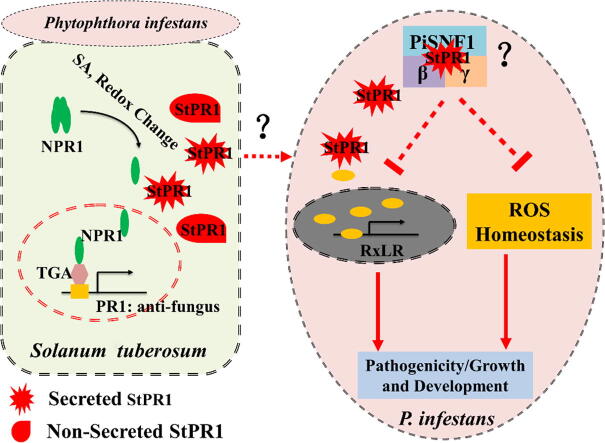
Graphical abstract
The potato-Phytophthora infestans system was used as a model to investigate how PR1 proteins protect plants against oomycete pathogens. Results showed that upon pathogen infection, PR1 proteins were induced, and secreted PR1 translocated into pathogen cells to target AMPK kinase complex, thus inhibiting vegetative growth and pathogenicity. Question marks and dotted lines represent scientific problems which need more detailed proofs. NPR1, nonexpressor of pathogenesis-related genes 1; TGA, TGACG-binding factor; SNF1, Sucrose non-fermenting 1; RxLR, the amino acid (Arg-x-Leu-Arg) of conserved motif in effector, where x is any amino acid.
Keywords: Pathogenesis-related protein 1, AMPK kinase complex, Phytophthora infestans, Potato late blight, Host-pathogen interaction, Cross-kingdom translocation
Highlights
-
•
Upon infecting by P. infestans, StPR1 expression was induced in host and secretory StPR1 proteins translocated into pathogen.
-
•
The translocated PR1 proteins target AMPK complex in P. infestans, and impaired the AMPK activation to downstream targets.
-
•
StPR1 prevented ROS homeostasis and inhibited the expression of RxLR effector-encoding genes in P. infestans.
Abstract
Introduction
During the arms race between plants and pathogens, pathogenesis-related proteins (PR) in host plants play a crucial role in disease resistance, especially PR1. PR1 constitute a secretory peptide family, and their role in plant defense has been widely demonstrated in both hosts and in vitro. However, the mechanisms by which they control host-pathogen interactions and the nature of their targets within the pathogen remain poorly understood.
Objectives
The present study was aimed to investigate the anti-oomycete activity of secretory PR1 proteins and elaborate their underlying mechanisms.
Methods
This study was conducted in the potato-Phytophthora infestans pathosystem. After being induced by the pathogen infection, the cross-kingdom translocation of secretory PR1 was demonstrated by histochemical assays and western blot, and their targets in P. infestans were identified by yeast-two-hybrid assays, bimolecular fluorescence complementation assays, and co-immunoprecipitation assay.
Results
The results showed that the expression of secretory PR1-encoding genes was induced during pathogen infection, and the host could deliver PR1 into P. infestans to inhibit its vegetative growth and pathogenicity. The translocated secretory PR1 targeted the subunits of the AMPK kinase complex in P. infestans, thus affecting the AMPK-driven phosphorylation of downstream target proteins, preventing ROS homeostasis, and down-regulating the expression of RxLR effectors.
Conclusion
The results provide novel insights into the molecular function of PR1 in protecting plants against pathogen infection, and uncover a potential target for preventing pre- and post-harvest late blight.
Introduction
Potato late blight is a devastating disease caused by the oomycete Phytophthora infestans, resulting in stem death and tuber rot, and causing severe economic losses reaching $12 billion annually [1]. During plant-pathogen interactions, the RxLR (amino acid of conserved motif Arg-x-Leu-Arg) effectors from P. infestans and the resistance-related proteins (R) from Solanum tuberosum are the two key factors determining whether the disease will prevail or not. Besides R proteins, host plants also secrete cross-kingdom proteins that target pathogens to inhibit their growth, development, metabolism, and pathogenicity, including the pathogenesis-related proteins (PR). PR proteins are antimicrobial, low molecular weight, secretory or vacuolar-targeted proteins encoded by host plants, and they are induced by pathological conditions during plant-pathogen interactions [2]. Inducible PR proteins were first identified in the 1970 s in Nicotiana tabacum infected with tobacco mosaic virus and they were later reported in different plant species that had previously been infected by pathogenic oomycetes, fungi, bacteria, and viruses [3]. Among them, PR1 is an inherent component of innate immunity in plants, and has been used as markers for enhanced disease resistance for a long time. Its biochemical activity and mode of action remain elusive, being the only PR without functional annotations [4]. The PR1 family is highly conserved among plants, and homologs have also been found in fungi, insects, and vertebrates, including humans. The cysteine-rich secretory protein (CRISP), antigen 5, and PR1 proteins form a superfamily of secreted proteins named CAP together. In plants, PR1 proteins have unique resistance to oomycetes. The anti-oomycete activity of PR1 proteins from tomato and tobacco was directly demonstrated to inhibit zoospore germination of P. infestans in vitro and restricted P. infestans colonization on tomato surfaces in vivo [5]. Furthermore, the heterologous expression of the PR1 proteins P14c (from Solanum lycopersicum) and PR-1a (from N. tabacum) inhibited the growth of the oomycete pathogen P. brassicae at concentrations of 20 μg/mL [6] and restricted zoospore germination of P. infestans [7]. However, the antifungal mechanism of PR1 proteins was a mystery until a CAP protein of yeast was found to bind sterols [8]. Since then, the inhibitory effect of PR1 on pathogens was demonstrated to sequester sterol from pathogens [6]. Sterol-auxotroph pathogens, such as oomycete Phytophthora, are particularly sensitive to PR1; and when the sterol biosynthesis was restricted, the sterol-prototroph fungal pathogens became highly sensitive to PR1, indicating a critical relationship between sterol binding and anti-fungal property, and suggesting some other mechanisms may contribute to the anti-oomycete activity [6].
Besides sterol sequestration, PR1 inhibited programmed cell death at the pathogen infection sites [9], and induced the expression of host defense genes by releasing a defense signaling peptide called CAPE1 (for CAP-derived peptide1) [10]. CAPE1 originates from the last 11 amino acids at C-terminus derived from the tomato PR1 protein P14c, and contributes to broad-spectrum antibacterial activity against Pseudomonas syringae DC3000 and larvae of Spodopteralitura by inducing the expression of defense-related genes [11]. The importance of PR1 proteins in plant defense is further evidenced by an increasing number of studies showing that they are targeted by effector proteins from multiple fungal pathogens [9], such as the TaPR1-5 that is targeted by the ToxA effector from wheat pathogens Parastagonospora nodorum and Pyrenophora tritici-repentis [12], [13], the PR1a and PR1b that are targeted by barley powdery mildew effector CSEPP0055 [12], [14], and the PR1 in Arabidopsis thaliana that is targeted by the virulence factor SsCP1 (cerato-platanin protein) from Sclerotinia sclerotiorum [15].
Although sterol sequestration and releasing host defense signaling peptides are two important factors that contributing to the antifungal activity of PR1 proteins, our knowledge regarding the molecular function of PR1 proteins is still incomplete, especially in pathogen cells. Can host secretory PR1 proteins translocate into pathogen cells, thus inhibiting the vegetative growth and pathogenicity by targeting key intracellular proteins or signaling pathways? Given the sterol-defect and high sensitivity of P. infestans to PR1 proteins, the S. tuberosum-P. infestans pathosystem was used to investigate whether host secretory PR1 proteins mediate cross-kingdom anti-oomycete activity. Additionally, we aimed to identify the molecular targets of PR1 proteins in pathogens. Our research provides new insights into the anti-oomycete mechanisms of PR1 proteins. Moreover, the functional analysis of PR1 proteins provides a theoretical basis for breeding resistant varieties, and the identification of their molecular targets in pathogens supply potential target resources for biocontrol against the potato late blight.
Materials and methods
Strains and plant materials
The international aggressive oomycete pathogen P. infestans strain T30-4 was used for the pathogenicity and sensitivity tests, and for generating mutants. It was cultured on rye agar medium supplemented with 2% (w/v) sucrose (RSA) or in a liquid rye sucrose broth at 20 °C. Agrobacterium GV3101 was used for the transient expression and genetic transformations, and was grown in Luria-Bertani (LB) agar/liquid medium. S. cerevisiae strain Y2HGold was used for yeast transformation. For S. tuberosum transformations, the susceptible potato variety ‘Desiree’ and the resistant variety ‘E3′ were used. N. benthamiana was used for transient expression and protein interaction assays. All plants were grown at 25 °C with a 16-h photoperiod and 55% relative humidity.
Vector construction
The RNAi and overexpression plasmids were constructed using the Gateway cloning system as previously described by Xiong et al. [16] (presented in supplemental materials and methods in detail). For the yeast-two-hybrid (Y2H) assays, all StPR1 and mutations without signal peptide were cloned into pGADT7, and the subunits of PiAMPK were cloned into pGBKT7 through in-fusion HD Cloning kit (Cat No. 639648; Takara Bio USA, Inc). Meanwhile, the StPR1 genes were cloned into pGBKT7 and PiAMPK was cloned into pGADT7 vectors as a swap. For bimolecular fluorescence complementation (BiFC) assays, all the StPR1 and mutations were cloned into pAB862, while the subunits of PiAMPK were cloned into pAB855. pColdTF (Cat No. 3365; TaKaRa) was used for prokaryotic expression. All constructs were sequenced at TSINGKE (Chongqing, China). All primers used for the construction of plasmids are listed in Supplementary Table 1.
Y2H assays
To screen for the StPR1 targets, key kinases involved in the TOR signaling pathway in P. infestans were selected as potential targets. Y2H assays were performed using the MatchmakerTM Gal4 Two-Hybrid System 3 (Clontech, Palo Alto, CA, USA), as described in the manual. The plasmid pGBKT7-target gene with the empty vector pGADT7 was co-transformed into S. cerevisiae to exclude the false positives caused by autoactivation. The same procedure was performed in the pGADT7-target gene with pGBKT7. Yeast transformants were screened on SD/-Leu/-Trp and further confirmed on SD-Leu/-Trp/-His/-Ade medium with 20 μg/mL X-α-gal and 50 ng/mL AbA.
