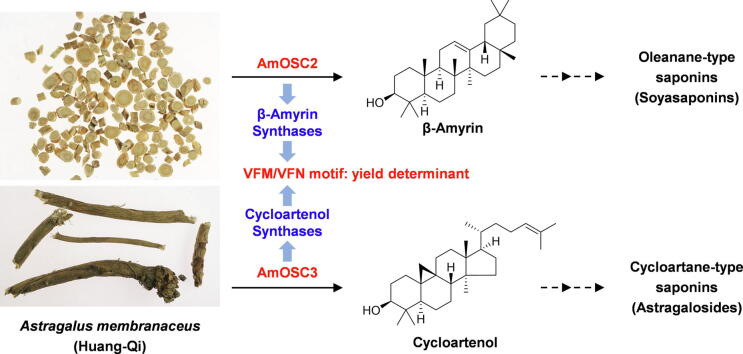
Graphical abstract
Oxidosqualene cyclases AmOSC2 and AmOSC3 participated in the biosynthesis of medicinally important soyasaponins and astragalosides in A. membranaceus. Conserved motifs VFM/VFN were found as functional signatures and yield determinants for β-amyrin/cycloartenol synthases.
Keywords: Astragalus membranaceus, Oxidosqualene cyclase (OSC), β-Amyrin, Cycloartenol, Biosynthesis, Conserved motif
Highlights
-
•
β-Amyrin and cycloartenol synthases in the medicinal plant A. membranaceus were characterized.
-
•
They contribute to the biosynthesis of medicinally important astragalosides and soyasaponins in Huang-Qi.
-
•
Conserved motifs VFM/VFN are critical for the function of β-amyrin and cycloartenol synthases.
Abstract
Introduction
Triterpenoids and saponins have a broad range of pharmacological activities. Unlike most legumes which contain mainly oleanane-type scaffold, Astragalus membranaceus contains not only oleanane-type but also cycloartane-type saponins, for which the biosynthetic pathways are unknown.
Objectives
This work aims to study the function and catalytic mechanism of oxidosqualene cyclases (OSCs), one of the most important enzymes in triterpenoid biosynthesis, in A. membranaceus.
Methods
Two OSC genes, AmOSC2 and AmOSC3, were cloned from A. membranaceus. Their functions were studied by heterologous expression in tobacco and yeast, together with in vivo transient expression and virus-induced gene silencing. Site-directed mutagenesis and molecular docking were used to explain the catalytic mechanism for the conserved motif.
Results
AmOSC2 is a β-amyrin synthase which showed higher expression levels in underground parts. It is associated with the production of β-amyrin and soyasaponins (oleanane-type) in vivo. AmOSC3 is a cycloartenol synthase expressed in both aerial and underground parts. It is related to the synthesis of astragalosides (cycloartane-type) in the roots, and to the synthesis of cycloartenol as a plant sterol precursor. From AmOSC2/3, conserved triad motifs VFM/VFN were discovered for β-amyrin/cycloartenol synthases, respectively. The motif is a critical determinant of yield as proved by 10 variants from different OSCs, where the variant containing the conserved motif increased the yield by up to 12.8-fold. Molecular docking and mutagenesis revealed that Val, Phe and Met residues acted together to stabilize the substrate, and the cation-π interactions from Phe played the major role.
Conclusion
The study provides insights into the biogenic origin of oleanane-type and cycloartane-type triterpenoids in Astragalus membranaceus. The conserved motif offers new opportunities for OSC engineering.
Introduction
Triterpenes are one of the largest and most structurally diverse classes of plant natural products. They protect plants from pathogens, and determine agronomically important traits such as flavor [1], [2]. They also have a broad range of pharmacological activities. Notably, many plant-derived triterpenes and saponins (glycosylated triterpenes) are used as medicines or dietary supplements. Examples include glycyrrhizin from Glycyrrhiza uralensis for hepatoprotection, anti-cancer ginsenosides from Panax ginseng, and the vaccine adjuvant QS-21 from Quillaja Saponaria [3], [4], [5]. More than 20,000 triterpenes have been reported [2]. These compounds can be categorized according to their structural backbones, the most common being β-amyrin.
Legumes (members of the Fabaceae family) produce various triterpenoids, most of which have the pentacyclic β-amyrin backbone [6], [7]. For example, the major triterpenoids found in barrel clover (Medicago truncatula), licorice (Glycyrrhiza glabra), and soybean (Glycine max) are derived from the β-amyrin-based scaffolds medicagenic acid, glycyrrhetinic acid, and soyasapogenol B, respectively (Fig. S1) [3], [8]. For the genus Astragalus, besides β-amyrin backbones, there are a lot of triterpenoids based on the tetracyclic skeleton, cycloartenol. Cycloartenol is well known as a precursor for the biosynthesis of essential plant sterols [9], as well as for steroidal alkaloids such as tomatine [10]. It could also serve as a precursor of specialized triterpenes from genus Astragalus (Fabaceae) and Cimicifuga (Ranunculaceae) [11], [12]. The roots of Astragalus membranaceus and A. membranaceus var. mongholicus are used as a tonic herb Huang-Qi in traditional Chinese medicine. Triterpenoids are the major effective constituents [13]. In total 77 triterpenoids have been purified from these two plants, including 51 cycloartane-type and 24 oleanane-type [14]. Representative compounds such as astragaloside IV and soyasaponin I, has been reported for cardiovascular protective effects and anti-inflammatory activities, respectively [14], [15].
Triterpene scaffolds such as β-amyrin and cycloartenol are formed by cyclization of the linear precursor 2,3-oxidosqualene, a process that is catalyzed by enzymes known as oxidosqualene cyclases (OSCs). Different OSCs generate different triterpene backbones through a cascade mechanism involving substrate folding, protonation, cyclization/rearrangement, and deprotonation/water capture [2], [16], [17]. Plant OSCs share several conserved motives including DCTAE, MXCXCR, and QXXXXXW, which contribute to reaction initiating, substrate binding, product specificity, and carbon cation stabilization [16]. Around 170 plant OSCs have now been characterized, collectively synthesizing more than 60 different types of triterpene backbones [16]. Most legume species investigated so far each have multiple OSCs, primarily β-amyrin synthases (βASs), cycloartenol synthases (CASs), and lupeol synthases [2], [16], [18], [19]. In order to explore the biosynthesis of oleanane-type and cycloartane-type triterpenoids in Astragalus species, especially why unlike other legumes, Astragalus species produce both cycloartane-type and oleanane-type triterpenoids, it is necessary to firstly investigate the nature and functions of their OSCs.
In this study, we discovered two OSCs in A. membranaceus. Their functions were characterized by heterologous expression in N. benthamiana and yeast, and by in vivo experiments including transient expression, virus-induced gene silencing, and RNA interference. We show that AmOSC2 and AmOSC3 make primarily the triterpenoid scaffolds β-amyrin and cycloartenol, and contribute to the biosynthesis of oleanane-type and cycloartane-type triterpenoids in vivo, respectively. We also discovered that most βAS and CAS have a conserved amino acid motif VFM/VFN. This motif is not conserved in AmOSC2 and led to a lower yield of β-amyrin. Mutagenesis and molecular docking are carried out to verify the role of this conservative region. Our results provide insights into triterpene biosynthesis in A. membranaceus, and also a conserved motif to improve the yield for OSCs.
Material and methods
Plant material and triterpenoid reference standards
Seeds were purchased from Anguo FengHua Seed Station (Hebei, China) and identified as Astragalus membranaceus based on psbA-trnH fragments [20]. The seeds were sown in mixotrophic soil and cultivated in 25 °C (16 h/8 h-light/dark). These plants were used for chemical analysis (3-month-old), transient expression and VIGS (2-month-old). For gene cloning, expression analysis and RNAi (2-week-old), the seedlings were sterilized and germinated as described in Supporting Information. In addition, a 3-year-old A. membranaceus plant was collected from Daxing’anling (Heilongjiang, China). Reference standards were purchased or purified in our group [21] as described in the Supporting Information.
