. 4D–F). However, in contrast to PDXA, we also observe reduced tumor growth and reduced levels of the plasmid in tumors of Ctr+Dox group, but there was no significant difference between Ctr groups with and without doxycycline. This might be a result of the prolonged doxycycline treatment compared with PDXA. Continued induction of Ctr shRNA might have a weak but continuous effect on cell viability.
Importantly, for tumors in mice from Huh7 cells transduced with the Chimera, the induction of the shRNA P eliminated expression of the chimera but had no effect on tumor growth (Fig. 5A–C). This in vivo observation corroborates the cytotoxic specificity of the shRNA against FLC seen in vitro. This rules out off-target toxic effects. Furthermore, by demonstrating that the dependence on DNAJB1-PRKACA is specific to FLC, it rules out on-target toxic effects from engaging the right target, which were not related to the dependence of the cell to the target.
Figure 5.
Expression of shRNA P in vivo does not affect Huh7-Chimera xenograft growth. A, Tumor volume over time of Huh7-Chimera xenografts transduced with inducible Ctrl shRNA or Chimera targeting shRNA P. The shRNA P group was feed with doxycycline chow. N = 3 for all groups. B, Images of extracted xenografts from A. C, Western blot analysis of tumors from A and B.
Discussion
Many pediatric cancers are driven by fusion genes, but only a few have experimental evidence demonstrating the oncogene to be indispensable for tumor growth (22). Studies in KRAS, MYC, or FGFR1 driven tumor models have demonstrated that these can develop independence from the triggering oncogene (11–15). Therefore, it is crucial to determine whether DNAJB1-PRKACA is a promising therapeutic target.
Here, we show that FLC tumors require the continued expression of the chimeric DNAJB1-PRKACA to grow. Thus, this oncokinase is not only a trigger for FLC but is needed for continued transformation. Furthermore, upon loss of the DNAJB1-PRKACA, FLC cells do not revert back to the phenotype of non-transformed cells but instead undergo apoptosis demonstrating an oncogenic addiction.
We screened shRNAs tiling over the fusion junction and identified several potent and specific candidates. We used a HCC cell line, Huh7, transduced with the chimera to select shRNAs that specifically knock down DNAJB1-PRKACA. We did not observe any adverse effects on Huh7-Chimera cells expressing the shRNA. This demonstrates that the effects of shRNA are not the result of either cytotoxic off-target effects, hitting sites other than the DNAJB1-PRKACA transcript, or the result of cytotoxic “on-target effects,” potentially triggered by formation of a double-stranded RNA. Instead, it demonstrates that in FLC, the cytotoxic effects of the shRNA are specific to a tumor that has developed an oncogenic addiction to DNAJB1-PRKACA. This is consistent with our recent observations that even after multiple passages, PDX of FLC never show loss of expression of DNAJB1-PRKACA (5). Using in vitro and in vivo models of FLC we show that knockdown of the chimeric transcript inhibits tumor growth and results in cell death. This validates DNAJB1-PRKACA as a target for therapeutic development.
We note that a murine knockout of DNAJB1 is not lethal, with only subtle effects on macrophages (23). Thus, exon 1 of DNAJB1 might also be tolerated as a potential therapeutic target. In contrast, it is critical that native PRKACA levels are unaffected as knockout is embryonically lethal and postnatally PRKACA is essential for many cellular functions across different tissues (24). In general, approaches targeting the fusion junction should offer high specificity and low toxicity as the fusion is only present in cancerous cells. Previous studies demonstrated the therapeutic effect of siRNA targeting the fusion junction of RET/PTC1 in papillary thyroid cancer, but delivery has been a critical bottleneck for clinical translation (25). However, recent advances in medicinal chemistry and delivery approaches of antisense oligonucleotides and siRNAs to the liver might allow translation of our findings targeting the fusion junction in FLC (26).
