Abstract
Polycystic ovary syndrome (PCOS) is characterized by hyperandrogenemia of ovarian theca cell origin. We report significant association of androgen production with 15 single nucleotide variants (SNVs) identified by exome sequencing of theca cells from women with PCOS and normal ovulatory women. Ten SNVs are located within a 150 kbp region on 12q13.2 which encompasses loci identified in PCOS genome-wide association studies (GWAS) and contains PCOS candidate genes ERBB3 and RAB5B. The region also contains PA2G4 which encodes a transcriptional corepressor of androgen receptor and androgen receptor-regulated genes. PA2G4 has not previously been recognized as related to PCOS in published GWAS studies. Two of the SNVs are predicted to have functional consequences (ERBB3 missense SNV, PA2G4 promoter SNV). PA2G4 interacts with the ERBB3 cytoplasmic domain containing the missense variant, suggesting a potential signaling pathway disruption that could lead to the PCOS ovarian phenotype. Single cell RNA sequencing of theca cells showed significantly less expression of PA2G4 after forskolin treatment in PCOS cells compared to normal cells (padj = 3.82E-30) and in cells heterozygous for the PA2G4 promoter SNV compared to those without the SNV (padj = 2.16E-11). This is consistent with a functional effect of the PA2G4 promoter SNV. No individual SNV was significantly associated with PCOS in an independent family cohort, but a haplotype with minor alleles of three SNVs was found preferentially in women with PCOS. These findings suggest a functional role for 12q13.2 variants in PCOS and implicate variants in ERBB3 and PA2G4 in the pathophysiology of PCOS.
Keywords: polycystic ovary syndrome, theca cells, family cohort, single nucleotide variants, candidate genes
1. Introduction
Polycystic ovary syndrome (PCOS) is the most common endocrine disorder of women of reproductive age. PCOS is characterized by hyperandrogenemia of ovarian origin, anovulatory infertility, and metabolic disturbances (Diamanti-Kandarakis and Dunaif, 2012). It is a complex genetic disorder, and approximately 20 susceptibility loci have been reproducibly associated in genome wide association studies (GWAS) (Dapas and Dunaif, 2022). However, the functional significance of many of the genes in these loci with respect to PCOS phenotypes is largely unknown (Dapas and Dunaif, 2022).
We examined the potential association of genes implicated in PCOS, focusing on those genes that could influence androgen production, since hyperandrogenemia is a cardinal phenotype of PCOS (Legro et al., 1998). The rationale for selecting theca cells for examination is that they produce the excess androgen characteristic of PCOS. For discovery of SNVs, we performed whole exome sequencing (WES) on DNA collected from theca cells isolated from size-matched follicles removed from ovaries of women diagnosed with PCOS or normal ovulatory women. PCOS status was directly related to excessive androgen synthesis by the respective theca cell preparations, especially when cells were stimulated with forskolin, which mimics the action of the gonadotropin, luteinizing hormone. We further examined the theca cells through single cell RNA sequencing and differential expression analyses of genes identified by our WES as associated with androgen levels. For validation, those SNVs identified as associated with forskolin-stimulated androgen production by the theca cells were examined in an independent family cohort with one or more daughters with PCOS. Here we describe SNVs and a haplotype located in a 150 kbp region of chromosome 12q13.2 that contains plausible PCOS genes (ERBB3, PA2G4, an RAB5B), and variants that could have a causal role in promoting PCOS ovarian phenotypes including excessive thecal androgen production.
