Abstract
Within Streptococcus suis serotype 2, pathogenic, weakly pathogenic, and nonpathogenic strains can be found. We introduced a genomic library of a pathogenic strain into a weakly pathogenic strain. After infection of the library into young piglets pathogenic transformants were selected. One specific transformant containing a 3-kb fragment of the pathogenic strain appeared to be dominantly enriched in diseased pigs. The observed enrichment was not tissue specific. The selected fragment, when introduced into two different weakly pathogenic strains, increased the virulence of these strains considerably. In contrast, introduction of the corresponding fragment of a weakly pathogenic strain had only minor effects on virulence. Nucleotide sequence analysis of the selected fragment of the pathogenic strain revealed the presence of two potential open reading frames, both of which were found to be mutated in the corresponding fragment of the weakly pathogenic strain. These data strongly suggest that the selected fragment contains determinants important for virulence.
Streptococcus suis is an important cause of meningitis, septicemia, arthritis, and sudden death in young pigs (6, 32). It can also cause meningitis in humans (1). Attempts to control the disease are still hampered by the lack of sufficient knowledge about the pathogenesis of the disease and the lack of effective vaccines and sensitive diagnostic methods.
So far, 35 serotypes of S. suis have been described (8–10). Virulence of S. suis can differ within and among serotypes (30, 31, 33, 34). Worldwide, S. suis serotype 2 is the most frequently isolated serotype. Within S. suis serotype 2, pathogenic, weakly pathogenic, and nonpathogenic strains can be found (33, 34). The pathogenic strains cause severe clinical signs of disease in pigs, and large numbers of bacteria can be reisolated from the central nervous system (CNS) and the joints after experimental infection (33, 34). The weakly pathogenic strains cause only mild clinical signs of disease, and only infrequently can bacteria be reisolated from the CNS and the joints after experimental infection (33, 34). The nonpathogenic strains are completely avirulent in young pigs after experimental infection (33, 34).
The 136-kDa muramidase-related protein and the 110-kDa extracellular factor are generally considered important virulence markers for S. suis serotype 2 strains isolated in Europe and the United States (2, 7, 17, 22, 29, 36). However, differences in virulence between pathogenic, weakly pathogenic, and nonpathogenic strains cannot be explained exclusively by differences in their muramidase-related protein and extracellular factor expression patterns (27). In addition, it is known that the capsule of S. suis serotype 2 is an important virulence factor (24). However, since both pathogenic, weakly pathogenic, and nonpathogenic strains seem to be fully encapsulated after growth in vitro and in vivo (H. E. Smith and H. J. Wisselink, unpublished data), it is not likely that the level of encapsulation of these strains is associated with the difference in virulence. To gain insight into the differences between pathogenic, weakly pathogenic, and nonpathogenic strains that determine the differences in virulence, we applied an in vivo complementation system.
It was previously shown, by ribotyping and random amplified polymorphic DNA analysis assays, that pathogenic and weakly pathogenic strains of S. suis serotype 2 are genetically closely related, whereas nonpathogenic strains showed a high degree of genetic heterogeneity (4, 25). Therefore, we envisaged the possibility that the introduction of DNA fragments of a pathogenic strain into a weakly pathogenic strain could increase its virulence. To challenge this hypothesis we constructed a genomic library of the pathogenic S. suis strain 10 in a plasmid and introduced the plasmid library into the weakly pathogenic reference strain of S. suis serotype 2, strain S735 (35). Pigs were inoculated intravenously with the recombinants, and bacteria were recovered from the CNS and the joints of diseased pigs. The reisolated bacteria were subsequently analyzed for their plasmid content and virulence.
Complementation system.