BiFC and co-localization assays
BiFC was performed using the protocols described previously [17]. Target genes were cloned into binary vectors pAB855 tagged with cYFP and pAB862 tagged with nYFP. These vectors were transiently co-expressed in 4-week-old N. benthamiana, and fluorescence was observed 3 days after agroinfiltration. As controls, only the YFP halves as well as single proteins fused to cYFP and nYFP alone were co-transformed into N. benthamiana. The agroinfiltration buffer consisted of 10 mM MES, 10 mM MgCl2, 200 μM AS. Agrobacterium strain with final OD600 values of 0.1 in agroinfiltration buffer was used. For YFP fluorescence images, cells were analyzed using an excitation wavelength of 514 nm and an emission wavelength of 527 nm with a Leica confocal fluorescence microscope (Leica TCS SP8, Germany). For the co-localization assay, p35S-StPR1GFP-t35S and pHam34-PiSNF1RFP-tHam34 fragments were cloned into modified multigene assembly vectors p35S-multi-8GWN through isocaudarner AsisI/AscI and PacI/AscI enzyme digestions, respectively. Co-localization assays were performed in N. benthamiana as described above.
Genetic transformation of potato lines and P. Infestans
For S. tuberosum transformations, the susceptible potato variety ‘Desiree’ was used to generate StPR1 overexpression lines, and the resistant variety ‘E3′ was used to generate RNAi transgenic lines. The transformation of potato was mediated by A. tumefaciens GV3103, and the detailed procedure was performed as described previously by Millam [18]. A. tumefaciens-mediated transformation was used to acquire P. infestans mutants according to the procedures described by Wu et al. [19] with some modifications. Detailed information is provided in supplemental materials and methods.
Pathogenicity assays
Pathogenicity assays were conducted according to the method of Whisson et al. [20] with some modifications. The sporangia (1 × 107 sporangia/mL) of P. infestans were harvested from 15-day-old cultures by washing with sterile water and were prepared as inoculum. Then, 10 μL of the inoculum were dropped onto the back of detached leaves and tubers of wild-type or transgenic lines. Owing to the inhibited sporangia formation of P. infestans mutants, oomycete disks (7-mm-diameter) were used as the inoculum on detached leaves of ‘Desiree’. All leaves and tubers were incubated at 20 °C with a 12-h/12-h light/dark cycle, and the relative humidity was greater than 90%. The disease index (DI) of the leaves was assessed at 5 days post-inoculation (dpi), and the size of necrotic patches on tubers was measured at 10 dpi. The disease grade and DI were determined as described previously by Wu et al. [21]. The pathogenicity tests were repeated three times, and each replication contained at least 10 leaves or tubers.
Prokaryotic expression and purification
The vector used for constructing prokaryotic expression plasmid was pColdTF (Cat No. 3365; TaKaRa), which was engineered to enhance the production of disulphide-bonded proteins, thus improving the intracellular yield, purity and solubility of the recombinant proteins. PR1 genes lacking N-terminal signal peptides were cloned into the pColdTF through in-fusion HD Cloning kit (Cat No. 639648; Takara Bio USA, Inc), and recombinant vectors were transformed into Escherichia coli BL21 competent cells. The protein expression was induced by 0.5 mM IPTG at 16 °C for 12 h. For removing proteins improperly folded, only soluble proteins of interest were collected and separated from the high-affinity Ni-charged resin (Qiagen, Valencia, CA, USA), and were visualized by coomassie-stained SDS-PAGE. The elution was dialyzed overnight to appropriate concentrations for further analysis. The expression of empty vector and GUS were used as a control.
Cross-kingdom translocation of StPR1 and its inhibition on P. Infestans in vitro
To test whether the secretory StPR1 proteins could translocate from potato to P. infestans, heterogenous StPR1 and its mutations (StPR1ΔSP, StPR1ΔSPΔTM, and StPR1ΔSPΔGH) tagged with GUS were purified and co-incubated with mycelia and sporangia of P. infestans in vitro. The mycelia that co-incubated with GUS only were used as control. The mycelia and sporangia of P. infestans were washed three times and then used for GUS histochemical staining after co-incubation with purified StPR1 and its mutations for 12 h. To test the cross-kingdom translocation of StPR1 in vivo, the intact StPR1 overexpressing potato lines (35S::StPR1GUS) were infected with a mixture of mycelia and sporangia for four days. The plants used for the inoculation were from the same line, and GUS histochemical assays were conducted every day. Those incubated with 35S::GUS were set as a control. At 2 dpi, the mycelia in/on the plants were also isolated for GUS histochemical assays. To measure the influence of StPR1 on sporangia germination, the 15-day-old sporangia (1 × 107 sporangia/mL) were co-incubated with heterogenous StPR1 at 20 °C, and the sporangia germination was calculated at 24 h post-inoculation. Each experiment was repeated three times.
Measurement of H2O2 and determination of enzyme activity
After being cultured in a liquid medium for 15 days, sporangia and mycelium mixtures of WT and mutants were harvested to measure H2O2 content and the activity of superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT). H2O2 content, and SOD, POD, CAT activities were determined according to procedures specified by the kit manufacturers (Cat No. TO1076, TE0720, TE0423, TE0742; Leagene). Each sample was tested in biological triplicate.
Co-immunoprecipitation assay and western blot analysis
To demonstrate the interaction of PiSNF1 and StPR1, total protein was extracted from S. cerevisiae expressing PiSNF1Myc and/or StPR1HA according to the instructions of Yeast Total Protein Extraction kit (Cat No. KGP650; KeyGEN, Nanjing, China). The total protein was incubated with anti-Myc and protein A/G sepharose (Abcam, ab193262) for 3 h at 4 °C. The immunoprecipitation complexes were washed three times in phosphate-buffered saline as described previously by Cai et al. [22]. The HA- or Myc-tagged proteins were detected using western blot with anti-HA (Cell Signaling Technology, 3724S) and anti-Myc antibodies (Cell Signaling Technology, 9402S), respectively.
For western blot, total protein was extracted using lysis buffer (25 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, and 0.5% NP-40) plus 2% (w/v) polyvinylpolypyrrolidone, 1 mM dithiothreitol, and a protease inhibitor cocktail. Primary antibodies were diluted in 5% BSA as follows: 1:1000 for anti-pAMPKThr172 and anti-APMKα (Cell Signaling Technology, 2535 and 2532, respectively), 1:2000 for anti-yeast beta Actin (Engibody Biotechnology, AT0014), anti-His (Solarbio, M1001020), anti-plant Actin (Abbkine, A01050), and anti-Gus (Agrisera, AS16 3689).
In vitro and in vivo phosphorylation assay
LKB1 is the upstream kinase of AMPK. For in vitro phosphorylation assay, PiSNF1His was incubated with active PiLKB1 in a kinase buffer (25 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 50 mM NaCl, 1 mM DTT, 50 µM ATP and 0.5 µCi of [32P]-ATP). StPR1 proteins (StPR1ΔSP, StPR1ΔSPΔTM, and StPR1ΔSPΔGH) were added into the system to check the influence of StPR1 on PiSNF1 activity. The system without the introduction of PR1 was used as a control. The reaction mixtures were incubated at 30 °C for 30 min, and then for western blot to analyze the phosphorylation level of AMPKα. For in vivo phosphorylation assay, total protein was extracted from the wild-type P. infestans and mutants using lysis buffer, and then for western blot and ELISA analysis. According to the manual protocol, PathScan® Phospho-AMPKα (Thr172) Sandwich ELISA kit (CST, 7955S) was used to detect the phosphorylation level of AMPK; PathScan® Total Acetyl-CoA Carboxylase (ACC) Sandwich ELISA kit (CST, #7996C) and PathScan® Phospho-ACC (Ser79) Sandwich ELISA kit (CST, #7986C) were used to detect the phosphorylation level of ACC.
qRT-PCR analysis
Total RNA was extracted using the EZNA® Total RNA kit I (Omega Bio-tek, Inc. Norcross, GA, R6834-01). The first strand of cDNA was synthesized according to the protocol of the PrimeScript™ RT reagent kit with gDNA Eraser (TaKaRa, Otsu, Japan). StEF1α was used as a reference gene for the normalization of S. tuberosum, and PiActin was used as a reference gene for the normalization of P. infestans. The reaction systems had a volume of 20 μL and were assessed using a CFX96 Real-Time System (Bio-Rad, USA) with TB Green® Premix Ex TaqTM II (Tli RNaseH Plus). The gene-specific primers designed for the qRT-PCR are listed in Supplementary Table 1.
Bioinformatics and statistical analysis
The structures and topology of PR1 were predicted using Protter (http://molbiol-tools.ca/Protein_secondary_structure.htm). The conserved domains of proteins were analyzed online using Pfam (http://pfam.xfam.org/). Multiple nucleic acid and protein sequences were aligned using ClustalX v2.0. The phylogenic tree was constructed using MEGA6.0 with a neighbor-joining method, and there were 1000 bootstrap replicates. Statistical analyses were performed using IBM SPSS v25.0. The differences between control and treatment were determined by Dunnett’s t-test, and the differences among independent groups were determined by One-way ANOVA. The significance levels were *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
Results
PR1 proteins are widely distributed in S. Tuberosum
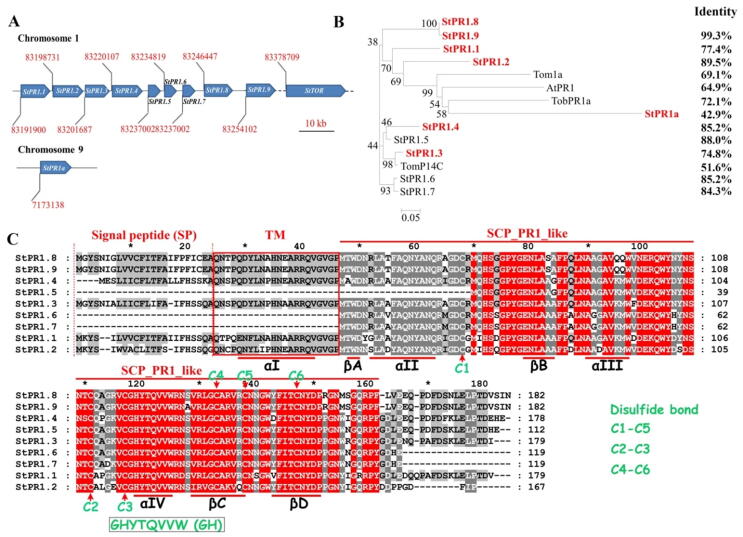
PR1 is a multigene (10 genes) family with StPR1.1–1.9 clustered on chromosome 1, and StPR1a on chromosome 9 (Fig. 1A). The gene information is provided in Supplementary Table 2. StPR1.1–1.4, StPR1.8–1.9, and StPR1a are predicted to be secretory proteins with a signal peptide (SP), while StPR1.5–1.7 are non-secretory proteins (Supplementary Fig. 1A). The nucleotide and protein sequence identity between StPR1.6 and StPR1.7, and that between StPR1.8 and StPR1.9 were over 99%, while the identity between StPR1a and other StPR1 proteins was less than 50% (Fig. 1B). Sequence alignment showed that the conserved GHYTQVVW motif (GH), four highly conserved α-helices (αI-IV) and β-strands (βA-D) were evident in different PR1 proteins. It is worth noting that the αI amino acid sequence, with a conserved motif of LxxHNxARxxVGV (TM, type-specific motif), was evident in secretory but not in non-secretory PR1 proteins (Fig. 1C). Additionally, all PR1 proteins contain six conserved cysteine residues to form disulfide bridges and show a high level of sequence conservation throughout different plant species (Supplementary Fig. 1B). During P. infestans infection, except for PR1.4 and PR1.6/1.7 genes in the sensitive variety ‘Desiree’, the expression of all PR1 genes was up-regulated (Supplementary Fig. 2). Moreover, the transcript levels for all PR1 genes were higher in resistant variety ‘E3′ than in ‘Desiree’ (Supplementary Fig. 2). Among the examined genes, StPR1.2, StPR1.3, and StPR1.8 were up-regulated in both ‘Desiree’ and ‘E3′ during P. infestans infection, suggesting their role in protecting plants against P. infestans (Supplementary Fig. 2). Hence, these genes were selected for further functional characterization.