2. Materials and Methods
2.1. Theca cell preparations and culture
Human theca interna tissue was obtained from follicles of women undergoing hysterectomy, following informed consent under a protocol approved by the Institutional Review Board of The Pennsylvania State University College of Medicine. As a standard of care, oophorectomies were performed during the luteal phase of the cycle. Theca cells from normal cycling and PCOS follicles were isolated and grown as we have as previously reported in detail(Nelson-Degrave et al., 2005; Wickenheisser et al., 2012, 2005). PCOS and normal ovarian tissue came from age-matched women, 38–41 years old. The diagnosis of PCOS was made according to National Institutes of Health (NIH) consensus guidelines (Azziz et al., 2016; Legro et al., 2013) which include hyperandrogenemia/hyperandrogenism and oligo-ovulation and the exclusion of other causes of hyperandrogenemia (e.g. 21-hydroxylase deficiency, Cushing’s syndrome, and adrenal or ovarian tumors). All of the PCOS theca cell preparations studied came from ovaries of women with fewer than six menses per year and elevated serum total testosterone or bioavailable testosterone levels (Nelson et al., 1999; Nelson-DeGrave et al., 2004; Wickenheisser et al., 2004, 2000). Each of the PCOS ovaries contained multiple subcortical follicles of less than 10 mm in diameter. The control (normal) theca cell preparations came from ovaries of fertile women with normal menstrual histories, menstrual cycles of 21–35 days, and no clinical signs of hyperandrogenism. Neither PCOS nor normal subjects were receiving hormonal medications at the time of surgery. Indications for surgery were dysfunctional uterine bleeding, endometrial cancer, and pelvic pain. Experiments comparing PCOS and normal theca were performed using fourth-passage (31–38 population doublings) theca cells isolated from individual size-matched follicles obtained from age-matched subjects, in the absence of in vivo stimulation. The use of fourth-passage cells allowed us to perform multiple experiments from the same patient population, and were propagated from frozen stocks of second passage cells in the media described above. The passage conditions and split ratios for all normal and PCOS cells were identical. These studies were approved by the Human Subjects Protection Offices of Virginia Commonwealth University) and Penn State College of Medicine.
Nine cell preparations obtained from women with PCOS (MC01, MC21, MC09_B, MC03, MC10, MC16, MC26, MC27, MC190) and seven from normal ovulating women (MC62_B, MC02, MC06, MC31, MC38, MC40, MC50) were studied. All subjects were unrelated and of European ancestry. The cells were characterized by their production of dehydroepiandrosterone (DHEA), the major androgen synthesized by these cells, under basal conditions or stimulated with forskolin (20 μM) for 16 h. DHEA was quantified by ELISA assays (DRG, Springfield,NJ) and production (pmol) was normalized to cell number (106 cells) determined at the end of the culture period.
2.2. Whole Exome Sequence Analysis of Normal and PCOS Theca Cells DNA
Theca cell DNA was extracted from flash frozen cultured cells using a DNAeasy Blood and Tissue Kit (Qiagen, Germantown Maryland). The DNA samples were subjected to whole exome sequencing at 100 millions reads providing 100× coverage using the Agilent SureSelect 51M capture kit with Illumina HiSeq 2000 sequencing, in conjunction with BGI Americas. Raw sequence data for each individual were mapped to the human reference genome (build GRCh37/hg19) using the BWA-MEM algorithm of Burrows-Wheeler Aligner (v 0.7.12) (H. Li, 2013). This was followed by a series of pre-processing steps–marking duplicates, realignment around indels and base quality recalibration. PCR duplicates were marked within the aligned reads using Picard tools. (http://picard.sourceforge.net) Next, mapping artifacts around indels were cleaned up using the RealignerTargetCreator, the IndelRealigner and the LeftAlignIndels walkers of the Genome Analysis ToolKit (GATK) (Depristo et al., 2011; McKenna et al., 2010). Inaccurate / biased base quality scores were recalibrated using the BaseRecalibrator, the AnalyzeCovariates and the PrintReads walkers of GATK, which use machine learning to model these errors empirically and adjust the quality scores accordingly.
2.3. Linkage disequilibrium
LDlink (https://analysistools.cancer.gov/LDlink/?tab=home) was used to identify haplotypes in Europeans. LDlink accesses data from 1000 Genomes in a suite of tools that allows determination of linkage disequilibrium (LD) and haplotypes. We used the LDlink SNPclip tool to examine LD among the 9 SNVs associated with DHEA response, using an r2 cutoff of 0.5. This identified two LD groups, which were then used to identify haplotypes using LDhap.
