Graphical abstract
Hyperlipidemia-associated cytokines and disturbed flow upregulate SOX4 expression in ECs, where SOX4 promotes EndoMT and atherogenesis. Metformin pharmacologically suppresses SOX4 level in ECs. EC, endothelial cell; IL-1β, interleukin-1β; TGF-β1, transforming growth factor β1.
Keywords: Atherosclerosis, EndoMT, Endothelial cells, Shear stress, Single-cell RNA sequencing
Highlights
-
•
Hyperlipidemia enriched mesenchymal markers in EC cluster of atherosclerotic mice.
-
•
EC cluster can be partitioned into ‘endothelial-like’ and ‘mesenchymal-like’ subpopulations.
-
•
SOX4 was upregulated in mouse and human atherosclerotic tissues.
-
•
SOX4 upregulation was observed in human aneurysmal tissues.
-
•
EC-specific SOX4 overexpression promoted EndoMT, endothelial dysfunction and atherogenesis.
-
•
Hyperlipidemia-related cytokines and disturbed flow upregulated endothelial SOX4 level.
Abstract
Introduction
Atherosclerotic complications represent the leading cause of cardiovascular mortality globally. Dysfunction of endothelial cells (ECs) often initiates the pathological events in atherosclerosis.
Objectives
In this study, we sought to investigate the transcriptional profile of atherosclerotic aortae, identify novel regulator in dysfunctional ECs and hence provide mechanistic insights into atherosclerotic progression.
Methods
We applied single-cell RNA sequencing (scRNA-seq) on aortic cells from Western diet-fed apolipoprotein E-deficient (ApoE−/−) mice to explore the transcriptional landscape and heterogeneity of dysfunctional ECs. In vivo validation of SOX4 upregulation in ECs were performed in atherosclerotic tissues, including mouse aortic tissues, human coronary arteries, and human renal arteries. Single-cell analysis on human aortic aneurysmal tissue was also performed. Downstream vascular abnormalities induced by EC-specific SOX4 overexpression, and upstream modulators of SOX4 were revealed by biochemical assays, immunostaining, and wire myography. Effects of shear stress on endothelial SOX4 expression was investigated by in vitro hemodynamic study.
Results
Among the compendium of aortic cells, mesenchymal markers in ECs were significantly enriched. Two EC subsets were subsequently distinguished, as the ‘endothelial-like’ and ‘mesenchymal-like’ subsets. Conventional assays consistently identified SOX4 as a novel atherosclerotic marker in mouse and different human arteries, additional to a cancer marker. EC-specific SOX4 overexpression promoted atherogenesis and endothelial-to-mesenchymal transition (EndoMT). Importantly, hyperlipidemia-associated cytokines and oscillatory blood flow upregulated, whereas the anti-diabetic drug metformin pharmacologically suppressed SOX4 level in ECs.
Conclusion
Our study unravels SOX4 as a novel phenotypic regulator during endothelial dysfunction, which exacerbates atherogenesis. Our study also pinpoints hyperlipidemia-associated cytokines and oscillatory blood flow as endogenous SOX4 inducers, providing more therapeutic insights against atherosclerotic diseases.
Nomenclature
- AA
Aortic arch
- ApoE
Apolipoprotein E
- ATAA
Ascending thoracic aortic aneurysm
- EC
Endothelial cell
- EndoMT
Endothelial-to-mesenchymal transition
- FB
Fibroblast
- FN1
Fibronectin
- GSEA
Gene set enrichment analysis
- HAEC
Human aortic endothelial cells
- HUVEC
Human umbilical vein endothelial cells
- IL-1β
Interleukin-1β
- LSS
Laminar shear stress
- MP
Macrophage
- ND
Normal diet
- OSS
Oscillatory shear stress
- RBC
Red blood cell
- scRNA-seq
Single-cell RNA sequencing
- siRNA
Small interfering RNA
- TA
Thoracic aorta
- TF
Transcription factor
- TGF-β
Transforming growth factor β
- UMAP
Uniform manifold approximation and projection
- VIM
Vimentin
- VSMC
Vascular smooth muscle cell
- WD
Western diet
Introduction
Atherosclerosis and its clinical complications, such as aneurysm and myocardial infarction, remain the leading cause of mortality worldwide [1]. Atherosclerosis is a progressive vascular disorder characterized by plaque formation due to accumulation of lipids, fibrous elements and cellular components underneath the thickened arterial intima [2]. Sharing similar risk factors like hypercholesterolemia and hypertension, atherosclerosis may predispose patients to aortic aneurysm [3], the weakening of arterial wall and subsequent formation of outward bulge. The monolayer of endothelial cells (ECs) constitutes the inner lining of blood vessels, responsible for transduction of biomechanical signal and regulation of vascular tone [4]. Dysfunction of ECs, characterized by imbalanced endothelium-dependent vasodilatation and vasoconstriction, signifies atherosclerotic lesion initiation [5]. However, detailed transcriptional landscape, regulatory network and molecular signature of ECs during atherosclerosis are not comprehensively understood.
Different cells are present in the vasculature, including ECs, vascular smooth muscle cells (VSMCs), fibroblasts (FBs), and macrophages (MPs). Previous efforts have been made to uncover transcriptional profiles of aortic cells during cardiovascular complications. In 2018, Winkels et al. and Cochain et al. applied single-cell RNA sequencing (scRNA-seq) to explore the heterogeneity of aortic leukocytes and macrophages in murine atherosclerosis, respectively [6], [7]. Recently, some studies have interrogated endothelial heterogeneity at single-cell resolution. In 2019, Kalluri et al. successfully identified transcriptionally distinct EC subpopulations in normal mouse aortae [8]. In 2020, He et al. comprehensively profiled spatial heterogeneity of vascular cells along mouse aortae [9]. Enlightened by these important recent findings, we aim to further deepen the understanding towards the role of ECs during atherosclerotic progression.
Chronic vascular inflammation provides a pro-atherogenic microenvironment that continually stimulates ECs while this unfavorable condition is associated with increased consumption of Western diets [10]. The presence of secreted cytokines, like interleukin-1β (IL-1β) and transforming growth factor β (TGF-β) in such microenvironment prompts the phenotypic shift of ECs [11]. This phenotypic alteration, termed endothelial-to-mesenchymal transition (EndoMT), aggravates atherosclerosis [12], by which mesenchymal-like ECs gradually express VSMC markers, thus worsening arterial stiffening [13].
Furthermore, vascular ECs are continually exposed to mechanical stimuli produced by blood flow (i.e. shear stress), where different shear patterns trigger distinct cellular responses. Steady laminar shear stress (LSS) in straight region of vasculature elicits anti-inflammatory and anti-atherogenic effects, whereas oscillatory shear stress (OSS) caused by disturbed flow in branched vascular region corresponds to a pro-inflammatory and pro-atherogenic factor [14]. However, very few studies have compared the genetic hallmarks of mature and functional ECs with mesenchymal-like ECs, particularly in the context of atherosclerotic disease.
By respectively comparing the genetic profiles of functional ECs and dysfunctional ECs in normal and atherosclerotic aortae from murine of the same genetic background (e.g. the well-established ApoE−/− mouse model [15]), we might identify novel molecular targets to provide diagnostic and therapeutic insights into atherosclerosis. By feeding ApoE−/− mice with either a normal chow, a standard diet of low cholesterol (0.02–0.03%) and fat (5–6%) contents, or a Western diet, the ‘humanized diet’ containing higher cholesterol (0.15%) and fat (∼21%) contents, normal and atherosclerotic aortae could be experimentally generated for genetic assays [16].
In this study, we applied scRNA-seq approach to (i) explore transcriptional landscape and heterogeneity of aortic ECs during atherosclerosis; (ii) identify novel and druggable molecular targets in dysfunctional ECs; (iii) link the newly identified molecular target to pathological conditions in vasculature; and (iv) correlate findings in experimental atherosclerosis to arteries and ECs in clinical specimens of human atherosclerotic diseases. Our findings shall open up new opportunities for identifying alternative therapeutic strategies against atherosclerotic diseases.
Materials and methods
Materials and reagents
ACh, hyaluronidase, metformin, NG-nitro-L-arginine methyl ester (L-Name), Phe, SNP, and TGF-β1 were purchased from Sigma-Aldrich. IL-1β was purchased from R&D Systems (Minneapolis, USA). Collagenase type I and DNase I were ordered from Worthington Biochemical Corporation (New Jersey, USA). Meanwhile, primer pairs were ordered from Tech Dragon Limited (Hong Kong, China).
Ethics statement
All experiments involving animals were conducted according to the ethical policies and procedures approved by the Animal Experimentation Ethics Committee of The Chinese University of Hong Kong (approval No. 18–114-TBR). The use of human arterial tissues was approved by the ethics committee of clinic research, The Chinese University of Hong Kong (approval No. 2014.468 and 2018.055); and the Hospital Human Subjects Review Committee of Tri-Service General Hospital in Taipei (approval NO. TSGHIRB-110305023). All participants signed an informed consent before inclusion into the study.
Animal experimentation
All animal experiments were conducted in compliance with the ARRIVE guidelines [17], and the Guide for the Care and Use of Laboratory Animals issued by the National Institutes of Health. Male ApoE−/− mice (10 weeks old, 23–26 g) and C57BL/6 mice (8 weeks old, 22–25 g) were supplied by the CUHK Laboratory Animal Services Center. Some male ApoE−/− mice were fed a normal diet (ND) or Western diet (WD; D12336; Research Diets, Inc., New Jersey, USA) for 20 weeks to develop advanced stage of atherosclerosis. Some ApoE−/− mice were subjected to tail vein injection of AAV-Vector or AAV-SOX4 (AAV1-ICAM2-SOX4; Vigene Biosciences, Maryland, USA) for EC-specific SOX4 overexpression, prior to WD feeding. Body weights and lipid profiles of ApoE−/− mice were recorded (Fig. S2 and Fig. S13). Mice were anaesthetized by intraperitoneal injection of a mixture of xylazine (10 mg/kg) and ketamine (80 mg/kg). Some C57BL/6 mice were sacrificed by CO2 suffocation to dissect thoracic aortae (TA) and aortic arches (AA) for en face immunofluorescence staining and Western blotting. Some ApoE−/− mice were sacrificed by CO2 suffocation to dissect aortae for ex vivo organ culture, Western blotting and immunofluorescence staining. All mice were housed in individually well-ventilated caging systems at constant temperature (23 ± 1 °C) and humidity (55% ± 5%) under a 12 h light:12 h dark cycle. Free access to laboratory food pellets and water were guaranteed.