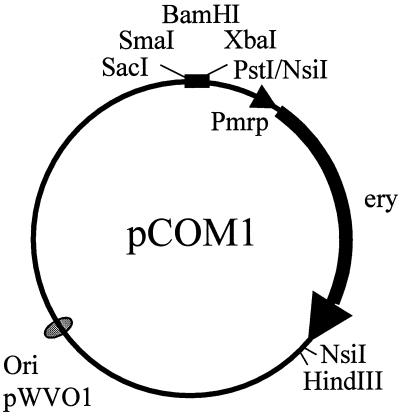
A genomic library of the pathogenic S. suis strain 10 (34) was constructed into the weakly pathogenic strain S735 (35). To do this, we used the plasmid pCOM1 (Fig. 1). pCOM1 is based on the replication functions of pWVO1 (13), which functions in Escherichia coli as well as in S. suis (28). Moreover, the vector contained the erythromycin resistance gene of pE194 (11) preceded by the promoter region of the mrp gene (26) as well as the SacI-PstI part of the multiple cloning site of pKUN19 (14). Sau3AI partial digests (23) of the DNA of the pathogenic S. suis serotype 2 strain 10 were size fractionated (>3 kb) by precipitation with 4.6% of polyethylene glycol 6000 (BDH Chemicals) (20). The fragments were ligated to BamHI-digested pCOM1 (Fig. 1), and the ligation mixtures were transformed into E. coli XL2-blue cells (Stratagene). Erythromycin-resistant colonies were selected on Luria broth (19) containing 1.5% (wt/vol) agar and 200 μg of erythromycin per ml. About 17,000 independent E. coli clones were obtained. Analysis of 55 of the transformants showed that 64% contained an insert of >3 kb (results not shown). From the pool of E. coli transformants, plasmid DNA was isolated and was subsequently used for the electrotransformation of the weakly pathogenic S. suis strain S735 (28). S. suis transformants were selected on Columbia agar blood base (code CM331; Oxoid) plates containing 6% (vol/vol) horse blood and 1 μg of erythromycin per ml. This resulted in approximately 30,000 independent S. suis transformants. The S. suis library was designated S735(pCOM-L). As determined by analysis of 24 randomly selected transformants, more than 30% of the S735(pCOM-L) transformants contained an insert of >3 kb (results not shown). The transformants were pooled and stored at −80°C.
FIG. 1.
pCOM1 vector used in this study. pCOM1 contains the replication functions of pWVO1 (Ori) (13) and the erythromycin resistance gene (ery) of pE194 (11) preceded by the promoter region of the mrp gene (Pmrp) (26) as well as the SacI-PstI part of the multiple cloning site of pKUN19 (14).
Selection of genomic fragments associated with virulence.
To select for genetic determinants of the pathogenic S. suis strain 10 that could increase the virulence of the weakly pathogenic strain S735, 1-week-old pigs were inoculated intravenously with the S. suis library S735(pCOM-L) as described before (31, 34). We used a dose of either 107 or 108 CFU, and the pigs received erythromycin twice a day orally (erythromycin stearate, 40 mg/kg of body weight; Abbott B.V., Amstelveen, The Netherlands). Two hours after the infection, the pigs were treated with erythromycin for the first time. To score disease, we measured the body temperature of the pigs, the number of polymorphonuclear leukocytes in blood, and clinical signs of disease such as signs of nervousness and lameness. Moreover, to monitor infection with S. suis we collected swabs of the nasopharynx and feces daily. The swabs were plated directly onto Columbia agar containing 6% horse blood. After the pigs were killed, they were examined for pathological changes. Moreover, tissue specimens were collected from the CNS, serosae, joints, lungs, liver, kidneys, spleen, heart, and tonsils. The tissues were homogenized in the presence of Todd-Hewitt medium (code CM189; Oxoid) by using an Ultra-Turrax tissuemizer (Omni International, Waterbury, Conn.) and were centrifuged for 5 min at 1,200 × g, and the supernatants were plated onto Columbia agar containing 6% horse blood.
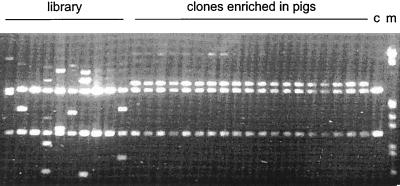
All pigs showed specific S. suis symptoms (Table 1) 3 to 7 days after the infection, and except for one, all pigs died during the course of the experiment. From five of the pigs bacteria could be reisolated from the CNS, and from two other pigs bacteria were isolated from the joints (Table 1). In previous experiments in which pigs were inoculated with weakly pathogenic strains, specific S. suis symptoms were observed at a very low frequency (34, 35). In addition, from those pigs, bacteria could never be reisolated from the CNS or from the joints. Therefore, these data indicate that, compared to the virulence of strain S735, bacteria isolated from pigs inoculated with the S. suis library S735(pCOM-L) are more virulent due to the presence of a DNA fragment of the pathogenic strain 10. The plasmid content of 90 randomly selected clones isolated from the CNS or the joints of the seven diseased pigs was analyzed by PCR and restriction analysis. The results showed that 88 of the 90 clones analyzed (19 of which are shown in Fig. 2) contained an insert of about 3 kb and had identical restriction patterns. Moreover, the inserts of 10 randomly selected clones having identical restriction patterns also showed identical DNA sequences (results not shown). Plasmid DNA of 10 randomly selected clones from the original S735(pCOM-L) library showed 10 different restriction patterns (Fig. 2). These data suggest that one specific clone, which was designated S735(pCOM-V10), was greatly enriched in seven different pigs. Moreover, this particular clone was isolated from the CNS as well as from the joints of the various pigs, indicating that the observed enrichment was not tissue specific.
