Abstract
Purpose:
Follicular lymphoma (FL) is the most frequent indolent non-Hodgkin lymphoma. Around 20% of patients suffer early disease progression within 24 months (POD24) of diagnosis. This study examined the significance of circulating tumor DNA (ctDNA) in predicting response to therapy and POD24 in patients with FL.
Experimental Design:
We collected 100 plasma samples, before and during the treatment, from 36 patients with FL prospectively enrolled in 8 Spanish hospitals. They were treated with a chemotherapy-rituximab regimen and followed up for a median of 3.43 years. We performed targeted deep sequencing in cell-free DNA (cfDNA) and tumor genomic DNA from 31 diagnostic biopsy samples.
Results:
Of the alterations detected in the diagnostic tissue samples, 73% (300/411) were also identified in basal cfDNA. The mean numbers of alterations per basal cfDNA sample in patients who suffered progression of disease within 24 months (POD24-pos) or did not achieve complete response (non-CR) were significantly higher than in POD24-neg or CR patients (unpaired samples t test, P = 0.0001 and 0.001, respectively). Pretreatment ctDNA levels, as haploid genome equivalents per milliliter of plasma, were higher in patients without CR (P = 0.02) and in POD24-pos patients compared with POD24-neg patients (P < 0.001). Dynamic analysis showed that ctDNA levels decreased dramatically after treatment, although the reduction was more significant in patients with CR and POD24-neg patients.
Conclusions:
Basal ctDNA levels are associated with the risk of early progression and response to treatment in FL. cfDNA monitoring and genotyping during treatment and follow-up predict response to treatment and early progression.
Translational Relevance.
Circulating tumor DNA (ctDNA) levels are associated with tumor volume and minimal residual disease in lymphoid malignancies, mainly in Hodgkin lymphoma and non-Hodgkin aggressive lymphomas. However, few studies have studied ctDNA in indolent subtypes such as follicular lymphoma that, despite being considered an indolent disease, remains incurable for most patients. Identifying clinical or molecular markers that could identify patients at higher risk of early progression at diagnosis is clinically relevant.
Our study suggests that pretreatment ctDNA levels could be useful to predict response to treatment and early progression as an independent predictive biomarker or in combination with the Follicular Lymphoma International Prognostic Index. Our pilot study also demonstrates that ctDNA levels and cell-free DNA genotyping at mid-treatment and end-of-treatment are associated with treatment response and early progression. Therefore, we propose that ctDNA measurement at diagnosis and follow-up is a valuable tool that might feasibly be used in clinical trials and, eventually, in daily clinical practice.
Introduction
Follicular lymphoma (FL) is the second most common type of B-cell non-Hodgkin lymphoma, with an annual incidence of 3 to 5 per 100,000 habitants in the United States and Europe (1, 2). FL is a heterogeneous though generally indolent disease. It is characterized by slow progression, high response rates to therapy, and high overall survival (OS) but remains an incurable disease (3, 4). Early FL relapse, defined as recurrence or progression of the disease within 24 months of diagnosis (POD24), is associated with inferior outcomes as POD24 patients had a 5-year OS of 34% to 50% compared with a 5-year OS of 90% to 94% for patients without POD24 (4). Recent analysis of data from more than 5,000 patients with FL enrolled in 13 clinical trials showed POD24 to be a robust indicator of poor FL survival (5). Around 20% to 30% of patients will die from refractory disease or transformation to high-grade lymphoma, most frequently to a diffuse large B-cell lymphoma (DLBCL; ref. 6). The transformation event has been associated with treatment resistance and a worse prognosis (6–8). The use of the Follicular Lymphoma International Prognostic Index (FLIPI) and PET has improved patient stratification, but both tools have limitations, meaning that alternative, more accurate methods for predicting patient outcomes are needed (9–12). In recent years, high-throughput sequencing technology has increased our knowledge of the FL mutational landscape (13–18), and the inclusion of predictors incorporating clinical and genetic features, such as m7-FLIPI or POD24-PI, has been proposed as means of improving the identification of patients with poor clinical outcomes (11, 19). In addition, alterations in several genes, including TP53, MYC, CCND3, GNA13, POU2AF1, NOTCH2, DTX1, UBE2A, and HISTIH1E, among others, have been associated with transformation (20–24).
Identifying genetic alterations in mature B-cell tumors has made it possible to detect genetic subtypes with a higher risk of treatment resistance or progression, track clonal evolution-driven resistance upon treatment in real time, and generate maps of genetic alterations for targeted therapies (25, 26). The difficulties of serial sampling during patient follow-up for monitoring disease evolution make this approach difficult to use in daily clinical practice.
In recent years, the analysis of cell-free DNA (cfDNA), DNA fragments released into the bloodstream from apoptotic normal and tumor cells, and the detection of circulating tumor DNA (ctDNA), has made it possible to monitor disease with a minimally invasive approach. ctDNA levels have been quantified to determine tumor volume and minimal residual disease (MRD). The application of high-throughput sequencing technology in cfDNA has emerged as a valuable tool for identifying and following tumor genetic alterations, and monitoring the clonotypic immunoglobulin gene rearrangement for MRD in hematologic diseases. cfDNA has proven useful for identifying tumor alterations at diagnosis with robust sensitivity in Hodgkin lymphoma, DLBCL, NK/T-cell lymphoma, T-cell lymphoblastic lymphoma, angioimmunoblastic T-cell lymphoma, mantle cell lymphoma, and chronic lymphocytic leukemia (25–35), and high levels of bloodstream ctDNA have been associated with shorter progression-free survival (27, 29, 36). In addition, sensitivity to detecting ctDNA has been improved by identifying phased variants (PVs). PVs are present in several types of cancers, but are particularly frequent in B-cell malignancies due to aberrant somatic hypermutation (aSHM) in genes relevant to B-cell biology. The detection of somatic PVs in plasma samples has been shown to improve the detection sensitivity of MRD in B-cell lymphomas (37, 38). Although some cfDNA studies have included patients with FL (30, 32, 39–41), its usefulness in this indolent lymphoma has not yet been fully resolved.
This study shows that basal and dynamic analyses of cfDNA in patients with FL are of predictive and prognostic value. Using a targeted deep-sequencing approach, we assessed the predictive and prognostic value of ctDNA in a prospective, real-life series of patients with FL, at diagnosis and during treatment and/or follow-up. For this purpose, we evaluated the role of pretreatment ctDNA levels as a prognostic and predictive biomarker of POD24 and response to treatment, compared pretreatment ctDNA and tissue biopsy genotyping, and addressed the predictive value of the dynamic levels of ctDNA during treatment and follow-up. To our knowledge, this is the first analysis of both basal and dynamic changes in ctDNA levels that employ longitudinal cfDNA genotyping in a multicenter FL cohort.
Materials and Methods
Patients and sample collection
This is a prospective, observational, nonintervention study. Thirty-six patients diagnosed with FL were enrolled in 8 Spanish hospitals between April 2017 and November 2020. Three more patients who suffered histologic transformation to DLBCL before recruitment were included. We collected 110 plasma samples, formalin-fixed, paraffin-embedded tissue (FFPEt) from diagnostic biopsies of 31/39 patients to identify tumor somatic mutations, and germline genomic DNA from the oral mucosa of 36/39 patients (Supplementary Fig. S1; see Supplementary for detailed methods). According to clinical guidelines, responses were assessed by PET 4 ± 1 weeks after end-of-treatment (EOT; ref. 42). PET/CTs were reviewed locally by the corresponding nuclear medicine services. Additional cohort details are presented in Table 1, Supplementary Fig. S2, and the Supplementary information (Methods and Supplementary Table S1).
Table 1.