We believe that inducible shRNAs specific to the junction of gene fusions can be useful for investigation of other fusion-driven cancers. Specifically, as in the current investigation, shRNA knockdown can be used to determine whether tumor cells are addicted to the oncogene, rather than merely initiated by it. Addiction is a precondition to qualifying the fusion product as a therapeutic target. While adult tumors often harbor multiple mutations, pediatric cancers show low mutational burden with often only a single genetic driver mutation (27, 28). Commonly, this mutation is a deletion event resulting in formation of oncogenic fusion proteins, such as EWS-FLI or SS18-SSX in sarcomas. In the case of FLC, we know the oncogenic event is formation of the fusion gene, not the loss of genes in between the breakpoints (10). For many of these fusion genes, oncogene addiction has not been demonstrated and therapeutics focus on other vulnerabilities independent of the fusion gene. For instance, CRISPR knockout screens have identified new dependencies in pediatric cancers that can be harnessed for therapeutic development (29). However, CRISPR-mediated knockout of the fusion proteins themselves are complicated, as the junction region provides only a narrow window for gRNAs that do not also target the native fusion partners. Some progress has been made for interchromosomal fusion genes such as EWSR1-FLI1 (30). In the case of DNAJB1-PRKACA, CRISPR approaches are complicated because of the high CG content in exon 1 of DNAJB1 and the essential function of PRKACA, which prohibits targeting exons 2–10 of the fusion. Targeting the enzymatic activity of PRKACA with small molecules would likely result in significant systemic toxicities and thus is a risky therapeutic approach. Inducible shRNAs offer an elegant system to study oncogene addiction of fusion genes which can be applied broadly across tumor types. Precision medicines targeting the unique fusion junction should mitigate toxic side effects and provide a wider therapeutic window. We anticipate this study will spur the development of targeted therapies for FLC.
Acknowledgments
We would like to thank all of the fibrolamellar patients, and their caregivers, through their contributions to The Fibrolamellar Registry, to our Fibrolamellar Tissue Repository, through work at the bench and contributions in too many ways to enumerate. We would also like to thank for their financial support, critical seed funds from private foundations, and the NIH/NCI R01CA248507 (C. Neumayer, S.M. Simon); NIH/NCI P50CA210964 (D. Ng, G. Lalazar, S.M. Simon); NIH/NCI U54CA243126 (D. Ng, G. Lalazar, S.M. Simon); Robertson Therapeutic Development Fund (S.M. Simon); Center for Basic and Translational Research on Disorders of the Digestive System through the generosity of the Leona M. and Harry B. Helmsley Charitable Trust (G. Lalazar, S.M. Simon); The Sohn Conference Foundation (S.M. Simon); The Rally Foundation (S.M. Simon); The Bear Necessities (S.M. Simon); The Truth365 (S.M. Simon). Supported in part by grant NIH/NCATS #UL1 TR001866 from a Clinical and Translational Science Award (CTSA).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authors' Disclosures
C. Neumayer, D. Ng, and S.M. Simon report grants from NIH during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
C. Neumayer: Conceptualization, resources, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. D. Ng: Formal analysis, investigation, visualization, methodology, writing–review and editing. C.S. Jiang: Formal analysis, methodology, writing–review and editing. A. Qureshi: Formal analysis, methodology, writing–review and editing. G. Lalazar: Conceptualization, investigation, writing–review and editing. R. Vaughan: Formal analysis, supervision, methodology, writing–review and editing. S.M. Simon: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, visualization, methodology, writing–original draft, writing–review and editing.
References
- 1. O'Neill AF, Church AJ, Perez-Atayde AR, Shaikh R, Marcus KJ, Vakili K. Fibrolamellar carcinoma: an entity all its own. Curr Probl Cancer 2021:100770. [DOI] [PubMed] [Google Scholar]
- 2. Lalazar G, Simon SM. Fibrolamellar carcinoma: recent advances and unresolved questions on the molecular mechanisms. Semin Liver Dis 2018;38:51–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Torbenson M. Fibrolamellar carcinoma: 2012 update. Scientifica 2012;2012:743790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fritz AG. International classification of diseases for oncology: ICD-O. Geneva: World Health Organization; 2013.viii, p. 242. [Google Scholar]