Aortic CD45− cell isolation
To prepare a single-cell suspension of aortic cells, a bead-based strategy to screen viable CD45− cells was applied. The whole mouse aortae (n = 3) of ND and WD groups of ApoE−/− mice were isolated after intracardiac perfusion with ice-cold PBS. The finely cut aortae were incubated in 1x aortic dissociation enzyme cocktail (450 U/mL collagenase type I, 60 U/mL DNase I, and 60 U/mL hyaluronidase) at 37 °C for 1 h. The digestion was halted by adding FACS buffer (2 mM EDTA, 2% FBS). The cell suspension was transferred through 50 μm cell strainer for subsequent incubation with dead cell removal microbeads (Miltenyi Biotec, North Rhine-Westphalia, Germany) at room temperature for 15 min. Viable cells were collected by transferring cell suspension through a MS column (Miltenyi Biotec) on a magnet. Later, the viable cells were incubated with CD45 microbeads (Miltenyi Biotec) on ice for 15 min for negative selection through a LS column (Miltenyi Biotec) on a magnet. The flow-through cells were centrifuged and resuspended in 0.04% BSA-PBS for droplet-based scRNA-seq.
Droplet-based scRNA-seq
Single-cell suspension was made into sequencing library using the Chromium Single Cell 3′ Reagent Kits (v2 Chemistry) on a Chromium Controller (10x Genomics, California, USA) following the manufacturer’s instructions. In brief, 3500 sorted single cells (0.04% BSA-PBS) were added to each channel for partition into gel beads in emulsion within the Chromium Controller. Cell lysis, barcoded reverse transcription of RNA, amplification, fragmentation, adaptor ligation and indexing were accomplished sequentially. Libraries were consequently sequenced on an Illumina HiSeq 2500 instrument (Macrogen, Seoul, Korea).
scRNA-seq data processing and functional analysis
Raw scRNA-seq data from individual samples of ND and WD groups, were processed by using Cell Ranger (3.0.2) with the mouse reference genome and annotation (refdata-cellranger-mm10-3.0.0) (10x Genomics). The filtered feature barcode matrices generated by Cell Ranger were then passed to Seurat (3.1.5) [18]. Only cells featured by threshold gene numbers (>200 genes and < 5000 genes) and mitochondrial reads (<10%) were sorted for further analyses in order to remove potential empty droplets and doublets. DoubletFinder was used to predict doublets in the pre-processed data of ND and WD groups, respectively, following the authors’ example (https://github.com/chris-mcginnis-ucsf/DoubletFinder) [19]. The first 14 principal components were used for UMAP dimension reduction. In the step for estimating doublet rate using the doubletFinder_v3 function, the doublet rate was assumed to be 1%, as suggested by 10x Genomics (https://kb.10xgenomics.com/hc/en-us/articles/360001378811-What-is-the-maximum-number-of-cells-that-can-be-profiled-), pN was set at 0.05, pK was set at 0.1 and 0.01 for the ND and WD groups, respectively. The doublet label predicted by DoubletFinder would be overlaid with the integrated data in the following step. Before integration of the two samples, normalization and variable feature identification of individual samples were performed. The first 30 dimensions of the canonical correlation vectors were used for anchoring. The cell cycle scores, for both S phase and G2/M phase, were calculated, based on a previously reported list of cell cycles markers [20]. Total RNA count and percentage of reads, together with the cell cycle scores, were used to scale the expression data. After data scaling, principal component analysis was performed and the first 14 principal components were selected for further dimension reduction by uniform manifold approximation and projection (UMAP). Cell clustering was performed using the first 14 principal components and at a resolution of 0.1, using the “FindClusters” function, while the marker genes were identified, using the “FindAllMarkers” function with both “min.pct” and “logfc.threshold” set at 0.5. Differential gene expression analysis was performed between ND and WD groups for each cluster, using the “FindMarkers” function with “min.pct” set at 0.5. These lists of differentially expressed genes would be used for gene regulatory network analysis in later steps. In addition, the fold-changes of all genes between ND and WD groups for each cluster were calculated, using the “FindMarkers” function with both “min.pct” and “logfc.threshold” set at 0. These complete lists of genes with their fold-changes between ND and WD groups were used for gene set enrichment analysis (4.0.3) [21], in combination with the Molecular Signature Database hallmark gene set collection [22].
Gene regulatory network analysis for EC cluster
The list of differentially expressed genes of the EC cluster generated by Seurat was used for gene regulatory network analysis by using QIAGEN IPA (QIAGEN Inc., https://digitalinsights.qiagen.com/IPAQiagen). Briefly, the gene list with the average log (fold-change) and adjusted p-value were imported. The “Core Analysis” function was used for genes meeting the criteria of |log (fold-change)| > 0.5 and adjusted p-value < 0.05. The output gene networks were merged and manually curated to retain the differentially expressed transcription factors and a minimal set of differentially expressed regulatees, generating the gene regulatory network of EC cluster.
Gene interaction network between EC and VSMC clusters
The lists of differentially expressed genes of the EC and VSMC clusters generated by Seurat were used for gene interaction network analysis, again, using QIAGEN IPA. Briefly, the gene lists with the average log (fold-change) and adjusted p-value were imported. The “Core Analysis” function was used for genes meeting the criteria of |log (fold-change)| > 0.5 and adjusted p-value < 0.05. The differentially expressed genes annotated as locating in extracellular space were compared between the two clusters. After that, differentially expressed genes locating in extracellular space, which were unique to the VSMC cluster, were combined with differentially expressed non-extracellular space locating genes of the EC cluster, and this newly generated list were used for “Core Analysis”. The output gene networks were manually curated to output the gene interaction network containing at least one extracellular space located gene of the VSMC cluster, and at least one differentially expressed transcription factor of the EC cluster.
EC subclustering
In order to further identify different subpopulations of ECs, the entire EC cluster was extracted for subclustering using Seurat. Principal component analysis was performed and the first 14 components were selected for further dimension reduction using UMAP. Cell clustering was performed using the first principal components and at a resolution of 0.2, using the “FindClusters” function, while the marker genes were identified, using the “FindAllMarkers” function with both “min.pct” and “logfc.threshold” set at 0.5. The fold-changes of all genes between ND and WD groups for each EC cluster were calculated, using the “FindMarkers” function with both “min.pct” and “logfc.threshold” set at 0. These complete lists of genes with their fold-changes between ND and WD groups were used for gene set enrichment analysis, in combination with the Molecular Signature Database hallmark gene set collection.
Human arteries
Human diseased coronary arteries (n = 7) and control internal thoracic arteries (n = 7) were harvested from patients with end-stage heart failure, undergoing heart transplantation at Tri-Service General Hospital in Taipei. Human renal arteries (n = 11) were collected from patients subjected to nephrectomy. All experiments conformed to the principles set out in the 1964 Declaration of Helsinki and the Department of Health and Human Services Belmont Report. These samples were from Department of Surgery in Prince of Wales Hospital, CUHK. The samples were fixed in paraformaldehyde solution (4% in PBS) and later embedded in paraffin. The paraffin-embedded specimen were sliced into 5 μm cross sections for Masson’s trichrome staining, immunohistochemistry staining and immunofluorescence staining. Some human renal arteries were freshly subjected to ex vivo IL-1β treatment (10 ngml−1, 48 h) prior to immunohistochemistry and Western blotting.
Human aortic aneurysm scRNA-seq analysis
scRNA-seq data from ascending aortic samples of control individuals and patients with ascending thoracic aortic aneurysm (ATAA) were extracted from a publicly available dataset at NCBI BioProject with accession number PRJNA649846 [23]. This scRNA-seq data were processed and re-analyzed for cell type clustering following the procedure described in section scRNA-seq data processing and functional analysis, with the following modifications: (1) no cell filtering was performed as the data was clean, (2) the first 18 principal components were used for dimension reduction, and (3) the resolution for cell clustering was set at 0.09. Similarly, EC sub-clustering was performed following procedure described in section EC sub-clustering, with the following modifications: (1) the first six principal components were used for dimension reduction and (2) the resolution for clustering ECs was set at 0.1.
Cell culture
Human umbilical vein endothelial cells (HUVECs; CC-2519; Lonza, MD, USA) were cultured in DMEM/F12 medium, supplemented with 10% FBS, EC growth supplement (50 μgmL−1; Lonza) and antibiotics, until confluency. Human aortic endothelial cells (HAECs; C-006-5C; Invitrogen) were cultured in medium 200 (M200500; Gibco) in the presence of low serum growth supplement kit (S003K; Thermo Fisher Scientific). Cells at passages four to seven were used for experiments. For adenoviral infection, HUVECs and HAECs were transfected with Ad-Vector or Ad-CMV-SOX4 (Vigene Biosciences) for 48 h. Morphology of HUVECs and HAECs were monitored by brightfield microscopy. For pharmacological experiments, HUVECs were incubated with metformin (200 μmolL−1, 72 h), IL-1β (10 ngml−1, 48 h) and TGF-β1 (10 ngml−1, 48 h) prior to Western blotting.
Aortic CD31+ cell isolation
To study gene expression in aortic ECs, a bead-based strategy to positively select CD31+ cells was applied. The whole mouse aortae of ND, WD, AAV-Vector and AAV-SOX4 groups were isolated after intracardiac perfusion with ice-cold PBS. The finely cut aortae were incubated in 1x aortic dissociation enzyme cocktail (450 U/mL collagenase type I, 60 U/mL DNase I, and 60U/mL hyaluronidase) at 37 °C for 1 h. The digestion was stopped by adding FACS buffer (2 mM EDTA, 2% FBS). The cell suspension was transferred through 50 μm cell strainer for subsequent incubation with CD31 microbeads (Miltenyi Biotec) on ice for 15 min. The CD31+ cells were sorted by using a LS column (Miltenyi Biotec) on a magnet, in which the column was flushed with FACS buffer for several times before pushing the syringe plug for cell collection. The isolated cells were later subjected to RNA or protein isolation.