Transcriptome and phylogenetic analysis
Four transcriptome datasets (NCBI SRA database numbers SRR923811, ERR706814, SRR5343992 and SRR5343993, Table S1) were used in this study. The data were assembled using Trinity software. Candidate OSC genes were identified using tBLASTn searches with GgbAS1 (Genbank AB037203) [22] and GgCAS1 (Genbank AB025968) [23] as templates. For phylogenetic analysis of OSCs, a total of 98 amino acid sequences, including β-amyrin synthase, cycloartenol synthase, and other functional OSCs were used (Table S2). Sequences were aligned using ClustalW and evolutionary distances computed through MEGA6 using the Maximum Likelihood method with default parameters while bootstrap = 1000 [24], [25]. All positions containing gaps and missing data were eliminated.
Molecular cloning and construct generation
Total RNA was extracted from roots of 2-week-old A. membranaceus plants and the cDNA was synthesized (Supporting Information). PCR was performed using Phusion High Fidelity DNA polymerase (New England BioLabs, US) or TransScript KD Plus DNA polymerases (Transgen Biotech, CN). The coding regions of AmOSC2 and AmOSC3 were amplified using primers in Table S3, and were then cloned into pDONR207 vectors following the Gateway Technology manual (Invitrogen). After verification of the sequences, the genes were transferred into pEAQ-HT vectors [26] using for transient expression in N. benthamiana and A. membranaceus.
For heterologous expression in yeast, genes were amplified using primers shown in Table S4. The PCR products were inserted into pYES2 vector (Invitrogen) using Quick-Change method [27]. Other known OSC genes (TcOSC1, TkOSC6, GuOSC, PgPNY1, PgPNY2, PgPNX1, LjOSC3, AtPEN1) were constructed into pYES2 vectors using the same method (Table S4 and Table S5). Plasmids for site-directed mutagenesis of OSCs were constructed using Fast Mutagensis System kit (TransGen Biotech, CN) using the primers in Table S6. All plasmids were confirmed by sequencing and then transformed into the GIL77 yeast strain [28] using Frozen-EZ Yeast Transformation II Kit (Zymoresearch, US) for expression.
For VIGS and RNAi experiment, specific fragments were cloned from AmOSC2 and AmOSC3 with primers shown in Table S7 and Table S8, respectively. The fragments were then inserted into the pTRV2 vector using pEASY-Uni Seamless Cloning and Assembly Kit (TransGen Biotech, CN) for VIGS, or inserted into pDONR207 and pK7WGIGW2R vectors using Gateway Technology [29].
Gene expression analysis
Total RNA was extracted from 0-day to 2-week A. membranaceus seedlings, and the cDNA was synthesized following the Supporting Information. Gene expressions were determined by real-time qPCR using gene-specific primers (Table S9) and TransStart Green qPCR SuperMix (Transgen Biotech, CN) on an Agilent MX3005P real-time PCR system (Agilent Technologies, US). All qPCR data were normalized to a 217-bp fragment of the A. membranaceus 18S RNA [30].
Transient expression in Nicotiana benthamiana and Astragalus membranaceus
Transient expression was carried out using the pEAQ-HT-DEST1 expression constructs in Agrobacterium tumefaciens strain LBA4404 following our previous report [31]. The A. tumefaciens cell suspensions were infiltrated into the undersides of leaves of 4-week-old N. benthamiana plants, and the leaves were harvested 5 days after agro-infiltration and freeze-dried. For transient expression in A. membranaceus, 2-month-old plants were used, and other steps were consistent with N. benthamiana. Scaled-up heterologous expression in N. benthamiana was described in the Supporting Information.
Virus-induced gene silencing (VIGS)
pTRV1, pTRV2, pTRV2/PDS (phytoene desaturase as a positive control), pTRV2/AmOSC2793-1101, and pTRV2/AmOSC31553-1893 constructs were introduced separately into A. tumefaciens strain GV3101 by electroporation. A 40-mL culture of each strain was incubated at 28 °C in LB broth (50 μg/mL kanamycin and 100 μg/mL rifampicin) until OD600 = 0.6. After centrifugation, bacteria were re-suspended with MMA buffer to OD600 = 1. Agrobacterium cultures containing pTRV1 (encodes proteins for viral replication and movement) and different pTRV2 (encodes the coat protein and harbors the sequence used for VIGS) constructs were mixed in 1:1 ratio and kept in dark at room temperature for 2 h. The mixture was infiltrated into the underside of the leaves of 2-month-old A. membranaceus plants with a 1-mL syringe [32], [33]. After one week, leaves were harvested for GC/MS analysis.
RNA interference (RNAi)
The recombinant plasmids pK7WGIGW2R containing RNAi fragments of AmOSCs and an empty vector pK7WG2R-EV as negative control were transformed into Agrobacterium rhizogenes A4 by electroporation respectively. Positive transformants were cultured in LB medium supplemented with spectinomycin (50 mg/L) and kanamycin (50 mg/L) at 28 °C until OD600 = 0.6. Ten milliliters of the bacteria were collected and resuspended using 2 mL of sterilized water containing 50 µM acetosyrengone. Two-week-old A. membranaceus aseptic seedlings were infected with the bacterial solution. The infected seedlings were co-cultured on B5 solid media for one week and transferred onto B5 solid media containing 3% sucrose and 500 mg/L cefotaxime (Sangon Biotech, CN) for about 4 weeks at 25 °C (12 h light/12 h dark). Positive hairy roots were screened out according to the fluorescence of dsRed protein (554 nm for exciting and 586 nm for emitting light), and were then transferred onto a new B5 solid media containing 3% sucrose and 500 mg/L cefotaxime at 25 °C (24 h dark). After 5 weeks, the positive hairy roots were harvested.
Heterologous expression in yeast
Yeast expression was carried out in strain GIL77 (gal2 hem3-6 erg7 ura3-167) following our previous study [21]. Briefly, pYES2 plasmids containing AmOSC2 and AmOSC3 and an empty pYES2 vector were transformed into GIL77. The yeast strains were grown at 30 °C in 40 mL cultures in selective medium [SC-URA + 2% (w/v) glucose + supplements] for 48 h with shaking (200 rpm). The supplements used were as follows: ergosterol 20 µg mL−1; hemin 13 µg mL−1; and Tween 80 5 mg mL−1 (Yuanye Bio-Technology, CN). Cells were then pelleted (collected by centrifugation at a speed of 500 g for 5 min) and transferred to 40 mL induction medium [SC-URA + 2% (w/v) galactose + supplements], and incubated for 24 h at 30 °C. The purification of yeast microsomes was performed following previous reports [34], [35] and was described in the Supporting Information. Scaled-up heterologous expression in yeast was described in the Supporting Information.
Molecular simulation
For homology modeling of AmOSC2 and its mutant, human lanosterol synthase (PDB ID: 1W6J) [36] was used as a template. Simulated protein structures were generated using SWISS-MODEL (swissmodel.expasy.org). Molecular docking of protein models and intermediates were carried out by autodock [37]. The conformation result with the lowest binding energy was used for further study. Models were visualized by using AutoDocktools v1.5.6 and Pymol.
GC/MS analysis
An aliquot of 5 mg of dried plant samples were extracted with 0.5 mL of saponification reagent (10% (w/v) KOH in 90% (v/v) ethanol, containing 10 μg/mL of coprostanol or 1.5 μg/mL of betulin as the internal standard), and incubated at 70 °C for 2 h before extraction with 0.5 mL of ethyl acetate twice. The ethyl acetate extracts were combined and concentrated to 250 μL using a Speedvac (Hualida Co., CN). Yeast cells (from 40-mL culture) were extracted by ultrasonication for 1 h with 10 mL of 20% (w/v) KOH in 50% (v/v) ethanol, containing the same internal standards above. The mixtures were extracted three times with 10 mL hexane and the hexane extracts were concentrated to 1 mL. Aliquots of the plant extracts (50 μL) or yeast extracts (50 μL) were then dried down using a water bath at 70 °C and the residues were re-suspended in 50 μL of Tri-Sil Z reagent (Sigma-Aldrich, USA) or TMS-HT (TCI, JPN) before incubating at 70 °C for 30 min. An additional 50 μL of ethyl acetate or 200 μL of hexane was added into the derivative products for plant and yeast respectively. After centrifugation, the supernatant was used for GC/MS detection.