2.4. Statistical analysis
The Wilcoxon rank sum test was used to compare age, basal DHEA, and forskolin-stimulated DHEA production between the PCOS and control samples. The WES data were subjected to the following filters. We retained genetic variants having a unique combination of Gene ID, Chromosome, Position, Variant ID, Reference and Alternate Allele. Additionally, variants that were homogeneous across all samples (i.e., no sample displayed the minor allele or all samples displayed the minor allele (N=21)) were removed, leaving 441 variants for statistical analysis.
For each variant, a Wilcoxon rank sum test was used to compare those with and without the variant with respect to forskolin-stimulated DHEA production. In order to apply a statistical comparison, a minimum of two samples per group were required so that the set of variants was restricted to 252 variants. P values of <0.05 were considered significant. We did not apply Bonferroni correction because we are testing a restricted set of genetic variants in robust loci for PCOS.
2.5. Single Cell RNA Sequencing of Normal and PCOS Theca Cells
Single cell RNA sequencing was performed using 4th passage normal (MC02, MC06, MC31, MC40, MC50) and PCOS (MC03, MC10, MC16, MC26, MC27), that were grown until subconfluent, transferred into serum free medium with and without 20 μM forskolin for 24 h. Following treatment, the theca cells were rinsed in PBS and harvested following trypsinization, and centrifugation at 500 × g at 4°C to pellet the cells. The cell pellet was subsequently rinsed in PBS, and the cells were gently resuspend in ice cold cryopreservation solution containing 50% FBS/40% growth media/10% DMSO at a concentration of 4 million cells/mL aliquoted in 2mL Corning cryopreservation tubes. The cells were then frozen using a −80 freezing device for 24 h, and stored in liquid nitrogen prior to shipment and processing at Active Motif (Carlsbad, CA). Following slow thawing in 37 degree 10% FBS in DME/F12, single cell libraries were generated using the 10X Genomics Chromium platform followed by sequencing at a minimun 250 Million reads, and 50,000 read pairs per cell on the Illumina platform.
2.6. Analysis of Single Cell RNA Sequencing Data
The 10X CellRanger software was used for alignment, filtering, barcode counting, and UMI counting of the sequencing data. CellRanger processed files were loaded into the Seurat v. 4.1.1 R package (Hao et al., 2021). Each sample was filtered for cells with nFeature_RNA > 200 and percent mitochondria < 5% followed by normalization (NormalizeData function) and identification of variable features (FindVariableFeatures function). The Seurat FindMarkers function was used for transcriptome wide differential expression analyses based on the Wilcoxon rank sum test with Bonferroni correction. FindMarkers avg_log2FC is the log fold-change of the average expression between the two groups with positive values indicating the feature is more highly expressed in the first group. FindMarkers was ran with default setting except for logfc.threshold = 0.1. Differential expression analyses were performed based solely on PCOS affection or forskolin treatment status and also among combinations of affection and treatment status (normal untreated vs normal treated; PCOS untreated vs PCOS treated; normal untreated vs PCOS untreated; normal treated vs PCOS treated). Differential expression analyses were also used to compare samples heterozygous for rs773121 (MC03 and MC27) to those without the SNV (MC02, MC06, MC31, MC40, MC50, MC10, MC16, MC26). The Seurat AverageExpression function was used to calculate average expression levels of genes in the groups.
2.7. SNV analyses of a PCOS cohort
We analyzed the 10 SNVs on chromosome 12 that were significantly associated with thecal cell androgen production in whole genome sequencing data from a family based PCOS cohort (Dapas et al., 2019). The study was approved by the Institutional Review Boards of Northwestern University Feinberg School of Medicine, Penn State Health Milton S. Hershey Medical Center, and Brigham and Women’s Hospital. Written informed consent was obtained from all subjects prior to the study. The cohort consisted of 318 individuals of European ancestry from 77 families with one or more daughters with PCOS. Among the index cases and sisters (n=171), the following phenotypes were identified: PCOS (T>58 ng/dl and/or uT>15 ng/dl and ≤ 8 menses/year) (n=90); Hyperandrogenemic (HA) (T>58 ng/dl and/or uT>15 ng/dl and regular menses (every 27–35 days)) (n=5); Unaffected (n=76). The women were ages 14 to 49 years. Women were assigned affected status if they fulfilled criteria for PCOS or HA, as we have done in our previous family-based genetic analyses (Urbanek et al., 1999).