Quantitative RT-PCR
Total RNA from isolated ECs of ApoE−/− mouse aortae and HUVECs were extracted by TRIzol reagent (Invitrogen). cDNA was synthesized by using iScriptTM cDNA synthesis kit (Bio-Rad). Quantitative RT-PCR was conducted by SYBR Premix ExTaq (TaKaRa, Shiga Prefecture, Kusatsu, Japan) in the ABI ViiA7 system (Applied Biosystems, Massachusetts, USA). GAPDH was selected as an endogenous control. Gene expression was quantified by the C(t) (ΔΔCt) method. The primer pairs used in this study were displayed in Table S5. The mRNA expressions were normalized to the mean of ND group and were expressed as the relative fold change as a general practice for RT-PCR analysis. The samples of ND group retained their individual fold change.
Western blotting
Isolated ECs of mouse aortae, mouse aortae (whole, TA and AA), human ECs (HUVECs and HAECs) and human renal arteries were homogenized in ice-cold 1X RIPA lysis buffer supplemented with Complete Protease Inhibitor cocktail (Sigma-Aldrich) and phosSTOP phosphatase inhibitor (Roche, Basel, Switzerland). Protein concentrations were determined by a BCA protein assay kit (Pierce Biotechnology, Rockford, USA). Equal amount of proteins were loaded and resolved by a 10% SDS-polyacrylamide gel and transferred onto an Immobilon-P polyvinylidene difluoride membrane (Millipore Corp., Bedford, MA, USA). Nonspecific binding sites were blocked by 3% BSA in TBS containing 0.05% Tween-20. The blots were incubated overnight at 4 °C with primary antibodies: anti-SOX4 (1:200; sc-130633; Santa Cruz Biotechnology, Texas, USA), anti-eNOS (1:1000; 610297; BD Transduction Laboratory, San Diego, USA), anti-PECAM-1 (1:500; sc-1506-R; Santa Cruz Biotechnology), anti-VIM (1:500; sc-6260; Santa Cruz Biotechnology) and anti-GAPDH (1:1000; 2118S; Cell Signaling Technology, Massachusetts, USA). Bound antibodies were incubated with horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology) at room temperature for 1 h. Protein bands were later visualized by enhanced chemiluminescence (Cell Signaling Technology) with the aid of ChemiDoc™ Imaging System (Bio-Rad).
Oil red O staining
Hearts of ApoE−/− mice were harvested in ice-cold PBS, followed by an overnight fixation in paraformaldehyde solution (4% in PBS). The hearts were then embedded in optimum cutting temperature compound (OCT; Sakura Finetek, Flemingweg, Holland) for later cryosection into 10 µm sections. The sections were stored at −80 °C for later Oil Red O staining, immunofluorescence staining and immunohistochemical staining. Meanwhile, aortae of ApoE−/− mice were dissected and cut longitudinally to expose atherosclerotic plaques on en face aortae. The exposed aortae were pinned in a matric gel for overnight fixation in 4% paraformaldehyde solution at 4 °C. To examine atherosclerotic lesions, frozen sections of aortic roots and en face aortae were firstly rinsed in water for 10 min, and subsequently in 60% isopropanol. After The staining with Oil Red O solution with gentle shaking for 15 min, the samples were rinsed in 60% isopropanol again and then thrice in water. For en face aortae, the aortic tissues were fixed on cover slides with endothelium facing upwards. Images were taken by brightfield microscopy (aortic root) or by a digital camera (en face aortae). The lesion areas were determined by ImageJ software (NIH) by calculating the plaque area with relative to total vascular area [24].
Masson’s trichrome staining
Collagen content in the paraffin sections of human aortic tissues were determined by Masson’s trichrome staining following standard protocols. In brief, the samples were deparaffinized and rehydrated before staining in Weigert's iron hematoxylin working solution for 10 min. After that, the samples were stained in Biebrich scarlet-acid fuchsin solution for 10 min, and then differentiated in phosphomolybdic–phosphotungstic acid solution for 10 min or until the collagen was not red. Before mounting, the samples were rinsed in aniline blue solution for 5 min, followed by a 2-min differentiation in 1% acetic acid solution.
Immunofluorescence staining
Paraffin sections of human coronary, control internal thoracic and renal arteries, frozen sections of mouse aortic roots and aortic cross sections, en face mouse aortae (TA and AA), and cultured HUVECs were fixed in paraformaldehyde solution (4% in PBS). The samples were blocked by 5% BSA at room temperature for 2 h, followed by a 4 °C overnight incubation with primary antibodies: anti-PECAM-1 (1:200; sc-1506-R; Santa Cruz Biotechnology), anti-PRSS23 (1:200; A17092; ABclonal), anti-SOX4 (1:100; sc-130633; Santa Cruz Biotechnology), anti-VIM (1:200; sc-6260; Santa Cruz Biotechnology), and/or isolectin IB4-conjugated Alexa Fluor 647 (1:1000; I21414; Thermo Fisher). The samples were later incubated with AlexaFluor secondary antibodies (1:500) at room temperature for 2 h in the dark. Nuclei were subsequently stained with DAPI (Invitrogen) in PBS for 5 min. Fluorescent signals were detected by Fluoview FV1000 laser scanning confocal system (Olympus, Tokyo, Japan).
Immunohistochemical staining
Paraffin sections of human renal arteries and frozen sections of mouse aortic roots were used for the immunohistochemistry for SOX4. Antigen retrieval was performed in 0.01 M citrate buffer at pH 6.0. After blocking by 5% BSA at room temperature for 2 h, the tissue sections were incubated with the primary antibody anti-SOX4 (1:100; sc-130633; Santa Cruz Biotechnology) at 4 °C overnight. The signals were developed by using UltraSensitiveTM SP kit (Fuzhou MAXIM Biological Technology Development Co., Ltd., China) according to the manufacturer’s instructions. The nuclei were counterstained in hematoxylin.
Ex vivo organ culture
After C57BL/6 and ApoE−/− mice were sacrificed by CO2 suffocation, the thoracic aortae were dissected out and adhering connective tissues were removed in sterile PBS. The aortae of C57BL/6 mice were cut into ring segments of ∼2 mm in length and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies, Paisley, UK) supplemented with 10% FBS and antibiotics. For ex vivo viral infection, the segments were incubated with AAV-Vector or AAV-ICAM2-SOX4 (Vigene Biosciences) for 48 h before functional assays on wire myograph. For pharmacological experiments, aortae of ApoE−/− mice were incubated with IL-1β (10 ngml−1, 48 h) in DMEM prior to Western blotting.
Wire myography
The aortic segments (∼2mm) of C57BL/6 mice were transferred to Krebs solution containing (in mmol·L−1): 119 NaCl, 4.7 KCl, 1 MgCl2, 2.5 CaCl2, 1.2 KH2PO4, 25 NaHCO3, and 11 D-glucose, and mounted individually on the Multi Wire Myograph System (Danish Myo Technology, Aarhus, Denmark) for functional assays. Each segment was initially stretched to an optimal baseline tension (3 mN) and was allowed to equilibrate for 1 h at 37 °C under continuous oxygenation (95% O2, 5% CO2) in Krebs solution. The segments were precontracted by 60 mmolL−1 KCl and later rinsed thrice in Krebs solution. Phenylephrine (Phe; 3 μmolL−1) was subsequently added to precontract the segments, followed by the induction of endothelium-dependent relaxations by cumulative additions of acetylcholine (ACh; 3 nmol·L−1 to 10 μmol·L−1) [25]. Endothelium-independent relaxations were also determined by cumulative additions of sodium nitroprusside (SNP; 1 nmol·L−1 to 10 μmol·L−1) into bathing solution. Changes in isometric tension were recorded by PowerLab LabChart 7.0 system (AD Instruments, NSW, Australia).
Serum lipid profile
Blood of ApoE−/− mice were collected via celiac vein, and serum was obtained by centrifugation at 3000 rpm at room temperature for 10 min. The serum lipid profile was evaluated by a commercially available assay kit (Stanbio, Boerne, Texas, USA) specialized for serum total cholesterol (TC), triglycerides (TG) and high-density lipoprotein (HDL) cholesterol. To separate HDL from the whole serum, HDL precipitating reagent (Stanbio) was added to serum (1:10), prior to a centrifugation at 1000g for 10 min. Lipid profile was determined according to the manufacturer’s protocol and lipid levels were measured on a plate reader (Bio-Rad) by detecting the absorbance at 500 nm. Levels of non-HDL cholesterol was calculated from the following formula: non-HDL cholesterol = TC-HDL-(TG/5).
Small interfering RNA (siRNA)-mediated genetic knockdown
After reaching ∼80% confluency, HUVECs were transfected by either scrambled siRNA (Invitrogen) or siRNA targeting SOX4 (siSOX4; Invitrogen) via electroporation by the Amaxa Basic Nucleofector Kit specialized for mammalian ECs (Lonza). Briefly, HUVECs were first trypsinized and washed twice in PBS. HUVECs were subsequently resuspended in 100 μL basic Nucleofector solution and transferred to an electroporation vial containing 50 pmol scrambled siRNA or siSOX4. HUVECs were then electroporated by the Amaxa NucleofectorTM apparatus and cultured in six-well plates containing FBS-free DMEM [26]. After 12 h, the medium was changed to EGM, and HUVECs were exposed to IL-1β (10 ngml−1, 48 h) and TGF-β1 (10 ngml−1, 48 h) prior to Western blotting.
In vitro hemodynamic study
LSS (12 dyn cm−2) and OSS (0.5 ± 6 dyn cm−2, 1 Hz) were generated by ibidi flow system (Gräfelfing, Germany), together with custom-built flow chambers (Fig. 7D). The assembly of flow system and computer control program were established following the manufacturer’s instructions. HUVECs (5 × 105 cells) were seeded on glass slides (75 mm × 38 mm; Corning, USA) coated with fibronectin (50 μgml−1, 24 h). After a 16-h attachment, HUVECs were either incubated in EGM supplemented with 2% FBS for LSS, or in EGM supplemented with 10% fatty-acid-free BSA for OSS. The slides were mounted into the flow chambers, which were subsequently connected to ibidi flow system [27]. The cells were exposed to either LSS or OSS for 24 h, prior to protein and RNA extraction.