For functional characterization of AmOSC genes, GC/MS analysis was performed on an Agilent GC–MSD (7890B-5977A) equipped with a Zebron™ ZB-5HT, GC Cap. Column (30 m × 0.25 mm × 0.10 µm, Phenomenex). An 1-μL aliquot was injected into the GC split/splitless inlet (250 °C) using a pulsed splitless mode (30 psi pulse pressure). The transfer line temperature was 280 °C. The oven temperature program was set to 170 °C for 2 min, followed by a ramp to 300 °C at 20 °C/min and held at 300 °C for 11.5 min to give a final run time of 20 min. MS scan range, m/z 60–800; scan time, 0.1184 s (per spectrum); inter-scan delay time, 0.0205 s (per spectrum); solvent delay, 8 min. For chemical analysis, transient expression, VIGS, and mutagenesis analysis, GC/MS analysis was performed on an ISQ 7000 Single Quadrupole GC–MS System (Thermo Scientific) using a similar program (Supporting Information).
LC/MS analysis
LC/MS analysis was performed on a Vanquish UHPLC system coupled with a Q-Exactive quadrupole-orbitrap mass spectrometer (ThermoFisher Scientific, USA) equipped with an Acquity UPLC HSS T3 column (100 mm × 2.1 mm, 1.8 µm, Waters, USA). Details were described in the Supporting Information.
Results
Chemical analysis of A. membranaceus
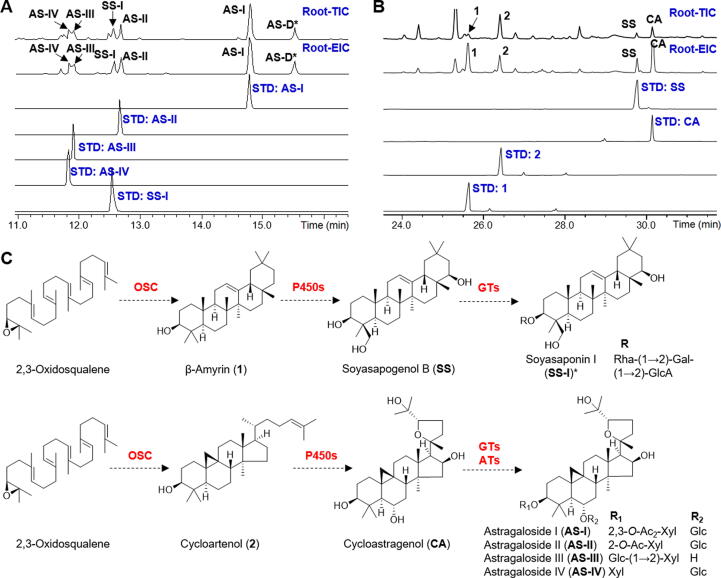
Saponins are major bioactive compounds in the roots of A. membranaceus [13], [14]. They mainly belong to cycloartane-type and oleanane-type [13], [14]. To confirm the chemical composition, the roots of A. membranaceus were analyzed by LC/MS and GC/MS respectively. As shown in Fig. 1A, cycloartane-type saponins such as astragaloside I, II, III, and IV were detected abundantly in 3-year-old roots. Meanwhile, oleanane-type saponins including soyasaponin I and astroolesaponin D were also found. To confirm the triterpene backbones, 3-month-old roots were extracted using 10% KOH and analyzed using GC/MS. As shown in Fig. 1B, β-amyrin (1) and cycloartenol (2), as well as their oxidized products soyasapogenol B (SS) and cycloastragenol (CA), were detected. All compounds above were unambiguously characterized by comparing with reference standards except for astroolesaponin D, which was identified by analyzing its high-resolution MS/MS data (Fig. S2A). The biosynthetic pathway of these saponins was proposed in Fig. 1C, where OSC genes should be present in A. membranaceus and dedicated to the biosynthesis of cycloartane-type and oleanane-type saponins.
Fig. 1.
Chemical analysis of A. membranaceus and the proposed biosynthetic pathways for its major saponins. (A) LC/MS analysis of a 3-year-old root. TIC, total ion chromatogram; EIC, extracted ion chromatogram. In EIC, m/z 913.48 ([AS-I + HCOO]-), m/z 871.47 ([AS-II + HCOO]-), m/z 829.46 ([AS-III + HCOO]-/[AS-IV + HCOO]-), m/z 941.51 ([SS-I-H]-), and m/z 953.47 ([AS-D-H]-) were extracted. (B) GC/MS analysis of a 3-month-old root. In EIC, m/z 218.2 (1), 339.3 (2), 306.3 (SS), and 215.2 (CA) were extracted. All compounds were identified by comparing with reference standards except for AS-D (identified by HR-MS/MS, Fig. S2A). (C) Proposed biosynthetic pathways of oleanane-type and cycloartane-type saponins in A. membranaceus. P450, cytochrome P450. GT, glycosyltransferase. AT, acyltransferase. Rha, rhamnose. Gal, galactose. GlcA, glucuronic acid. Glc, glucose. Xyl, xylose. Ac, acetyl.
Transcriptome mining and phylogenetic analysis
In total four sets of transcriptome data have been published for A. membranaceus (Table S1), including for hairy roots (Genbank SRR923811), shoots and roots (ERR706814), taproots (SRR5343992) and leaves (SRR5343993). Candidate OSC genes were identified by transcriptome mining of these four databases by BLASTn (e < 10-140 and alignment greater than 1700 bp), using the licorice OSCs GgbAS1 [22] and GgCAS1 [23] as probes. Three OSC genes were found and named as AmOSC1, AmOSC2, and AmOSC3.
The function of AmOSC1 was not characterized, so we reported AmOSC2 and AmOSC3 (Genbank Accession Nos. MT080938 and MT080939) in this study. Phylogenetic analysis of AmOSCs and 96 previously characterized plant OSCs revealed AmOSC2 and AmOSC3 grouped with other previously characterized β-amyrin synthases and cycloartenol synthases respectively (Fig. S3). In addition, AmOSCs were more closely related to OSCs from other legume plants, as expected.
Functional characterization in vitro
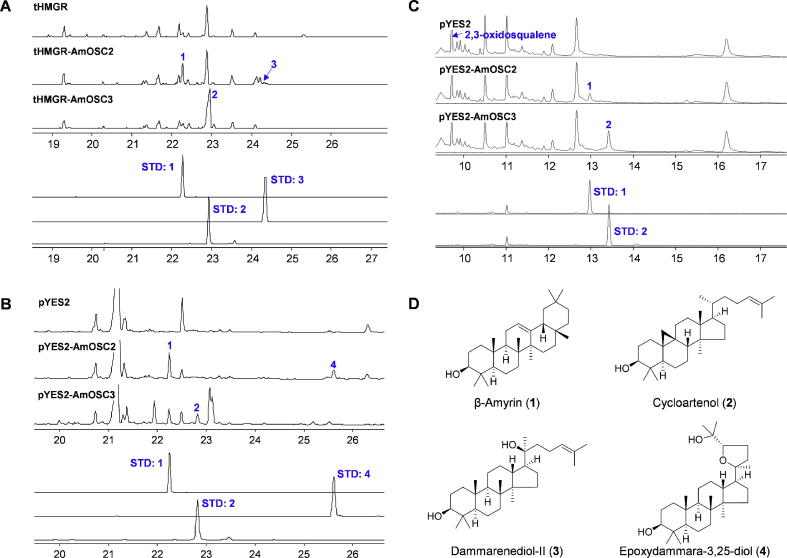
Transient expression of AmOSC 2 and 3 in N. benthamiana resulted in new products, although some peaks were minor (Fig. S4). When co-expressed with a truncated feedback-insensitive version of HMG-CoA reductase (tHMGR, KY28473) [31], new product peaks were clearly detected. AmOSC2 produced a major product β-amyrin (1) together with a minor product dammarenediol-II (3), which was an upstream intermediate of β-amyrin [2], [16]. AmOSC3 gave a single major product cycloartenol (2). Although it could be detected in the control group as a plant sterol precursor [9], expression of AmOSC3 gave a considerable increase in the amount of cycloartenol (Fig. 2A).