Sequencing of the cohort was performed using the Complete Genomics, Inc. platform. Sequence reads were aligned to the human reference genome (GRCh37/hg19) and variants were called using the CGI AssemblyPipeline version 2.0. The SNVs were analyzed individually using the PLINK v1.90 (Purcell et al., 2007) transmission disequilibrium test (TDT) based on PCOS affection status. An individual was considered affected if they had a phenotype of PCOS or hyperandrogenemia. The haplotypes containing the SNVs were analyzed using the Family-Based Association Tests (FBAT) v2.0.3 (Horvath et al., 2001) HBAT function which is the haplotype version of the association test. FBAT HBAT was performed using the PCOS affection status.
2.8. Functional Annotations
Potential functional consequences of the SNVs were examined using Combined Annotation-Dependent Depletion (CADD) v1.6 (Rentzsch et al., 2019), FATHMM-MKL (Shihab et al., 2015), FATHMM-XF (Rogers et al., 2018), PolyPhen-2 (Adzhubei et al., 2013), SIFT (Kumar et al., 2009; Ng and Henikoff, 2003), and GeneHancer (Fishilevich et al., 2017).
3. Results
3.1. Whole Exome Sequencing (WES) Identifies SNVs on Chromosome 12 Associated with Thecal Cell Androgen Production.
Theca cells isolated from size-matched follicles micro-dissected from ovaries of women diagnosed with PCOS (N=9) and normal ovulatory women (N=7) were utilized to discover variants in selected PCOS candidate genes and their association with theca cell androgen production. Table 1 lists the 18 PCOS candidate genes (Chen et al., 2011; Day et al., 2018, 2015; Hayes et al., 2015; Shi et al., 2012) chosen for examination from all genes interrogated in the whole exome sequencing study. These genes were selected from published GWAS and meta-analyses, representing genes in loci associated with PCOS in women of European ancestry.
Table 1:
PCOS Candidate Loci Interrogated.
| Locus | Population | GWAS Reference |
|---|---|---|
| C9orf3 | Han, European | Shi et al., 2012; Hayes et al., 2015 |
| DENND1A | Han, European | Chen et al., 2011; Shi et al., 2012; Day et al., 2018 |
| ERBB2 | European | Day et al., 2018 |
| ERBB3 | European | Day et al., 2018 |
| ERBB4 | European | Day et al., 2015, 2018 |
| FSHB | European | Day et al., 2015; Hayes et al., 2015 |
| GATA4/NEIL2 | European | Hayes et al., 2015 |
| KRR1 | European | Day et al., 2015, 2018 |
| MAPRE1 | European | Day et al., 2018 |
| PLGRKT | European | Day et al., 2018 |
| RAB5B/SUOX | Han, European | Shi et al., 2012; Day et al., 2018 |
| RAD50 | European | Day et al., 2015, 2018 |
| THADA | Han, European | Chen et al., 2011; Shi et al., 2012; Day et al., 2015, 2018 |
| TOX3 | Han, European | Shi et al., 2012; Day et al., 2018 |
| YAP1 | Han, European | Shi et al., 2012; Day et al., 2015, 2018 |
| ZBTB16 | European | Day et al., 2018 |
We used DHEA as the marker of theca cell androgen biosynthesis since it is the predominant androgen secreted by our theca cell preparations (Table 2). In addition to producing higher DHEA levels than normal theca cells, PCOS theca cells also secrete higher amounts of other androgens including androstenedione and testosterone under basal conditions and when challenged with forskolin (Nelson et al., 1999). Forskolin activates adenylate cyclase and mimics the action of luteinizing hormone (LH), the main gonadotropin regulating theca cell steroidogenesis. The variants identified by WES in the genes of interest are presented in Supplemental Table S1 along with SIFT (Kumar et al., 2009; Ng and Henikoff, 2003) and PolyPhen-2 HDIV and HVAR predictions (Adzhubei et al., 2013). The gene name, chromosome position, dbSNP accession, GenBank transcript accession, reference and alternate alleles, consequence, and number of minor alleles detected in each theca cell preparation are also provided. Table S2 presents the number of variants analyzed in each gene of interest after filtering to remove duplicate entries and sites that were not variable across the cell preparations analyzed. An allele-based Wilcoxon rank sum test was then performed on each of the filtered SNVs (N=252) to test for association with levels of forskolin-stimulated DHEA production, a measure of maximal androgen-producing capacity (Table 3). Among the 15 variants that were found to exhibit significant association at a P-value < 0.05, 10 were located on chromosome 12. This over representation of chromosome 12 was highly significant (Fisher’s exact test, P=0.000002).