Fig. 7.
Biomechanical inducers of SOX4 upregulation. (A) Schematic diagram on different flow patterns along the aorta. (B) Immunofluorescence staining on en face SOX4 expression of mouse thoracic aortae (TA) and aortic arches (AA) (n = 6 per group; Scale bar: 50 μm). (C) Western blotting on SOX4 level in mouse TA and AA (n = 6 per group). (D) Schematic diagrams on design of ibidi flow system and cross sections of cell chamber. (E) Western blotting on SOX4 level in HUVECs exposed to LSS or OSS for 24 h (n = 6 per group). Data are presented as mean ± SD. *P < 0.05 vs TA or LSS (unpaired t-test and nonparametric Mann-Whitney test). AA, aortic arch; HUVECs, human umbilical vein endothelial cell; LSS, laminar shear stress; OSS, oscillatory shear stress; TA, thoracic aorta.
Experimental blinding and randomization
Experimental blinding was used to reduce the risk of bias in this study whenever possible. The drugs used for in vitro and ex vivo experiments were prepared by laboratory members who did not conduct the experiments. Besides, all animals were randomized before diet feeding and viral injection.
Statistical analysis
Data obtained from experiments are presented as mean ± SD. Statistical analyses were conducted by using GraphPad Prism software (Version 8.0). Sample sizes were selected on the basis of previous experiments. Statistical significance were evaluated by unpaired t-test and nonparametric Mann-Whitney test for two-tailed comparison between two groups, and by Brown-Forsythe and Welch ANOVA followed by Games-Howell’s multiple comparisons test for comparison between multiple groups. A p value < 0.05 indicates statistical significance.
Results
Single-cell survey of murine atherosclerotic aorta
Due to the presence of a large proportion of immune cells, especially lesional MPs present in atherosclerotic plaque, suggested by previous investigations on CD45+ cells for scRNA-seq [6], [7], the EC percentage would be largely diluted for single-cell analysis. Therefore, we applied an opposite bead-based strategy to select CD45− cells for higher EC yield. Viable CD45− cells were enriched from the aortae of 3 male ApoE−/− mice fed either a normal diet (ND) or a western diet (WD) for 20 weeks, representing an advanced stage of atherosclerosis (Fig. 1A and Fig. S1) [28]. Body weights and lipid profiles of ApoE−/− mice were recorded (Fig. S2). CD31+ screening strategy was not applied for preparing single-cell suspension because this procedure would greatly decrease the abundance of non-EC cells, hindering further analysis of intercellular interactions. Conversely, CD45− sorting strategy ensured a relatively higher abundance of ECs. Sorted cells were sequenced at high depth (>150 000 reads per cell), with the median of detected genes per cell over 850 (Table S1). After quality control filtering, 1063 cells and 991 cells from normal and atherosclerotic aortae respectively, were included for single-cell analysis. The doublet rate was around 1% by estimation using DoubletFinder and the predicted doublets were clustered with other singlets (Fig. S3), suggesting that data preprocessing was effective in preventing any doublet specific cluster.
Fig. 1.
Identification of five distinct cell populations in atherosclerotic aorta. (A) Schematic overview of experimental design. (B, C) Uniform manifold approximation and projection (UMAP) visualization of single cells from normal (n = 1063) and atherosclerotic (n = 991) aortae of 3 ApoE−/− mice representing (B) cellular origin and (C) segregation into 5 distinct clusters. (D) Dotplots demonstrating the gene signatures of the 5 aortic cell types. Dot colors, expression levels; size, cell proportion with expression. EC, endothelial cell; FB, fibroblast; MP, macrophage; ND, normal diet; RBC, red blood cell; VSMC, vascular smooth muscle cell; WD, western diet. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Identification of distinct cell populations in atherosclerotic aorta
Based on the k-nearest neighbor algorithm, individual cells from pooled sample containing 3 aortae of both the ND and WD groups were clustered to 5 common cell clusters. The first two UMAP values were used for visualization in a lower dimension (Fig. 1B and 1C). To distinguish distinct clusters, we examined the expression levels of well-established marker genes in all clusters and compared their expression profiles to those in a published single-cell survey of normal mouse aortae [8]. We identified 5 major clusters including: ECs (enriched genes: Cytl1 and Pecam1), FBs (Dcn, Gsn and Serpinf1), MPs (Cd52, Coro1a and Rac2), red blood cells (RBCs; Hba and Hbb) and VSMCs (Acta2, Myl9 and Tagln), where their expression profiles of selected marker genes and heatmap of top signature genes (log(fold-change) > 2) were displayed (Fig. 1D, Fig. S4 and Table S2).
Transcriptional landscape of aortic cells in atherosclerotic aorta
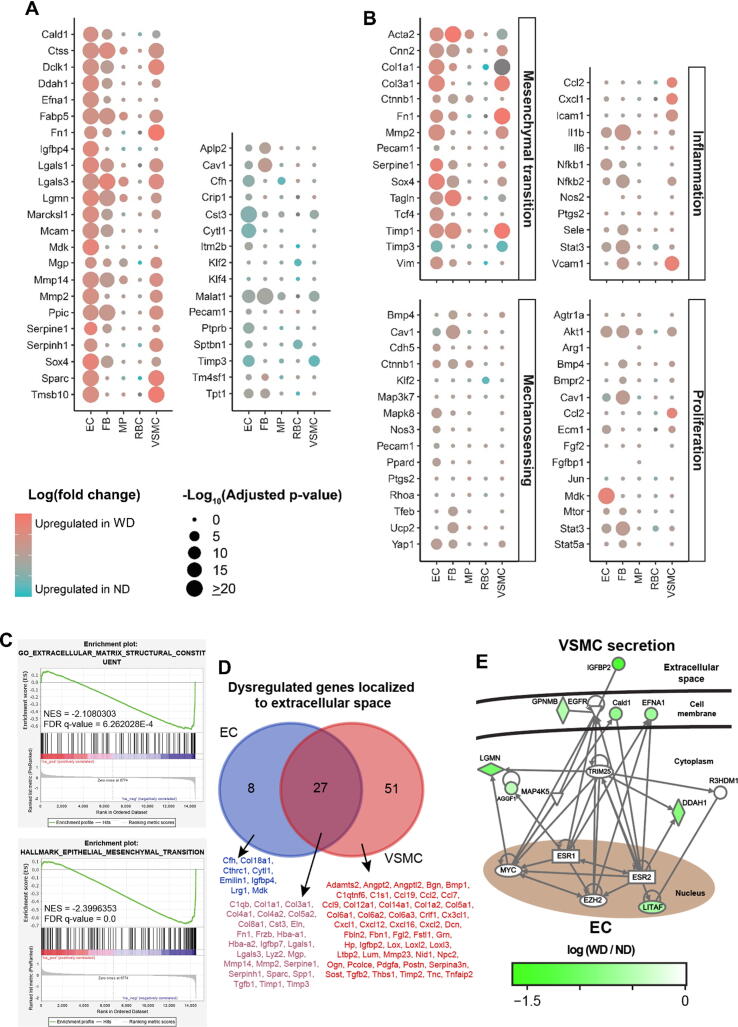
Transcriptional profiles of 5 clusters were studied accordingly. Up-regulated and down-regulated genes among clusters were selected to be displayed based on their expressional fold changes and biological correlation to atherosclerosis (Fig. 2A and Table S3). Atherosclerosis is generally associated with profound vascular remodeling, chronic vascular inflammation and VSMC proliferation [29]. Importantly, shear stress-induced mechanosignaling also contributes to the susceptibility of atherogenesis [30]. Therefore, the expression of key genes related to mesenchymal transition, inflammation, mechanosensing and proliferation were selected and outlined in dotplots (Fig. 2B and Table S4).
Fig. 2.
Transcriptional profiles of distinct cells in atherosclerotic aorta. (A) Dotplots overviewing selected up-regulated and down-regulated genes, and (B) highlighting expression of key genes in terms of mesenchymal transition, inflammation, mechanosening and proliferation. Dot colors, log (fold-change); size, adjusted p-value. (C) Gene set enrichment analysis (GSEA) on extracellular matrix structural constituent and mesenchymal transition of ECs between ND and WD groups. Upper plot: enrichment score distribution. Middle plot: black bars, genes of gene set; red and blue, downregulation and upregulation in WD group, respectively. Lower plot: ordered log (fold-change). Negative NES: enriched expression levels in WD group. (D) Venn-diagram on overlap of dysregulated genes localized to extracellular space of ECs and VSMCs, with the lists of common and unique genes. (E) Gene regulatory network analysis on the crosstalk between ECs and VSMCs during atherosclerosis. Scale bar: log (fold-change). EC, endothelial cell; FB, fibroblast; MP, macrophage; ND, normal diet; RBC, red blood cell; VSMC, vascular smooth muscle cell; WD, western diet. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Cells from the WD group showed marked upregulation of mesenchymal transition-related genes in ECs, FBs and VSMCs. Slight upregulation of inflammatory markers (e.g. Ccl2, Icam1, Il1b, Nfkb and Vcam1) and proliferation-related genes (e.g. Akt1 and Mdk) were observed in ECs or VSMCs (Fig. 2B). However, no significant alteration in mechanosensitive genes were noted that most genes were neither showing high fold-changes nor significant p-values, except that Klf2 was slightly upregulated in the RBCs of ND group (Fig. 2B). The expressional alterations (endothelial and mesenchymal markers) in EC cluster were verified by conventional RT-PCR on ECs isolated from atherosclerotic aortae (Fig. S5). Gene set enrichment analysis (GSEA) suggested that genes related to mesenchymal transition and structural constituent of extracellular matrix were enriched in ECs as reflected by negative enrichment scores (Fig. 2C and Fig. S6). Notably, dysregulation of mesenchymal transition-related genes was noted in both EC and VSMC clusters (Fig. 2A and 2B). Due to the close proximity and pathophysiological relationship between ECs and VSMCs in vasculature [31], we further investigated the overlapping of dysregulated genes and genetic interaction of these two clusters. Venn-diagram showed overlapping of 27 dysregulated genes in the extracellular space of ECs and VSMCs (Fig. 2D). Gene regulatory network analysis also implied a close crosstalk between ECs and VSMCs, via dysregulated genes in the extracellular space that were unique to VSMCs (e.g. Igfbp2 upregulation) (Fig. 2E).
