Graphical abstract
Keywords: Microplitis mediator, Lepidopteran pest, Odorant receptor, Sex pheromone, Z5-10:Ac, Host habitat location
Highlights
-
•
Female parasitoid wasps (Microplitis mediator) are attracted by the sex pheromone of the turnip moth (Agrotis segetum).
-
•
The female-biased odorant receptor MmedOR49 tuned to the Z5-10:Ac (the main sex pheromone component of the turnip moth) in vitro.
-
•
Z5-10: Ac strongly binds to MmedOR49 by the formation of hydrogen bonds with the key residues (His 80, Ile 81, and Arg 84).
-
•
The behavioral response of female M. mediator to Z5-10:Ac was strongly diminished when MmedOR49 was downregulated by RNAi.
-
•
MmedOR49 is involved in the locating of parasitic wasp host habitats using the perception of host sex pheromone.
Abstract
Introduction
The parasitoid wasp Microplitis mediator is an important natural enemy of the turnip moth Agrotis segetum and other Noctuidae pests. In our field observation, it was fortuitously discovered that sex pheromone traps used for A. segetum also attract female wasps, verified by a simulated field condition dual-choice laboratory assay. Therefore, it was hypothesized that olfactory recognition could be crucial in this process. In this regard, a female-biased odorant receptor of the wasp, MmedOR49, attracted our attention.
Objectives
To unravel the significance of the female-biased MmedOR49 regulating host pheromone recognition.
Methods
Expression analysis (fluorescence in situ hybridization; quantitative realtime PCR), in vitro (two-electrode voltage-clamp recordings) and in vivo (RNAi combined with behavioral assessments) functional studies, and bioinformatics (structural modeling and molecular docking) were carried out to investigate the characteristics of MmedOR49.
Results
MmedOR49 expression was detected in the antennae of females by FISH. Quantification indicated that the expression level of MmedOR49 increased significantly after adult emergence. In vitro functional study revealed that MmedOR49 was specifically tuned to cis-5-decenyl acetate (Z5-10:Ac), the major sex pheromone component of A. segetum. Molecular docking showed that Z5-10:Ac strongly bound to the key amino acid residues His 80, Ile 81, and Arg 84 of MmedOR49 through hydrogen bonding. Behavioral assays indicated that female wasps were significantly attracted by Z5-10:Ac in a three-cage olfactometer. RNAi targeting further confirmed that MmedOR49 was necessary to recognize Z5-10:Ac, as female wasps lost their original behavioral responses to Z5-10:Ac after down-regulation of the MmedOR49 transcript.
Conclusion
Although M. mediator is a larval endoparasitoid, female wasps have a behavioral preference for a sex pheromone component of lepidopteran hosts. In this behavior, for female M. mediator, MmedOR49 plays an important role in guiding the habitat of host insects. These data provide a potential target for enhancing natural enemy utilization and pest control.
Introduction
Parasitoid wasps, the largest group of hymenopterans, have been successfully exploited as natural enemies in green agricultural production against a diverse group of insect pests [1], [2], [3], [4], [5]. The parasitoid wasp Microplitis mediator (Hymenoptera: Braconidae) is a generalist endoparasitoid of larvae with a broad host range. It primarily parasitizes Noctuidae insect pests, including Helicoverpa armigera, Agrotis segetum, Agrotis ipsilon, and Argyrogramma agnata [6], [7]. In China, M. mediator has been artificially reared and utilized as part of integrated pest management strategies to control the cotton bollworm H. armigera and other insect pests in the field [8]. The turnip moth A. segetum is a destructive polyphagous pest that occurs throughout Asia, Europe, and Africa. Its larvae cause severe damage to crops, including grains and vegetables [9], [10], [11], [12], [13]. In our previous field observation, it was found that female M. mediator was also attracted by the sex pheromone traps set for A. segetum. Similar to other insects, M. mediator takes advantage of a sensitive olfactory system to discriminate host-associated semiochemicals for host location and spawning. Accordingly, olfactory perception may perform a crucial function in the process by which A. segetum sex pheromone traps attract female M. mediator.
Odorant receptors (ORs), the first group of chemoreceptors found in insects, have been extensively investigated [14], [15], [16]. Insect ORs are seven-transmembrane domain (TMD) proteins with an inverted topology that includes an intracellular N-terminus and extracellular C-terminus, unlike G protein-coupled receptors in vertebrates and nematodes [14], [15], [17], [18], [19]. Heterologous expression techniques, including Xenopus oocytes, cell lines, and the “empty neuron” of transgenic Drosophila, have been shown to be very effective in exploring the ligand specificities of insect ORs. In addition, recent advances in the development of in vivo analysis such as RNAi or CRISPR-mediated mutagenesis have helped to deorphanize the ORs of several model and non-model insects [19], [20], [21], [22], [23]. Insect ORs respond to specific types of odor molecules, whereas distinct ligands can activate multiple ORs [19], [24]. In lepidopterans, HarmOR42-orthologous ORs conservatively recognize plant floral cues [23]. In female Plutella xylostella, both OR35 and OR49 are necessary and sufficient for perceiving isothiocyanates and choosing host plants [25]. In the pea aphid Acyrthosiphon pisum, the alarm pheromone (E)-β-farnesene and some specific plant volatiles are detected by the receptors OR5 and OR4, respectively [22], [26]. General ORs (OR59, OR80) and sex pheromone ORs (OR4, OR33) have been identified and functionally characterized in mirid bugs [27], [28], [29], [30]. A large expansion of the OR gene family has been found in hymenopteran species. Gene gain and loss events are diverse and common in bees, wasps, and ants [31]. The size of the OR gene family in wasps reflects the complexity of the chemical cues in their habitats. The members of wasp OR families recognize distinct volatile components specifically and efficiently, and all ORs function together to identify multiple semiochemicals [19], [32], [33]. Recently, investigations of the ORs of parasitoid wasps have received increasing attention. The two major oviposition attractants β-caryophyllene and (E)-α-farnesene are detected by OR35 of Anastatus japonicus [34]. OR62 specifically tuned to cis-jasmone was confirmed to mediate the behavior of female Campoletis chlorideae wasps [35]. Moreover, odorant receptor co-receptor (Orco) channel composition has been elucidated by cryo-electron microscopy (cryo-EM) for the parasitic wasp Apocrypta bakeri [36].
In several preliminary studies, large sets of M. mediator proteins implicated in chemoreception, including olfactory receptors (166 ORs and 17 ionotropic receptors (IRs)), odorant carrier proteins (18 odorant binding proteins (OBPs), 3 chemosensory proteins (CSPs), and 2 Niemann-Pick type C2s (NPC2s)), and 2 sensory neuron membrane proteins (SNMPs) were identified [37], [38], [39], [40], [41], [42], [43], [44]. In addition, the expression profile of approximately 60 MmedORs has been characterized [45], [46]. However, none of their functions are known and all MmedORs remain orphans. In our ongoing project, female-biased MmedORs were functionally characterized by screening a large odorant panel, including plant volatiles, host volatiles, and host adult sex pheromone components. The MmedOR49 was selectively activated by cis-5-decenyl acetate (Z5-10:Ac), the main component of the A. segetum sex pheromone. Here, Xenopus expression coupled with two-electrode voltage-clamp (TEVC) recordings, quantitative real-time PCR (qPCR) measurement, fluorescence in situ hybridization (FISH), protein homology modeling, molecular docking, and RNAi combined with behavioral assays were applied to fully characterize the function of target MmedOR49 in response to Z5-10:Ac. Our findings provide valuable support for the development of a biological pest control strategy based on the regulation of parasitoid chemosensory behavior.
Materials and methods
Plants and insects
Cotton seeds (Gossypium hirsutum cv. CCRI12) were planted in plastic pots (height 14 cm, diameter 6 cm) and kept in a greenhouse under natural light at 29/25°C. Cotton plants at the 6–7 true leaf stage were used in experiments. M. mediator individuals were reared in a climate chamber under controlled conditions (28 ± 1°C, 60 ± 10% R. H., and 16L : 8D photoperiod). Antennal samples from 3-day-old female wasps were collected for RNA extraction. For expression analyses, antennae were dissected from females at different developmental stages including the red-eyed pupae, half-pigmented pupae, fully pigmented pupae, and adults one day after emergence, as well as from different physiological states, including 3-day-old virgins, 3-day-old mated wasps without parasitism experience, and 3-day-old mated wasps with parasitism experience. All antennal samples were directly frozen in liquid nitrogen and stored at −80°C for further use.