Summary of clinical features of the FL cohort (n = 36).
| Clinical features | Categories | N (%) |
|---|---|---|
| Age | ≥60 | 22 (61) |
| <60 | 14 (39) | |
| Sex | Female | 22 (61) |
| Male | 14 (39) | |
| POD24 | POD24-pos | 6 (17) |
| POD24-neg | 30 (83) | |
| Transformation | Transformation | 2 (6) |
| Non-transformation | 34 (94) | |
| Status | Alive with lymphoma | 12 (33) |
| Alive without lymphoma | 20 (56) | |
| Exitus | 4 (11) | |
| Watch-and-wait | No | 31 (86) |
| Yes | 5 (14) | |
| Grade | I | 9 (27) |
| II | 15 (46) | |
| IIIA | 9 (27) | |
| Bulky | No | 32 (89) |
| Yes | 4 (11) | |
| Stage | IA | 3 (8) |
| IIA | 2 (6) | |
| IIIA | 13 (36) | |
| IVA | 14 (39) | |
| IVB | 4 (11) | |
| B symptoms | No | 32 (89) |
| Yes | 4 (11) | |
| Extranodal involvement | No | 18 (50) |
| Yes | 18 (50) | |
| Affected areas | >1 | 12 (71) |
| 1 | 5 (29) | |
| Bone marrow involvement | No | 20 (57) |
| Yes | 15 (43) | |
| Hemoglobin | ≤12 | 6 (17) |
| >12 | 30 (83) | |
| LDH | Elevated | 9 (25) |
| Normal | 27 (75) | |
| ECOG | 0 | 20 (56) |
| 1 | 16 (44) | |
| β2-microglobulin | Elevated | 10 (30) |
| Normal | 23 (70) | |
| FLIPI | Low | 11 (31) |
| Intermediate | 15 (42) | |
| High | 10 (27) | |
| GELF criteria | Positive | 18 (50) |
| Negative | 18 (50) | |
| First line of treatment | R-Bendamustine | 21 (66) |
| R-CHOP | 6 (19) | |
| R-CVP | 2 (6) | |
| Rituximab | 3 (9) | |
| Response | Complete Response | 25 (74) |
| Partial Response | 8 (24) | |
| Non-response | 1 (2) | |
| Rituximab maintenance | Yes | 20 (56) |
| No | 16 (44) |
The study was approved by the Ethics Committee of the Hospital Universitario Puerta de Hierro-Majadahonda (PI-67/14), and was conducted following the principles of the Declaration of Helsinki. All participants gave their signed informed consent for inclusion. Samples were collected and clinical data were managed following standardized protocols to guarantee the quality of the samples and the confidentiality of donor data. After approval by the corresponding ethics committees, material and data from other centers were anonymously transferred to our laboratory in compliance with the current Spanish legislation (Ley 14/2007 de Investigación Biomédica and Real Decreto 1716/2011).
Targeted DNA sequencing
cfDNA and genomic DNA were extracted as described in Supplementary Data (Methods).
A targeted sequencing gene panel was designed that included coding exons, UTRs and splicing sites of 78 genes (Human Assembly GRCh38/hg38) that are recurrently mutated in B-cell lymphomas (FL and DLBCL), selected based on the findings of previous studies (24, 43). Coding and non-coding regions targeted by aSHM were also included (Supplementary Table S2). The bioinformatics pipeline is described in Supplementary Methods.
Plasma ctDNA concentrations were calculated in haploid genome equivalents per milliliter of plasma (hGE/mL), determined as the product of total cfDNA in pg/mL, and the mean variant allele frequency (VAF) divided by 3.3 pg per DNA molecule or hGE (44). The dynamics of ctDNA levels during treatment, measured as the log10 fold-change, were normalized to pretreatment levels.
Statistical analysis
Statistical analyses were performed using R 3.6.1 (https://www.R-project.org, R Foundation for Statistical Computing, Vienna, Austria). The differences between categories were evaluated with the Mann–Whitney U and Student t tests. ROC curve analysis was used to determine the optimal cut-off point for stratification.
Event-free survival (EFS) was calculated from diagnosis until relapse, progression or transformation, unplanned retreatment of lymphoma after initial management, or death due to any cause (45). OS was calculated from diagnosis until death. EFS and OS probabilities were estimated by the Kaplan–Meier method. Early lymphoma progression was defined as the progression/transformation of disease at 24 months from diagnosis (POD24; ref. 45).
Data availability
The data generated in this study are publicly available in the Sequence Read Archive database at PRJNA813747.
Results
Characteristics of the study cohort
The characteristics of the patient cohort and tumors are shown in Table 1 (Supplementary Fig. S1 and Supplementary Table S1). Thirty-six patients were diagnosed with FL at recruitment; 2 transformed during the follow-up. Samples from 3 additional patients with FL already transformed to DLBLC were also collected. The 36 patients with FL were treated with rituximab (R; 3 patients), R-CHOP (cyclophosphamide, hydroxydaunorubicin, vincristine, prednisolone; 6 patients), R-CVP (cyclophosphamide, vincristine, prednisolone; 2 patients), or R-bendamustine (21 patients), and radiotherapy (RT) only (1 patient; Table 1). Five patients did not require treatment at diagnosis, so a watch and wait (W&W) approach was adopted, but 2 were subsequently treated when disease symptoms appeared. Twenty-three patients had a complete response (CR) to first-line chemoimmunotherapy (CI) or RT, 8 patients had a partial response (PR), and 1 patient did not respond to treatment (NR). All the patients who did not achieve CR (PR and NR) were analyzed as a single group. According to the Groupe d'Etude des Lymphomes Folliculaires (GELF), half of the patients with FL (18/36) were positive for GELF criteria (GELF ≥ 1; Supplementary Table S1). All these patients received treatment, and ten (55.5%) achieved a CR. Twelve patients negative for GELF criteria (GELF = 0) were treated with CI or RT, and eleven (91.2%) achieved a CR. The mean follow-up time from diagnosis for alive patients was 43.6 months (8–137 months).
We collected 100 samples from the 36 patients with FL as follows: 31 samples at diagnosis or before treatment (basal or pretreatment samples), 25 samples at mid-treatment (one from additional lines of treatment), 22 samples at EOT (two of them after the second or third lines of treatment), 19 samples in follow-up visits (W&W patients, and revisions after treatment surveillance assessment every 6 months); three were collected at the time of progression or transformation.
Ten cfDNA samples were collected from the 3 patients who had suffered histologic transformation to DLBCL before recruitment (Supplementary Figures S1 and S2; Supplementary Table S3).
In total, 97/110 cfDNA samples (93%) had enough cfDNA (at least 10 ng of cfDNA corresponding to ∼1,000 genome equivalents) to be sequenced (ref. 32; Supplementary Figures S1 and S2; Supplementary Tables S3 and S4). The mean target coverage of the cfDNA samples was 1,969x (432–6, 466x) for those sequenced on the NovaSeq instrument. Due to the lower quality of some samples and to avoid compromising the sequencing quality of the others, six samples were sequenced on the MiSeq instrument. The mean coverage for these six samples was 254x (79–398x). The lower level of coverage may have influenced ctDNA detection in these samples, but we could still detect somatic mutations. Thirty-one tumor gDNA from paired diagnostic tissue samples and 36 germinal gDNA from paired oral mucosa were also sequenced. The mean target coverage was 270x (72–458x) for tumor gDNA, and 40x (4–206x) for germline-gDNA (Supplementary Table S5).
FL basal ctDNA and tissue biopsy genotyping
Twenty-eight of the 31 basal plasma samples from patients with FL had enough cfDNA to proceed with sequencing. We analyzed their mutation profile and at least one mutation was identified in 19/28 samples, considering synonymous and non-synonymous mutations in coding exons and splice site regions, with a mean of 6.71 mutations per cfDNA sample (range, 0–29). The most frequently mutated genes detected in more than 20% of the samples were KMT2D, BCL2, EZH2, IGLL5, CREBBP, and TNFSR14 (Fig. 1A; Supplementary Table S6).