Fig. 1.
Bioinformatic analysis of StPR1 in Solanum tuberosum. (A) Ten genes encode pathogenesis-related protein 1 (PR1) in the genome of S. tuberosum. StPR1.1–1.9 are clustered on chromosome 1 whereas StPR1a is located at chromosome 9. (B) Phylogenetic analysis of PR1 proteins from S. tuberosum was performed with MEGA6.0 using the neighbor-joining method. One thousand bootstrap replicates were used. The PR1 proteins in red represent secretory proteins in S. tuberosum, while those in black indicate non-secretory proteins. The identity between StPR1.8 and other PR1 proteins was analyzed using Clustal X. The amino acid sequences were collected from the following organisms: St, S. tuberosum; At, Arabidopsis thaliana; Tom, Solanum lycopersicum; Tob, Nicotiana benthamiana. (C) The analysis of conserved domains of StPR1.1–1.9 in S. tuberosum. The secretory PR1 proteins have four highly conserved α-helices (αI-IV) and β-strands (βA-D) to form a α-β-α sandwich structure, while the non-secretory PR1 proteins were absent from the amino acid sequence of αI. All the PR1 proteins have a conserved “GHYTQVVW” motif, and six conserved cysteine residues (Cys) to form three disulfide bonds. The red blank indicates a specific sequence between secretory and non-secretory PR1 proteins, which was abbreviated as TM (type-specific motif). The words in red background represent completely uniform sequences.
StPR1 is involved in plant defense in vivo and shows anti-oomycete activity in vitro
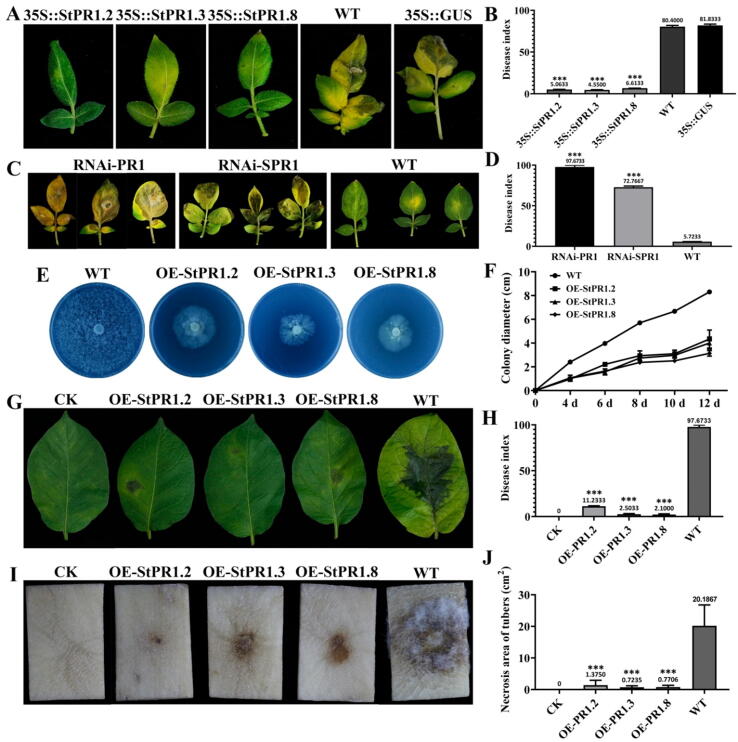
To investigate PR1 function in host defense against P. infestans in vivo, transgenic lines overexpressing StPR1.2, StPR1.3 and StPR1.8 as well as RNA interference lines (RNAi) were generated (Supplementary Fig. 3). No differences were found in terms of plant growth among wild-type (WT), control plants (CK), and transgenic lines (Supplementary Fig. 4). In overexpressed transgenic plants 35S::StPR1.2GUS, 35S::StPR1.3GUS and 35S::StPR1.8GUS, the disease resistance to P. infestans was enhanced, with the disease index (DI) being lower than 7 in all cases, while in WT and control (35S::GUS) plants, the disease symptoms were severe, with the DI reaching 80.4 and 80.8, respectively (Fig. 2A, B). To explore the functional differences between secretory and non-secretory PR1 proteins in disease resistance against P. infestans, either the expression of all PR1 genes (RNAi-PR1) or only that of all secretory PR1-encoding genes were knocked down (RNAi-SPR1). Pathogenicity tests showed that both RNAi-PR1 and RNAi-SPR1 transgenic lines were more susceptible to P. infestans infection compared with the WT, as the DI was 97.7 and 72.8 in the RNAi-PR1 and RNAi-SPR1, respectively; and only 5.7 in the WT (Fig. 2C, D). The disease symptoms and DI were more severe in RNAi-PR1 than those in RNAi-SPR1, suggesting that although both types of PR1 proteins are important, the secretory PR1 proteins may play a more crucial role in disease resistance. Furthermore, StPR1.2, StPR1.3 and StPR1.8 tagged with RFP (OE-StPR1) were heterologously expressed under the Ham34 promoter without signal peptide in P. infestans (Supplementary Fig. 5A-C). Fluorescence analysis showed that all PR1 proteins were expressed successfully in P. infestans, and the colony growth and sporulation were highly inhibited in PR1 overexpression mutants (Fig. 2E, F and Supplementary Fig. 5C, D). Moreover, the virulence of each mutant on leaves was significantly decreased, as the DIs of OE-StPR1.2, OE-StPR1.3 and OE-StPR1.8 were 11.2, 2.5 and 2.1, respectively; whereas the DI of wild-type P. infestans (WT) reached almost 100 (Fig. 2G, H). Additionally, the infection of PR1 overexpression mutants on tubers was also lower than WT (Fig. 2I, J).
Fig. 2.
StPR1 involved in plant defense against Phytophthora infestans in vivo. (A, B) Compared with the wild-type ‘Desiree’ (WT), the disease resistance to P. infestans in transgenic lines overexpressing StPR1.2, StPR1.3 and StPR1.8 was enhanced. 35S::GUS represent GUS in S. tuberosum under 35S promoter. (C, D) Compared with the wild-type resistant variety ‘E3′ (WT), the RNAi lines RNAi-PR1 and RNAi-SPR1 displayed enhanced susceptibility to P. infestans with higher DI, and the disease symptoms on RNAi-PR1 lines were more serious. RNAi-PR1 indicates that all the PR1 genes were knocked down, and the RNAi-SPR1 indicates that only the secretory PR1 genes were knocked down. The DIs of transgenic lines were measured after inoculating 1 × 107 conidia of wild-type P. infestans for 5 days. (E, F) Heterologous overexpression of StPR1.2, StPR1.3 and StPR1.8 in P. infestans inhibited the colony growth and decreased the growth rate of P. infestans. WT, wild-type P. infestans; OE-StPR1, P. infestans mutants overexpressing StPR1. (G, H) Reduced pathogenicity of OE-StPR1.2, OE-StPR1.3 and OE-StPR1.8 on ‘Desiree’ leaves with lower DIs after 5 days of inoculation. The leaves infected by WT were used as a control. (I, J) Heterologous expression of StPR1 in P. infestans decreased the infection on tubers after 10 days of inoculation. The tubers infected by WT were used as control. CK in G-J, leaves and tubers inoculated with sterile water. Each experiment has three biological replicates. Each replication of pathogenicity tests contained at least 10 leaves and tubers, respectively. Error bars indicate standard deviation (SD). Asterisks indicate significant differences (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; t-test).