TABLE 1.
Virulence of S. suis library and strains in germfree pigs
| Strain | No. of pigs | Dose of S. suis (CFU) | Mortalitya (%) | Mean no. of days until death | Morbidityb (%) | Clinical index of the group
|
Fever indexe | Leukocyte indexf | No. of pigs in which S. suis was isolated from:
|
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Specificc symptoms | Nonspecificd symptoms | CNS | Serosae | Joints | ||||||||
| S735(pCOM-L) | 4 | 107 | 100 | 4 | 100 | 69 | 91 | 25 | NAg | 3 | 2 | 3 |
| 4 | 108 | 75 | 7 | 100 | 50 | 69 | 20 | 17 | 2 | 1 | 2 | |
| S735(pCOM-V10) | 5 | 106 | 100 | 1 | 100 | 100 | 100 | 54 | 4 | 5 | 5 | 5 |
| S735(pCOM1) | 4 | 106 | 25 | 12 | 25 | 2 | 11 | 6 | 80 | 1 | 1 | 2 |
| S735(pCOM-V10) | 5 | 106 | 100 | 1 | 100 | 100 | 100 | 60 | NA | 5 | 5 | 5 |
| S735(pCOM-V735) | 5 | 106 | 20 | 15 | 100 | 40 | 26 | 17 | 52 | 1 | 1 | 1 |
| S735(pCOM1) | 5 | 106 | 20 | 16 | 60 | 11 | 9 | 11 | 20 | 1 | 0 | 0 |
| 24(pCOM-V10) | 5 | 106 | 100 | 2 | 100 | 50 | 66 | 42 | 29 | 3 | 3 | 5 |
| 24(pCOM-V735) | 4 | 106 | 25 | 15 | 100 | 40 | 30 | 17 | 18 | 1 | 0 | 0 |
| 24(pCOM1) | 5 | 106 | 20 | 15 | 20 | 2 | 14 | 6 | 21 | 1 | 0 | 0 |
Percentage of pigs that died due to infection or had to be killed for animal welfare reasons.
Percentage of pigs with specific symptoms.
Percentage of observations for the experimental group in which specific symptoms (ataxia, lameness of at least one joint, and/or stillness) were observed.
Percentage of observations for the experimental group in which nonspecific symptoms (loss of appetite and/or depression) were observed.
Percentage of observations for the experimental group of a body temperature of >40°C.
Percentage of blood samples for the experimental group in which the concentration of granulocytes was >1010/liter.
NA, not applicable.
FIG. 2.
Plasmids digested with SmaI and XbaI on a 0.8% agarose gel. Library, plasmids isolated from 10 randomly selected clones of the original library; clones enriched in pigs, plasmids isolated from 19 independently selected clones enriched in pigs; c, pCOM1; m, molecular size marker.
Virulence-associated properties of the selected fragment, V10.
To further analyze the virulence properties of strain S735(pCOM-V10), pigs were inoculated intravenously with 106 CFU of strain S735(pCOM1) or strain S735(pCOM-V10). The results (Table 1) clearly show that, compared to the virulence of strain S735(pCOM1), the virulence of strain S735(pCOM-V10) was greatly enhanced. All pigs inoculated with strain S735(pCOM-V10) showed specific S. suis symptoms and died within 1 day of infection. In contrast, except for one, none of the pigs inoculated with the control strain S735(pCOM1) showed specific clinical symptoms, and these pigs survived until the end of the experiment (15 days after infection). These data proved that the introduction of fragment V10 of strain 10 into S735 transformed the weakly pathogenic strain S735 into a highly pathogenic strain. This strongly suggests that the protein(s) encoded by V10 is an important virulence determinant and plays an important role in the pathogenesis of S. suis serotype 2 infections in pigs.