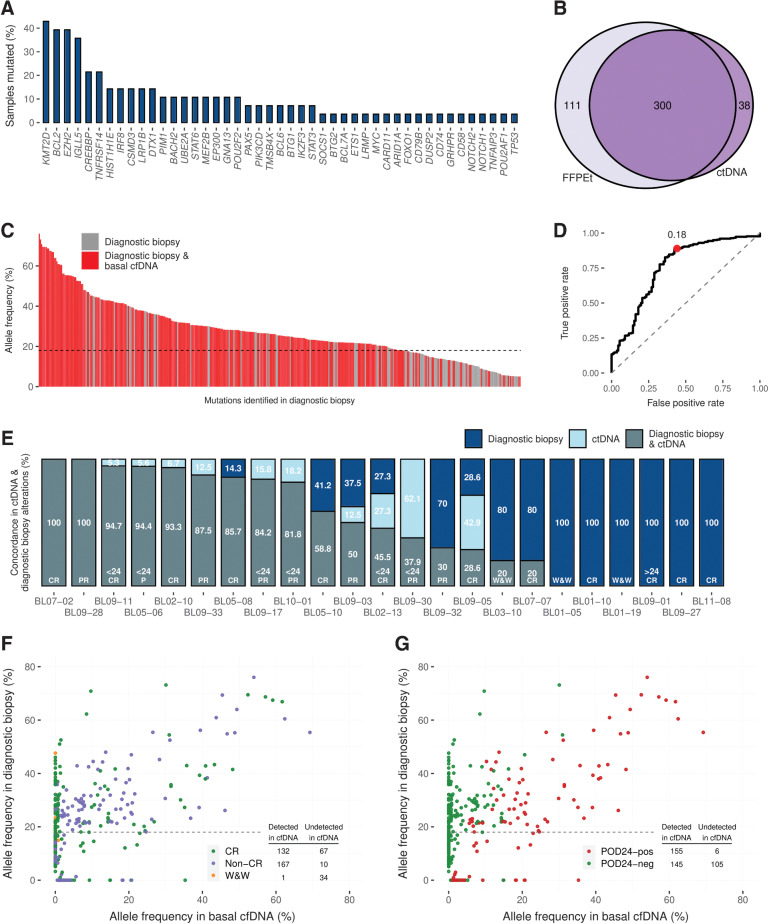
Figure 1.
Frequency of somatic mutations detected in basal plasma ctDNA and concordance between mutations detected in pretreatment plasma cfDNA and matched tumor gDNA genotyping (A) Percentage of samples with mutated genes. B, The Venn diagram shows the number of mutations detected in tumor biopsy gDNA and cfDNA. C, The bar graph shows the VAF of the mutations identified in tumor gDNA found in pretreatment cfDNA (red bars) or were missed (grey bars). The dashed line tracks the 18% VAF threshold calculated by the ROC analysis. D, ROC analysis shows the ability of pretreatment cfDNA genotyping to detect diagnostic biopsy tumor mutations according to the VAF. E, The bar graph shows the percentage of mutations by case. Mutations are coded by color (see figure box legend). The scatter plots represent the VAF in pretreatment cfDNA compared with that in diagnostic biopsy for each variant identified. Each variant is represented by the color code indicated in the figure: (F) according to response to treatment and (G) POD24. gDNA, genomic DNA.
We then evaluated paired basal cfDNA and tumor gDNA from 23 patients with FL with matched basal cfDNA and FFPEt diagnostic samples. At least one somatic mutation was detected in all tissue biopsies with a mean of 14.54 mutations per FFPEt sample (range, 5–34), whereas we only identified alterations in 17/23 basal cfDNA samples. Four hundred and eleven mutations were detected in tissue biopsies, 300 of which (73%) were also identified in cfDNA (Figs. 1B and C). In addition, 38 alterations were identified in cfDNA but not in tissue biopsies. All 38 mutations passed the quality filters (see Supplementary for detailed methods), and seven were identified in other follow-up cfDNA samples. The ROC curve analysis (Fig. 1D) showed that the sensitivity for detecting mutations in cfDNA increased for those with VAFs ≥ 18% in tumor gDNA. If we considered these alterations with VAF ≥ 18% in tumor biopsy, 84.84% were also detected in cfDNA. On the other hand, for those with a VAF of < 18%, only 36.63% were identified in cfDNA.
Of the 6/23 patients in which we did detect mutations in tumor gDNA but not in paired basal cfDNA, all were POD24-neg; 4 achieved CR after treatment, and 2 did not require treatment (W&W; Fig. 1E). The median read coverage for these samples was 1,253x. On the other hand, at least one genetic alteration was detected in every patient who did not achieve CR (8 patients) or who was POD24-pos (6 patients; Fig. 1E).
Several mutations (111/411, 27%) were detected in tumor gDNA but not in paired basal cfDNA. Most of them (represented by green points in Figs. 1F and G) corresponded to patients with a favorable outcome: CR or W&W (101/111, 91%) and POD24-neg (105/111, 94.6%).
The sensitivity to detecting alterations in basal cfDNA samples was higher in patients who did not achieve complete response (non-CR; 94.4%) and POD24-pos patients (96.3%) than in patients who achieved CR (66.3%) or who were POD24-neg (58%; Supplementary Figures S3A and S3B). If we considered only mutations with VAF ≥ 18%, the sensitivity increased to 99.4% in non-CR and 99.3% in POD24-pos patients.
The number of mutations identified in basal cfDNA samples was associated with response to treatment and POD24: the mean numbers of alterations per basal cfDNA sample in POD24-pos (18; range 11–29) and non-CR patients (14.2; 3–29) were significantly higher than in POD24-neg (4.7; 0–19) or CR patients (3.9; 0–17) (unpaired samples t test, P = 0.0001 and 0.001, respectively; Figs. 2A and B). However, no significant differences were found when we examined the correlation with the number of mutations detected in diagnostic tissue biopsies with the same endpoints (Supplementary Figures S4A and S4B).
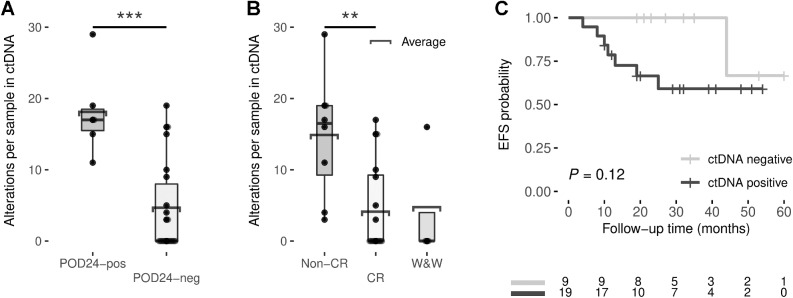
Figure 2.
Box plots showing the number of alterations per basal cfDNA sample by POD24 (A) or response to treatment (B). C, Kaplan–Meier estimates of EFS based on risk groups determined by pretreatment ctDNA detection. EFS differences between risk groups were tested using the log-rank test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Recent studies have used the analysis of PVs to increase the sensitivity of ctDNA detection (37). Therefore, as previously proposed, we examined the value of analyzing PVs to improve ctDNA detection sensitivity (37). The sequencing panel used in this study included known target regions of aSHM (Supplementary Table S2), allowing us to enrich PV detection in these samples. The aSHM regions of IGLL5 and BCL2 and, to a lesser extent MYC and BCL7A, have a high proportion of PVs (Supplementary Table S7 and Supplementary Figures S5A). A total of 138 PVs were identified in diagnostic tissue biopsy samples (Supplementary Figures S5B). ctDNAtools software, described by Meriranta and colleagues (38), was used to look for the 138 PVs, previously identified in tumoral gDNA, in the cfDNA samples. Twenty-one of 23 patients with FL with matched basal cfDNA and FFPEt diagnostic samples had at least one PVs in the FFPEt diagnostic sample, and 11 of 21 patients had at least one PVs detected in the basal cfDNA sample. The PVs analyses of basal cfDNA samples from the five cases without paired FFPEt identified ctDNA in the BL09–19 patient, previously reported as ctDNA-positive due to detection of other somatic alterations. The analysis allowed us ctDNA detection in samples reported as ctDNA-negative when considering only somatic alterations, such as BL11–08–02, BL11–08–03, BL11–08–04, and BL12–13–02 (Supplementary Fig. S2).
ctDNA detection combining somatic mutations and PVs (n = 28) showed that patients with at least one abnormality identified in basal cfDNA presented a shorter EFS trend than patients without abnormalities (66.5 vs. 100%, log-rank test, P = 0.12, by the Kaplan–Meier analysis; Fig. 2C). No significant difference was found for OS (Supplementary Fig. S6A).