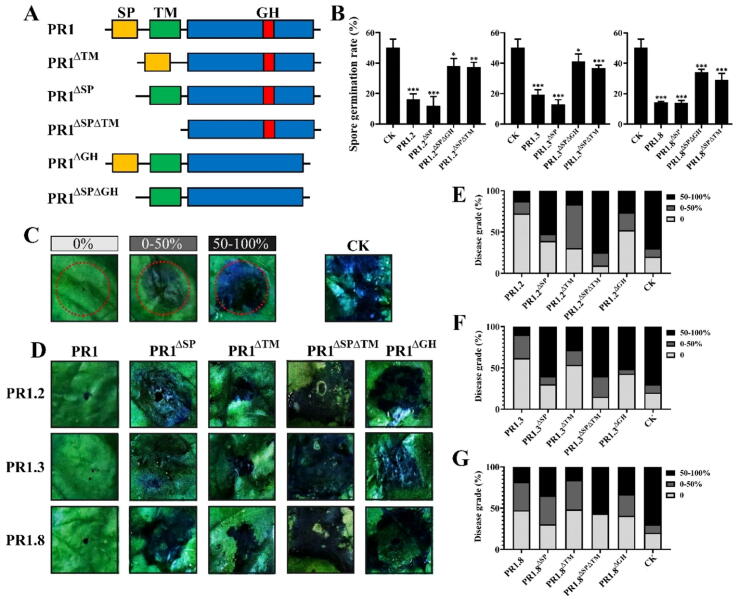
To test the role of SP, TM and GH motifs in StPR1, five types of StPR1 mutations were generated in E. coli or N. benthamiana (Fig. 3A). In vitro tests showed that 1 mM StPR1.2, StPR1.2ΔSP, StPR1.2ΔSPΔGH and StPR1.2ΔSPΔTM could inhibit sporangiospore germination significantly; however, the germination rates of StPR1.2ΔSPΔGH and StPR1.2ΔSPΔTM were higher than those of StPR1.2 and StPR1.2ΔSP (Fig. 3B and Supplementary Fig. 6). A similar inhibition pattern was also observed in StPR1.3, StPR1.8, and their mutants (Supplementary Fig. 6). Further pathogenicity assays for P. infestans were conducted in N. benthamiana leaves that transiently expressed StPR1 and StPR1 mutations tagged with GFP. The abundance of target proteins was measured according to the GFP fluorescence intensity (Supplementary Fig. 7). To precisely measure the severity of necrosis, three necrosis grades for the treated N. benthamiana leaves were considered as defined by Li et al. [23] (Fig. 3C). Data showed that compared with StPR1, the necrosis was higher in StPR1ΔSP, StPR1ΔTM, StPR1ΔSPΔTM and StPR1ΔGH at 3 dpi (P ≤ 0.001), but lower compared with control plants (P ≤ 0.001) (Fig. 3D-G). The results indicate that both TM and GH are critically important for the function of StPR1 proteins in conferring disease resistance upon late blight infection.
Fig. 3.
The GH and TM motifs are important for the anti-oomycete activity of StPR1 against Phytophthora infestans. (A) Overview of five types of StPR1 mutations (StPR1ΔSP, StPR1ΔTM, StPR1ΔSPΔTM, StPR1ΔGH and StPR1ΔSPΔGH) that were changed at the SP, GH and TM motifs. (B) The significant inhibition of prokaryotic expressed StPR1, StPR1ΔSP, StPR1ΔSPΔGH and StPR1ΔSPΔTM on sporangia germination of P. infestans. The sporangia germination rates treated with StPR1ΔSPΔGH and StPR1ΔSPΔTM were higher than those treated with StPR1 and StPR1ΔSP. The sporangia germination treated with GUS was used as a control (CK). (C) Three necrosis grades are defined according to the percentage of the necrosis area developed in the pathogen inoculation area. (D) Phenotypes of plant necrosis triggered by P. infestans in Nicotiana benthamiana leaves transiently expressing PR1, PR1ΔSP, PR1ΔTM, PR1ΔSPΔTM, and PR1ΔGH indicate that the deletion of SP, GH and TM motifs decreased the anti-oomycete activity of StPR1. The necrosis caused by P. infestans in N. benthamiana expressing GFP was used as a control (CK). (E-G) The disease grades of plant necrosis triggered by P. infestans in N. benthamiana expressing StPR1 and mutations. One-tailed t-tests were used to assess statistical significance between means. Error bars indicate standard deviation (SD). In (B, E, F, and G): *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Secretory verification of StPR1
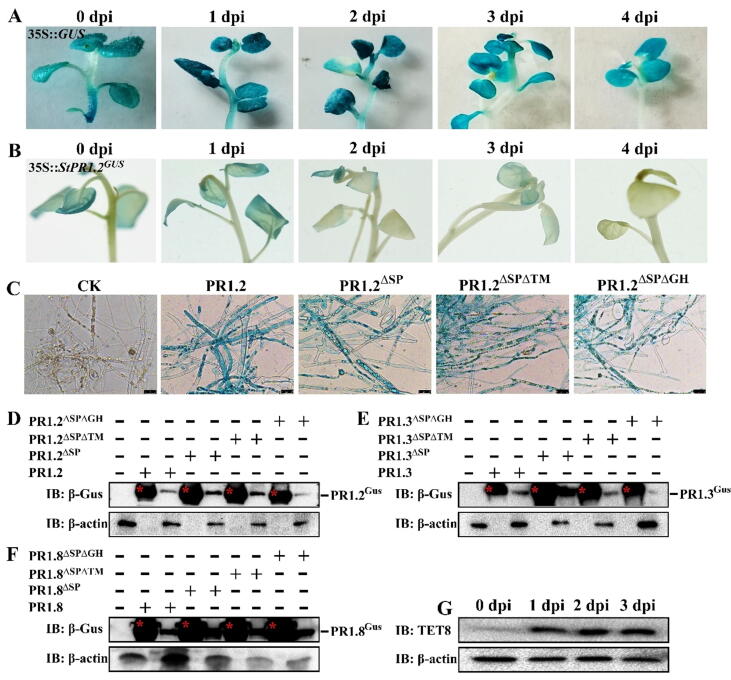
Co-incubation analysis showed that, in accordance with the secreted characteristics of StPR1, GUS staining was attenuated in the 35S::StPR1.2GUS plants as the inoculation day progressed (Fig. 4B), while there was no decrease in the control plant 35S::GUS (Fig. 4A), suggesting that StPR1 was secreted into the extracellular environment during P. infestans infection. Interestingly, GUS histochemical assays showed that StPR1.2GUS could be taken up by the hyphae and germ tube in or around the 35S::StPR1.2GUS plants after 2 days of co-incubation, while the hyphae and sporangia from the control plant 35S::GUS were unstained (Supplementary Fig. 8A-F), implying that the host StPR1 can translocate into the oomycetes. Consistent with the in vivo results, the hyphae co-incubated with StPR1.2, StPR1.2ΔSP, StPR1.2ΔSPΔTM, and StPR1.2ΔSPΔGH in vitro were stained in blue, while those co-incubated only with GUS were unstained (Fig. 4B). Western blot analysis further confirmed the presence of the PR1 protein inside the hyphae (Fig. 4C). StPR1.3/1.8, StPR1.3/1.8ΔSP, StPR1.3/1.8ΔSPΔTM, and StPR1.3/1.8ΔSPΔGH tagged with GUS were also detected in the P. infestans after 12 h of co-incubation (Fig. 4D, E and Supplementary Fig. 8H, I). Secretory verification of StPR1 in vivo and in vitro indicated that the host secretory StPR1 could efficiently translocate into P. infestans cells; moreover, the StPR1ΔSPΔTM could also be taken up by P. infestans, suggesting that once secreted into the external environment, the secretory PR1-specific TM motif was not necessary anymore for the cross-kingdom translocation. Extracellular vesicles (EVs) represent an important mode of intercellular communication by serving as vehicles for transferring proteins, lipids, and RNA [24]. Tetraspanins, such as AtTET8 and AtTET9, are specific EVs markers. S. tuberosum has 20 TETRASPANIN (TET)-like genes, including the StTET8, which is closely related to AtTET8 (Supplementary Fig. 9A). The transcript levels of StTET8 (Supplementary Fig. 9B) and the amount of its corresponding protein increased during P. infestans infection (Fig. 4F). However, it is unclear whether the PR1 proteins are transported through EVs. Hence, the positive correlation between StPR1 and EVs requires further investigation.
Fig. 4.
Cross-kingdom translocation of StPR1 from host to Phytophthora infestans in vivo and in vitro. (A, B) The GUS histochemical assays of 35S::StPR1.2GUS transgenic plants during P. infestans infection (B). The assays in 35S::GUS were used as control (A). The GUS staining was attenuated in 35S::StPR1.2GUS as the inoculation day progressed, while that was not in the 35S::GUS. (C) GUS histochemical assays showed that the prokaryotic expressed StPR1.2 and its mutations tagged with GUS could translocate into P. infestans cells after 12 h of co-incubation in vitro. The hyphae co-incubated with GUS were used as control. The scale bars represent 25 μm. (D, F) Western blot analysis further confirmed the presence of StPR1.2 (D), StPR1.3 (E), StPR1.8 (F) proteins and their mutations in the hyphae of P. infestans cells after 12 h of co-incubation. Purified prokaryotic expressed StPR1 and mutations are marked with red asterisks. Total proteins of P. infestans used for western blot analysis were extracted after 12 h of co-incubation with indicated proteins. β-actin was used as a loading control. (G) The specific marker protein of extracellular vesicles (EVs), TET8, was detected to accumulate significantly during P. infestans infection by western blot analysis. β-actin was used as a loading control.
PiAMPK complex is the target of StPR1 in P. Infestans
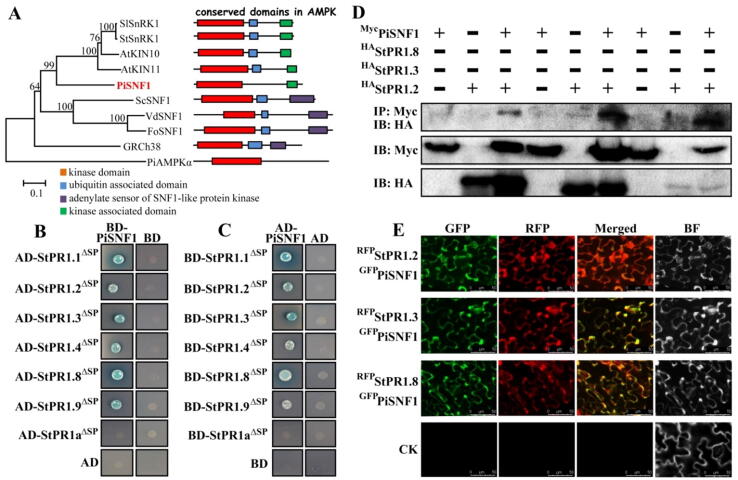
The above results indicate that the secretory StPR1 proteins have an anti-oomycete activity and can translocate in a cross-kingdom manner between S. tuberosum and P. infestans cells. Through multidimensional analysis, target of rapamycin (TOR) signaling pathway was affected mostly by StPR1 proteins, which is a core regulator of cell growth, development, proliferation and death in eukaryotes [25]. Taking the S. cerevisiae as a reference, the corresponding genes of the TOR signaling pathway in P. infestans were firstly identified (Supplementary Table 3). To determine the target proteins of StPR1 in the TOR signaling pathway in P. infestans, StPR1.2 was used as a bait in Y2H assays. Among the examined target proteins, only PiSNF1 could interact with StPR1.2 (Supplementary Fig. 10A). PiSNF1 (PITG_14707) and PiAMPKα (PITG_07910) are two genes encoding the α-catalytic subunit of AMPK in P. infestans. Phylogenetic analysis showed that PiSNF1 belonged to the same clade of the plant catalytic subunit of AMPK with a kinase and kinase associated domain, while PiAMPKα was independent from the clade of catalytic subunits of AMPK from plants and fungi (Fig. 5A). According to Y2H assays, all StPR1ΔSP, except for StPR1aΔSP, interacted with PiSNF1 (Fig. 5B), but not with PiAMPKα (Supplementary Fig. 10B), indicating that StPR1ΔSP can bind specifically with PiSNF1. The same results were also obtained when using the target proteins as baits (Fig. 5C). The interactions of StPR1.2ΔSP, StPR1.3ΔSP, and StPR1.8ΔSP with PiSNF1 were further confirmed by co-immunoprecipitation when co-expressed in S. cerevisiae (Fig. 5D). To investigate whether StPR1 proteins and PiSNF1 exhibited similar subcellular distributions, StPR1.2RFP, StPR1.3RFP, and StPR1.8RFP with PiSNF1GFP were transiently co-expressed in N. benthamiana. Confocal microscopy showed that StPR1.2, StPR1.3, and StPR1.8 displayed the same distribution as PiSNF1 at 3 dpi (Fig. 5E), consistent with the observation that they interact with each other.