Table 2: Androgen production by PCOS and normal thecal cells employed in this study.
(A) DHEA production by normal and PCOS theca cell preparations employed in this study and summary statistics. Cells were cultured for 16 h with or without forskolin (20 μM) and DHEA production was assessed by immunoassay normalized to cell number. Values presented are means (S.D.) from triplicate cultures for each preparation. (B) Median and range (minimum, maximum) with P-value from Wilcoxon rank sum test.
| A | |||
|---|---|---|---|
| Sample | Age | Basal DHEA pmol/106 cells | Forskolin-DHEA pmol/106 cells |
| Normal | |||
| MC02 | 41 | 40.37 ± 2.37 | 183.32 ± 2.57 |
| MC06 | 38 | 45.23 ± 3.99 | 304.90 ± 30.22 |
| MC31 | 37 | 24.50 ± 2.09 | 139.64 ± 15.94 |
| MC38 | 36 | 52.87 ± 22.60 | 578.52 ± 5.34 |
| MC40 | 41 | 46.95 ± 3.60 | 156.40 ± 14.57 |
| MC50 | 37 | 35.84 ± 2.65 | 147.88 ± 8.11 |
| MC62 | 37 | 31.95 ± 0.58 | 273.46 ± 35.13 |
| PCOS | |||
| MC01 | 39 | 386.71 ± 49.65 | 6504.54 ± 358.27 |
| MC03 | 30 | 1281.66 ± 214.17 | 6412.01 ± 558.38 |
| MC09 | 37 | 799.64 ± 29.87 | 5326.77 ± 328.37 |
| MC10 | 31 | 196.43 ± 10.69 | 2005.88 ± 105.57 |
| MC16 | 33 | 148.43 ± 9.76 | 2714.01 ± 100.84 |
| MC21 | 34 | 814.00 ± 21.21 | 5350.00 ± 212.13 |
| MC26 | 30 | 439.96 ± 66.17 | 1928.96 ± 120.18 |
| MC27 | 35 | 837.22 ± 78.87 | 5880.64 ± 1131.21 |
| MC190 | 41 | 128.60 ± 11.25 | 3749.16 ± 365.41 |
| B | |||
|---|---|---|---|
| Normal | PCOS | p | |
| N | 7 | 9 | |
| Age | 37 (36, 41) | 34 (30, 41) | 0.101 |
| Basal DHEA production | 40.37 (24.5, 52.87) | 439.96 (128.6, 1281.66) | <0.001 |
| Forskolin-stimulated DHEA production | 183.32 (139.64, 578.52) | 5326.77 (1928.96, 6504.54) | <0.001 |
Table 3.