We then calculated ctDNA levels in hGE/mL, as described in the Experimental Design section, and associated the pretreatment ctDNA with FL patients' clinical outcomes. No alterations were identified in 9/28 basal cfDNA samples. The mean coverage of these nine samples was 1,300x (356–2, 549x), whereas that of the whole basal cohort was 1,900x (180–6, 466x). The mean level of ctDNA, measured as hGE/mL, was 744, based on all cases.
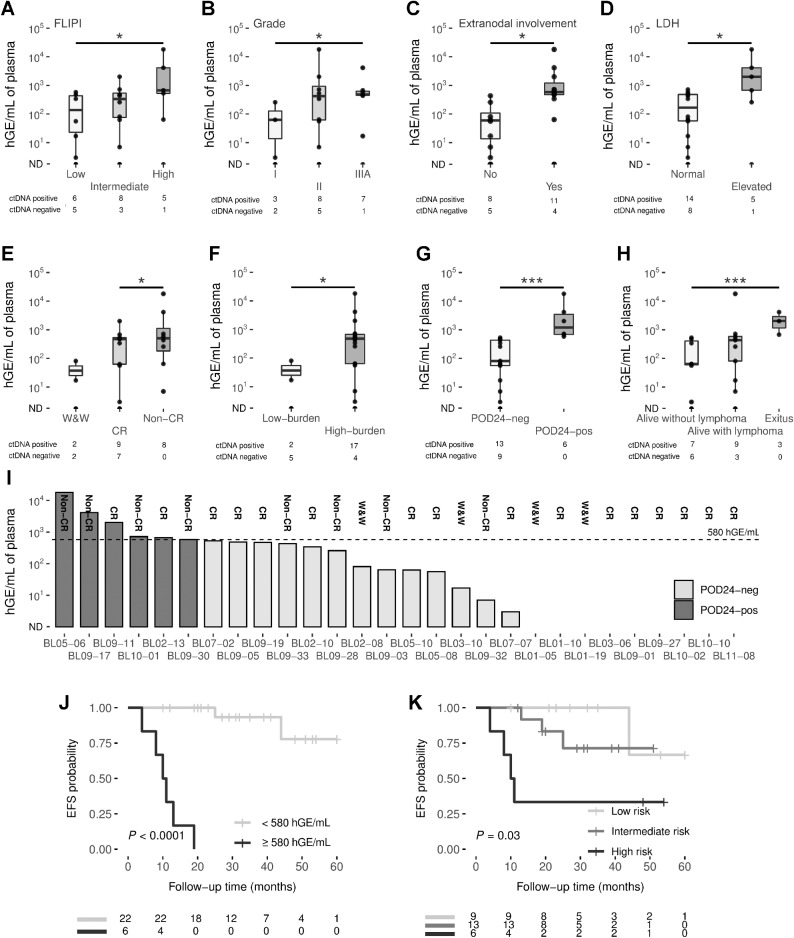
Basal ctDNA levels correlated with clinical characteristics at the time of diagnosis. Higher ctDNA levels were significantly associated with high-risk FLIPI patients (P = 0.02), higher tumor grade (P = 0.04), extranodal involvement (P = 0.02) and elevated levels of LDH (P = 0.03; Figs. 3A–C and D). No significant differences were observed between stage categories, patient Eastern Cooperative Oncology Group (ECOG), β2-microglobulin levels and GELF criteria (Supplementary Figures S7A–S7D).
Figure 3.
A, Box plots showing significant correlations of pretreatment ctDNA levels with FLIPI (A); grade (B); extranodal involvement (C); LDH (D); response to treatment (E); tumor burden (F); progression of disease within 24 months (G); and patient status (H). Group differences were assessed with the Mann–Whitney U test. The box plot represents the quartiles and the median for the hGE/mL values. I, The bar graph shows the levels of ctDNA in pretreatment cases, measured by hGE/mL. POD24 status is coded by color (see figure box legend). J, Kaplan–Meier estimates of EFS based on risk groups determined by hGE/mL levels, and (K) the combination of FLIPI and ctDNA levels. EFS differences between risk groups were tested using the log-rank test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. LDH, lactate dehydrogenase; ND, non-detected.
Basal ctDNA levels also correlated with response to therapy, being significantly higher in non-CR patients than in those who achieved CR (Mann–Whitney U, P = 0.02; Fig. 3E). The low-burden patients that were not treated (W&W) or treated with rituximab in monotherapy, had lower basal ctDNA levels than patients requiring treatment (high-burden patients; P = 0.01; Fig. 3F).
We also estimated the correlation between basal ctDNA levels with early lymphoma progression/transformation or POD24. Patients with POD24 (POD24-pos) had significantly higher levels of ctDNA than those that did not progress (POD24) (P < 0.001; Fig. 3G); deceased patients had higher basal ctDNA levels than those who remained alive without lymphoma (P < 0.001; Fig. 3H). This significant difference was conserved when we analyzed patients separately according to the GELF criteria (11 GELF-neg and 17 GELF-pos): pretreatment ctDNA levels were higher in POD24-pos compared with POD24-neg patients in both GELF groups (Mann–Whitney U, P = 0.04 and P = 0.004 for patients negative and positive for GELF criteria, respectively; Figures S7E and S7F).
We established the best threshold for stratifying patients into two POD24 risk groups (Fig. 3I) by ROC curve analysis based on basal ctDNA levels. Using a threshold of 580 hGE/mL, patients with higher levels had significantly inferior rates of EFS at 24 months than those with low levels (log-rank test, for EFS, P < 0.0001 and OS, P = 0.029, by the Kaplan–Meier analysis; Fig. 3J; Supplementary Fig. S6B).
FLIPI is the most widely used clinical tool for stratifying patients with FL and predicting their clinical outcomes (10). In this cohort, although patients with high-risk FLIPI had a lower EFS and OS probability than those with low-risk FLIPI, the relationship was not significant (log-rank test, for EFS, P = 0.078 and OS, P = 0.19; Supplementary Fig. S8A and S8D). Several attempts have been made to improve the tool's predictive and prognostic power by combining clinical and genetic data, such as m7-FLIPI and the POD24 Prognostic Index (POD24-PI; refs. 11, 12, 19). However, when we tested them in our series, neither identified significant differences in EFS and OS probability by Kaplan–Meier analysis (Supplementary Figures S8B, S8C, S8E, and S8F). We did not find any significant association of stage, ECOG or grade with progression by a multivariate Cox analysis, probably due to the small number of patients (Supplementary Table S8).
Therefore, we examined whether incorporating ctDNA levels could improve FLIPI predictive capacity. When we combined the two, the Kaplan–Meier analysis showed a significant association for EFS (log-rank test for EFS, P = 0.03) but not for OS (Fig. 3K; Supplementary Fig. S6C). Furthermore, multivariate Cox analysis showed ctDNA levels to have an independent predictive capacity to predict progression compared with FLIPI, m7-FLIPI, POD24-PI, and GELF criteria (Supplementary Table S8).