Simulated field condition behavioral trial
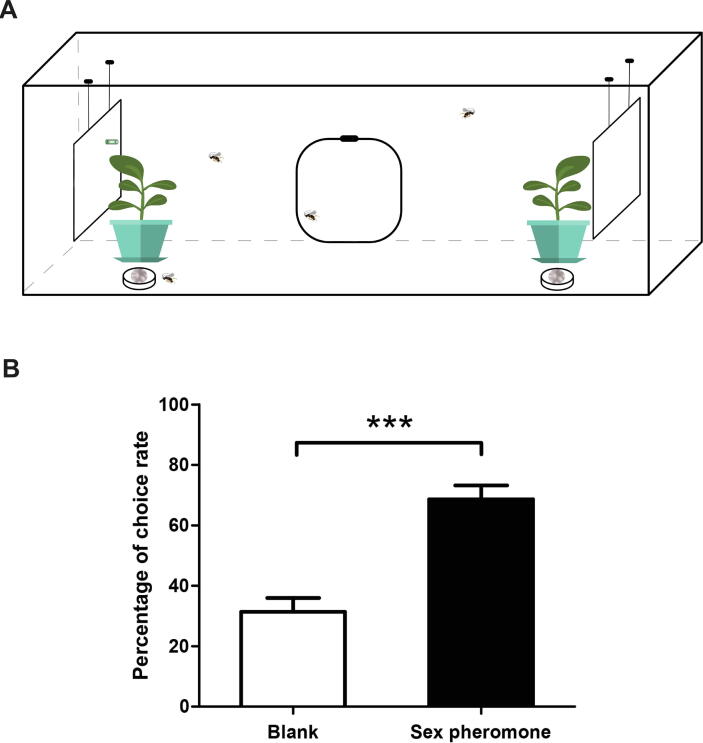
To verify our preliminary field observation of the attraction of M. mediator females to moth sex pheromone components, a two-choice trial imitating the field conditions was performed in nylon net cages (50 × 40 × 120 cm, 100 mesh). A single pot containing one cotton plant with 6–7 leaves was placed on each side of the cage. One white adhesive card (20 × 20 cm) was hung vertically 5 cm from each cotton plant. The A. segetum sex pheromone components (decyl acetate (10:AC) : Z5-10:AC : cis-7-dodecenyl acetate (Z7-12:AC) : cis-9-tetradecenyl acetate (Z9-14:AC) = 0.6 : 1 : 5 : 2.5) were diluted to 5 ng/μL (Z7-12:AC) in hexane [47], [48]. A 100 µL aliquot of pheromone solution was loaded into a green rubber lure, then fixed to the center of the card. One rubber lure containing 100 μL of hexane fixed on a card at the opposite end of the net cage was used as a control (Fig. 1A). The cages were placed in a dark room at 28 ± 1°C and 60 ± 10% R. H. Fifty 1–2-day-old female wasps were released in the center of the nets at 8:00 a.m. The numbers of wasps on each sticky trap were counted at 4:00 p.m. Two Petri dishes with 10% sucrose solution were placed on the sides of the cage as food sources for wasps. This assay was repeated five times.
Fig. 1.
Behavior choices of female M. mediator to sex pheromones of A. segetum. (A) Schematic diagram of the behavior test. (B) Behavioral tendency of female wasps to a blend of A. segetum sex pheromones. “***” indicates extremely significant differences (P < 0.001).
RNA isolation and cDNA synthesis
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The integrity of the RNA templates was examined on a 1.2% agarose gel and a NanoDrop 2000 spectrophotometer (NanoDrop, Wilmington, MA, USA). First-strand cDNA was synthesized from 1 μg of total RNA using the FastKing gDNA Dispelling RT SuperMix (TIANGEN BIOTECH, Beijing, China).
Sequence analysis of MmedORs
A total of 515 ORs with a minimum length of 200 amino acid residues from hymenopteran insects were used in the phylogenetic analysis. Sequence alignment was conducted using MAFFT (version 7.475) with the auto-option strategy [49]. To construct a maximum-likelihood tree, aligned sequences were analyzed with IQ-TREE (version 1.6.12), which runs automatic model testing and selection using the ModelFinder function [50]. According to the Bayesian Information Criterion (BIC), JTT + F + R9 was selected as the best-fit model. Tree branch support was determined using 2000 UltraFast bootstraps. The resulting tree was further edited using the Interactive Tree of Life (iTOL, https://itol.embl.de).
Six MmedORs, with full-length sequences and high sequence similarity in the same branch as MmedOR49, were aligned using PRALINE (http://www.ibi.vu.nl/programs/pralinewww/). The OR sequences employed in this analysis are provided in the Supplementary Material. The TOPCONS online server (http://topcons.cbr.su.se) was utilized to predict TMDs of MmedOR49. The model that best fitted the OR structure was selected and illustrated using Protter (http://wlab.ethz.ch/protter/) [51], [52].
qPCR measurement
The expression of MmedOR49 was assessed by performing qPCR on an ABI Prism 7500 Fast Detection System (Applied Biosystems, Carlsbad, CA, USA). β-actin (accession number KC193266.1) was used as an internal reference gene to normalize the transcript level of MmedOR49 and correct the sample variation. Previously designed primers for MmedOR49 and β-actin were used in this study [46]. The productivity of amplification was evaluated using a standard curve generated from a five-fold cDNA dilution series. The qPCR mixture (20 μL) was 10 μL SuperReal PreMix Plus (TIANGEN), 1 μL template cDNA (200 ng), 0.6 μL forward and reverse primers (10 μmol/L), 0.4 μL Rox reference dye, and 7.4 μL sterile H2O. The qPCR conditions were set as follows: 15 min at 95°C, 10 s at 95°C (40 cycles), 30 s at 55°C, and 32 s at 72°C; the melt curve was 15 s at 95°C, 1 min at 60°C, and 15 s at 95°C. Three independent biological replicates were tested with three technical replicates for each reaction. The comparative 2−ΔΔCT formula was used to estimate the relative expression levels of the target gene in different samples [53].
Fluorescence in situ hybridization
FISH was conducted to investigate the localization of MmedOR49 in the antennae of female wasps. The full-length sequence of MmedOR49 was PCR-amplified using specific primers (Table S1). The recombinant plasmid for probe synthesis was constructed using PCR product. The biotin-labeled antisense RNA probe was transcribed from linearized plasmid using the Biotin RNA Labeling Mix kit (Roche, Mannheim, Germany). The probe was digested to approximately 400-base fragments using carbonate buffer (80 mM NaHCO3, 120 mM Na2CO3, pH 10.2) as previously described [54]. Freshly dissected female antennae were embedded immediately in Tissue-Tek optimal cutting temperature (O.C.T.) compound (Sakura Finetek, Torrance, CA, USA) at −26°C. The samples were excised into 12 μm slices using a CryoStar™ NX50 microtome (ThermoFisher, Waltham, MA, USA) and fixed onto super-frost plus microscope slides (ThermoFisher). Slides were stored at −80°C until use.
Hybridization was performed as previously described [55], [56], [57]. Briefly, slides stored at −80°C were taken out, incubated at room temperature (∼25°C) for approximately 30 min, and then fixed in 4% paraformaldehyde for 30 min at 4°C. Subsequently, slides were treated with 0.2 M HCl (10 min) and washed with 1 × PBS buffer. For pre-hybridization, slides were immersed in formamide (50%) with 2 × SSC (0.3 M NaCl and 0.03 M sodium citrate) for 1 h. A 100 μL aliquot of hybridization buffer containing the labeled probe of MmedOR49 was added to each slide, followed by incubation at 60°C for at least 16 h. After hybridization, slides were washed in 0.1 × SSC at 60°C (three times, 20 min each) and immediately treated with 1% blocking solution (Roche) diluted in TBS buffer (100 mM Tris, 150 mM NaCl, pH 7.5) with Triton X-100 (0.03%) at RT for 30 min. Finally, streptavidin-HRP and a TSA Kit (Perkin Elmer, Waltham, MA, USA) were used to detect the biotin-labeled probe. Hybridization signals were visualized using a Zeiss LSM 880 confocal microscope (Zeiss, Jena, Germany) and refined using ZEN 2012 software (blue edition) (Zeiss).
Heterologous expression and TEVC recording
As described by Wang et al. (2010), Xenopus oocyte expression and TEVC recording were performed [58]. To construct the expression vector, the entire coding regions of MmedOR49 and MmedOrco were amplified using specific primers (Table S1) and cloned into the pT7TS expression plasmid. Plasmids were linearized by the restriction endonuclease and purified using the phenol-chloroform-isoamyl alcohol extraction method. The cRNAs of MmedOR49 and MmedOrco were transcribed from purified products using the mMESSAGE mMACHINE® Kit (Invitrogen).