Fig. 2.
Functional characterization of AmOSC2 and AmOSC3 in vitro. (A) Transient expression of AmOSC genes in N. benthamiana. (B) Heterologous expression of AmOSC genes in yeast. (C) Incubation reaction with microsomes from yeast harboring AmOSC genes. (D) Structures of compounds 1–4.
To confirm the functions of AmOSC 2 and 3, we next expressed them in the yeast strain GIL77 [28]. AmOSC2 yielded β-amyrin (1) as a major product as expected, and also a minor product epoxydammara-3,25-diol (4), which might be derived from another substrate dioxidosqualene [21]. The yeast strain harboring AmOSC3 produced cycloartenol (2), which was consistent with its function in N. benthamiana (Fig. 2B). Additional peaks were also detected, which may represent yeast-modified metabolites according to previous reports [38], [39]. Compounds 1, 2, and 4 were purified and characterized by NMR (Supporting Information). Compounds 1–4 were identified by comparing the retention times and mass spectra (Fig. S5) with those of reference standards.
To further clarify their functions, microsomes were extracted from GIL77 strains harbouring AmOSC2 and AmOSC3 genes [35]. When 2,3-oxidosqualene was added as a substrate, AmOSC2 and AmOSC3 produced β-amyrin (1) and cycloartenol (2), respectively (Fig. 2C). Thus the functions of AmOSC 2 and 3 were fully confirmed.
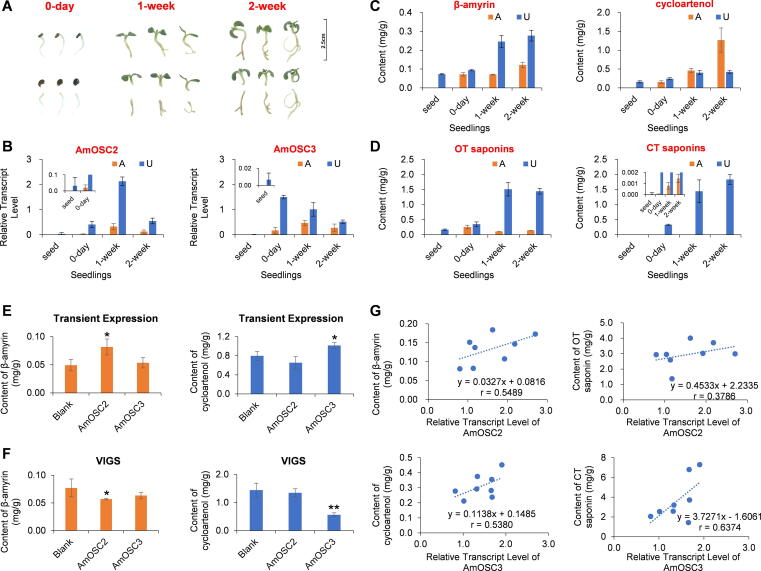
Expression patterns of AmOSCs
We then determined the expression levels of the two OSC genes and the concentrations of their products in A. membranaceus seedlings. In RT-qPCR analysis, AmOSC2 showed much higher expression levels in underground parts than aerial parts (Fig. 3B). β-Amyrin and its downstream oleanane-type saponins (OT saponins, including soyasaponin I, astroolesaponin D, and robinioside B) also showed higher content in underground parts (Fig. 3CD, Fig. S6A). AmOSC3 was expressed in all samples though the expression level was higher in underground parts. Accordingly, cycloartenol was detected in seeds and seedlings (Fig. 3C). However, the downstream product cycloartane-type saponins (CT saponins, namely astragaloside I/II/III/IV) specifically existed in the underground parts (Fig. 3D, Fig. S6B).
Fig. 3.
Functional characterization of AmOSC2 and AmOSC3 in vivo. (A-D) AmOSC expression and metabolite accumulation in A. membranaceus seedlings. (A) Pictures of seedlings of different ages. The upper panel showed complete seedlings while lower showed the aerial and underground parts. (B) RT-qPCR analysis of the transcript levels for AmOSC2/3. A, aerial parts; U, underground parts. (C) Contents of β-amyrin and cycloartenol in the seedlings determined by GC/MS. (D) Contents of OT saponins (oleanane-type saponins, including SS-I, AS-D, and RS-B, Fig. S6A) and CT saponins (cycloartane-type saponins, including AS-I/II/III/IV, Fig. S6B) determined by LC/MS. All compounds were identified by comparing with reference standards except for AS-D and RS-B (identified by HR-MS/MS, Fig. S2). (E) Transient expression of AmOSC genes in A. membranaceus. Blank, AmOSC2, and AmOSC3 groups were infected with A. tumefaciens strains LBA4404 carrying pEAQ-HT/AstHMGR, pEAQ-HT/AstHMGR + pEAQ-HT/AmOSC2, and pEAQ-HT/AstHMGR + pEAQ-HT/AmOSC3 plasmids respectively. (F) VIGS of AmOSC genes in A. membranaceus. Blank, AmOSC2, and AmOSC3 groups were infected with A. tumefaciens strains GV3101 carrying pTRV1 + pTRV2, pTRV1 + pTRV2-AmOSC2793-1101, and pTRV1 + pTRV2-AmOSC31553-1893, respectively. (G) Correlation between gene expression levels and triterpenoid contents. OT saponins, oleanane-type saponins (SS-I, AS-D, RS-B); CT saponins, cycloartane-type saponins (AS-I/II/III/IV). For all groups in B-F, n = 3. * P < 0.05 and ** P < 0.01.
Functional characterization in vivo
To study the in vivo function of AmOSC2 and AmOSC3, transient expression, virus-induced gene silencing (VIGS), and RNA interference (RNAi) experiments were carried out. When AmOSC2 and AmOSC3 were transiently overexpressed in A. membranaceus, the contents of β-amyrin and cycloartenol significantly increased by 65.7% and 27.1%, respectively (Fig. 3E). Meanwhile, in the VIGS experiment (Fig. S7), when AmOSC2 and AmOSC3 were suppressed, the contents of β-amyrin and cycloartenol significantly decreased by 26.3% and 61.0%, respectively (Fig. 3F). These results indicated that AmOSC2 and AmOSC3 are associated with the synthesis of β-amyrin and cycloartenol in A. membranaceus, respectively.
To further study the correlation between AmOSC2/3 and the saponin contents, RNAi was carried out using the hairy roots of A. membranaceus (Fig. S8). Regretfully, according to more than 30 lines generated, silencing of the sterol biosynthetic gene AmOSC3 were more likely to be detrimental, and compensational increase of AmOSC3 expression was observed when AmOSC2 was interfered. Thus we correlated the gene expression levels with the contents of triterpenes. As shown in Fig. 3G, the transcript level of AmOSC2 was higher in the roots that accumulating more β-amyrin or more oleanane-type saponins (soyasaponin I, astroolesaponin D and robinioside B, Table S10) (Pearson correlation coefficients r = 0.55 and 0.38, respectively). Meanwhile, the transcript level of AmOSC3 was higher in the roots that accumulating more cycloartenol or more cycloartane-type saponins (astragalosides I, II, III, and IV, Table S10) (r = 0.54 and 0.64, respectively). These results further proved that AmOSC2 and AmOSC3 are associated with the synthesis of oleanane-type and cycloartane-type saponins in the roots of A. membranaceus, respectively.