To find out whether the observed increase of virulence by fragment V10 was specific for strain S735, we introduced pCOM1 and pCOM-V10 into another weakly pathogenic strain, strain 24 (34). Subsequently, we determined the virulence properties of the strains 24(pCOM1) and 24(pCOM-V10). As shown in Table 1, similar effects of V10 on the virulence of strains S735 and 24 were observed. Both strains 24(pCOM-V10) and S735(pCOM-V10) were highly pathogenic for young piglets, whereas strains 24(pCOM1) and S735(pCOM1) were shown to be only weakly pathogenic (Table 1). This strongly indicates that V10 has a more general ability to transform weakly pathogenic serotype 2 strains into highly pathogenic strains.
Because we used a plasmid system for the complementation approach, gene-dose effects cannot be excluded. Plasmid pCOM1 is based on the replication functions of pWVO1. In gram-positive bacteria the latter plasmid has a copy number between 3 and 6 (13). To find out whether copy number effects play a role, we cloned the genomic region of strain S735 homologous to fragment V10 of strain 10 (see below) into plasmid pCOM1. This plasmid was designated pCOM-V735. The virulence of strains S735(pCOM-V735) and 24(pCOM-V735) was subsequently compared to that of S735(pCOM-V10), S735(pCOM1), 24(pCOM-V10), and 24(pCOM1). The results (Table 1) clearly show that, in contrast to pCOM-V10, the plasmid pCOM-V735 did not carry virulence-enhancing activity. Pigs infected with strains S735(pCOM-V10) and 24(pCOM-V10) died within 1 or 2 days after infection, whereas most of the pigs infected with strains S735(pCOM-V735), 24(pCOM-V735), S735(pCOM1), and 24(pCOM1) survived until the end of the experiment (17 days after infection). Compared to pigs infected with strains containing pCOM1, pigs infected with strains containing pCOM-V735 developed more general and specific signs of disease, but much less so than pigs infected with strains containing pCOM-V10 (Table 1). From these data we concluded that the differences in virulence observed between the strains containing pCOM-V10 and the strains containing pCOM-V735 are caused by differences between the fragments V10 and V735 (see below). The differences in virulence observed between the strains containing pCOM1 and the strains containing pCOM-VS735 may be due to gene-dose effects.
In previous experiments it was found that pigs infected with weakly pathogenic strains showed only mild clinical signs of disease and that bacteria could never be reisolated from the CNS or the joints (34, 35). Surprisingly, in the experiments described in this paper, in which we used weakly pathogenic strains containing the control plasmid pCOM1, bacteria could (with a low frequency) be reisolated from the CNS as well as from the joints. There are several possible explanations for these observed differences. One explanation is that the presence of the plasmid somehow affects the virulence properties of the strains. Another possibility is that the treatment of the pigs with erythromycin makes the pigs more sensitive to S. suis infections, and a third possibility is that compared to the pigs used previously, the pigs used for the current experiments were more sensitive to S. suis infections.
Previously, Charland and coworkers (5) tested the virulence of strain S735-SM in SPF piglets. In these experiments they showed that animals infected with strain S735-SM had signs of meningitis and arthritis. These data did not seem to be in accord with our data concerning the virulence of strain S735. However, strain S735-SM is a streptomycin-resistant variant of strain S735 that was selected by serial passages of strain S735 on broth containing increasing concentrations of streptomycin. In their experiments the authors did not compare the virulence of strain S735-SM to that of the original S735 strain. Moreover, the animal model used by these authors differed considerably from the model used in our experiments. Charland and coworkers (5) used 6- to 7-week-old specific-pathogen-free piglets that were infected intravenously with 108 CFU of S. suis, whereas we used 1-week-old germfree piglets that were infected intravenously with 106 CFU. These differences will probably explain the different results obtained.
Sequence analysis of fragments V10 and V735.