Stage V-VI Xenopus oocytes were excised surgically and treated with 1.3 mg/mL collagenase type I (Life Technologies, NY, USA) in 1 × Ringer’s solution (pH 7.6) at RT for 1 h, then incubated overnight in 1 × Ringer’s buffer supplemented with dialyzed horse serum (5%), tetracycline (50 μg/mL), streptomycin (100 μg/mL), and sodium pyruvate (550 μg/mL) at 18°C. The next day, oocytes were injected with a mixture of MmedOR49 and MmedOrco cRNA (27.6 ng each). After injection, oocytes were further incubated for 2–3 days under the same conditions for target protein expression. A panel of 118 test compounds (diluted to 1 M stock solution in DMSO, Table S2) were prepared and stored at −20°C; work solutions (10−3 M) were prepared on the day of recording. Each compound was delivered to the oocyte holding chamber through a flow rate control system (2 mL/min, 15 s). Oocytes were washed with 1 × Ringer’s buffer between each delivery, allowing the current to return to the baseline. Ligand-stimulated currents from the injected oocytes were amplified using an OC-725C amplifier (Warner Instruments, Hamden, CT, USA) at a holding potential of −80 mV.
Data acquisition and analysis were carried out using Digidata 1440A and pCLAMP 10.2 software (Axon Instruments Inc., CA, USA). The Orco antagonist VUAA1 was used as a positive control. For dose-response assays, oocytes were exposed to ligand with a series of ascending concentrations from 10−8 to 10−3 M to measure the concentration for 50% of maximal effect (EC50). Data were analyzed using PRISM 7 software (GraphPad, San Diego, CA, USA).
Structural analysis and molecular docking
The 3D structure of the MmedOR49 protein was modeled via the trRosetta web portal (https://yanglab.nankai.edu.cn/trRosetta/) using the Orco structure (PDB ID 6c70) of the parasitic fig wasp A. Bakeri, the only available structure for insect ORs [36], [59]. The stereochemical quality and residue profiles of the MmedOR49 model were validated using structural assessment tools (ERRAT, Verify3D, and PROCHECK) of the SAVES v6.0 structure validation server (https://saves.mbi.ucla.edu/). The verified model was subjected to MD simulations and energy minimization using the Gromacs 2020.1 package (http://www.gromacs.org/) prior to docking analysis. Based on the TEVC recordings, a key ligand was chosen for molecular docking. The structure data file (SDF) of the target compound was downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). The SDF was then converted to PDB format using OpenBabel v. 2.4.1 [60].
Docking was carried out using Vina integrated with AutoDock tools v 1.5.6 on Ubuntu 20.04.2.0 LTS [61]. The docking scores correspond to interaction energies (kcal/mol). The top 10 poses of the target ligand were ranked based on the lowest binding affinity. The interaction between receptor and ligand was analyzed and visualized using Discovery Studio Visualizer v 20.1.0.19295 and LigPlot+ to draw 3D and 2D pictures, respectively. DoGSiteScorer (https://proteins.plus) [62] and CASTp (http://cast.engr.uic.edu9) were utilized to identify putative binding pockets in the modeled structure of MmedOR49.
Behavioral trial
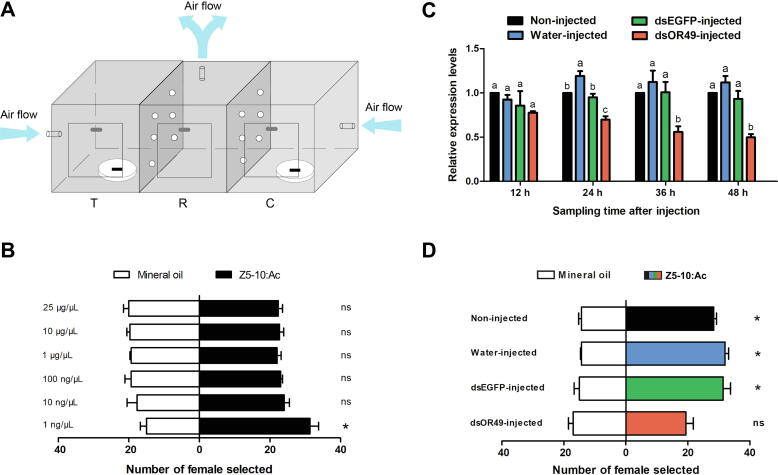
The behavioral tendency of female wasps to Z5-10:Ac was evaluated using an olfactometer made of three cages (each cage 25 × 25 × 25 cm), interconnecting with 5 cm holes between cages (Fig. 6A). The middle cage (R) was used to release wasps and the two lateral cages (T and C) contained treatment and control compounds. Z5-10:Ac was diluted in a series of concentrations (1, 10, and 100 ng/μL, 1, 10, and 25 μg/μL) in mineral oil. Mineral oil alone was used as the control. One hundred microliters of mineral oil or Z5-10:Ac was loaded onto the filter paper strip and placed into the open Petri dishes in the C or T cage. Clean air at 0.5 L/min was provided to the C and T cages and discharged from the R cage to ensure the flow of odors. Fifty 2–3-day-old female M. mediator adults were released into the R cage and left in the olfactometer from 8:00 a.m. to 4:00 p.m. Finally, the wasps moving into the T or C cages were counted. All assays were performed in five replicates under identical conditions. In addition, the olfactometer was cleaned between replicates and the positions of the T and C cages were exchanged to avoid contamination and orientation cues.
Fig. 6.
Behavioral tendency of female M. mediator to Z5-10:Ac and RNAi assessment of MmedOR49. (A) Three-cage olfactometer. C, control cage; T, treatment cage; R, release cage. (B) Behavior choice of females to different concentrations of Z5-10:Ac. “*” indicates significant differences (P < 0.05) and “ns” indicates no significant difference. (C) The expression of MmedOR49 12, 24, 36, and 48 h after injection of dsRNA. The significant differences of transcription levels among samples are indicated by different lowercase letters (P < 0.05). (D) Behavior choices of female adults to 1 ng/μL of Z5-10:Ac 36 h after dsRNA injection. “*” indicates significant differences (P < 0.05) and “ns” represents no significant difference.
RNAi assessment
To further investigate the role of MmedOR49 in vivo, RNAi of the target MmedOR49 was conducted. MmedOR49 and EGFP fragments (approximately 400 bp) were amplified using specific primers (Table S1). Double-stranded RNA (dsRNA) from MmedOR49 and EGFP were transcribed in vitro using the T7 RiboMAX™ Express RNAi System (Promega, Madison, WI, USA). About 0.7 μg dsRNA or 200 nL of sterile water were injected into the abdomen of newly emerged female adults of M. mediator. The treatment groups were non-injected (non-injected), injected with sterilized water (water-injected), injected with dsRNA of EGFP (dsEGFP-injected), and injected with dsRNA of MmedOR49 (dsOR49-injected). Each treatment was repeated three times with 30 injected wasps per replicate. Injected wasps were reared in a climate incubator and fed a 10% sucrose solution. At 12, 24, 36, and 48 h after injection, antennae were collected for qPCR. A behavioral tendency assay using a three-cage olfactometer was also employed to assess the effects of RNAi.
Statistical analysis
All analyses were performed using SPSS Statistics software (version 17.0; SPSS Inc., Chicago, IL, USA). Data are presented as the mean ± standard error of the mean (SEM), and if needed, were transformed before analysis. Differences between treatments in the expression of MmedOR49, TEVC recording, and RNAi data were analyzed using one-way analysis of variance (ANOVA) followed by Duncan’s multiple-range test (P < 0.05). For behavioral assays, a Chi-squared test (50 : 50 distribution) was used to assess the preferences of female wasps between controls and treatments.
Results
Female wasps are attracted to A. segetum sex pheromones in the laboratory
In the field, female wasps were found to be attracted to sex pheromone traps of A. segetum. Hence, an indoor behavioral experiment, similar to what was observed in the field, was designed to verify the above findings (Fig. 1A). The results showed that the mixture of sex pheromone components of A. segetum significantly attracted female M. mediator. After 8 h, 68.61% of the parasitoids were found on the sex pheromone sticky cards, which was significantly higher than that on the controls (31.39%, Fig. 1B).