Fig. 5.
PiSNF1 is the target of StPR1. (A) Phylogenetic relationship among catalytic subunits of AMPK from different species. The phylogenetic tree was constructed using MEGA6.0 based on an alignment generated with ClustalX. Sl: Solanum lycopersicum, St: Solanum tuberosum, At: Arabidopsis thaliana, Pi: Phytophthora infestans, Sc: Saccharomyces cerevisiae, Vd: Verticillium dahliae, Fo: Fusarium oxysporum, GRCH: human. (B) StPR1 proteins interacts with PiSNF1 as determined by Y2H assays. PiSNF1 on pGBKT7 (BD) vector was used to confirm the interactions with secretory StPR1ΔSP cloned into pGADT7 (AD). Yeast transformants were grown on SD/-Leu/-Trp/-His/-Ade medium with 40 g/mL X-α-gal, and the blue colony indicate interactions. Pictures were taken after 3 d of culture. (C) Y2H confirmed the interaction between PiSNF1 and StPR1ΔSP using the target proteins as baits. (D) Co-IP assays demonstrating the interactions of StPR1.2, StPR1.3, and StPR1.8 with PiSNF1. The proteins used for Co-IP were derived from S. cerevisiae co-expressing PiSNF1Myc with StPR1.2HA, StPR1.3HA, and StPR1.8HA. (E) Co-localization of RFP-labeled StPR1.2, StPR1.3, StPR1.8 and GFP-labeled PiSNF1. The fusion constructs were transiently co-expressed in N. benthamiana and the expressions were analyzed by confocal microscopy at 3 dpi. Bar = 50 μm. The N. benthamiana infiltrated with MMA buffer was used as control.
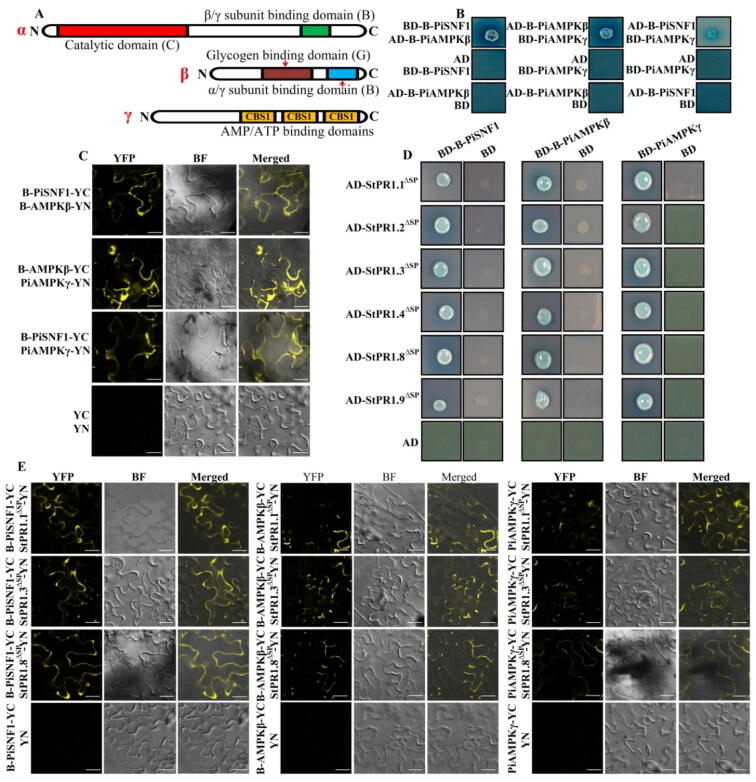
In AMPK, the catalytic subunit (α) and two regulatory subunits (β and γ) interact to form a heterotrimer. In the catalytic subunit of PiSNF1, the kinase catalytic domain (C) was located at 11–263 aa, and the β/γ subunit binding domain (B) was at 493–535 aa; in the β subunit PiAMPKβ (PITG_14586), the glycogen binding domain (G) was found to be located at 98–181 aa, and the α/γ subunit binding domain (B) was located at 211–279 aa. Furthermore, in the γ subunit PiAMPKγ (PITG_17395), there were three AMP/ATP binding domains (CBS) (Fig. 6A). Y2H and BiFC assays showed that B-PiSNF1, B-PiAMPKβ and PiAMPKγ interacted with each other to form a heterotrimer, as in plants and animals (Fig. 6B, C), while the kinase domains in PiSNF1 and glycogen binding domain in PiAMPKβ were not involved in the interaction (Supplementary Fig. 10C). Moreover, the secretory StPR1ΔSP, except for StPR1aΔSP, also interacted with these three subunits through binding domains in Y2H assays (Fig. 6D), implying that the β/γ subunit binding domain is the binding site of PiSNF1 with StPR1, and the α/γ subunit binding domain is the interaction domain of PiAMPKβ with StPR1. The interaction of StPR1ΔSP (StPR1.2ΔSP-nYFP, StPR1.3ΔSP-nYFP, StPR1.8ΔSP-nYFP) with the AMPK subunits (B-PiSNF1-cYFP, B-PiAMPKβ-cYFP, PiAMPKγ-cYFP) was further confirmed by BiFC in N. benthamiana, while there was no fluorescence detected in the controls (Fig. 6E). It is worth noting that StPR1.5, StPR1.6 and StPR1.7 could also interact with PiSNF1 (Supplementary Fig. 10D, F). Further Y2H assays confirmed that StPR1ΔSPΔGH could interact with PiAMPKβ and PiAMPKγ but not PiSNF1, and that StPR1ΔSPΔTM could not interact with PiAMPKγ (Supplementary Fig. 11), suggesting that GH was the binding motif of StPR1 with PiSNF1, and TM was the binding motif of StPR1 with PiAMPKγ.
Fig. 6.
AMPK kinase complex in Phytophthora infestans is the target of StPR1. (A) Structures of the AMPK complex. AMPK exists as a heterotrimer consisting of a catalytic subunit (α) and two regulatory subunits (β and γ). (B, C) Y2H (B) and bimolecular fluorescence complementation (BiFC) (C) assays showed that B-PiSNF1, B-PiAMPKβ and PiAMPKγ interact with each other to form a heterotrimer as those in plant and animal. (D) B-PiSNF1, B-PiAMPKβ and PiAMPKγ interact with secretory StPR1ΔSP in Y2H assays. (E) The AMPK subunits interact with StPR1.2ΔSP, StPR1.3ΔSP, and StPR1.8ΔSP in BiFC assays. The StPR1.2ΔSP, StPR1.3ΔSP, and StPR1.8ΔSP fused with the N terminus of YFP, and AMPK subunits fused with the C terminus of YFP. The constructs were transiently co-expressed in N. benthamiana and examined by confocal microscopy at 3 d post-infiltration (dpi). Only the nYFP halves and single proteins fused to cYFP alone were used as control. The complementation of fluorescence indicates interaction between assayed proteins.
StPR1 affected the PiAMPK phosphorylation of downstream target proteins
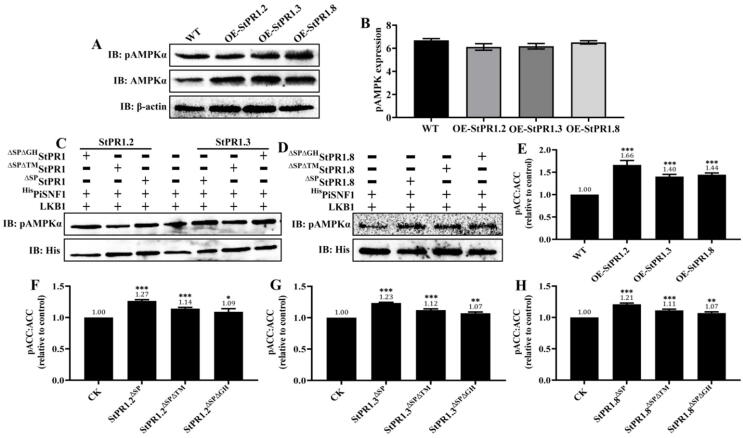
To assess the influence of StPR1 on PiAMPK kinase activity, the total protein of P. infestans mutants OE-StPR1.2, OE-StPR1.3 and OE-StPR1.8 were collected for western blot analysis to measure the phosphorylation levels of AMPK catalytic subunit (pAMPKα) in vivo. Surprisingly, overexpression of StPR1ΔSP in P. infestans did not change the pAMPKα in vivo (Fig. 7A and Supplementary Fig. 12A), which was further confirmed by ELISA analysis (Fig. 7B). To demonstrate whether the TM or GH motif of StPR1 proteins is necessary to affect the pAMPKα, we performed phosphorylation assays in vitro and found that the pAMPKα treated with StPR1.2ΔSP and its mutations (StPR1ΔSPΔTM, StPR1ΔSPΔGH) were not significantly different to that of the control (Fig. 7C and Supplementary Fig. 12B). The unchanged levels of pAMPKα were also observed in StPR1.3ΔSP, StPR1.8ΔSP, and their mutations (Fig. 7C, D and Supplementary Fig. 12C, D), suggesting that the TM and GH motifs of StPR1 proteins were not necessary to alter the AMPKα kinase activity.
Fig. 7.