Variants that are significant and their observed p-values from the Wilcoxon rank sum test when comparing forskolin-stimulated DHEA between samples having the minor allele to those having only the major allele.
| Gene | Chromosome | GRCh37 Position | GRCh38 Position | dbSNP ID | Major Allele (n) | Minor Allele (n) | P-value |
|---|---|---|---|---|---|---|---|
| THADA | chr2 | 43519977 | 43292838 | rs35720761 | C (11) | T (5) | 0.013278 |
| THADA | chr2 | 43736171 | 43509032 | rs6544669 | T (12) | C (4) | 0.007692 |
| ERBB4 | chr2 | 212530466 | 211665741 | rs6725181 | C (12) | T (4) | 0.02967 |
| ERBB4 | chr2 | 212587321 | 211722596 | rs13002712 | G (2) | A (14) | 0.033333 |
| DENND1A | chr9 | 126304695 | 123542416 | rs748994 | T (13) | G (3) | 0.039286 |
| RAB5B | chr12 | 56374318 | 55980534 | rs67594137 | C (13) | T (3) | 0.003571 |
| RAB5B | chr12 | 56374803 | 55981019 | rs11171713 | G (13) | A (3) | 0.003571 |
| RAB5B | chr12 | 56386076 | 55992292 | rs11550558 | A (13) | G (3) | 0.003571 |
| SUOX | chr12 | 56395689 | 56001905 | rs7963590 | G (13) | A (3) | 0.003571 |
| ERBB3 | chr12 | 56473808 | 56080024 | rs7297175 | T (7) | C (9) | 0.041783 |
| ERBB3 | chr12 | 56478607 | 56084823 | rs12817471 | G (13) | A (3) | 0.003571 |
| ERBB3 | chr12 | 56487201 | 56093417 | rs2229046 | T (13) | C (3) | 0.003571 |
| ERBB3 | chr12 | 56494998 | 56101214 | rs773123 | A (13) | T (3) | 0.003571 |
| ERBB3 | chr12 | 56495306 | 56101522 | rs812826 | C (13) | T (3) | 0.003571 |
| ERBB3 | chr12 | 56498241 | 56104457 | rs773121 | G (13) | A (3) | 0.003571 |
The ten variants on chromosome 12 showing significant effects include 6 in the ERBB3 gene (rs7297125, rs12817471,S rs2229046, rs773123, rs812826, rs773121), and 4 (rs67594137, rs11171713, rs11550558, rs7963590) in RAB5B/SUOX. The latter genes overlap each other on opposite DNA strands. Notably, three PCOS theca cell preparations (MC01, MC03, MC27) each derived from a different subject and representing one third of the PCOS sample, contained the minor allele of these SNVs in a heterozygous state, suggesting linkage disequilibrium (LD) and a large effect size. The LD was not unexpected since the variants are located within a 150 kb stretch of chromosome 12q13.2. Moreover, using LDlink programs to investigate linkage disequilibrium in European populations, we found that one SNV, rs7297175, is independent of the other 9, while 6 SNVs (rs67594137, rs11171713, rs11550558, rs7963590, rs12817471, rs2229046) formed one LD group (r2 0.77 to 0.97 among the SNVs) and 3 SNVs (rs773123, rs812826, rs773121) formed another LD group (r2 1.0) (Figure 1). We then used LDlink to identify the haplotypes present in the two haplotype blocks, finding two haplotypes in each block, one consisting of the major allele of the constituent SNVs and the other containing the minor alleles (Figure 2).
Fig 1. Linkage disequilibrium among chromosome 12 SNVs in Individuals of European Ancestry.