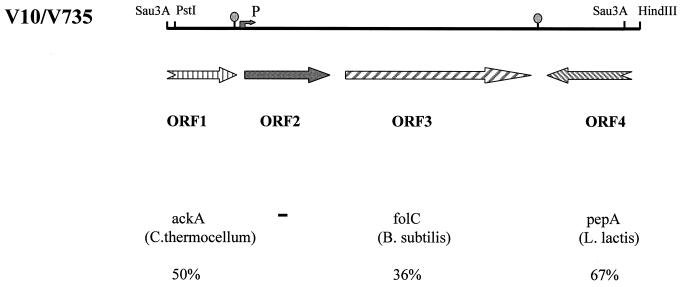
Large differences were observed between the effects of the selected fragment, V10, of the pathogenic strain 10 and the corresponding fragment, V735, isolated from the weakly pathogenic strain S735, on virulence. In contrast to V10, which had a strong virulence-enhancing effect on weakly pathogenic strains, V735 showed only minor effects. Therefore, differences between these two fragments should be responsible for the observed differences in virulence. To analyze the differences between fragments V10 and V735, we cloned fragment V735 and determined the nucleotide sequences of fragments V10 and V735. A 3.1-kb PstI-HindIII fragment of strain S735 (V735) was identified by using the fragment V10 as a probe, and this fragment was subsequently cloned into pCOM1 (Fig. 3). DNA sequences were determined with a 373A DNA Sequencing System (Applied Biosystems, Warrington, United Kingdom), and samples were prepared by use of an ABI/PRISM dye-terminator cycle-sequencing ready reaction kit (Applied Biosystems). The sequence of V10 revealed two complete and two incomplete open reading frames (ORFs) (Fig. 3). ORF 1 (nucleotides 1 to 461) codes for a polypeptide of 153 amino acids. This protein shows homology (49% identity) to the C-terminal region of acetate kinase of Clostridium thermocellum (accession number AF041841) and various other bacterial species (12). ORF 2 (nucleotides 625 to 1327) codes for a protein of 233 amino acids. No significant similarities were found between the predicted amino acid sequence of this protein and proteins present in the data libraries. ORF 3 (nucleotides 1382 to 2639) codes for a protein of 418 amino acids. This protein shows homology (36% identity) to folylpolyglutamate synthetase (FolC) of Bacillus subtilis (16, 18). Compared to the other ORFs, ORF 4 is transcribed in the opposite direction. ORF 4 (nucleotides 2684 to 2972) codes for a polypeptide of 96 amino acids. This polypeptide shows homology (67% identity) to the C-terminal part of glutamyl aminopeptidase (PepA) of Lactococcus lactis (15). ORFs 2 and 3 both possess putative initiation codons and ribosome-binding sites. Putative −35 (TGGACA) and −10 (TACAAT) sequences, which may function as promoter sequences, were found preceding ORF 2. ORFs 2 and 3 are separated by 55 nucleotides. In this region no putative promoter sequences could be observed. This could indicate that ORFs 2 and 3 are cotranscribed. Downstream of ORFs 1 and 3 we found regions of extended dyad symmetry, which probably function as transcription termination signals.
FIG. 3.
Schematic representation of fragments V10 and V735. The arrows indicate the potential ORFs. P, position of the potential promoter sequence; ○, positions of the potential transcription regulator sequences. Homologies (% identities) between the potential proteins encoded by the ORFs and proteins present in the data libraries are indicated.
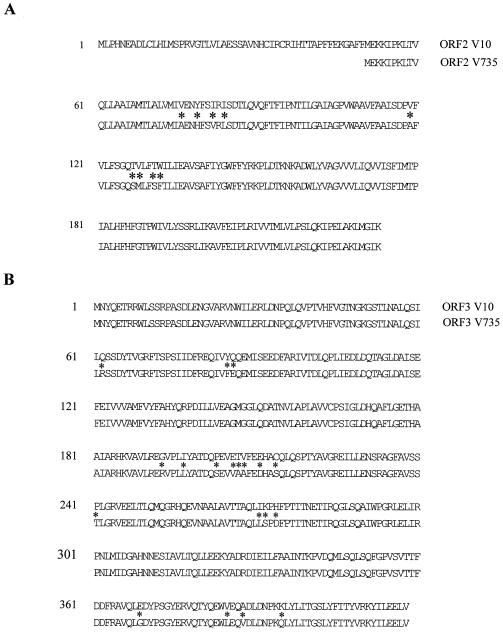
The sequence of fragment V735 was determined and compared to the sequence of fragment V10. No major deletions or insertions were found between the sequenced regions. ORFs 1, 3, and 4 of strains 10 and S735 were highly homologous. The putative protein fragments encoded by the ORF 1's differ in two (1.3%) amino acids, whereas the putative protein fragments of the ORF 4's are identical. The putative proteins encoded by the ORF 3's are highly homologous and differ in only 19 (4.5%) amino acids (Fig. 4B). The proteins encoded by the ORF 3's show homology to FolC of various pro- and eucaryotic organisms. FolC catalyzes the conversion of folates to polyglutamate derivatives (3). Bacteria require folates for the biosynthesis of glycine, methionine, formylmethionine, thymidine, purines, and pantothenate (3). Whether the FolC proteins encoded by fragments V10 and V735 have different enzymatic activities or different substrate specificities is unknown so far. In E. coli, a folC mutant is methionine deficient (3); however, so far a role of FolC in virulence has not been described.