Sequence analysis of AmOSCs and other OSC proteins
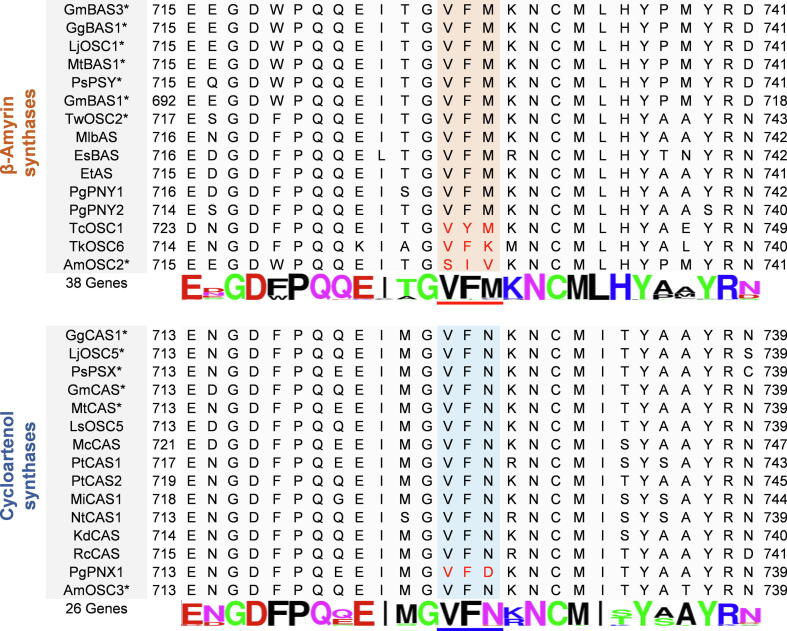
In a small-scale phylogenetic analysis, AmOSC3 grouped with legume cycloartenol synthases, while AmOSC2 formed a single branch from other legume OSCs (Fig. S9). To reveal the specific feature of AmOSC2, we used sequence alignment to find that AmOSC2 has a mutation on a triad motif. The triad is close to the C-terminus, and conserved as VFM in 33 out of 38 β-amyrin synthases (Fig. 4, Fig. S10). Besides AmOSC2 (SIV), exceptions were also found in TcOSC1 (VYM) (from Taraxacum coreanum) [40] and TkOSC6 (VFK) (from Taraxacum kok-saghyz) [41]. To evaluate the consistency of the triad in other OSCs, 26 cycloartenol synthases including AmOSC3 were aligned using the same method. As a result, the triad motif was conserved as VFN in 24 out of 26 cycloartenol synthases (Fig. S10), except for PgPNX1 (VFD) (from Panax ginseng) [28] and LcCAS1 (IFN) (from Luffa cylindrica) [42]. These results suggested that the VFM/VFN triad might be signatures for βASs/CASs, respectively. This motif has rarely been studied before, and its function warrants further investigation.
Fig. 4.
Sequence alignments of β-amyrin synthases and cycloartenol synthases. Conserved motifs VFM and VFN were marked in orange and blue, respectively. The images representing 38 or 26 OSC genes were created using WebLogo (https://weblogo.berkeley.edu/) and the genes were listed in Fig. S10. *, Genes from legumes.
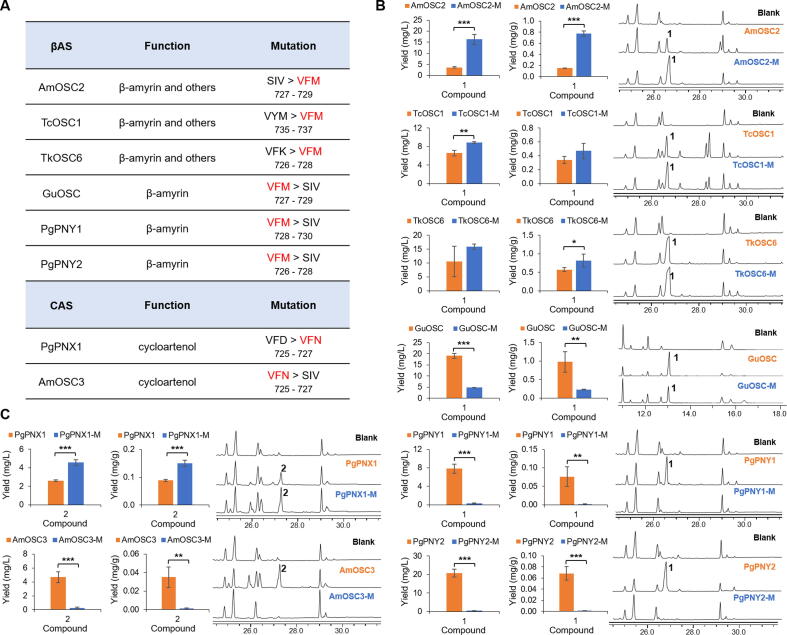
Site-directed mutagenesis of OSCs
To test the functions of the VFM/VFN motif, we carried out site-directed mutagenesis to replace the triad for different OSCs including AmOSC2/3, TcOSC1, TkOSC6 and PgPNX1 mentioned in the last paragraph (Fig. 5A, Fig. S11). For example, an AmOSC2 mutant was constructed by replacing SIV with VFM. The AmOSC2SIV727-729VFM mutant showed a 4.4-fold increase in β-amyrin (1) production compared to the wild-type when expressed in yeast (Fig. 5B). Similar results were observed in N. benthamiana, where the AmOSC2SIV727-729VFM mutant increased the production of β-amyrin (1) by 12.8-fold (Fig. S12). Similarly, for β-amyrin synthases TcOSC1 and TkOSC6, the TcOSC1VYM735-737VFM and TkOSC6VFK726-728VFM variants could increase the yield of β-amyrin by 34.7% and 50.0%, respectively. For TcOSC1, we also observed the decreased yield of by-products (Fig. S13). On the contrary, when the VFM triad was replaced by SIV, the activity of GuOSC (from Glycyrrhiza uralensis) decreased by 4.0-fold, while the activities of PgPNY1 and PgPNY2 (from P. ginseng) were almost abolished. For cycloartenol synthases, mutation of VFN diminished the activity of AmOSC3, while mutation of VFD to VFN increased the activity of the PgPNX1 by 75.3% (Fig. 5C). These results indicate that the VFM and VFN motifs are important for the function of β-amyrin synthases and cycloartenol synthases. The triad motif is also important for other OSCs, as exemplified by LjOSC3 (lupeol synthase from Lotus japonicus) [18] and AtPEN1 (arabidiol synthase from Arabidopsis thaliana) [43] (Table S5). Mutation of the corresponding residues also significantly decreased their activities (Fig. S14).
Fig. 5.
Site-directed mutagenesis of OSCs. (A) β-amyrin synthases (βAS) and cycloartenol synthases (CAS) studied by mutagenesis (accession numbers see Table S5). (B) Comparison of β-amyrin (1) yields and GC/MS chromatograms for β-amyrin synthases and their mutants. (C) Comparison of cycloartenol (2) yields and GC/MS chromatograms for cycloartenol synthases and their mutants. Mutant of each OSC was labeled as OSC-M, and blank refers to the results from yeast carrying an empty pYES2 vector. For all groups, n = 3. * P < 0.05, ** P < 0.01, and *** P < 0.001. Protein sequences alignment for these OSCs were shown in Fig. S11.