StPR1 did not change the levels of pAMPKα, while affected the AMPK phosphorylation to downstream target proteins. (A) Western blot of AMPK and phosphorylation of AMPKα (pAMPKα) in wild-type (WT) and PR1 overexpression mutants of P. infestans (OE-StPR1.2, OE-StPR1.3 and OE-StPR1.8). β-actin was used as a loading control. The AMPK expression and pAMPKα level were not different between WT and mutants. (B) Quantification of pAMPKα from WT, OE-StPR1.2, OE-StPR1.3 and OE-StPR1.8 (n = 3) through ELISA analysis. The pAMPKα levels were not significantly different among them. (C, D) Phosphorylation of AMPKα under the involvement of StPR1.2ΔSP, StPR1.3ΔSP, StPR1.8ΔSP, and their mutations through phosphorylation assay in vitro. StPR1.2ΔSP, StPR1.3ΔSP, StPR1.8ΔSP, and their mutations had no influence on the phosphorylation levels of AMPKα. StPR1ΔSP: deleting the signal peptide; StPR1ΔSPΔTM: deleting the signal peptide and TM motif; StPR1ΔSPΔGH: deleting the signal peptide and GH motif. (E) Quantification of pACC:ACC ratio from WT, OE-StPR1.2, OE-StPR1.3 and OE-StPR1.8 (n = 3) through ELISA analysis. The pACC:ACC ratio was increased significantly in OE-StPR1.2, OE-StPR1.3 and OE-StPR1.8. (F, H) Quantification of pACC:ACC ratio after 48 h of treatment with StPR1ΔSP, StPR1ΔSPΔTM, and StPR1ΔSPΔGH (n = 3) through ELISA analysis. The analysis of pACC:ACC ratio of P. infestans without PR1 treatment was used as a control. Results that were significantly different from control (CK) were indicated by asterisks. One-tailed t-tests were used to assess significances. Error bars indicate standard deviation (SD). In E-H: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Even though the in vitro and in vivo phosphorylation assays demonstrated that StPR1 proteins have no influence on the AMPK kinase activity, it was subsequently found that StPR1 proteins altered the AMPK activation to downstream targets. Acetyl-CoA Carboxylase (ACC) is the conserved target substrate of AMPK kinase and is an appropriate choice for detecting the AMPK kinase activity, which is negatively regulated by AMPK at the site of Ser79 in ACC1 [26]. Compared with the wild-type strain, the pACC:ACC ratios were increased by 66.2%, 40.3%, and 44.5% in OE-StPR1.2, OE-StPR1.3 and OE-StPR1.8 mutants, respectively (P ≤ 0.001) (Fig. 7E). Further ELISA analysis showed that StPR1.2ΔSP, StPR1.2ΔSPΔTM, and StPR1.2ΔSPΔGH increased the levels of pACCSer79 and the pACC:ACC ratio in vitro, while the influence of StPR1.2ΔSP on pACC:ACC ratio was stronger than that of StPR1.2ΔSPΔTM and StPR1.2ΔSPΔGH (Fig. 7F). The same results were obtained in P. infestans treated with StPR1.3ΔSP, StPR1.8ΔSP, and their mutations (Fig. 7G, H), implying that TM and GH motifs are necessary for StPR1 to change the AMPK phosphorylation to its downstream targets.
AMPK inhibition in P. Infestans restricted vegetative growth and decreased pathogenicity
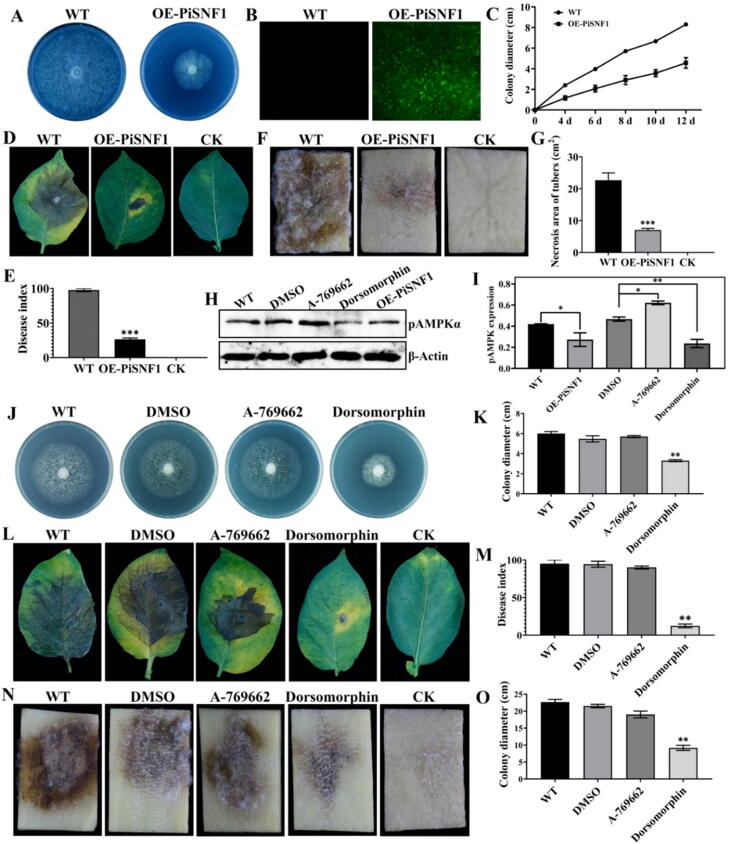
To gain insights into the function of AMPK activity, overexpression mutants of the catalytic subunit encoding gene (PiSNF1) fused with GFP (OE-PiSNF1) were generated (Fig. 8A, B and Supplementary Fig. 13). The overexpression of PiSNF1 decreased the colony growth with aerial hyphae growing almost 50% less compared with the wild-type (Fig. 8C). Likewise, compared with wild-type P. infestans (WT), the pathogenicity of OE-PiSNF1 was lower on leaves and tubers (Fig. 8D-G). Additionally, the phenotype of OE-PiSNF1 resembled that of the P. infestans treated with the AMPK kinase inhibitor dorsomorphin (800 μM; Selleck, cat no. S7306) (Fig. 8J, K). Western blot analysis revealed that the pAMPKα level of OE-PiSNF1 was lower than that of WT, while the pAMPKα level with dorsomorphin treatment was lower than that with DMSO treatment (Fig. 8H, I). However, the phenotype of P. infestans treated with the AMPK activator A-769662 (800 μM; Selleck, cat no. S2697) did not differ from the WT and the DMSO-treated P. infestans (Fig. 8L-O). Overall, these results indicate that the inhibition of AMPK activity decreased the vegetative growth and pathogenicity of P. infestans.
Fig. 8.
Functional characterization of PiSNF1 and the influence of AMPK activation and inhibition on the colony growth and pathogenicity of Phytophthora infestans. (A) The colony phenotype of PiSNF1 overexpression mutant (OE-PiSNF1). (B) GFP fluorescence observation of OE-PiSNF1 under a fluorescence microscope. (C) Overexpression of PiSNF1 decreased the colony growth rate of P. infestans. (D, E) Reduced pathogenicity of OE-PiSNF1 on ‘Desiree’ leaves with lower DI after inoculating for 5 days. (F, G) The pathogenicity of OE-PiSNF1 on tubers was decreased. (H) Western blot analysis of pAMPKα in wild-type P. infestans (WT), wild-type P. infestans treated with DMSO (DMSO), wild-type P. infestans treated with AMPK activator A-769662 (A-769662), wild-type P. infestans treated with AMPK kinase inhibitor dorsomorphin (Dorsomorphin), and OE-PiSNF1. (I) Quantification of pAMPKα in OE-PiSNF1 mutant and under AMPK activator/inhibitor treatment. (J, K) Reduced colony growth of P. infestans following dorsomorphin treatment. The colony growth of P. infestans under the treatment of A-769662 and DMSO was not different from that of wild-type. (L, M) Reduced pathogenicity of Dorsomorphin on ‘Desiree’ leaves with lower DI after 5 days of inoculation. (N, O) Dorsomorphin decreased the infection of P. infestans on tubers. In pathogenicity tests, the leaves or tubers infected by wild-type P. infestans were used as positive control (WT). CK, leaves or tubers inoculated with sterile water. Each pathogenicity test contained at least 10 leaves or tubers, respectively. Each experiment was repeated three times. Asterisks indicate significant differences (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; t-test).
StPR1-PiAMPK disrupted ROS homeostasis and inhibited the expression of RxLR effector-encoding genes in P. Infestans
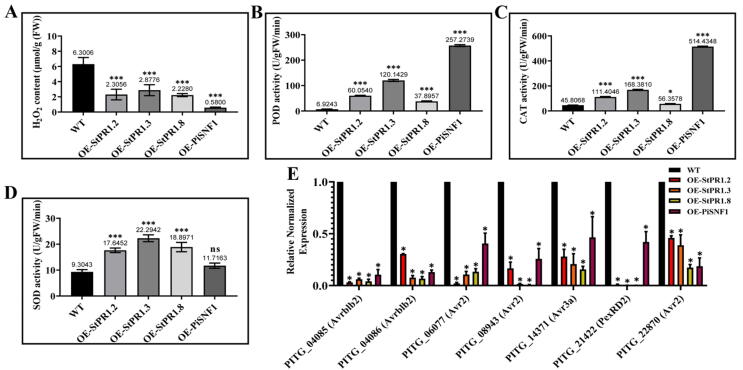
Oxidative stress is critical for establishing pathogenesis [27], [28]. The H2O2 content was significantly lower in OE-StPR1.2, OE-StPR1.3, OE-StPR1.8, and OE-PiSNF1 than in WT (Fig. 9A); while compared with WT, the enzyme activity of ROS scavenging enzymes SOD, POD and CAT was higher in the mutants (Fig. 9B-D). Apart from the changes in ROS homeostasis, the transcript levels of the RxLR effector-encoding genes were down-regulated in the mutants. Ten effectors have been published in P. infestans, including cystatin-like protease inhibitors (EPIC1, EEY55256; EPIC2, A1L016; EPIC2B, D0NBV3), extracellular serine protease inhibitors (EPI1, G8FQ60; EPI10, Q6PQG3), RxLR cytoplasmic effectors (AVRBLB1, D0P3S7; AVRBLB2, D0P1B2; AVR2, D0NN59; AVR3a, A5YTY8), and CRN cytoplasmic effectors (CRN8, Q2M405). Among them, PiAvr3a, a typical RxLR effector protein, targets and stabilizes the E3 ubiquitin ligase CMPG1 in plant cells to inhibit INF-induced necrosis [29]; PiPexRD2, interacts with MAPKKKε and inhibits its kinase activity to obstruct the transmission of the MAPK immune signaling pathway [30]; PiAvr2 could activate the lipid signal transduction pathway and inhibit the PTI defense response induced by INF1 [31]; PiAvrblb2 can block the secretion of C14, a disease resistance protein belonging to cysteine protease [32]. Owing to their role in pathogenicity, the RxLR effector-encoding genes Avrblb2 (PITG_04085, PITG_04086), Avr2 (PITG_06077, PITG_08943, PITG_22870), Avr3a (PITG_14371), and PexRD2 (PITG_21422) were selected for quantitative analysis. qRT-PCR analysis showed that compared with WT, all the examined genes were down-regulated by more than 2-fold in the OE-StPR1.2, OE-StPR1.3, OE-StPR1.8, and OE-PiSNF1 (Fig. 9E), indicating that inhibiting the expression of effectors was one strategy employed by PR1 proteins to protect plants against pathogen infection. In turn, AVR3a was found to bind with StPR1 through Y2H assays (Supplementary Fig. 14).