Dynamic analysis of ctDNA
We next tested the effect of the treatment on ctDNA dynamics and its predictive value. The plasma ctDNA was monitored during the patients' treatment and follow-up.
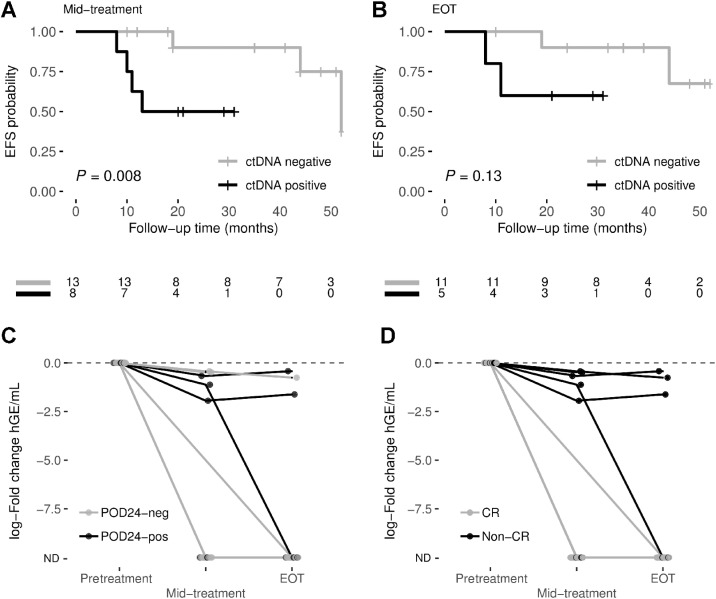
First, we analyzed the alterations in paired cfDNA and FFPEt. At mid-treatment, the sensitivity to detecting at least one mutation in cfDNA was higher in patients who did not achieve CR (45.5%) than in those who did achieve it (7.0%; Supplementary Fig. S9A). The sensitivity increased from 45.5 to 52.3% in non-CR patients when only the mutations with VAF ≥ 18% in the tumor gDNA were considered.
At EOT, the sensitivity to detecting alterations in non-CR patients was 83.1%, and 87.0% for alterations with VAF ≥ 18% in tumor gDNA (Supplementary Fig. S9B). EOT cfDNA samples without tumor alterations showed a mean target coverage per position above 2,000x (mean: 2,079 range: 398–3,743x).
Despite the somewhat low level of sensitivity, we were still able to observe inferior EFS in those patients with at least one mutation in mid-treatment cfDNA (including SNVs and PVs; 50% at 24 months) than in patients without mutations (90%; log-rank test, P = 0.008; Fig. 4A). No significant differences were observed for OS (Supplementary Fig. S10A). Of the cfDNA samples at mid-treatment (13) in which we did not detect alterations, the mean target coverage per position was 1,500x (range: 551–3,238x).
Figure 4.
Prognostic value of dynamic analysis of ctDNA. Kaplan–Meier estimates of EFS probabilities are shown for ctDNA-positive (at least one mutation detected in cfDNA) and ctDNA-negative patients at (A) mid-treatment, and (B) end-of-treatment. EFS differences between groups were tested using the log-rank test. The dynamic of ctDNA levels, measured as log10 fold-change, grouped by their (C) POD24 status and (D) response to treatment, is represented for each patient. ND, non-detected.
Patients with at least one identified ctDNA mutation at EOT showed a worse EFS trend than patients without (60 vs. 90% at 24 months, respectively; Fig. 4B). No significant differences were observed for OS (Supplementary Fig. S10B). Three patients showed different responses when comparing PET and ctDNA analysis response information at EOT. Patient BL09–03, with a partial response (non-CR), was ctDNA-negative; however, BL03–07 and BL11–08 patients had CR by PET but were ctDNA-positive. Despite the differences, the EFS trend by PET response at EOT (50 vs. 90.9% at 24 months; Supplementary Fig. S10C) was similar to that found for ctDNA detection (Fig. 4B) by the Kaplan–Meier method, although neither reached significance possibly due to the small number of samples analyzed at EOT.
We then evaluated the changes in ctDNA levels. ctDNA levels decreased dramatically after treatment for every patient, although the reduction was more significant in patients with favorable outcomes. In POD24-neg patients, the reduction in ctDNA levels was more marked at both mid-treatment (86.6% of POD24-neg cases showed non-detected ctDNA vs. 40% of POD24-pos cases) and EOT (92.3% of POD24-neg cases showed non-detected ctDNA vs. 60% of POD24-pos cases; Fig. 4C). Similarly, when we analyzed the response to treatment, a significant decline in ctDNA levels was observed at mid-treatment and EOT in patients with CR compared with those who did not achieve CR (Fisher exact test P = 0.002 at mid-treatment and P = 0.01 at EOT; Fig. 4D). Moreover, comparing the absolute ctDNA levels (hGE/mL) showed lower values for POD24-neg and CR patients at mid-treatment and EOT (Supplementary Fig. S11).
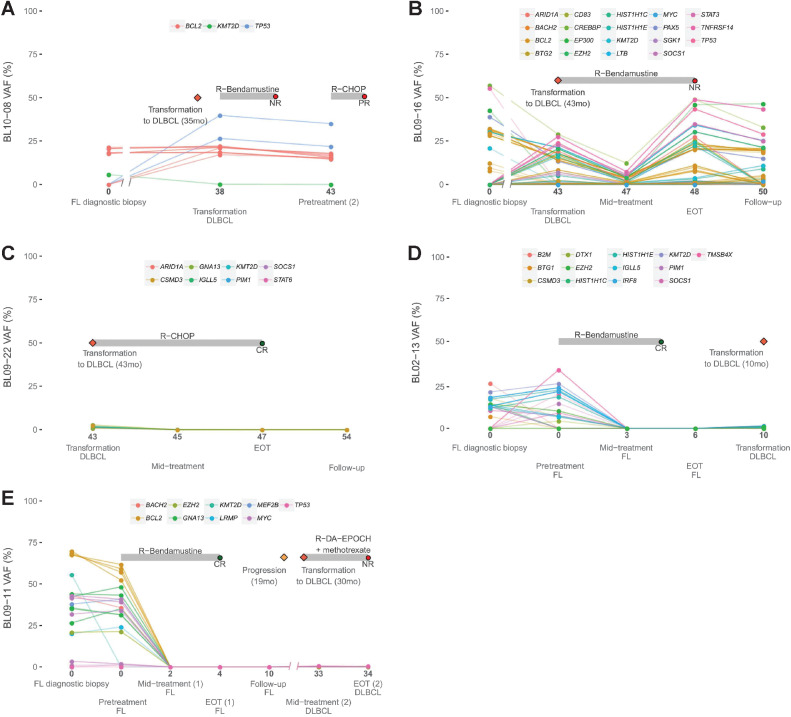
While recruiting patients for the study, we collected samples from five patients with histologic transformation to DLBLC. Three of them had transformed before recruitment (BL10–08, BL09–16, and BL09–22; (Figs. 5A–C), and the other two (BL02–13 and BL09–11) transformed during the study (Figures D and E). We compared the cfDNA samples after transformation with the paired pre-transformed FL biopsy and observed the appearance of novel genetic mutations of TP53 in BL10–08 and BL09–11 (Figs. 5A and E). In BL09–16, we identified mutations in TNFRSF14, TP53, SOCS1, HIST1H1C, HIST1H1E, STAT3, MYC, SGK1, EZH2, and BTG2 genes in the cfDNA after transformation (Fig. 5B), many of which had previously been found to be associated with transformation (20–24). However, mutations in KMT2D, EP300, and TNFRSF14, present in the FL diagnostic biopsy, were not detected in the paired cfDNA DLCBL sample. Something similar occurred in BL02–13, in which a novel alteration in EZH2 was detected in the DLBCL sample that was not present in the FL diagnostic biopsy. However, other alterations in genes such as TNFRSF14, SOCS1, B2M, BTG1, CSMD3, PIM1, or DTX1, present in the FL diagnostic biopsy, disappeared in the DLBCL cfDNA sample (Fig. 5D). No alterations were detected in the mid-treatment, EOT and follow-up cfDNA samples from the transformed DLCBL BL09–22 patient, consistent with the CR to first-line R-CHOP therapy (Fig. 5C).