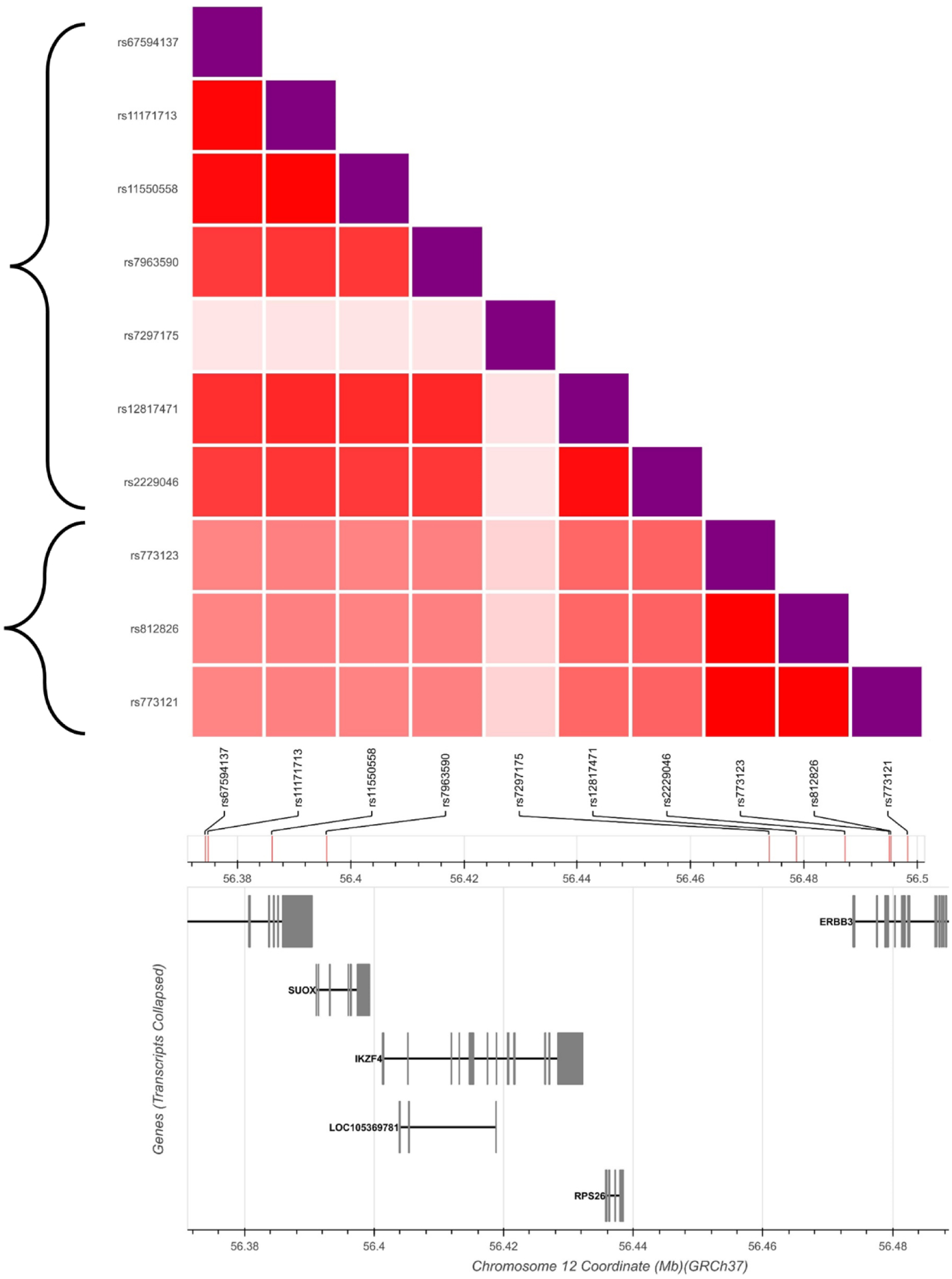
The plot displays linkage disequilibrium as r2 between each pair of SNVs. The brackets outline the two linkage disequilibrium groups (r2 > 0.5). Note that rs7297175 is independent of the other 9 SNVs and is not included in either haplotype block. Darker red indicates higher linkage disequilibrium. SNV locations and genes in the region are displayed at bottom.
Fig 2. Haplotypes in the chromosome 12 region of interest.