MmedOR49/Orco is activated by the main sex pheromone component from A. segetum
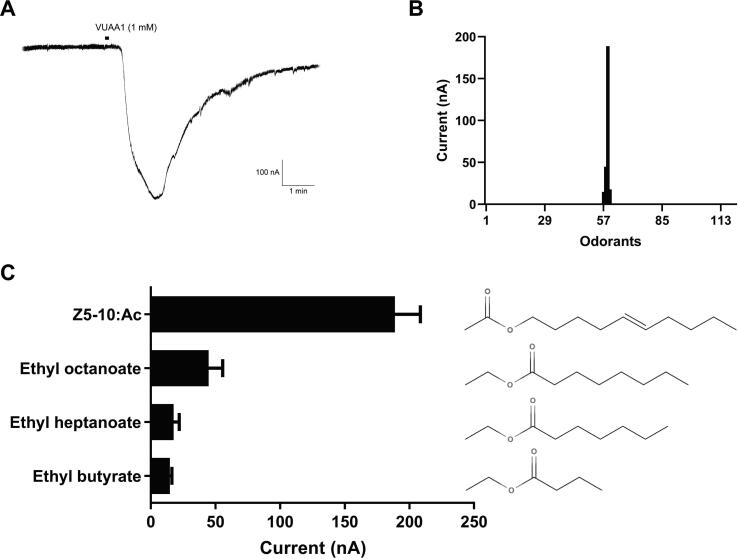
TEVC recordings demonstrated the activation of MmedOR49/Orco by Z5-10:AC. Notably, of the 4 A. segetum sex pheromone components (10:AC, Z5-10:AC, Z7-12:AC, and Z9-14:AC) and 10 sex pheromone components from other lepidopteran insects, Z5-10:AC was recorded as the unique ligand of MmedOR49 (Fig. S2A). The dose-dependency of activation by Z5-10:AC demonstrated that the MmedOR49 response was concentration-dependent (EC50: 3.61 × 10−4 M; Fig. S2B, C). Moreover, the binding affinities of MmedOR49 with the remaining 104 compounds, including habitat cue compounds of wasps and their analogs, were examined. MmedOR49 also narrowly recognized three esters: ethyl octanoate, ethyl heptanoate, and ethyl butyrate. However, Z5-10:AC was the most dynamic ligand (Fig. 2).
Fig. 2.
Binding characteristics of the MmedOR49/Orco complex to active ligands. (A) Current trace to VUAA1. (B) Tuning profile of the MmedOR49/Orco complex to an odor library including 118 odorants. (C) Response of the MmedOR49/Orco complex to active ligands. The chemical formula of corresponding ligands are shown on the right. Data are shown as means ± SEM (n = 5–6).
MmedOR49 sequence analysis
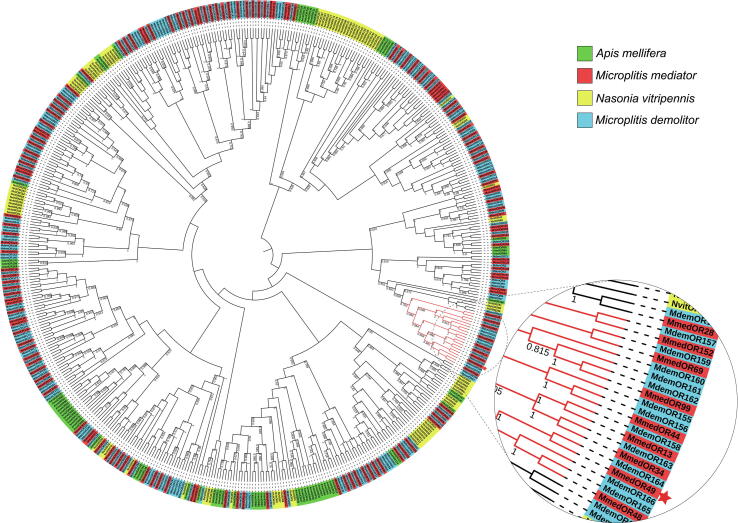
The whole ORF of MmedOR49 contained 1182 bp nucleotides encoding a 393-residue protein with a predicted molecular weight of 45.21 kDa and an isoelectric point of 9.12. Phylogenetic analysis showed that ORs from different hymenopteran species had a relatively divergent distribution, forming several monophyletic clades (Fig. 3). However, M. mediator ORs were clustered in the same branch as those of its relative M. demolitor. In the phylogenetic tree, MmedOR49 was clustered with 7 MmedORs and 12 MdemORs on one branch. Seven MmedORs, including MmedOR49, with full-length sequences and high sequence similarity in the same branch, were further aligned. The multiple sequence alignment results demonstrated that aligned ORs share 62.91% amino acid identity (Fig. S1).
Fig. 3.
Phylogenetic analysis of MmedOR49. Numbers at the nodes are bootstrap values (only > 0.5 are displayed). The branch where MmedOR49 is located is marked in red and displayed as an enlarged view. The red star indicates the position of MmedOR49.
Expression characteristic of MmedOR49 in female antennae
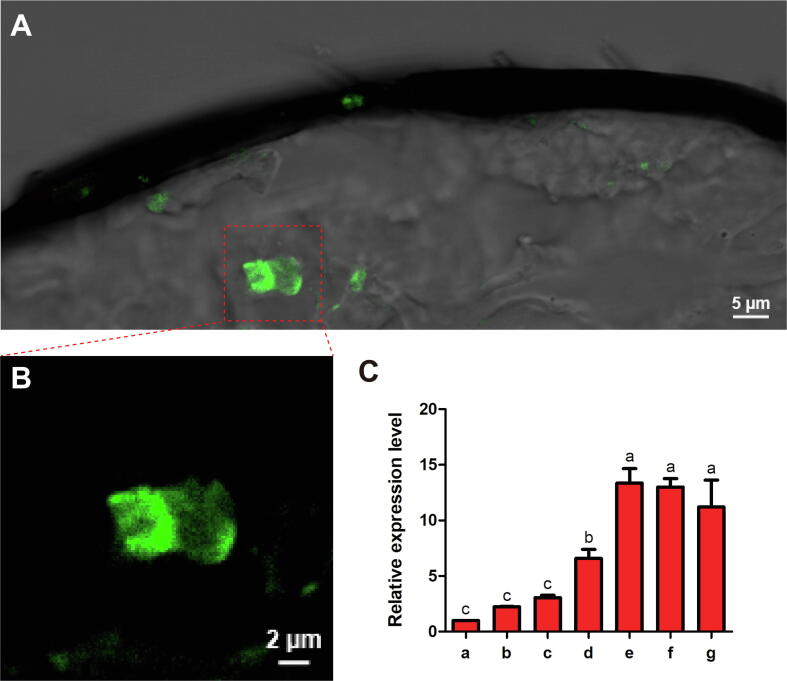
In the FISH assay, the fluorescence signals of MmedOR49 were detected in the antennae of female M. mediator (Fig. 4A, B). MmedOR49 transcripts were mainly distributed in olfactory sensory neurons (OSNs) of the antennae. Quantification by qPCR indicated that transcription of MmedOR49 is developmentally regulated. At the pupal stage, low expression was observed a few days before emergence, whereas expression increased remarkably after eclosion. However, parasitism and mating experience had no significant effect on MmedOR49 transcription (Fig. 4C).
Fig. 4.
The expression profile of MmedOR49 in the antennae of female M. mediator. (A) Location of MmedOR49 in the antennae of female M. mediator. The biotin-labeled antisense RNA probe for MmedOR49 was labeled in the olfactory sensory neurons (OSNs) of the antennal flagella of the females. (B) Amplification of the labeled OSNs. (C)MmedOR49 expressions in female antennae at different developmental stages and physiological states. (a) Red-eyed pupae, (b) half-pigmented pupae, (c) fully pigmented pupae, (d) newly emerged adult, (e) unmated adult, (f) mated adult without parasitism experience, (g) mated adult with parasitism experience. The significant differences of transcription levels among samples are indicated by different lowercase letters (P < 0.05).
Modeling of Z5-10:Ac binding to MmedOR49
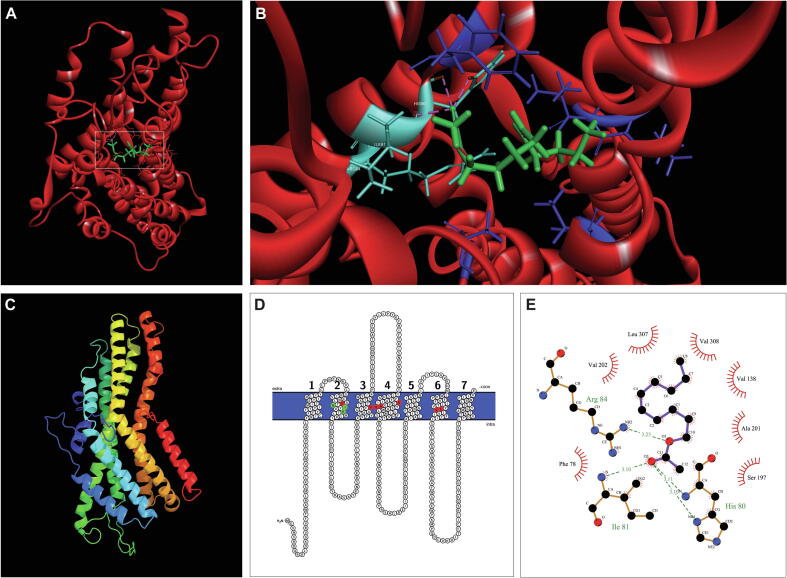
To understand the interaction of Z5-10:Ac with MmedOR49 and predict the key binding sites of MmedOR49 to the main ligand, protein homology modeling and molecular docking simulations were performed. The three-dimensional model of MmedOR49 is shown in Fig. 5C with helical and loop structures. Validation assessments using SAVES 6.0 demonstrated that the MmedOR49 model surpassed the basic prerequisite; therefore, it was used in further analyses. ERRAT showed an overall quality factor of 93.5% (Fig. S3). VERIFY3D showed that 79% of the model’s residues had a score higher than 0.2 in the 3D/1D profile (Fig. S4). A Ramachandran plot created using PROCHECK indicated that 98% of residues were in the most favored region (Fig. S5). Molecular docking indicated that Z5-10:Ac bound tightly to MmedOR49 with a G value of −5.7 kcal/mol (Table S3). Z5-10:Ac interacted via hydrogen bonds to His 80 with 3.10 and 3.11 Å bond lengths, Ile 81 with 3.10 Å bond length, and Arg 84 with 3.23 Å bond length (Fig. 5A, B). These three residues, which played key roles in Z5-10:Ac binding, were in the second transmembrane region of MmedOR49. An additional seven residues, Phe 78, Val 138, Ser 197, Ala 201, Val 202, Leu 307, and Val 308, with the above three key residues together formed the binding site (Fig. 5B, D, E). Pocket-finding analyses yielded 66 and 15 pockets from CASTp and DoGSiteScorer, respectively. The main active site (top pocket in the ranking) (Fig. S6) predicted by DoGSiteScorer and CASTp included 39 and 35 residues, respectively (Table S4).