Figure 5.
A–E, Longitudinal monitoring of cfDNA genotyping during disease evolution for patients with FL that suffered transformation to DLBLC. Time in months of cfDNA sample collection after diagnosis is indicated. NR, non-response; P, progression.
Discussion
In this study, we assessed the utility of cfDNA profiling by targeted deep-sequencing for FL prognosticating. ctDNA is already known to be of clinical utility in other malignancies, including lymphomas, and its presence has been correlated with clinical characteristics and can be used to track MRD (25, 26, 33, 36, 46, 47). On the other hand, several studies confirm the increased mortality risk in patients with POD24 (4, 5, 19), but their assessment at diagnosis is still inaccurate, and it is not currently possible in clinical practice to identify patients with FL with a higher risk of early relapse before it happens (5, 48). In this real-life, observational, multicenter, prospective-based population study, we show the utility of pretreatment and dynamic ctDNA analysis for predicting outcomes and monitoring patients with FL. This pilot study shows that ctDNA levels might be a prognostic biomarker of early progression at diagnosis or before treatment. We also show that cfDNA monitoring and genotyping during treatment and follow-up predict response to treatment and early progression or relapse.
Pretreatment ctDNA levels are correlated with some clinical characteristics such as grade, FLIPI, extranodal involvement and LDH, suggesting that it might be a putative biomarker of tumor burden in FL. We have also explored the significance of measuring ctDNA levels to predict patient response to treatment, and we found that the detection of ctDNA in plasma correlated with non-CR and early progression.
Identifying patients at higher risk of POD24 at diagnosis is a clinically relevant problem. Our results show that pretreatment ctDNA levels are significantly higher in POD24-pos patients. In addition, although we have not found differences in basal ctDNA levels in patients according to GELF criteria, the fact is that two thirds of the patients negative for GELF criteria were treated with immunochemotherapy. However, if we analyzed basal ctDNA levels in treated patients versus those that did not receive treatment or were treated only with rituximab, we did find significantly higher ctDNA levels at diagnosis in treated patients. Finally, diagnostic ctDNA levels were higher in POD-pos patients in both GELF-pos (P = 0.004) and GELF-neg (P = 0.04) groups. Furthermore, our data demonstrate that pretreatment ctDNA level might be a more robust predictor of POD24 than m7-FLIPI, POD24-PI or GELF criteria (11, 19). In this context, the non-detection of pretreatment ctDNA may be used to identify patients who would achieve CR and/or not suffer POD24. These results suggest that ctDNA analysis, combined with patients' clinical features, could improve their risk stratification and be used as a prognostic biomarker at diagnosis. More studies and further validation in larger cohorts are needed to develop the standardization required for its use in monitoring FL.
Comparative analysis of diagnostic tissue biopsy with pretreatment cfDNA showed that the sensitivity to detecting tumor alterations in cfDNA, which were also identified in the tumor gDNA, was lower (73%) than the values found in other studies of more aggressive B-cell lymphomas (25, 26, 32, 49). This apparently lower sensitivity is due to the absence of ctDNA (measured as hGE/mL) in plasma samples from patients with good-prognosis FL (W&W, CR, and POD24-neg patients). There were fewer alterations per basal cfDNA sample in these patients than in those with worse clinical outcomes, but the read sequencing coverage data in cfDNA from these patients were enough (median, 1,253x) not to preclude the detection of alterations. FL is considered an indolent lymphoma, and plasma samples from patients with FL are expected to have around one tenth of the ctDNA calculated in patients with DLBLC (44, 50). This lesser amount of ctDNA reflects the smaller tumor volume, clinical stage and more indolent characteristics of this type of lymphoma. Sensitivity would be improved by using larger plasma volumes and more sensitive methods, such as PV detection. However, the sensitivity of our method for detecting tumor alterations in pretreatment cfDNA from patients who did not achieve CR or POD24-pos was higher (94.4 and 96.3%, respectively) and similar to that found in other studies (25, 26, 49). These results suggest that detecting somatic mutations in pretreatment cfDNA is favored by lower response rates to first-line CI, a higher risk of early progression, and greater FL aggressiveness. From a different perspective, the non-detection of ctDNA in pretreatment plasma samples could be a biomarker of good prognosis.
We also observed that the sensitivity for detecting mutations in cfDNA decreases considerably for mutations with VAF < 18% in the diagnostic tissue biopsy. For this reason, the approach could be compromised in those cases with alterations below the detection limit, as other studies have shown (26). Longitudinal genotyping of cfDNA samples showed that a large proportion of the mutations identified in the diagnostic tissue biopsy and/or basal cfDNA disappear by mid-treatment. In unfavorable cases, the number of mutations detected increased by the EOT. These findings were correlated with the dynamic ctDNA analysis, in which we found a decrease in ctDNA levels at mid-treatment and EOT in all patients, with higher ctDNA levels found in non-CR or POD24-pos patients. The drop in ctDNA at mid-treatment and EOT was also associated with their clinical outcomes, highlighting the value of ctDNA as a putative predictive biomarker in FL.
PVs have been shown to improve the detection limit of ctDNA in B-cell lymphomas (37). In our cohort, PVs detection allowed the presence of ctDNA to be detected in samples that were otherwise reported as being ctDNA-negative, although our results may have been limited by the absence of the immunoglobulin genes regions in the sequencing panel (37). However, the detection of somatic PVs in cfDNA provides us with a promising tool for improving ctDNA detection sensitivity that would be especially useful in relation to indolent lymphomas, such as FL, that contain smaller amounts of ctDNA than do more aggressive lymphomas.
In conclusion, our data suggest that the use of ctDNA could be useful to identify patients at high risk of POD24, or who would not achieve CR. This study illustrates the utility of ctDNA analysis (quantifying pretreatment and follow-up levels and longitudinal genotyping) in FL. More studies in the context of clinical trials and further validation in larger cohorts are needed to develop the standardization required for its use in monitoring FL. However, the results of this pilot study, based on a multicenter, prospective cohort, support the notion that basal and dynamic ctDNA analysis in FL, in combination with clinical characteristics, would improve patient stratification and help predict the response to CI and the risk of early progression of the disease.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the Hospital Universitario Puerta de Hierro-Majadahonda (PI-67/14), and was conducted following the principles of the Declaration of Helsinki. All participants gave their signed informed consent for inclusion.
Supplementary Material
Supplementary Data 1
Supplementary Data 2
Acknowledgments
We are indebted to the patients who contributed to this study, and to the members of GOTEL (Grupo Oncológico para el Tratamiento y el Estudio de los Linfomas). We are especially thankful to E. Ramil of the Sequencing Unit of the Instituto de Investigación Sanitaria Puerta de Hierro-Segovia de Arana (IDIPHISA). We acknowledge the Biobank of the Hospital Universitario Puerta de Hierro-Majadahonda, Biobank of Sistema Sanitario Público de Andalucía, and the Pathology Departments of the centers collaborating in the study.