Fig. 9.
StPR1-PiAMPK signaling pathway is involved in regulating ROS homeostasis and RxLR expression in Phytophthora infestans. (A) The H2O2 content was lower in OE-StPR1.2, OE-StPR1.3, OE-StPR1.8, and OE-PiSNF1 than in wild-type P. infestans (WT). (B-D) Compared with WT, the enzyme activities of SOD, POD, and CAT were significantly increased in OE-StPR1.2, OE-StPR1.3, OE-StPR1.8, and OE-PiSNF1 mutants. (E) qRT-PCR analysis showed that compared with WT, the transcript of RxLR effectors encoding genes were down-regulated in OE-StPR1.2, OE-StPR1.3, OE-StPR1.8, and OE-PiSNF1 mutants. Y-axis indicated the relative normalized expression. Error bars represent standard error from three independent replicates. Asterisks indicate significant differences (*P ≤ 0.05; ***P ≤ 0.001; t-test).
Discussion
PR1 is an indispensable component of innate immune responses in plants under biotic or abiotic stresses and is activated by salicylic acid (SA) application. Its accumulation has long been used as a marker for SA-mediated disease resistance [33]. Many researchers have reported that the PR1 proteins are involved in plant defenses against Phytophthora sp. [5], [6], while their function and mode of action are still unclear. During P. infestans infection, the ten PR1 genes in S. tuberosum were up-regulated and their transcript levels were higher in the resistant variety ‘E3′ than those in the susceptible variety ‘Desiree’ (Supplementary Fig. 2). Silencing of PR1 proteins increased the susceptibility to the pathogen, with the disease index caused by secretory PR1 proteins being higher than that caused by non-secretory PR1 proteins (Fig. 2C, D). Moreover, the overexpression of secretory PR1 proteins in potato enhanced its resistance to late blight, suggesting the critical role of secretory PR1 proteins in plant defense. In vitro analysis indicated that the secretory PR1 proteins inhibited the sporangiospore germination significantly (Supplementary Fig. 6), and the heterologous expression of secretory PR1 proteins in P. infestans inhibited its vegetative growth and pathogenicity (Fig. 2E-H and Supplementary Fig. 5C, D). These results agree with the findings of Niderman et al. [5] and Gamir et al. [6]. Even though there has been considerable interest in PR1 proteins for several decades, its necessary and cross-kingdom translocations in plant defenses remain poorly understood. Pecenková et al. [34] demonstrated that in A. thaliana, the PR1 (AtPR1) has an unconventional secretion pathway, being transported via phosphatidyl-inositol-3-phosphate (PI(3)P)-positive LE/MVB-like vesicles. Besides antimicrobial proteins, EVs also send host sRNAs into pathogens to silence virulent genes and inhibit their pathogenicity [22]. The EVs marker proteins TET8 and TET9 were up-regulated during P. infestans infection; however, further research is necessary to demonstrate the relationship between PR1 proteins and EVs, and to establish whether PR1 proteins are indeed transported via EVs
Since the identification of CAPE peptides and the establishment of their role in biotic and abiotic stress responses, there have been significant advances in our understanding in relation to the function of PR1 proteins [33]. Gamir et al. [6] demonstrated that PR1 proteins possess a sterol-binding activity, and their inhibitory effects on pathogen growth are attributed to the deprivation of sterols from pathogens, including the sterol-auxotroph oomycete pathogens. The CAP domain is responsible for the sterol-binding function. The conserved consensus motif of CAP-derived peptide 1 (CAPE1) is PxGNxxxxxPY, which is a 11 aa peptide cleaved from the C terminus of cysteine-rich secretory proteins PR1 and PR5 in various monocot and dicot plants [11], [35]. The bioactive CAPE1 can induce defense genes to produce immune responses against herbivores, pathogens, and abiotic stresses [11], [35]. It is essential for host defense responses by acting as a defense-signaling molecule rather than as a protein with a direct antimicrobe/herbivore function [33]. In our research, the secretory PR1 proteins were found to have a different function by acting as kinase regulators. In particular, they translocated from the host to pathogen cells to target the Ser/Thr protein kinase AMPK in P. infestans, leading to changes in AMPK phosphorylation of downstream target proteins. The TM and GH motifs are key domains for interacting with AMPK, while the CAP domain is not necessary for the interaction, which probably explains why secretory PR1 proteins have a direct antimicrobial function.
AMPK is a Ser/Thr protein kinase that is widely distributed in eukaryotic cells. It is a highly conserved sensor of energy and metabolism and is one of the key regulators of growth and development. In most species, AMPK exists as an obligate heterotrimer, containing a catalytic subunit (α) and two regulatory subunits (β and γ) [26]. Phosphorylation of Thr172 in the activation loop of AMPK is required for AMPK activation, which is directly mediated by the Ser/Thr kinase LKB1 [36]. Additionally, AMPK can also be phosphorylated on Thr172 in response to a calcium flux, independent of LKB1, by CAMKK2 (also known as CAMKKβ) kinase [37]. According to the alignment sequences of the established physiological substrates, the conserved phosphorylation motif øβxxx(S/T) is the most well-defined substrate recognition motif by AMPK [38]. In S. cerevisiae, the α and γ subunits are annotated as SNF1 and SNF4, respectively [39]; and in fungi, SNF1 is closely related to vegetative growth and pathogenicity [40]. However, the function of SNF1 in oomycetes has not been well established. By constructing SNF1 mutants and treating P. infestans with AMPK activators and inhibitors, AMPK was demonstrated to be involved in vegetative growth and pathogenicity, accompanying with regulating the expression of effector-encoding genes and ROS homeostasis. However, instead of affecting the AMPK activity through T172 phosphorylation, PR1 proteins bound to the three subunits of the AMPK complex, thus changed the AMPK-driven phosphorylation to downstream targets. For example, PR1 proteins strengthen the ACC phosphorylation level, which is negatively regulated by AMPK. It is a discovery with regards to the antimicrobial function of the secretory PR1 proteins.
To date, PR1 proteins are regarded as pathogen effector hubs. Three effectors from different fungal plant pathogens have been found to interact with host PR1 proteins, including ToxA, Tox3, and CSEP0055 [12], [13], [14]. ToxA is a universal effector in Parastagonospora nodorum, Pyrenophora tritici-repentis, and Bipolaris sorokiniana, while Tox3 is unique to P. nodorum [41]. They all interact with amino acids toward the C terminus of PR1, and the ToxA/Tox3-PR1 interaction facilitates pathogen infection of the host [12], [14]. The third effector CSEP0055 that interacted with PR1, was identified in powdery mildew fungus Blumeria graminis [14], but no further information has been reported to describe the biological effects of their interaction. In our work, independent Y2H assays showed that the RxLR effector Avr3a from P. infestans could interact with potato PR1 proteins without specificity, and this interaction in turn regulates the resistance or susceptibility of the host. As PR1 has a critical role in plant immunity, the identification of more effectors from various plant pathogens that target these proteins and associated pathways will be of great interest [33]. Additionally, PR1 reversely inhibited the expression of effectors. More detailed insights into PR-mediated defense responses will contribute to our understanding of pathogen-plant communication, and shed light on the putative roles of StPR1-PiAMPK for disease control.