Differential expression profiles of two EC subpopulations
Dysfunction of EC population precedes atherosclerotic lesion [32]. We therefore sought to unravel EC subpopulations and their corresponding features in atherosclerosis, which would provide structural and therapeutic insights for atherosclerosis research. Two EC subpopulations (EC1 and EC2) were segregated based on their transcriptome profiles (Fig. 3A and 3B). The selected and top (log(fold-change) > 1) upregulated signature genes of EC1 and EC2 were highlighted in the dotplot (Fig. 3C) and heatmap (Fig. S7), respectively. GSEA analyses suggested that genes related to mesenchymal transition were enriched in EC1 and EC2 of the WD group (Fig. S8). Notably, upregulation of certain endothelial markers (e.g. Icam2, Pecam1 and Vwf) were observed in EC1 (Fig. 3D), whereas higher levels of mesenchymal markers (e.g. Acta2, Prss23, Timp1 and Vim) were found in EC2 (Fig. 3E). These findings suggested that EC1 was more ‘endothelial-like’ while EC2 was more ‘mesenchymal-like’. Importantly, more mesenchymal-like ECs were observed in WD-induced atherosclerosis. Since Prss23 was mainly expressed by EC2 (Fig. 3E), the presence of EC1 and EC2 were verified by immunofluorescence staining on PRSS23 in ApoE−/− mouse aortae (Fig. 3F).
Fig. 3.
EC subpopulations in atherosclerotic aorta. (A, B) Uniform manifold approximation and projection (UMAP) plots on (A) cellular origin and (B) partition into two subpopulations (EC1 and EC2) of single ECs from normal and atherosclerotic aortae. (C) Dot plots showing the gene signatures of the two EC subpopulations. Dot colors, expression levels; size, cell proportion with expression. (D, E) Violin plots on the expression of selected (D) endothelial markers and (E) mesenchymal markers of 2 EC subpopulations. Dot: median. (F) Immunofluorescence staining on PRSS23 level in cross sections of ApoE−/− mouse aortae (n = 4 per group; Scale bar: 200 μm). EC, endothelial cell; ND, normal diet; WD, western diet.
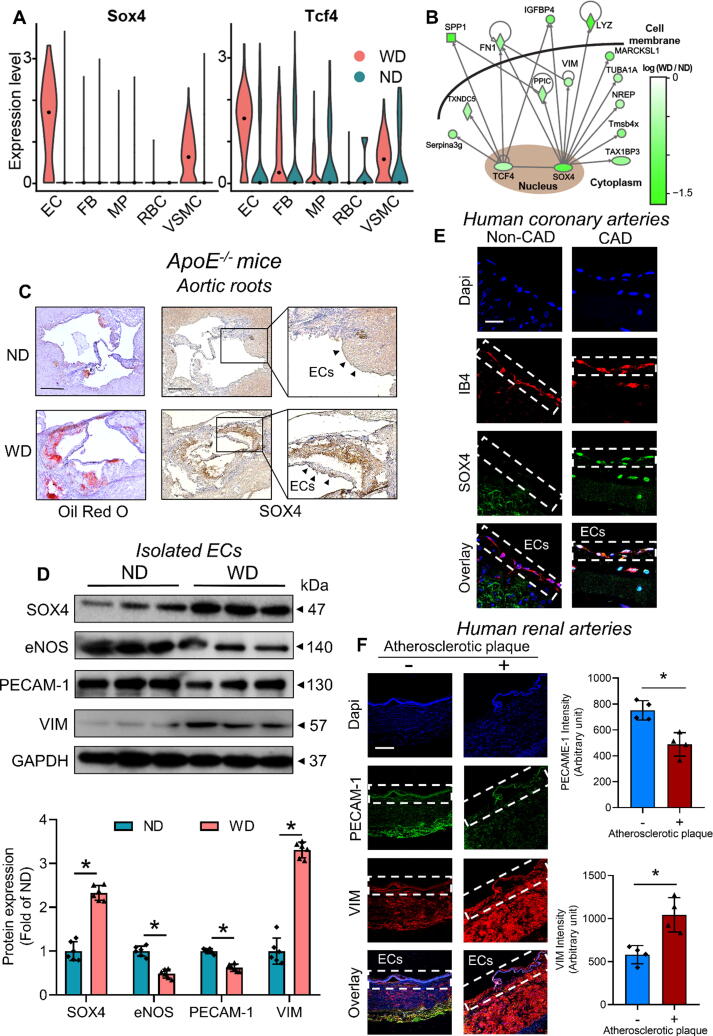
SOX4 upregulation in atherosclerotic aorta
Since transcription factors (TFs) represent the point of convergence of various signaling pathways in eukaryotic cells, direct modulation of TF function may provide novel therapeutic insights against different diseases [33], including atherosclerosis. In atherosclerotic aortae, upregulation of the TF Sox4 was detected solely in EC and VSMC clusters (Fig. 4A), consistent with RT-PCR results of isolated ECs (Fig. S5). Notably, Sox4 upregulation was observed in both EC subpopulations (EC1 and EC2) of the WD group (Fig. S9), along with the enriched genes related to mesenchymal transition (Fig. S8). Although Tcf4 upregulation was also noted in ECs of atherosclerotic aortae (Fig. 4A), its complex expression alterations in multiple cell clusters implied that Tcf4 might be involved in multiple cellular processes but not solely involved in EC function during atherosclerosis. To unravel the effector genes downstream to these TFs, we thereby conducted an interactome analysis. Furthermore, SOX4 is a direct upstream regulator of certain mesenchymal markers (i.e. VIM and FN1) as shown in the gene regulatory network analysis (Fig. 4B). In aortic roots of WD-fed ApoE−/− mice, and human atherosclerotic coronary and renal arteries, SOX4 upregulation was observed in plaque regions and EC layer, respectively (Fig. 4C, 4E and Fig. S10). Such upregulation was accompanied by the loss of endothelial markers (i.e. eNOS and PECAM-1) and gain of mesenchymal marker (i.e. VIM) in isolated ECs of mouse aortae, and human atherosclerotic renal arteries (Fig. 4D and 4F).
Fig. 4.
Endothelial SOX4 upregulation in human and mouse atherosclerotic aortae. (A) Violin plots on the expression of the transcription factors Sox4 and Tcf4 among different aortic cells. Dot: median. (B) Gene regulatory network analysis on the interactome between SOX4 and other proteins. Scale bar: log (fold-change). (C) Oil red O staining on atherosclerotic lesions and immunohistochemical staining on SOX4 expression in aortic roots of ApoE-/- mice (n = 6 per group; Scale bar: 500 μm). (D) Western blotting on expression levels of SOX4, endothelial markers (i.e. eNOS and PECAM-1) and mesenchymal marker (i.e. VIM) of isolated ECs from ApoE−/− mice (n = 6 per group). (E) Immunofluorescence staining on SOX4 level in endothelial cells of human diseased coronary arteries and control internal thoracic arteries (n = 7 per group; Scale bar: 20 μm). (F) Immunofluorescence staining on the levels of endothelial (PECAM-1) and mesenchymal (VIM) markers in human renal arteries (n = 4 per group; Scale bar: 200 μm). Data are presented as mean ± SD. *P < 0.05 vs ND or Non-atherosclerotic artery. CAD, coronary artery disease; EC, endothelial cell; FB, fibroblast; MP, macrophage; ND, normal diet; RBC, red blood cell; VSMC, vascular smooth muscle cell; WD, western diet. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
SOX4 expression and EC subpopulations in human aortic aneurysm
Aortic aneurysm may be an adverse consequence of advanced atherosclerosis [3]. To further correlate our experimental findings to clinical specimens, we extracted scRNA-seq data from a previous study on ascending aortic samples of control individuals and ATAA patients (Fig. S11A) [23]. SOX4 expression was identified in EC cluster of most ATAA samples (Fig. S11B). Consistent to our scRNA-seq data on atherosclerosis, SOX4 expression was mainly observed in structural cells of vasculature (e.g. ECs and VSMCs), rather than immune cells (e.g. MPs) (Fig. S12). For structural and therapeutic insights, we also evaluated EC subpopulations in human aneurysmal aortic tissues. Two EC subpopulations (EC1 and EC2) were segregated based on their transcriptome profiles (Fig. S11C). The upregulated signature genes of EC1 and EC2 were highlighted in dotplots (Fig. S11D). Notably, certain endothelial markers (e.g. CD9, ICAM2, and TIE1) were upregulated in EC1 (Fig. S11E), while higher levels of mesenchymal markers (e.g. PRSS23, TIMP1 and VIM) were noted in EC2 (Fig. S11F). These findings might imply that EC1 was more ‘endothelial-like’, whereas EC2 was more ‘mesenchymal-like’.