Supported by: Fundación GILEAD (GLD18/00019), Spanish Ministry of Economy and Competence (MINECO) and Instituto de Salud Carlos III (ISCIII), ISCIII-MINECO AES-FEDER (PI14/00221, DTS17/00039, PI17/00272, PI20/00591, CIBERONC CB16/12/00291) and by Dirección General de Universidades e Investigación de la Consejería de Educación e Investigación de la Comunidad de Madrid (CAM) (B2017/BMD-3778). Ismael Fernández-Miranda is supported by B2017/BMD-3778 and Fundación de Investigación Biomédica HU Puerta de Hierro-Majadahonda. Lucía Pedrosa is recipient of iPFIS predoctoral fellowship (IFI18/0004, by ISCIII-MINECO AES-FEDER, Plan Estatal I+D+I 2017–2020). Natalia Yanguas-Casás is supported by the Asociación Española Contra el Cáncer. Marta Navarro and Beatriz Horcajo are supported by Plan de empleo juvenil de la CAM (PEJ-2020-AI/BMD-19527 and PEJ-2020-TL/BMD-19530, respectively). Margarita Sánchez-Beato was a beneficiary of a Miguel Servet II contract (CPII16/00024), supported by ISCIII-MINECO AES-FEDER (Plan Estatal I+D+I 2013–2016) and Fundación de Investigación Biomédica Puerta de Hierro.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
L. Pedrosa reports grants from Instituto de Salud Carlos III during the conduct of the study. J. Gumá reports grants from Amgen, Pfizer, Ipsen, Clovis Oncology, Roche, Janssen, MSD, Merck, BMS, EUSA Pharma, Lilly, Gilead, Astellas, and Bayer outside the submitted work. V. Calvo reports personal fees from Roche, BMS, MSD, AstraZeneca, Takeda, Pfizer, LILLY, and Boehringer Ingelheim outside the submitted work. A. Rueda-Domínguez reports other support from BMS/Celgene, Roche, MSD, Takeda, and Merck outside the submitted work. M. Provencio reports grants and personal fees from BMS, Takeda, and Janssen outside the submitted work; and grants, personal fees, and nonfinancial support from MSD, Roche, and AstraZeneca. M. Sánchez-Beato reports grants from Fundación Gilead, Spain, ISCIII-MINECO AES- FEDER, Dirección General de Universidades e Investigación de la Consejería de Educación e Investigación de la Comunidad de Madrid; and personal fees from ISCI II during the conduct of the study. No disclosures were reported by the other authors.
Disclaimer
The funders had no role in the study design, data collection and analysis, the decision to publish, or the preparation of the manuscript.
Authors' Contributions
I. Fernández-Miranda: Data curation, formal analysis, supervision, methodology, writing–original draft, writing–review and editing. L. Pedrosa: Methodology, writing–review and editing. M. Llanos: Resources, writing–review and editing. F.F. Franco: Resources, writing–review and editing. S. Gómez: Methodology, writing–review and editing. P. Martín-Acosta: Resources, writing–review and editing. F.R. García-Arroyo: Resources, writing–review and editing. J. Gumá: Resources, writing–review and editing. B. Horcajo: Methodology, writing–review and editing. A.K. Ballesteros: Methodology, writing–review and editing. L. Gálvez: Resources, writing–review and editing. N. Martínez: Resources, writing–review and editing. M. Marín: Resources, writing–review and editing. S. Sequero: Resources, writing–review and editing. M. Navarro: Methodology, writing–review and editing. N. Yanguas-Casás: Methodology, writing–review and editing. V. Calvo: Resources, writing–review and editing. A. Rueda-Domínguez: Resources, writing–review and editing. M. Provencio: Conceptualization, resources, formal analysis, writing–review and editing. M. Sánchez-Beato: Conceptualization, data curation, formal analysis, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Sant M, Allemani C, Tereanu C, De Angelis R, Capocaccia R, Visser O, et al. Incidence of hematologic malignancies in Europe by morphologic subtype: results of the HAEMACARE project. Blood 2010;116:3724–34. [DOI] [PubMed] [Google Scholar]
- 2. Kanas G, Ge W, Quek RGW, Keeven K, Nersesyan K, Arnason JE. Epidemiology of diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma (FL) in the United States and Western Europe: population-level projections for 2020–2025. Leuk Lymphoma. 2022;63:54–63. [DOI] [PubMed] [Google Scholar]
- 3. Carbone A, Roulland S, Gloghini A, Younes A, von Keudell G, López-Guillermo A, et al. Follicular lymphoma. Nat Rev Dis Primers 2019;5:83. [DOI] [PubMed] [Google Scholar]
- 4. Casulo C, Byrtek M, Dawson KL, Zhou X, Farber CM, Flowers CR, et al. Early relapse of follicular lymphoma after rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone defines patients at high risk for death: an analysis from the national LymphoCare study. J Clin Oncol 2015;33:2516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Casulo C, Dixon JG, Le-Rademacher J, Hoster E, Hochster HS, Hiddemann W, et al. Validation of POD24 as a robust early clinical end point of poor survival in FL from 5225 patients on 13 clinical trials. Blood 2022;139:1684–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wagner-Johnston ND, Link BK, Byrtek M, Dawson KL, Hainsworth J, Flowers CR, et al. Outcomes of transformed follicular lymphoma in the modern era: a report from the National LymphoCare Study (NLCS). Blood 2015;126:851–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Federico M, Barrigón MDC, Marcheselli L, Tarantino V, Manni M, Sarkozy C, et al. Rituximab and the risk of transformation of follicular lymphoma: a retrospective pooled analysis. Lancet Haematol 2018;5:e359–67. [DOI] [PubMed] [Google Scholar]
- 8. Sarkozy C, Trneny M, Xerri L, Wickham N, Feugier P, Leppa S, et al. Risk factors and outcomes for patients with follicular lymphoma who had histologic transformation after response to first-line immunochemotherapy in the PRIMA trial. J Clin Oncol 2016;34:2575–82. [DOI] [PubMed] [Google Scholar]
- 9. Cottereau AS, Versari A, Luminari S, Dupuis J, Chartier L, Casasnovas R-O, et al. Prognostic model for high-tumor-burden follicular lymphoma integrating baseline and end-induction PET: a LYSA/FIL study. Blood 2018;131:2449–53. [DOI] [PubMed] [Google Scholar]
- 10. Buske C, Hoster E, Dreyling M, Hasford J, Unterhalt M, Hiddemann W, et al. The follicular lymphoma international prognostic index (FLIPI) separates high-risk from intermediate- or low-risk patients with advanced-stage follicular lymphoma treated front-line with rituximab and the combination of cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) with respect to treatment outcome. Blood 2006;108:1504–8. [DOI] [PubMed] [Google Scholar]
- 11. Pastore A, Jurinovic V, Kridel R, Hoster E, Staiger AM, Szczepanowski M, et al. Integration of gene mutations in risk prognostication for patients receiving first-line immunochemotherapy for follicular lymphoma: a retrospective analysis of a prospective clinical trial and validation in a population-based registry. Lancet Oncol 2015;16:1111–22. [DOI] [PubMed] [Google Scholar]
- 12. Federico M, Bellei M, Marcheselli L, Luminari S, Lopez-Guillermo A, Vitolo U, et al. Follicular lymphoma international prognostic index 2: a new prognostic index for follicular lymphoma developed by the international follicular lymphoma prognostic factor project. J Clin Oncol 2009;27:4555–62. [DOI] [PubMed] [Google Scholar]
- 13. Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 2011;476:298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 2011;471:189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 2010;42:181–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Okosun J, Bödör C, Wang J, Araf S, Yang C-Y, Pan C, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet 2014;46:176–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Green MR. Chromatin modifying gene mutations in follicular lymphoma. Blood 2018;131:595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Devan J, Janikova A, Mraz M. New concepts in follicular lymphoma biology: from BCL2 to epigenetic regulators and noncoding RNAs. Semin Oncol 2018;45:291–302. [DOI] [PubMed] [Google Scholar]