Fig. 5.
Three-dimensional model and molecular docking of MmedOR49. (A) Binding model of Z5-10:Ac with MmedOR49. The helix and loop structure of MmedOR49 is indicated by the red color. The Z5-10:Ac binding conformation is indicated by the green color. (B) The main region of MmedOR49 binding with Z5-10:Ac. Light blue represents the key binding sites and dark blue represents the hydrophobic residues. (B) displays the enlargement of the white box area of (A). (C) Three-dimensional structure of MmedOR49. The image showing rainbow colors indicates the helix and loop structure. (D) Predicted protein topology of MmedOR49. The numbers indicate the seven transmembrane domains. The green amino acids are the key binding residues of MmedOR49 to Z5-10:Ac, and the binding pocket residues are highlighted in red. (E) The interaction of MmedOR49 with Z5-10:Ac visualized by LigPlot+. The green dashes and corresponding values represent hydrogen bonds and distances, respectively. The residues with hydrophobic interactions are shown as arcs with spokes radiating towards the ligand.
Preference of female wasps for Z5-10:Ac
In the three-cage olfactometer trial, the attraction of female adults to 1 ng/μL of Z5-10:Ac was significantly higher than that to the control (Fig. 6A). In contrast, 10 and 100 ng/μL and 1, 10, and 25 μg/μL of Z5-10:Ac failed to trigger the choice behavior of female wasps (Fig. 6B).
MmedOR49 knockdown
To further investigate the function of MmedOR49 in vivo, the RNAi coupled with behavior assay strategy was conducted. At 12 h following MmedOR49 dsRNA injection, no obvious change was observed in the level of MmedOR49 mRNA, but it was significantly decreased at 24, 36, and 48 h after injection (Fig. 6C). Consequently, 36 h-injected individuals were chosen for behavioral assays, which showed that the wasp’s original behavioral tendency to 1 ng/μL of Z5-10:Ac was eliminated by the knockdown (Fig. 6D).
Discussion
In both field and laboratory conditions, M. mediator females were attracted by the sex pheromone components of A. segetum. It was previously reported that the antennae of the female parasitoid have a strong electrophysiological response to Z9-14:Ald, the sex pheromone component of its main host, H. armigera, and the olfactory receptor IRs are involved in this semiochemical recognition [43]. These two phenomena suggest that M. mediator olfaction plays an important role in recognizing host sex pheromone components. Subsequently, TEVC recording was used to confirm the specificity of MmedOR49 for Z5-10:Ac, the main sex pheromone component of A. segetum. Here, the roles of MmedOR49 in vitro and in vivo were thoroughly characterized. Our research provides a basis for understanding the underlying mechanisms and the ecological significance of parasitoid wasp-host interactions in the process of sex pheromone perception.
Ecological role of MmedOR49 recognition of Z5-10:Ac
The ligand specificities of OR repertoires have been studied in several insects. Some insect ORs are activated by a narrow range of semiochemicals, whereas most respond to multiple ligands [23], [33], [58], [63]. However, both the deorphanized ORs reported in parasitoids respond to a narrow range of ligands, such as β-caryophyllene, (E)-α-farnesene, and cis-jasmone [34], [35]. Here, using Xenopus oocyte expression coupled with TEVC recording, MmedOR49 responded to four compounds, all of which belong to the ester class of chemicals. This is similar to the odorant range of ORs studied in other insects. In D. melanogaster, the majority of OR ligand clusters were esters and alcohols, which stimulated no or low responses of IR repertoires (another olfactory receptor family in insects). Instead, IR ligands are primarily carboxylic acids and amines, which trigger little or no response from ORs [64]. Consistently, two homologous MmedIRs primarily responded to acids and aldehydes, suggesting that the tuning spectra of ORs and IRs are complementary to assist parasitoids in processing habitats and hosts information in an energy-saving and efficient manner [43].
Z5-10:Ac was the main ligand of MmedOR49, producing the largest response among a panel of 118 odorants, including lepidopteran sex pheromones, plant volatiles, and other host-associated semiochemicals. Z5-10:AC, a general component of lepidopteran sex pheromones, both alone and blended with other sex pheromone compounds triggers significant electrophysiological and behavioral responses in males [47], [48], [65], [66], [67], [68]. Notably, Z5-10:AC is also a vital sex pheromone component of the turnip moth A. segetum and black cutworm moth A. ipsilon (Lepidoptera: Noctuidae). Both are destructive pests that damage a large number of crops worldwide and are also important hosts for M. mediator [47], [69], [70]. Thus, the female-biased MmedOR49 might be involved in recognizing the turnip moth sex pheromone.
Similarly, MmedIR64a2 has been shown to be activated by Z9-14:Ald [43], the main sex pheromone component of H. armigera [71], [72], [73], which is another host for M. mediator. These data suggest that different olfactory receptor families and multiple receptor proteins are activated by distinct and general or specific host pheromone components in wasps. M. mediator, a generalist larval endoparasitoid with a wide host range of Noctuidae pests [6], [7], [8], may use host pheromone components to recognize different host signals in a combinatorial coded pattern. Moreover, ethyl octanoate, ethyl heptanoate, and ethyl butyrate emitted from the H. armigera larvae body surface or fruits [74], [75], [76], [77] were recorded as secondary ligands of MmedOR49. The sensitivity of MmedOR49 to these three chemicals could be due to their structural similarities to Z5-10:Ac. As an inclusive characteristic of insect ORs, it also has been demonstrated that compounds with similar structures can be detected by distinct ORs. For example, RferOR1, a pheromone receptor in the red palm weevil (Rhynchophorus ferrugineus), can be activated by the main components of the aggregation pheromone and five other structurally related compounds [78]. Therefore, here, the effect of Z5-10:AC, the most important and special ligand of MmedOR49 on wasps, was focused on.
In this study, a dual choice behavior assay was carried out to test the behavioral activity of Z5-10:Ac against female wasps. A certain concentration of Z5-10:Ac attracted female adults, but the response was attenuated by MmedOR49 RNAi knockdown. Thus, MmedOR49 mediates the behavioral tendency of female wasps via the response to Z5-10:Ac. Coincidentally, Z9-14:Ald, the ligand of MmedIR64a2, also elicited electrophysiological responses in adults antennae [43]. These results suggest that not only the volatiles from host larvae or host plants are directly utilized by M. mediator in the host-locating process, but also the sex pheromone components of host adults are substantial cues for wasp host-locating behavior. Taken together, these results suggest that female parasitoids intelligently exploit the pheromone communication of their host adults to identify the habitat of host larvae. This complex procedure is assumed to be operated via an abundant number of chemosensory-related receptors that arose during the co-evolution of parasitoid-host interactions.
Increased expression of MmedOR49 in adult females
Quantification by qPCR showed that MmedOR49 expression is regulated during development. MmedOR49 expression was significantly increased in adult antennae compared to that in the pupae, but parasitism and mating experience had no significant effect on it. Therefore, it could be hypothesized that female wasps increase the expression of MmedOR49 immediately after emergence to complement their host-seeking behavior. Similarly, the expression of AlucOR28 in the antennae of Apolygus lucorum, mediating the behavioral activities of both sexes towards host plant volatiles, dramatically increased after adult emergence and reached its highest level 4 days after emergence [79]. In the malaria vector Anopheles coluzzii, the expression of four functional ORs significantly increased in the host-seeking female mosquitoes 4 days after emergence [80]. Increases in the transcript levels of olfactory-related genes primarily expressed in the antennae after adult emergence are consistent with the development of the insect olfactory system, demonstrating their critical functions in chemical communication in adults.