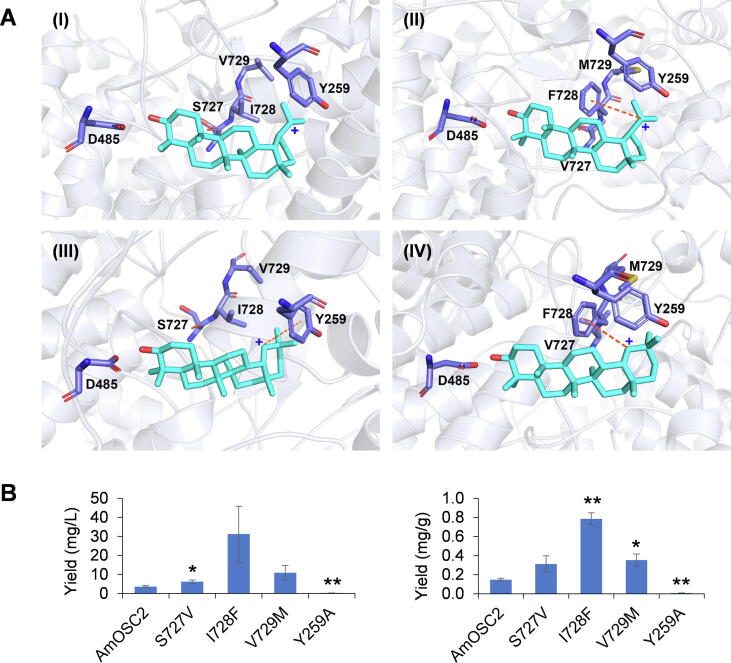
Homology modeling and molecular docking for AmOSC2
To further explain the mechanism of function for the triad motif, we used homology modeling and molecular docking to simulate the interactions between AmOSC2 and its intermediates. The structure models of AmOSC2 and AmOSC2SIV727-729VFM mutant were generated using human lanosterol synthase (PDB ID: 1W6J) as a template [36]. It showed amino acids sequence identity of 35.15% with AmOSC2. According to the cyclization process of 2,3-oxidosqualene to form β-amyrin, high-energy intermediates [44] including lupenyl cation and germanicyl cation were used as ligands [45], [46]. To validate the model, a key amino acid Y259 was discovered as consistent with previous reports [45]. Y259 plays an important role in stabilizing the C-19 germanicyl cation intermediate through cation-π interaction. This was further proved by the Y259A mutant, whose function was abolished (Fig. 6B). These data confirmed the accuracy of the model.
Fig. 6.
Molecular docking and site-directed mutagenesis of AmOSC2. (A) Molecular docking of AmOSC2 with lupenyl/germanicyl cation (I/III), and AmOSC2SIV727-729VFM mutant with lupenyl/germanicyl cation (II/IV). (B) Yields of β-amyrin from AmOSC2 and its S727V, I728F, V729M, and Y259A mutants. For all groups, n = 3. * P < 0.05 and ** P < 0.01 compared to the wild-type AmOSC2.
When the SIV residues at 727–729 were replaced by the conserved VFM, both lupenyl and germanicyl cation could be more stable due to the emerging cation-π interaction between the phenyl of F728 and the cation intermediate (Fig. 6A). In addition, when V729 was mutated to Met (M), the locations of the intermediates were altered probably due to steric bulk from the longer side chain [46]. To further confirm the docking results, single mutants were constructed for AmOSC2. The I728F mutant significantly increased the β-amyrin yield, while S727V and V729M mutants slightly improved the yield (Fig. 6B). These results indicated that V727, F728 and M729 residues acted together to provide the catalytic activity, and the cation-π interactions from Phe played the major role.
Discussion
AmOSC2 is the first β-amyrin synthase characterized from A. membranaceus. Transient expression or silencing of AmOSC2 in leaves could increase or decrease the yield of β-amyrin, respectively. In hairy roots, the transcript level of AmOSC2 showed positive correlations to β-amyrin and to oleanane-type saponins. In A. membranaceus seedlings, AmOSC2 showed higher expression levels in underground parts where β-amyrin and its downstream soyasaponins were accumulated. These data suggested that AmOSC2 is associated with the production of β-amyrin and oleanane-type saponins such as soyasaponin I in vivo. Compared with other βASs, AmOSC2 lacks of the conserved VFM motif and instead has SIV in this position. Our results indicated that variation in this motif is associated with lower yield of β-amyrin. This difference may affect the contents of oleanane-type triterpenoids in A. membranaceus.
AmOSC3 is the first biochemically characterized cycloartenol synthase from A. membranaceus. This enzyme is likely to contribute to both the biosynthesis of essential sterols and cycloartane-type triterpenes [9], [14]. Neither the expression of AmOSC3 nor the distribution of cycloartenol showed tissue-specificity in the seedlings. However, the downstream product astragalosides I/II/III/IV specifically existed in the underground parts. Transient expression or silencing of AmOSC3 in A. membranaceus leaves could increase or decrease the yield of cycloartenol, respectively. In hairy roots, the transcript level of AmOSC3 also showed positive correlations to cycloartenol and cycloartane-type saponins (astragalosides). Presumably, AmOSC3 is related to the synthesis of cycloartenol, which is synthesized in all tissues as a plant sterol precursor [9] and is consumed to form astragalosides underground.
Astragalosides I-IV are the major bioactive compounds in Astragalus roots (Huang-Qi) with cardiovascular protective effects [14]. They specifically accumulated in the roots of A. membranaceus. The conversion of cycloartenol to astragaloside is likely to involve several enzymes belonging to different families (Fig. 1C). Our recent work has revealed the glycosyltransferases that catalyzed the 3-/6-/2′-O-glycosylation of cycloastragenol [47]. Future work will focus on the identification of other downstream pathway enzymes by mining for candidates in root transcriptome, with the ultimate aim of elucidating and reconstituting the pathway for astragalosides in a heterologous expression system (yeast and/or N. benthamiana).
Several conserved motives have been reported for OSCs, including DCTAE which initiates the polycyclization cascade, MXCXCR which influences the product specificity, and the repeated QXXXXXW motifs which stabilize the carbocations during cyclization [16]. Here we have discovered the amino acid triads VFM/VFN, which are proposed as signatures for β-amyrin synthases and cycloartenol synthases, respectively. Functional analysis suggested these triad motifs as critical determinants of yield in the 10 OSCs tested. In the case of TcOSC1, an alteration of product selectivity has also been observed. Function of this triad has not been reported except for F728 in EtAS, which was found to be critical for its activity [48]. Our results showed that the consecutive residues Val727, Phe728 and Met729 in AmOSC2 acted together to stabilize the substrate intermediate. Improving the enzyme activity and altering the relative product selectivity open up new opportunities to engineer OSC function in the future.
Catalytic activity of OSCs and their mutants were compared using the yields of β-amyrin or cycloartenol. The yields were calculated as mg/L of yeast culture, or mg/g of yeast cells (Fig. 5BC) following general methods in the literature [19], [21], [49]. Since the mutation might also alter other features of the target protein, we studied the protein expression level and protein stability for 4 OSCs (AmOSC2, GuOSC, PgPNX1, AmOSC3) and their mutants (Supporting Information). The OSC protein abundance were determined in yeast microsome/protein extracts using LC/MS. The expression levels changed slightly (0.7–1.3 folds) after mutation. The protein stability was calculated using molecular dynamics. The proteins showed similar stable states after mutation except for GuOSC. More importantly, the changes in protein expression or structural stability were not correlated with the changes in catalytic activity. For example, the mutant for PgPNX1 showed an increased activity of 175.3%, though the protein expression level decreased for 31.0%. Similarly, the mutant of GuOSC showed higher stability than the wild-type though its activity decreased by 75.0% (Fig. S15). These results ruled out the influence of site-directed mutagenesis to protein expression or protein stability, which further supported the interaction between the OSC triad motif and the substrate.
Interestingly, A. membranaceus owns a βAS mutated in the triad motif, and contains both oleanane-type and cycloartane-type saponins. Compared to other legume plants, Glycyrrhiza uralensis and Glycine max has βASs conserved in the triad motif and they contain mainly oleanane-type saponins [6], [7]. Similar phenomenon was also observed in Taraxacum plants, where TcOSC1 (T. coreanum) and TkOSC6 (T. kok-saghyz) are mutated in the triad motif, and the corresponding plants contain both oleanane-type and taraxarane-type saponins [40], [41]. Generally, one plant could contain several different species of OSC genes, but not all of them could function to synthesize the specialized metabolites [16]. Correlation between mutated βAS and the triterpene backbone diversity in plants still warrants further investigation.