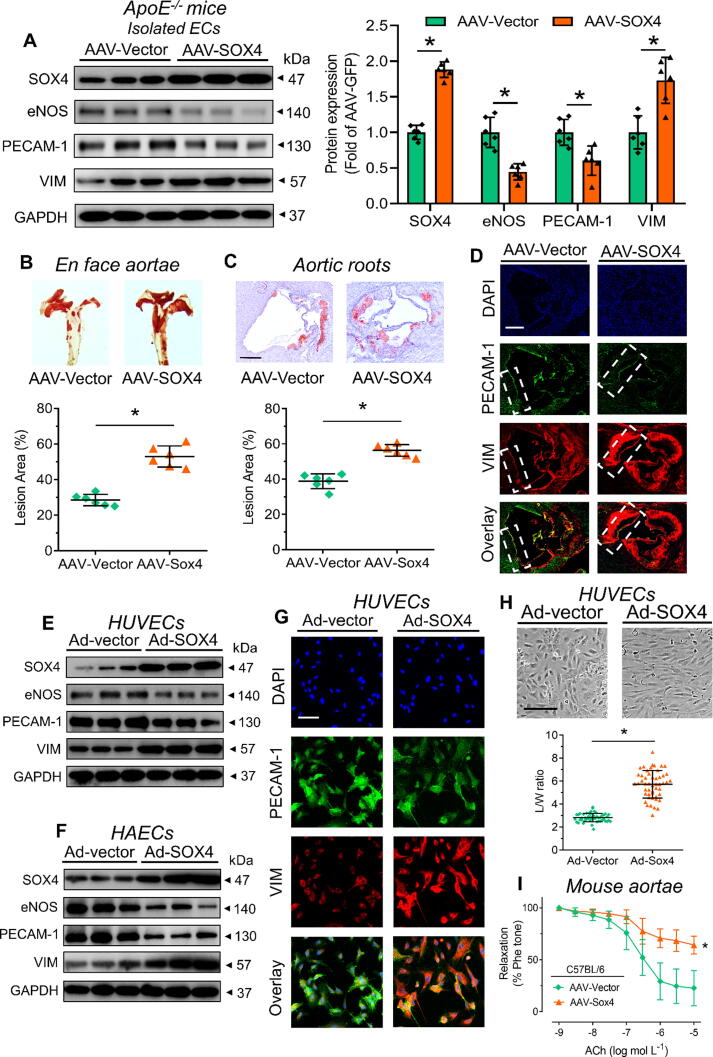
Pro-atherogenic and pro-EndoMT effects of SOX4 overexpression
To understand the role of SOX4 in atherosclerosis, endothelial-specific AAV-SOX4 was injected into ApoE−/− mice via tail vein prior to WD feeding. Body weights and lipid profiles of injected ApoE−/− mice were recorded (Fig. S13). SOX4 overexpression increased SOX4 protein expression in aorta of ApoE−/− mice (Fig. 5A). SOX4 overexpression increased lesion areas in en face aortae and aortic roots of ApoE−/− mice (Fig. 5B and 5C). Furthermore, SOX4 overexpression caused loss of endothelial markers (i.e. eNOS and PECAM-1) and gain of mesenchymal marker (i.e. VIM) in isolated ECs and aortic roots (Fig. 5A and 5D). Accordingly, adenovirus-mediated SOX4 overexpression caused similar alterations in human umbilical vein endothelial cells (HUVECs) and human aortic endothelial cells (HAECs) (Fig. 5E-5G). Consistent with the regulatory network (Fig. 4B), SOX4 overexpression enhanced VIM level in ECs. Moreover, we investigated the effects of SOX4 overexpression on EC morphology and function. Such overexpression triggered elongation of HUVECs and HAECs (Fig. 5H and Fig. S14), and impairment on endothelium-dependent relaxations of C57BL/6 mouse aortae measured by wire myograph (Fig. 5I and Fig. S15), signifying structural modification and endothelial dysfunction. SOX4 overexpression did not alter sodium nitroprusside-induced endothelium-independent relaxations of mouse aortae (Fig. S16). These evidence implied the pro-atherogenic effect of SOX4, which is associated with SOX4-mediated EndoMT.
Fig. 5.
SOX4 overexpression in endothelial cells. (A) Western blotting on isolated ECs from ApoE−/− mice, previously subjected to tail vein injection of endothelial-specific AAV-SOX4 (n = 6 per group). (B, C) Oil red O staining on atherosclerotic lesions of (B) en face aortae and (C) aortic roots of AAV-SOX4-injected ApoE−/− mice (n = 6 per group; Scale bar: 500 μm). (D) Immunofluorescence staining on endothelial and mesenchymal markers of mouse aortic roots (n = 6 per group; Scale bar: 500 μm). (E, F) Western blotting on Ad-SOX4-treated (E) HUVECs and (F) HAECs (n = 6 per group). (G) Immunofluorescence staining on endothelial and mesenchymal markers of Ad-SOX4-infected HUVECs (n = 6 per group; Scale bar: 50 μm). (H) Morphology of Ad-SOX4-treated HUVECs (L/W ratio: Feret value/Min Feret value; Scale bar: 100 μm). (I) Functional assay on endothelium-dependent relaxation of Ad-SOX4-infected C57BL/6 mouse aortae by wire myograph (n = 6 per group). Data are presented as mean ± SD. *P < 0.05 vs AAV-Vector or Ad-Vector (unpaired t-test and nonparametric Mann-Whitney test). ACh, acetylcholine; HAEC, human aortic endothelial cell; HUVEC, human umbilical vein endothelial cell; Phe, phenylephrine. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Metformin-mediated suppression on cytokine-upregulated SOX4
Next, we aimed to uncover potential inducer and suppressor of SOX4. Notably, a Western dietary pattern often correlates to higher levels of circulatory cytokines (e.g. IL-1β) [34], and both IL-1β and TGF-β1 are EndoMT-inducing cytokines during atherosclerosis [13]. Moreover, SOX4 was previously shown to be a downstream target of these two cytokines in immune cells (e.g. T cells) [35]. Hence, we wondered whether IL-1β and TGF-β1 act as upstream initiators of SOX4-mediated EndoMT in endothelial cells. Indeed, ex vivo incubation with IL-1β (10 ng ml−1) induced SOX4 upregulation and EndoMT in human renal arteries and ApoE−/− mouse aortae (Fig. 6A-6C). We further verified the results in HUVECs exposed to IL-1β and TGF-β1, another EndoMT-inducing cytokine. Co-treatment of both cytokines markedly increased SOX4 protein expression and EndoMT in HUVECs (Fig. 6D). More importantly, treatment with the antidiabetic drug metformin, previously reported to be a SOX4 suppressor in carcinoma [36], reversed cytokine-induced elevation of SOX4 expression and EndoMT in HUVECs (Fig. 6D). More importantly, siRNA-mediated knockdown of SOX4 also reversed EndoMT induced by cytokines in HUVECs (Fig. 6E).
Fig. 6.
Biochemical inducers of SOX4 upregulation. (A) Immunohistochemical staining on SOX4 expression of IL-1β-treated human renal arteries (n = 3 per group; Scale bar: 200 μm). (B) Western blotting on IL-1β-treated human renal arteries (n = 3 per group). (C) Western blotting on IL-1β-treated ApoE−/− mouse aortae (n = 6 per group). (D) The effects of metformin in HUVECs co-treated by IL-1β and TGF-β1 (n = 6 per group). (E) The effects of siRNA-mediated SOX4 knockdown in HUVECs co-treated by IL-1β and TGF-β1 (n = 6 per group). Data are presented as mean ± SD. *P < 0.05 vs empty Ctl or empty Scr; #P < 0.05 vs IL-1β + TGF-β1 Ctl or IL-1β + TGF-β1 Scr (Brown-Forsythe and Welch ANOVA and Games-Howell’s multiple comparisons test). HUVECs, human umbilical vein endothelial cell; IL-1β, interleukin-1β; Met, metformin; TGF-β1, transforming growth factor β1.
OSS-induced SOX4 upregulation
Atherosclerotic plaques and aneurysms tend to develop at vascular regions exposed to disturbed flow [14]. Due to the high SOX4 expression in OSS-experiencing regions (e.g. mouse aortic roots) (Fig. 4C), we postulated that SOX4 might be sensitive to flow-induced mechanical stimuli, additional to cytokine-mediated biochemical stimuli. Since the relatively straight TA and the curved AA correspond to LSS and OSS respectively (Fig. 7A), we examined SOX4 level at these vascular regions. SOX4 expression was increased in ECs residing in the inner curvature of AA (Fig. 7B), meanwhile the overall SOX4 expression was also higher in AA (Fig. 7C). We further verified the results in HUVECs subjected to hemodynamic forces in vitro (Fig. 7D). A 24-h exposure to OSS transcriptionally and translationally increased SOX4 level in HUVECs (Fig. 7E and Fig. S17), consistent with in vivo results.
Discussion
Endothelial dysfunction serves as the antecedent marker of atherosclerosis. The current study reported the detailed transcriptional profile, regulatory network and molecular signature of dysfunctional ECs during atherosclerotic diseases. Within the enriched aortic cells of ApoE−/− mice, we generated a compendium of 5 distinct cell clusters, including ECs, FBs, MPs, RBCs and VSMCs, which generally resembled those previously identified from C57BL/6 mice [8]. From the transcriptional profiles of 5 clusters, the expression of remodeling-related genes in the structural units (e.g. ECs, FBs and VSMCs) were significantly altered in WD group. Both GSEA and top-ranked genes of EC cluster, including phenotypic markers (e.g. Acta2, Cnn2, Tagln and Vim), remodeling genes (e.g. Fn1, Mmp2 and Timp1) and collagens (e.g. Col1a1 and Col3a1), indicate EndoMT in WD group. Upregulation of proliferation-related genes (e.g. Mdk and Akt1) and inflammatory genes (e.g. Icam1, Cxcl1, Nfkb1 and Vcam1) highlighted enhanced proliferation and inflammation in WD-induced atherosclerosis.
ECs at straight and branched regions of blood vessels are subjected to unidirectional and oscillatory shear stress respectively [27]. Size of atherosclerotic lesions depend on the patterns of fluidic shear stress, where the oscillatory pattern increases lesion size [37]. We initially hypothesized that shear stress-regulated genes might be more strikingly altered in ECs of WD-fed mice, but only slight alterations were eventually noted. Since the transcriptome was generated from the whole aorta of ApoE−/− mice, spatial heterogeneity of vasculature could not be comprehensively revealed. Significant alterations would be expected if the transcriptional landscape at straight and curved regions of vasculature are separately investigated.
The EC cluster was further partitioned into two subpopulations (EC1 and EC2) based on different molecular signatures. Particularly, the expression of endothelial markers (e.g. Pecam1, Icam2 and Vwf) was lower, while that of mesenchymal markers (e.g. Acta2, Serpine1 and Vim) was higher in EC2. Prss23, essential to Snail-dependent EndoMT [38], was exclusively expressed in EC2. These evidence suggested that EC2 is more ‘mesenchymal-like’. We therefore proposed that EC2 might be an intermediate state before ECs completely lose the endothelial phenotype to become mesenchymal cells, where the presence of two EC subpopulations were verified by immunofluorescence staining on ApoE−/− mouse aortae. Compared to a previous single-cell analysis which categorized ECs from normal aortae into three subsets [8], our findings, for the first time, correlated the two unique EC subpopulations to phenotypic and structural implications. Subtype switching of vascular cells might correlate with the severity of cardiovascular complications [39]. It would therefore be interesting to uncover molecular mediators participating in EC1-EC2 transition, and whether such transition is reversible.