Conclusion
In conclusion, we found that plant extracellular PR1 play a crucial role in the cross-kingdom trafficking between S. tuberosum and the oomycete pathogen P. infestans. Cross-kingdom trafficking events have been reported in Arabidopsis, which result in the secretion of exosomes to deliver host sRNAs into fungal cells and silence pathogenicity-related genes in the fungal pathogen B. cinerea [22]. However, the same phenomenon was rarely reported in antifungal proteins, such as PR1. Functional analysis showed that secretory PR1 proteins have anti-oomycete activity and can translocate from the host to the pathogen. The translocated PR1 proteins target Ser/Thr protein kinase AMPK in P. infestans, thereby reducing AMPK-driven phosphorylation of downstream proteins, and inhibiting the vegetative growth and pathogenicity. The binding mechanisms of PR1 with AMPK could be a potential target for developing new biofungicide, while the identification of the molecular function of PR1 could provide effective strategies to precisely control plant diseases in various crops, both at pre- and post-harvest stages.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
CRediT authorship contribution statement
Xiumei Luo: Funding acquisition, Investigation, Data curation, Formal analysis, Writing – original draft. Tingting Tian: Data curation, Investigation. Li Feng: Investigation. Xingyong Yang: Writing – review & editing. Linxuan Li: Data curation. Xue Tan: Data curation, Investigation. Wenxian Wu: Investigation. Zhengguo Li: Writing – review & editing. Haim Treves: Writing – review & editing. Francois Serneels: Writing – review & editing. I-Son Ng: Writing – review & editing. Kan Tanaka: Writing – review & editing. Maozhi Ren: Funding acquisition, Conceptualization, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work is funded by the National key R&D program of China (2017YFE0115500, 2020YFA0908002), the National Natural Science Foundation of China (31801911, 31972469, 32002105, U1804231), Central Public-Interest Scientific Institution Basal Research Fund (Y2021XK05), Chengdu Agricultural Science and Technology Center local financial special fund project (NASC2019TI13). Thanks for the support of the Science and Technology Innovation Project of the Chinese Academy of Agricultural Sciences (No.34-IUA-02).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.02.002.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Haverkort A.J., Struik P.C., Visser R.G.F., Jacobsen E. Applied biotechnology to combat late blight in potato caused by Phytophthora infestans. Potato Res. 2009;52(3):249–264. [Google Scholar]
- 2.Dong X. NPR1, all things considered. Curr Opin Plant Biol. 2004;7(5):547–552. doi: 10.1016/j.pbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Van Loon L.C., Van Kammen A. Polyacrylamide disc electro-phoresis of the soluble leaf proteins from Nicotiana tabacum var. 'Samsun' and 'Samsun NN'. II. Changes in protein constitution after infection with tobacco mosaic virus. Virology. 1970;40(2):199–211. doi: 10.1016/0042-6822(70)90395-8. [DOI] [PubMed] [Google Scholar]
- 4.van Loon L.C., Rep M., Pieterse C.M.J. Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol. 2006;44(1):135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- 5.Niderman T., Genetet I., Bruyere T., Gees R., Stintzi A., Legrand M., et al. Pathogenesis-Related PR-1 proteins are antifungal (isolation and characterization of three 14-kilodalton proteins of tomato and of a basic PR-1 of tobacco with inhibitory activity against Phytophthora infestans) Plant Physiol. 1995;108(1):17–27. doi: 10.1104/pp.108.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gamir J., Darwiche R., van't Hof P., Choudhary V., Stumpe M., Schneiter R., et al. The sterol-binding activity of Pathogenesis-Related Protein 1 reveals the mode of action of an antimicrobial protein. Plant J. 2017;89(3):502–509. doi: 10.1111/tpj.13398. [DOI] [PubMed] [Google Scholar]
- 7.Li J., Du L.F., Liu X., Du S.W., Huang X.H., Jiang J.H. Expression and purification of tobacco PR-1a protein tor function analysis. Asian J Chem. 2009;21:3697–3707. [Google Scholar]
- 8.Choudhary V., Schneiter R. Pathogen-Related Yeast (PRY) proteins and members of the CAP superfamily are secreted sterol-binding proteins. P Natl Acad Sci USA. 2012;109(42):16882–16887. doi: 10.1073/pnas.1209086109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lincoln J.E., Sanchez J.P., Zumstein K., Gilchrist D.G. Plant and animal PR1 family members inhibit programmed cell death and suppress bacterial pathogens in plant tissues. Mol Plant Pathol. 2018;19(9):2111–2123. doi: 10.1111/mpp.12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sung Y.-C., Outram M.A., Breen S., Wang C., Dagvadorj B., Winterberg B., et al. PR1-mediated defence via C-terminal peptide release is targeted by a fungal pathogen effector. New Phytol. 2021;229(6):3467–3480. doi: 10.1111/nph.17128. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y.-L., Lee C.-Y., Cheng K.-T., Chang W.-H., Huang R.-N., Nam H.G., et al. Quantitative peptidomics study reveals that a wound-induced peptide from PR-1 regulates immune signaling in tomato. Plant Cell. 2014;26(10):4135–4148. doi: 10.1105/tpc.114.131185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breen S., Williams S.J., Winterberg B., Kobe B., Solomon P.S. Wheat PR-1 proteins are targeted by necrotrophic pathogen effector proteins. Plant J. 2016;88:13–25. doi: 10.1111/tpj.13228. [DOI] [PubMed] [Google Scholar]
- 13.Lu S., Faris J.D., Sherwood R., Friesen T.L., Edwards M.C. A dimeric PR-1-type pathogenesis-related protein interacts with ToxA and potentially mediates ToxA-induced necrosis in sensitive wheat. Mol Plant Pathol. 2014;15(7):650–663. doi: 10.1111/mpp.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W.J., Pedersen C., Kwaaitaal M., Gregersen P.L., Mørch S.M., Hanisch S., et al. Interaction of barley powdery mildew effector candidate CSEP0055 with the defence protein PR17c. Mol Plant Pathol. 2012;13(9):1110–1119. doi: 10.1111/j.1364-3703.2012.00820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang G., Tang L., Gong Y., Xie J., Fu Y., Jiang D., et al. A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 2018;217(2):739–755. doi: 10.1111/nph.14842. [DOI] [PubMed] [Google Scholar]
- 16.Xiong F., Zhang R., Meng Z., Deng K., Que Y., Zhuo F., et al. Brassinosteriod Insensitive 2 (BIN2) acts as a downstream effector of the Target of Rapamycin (TOR) signaling pathway to regulate photoautotrophic growth in Arabidopsis. New Phytol. 2017;213(1):233–249. doi: 10.1111/nph.14118. [DOI] [PubMed] [Google Scholar]
- 17.Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40(3):428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 18.Millam S. Potato (Solanum tuberosum L.). In: Wang K. (eds) Agrobacterium protocols Volume 2. Methods in Molecular Biology. Springer, AG, Switzerland. 2006; 344:25-35. [DOI] [PubMed]
- 19.Wu D., Navet N., Liu Y., Uchida J., Tian M. Establishment of a simple and efficient Agrobacterium-mediated transformation system for Phytophthora palmivora. BMC Microbiol. 2016;16:204. doi: 10.1186/s12866-016-0825-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whisson S.C., Boevink P.C., Moleleki L., Avrova A.O., Morales J.G., Gilroy E.M., et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450(7166):115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 21.Wu X., Gu B., Liu H., Zhu Q. China standard press; Beijing: 2000. Pesticide-Guidelines for the field efficacy trials (I)—fungicides against late blight of potato; pp. 483–486. [Google Scholar]
- 22.Cai Q., Qiao L., Wang M., He B., Lin F.M., Palmquist J., et al. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science. 2018;360(6393):1126–1129. doi: 10.1126/science.aar4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li T, Wang Q, Feng R, Li L, Ding L, Fan G, et al. Negative regulators of plant immunity derived from cinnamyl alcohol dehydrogenases are targeted by multiple Phytophthora Avr3a-like effectors. New Phytol 2019; doi: 10.1111/nph.16139. [DOI] [PubMed]
- 24.Raposo G., Stoorvogel W. Extracellular vesicles: Exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eltschinger S., Loewith R. TOR complexes and the maintenance of cellular homeostasis. Trends Cell Biol. 2016;26(2):148–159. doi: 10.1016/j.tcb.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Mihaylova M.M., Shaw R.J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13(9):1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mqa B., Hxa C., Kang H.A., Maao A., Dh D., An E., et al. Host-pathogen interaction between asian citrus psyllid and entomopathogenic fungus (cordyceps fumosorosea) is regulated by modulations in gene expression, enzymatic activity and hlb-bacterial population of the host. Comp Biochem Phys C. 2012;248 doi: 10.1016/j.cbpc.2021.109112. [DOI] [PubMed] [Google Scholar]
- 28.Qasim M., Lin Y., Dash C.K., Bamisile B.S., Ravindran K., Islam S.U., et al. Temperature-dependent development of asian citrus psyllid on various hosts, and mortality by two strains of isaria. Microb Pathog. 2018;119:109–118. doi: 10.1016/j.micpath.2018.04.019. [DOI] [PubMed] [Google Scholar]
- 29.Bos J.I.B., Armstrong M.R., Gilroy E.M., Boevink P.C., Hein I., Taylor R.M., et al. Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. P Natl Acad Sci USA. 2010;107(21):9909–9914. doi: 10.1073/pnas.0914408107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King S.R.F., McLellan H., Boevink P.C., Armstrong M.R., Bukharova T., Sukarta O., et al. Phytophthora infestans RXLR effector PexRD2 interacts with host MAPKKK epsilon to suppress plant immune signaling. Plant Cell. 2014;26:1345–1359. doi: 10.1105/tpc.113.120055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turnbull D., Yang L., Naqvi S., Breen S., Welsh L., Stephens J., et al. RXLR effector AVR2 up-regulates a brassinosteroid-responsive bHLH transcription factor to suppress immunity. Plant Physiol. 2017;174(1):356–369. doi: 10.1104/pp.16.01804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bozkurt T.O., Schornack S., Win J., Shindo T., Ilyas M., Oliva R., et al. Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. P Natl Acad Sci USA. 2011;108(51):20832–20837. doi: 10.1073/pnas.1112708109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breen S., Williams S.J., Outram M., Kobe B., Solomon P.S. Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 2017;22(10):871–879. doi: 10.1016/j.tplants.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 34.Pečenková T., Pleskot R., Žárský V. Subcellular localization of Arabidopsis pathogenesis-related 1 (PR1) protein. Int J Mol Sci. 2017;18(4):825. doi: 10.3390/ijms18040825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chien P.S., Nam H.G., Chen Y.R. A salt-regulated peptide derived from the CAP superfamily protein negatively regulates salt-stress tolerance in Arabidopsis. J Exp Bot. 2015;66(17):5301–5313. doi: 10.1093/jxb/erv263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw R.J., Kosmatka M., Bardeesy N., Hurley R.L., Witters L.A., DePinho R.A., et al. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. P Natl Acad Sci USA. 2004;101(10):3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hurley R.L. The Ca2+/Calmodulin-dependent orotein kinase kinases are AMP-activated protein kinase kinases. J Biol Chem. 2005;280:29060–29066. doi: 10.1074/jbc.M503824200. [DOI] [PubMed] [Google Scholar]
- 38.Scott J.W., Norman D.G., Hawley S.A., Kontogiannis L., Hardie D.G. Protein kinase substrate recognition studied using the recombinant catalytic domain of AMP-activated protein kinase and a model substrate. J Mol Biol. 2002;317(2):309–323. doi: 10.1006/jmbi.2001.5316. [DOI] [PubMed] [Google Scholar]
- 39.Amodeo G.A., Rudolph M.J., Tong L. Crystal structure of the heterotrimer core of Saccharomyces cerevisiae AMPK homologue SNF1. Nature. 2007;449(7161):492–495. doi: 10.1038/nature06127. [DOI] [PubMed] [Google Scholar]
- 40.Tzima A.K., Paplomatas E.J., Rauyaree P., Ospina-Giraldo M.D., Kang S. VdSNF1, the sucrose nonfermenting protein kinase gene of Verticillium dahliae, is required for virulence and expression of genes involved in cell-wall degradation. Mol Plant Microbe In. 2011;24(1):129–142. doi: 10.1094/MPMI-09-09-0217. [DOI] [PubMed] [Google Scholar]
- 41.McDonald M.C., Ahren D., Simpfendorfer S., Milgate A., Solomon P.S. The discovery of the virulence gene ToxA in the wheat and barley pathogen Bipolaris sorokiniana. Mol Plant Pathol. 2018;19(2):432–439. doi: 10.1111/mpp.12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.