Conclusion
In this study, two OSC genes, AmOSC2 and AmOSC3, were cloned from A. membranaceus, the original plant of the herb Huang-Qi. Their functions were investigated through expression in N. benthamiana and yeast, together with transient expression, VIGS, and gene expression-product content correlation in vivo. AmOSC2/3 are β-amyrin synthase and cycloartenol synthase, respectively, which contribute to the biosynthesis of medicinally important soyasaponins and astragaosides in Huang-Qi, respectively. In addition, conserved amino acid motifs VFM/VFN were discovered for β-amyrin/cycloartenol synthases. Site-directed mutagenesis and molecular docking revealed that the triad could significantly affect the enzymatic activity by stabilizing the cation intermediates, mainly via cation-π interaction. The study provides insights for the biosynthesis of oleanane-type and cycloartane-type triterpenoids in A. membranaceus, and the newly discovered conserved motif could contribute to the engineering of OSC enzymes.
Compliance with Ethics Requirements
There was no animal or human experiment in this study.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are grateful to Prof. George Lomonossoff at John Innes Centre for providing the pEAQ-HT vectors. We thank Prof. Zhihua Liao and Dr. Fangyuan Zhang at Southwest University for providing VIGS vectors. We also wish to thank Prof. Qing Zhao and Dr. Mengying Cui at Shanghai Chenshan Plant Science Research Center of the Chinese Academy of Sciences for their technical help in the RNAi experiments. This work was supported by National Natural Science Foundation of China (82122073, 81973448, 81725023) and Beijing Natural Science Foundation (JQ18027). A.O.’s program is supported by the BBSRC Institute Strategic Programme Grant ‘Molecules from Nature – Products and Pathways’ (BBS/E/J/000PR9790) and the John Innes Foundation.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.03.014.
Contributor Information
Anne Osbourn, Email: anne.osbourn@jic.ac.uk.
Xue Qiao, Email: qiaoxue@bjmu.edu.cn.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Osbourn A., Goss R.J.M., Field R.A. The saponins - polar isoprenoids with important and diverse biological activities. Nat Prod Rep. 2011;28(7):1261. doi: 10.1039/c1np00015b. [DOI] [PubMed] [Google Scholar]
- 2.Thimmappa R., Geisler K., Louveau T., O'Maille P., Osbourn A. Triterpene biosynthesis in plants. Annu Rev Plant Biol. 2014;65(1):225–257. doi: 10.1146/annurev-arplant-050312-120229. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Q.Y., Ye M. Chemical analysis of the Chinese herbal medicine Gan-Cao (licorice) J Chromatogr A. 2009;1216:1954–1969. doi: 10.1016/j.chroma.2008.07.072. [DOI] [PubMed] [Google Scholar]
- 4.Yang W.Z., Hu Y., Wu W.Y., Ye M., Guo D.A. Saponins in the genus Panax L. (Araliaceae): A systematic review of their chemical diversity. Phytochemistry. 2014;106:7–24. doi: 10.1016/j.phytochem.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 5.Ragupathi G., Gardner J.R., Livingston P.O., Gin D.Y. Natural and synthetic saponin adjuvant QS-21 for vaccines against cancer. Expert Rev Vaccines. 2011;10(4):463–470. doi: 10.1586/erv.11.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukushima E.O., Seki H., Sawai S., Suzuki M., Ohyama K., Saito K., et al. Combinatorial biosynthesis of Legume natural and rare triterpenoids in engineered yeast. Plant Cell Physiol. 2013;54(5):740–749. doi: 10.1093/pcp/pct015. [DOI] [PubMed] [Google Scholar]
- 7.Parente J.P., Silva B.P. Bioactive complex triterpenoid saponins from the leguminosae family. Nat Prod Commun. 2009;4:143–155. [PubMed] [Google Scholar]
- 8.Tava A., Scotti C., Avato P. Biosynthesis of saponins in the genus Medicago. Phytochem Rev. 2011;10(4):459–469. [Google Scholar]
- 9.Benveniste P. Biosynthesis and accumulation of sterols. Annu Rev Plant Biol. 2004;55(1):429–457. doi: 10.1146/annurev.arplant.55.031903.141616. [DOI] [PubMed] [Google Scholar]
- 10.Sonawane P.D., Pollier J., Panda S., Szymanski J., Massalha H., Yona M., et al. Plant cholesterol biosynthetic pathway overlaps with phytosterol metabolism. Nat Plants. 2017;3:1–13. doi: 10.1038/nplants.2017.101. [DOI] [PubMed] [Google Scholar]
- 11.Yang L.P., Shen J.G., Xu W.C., Li J., Jiang J.Q. Secondary metabolites of the genus Astragalus: Structure and biological-activity update. Chem Biodivers. 2013;10:1004–1054. doi: 10.1002/cbdv.201100444. [DOI] [PubMed] [Google Scholar]
- 12.Li H.X., Yu Z.Y. Cimicifugae rhizoma: From origins, bioactive constituents to clinical outcomes. Curr Med Chem. 2006;13:2927–2951. doi: 10.2174/092986706778521869. [DOI] [PubMed] [Google Scholar]
- 13.Ionkova I., Shkondrov A., Krasteva I., Ionkov T. Recent progress in phytochemistry, pharmacology and biotechnology of Astragalus saponins. Phytochem Rev. 2014;13(2):343–374. [Google Scholar]
- 14.Su H.-F., Shaker S., Kuang Y.i., Zhang M., Ye M., Qiao X. Phytochemistry and cardiovascular protective effects of Huang-Qi. Med Res Rev. 2021;41(4):1999–2038. doi: 10.1002/med.21785. [DOI] [PubMed] [Google Scholar]
- 15.Chen J.B., Ullah H., Zheng Z., Gu X.F., Su C.H., Xiao L.Y., et al. Soyasaponins reduce inflammation by downregulating MyD88 expression and suppressing the recruitments of TLR4 and MyD88 into lipid rafts. BMC Complement Med Ther. 2020;20:1–16. doi: 10.1186/s12906-020-2864-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen K., Zhang M., Ye M., Qiao X. Site-directed mutagenesis and substrate compatibility to reveal the structure-function relationships of plant oxidosqualene cyclases. Nat Prod Rep. 2021;38(12):2261–2275. doi: 10.1039/d1np00015b. [DOI] [PubMed] [Google Scholar]
- 17.Stephenson M.J., Field R.A., Osbourn A. The protosteryl and dammarenyl cation dichotomy in polycyclic triterpene biosynthesis revisited: has this 'rule' finally been broken? Nat Prod Rep. 2019;36:1044–1052. doi: 10.1039/c8np00096d. [DOI] [PubMed] [Google Scholar]
- 18.Sawai S., Shindo T., Sato S., Kaneko T., Tabata S., Ayabe S.-I., et al. Functional and structural analysis of genes encoding oxidosqualene cyclases of Lotus japonicus. Plant Sci. 2006;170(2):247–257. [Google Scholar]
- 19.Srisawat P., Fukushima E.O., Yasumoto S., Robertlee J., Suzuki H., Seki H., et al. Identification of oxidosqualene cyclases from the medicinal legume tree Bauhinia forficata: a step toward discovering preponderant alpha-amyrin-producing activity. New Phytol. 2019;224:352–366. doi: 10.1111/nph.16013. [DOI] [PubMed] [Google Scholar]
- 20.Zheng S.H., Liu D.W., Ren W.G., Fu J., Huang L.F., Chen S.L. Integrated analysis for identifying Radix Astragali and its adulterants based on DNA barcoding. Evid-Based Compl Alt. 2014;2014:1–11. doi: 10.1155/2014/843923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salmon M., Thimmappa R.B., Minto R.E., Melton R.E., Hughes R.K., O’Maille P.E., et al. A conserved amino acid residue critical for product and substrate specificity in plant triterpene synthases. Proc Natl Acad Sci USA. 2016;113(30) doi: 10.1073/pnas.1605509113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi H., Huang P., Kirakosyan A., Inoue K., Hiraoka N., Ikeshiro Y., et al. Cloning and characterization of a cDNA encoding beta-amyrin synthase involved in glycyrrhizin and soyasaponin biosyntheses in licorice. Biol Pharm Bull. 2001;24(8):912–916. doi: 10.1248/bpb.24.912. [DOI] [PubMed] [Google Scholar]