Importantly, ECs and VSMCs of WD group expressed a higher level of Sox4, shown to be upstream to mesenchymal markers (VIM and FN1) by gene regulatory network analysis. Although such analysis could not reflect the most comprehensive gene regulatory network with relevance to immune regulation after depletion of majority of CD45+ immune cells, it could still provide mechanistic insights to phenotypic shift downstream to Sox4. As a TF and a marker in various cancers, SOX4 promotes epithelial-to-mesenchymal transition during tumorigenesis, enhancing anti-apoptotic, invasive and migratory properties of cancer cells [40]. Both tumorigenesis and atherosclerosis involve the acquiring of mesenchymal phenotype in corresponding cell types, exacerbating disease progression [41]. Due to commonalities in cellular plasticity among two complications [42], it is reasonable to hypothesize that similar mediators (e.g. SOX4) participate in cellular remodeling during atherogenesis.
Our study, for the first time, reported SOX4 to be a novel regulator in atherosclerosis, beyond its established role in cancer development. As shown by histological and biochemical analysis, SOX4 was upregulated in plaque regions and aortic ECs of ApoE−/− mice, and in endothelium of different human atherosclerotic arteries (e.g. coronary and renal arteries). Upregulated SOX4 level in plaque region might attribute to the high abundance of VSMCs in atherosclerotic plaque [43], coherent with our scRNA-seq finding that SOX4 was upregulated in both EC and VSMC clusters. From scRNA-seq analysis, higher SOX4 levels were observed in both EC1 and EC2 subpopulations of WD group. Such observation implies that SOX4 might participate in the entire process of EC1-EC2-mesenchymal transition, pushing both EC1 and EC2 towards a more mesenchymal state.
To improve the clinical relevance, we further extracted scRNA-seq data on human ascending aortic tissues from control individuals and ATAA patients. Sharing similar risk factors like hyperlipidemia, atherosclerosis and aneurysm may share similar pathological events, such as endothelial dysfunction and EndoMT [44]. Coherent to aortic tissues of WD-fed mice, SOX4 was upregulated in those of ATAA patients and its expression was mainly observed in structural units of vasculature (e.g. ECs and VSMCs). Two EC subpopulations were also found in human specimens, in which EC1 and EC2 correspond to a more ‘endothelial-like’ and ‘mesenchymal-like’ genotype respectively. Notably, PRSS23, the positive EndoMT regulator, was also upregulated in EC2, consistent with murine scRNA-seq data. These findings implied the involvement of SOX4 in atherosclerotic diseases. However, since aneurysm represents a later stage or a parallel event of atherosclerotic diseases [45], the scRNA-seq data on human aneurysm is not fully comparable to those obtained from atherosclerotic aortae of ApoE−/− mice.
We further examined the downstream events mediated by SOX4. On top of WD, AAV-mediated overexpression further boosted SOX4 in ECs of ApoE−/− mice. In vivo, ex vivo and in vitro findings supported that SOX4 aggravated EndoMT, endothelial dysfunction and atherogenesis. Since EC2 is associated with Vim upregulation and Pecam-1 downregulation, we reasoned that SOX4 might serve as one of the accelerators during EC1-EC2 transition. However, detailed mechanism on how SOX4 suppresses endothelial markers remains elusive. Several factors downstream of SOX4 could be involved. For instance, IGFBP-4, previously shown to be anti-angiogenic in cancer [46], might account for the loss of endothelial phenotype. Previously, SOX4 was shown to interact with other TFs, such as SMAD3 and β-catenin, to directly regulate epithelial-to-mesenchymal transition via EZH2 and other targets during tumorigenesis [47]. However, whether SOX4 mediates EndoMT in ECs through similar mechanisms during atherosclerosis requires further study.
In this study, we identified the upstream modulators of SOX4 during atherosclerosis. Ex vivo and in vitro findings suggested that EndoMT-inducing cytokines (i.e. IL-1β and TGF-β1) were upstream to SOX4. These results suggested the mechanism of SOX4 upregulation induced by cytokines present in atherosclerotic plaques. We next sought to identify pharmacological suppressor of SOX4. Importantly, the antidiabetic drug metformin reversed SOX4 upregulation and EndoMT in HUVECs, consistent with a previous report that metformin inhibits SOX4 in lung carcinoma cells [36]. Metformin was clinically shown to improve endothelial function [48]. Additional to anti-diabetic and anti-cancer effects, the present study extended the cardiovascular benefits of metformin as a potential anti-atherogenic agent. Furthermore, metformin suppressed EndoMT, suggesting that metformin may potentially reverse the EC1-EC2 transition.
High SOX4 expression at vascular regions of low shear stress (e.g. mouse aortic roots and human aneurysmal aortic tissues) implied that SOX4 might be a flow-sensitive gene. In vivo and in vitro results of this study confirmed that OSS could increase SOX4 level, additional to biochemical stimuli mediated by cytokines. Our findings extended the underlying mechanism of disturbed flow-induced EndoMT in ECs [49], where our findings connected physiological mechanical stimulus to SOX4-induced EndoMT in vascular abnormalities. In summary, the present study revealed the change of transcriptional landscape of aortic atherosclerosis, connected a previously unrecognized transition of endothelial cells to atherosclerosis, and identified SOX4 as a novel TF and a target for potential therapeutic intervention of atherosclerotic cardiovascular diseases.
Conclusion
In summary, the present study uncovers the cellular composition and transcriptional profile of murine atherosclerotic aortae, and categorized ECs into two subpopulations based on transcriptional and cellular heterogeneity. Furthermore, SOX4 was successfully identified as a novel marker in both human and murine atherosclerosis. Hyperlipidemia-associated cytokines and fluidic shear stress were identified as the upstream stimuli that mediate SOX4 upregulation, thus providing mechanistic insights into SOX4-accelerated atherosclerotic progression (Graphical abstract). Metformin was shown to suppress cytokine-induced SOX4 upregulation, opening up new therapeutic opportunities against atherosclerotic diseases.
Compliance with ethics requirements
For experimental procedures involving animals, all Institutional and National Guidelines for the care and use of animals (fisheries) were followed. For experiments involving human samples, all procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008 (5). Informed consent was obtained from all patients for being included in the study.
CRediT authorship contribution statement
Chak Kwong Cheng: Conceptualization, Methodology, Investigation, Writing – original draft, Visualization. Xiao Lin: Methodology, Software, Formal analysis, Investigation, Data curation, Writing – original draft. Yujie Pu: Methodology, Validation, Investigation. Joyce Ka Yu Tse: Software, Formal analysis. Yu Wang: Methodology, Investigation. Cheng-Lin Zhang: Methodology, Investigation. Xiaoyun Cao: Investigation. Chi Wai Lau: Investigation. Juan Huang: Investigation. Lei He: Investigation. Jiang-Yun Luo: Writing – review & editing. Yu-Tsung Shih: Investigation. Song Wan: Resources. Chi Fai Ng: Resources. Li Wang: Writing – review & editing. Ronald Ching Wan Ma: Writing – review & editing, Funding acquisition. Jeng-Jiann Chiu: Resources, Funding acquisition. Ting Fung Chan: Resources, Writing – review & editing, Funding acquisition. Xiao Yu Tian: Conceptualization, Supervision, Funding acquisition. Yu Huang: Conceptualization, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This present study was supported by grants from National Natural Science Foundation of China (91939302, 81922078), Hong Kong RGC-Collaborative Research Fund (C4024-16W), Hong Kong RGC-Senior Research Fellow Scheme (SRFS2021-4S04), Hong Kong RGC-General Research Fund (14109519, 14105321), and Hong Kong RGC-Research Impact Fund (R4012-18). This study was also supported by Taiwan Ministry of Science and Technology grants (MOST 109-2326-B-400-006 and MOST 109-2320-B-400-010-MY3), an Innovation and Technology Fund from the Innovation and Technology Commission, Hong Kong Government to the State Key Laboratory, and a generous donation from Mr. and Mrs. Sunny Yang. The authors thank the members of core laboratory facilities (CUHK) for assistance.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.02.017.
Contributor Information
Xiao Yu Tian, Email: xytian@cuhk.edu.hk.