FIG. 4.
Homology between ORFs 2 (A) and 3 (B) encoding proteins of fragments V10 and V735. Asterisks indicate nonidentical amino acids.
Major differences were observed between the ORF 2's of strains 10 and S735. In the pathogenic strain 10 an ORF of 699 bases was found, predicting a protein product of 233 amino acids. In contrast, due to a frameshift mutation, in the weakly pathogenic strain S735 an ORF of 569 bases, coding for a polypeptide of 183 amino acids, was found. Compared to the putative protein encoded by strain 10, the putative protein encoded by strain S735 lacks the N-terminal 50 amino acids (Fig. 4A). In strain S735 a strong ribosome-binding site precedes the methionine start codon of ORF 2. In contrast, however, in strain 10 the sequence did not indicate the presence of a strong ribosome-binding site preceding the methionine start codon of ORF 2. Therefore, although ORF 2 of strain 10 is extended compared to ORF 2 of strain S735, it is not clear whether the proteins expressed by these two ORFs differ in length. Future experiments will be required to analyze the expressed proteins in detail. Besides these N-terminal differences, the putative proteins differ at nine amino acid positions (4.9%). Except for one, these amino acid substitutions are clustered at two different positions in the putative protein. The function of the ORF 2 protein is unknown so far. No homologies were found between the ORF 2 protein sequences and protein sequences present in the data libraries. Hydrophobicity profiles showed that the ORF 2-encoded proteins are very hydrophobic. A role of the ORF 2 protein in the cellular membrane is therefore suggested. In addition, the putative −35 regions that may be part of the promoter sequences involved in the expression of ORFs 2 and 3 differed between the two strains. A TGGACA sequence was found in strain 10, whereas a TGGTCA sequence was found in strain S735. The sequence data suggest that the differences in the virulence-enhancing effects of fragments V10 and V735 may be the results of functional differences between the putative proteins expressed by ORFs 2 and/or 3 and/or by differences in their levels of expression.
In the present paper we describe the development and the successful application of an in vivo complementation approach for the identification of important molecular determinants that determine the differences in virulence between pathogenic and weakly pathogenic strains of S. suis serotype 2. The strategy to identify genetic determinants by in vivo complementation requires two genetically related strains which can be distinguished by their ability to cause disease as well as a gene transfer system. Therefore, it is likely that the system is more generally applicable for the identification of virulence determinants in various bacterial species. Previously, a similar approach was used to identify virulence genes in Mycobacterium tuberculosis (21). A major difference between the two systems is that the M. tuberculosis virulence genes were selected by using a mouse model. In contrast, we used the natural host (pigs). Moreover, in the system described for M. tuberculosis an integrating cosmid vector was used to introduce the virulence genes instead of the plasmid system that was used here.
Nucleotide sequence accession numbers.
The nucleotide sequences for fragments V10 and V735 have been deposited into the National Center for Biotechnology Information under accession numbers AF306940 and AF306941, respectively.
Acknowledgments
We thank D. Mevius for helpful discussions concerning the treatments of the pigs with erythromycin.