- 5. Lalazar G, Requena D, Ramos-Espiritu L, Ng D, Bhola PD, de Jong YP, et al. Identification of novel therapeutic targets for fibrolamellar carcinoma using patient-derived xenografts and direct-from-patient screening. Cancer Discov 2021;11:2544–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Graham RP, Lackner C, Terracciano L, Gonzalez-Cantu Y, Maleszewski JJ, Greipp PT, et al. Fibrolamellar carcinoma in the Carney complex: PRKAR1A loss instead of the classic DNAJB1-PRKACA fusion. Hepatology 2018;68:1441–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Honeyman JN, Simon EP, Robine N, Chiaroni-Clarke R, Darcy DG, Lim II, et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science 2014;343:1010–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Graham RP, Jin L, Knutson DL, Kloft-Nelson SM, Greipp PT, Waldburger N, et al. DNAJB1-PRKACA is specific for fibrolamellar carcinoma. Mod Pathol 2015;28:822–9. [DOI] [PubMed] [Google Scholar]
- 9. Engelholm LH, Riaz A, Serra D, Dagnaes-Hansen F, Johansen JV, Santoni-Rugiu E, et al. CRISPR/Cas9 engineering of adult mouse liver demonstrates that the dnajb1-prkaca gene fusion is sufficient to induce tumors resembling fibrolamellar hepatocellular carcinoma. Gastroenterology 2017;153:1662–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kastenhuber ER, Lalazar G, Houlihan SL, Tschaharganeh DF, Baslan T, Chen CC, et al. DNAJB1-PRKACA fusion kinase interacts with beta-catenin and the liver regenerative response to drive fibrolamellar hepatocellular carcinoma. Proc Nat Acad Sci U S A 2017;114:13076–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Abdulla H, Vo A, Shields BJ, Davies TJ, Jackson JT, Alserihi R, et al. T-ALL can evolve to oncogene independence. Leukemia 2021;35:2205–19. [DOI] [PubMed] [Google Scholar]
- 12. Rajbhandari N, Lin WC, Wehde BL, Triplett AA, Wagner KU. Autocrine IGF1 signaling mediates pancreatic tumor cell dormancy in the absence of oncogenic drivers. Cell Rep 2017;18:2243–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell 2014;158:185–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 2014;514:628–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Acevedo VD, Gangula RD, Freeman KW, Li R, Zhang Y, Wang F, et al. Inducible FGFR-1 activation leads to irreversible prostate adenocarcinoma and an epithelial-to-mesenchymal transition. Cancer Cell 2007;12:559–71. [DOI] [PubMed] [Google Scholar]
- 16. Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 2006;124:1283–98. [DOI] [PubMed] [Google Scholar]
- 17. Wiederschain D, Susan W, Chen L, Loo A, Yang G, Huang A, et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle 2009;8:498–504. [DOI] [PubMed] [Google Scholar]
- 18. Harrison XA, Donaldson L, Correa-Cano ME, Evans J, Fisher DN, Goodwin CED, et al. A brief introduction to mixed effects modelling and multi-model inference in ecology. PeerJ 2018;6:e4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res 1982;42:3858–63. [PubMed] [Google Scholar]
- 20. Ahler E, Sullivan WJ, Cass A, Braas D, York AG, Bensinger SJ, et al. Doxycycline alters metabolism and proliferation of human cell lines. PLoS One 2013;8:e64561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dijk SN, Protasoni M, Elpidorou M, Kroon AM, Taanman J-W. Mitochondria as target to inhibit proliferation and induce apoptosis of cancer cells: the effects of doxycycline and gemcitabine. Sci Rep 2020;10:4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shao L, Tekedereli I, Wang J, Yuca E, Tsang S, Sood A, et al. Highly specific targeting of the TMPRSS2/ERG fusion gene using liposomal nanovectors. Clin Cancer Res 2012;18:6648–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Uchiyama Y, Takeda N, Mori M, Terada K. Heat shock protein 40/DjB1 is required for thermotolerance in early phase. J Biochem 2006;140:805–12. [DOI] [PubMed] [Google Scholar]
- 24. Skalhegg BS, Huang Y, Su T, Idzerda RL, McKnight GS, Burton KA. Mutation of the Calpha subunit of PKA leads to growth retardation and sperm dysfunction. Mol Endocrinol 2002;16:630–9. [DOI] [PubMed] [Google Scholar]
- 25. de Martimprey H, Bertrand JR, Fusco A, Santoro M, Couvreur P, Vauthier C, et al. siRNA nanoformulation against the ret/PTC1 junction oncogene is efficient in an in vivo model of papillary thyroid carcinoma. Nucleic Acids Res 2008;36:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roberts TC, Langer R, Wood MJA. Advances in oligonucleotide drug delivery. Nat Rev Drug Discov 2020;19:673–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dupain C, Harttrampf AC, Urbinati G, Geoerger B, Massaad-Massade L. Relevance of fusion genes in pediatric cancers: toward precision medicine. Mol Ther Nucleic Acids 2017;6:315–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grobner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA, et al. The landscape of genomic alterations across childhood cancers. Nature 2018;555:321–7. [DOI] [PubMed] [Google Scholar]
- 29. Dharia NV, Kugener G, Guenther LM, Malone CF, Durbin AD, Hong AL, et al. A first-generation pediatric cancer dependency map. Nat Genet 2021;53:529–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martinez-Lage M, Torres-Ruiz R, Puig-Serra P, Moreno-Gaona P, Martin MC, Moya FJ, et al. In vivo CRISPR/Cas9 targeting of fusion oncogenes for selective elimination of cancer cells. Nat Commun 2020;11:5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data were generated by the authors and are available on request from the corresponding author.