Molecular docking of MmedOR49 to Z5-10:Ac
Recently, molecular docking has been employed to predict binding sites and binding strengths of ORs with their candidate ligands. Moreover, putative binding pockets in protein structures can be identified using multiple bioinformatics tools. In the spruce bark beetle Ips typographus, homology modeling and molecular docking were used to predict OR46 and OR49 binding sites that were confirmed by site-directed mutagenesis and functional assays. Substitutions of key residues resulted in reduced or no odorant detection [81]. The binding affinity of the pheromone receptor for sex pheromone components has been verified via in silico docking in the red palm weevil R. ferrugineus [78]. The binding of OR27 to host plant volatiles has also been predicted in dark black chafer Holotrichia parallela [82]. In this study, the MmedOR49 − Z5-10:Ac interaction decoded using in silico docking predicted high binding affinity through hydrogen bonding between the receptor and ligand, which was in good agreement with the results of TEVC recording. Putative pocket predictions using two different tools (CASTp and DoGSiteScorer) identified similar active sites (pockets) based on residues, size, and surface area. Notably, comparing the results of putative pocket prediction and docking analyses revealed that Z5-10:Ac interacted with MmedOR49 residues in the identified active site. All of the key residues predicted to be responsible for interacting with the target ligand were shown to be part of the identified active site, suggesting the accuracy of the analysis. However, the key residues associated with the formation of the MmedOR49 − Z5-10:Ac complex need to be verified through further experiments, such as site-directed mutagenesis, to prove the functionality of the predicted residues. Furthermore, the key residues, such as His 80, Ile 81, and Arg 84, in ligand-binding sites should be investigated in future to design more active and effective chemicals for eco-friendly pest control. Along with the Orco cryo-EM structure [36], the discovery of the intact structure of insect ORs will provide a foundation that will be useful in the screening of unknown ligands for orphan receptors.
Conclusion
In the current study, it was proved that the female M. mediator could be attracted by the sex pheromone blend of A. segetum. The female-biased MmedOR49 specifically was tuned to Z5-10:Ac, a vital sex pheromone component of A. segetum and other Noctuidae pests. This component could trigger the behavioral response of female wasps in the olfactometer assay in the laboratory. Moreover, the knockdown assay using RNAi showed that MmedOR49 was necessary for the recognition of Z5-10:Ac. The ecological significance of these findings could be rationalized by the following conjecture: the parasitoid wasp recognizes the specific sex pheromone of the host adult using a certain OR to find potential host larval habitat and further accurately locates the host through direct host signals. Based on this study, deeper research is required to verify this hypothesis. The results of the current research will enrich the understanding of parasitoid ORs and help to comprehend the olfactory mechanism of parasitoid host localization. Furthermore, MmedOR49 and Z5-10:Ac could be used as potential molecular targets for designing efficient biological control strategies.
Compliance with ethics requirements
-
•
For studies on the insect species which has been used in our research works all the institutional and national guidelines for the care and use of laboratory animals were followed.
-
•
This article does not contain any studies with human subjects.
CRediT authorship contribution statement
Shuang Shan: Conceptualization, Methodology, Investigation, Data curation, Formal analysis, Writing - original draft, Writing - review & editing, Fund acquisition. Xuan Song: Methodology, Investigation. Adel Khashaveh: Conceptualization, Methodology, Investigation, Writing - original draft, Writing - review & editing. Shan-Ning Wang: Data curation, Formal analysis, Validation, Supervision. Zi-Yun Lu: Data curation, Formal analysis. Khalid Hussain Dhiloo: Software, Visulalization. Rui-Jun Li: Software, Visulalization. Yong-Jun Zhang: Conceptualization, Writing - original draft, Writing - review & editing, Validation, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31772176, 31972338), and the National Key Research and Development Program of China (2019YFD0300100), the China Postdoctoral Science Foundation (2020M670551) and Beijing Natural Science Foundation (6214044).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.03.006.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Beckage N.E., Gelman D.B. Wasp parasitoid disruption of host development: implications for new biologically based strategies for insect control. Annu Rev Entomol. 2004;49:299–330. doi: 10.1146/annurev.ento.49.061802.123324. [DOI] [PubMed] [Google Scholar]
- 2.Guo H., Wang C.Z. The ethological significance and olfactory detection of herbivore-induced plant volatiles in interactions of plants, herbivorous insects, and parasitoids. Arth-Plant Int. 2019;13(2):161–179. doi: 10.1007/s11829-019-09672-5. [DOI] [Google Scholar]
- 3.Russell M. A meta-analysis of physiological and behavioral responses of parasitoid wasps to flowers of individual plant species. Biol Control. 2015;82:96–103. doi: 10.1016/j.biocontrol.2014.11.014. [DOI] [Google Scholar]
- 4.Qasim M., Xiao H.M., He K., Omar M.A.A., Hussain D., Noman A., et al. Host-pathogen interaction between Asian citrus psyllid and entomopathogenic fungus (Cordyceps fumosorosea) is regulated by modulations in gene expression, enzymatic activity and HLB-bacterial population of the host. Comp Biochem Physiol C Toxicol Pharmacol. 2021;248:109112. doi: 10.1016/j.cbpc.2021.109112. [DOI] [PubMed] [Google Scholar]
- 5.Qasim M., Lin Y., Dash C.K., Bamisile B.S., Ravindran K., Islam S.U., et al. Temperature-dependent development of Asian citrus psyllid on various hosts, and mortality by two strains of Isaria. Microb Pathog. 2018;119:109–118. doi: 10.1016/j.micpath.2018.04.019. [DOI] [PubMed] [Google Scholar]
- 6.Khan S.M. Effectiveness of Microplitis mediator (HYM.: Braconidae) against its hosts Agrotis segetum and A. ipsilon (Lepidoptera: noctuidaeì) Pak J Biol Sci. 1999;2(2):344–346. [Google Scholar]
- 7.Lauro N., Kuhlmann U., Mason P.G., Holliday N.J. Interaction of a solitary larval endoparasitoid, Microplitis mediator, with its host, Mamestra brassicae: host acceptance and host suitability. J Appl Entomol. 2005;129(9-10):567–573. doi: 10.1111/j.1439-0418.2005.01011.x. [DOI] [Google Scholar]
- 8.Li J.C., Yan F.M., Coudron T.A., Pan W.L., Zhang X.F., Liu X.X. Field release of the parasitoid Microplitis mediator (Hymenoptera: braconidae) for control of Helicoverpa armigera (Lepidoptera: noctuidae) in cotton fields in northwestern China's xinjiang province. Environ Entomol. 2006;35:694–699. doi: 10.1603/0046-225X-35.3.694. [DOI] [Google Scholar]
- 9.Hülbert D., Süss A. Biologie und wirtschaftliche Bedeutung der Wintersaateule, Scotia (Agrotis) segetum Schiffermüller (Lepidoptera: Noctuidae) Beitr Ent. 1983;33:383–438. doi: 10.21248/contrib.entomol.33.2.383-438. [DOI] [Google Scholar]
- 10.Anonymous. Cotton. EPPO Bull 2004;34:57-63. doi: https://doi.org/10.1111/j.1365-2338.2004.00699.x.