REFERENCES
- 1.Arends J P, Zanen H C. Meningitis caused by Streptococcus suis in humans. Rev Infect Dis. 1988;10:131–137. doi: 10.1093/clinids/10.1.131. [DOI] [PubMed] [Google Scholar]
- 2.Awad-Masalmeh M, Köfer J, Schuh M, Hinterdorfer F. Serotypen, virulenzfaktoren und empfindlichkeit gegenuber antibiotika von Streptococcus suis stämmer isoliert aus klinisch gesunden und erkrankten schweinen in Österreich. Wein Tieräerztl Monschr. 1999;86:262–269. [Google Scholar]
- 3.Bogner A L, Osborne C, Shane B. Primary structure of the Escherichia coli folC gene and its folylpolyglutamate synthetase-dihydrofolate synthetase product and regulation of expression by an upstream gene. J Biol Chem. 1987;262:12337–12343. [PubMed] [Google Scholar]
- 4.Chalettier S, Gottschalk M, Higgins R, Brousseau R, Harel J. Relatedness of Streptococcus suis serotype 2 isolates from different geographic origins as evaluated by molecular fingerprinting and phenotyping. J Clin Microbiol. 1999;37:362–366. doi: 10.1128/jcm.37.2.362-366.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charland N, Harel J, Kobisch M, Lacasse S, Gottschalk M. Streptococcus suis serotype 2 mutants deficient in capsular expression. Microbiology. 1998;144:325–332. doi: 10.1099/00221287-144-2-325. [DOI] [PubMed] [Google Scholar]
- 6.Clifton-Hadley F A. Streptococcus suis type 2 infections. Br Vet J. 1983;139:1–5. doi: 10.1016/s0007-1935(17)30581-x. [DOI] [PubMed] [Google Scholar]
- 7.Galina L, Vecht U, Wisselink H J, Pijoan C. Prevalence of various phenotypes of Streptococcus suis isolated from swine in the U.S.A. based on the presence of muramidase-released protein and extracellular factor. Can J Vet Res. 1996;60:72–74. [PMC free article] [PubMed] [Google Scholar]
- 8.Gottschalk M, Higgins R, Jacques M, Mittal R K, Henrichsen J. Description of 14 new capsular types of Streptococcus suis. J Clin Microbiol. 1989;27:2633–2636. doi: 10.1128/jcm.27.12.2633-2636.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottschalk M, Higgins R, Jacques M, Beaudain M, Henrichsen J. Characterization of six new capsular types (23–28) of Streptococcus suis. J Clin Microbiol. 1991;29:2590–2594. doi: 10.1128/jcm.29.11.2590-2594.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higgins R, Gottschalk M, Jacques M, Beaudain M, Henrichsen J. Description of six new capsular types (29–34) of Streptococcus suis. J Vet Diagn Investig. 1995;7:405–406. doi: 10.1177/104063879500700322. [DOI] [PubMed] [Google Scholar]
- 11.Horinouchi S, Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibiotics. J Bacteriol. 1982;150:804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kakuda H, Honoso K, Shiroishi K, Ichihara S. Identification and characterization of the ack (acetate kinase A)-pta (phosphotransacetylase) operon and complementation analysis of acetate utilization by an ackA-pta deletion mutant of Escherichia coli. J Biochem. 1994;116:916–922. doi: 10.1093/oxfordjournals.jbchem.a124616. [DOI] [PubMed] [Google Scholar]
- 13.Kok J, van der Vossen J M B M, Venema G. Construction of plasmid cloning vectors for lactic acid streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl Environ Microbiol. 1984;48:726–731. doi: 10.1128/aem.48.4.726-731.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konings R N H, Verhoeven E J M, Peeters B P H. pKUN vectors for the separate production of both DNA strands of recombinant plasmids. Methods Enzymol. 1987;153:12–34. doi: 10.1016/0076-6879(87)53045-2. [DOI] [PubMed] [Google Scholar]
- 15.l'Anson K J, Movahedi S, Griffin H G, Gasson M J, Mulholland F. A non-essential glutamyl aminopeptidase is required for optimal growth of Lactococcus lactis MG1363 in milk. Microbiology. 1995;141:2873–2881. doi: 10.1099/13500872-141-11-2873. [DOI] [PubMed] [Google Scholar]
- 16.Luo D, Leautey J, Grunberg-Manago M, Putzer H. Structure and regulation of expression of the Bacillus subtilis valyl-tRNA synthetase gene. J Bacteriol. 1997;179:2472–2478. doi: 10.1128/jb.179.8.2472-2478.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luque I, Tarradas C, Astorga R, Perea A, Wisselink H J, Vecht U. The presence of muramidase released protein and extracellular factor protein in various serotypes of Streptococcus suis isolated from diseased and healthy pigs in Spain. Res Vet Sci. 1998;66:69–72. doi: 10.1053/rvsc.1998.0242. [DOI] [PubMed] [Google Scholar]