- 23.Hayashi H., Hiraoka N., Ikeshiro Y., Kushiro T., Morita M., Shibuya M., et al. Molecular cloning and characterization of a cDNA for Glycyrrhiza glabra cycloartenol synthase. Biol Pharm Bull. 2000;23(2):231–234. doi: 10.1248/bpb.23.231. [DOI] [PubMed] [Google Scholar]
- 24.Jones D.T., Taylor W.R., Thornton J.M. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis Version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sainsbury F., Thuenemann E.C., Lomonossoff G.P. pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol J. 2009;7:682–693. doi: 10.1111/j.1467-7652.2009.00434.x. [DOI] [PubMed] [Google Scholar]
- 27.Bok J.W., Keller N.P. Fast and easy method for construction of plasmid vectors using modified quick-change mutagenesis. Methods Mol Biol. 2012;944:163–174. doi: 10.1007/978-1-62703-122-6_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kushiro T., Shibuya M., Ebizuka Y. beta-Amyrin synthase - Cloning of oxidosqualene cyclase that catalyzes the formation of the most popular triterpene among higher plants. Eur J Biochem. 1998;256(1):238–244. doi: 10.1046/j.1432-1327.1998.2560238.x. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Q., Zhang Y., Wang G., Hill L., Weng J.K., Chen X.Y., et al. A specialized flavone biosynthetic pathway has evolved in the medicinal plant, Scutellaria baicalensis. Sci Adv. 2016;2:1–15. doi: 10.1126/sciadv.1501780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu R.-Y., Nan P., Pan H., Zhou T., Chen J. Molecular cloning, characterization and expression of a chalcone reductase gene from Astragalus membranaceus Bge. var. mongholicus (Bge.) Hsiao. Mol Biol Rep. 2012;39(3):2275–2283. doi: 10.1007/s11033-011-0977-x. [DOI] [PubMed] [Google Scholar]
- 31.Reed J., Stephenson M.J., Miettinen K., Brouwer B., Leveau A., Brett P., et al. A translational synthetic biology platform for rapid access to gram-scale quantities of novel drug-like molecules. Metab Eng. 2017;42:185–193. doi: 10.1016/j.ymben.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li R., Reed D.W., Liu E., Nowak J., Pelcher L.E., Page J.E., et al. Functional genomic analysis of alkaloid biosynthesis in Hyoscyamus niger reveals a cytochrome P450 involved in littorine rearrangement. Chem Biol. 2006;13(5):513–520. doi: 10.1016/j.chembiol.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 33.Geng C., Zhao T., Yang C., Zhang Q., Bai F., Zeng J., et al. Metabolic characterization of Hyoscyamus niger root-specific putrescine N-methyltransferase. Plant Physiol Biochem. 2018;127:47–54. doi: 10.1016/j.plaphy.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 34.Ito R., Masukawa Y., Hoshino T. Purification, kinetics, inhibitors and CD for recombinant beta-amyrin synthase from Euphorbia tirucalli L and functional analysis of the DCTA motif, which is highly conserved among oxidosqualene cyclases. FEBS J. 2013;280:1267–1280. doi: 10.1111/febs.12119. [DOI] [PubMed] [Google Scholar]
- 35.He J., Dong Z., Hu Z., Kuang Y.i., Fan J., Qiao X., et al. Regio-specific prenylation of pterocarpans by a membrane-bound prenyltransferase from Psoralea corylifolia. Org Biomol Chem. 2018;16(36):6760–6766. doi: 10.1039/c8ob01724g. [DOI] [PubMed] [Google Scholar]
- 36.Thoma R., Schulz-Gasch T., D'Arcy B., Benz J., Aebi J., Dehmlow H., et al. Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase. Nature. 2004;432(7013):118–122. doi: 10.1038/nature02993. [DOI] [PubMed] [Google Scholar]
- 37.Trott O., Olson A.J. Software news and update AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiang T., Shibuya M., Katsube Y., Tsutsumi T., Otsuka M., Zhang H., et al. A new triterpene synthase from Arabidopsis thaliana produces a tricyclic triterpene with two hydroxyl groups. Org Lett. 2006;8(13):2835–2838. doi: 10.1021/ol060973p. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z., Yeats T., Han H., Jetter R. Cloning and characterization of oxidosqualene cyclases from Kalanchoe daigremontiana. J Biol Chem. 2010;285(39):29703–29712. doi: 10.1074/jbc.M109.098871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han J.Y., Jo H.-J., Kwon E.K., Choi Y.E. Cloning and characterization of oxidosqualene cyclases involved in taraxasterol, taraxerol and bauerenol triterpene biosynthesis in Taraxacum coreanum. Plant Cell Physiol. 2019;60(7):1595–1603. doi: 10.1093/pcp/pcz062. [DOI] [PubMed] [Google Scholar]
- 41.Pütter K.M., van Deenen N., Müller B., Fuchs L., Vorwerk K., Unland K., et al. The enzymes OSC1 and CYP716A263 produce a high variety of triterpenoids in the latex of Taraxacum koksaghyz. Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-42381-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayashi H., Hiraoka N., Ikeshiro Y., Yazaki K., Tanaka S., Kushiro T., et al. Molecular cloning of a cDNA encoding cycloartenol synthase from Luffa cylindrica. Plant Physiol. 1999;121:1384. [Google Scholar]
- 43.Lodeiro S., Xiong Q., Wilson W.K., Kolesnikova M.D., Onak C.S., Matsuda S.P.T. An oxidosqualene cyclase makes numerous products by diverse mechanisms: A challenge to prevailing concepts of triterpene biosynthesis. J Am Chem Soc. 2007;129:11213–11222. doi: 10.1021/ja073133u. [DOI] [PubMed] [Google Scholar]
- 44.Hermann J.C., Ghanem E., Li Y., Raushel F.M., Irwin J.J., Shoichet B.K. Predicting substrates by docking high-energy intermediates to enzyme structures. J Am Chem Soc. 2006;128(49):15882–15891. doi: 10.1021/ja065860f. [DOI] [PubMed] [Google Scholar]
- 45.Kushiro T., Shibuya M., Masuda K., Ebizuka Y. Mutational studies on triterpene syntheses: Engineering lupeol synthase into beta-amyrin synthase. J Am Chem Soc. 2000;122:6816–6824. [Google Scholar]
- 46.Hoshino T. beta-Amyrin biosynthesis: catalytic mechanism and substrate recognition. Org Biomol Chem. 2017;15:2869–2891. doi: 10.1039/c7ob00238f. [DOI] [PubMed] [Google Scholar]
- 47.Zhang M., Yi Y., Gao B.-H., Su H.-F., Bao Y.-O., Shi X.-M., et al. Functional characterization and protein engineering of a triterpene 3-/6-/2’-O-glycosyltransferase reveal a conserved residue critical for the regiospecificity. Angew Chem Int Edit. 2022;61(8) doi: 10.1002/anie.202113587. [DOI] [PubMed] [Google Scholar]
- 48.Ito R., Hashimoto I., Masukawa Y., Hoshino T. Effect of cation-π interactions and steric bulk on the catalytic action of oxidosqualene cyclase: A case study of Phe728 of beta-amyrin synthase from Euphorbia tirucalli L. Chem-Eur J. 2013;19:17150–17158. doi: 10.1002/chem.201301917. [DOI] [PubMed] [Google Scholar]
- 49.Zhou J., Hu T., Gao L., Su P., Zhang Y., Zhao Y., et al. Friedelane-type triterpene cyclase in celastrol biosynthesis from Tripterygium wilfordii and its application for triterpenes biosynthesis in yeast. New Phytol. 2019;223(2):722–735. doi: 10.1111/nph.15809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.