Yu Huang, Email: yu.huang@cityu.edu.hk.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Libby P. Inflammation in atherosclerosis. Nature. 2002;420(6917):868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 2.Lusis A.J. Atherosclerosis. Nature. 2000;407(6801):233–241. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnsen S.H., Forsdahl S.H., Singh K., Jacobsen B.K. Atherosclerosis in Abdominal Aortic Aneurysms: A Causal Event or a Process Running in Parallel? The Tromsø Study. Arterioscler Thromb Vasc Biol. 2010;30(6):1263–1268. doi: 10.1161/ATVBAHA.110.203588. [DOI] [PubMed] [Google Scholar]
- 4.Baeyens N., Bandyopadhyay C., Coon B.G., Yun S., Schwartz M.A. Endothelial fluid shear stress sensing in vascular health and disease. J Clin Invest. 2016;126(3):821–828. doi: 10.1172/JCI83083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gimbrone M.A., García-Cardeña G. Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis. Circ Res. 2016;118(4):620–636. doi: 10.1161/CIRCRESAHA.115.306301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cochain C., Vafadarnejad E., Arampatzi P., Pelisek J., Winkels H., Ley K., et al. Single-Cell RNA-Seq Reveals the Transcriptional Landscape and Heterogeneity of Aortic Macrophages in Murine Atherosclerosis. Circ Res. 2018;122(12):1661–1674. doi: 10.1161/CIRCRESAHA.117.312509. [DOI] [PubMed] [Google Scholar]
- 7.Winkels H., Ehinger E., Vassallo M., Buscher K., Dinh H.Q., Kobiyama K., et al. Atlas of the immune cell repertoire in mouse atherosclerosis defined by single-cell RNA-sequencing and mass cytometry. Circ Res. 2018;122(12):1675–1688. doi: 10.1161/CIRCRESAHA.117.312513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalluri A.S., Vellarikkal S.K., Edelman E.R., Nguyen L., Subramanian A., Ellinor P.T., et al. Single-Cell Analysis of the Normal Mouse Aorta Reveals Functionally Distinct Endothelial Cell Populations. Circulation. 2019;140(2):147–163. doi: 10.1161/CIRCULATIONAHA.118.038362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He D., Mao A., Zheng C.-B., Kan H., Zhang K., Zhang Z., et al. Aortic heterogeneity across segments and under high fat/salt/glucose conditions at the single-cell level. Natl Sci Rev. 2020;7(5):881–896. doi: 10.1093/nsr/nwaa038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yurdagul A., Jr., Finney A., Woolard M.D., Orr A.W. The Arterial Microenvironment: The Where and Why of Atherosclerosis. Biochem J. 2016;473:1281. doi: 10.1042/BJ20150844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dejana E., Hirschi K.K., Simons M. The molecular basis of endothelial cell plasticity. Nat Commun. 2017;8:14361. doi: 10.1038/NCOMMS14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen P.-Y., Qin L., Baeyens N., Li G., Afolabi T., Budatha M., et al. Endothelial-to-mesenchymal transition drives atherosclerosis progression. J Clin Invest. 2015;125(12):4514–4528. doi: 10.1172/JCI82719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Souilhol C., Harmsen M., Evans P., Krenning G. Endothelial-mesenchymal transition in atherosclerosis. Cardiovasc Res. 2018;114:565–577. doi: 10.1093/CVR/CVX253. [DOI] [PubMed] [Google Scholar]
- 14.Cunningham K.S., Gotlieb A.I. The role of shear stress in the pathogenesis of atherosclerosis. Lab Invest. 2005;85(1):9–23. doi: 10.1038/labinvest.3700215. [DOI] [PubMed] [Google Scholar]
- 15.Getz G.S., Reardon C.A. Do the Apoe−/− and Ldlr−/− mice yield the same insight on atherogenesis? Arterioscler Thromb Vasc Biol. 2016;36(9):1734–1741. doi: 10.1161/ATVBAHA.116.306874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oppi S., Lüscher T.F., Stein S. Mouse Models for Atherosclerosis Research—Which Is My Line? Front Cardiovasc Med. 2019;6:46. doi: 10.3389/FCVM.2019.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilkenny C., Browne W., Cuthill I.C., Emerson M., Altman D.G. NC3Rs Reporting Guidelines Working Group. Animal research: Reporting in vivo experiments: The ARRIVE guidelines. Br J Pharmacol. 2010;160:1577–1579. doi: 10.1111/j.1476-5381.2010.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177(7):1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 2019;8(4):329–337.e4. doi: 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tirosh I., Izar B., Prakadan S.M., Wadsworth M.H., Treacy D., Trombetta J.J., et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mootha V.K., Lindgren C.M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J., et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 22.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J., Tamayo P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y., Ren P., Dawson A., Vasquez H.G., Ageedi W., Zhang C., et al. Single-Cell Transcriptome Analysis Reveals Dynamic Cell Populations and Differential Gene Expression Patterns in Control and Aneurysmal Human Aortic Tissue. Circulation. 2020;142(14):1374–1388. doi: 10.1161/CIRCULATIONAHA.120.046528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin Y., Bai L., Chen Y., Zhu N., Bai Y., Li Q., et al. Practical assessment of the quantification of atherosclerotic lesions in apoE-/- mice. Mol Med Rep. 2015;12(4):5298–5306. doi: 10.3892/mmr.2015.4084. [DOI] [PubMed] [Google Scholar]
- 25.Wong W.T., Tian X.Y., Xu A., Yu J., Lau C., Hoo R.C., et al. Adiponectin Is Required for PPARγ-Mediated Improvement of Endothelial Function in Diabetic Mice. Cell Metab. 2011;14(1):104–115. doi: 10.1016/j.cmet.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Cheng C.K., Luo J.-Y., Lau C.W., Cho W.-S., Ng C.F., Ma R.C.W., et al. A GLP-1 analog lowers ER stress and enhances protein folding to ameliorate homocysteine-induced endothelial dysfunction. Acta Pharmacol Sin. 2021;42(10):1598–1609. doi: 10.1038/s41401-020-00589-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L., Luo J.-Y., Li B., Tian X.Y., Chen L.-J., Huang Y., et al. Integrin-YAP/TAZ-JNK cascade mediates atheroprotective effect of unidirectional shear flow. Nature. 2016;540(7634):579–582. doi: 10.1038/nature20602. [DOI] [PubMed] [Google Scholar]
- 28.Tang J., Lobatto M.E., Hassing L., van der Staay S., van Rijs S.M., Calcagno C., et al. Inhibiting macrophage proliferation suppresses atherosclerotic plaque inflammation. Sci Adv. 2015;1(3) doi: 10.1126/sciadv.1400223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dzau V.J., Braun-Dullaeus R.C., Sedding D.G. Vascular proliferation and atherosclerosis: New perspectives and therapeutic strategies. Nat Med. 2002;8(11):1249–1256. doi: 10.1038/nm1102-1249. [DOI] [PubMed] [Google Scholar]
- 30.Hahn C., Schwartz M.A. The Role of Cellular Adaptation to Mechanical Forces in Atherosclerosis. Arterioscler Thromb Vasc Biol. 2008;28(12):2101–2107. doi: 10.1161/ATVBAHA.108.165951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M., Qian M., Kyler K., Xu J. Endothelial-Vascular Smooth Muscle Cells Interactions in Atherosclerosis. Front Cardiovasc Med. 2018;5 doi: 10.3389/FCVM.2018.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davignon J., Ganz P. Role of Endothelial Dysfunction in Atherosclerosis. Circulation. 2004;109(23_suppl_1) doi: 10.1161/01.CIR.0000131515.03336.f8. [DOI] [PubMed] [Google Scholar]
- 33.Papavassiliou K.A., Papavassiliou A.G. Transcription Factor Drug Targets. J Cell Biochem. 2016;117(12):2693–2696. doi: 10.1002/jcb.25605. [DOI] [PubMed] [Google Scholar]
- 34.Christ A., Günther P., Lauterbach M.A.R., Duewell P., Biswas D., Pelka K., et al. Western Diet Triggers NLRP3-Dependent Innate Immune Reprogramming. Cell. 2018;172(1-2):162–175.e14. doi: 10.1016/j.cell.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshitomi H., Kobayashi S., Miyagawa-Hayashino A., Okahata A., Doi K., Nishitani K., et al. Human Sox4 facilitates the development of CXCL13-producing helper T cells in inflammatory environments. Nat Commun. 2018;9(1) doi: 10.1038/s41467-018-06187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melnik S., Dvornikov D., Müller-Decker K., Depner S., Stannek P., Meister M., et al. Cancer cell specific inhibition of Wnt/β-catenin signaling by forced intracellular acidification. Cell Discov. 2018;4(1) doi: 10.1038/s41421-018-0033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng C., Tempel D., van Haperen R., van der Baan A., Grosveld F., Daemen M.J.A.P., et al. Atherosclerotic lesion size and vulnerability are determined by patterns of fluid shear stress. Circulation. 2006;113(23):2744–2753. doi: 10.1161/CIRCULATIONAHA.105.590018. [DOI] [PubMed] [Google Scholar]
- 38.Chen I.-H., Wang H.-H., Hsieh Y.-S., Huang W.-C., Yeh H.-I., Chuang Y.-J. PRSS23 is essential for the Snail-dependent endothelial-to-mesenchymal transition during valvulogenesis in zebrafish. Cardiovasc Res. 2013;97:443–453. doi: 10.1093/cvr/cvs355. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y., Yao F., Wang L., Li Z., Ren Z., Li D., et al. Single-cell analysis of murine fibroblasts identifies neonatal to adult switching that regulates cardiomyocyte maturation. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-16204-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lourenço A.R., Coffer P.J. SOX4: Joining the Master Regulators of Epithelial-to-Mesenchymal Transition? Trends Cancer. 2017;3(8):571–582. doi: 10.1016/j.trecan.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 41.Dongre A., Weinberg R.A. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20(2):69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 42.DiRenzo D., Owens G.K., Leeper N.J. “Attack of the Clones”: Commonalities Between Cancer and Atherosclerosis. Circ Res. 2017;120(4):624–626. doi: 10.1161/CIRCRESAHA.116.310091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Basatemur G.L., Jørgensen H.F., Clarke M.C.H., Bennett M.R., Mallat Z. Vascular smooth muscle cells in atherosclerosis. Nat Rev Cardiol. 2019;16(12):727–744. doi: 10.1038/s41569-019-0227-9. [DOI] [PubMed] [Google Scholar]
- 44.Zhao G., Chang Z., Zhao Y., Guo Y., Lu H., Liang W., et al. KLF11 protects against abdominal aortic aneurysm through inhibition of endothelial cell dysfunction. JCI Insight. 2021;6(5) doi: 10.1172/jci.insight.141673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Golledge J., Norman P.E. Atherosclerosis and abdominal aortic aneurysm: Cause, response or common risk factors? Arterioscler Thromb Vasc Biol. 2010;30(6):1075–1077. doi: 10.1161/ATVBAHA.110.206573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Contois L.W., Akalu A., Caron J.M., Tweedie E., Cretu A., Henderson T., et al. Inhibition of tumor-associated αvβ3 integrin regulates the angiogenic switch by enhancing expression of IGFBP-4 leading to reduced melanoma growth and angiogenesis in vivo. Angiogenesis. 2015;18(1):31–46. doi: 10.1007/s10456-014-9445-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moreno C.S. SOX4: The unappreciated oncogene. Semin Cancer Biol. 2020;67:57–64. doi: 10.1016/J.SEMCANCER.2019.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rena G., Lang C.C. Repurposing Metformin for Cardiovascular Disease. Circulation. 2018;137(5):422–424. doi: 10.1161/CIRCULATIONAHA.117.031735. [DOI] [PubMed] [Google Scholar]
- 49.Moonen J.-R.A.J., Lee E.S., Schmidt M., Maleszewska M., Koerts J.A., Brouwer L.A., et al. Endothelial-to-mesenchymal transition contributes to fibro-proliferative vascular disease and is modulated by fluid shear stress. Cardiovasc Res. 2015;108(3):377–386. doi: 10.1093/cvr/cvv175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.