- 11.Esbjerg P., Sigsgaard L. Phenology and pest status of Agrotis segetum in a changing climate. Crop Prot. 2014;62:64–71. doi: 10.1016/j.cropro.2014.04.003. [DOI] [Google Scholar]
- 12.Lemic D., Drmić Z., Bažok R. Population dynamics of noctuid moths and damage forecasting in sugar beet. Agr For Entomol. 2016;18(2):128–136. doi: 10.1111/afe.12145. [DOI] [Google Scholar]
- 13.Nowinszky L., Kiss M., Puskás J., Barta A. Light-trap catch of turnip moth (Agrotis segetum Denis et Schiffermüller, 1775) in connection with the night sky polarization phenomena. Global J Res Rev. 2017;4:22. doi: 10.21767/2393-8854.100022. [DOI] [Google Scholar]
- 14.Clyne P.J., Warr C.G., Freeman M.R., Lessing D., Kim J., Carlson J.R. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22(2):327–338. doi: 10.1016/S0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 15.Gao Q., Chess A. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics. 1999;60(1):31–39. doi: 10.1006/geno.1999.5894. [DOI] [PubMed] [Google Scholar]
- 16.Vosshall L.B. The molecular logic of olfaction in Drosophila. Chem Senses. 2001;26:207–213. doi: 10.1093/chemse/26.2.207. [DOI] [PubMed] [Google Scholar]
- 17.Robertson H.M., Warr C.G., Carlson J.R. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2003;100(suppl_2):14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lundin C., Käll L., Kreher S.A., Kapp K., Sonnhammer E.L., Carlson J.R., et al. Membrane topology of the Drosophila OR83b odorant receptor. FEBS Lett. 2007;581(29):5601–5604. doi: 10.1016/j.febslet.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleischer J., Pregitzer P., Breer H., Krieger J. Access to the odor world: olfactory receptors and their role for signal transduction in insects. Cell Mol Life Sci. 2018;75(3):485–508. doi: 10.1007/s00018-017-2627-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez D., Witzgall P., Walker W.B. Protocol for heterologous expression of insect odourant receptors in Drosophila. Front Ecol Evol. 2016;4:1–5. doi: 10.3389/fevo.2016.00024. [DOI] [Google Scholar]
- 21.Wang B., Liu Y., He K., Wang G.R. Comparison of research methods for functional characterization of insect olfactory receptors. Sci Rep. 2016;6:32806. doi: 10.1038/srep32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang R.B., Wang B., Grossi G., Falabella P., Liu Y., Yan S.C., et al. Molecular basis of alarm pheromone detection in aphids. Curr Biol. 2017;27(1):55–61. doi: 10.1016/j.cub.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 23.Guo M.B., Du L.X., Chen Q.Y., Feng Y.L., Zhang J., Zhang X.X., et al. Odorant receptors for detecting flowering plant cues are functionally conserved across moths and butterflies. Mol Biol Evol. 2021;38(4):1413–1427. doi: 10.1093/molbev/msaa300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hallem E.A., Ho M.G., Carlson J.R. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117(7):965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 25.Liu X.L., Zhang J., Yan Q., Miao C.L., Han W.K., Hou W., et al. The molecular basis of host selection in a crucifer-specialized moth. Curr Biol. 2020;30(22):4476–4482.e5. doi: 10.1016/j.cub.2020.08.047. [DOI] [PubMed] [Google Scholar]
- 26.Zhang R.B., Liu Y., Yan S.C., Wang G.R. Identification and functional characterization of an odorant receptor in pea aphid, Acyrthosiphon pisum. Insect Sci. 2019;26:58–67. doi: 10.1111/1744-7917.12510. [DOI] [PubMed] [Google Scholar]
- 27.An X.K, Khashaveh A., Liu D.F., Xiao Y., Wang Q., Wang S.N., et al. Functional characterization of one sex pheromone receptor (AlucOR4) in Apolygus lucorum (Meyer-Dür) J Insect Physiol. 2020;120:103986. doi: 10.1016/j.jinsphys.2019.103986. [DOI] [PubMed] [Google Scholar]
- 28.Khashaveh A., An X.K., Shan S., Xiao Y., Wang Q., Wang S.N., et al. Deorphanization of an odorant receptor revealed new bioactive components for green mirid bug Apolygus lucorum (Hemiptera: Miridae) Pest Manag Sci. 2020;76:1626–1638. doi: 10.1002/ps.5682. [DOI] [PubMed] [Google Scholar]
- 29.Xiao Y., An X.K., Khashaveh A., Shan S., Wang Q., Wang S.N., et al. Broadly tuned odorant receptor AlinOR59 involved in chemoreception of floral scent in Adelphocoris lineolatus. J Agric Food Chem. 2020;68(47):13815–13823. doi: 10.1021/acs.jafc.0c04434. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q., Xiao Y., An X.K., Shan S., Khashaveh A., Gu S.H., et al. Functional characterization of a candidate sex pheromone receptor AlinOR33 involved in the chemoreception of Adelphocoris lineolatus. J Agric Food Chem. 2021;69(24):6769–6778. doi: 10.1021/acs.jafc.1c01319. [DOI] [PubMed] [Google Scholar]
- 31.Zhou X.F., Rokas A., Berger S.L., Liebig J., Ray A., Zwiebel L.J. Chemoreceptor evolution in Hymenoptera and its implications for the evolution of eusociality. Genome Biol Evol. 2015;7(8):2407–2416. doi: 10.1093/gbe/evv149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Missbach C., Dweck H.K., Vogel H., Vilcinskas A., Stensmyr M.C., Hansson B.S., et al. Evolution of insect olfactory receptors. eLife. 2014;3:e02115. doi: 10.7554/eLife.02115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Fouchier A., Walker W.B., Montagné N., Steiner C., Binyameen M., Schlyter F., et al. Functional evolution of Lepidoptera olfactory receptors revealed by deorphanization of a moth repertoire. Nat Commun. 2017;8:15709. doi: 10.1038/ncomms15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y.L., Chen Q., Guo J.Q., Li J., Wang J.T., Wen M., et al. Molecular basis of peripheral olfactory sensing during oviposition in the behavior of the parasitic wasp Anastatus japonicus. Insect Biochem Mol Bio. 2017;89:58–70. doi: 10.1016/j.ibmb.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 35.Sun Y.L., Dong J.F., Ning C., Ding P.P., Huang L.Q., Sun J.G., et al. An odorant receptor mediates the attractiveness of cis-jasmone to Campoletis chlorideae, the endoparasitoid of Helicoverpa armigera. Insect Mol Biol. 2019;28(1):23–34. doi: 10.1111/imb.12523. [DOI] [PubMed] [Google Scholar]
- 36.Butterwick J.A., del Mármol J., Kim K.H., Kahlson M.A., Rogow J.A., Walz T., et al. Cryo-EM structure of the insect olfactory receptor Orco. Nature. 2018;560(7719):447–452. doi: 10.1038/s41586-018-0420-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang S., Zhang Y.J., Su H.H., Gao X.W., Guo Y.Y. Identification and expression pattern of putative odorant binding proteins and chemosensory proteins in antennae of the Microplitis mediator (Hymenoptera: Braconidae) Chem Senses. 2009;34(6):503–512. doi: 10.1093/chemse/bjp027. [DOI] [PubMed] [Google Scholar]
- 38.Zhang S., Chen L.Z., Gu S.H., Cui J.J., Gao X.W., Zhang Y.J., et al. Binding characterization of recombinant odorant-binding proteins from the parasitic wasp, Microplitis mediator (Hymenoptera: Braconidae) J Chem Ecol. 2011;37(2):189–194. doi: 10.1007/s10886-010-9902-3. [DOI] [PubMed] [Google Scholar]
- 39.Ma L., Gu S.H., Liu Z.W., Wang S.N., Guo Y.Y., Zhou J.J., et al. Molecular characterization and expression profiles of olfactory receptor genes in the parasitic wasp, Microplitis mediator (Hymenoptera: Braconidae) J Insect Physiol. 2014;60:118–126. doi: 10.1016/j.jinsphys.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 40.Wang S.N., Peng Y., Lu Z.Y., Dhiloo K.H., Zheng Y., Shan S., et al. Cloning and expression profile of ionotropic receptors in the parasitoid wasp Microplitis mediator (Hymenoptera: Braconidae) J Insect Physiol. 2016;90:27–35. doi: 10.1016/j.jinsphys.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 41.Peng Y., Wang S.N., Li K.M., Liu J.T., Zheng Y., Shan S., et al. Identification of odorant binding proteins and chemosensory proteins in Microplitis mediator as well as functional characterization of chemosensory protein 3. PLoS ONE. 2017;12(7):e0180775. doi: 10.1371/journal.pone.0180775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng Y., Wang S.N., Peng Y., Lu Z.Y., Shan S., Yang Y.Q., et al. Functional characterization of a Niemann-Pick type C2 protein in the parasitoid wasp Microplitis mediator. Insect Sci. 2018;25(5):765–777. doi: 10.1111/1744-7917.12473. [DOI] [PubMed] [Google Scholar]
- 43.Shan S., Wang S.N., Song X., Khashaveh A., Lu Z.Y., Dhiloo K., et al. Antennal ionotropic receptors IR64a1 and IR64a2 of parasitoid wasp Microplitis mediator (Hymenoptera: Braconidate) collaboratively perceive habitat/host cues in host-seeking. Insect Biochem Mol Biol. 2019;114 doi: 10.1016/j.ibmb.2019.103204. [DOI] [PubMed] [Google Scholar]