- 18.Margolis P S, Driks A, Losick R. Sporulation gene spoIIB from Bacillus subtilis. J Bacteriol. 1993;175:528–540. doi: 10.1128/jb.175.2.528-540.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 20.Paithankar K R, Prasad K S N. Precipitation of DNA by polyethylene glycol and ethanol. Nucleic Acids Res. 1991;19:1346. doi: 10.1093/nar/19.6.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pascopella L, Collins F M, Martin J M, Lee M H, Hatfull G F, Stover C K, Bloom B R, Jacobs W R., Jr Use of in vivo complementation in Mycobacterium tuberculosis to identify a genomic fragment associated with virulence. Infect Immun. 1994;62:1313–1319. doi: 10.1128/iai.62.4.1313-1319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salasia S I O, Lämmler C. Distribution of serotype, virulence markers and further characteristics of Streptococcus suis isolates from pigs. J Vet Med Ser B. 1995;42:78–83. doi: 10.1111/j.1439-0450.1995.tb00685.x. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Smith H E, Damman M, van der Velde J, Wagenaar F, Wisselink H J, Stockhofe-Zurwieden N, Smits M A. Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect Immun. 1999;67:1750–1756. doi: 10.1128/iai.67.4.1750-1756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith H E, Rijnsburger M, Stockhofe-Zurwieden N, Wisselink H J, Vecht U, Smits M A. Virulent strains of Streptococcus suis serotype 2 and highly virulent strains of Streptococcus suis serotype 1 can be recognized by a unique ribotype profile. J Clin Microbiol. 1997;35:1049–1053. doi: 10.1128/jcm.35.5.1049-1053.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith H E, Vecht U, Gielkens A L J, Smits M A. Cloning and nucleotide sequence of the gene encoding the 136-kilodalton surface protein (muramidase-released protein) of Streptococcus suis type 2. Infect Immun. 1992;60:2361–2367. doi: 10.1128/iai.60.6.2361-2367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith H E, Vecht U, Wisselink H J, Stockhofe-Zurwieden N, Biermann Y, Smits M A. Mutants of Streptococcus suis types 1 and 2 impaired in expression of muramidase-released protein and extracellular protein induce disease in newborn germfree pigs. Infect Immun. 1996;64:4409–4412. doi: 10.1128/iai.64.10.4409-4412.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith H E, Wisselink H J, Vecht U, Gielkens A L J, Smits M A. High-efficiency transformation and gene inactivation in Streptococcus suis type 2. Microbiology. 1995;141:181–188. doi: 10.1099/00221287-141-1-181. [DOI] [PubMed] [Google Scholar]
- 29.Staats J J, Plattner B L, Stewart G C, Chengappa M M. Presence of the Streptococcus suis suilysin gene and expression of MRP and EF correlates with high virulence in Streptococcus suis type 2 isolates. Vet Microbiol. 1999;70:201–211. doi: 10.1016/s0378-1135(99)00147-9. [DOI] [PubMed] [Google Scholar]
- 30.Stockhofe-Zurwieden N, Vecht U, Wisselink H J, van Lieshout H, Smith H E. Comparative studies on the pathogenicity of different Streptococcus suis serotype 1 strains. In: Monetti P G, Vignola G, editors. Proceedings of the 14th International Pig Veterinary Society Congress, Bologna, Italy. 1996. p. 299. [Google Scholar]
- 31.Vecht U, Arends J P, van der Molen E J, van Leengoed L A M G. Difference in virulence between two strains of Streptococcus suis type 2 after experimentally induced infection of newborn germfree pigs. Am J Vet Res. 1989;50:1037–1043. [PubMed] [Google Scholar]
- 32.Vecht U, van Leengoed L A M G, Verheyen E R M. Streptococcus suis infections in pigs in The Netherlands (part one) Vet Q. 1985;7:315–321. doi: 10.1080/01652176.1985.9694005. [DOI] [PubMed] [Google Scholar]
- 33.Vecht U, Wisselink H J, Jellema M L, Smith H E. Identification of two proteins associated with virulence of Streptococcus suis type 2. Infect Immun. 1991;59:3156–3162. doi: 10.1128/iai.59.9.3156-3162.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vecht U, Wisselink H J, van Dijk J E, Smith H E. Virulence of Streptococcus suis type 2 strains in newborn germfree pigs depends on phenotype. Infect Immun. 1992;60:550–556. doi: 10.1128/iai.60.2.550-556.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vecht U, Wisselink H J, Stockhofe-Zurwieden N, Smith H E. Characterization of virulence of the Streptococcus suis serotype 2 reference strain Henrichsen S735 in newborn germfree pigs. Vet Microbiol. 1995;51:125–136. doi: 10.1016/0378-1135(96)00028-4. [DOI] [PubMed] [Google Scholar]
- 36.Wisselink H J, Smith H E, Stockhofe-Zurwieden N, Peperkamp K, Vecht U. Distribution of capsular types and production of muramidase-released protein (MRP) and extracellular factor (EF) of Streptococcus suis strains isolated from diseased pigs in seven European countries. Vet Microbiol. 2000;74:237–247. doi: 10.1016/s0378-1135(00)00188-7. [DOI] [PubMed] [Google Scholar]