- 19. Jurinovic V, Kridel R, Staiger AM, Szczepanowski M, Horn H, Dreyling MH, et al. Clinicogenetic risk models predict early progression of follicular lymphoma after first-line immunochemotherapy. Blood 2016;128:1112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pasqualucci L, Khiabanian H, Fangazio M, Vasishtha M, Messina M, Holmes AB, et al. Genetics of follicular lymphoma transformation. Cell Rep 2014;6:130–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Davies AJ, Lee AM, Taylor C, Clear AJ, Goff LK, Iqbal S, et al. A limited role for TP53 mutation in the transformation of follicular lymphoma to diffuse large B-cell lymphoma. Leukemia 2005;19:1459–65. [DOI] [PubMed] [Google Scholar]
- 22. Kridel R, Chan FC, Mottok A, Boyle M, Farinha P, Tan K, et al. Histological transformation and progression in follicular lymphoma: a clonal evolution study. PLoS Med 2016;13:e1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Correia C, Schneider PA, Dai H, Dogan A, Maurer MJ, Church AK, et al. BCL2 mutations are associated with increased risk of transformation and shortened survival in follicular lymphoma. Blood 2015;125:658–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. González-Rincón J, Méndez M, Gómez S, García JF, Martín P, Bellas C, et al. Unraveling transformation of follicular lymphoma to diffuse large B-cell lymphoma. PLoS One 2019;14:e0212813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Scherer F, Kurtz DM, Newman AM, Stehr H, Craig AFM, Esfahani MS, et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci Transl Med 2016;8:364ra155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rossi D, Diop F, Spaccarotella E, Monti S, Zanni M, Rasi S, et al. Diffuse large B-cell lymphoma genotyping on the liquid biopsy. Blood 2017;129:1947–57. [DOI] [PubMed] [Google Scholar]
- 27. Hur JY, Kim YJ, Yoon SE, Son D-S, Park W-Y, Kim SJ, et al. Plasma cell-free DNA is a prognostic biomarker for survival in patients with aggressive non-Hodgkin lymphomas. Ann Hematol 2020;99:1293–302. [DOI] [PubMed] [Google Scholar]
- 28. Sun P, Chen C, Xia Y, Wang Y, Liu P-P, Bi X-W, et al. Mutation profiling of malignant lymphoma by next-generation sequencing of circulating cell-free DNA. J Cancer 2019;10:323–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hohaus S, Giachelia M, Massini G, Mansueto G, Vannata B, Bozzoli V, et al. Cell-free circulating DNA in Hodgkin's and non-Hodgkin's lymphomas. Ann Oncol 2009;20:1408–13. [DOI] [PubMed] [Google Scholar]
- 30. Wu J, Tang W, Huang L, Hou N, Wu J, Cheng X, et al. The analysis of cell-free DNA concentrations and integrity in serum of initial and treated of lymphoma patients. Clin Biochem 2019;63:59–65. [DOI] [PubMed] [Google Scholar]
- 31. Bohers E, Viailly P-J, Becker S, Marchand V, Ruminy P, Maingonnat C, et al. Noninvasive monitoring of diffuse large B-cell lymphoma by cell-free DNA high-throughput targeted sequencing: analysis of a prospective cohort. Blood Cancer J 2018;8:74.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bruscaggin A, di Bergamo LT, Spina V, Hodkinson B, Forestieri G, Bonfiglio F, et al. Circulating tumor DNA for comprehensive noninvasive monitoring of lymphoma treated with ibrutinib plus nivolumab. Blood Adv 2021;5:4674–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Spina V, Bruscaggin A, Cuccaro A, Martini M, Di Trani M, Forestieri G, et al. Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood 2018;131:2413–25. [DOI] [PubMed] [Google Scholar]
- 34. Yeh P, Hunter T, Sinha D, Ftouni S, Wallach E, Jiang D, et al. Circulating tumor DNA reflects treatment response and clonal evolution in chronic lymphocytic leukemia. Nat Commun 2017;8:14756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kurtz DM, Green MR, Bratman SV, Scherer F, Liu CL, Kunder CA, et al. Noninvasive monitoring of diffuse large B-cell lymphoma by immunoglobulin high-throughput sequencing. Blood 2015;125:3679–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roschewski M, Dunleavy K, Pittaluga S, Moorhead M, Pepin F, Kong K, et al. Circulating tumor DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: a correlative biomarker study. Lancet Oncol 2015;16:541–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kurtz DM, Soo J, Co Ting Keh L, Alig S, Chabon JJ, Sworder BJ, et al. Enhanced detection of minimal residual disease by targeted sequencing of phased variants in circulating tumor DNA. Nat Biotechnol 2021;39:1537–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Meriranta L, Alkodsi A, Pasanen A, Lepistö M, Mapar P, Blaker YN, et al. Molecular features encoded in the ctDNA reveal heterogeneity and predict outcome in high-risk aggressive B-cell lymphoma. Blood 2022;139:1863–77. [DOI] [PubMed] [Google Scholar]
- 39. Delfau-Larue M-H, van der Gucht A, Dupuis J, Jais J-P, Nel I, Beldi-Ferchiou A, et al. Total metabolic tumor volume, circulating tumor cells, cell-free DNA: distinct prognostic value in follicular lymphoma. Blood Adv 2018;2:807–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hatipoğlu T, Esmeray Sönmez E, Hu X, Yuan H, Danyeli AE, Şeyhanlı A, et al. Plasma concentrations and cancer-associated mutations in cell-free circulating DNA of treatment-naive follicular lymphoma for improved noninvasive diagnosis and prognosis. Front Oncol 2022;12:870487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sarkozy C, Huet S, Carlton VEH, Fabiani B, Delmer A, Jardin F, et al. The prognostic value of clonal heterogeneity and quantitative assessment of plasma circulating clonal IG-VDJ sequences at diagnosis in patients with follicular lymphoma. Oncotarget 2017;8:8765–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cheson BD, Fisher RI, Barrington SF, Cavalli F, Schwartz LH, Zucca E, et al. Recommendations for Initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: the Lugano classification. J Clin Oncol 2014;32:3059–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pedrosa L, Fernández-Miranda I, Pérez-Callejo D, Quero C, Rodríguez M, Martín-Acosta P, et al. Proposal and validation of a method to classify genetic subtypes of diffuse large B- cell lymphoma. Sci Rep 2021;11:1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bourbon E, Alcazer V, Cheli E, Huet S, Sujobert P. How to obtain a high quality ctDNA in lymphoma patients: preanalytical tips and tricks. Pharmaceuticals (Basel) 2021;14:617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Provencio M, Royuela A, Torrente M, Pollán M, Gómez-Codina J, Sabín P, et al. Prognostic value of event-free survival at 12 and 24 months and long-term mortality for non-Hodgkin follicular lymphoma patients: a study report from the Spanish lymphoma oncology group. Cancer 2017;123:3709–16. [DOI] [PubMed] [Google Scholar]
- 46. Frank MJ, Hossain NM, Bukhari A, Dean E, Spiegel JY, Claire GK, et al. Monitoring of circulating tumor DNA improves early relapse detection after axicabtagene ciloleucel infusion in large B-cell lymphoma: results of a prospective multi-institutional trial. J Clin Oncol 2021;39:3034–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shin S-H, Kim YJ, Lee D, Cho D, Ko YH, Cho J, et al. Analysis of circulating tumor DNA by targeted ultra-deep sequencing across various non-Hodgkin lymphoma subtypes. Leuk Lymphoma 2019;60:2237–46. [DOI] [PubMed] [Google Scholar]
- 48. Leonard JP. POD24 in follicular lymphoma: time to be “wise. Blood 2022;139:1609–10. [DOI] [PubMed] [Google Scholar]
- 49. Kurtz DM, Scherer F, Jin MC, Soo J, Craig AFM, Esfahani MS, et al. Circulating tumor DNA measurements as early outcome predictors in diffuse large B-cell lymphoma. J Clin Oncol 2018;36:2845–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huet S, Salles G. Potential of circulating tumor DNA for the management of patients with lymphoma. JCO Oncol Pract 2020;16:561–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data 1
Supplementary Data 2
Data Availability Statement
The data generated in this study are publicly available in the Sequence Read Archive database at PRJNA813747.