- 44.Shan S., Wang S.N., Song X., Khashaveh A., Lu Z.Y., Dhiloo K., et al. Molecular characterization and expression of sensory neuron membrane proteins SNMPs in the parasitoid Microplitis mediator (Hymenoptera: Braconidae) Insect Sci. 2020;27:425–439. doi: 10.1111/1744-7917.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang S.N., Peng Y., Lu Z.Y., Dhiloo K.H., Gu S.H., Li R.J., et al. Identification and expression analysis of putative chemosensory receptor genes in Microplitis mediator by antennal transcriptome screening. Int J Biol Sci. 2015;11(7):737–751. doi: 10.7150/ijbs.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang S.N., Shan S., Zheng Y., Peng Y., Lu Z.Y., Yang Y.Q., et al. Gene structure and expression characteristic of a novel odorant receptor gene cluster in the parasitoid wasp Microplitis mediator (Hymenoptera: Braconidae) Insect Mol Biol. 2017;26(4):420–431. doi: 10.1111/imb.12306. [DOI] [PubMed] [Google Scholar]
- 47.Löfstedt C., Van Der Pers J.N.C., Lofqvist J., Lanne B.S., Appelgren M., Bergström G., et al. Sex pheromone components of the turnip moth, Agrotis segetum. J Chem Ecol. 1982;8(10):1305–1321. doi: 10.1007/BF00987764. [DOI] [PubMed] [Google Scholar]
- 48.Löfstedt C., Linn C.E., Löfqvist J. Behavioral responses of male turnip moths, Agrotis segetum, to sex pheromone in a flight tunnel and in the field. J Chem Ecol. 1985;11(9):1209–1221. doi: 10.1007/BF01024109. [DOI] [PubMed] [Google Scholar]
- 49.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., et al. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37(5):1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Omasits U., Ahrens C.H., Müller S., Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014;30:884–886. doi: 10.1093/bioinformatics/btt607. [DOI] [PubMed] [Google Scholar]
- 52.Tsirigos K.D., Peters C., Shu N., Käll L., Elofsson A. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 2015;43(W1):W401–W407. doi: 10.1093/nar/gkv485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 54.Cox K.H., Deleon D.V., Angerer L.M., Angerer R.C. Detection of mRNAs in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol. 1984;101:485–502. doi: 10.1016/j.cub.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 55.Yang Y., Krieger J., Zhang L., Breer H. The olfactory co-receptor Orco from the migratory locust (Locusta migratoria) and the desert locust (Schistocerca gregaria): identification and expression pattern. Int J Biol Sci. 2012;8(2):159–170. doi: 10.7150/ijbs.8.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu H.Z., Guo M., Yang Y., You Y.W., Zhang L. Differential expression of two novel odorant receptors in the locust (Locusta migratoria) BMC Neurosci. 2013;14:50. doi: 10.1186/1471-2202-14-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu X., You Y.W., Zhang L. Localization of odorant receptor genes in locust antennae by RNA in situ hybridization. J Vis Exp. 2017;125 doi: 10.3791/55924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang G.R., Carey A.F., Carlson J.R., Zwiebel L.J. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci U S A. 2010;107(9):4418–4423. doi: 10.1073/pnas.0913392107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang J.Y., Anishchenko I., Park H., Peng Z.L., Ovchinnikov S., Baker D. Improved protein structure prediction using predicted interresidue orientations. Proc Natl Acad Sci U S A. 2020;117(3):1496–1503. doi: 10.1073/pnas.1914677117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O'Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: An open chemical toolbox. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Volkamer A., Kuhn D., Grombacher T., Rippmann F., Rarey M. Combining global and local measures for structure-based druggability predictions. J Chem Inf Model. 2012;52(2):360–372. doi: 10.1021/ci200454v. [DOI] [PubMed] [Google Scholar]
- 63.Carey A.F., Carlson J.R. Insect olfaction from model systems to disease control. Proc Natl Acad Sci U S A. 2011;108(32):12987–12995. doi: 10.1073/pnas.1103472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Silbering A.F., Rytz R., Grosjean Y., Abuin L., Ramdya P., Jefferis G.S.X.E., et al. Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J Neurosci. 2011;31(38):13357–13375. doi: 10.1523/JNEUROSCI.2360-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bestmann H.J., Vostrowsky O., Koschatzky K.H., Platz H., Brosche T., Kantardjiew I., et al. (Z)-5-Decenyl acetate, a sex attractant for the male turnip moth Agrotis segetum (Lepidoptera) Angew Chem. 1978;17(10):768. [Google Scholar]
- 66.Priesner E. (Z)-3-and (Z)-5-Decenyl acetates, Sex-attractant components for male Eustrotia uncula Cl. (Lepidoptera: Noctuidae) Zeitschrift für Naturforschung C. 1988;43:961–962. doi: 10.1515/znc-1988-11-1226. [DOI] [Google Scholar]
- 67.Priesner E. Two-component sexual attractant for male Batrachedra pinicolella (Zell.) (Lepidoptera: Batrachedridae) Zeitschrift für Naturforschung C. 1989;44:1061–1062. doi: 10.1515/znc-1989-11-1230. [DOI] [Google Scholar]
- 68.Suckling D.M., Gibb A.R., Gourlay H., Conant P., Hirayama C., Leen R. Sex attractant for the gorse biocontrol agent Agonopterix ulicetella (Oecophoridae) N Z Plant Prot. 2000;53:66–70. [Google Scholar]
- 69.Rings R.W., Arnold F.J., Johnson B.A. Host range of the black cutworm on vegetables: a bibliography. Bull E S A. 1975;21(4):229–234. doi: 10.1093/besa/21.4.229. [DOI] [Google Scholar]
- 70.Clement S.L., Show E.D., Way M.O. Black cutworm pheromone trapping in strawberries. Calif Agr. 1982;36:2021. [Google Scholar]
- 71.Kehat M., Gothilf S., Dunkelblum E., Greenberg S. Field evaluation of female sex pheromone components of the cotton bollworm Heliothis armgiera. Entomol Exp Appl. 1980;27:188–193. doi: 10.1111/j.1570-7458.1980.tb02963.x. [DOI] [Google Scholar]
- 72.Kehat M., Dunkelblum E. Behavioral responses of male Heliothis armigera (Lepidoptera: noctuidae) moths in a flight tunnel to combinations of components identified from female sex pheromone glands. J Insect Behav. 1990;3(1):75–83. doi: 10.1007/BF01049196. [DOI] [Google Scholar]
- 73.Chang H.T., Liu Y., Ai D., Jiang X.C., Dong S.L., Wang G.R. A pheromone antagonist regulates optimal mating time in the moth Helicoverpa armigera. Curr Biol. 2017;27(11):1610–1615.e3. doi: 10.1016/j.cub.2017.04.035. [DOI] [PubMed] [Google Scholar]
- 74.Alonzo G., Saiano F., Tusa N., Fatta Del Bosco S. Analysis of volatile compounds released from embryogenic cultures andsomatic embryos of sweet oranges by head space SPME. Plant Cell Tissue Organ Cult. 2001;66:31–34. doi: 10.1023/A:1010608417457. [DOI] [Google Scholar]
- 75.Izumi Y., Tian R., Sonoda S., Imayoshi Y., Iwabuchi H., Miyashita Y., et al. Analysis of peach fruit headspace volatiles and response by the fruit-piercing moth Oraesia excavata (Lepidoptera: Noctuidae) Appl Entomol Zool. 2015;50(2):231–238. doi: 10.1007/s13355-015-0330-2. [DOI] [Google Scholar]
- 76.Shi Q.X., Luo Q.H., Zhao L., Zhou Z.X., He G.Q., Wei W. Extraction and identification of maize volatiles and cuticular volatiles of larval Helicoverpa armigera (Lepidoptera: noctuidae) related to host habitat location and host location of parasitic wasp Microplitis mediator (Hymenoptera: braconidae) Acta Entomol Sin. 2015;58:244–255. doi: 10.16380/j.kcxb.2015.03.003. [DOI] [Google Scholar]
- 77.Lee S., Kim Y.J., Jones W.D. Central peptidergic modulation of peripheral olfactory responses. BMC Biol. 2017;15:35. doi: 10.1186/s12915-017-0374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Antony B., Johny J., Montagné N., Jacquin‐Joly E., Capoduro R., Cali K., et al. Pheromone receptor of the globally invasive quarantine pest of the palm tree, the red palm weevil (Rhynchophorus ferrugineus) Mol Ecol. 2021;30(9):2025–2039. doi: 10.1111/mec.15874. [DOI] [PubMed] [Google Scholar]
- 79.Yan S.W., Zhang J., Liu Y., Li G.Q., Wang G.R. An olfactory receptor from Apolygus lucorum (Meyer-Dur) mainly tuned to volatiles from flowering host plants. J Insect Physiol. 2015;79:36–41. doi: 10.1016/j.jinsphys.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 80.Omondi A.B., Ghaninia M., Dawit M., Svensson T., Ingell R. Age-dependent regulation of host seeking in Anopheles coluzzii. Sci Rep. 2019;9:9699. doi: 10.1038/2Fs41598-019-46220-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yuvaraj J.K., Roberts R.E., Sonntag Y., Hou X.Q., Grosse-Wilde E., Machara A., et al. Putative ligand binding sites of two functionally characterized bark beetle odorant receptors. BMC Biol. 2021;19:16. doi: 10.1186/s12915-020-00946-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang X., Wang S., Yi J.K., Li Y.S., Liu J.N., Wang J., et al. Three host plant volatiles, hexanal, lauric acid, and tetradecane, are detected by an antenna-biased expressed odorant receptor 27 in the dark black chafer Holotrichia parallela. J Agr Food Chem. 2020;68(28):7316–7323. doi: 10.1021/acs.jafc.0c00333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.