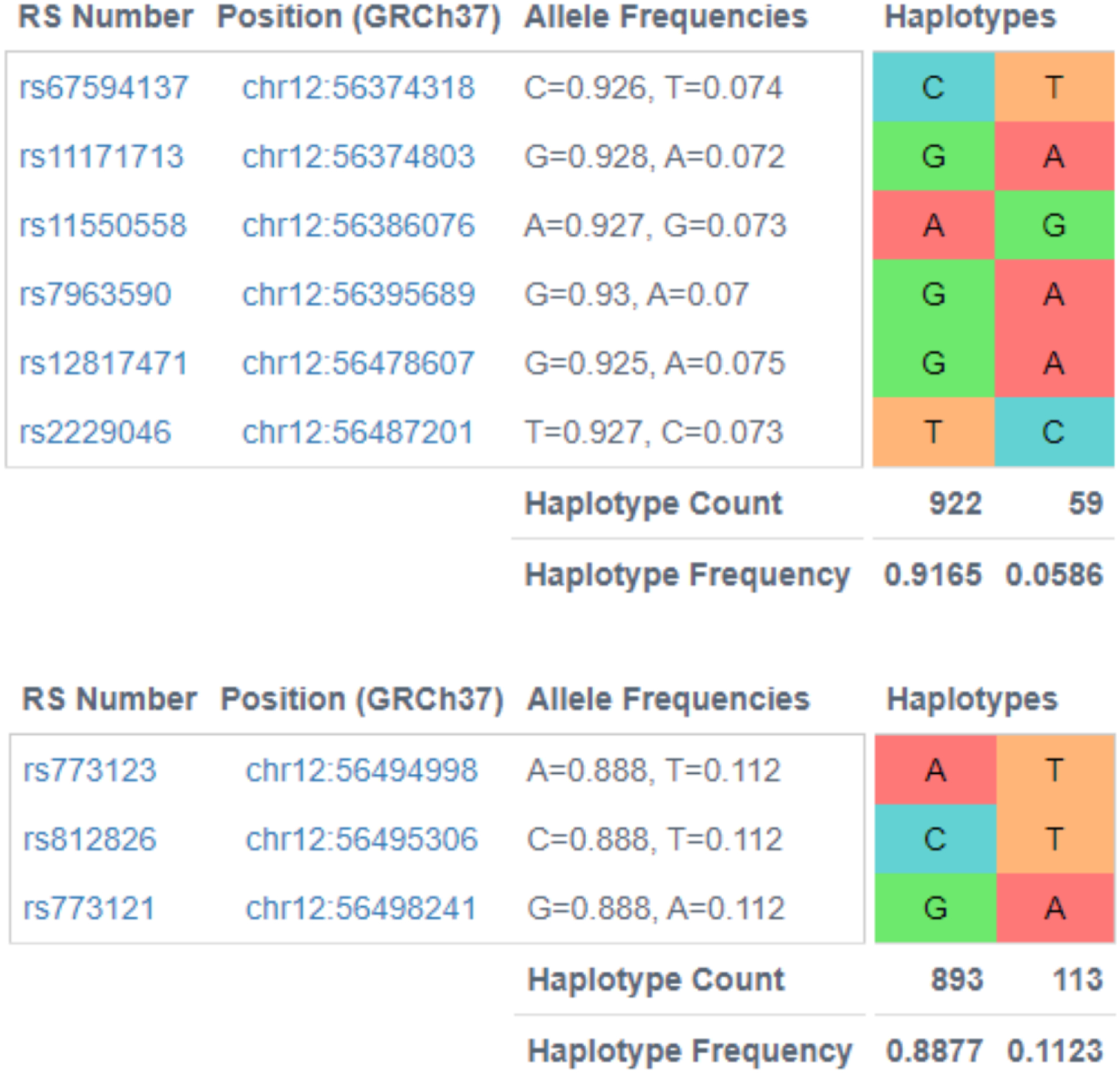
The first haplotype block consists of 6 SNVs and the second consists of 3 SNVs. Each block contains a common haplotype and a rare haplotype.
3.2. Single Cell RNA Sequencing of Theca Cells
We performed single cell RNA sequencing on normal (N=5) and PCOS (N=5) theca cells before and after forskolin treatment. Seurat (Hao et al., 2021) differential expression analyses were performed based solely on PCOS affection or forskolin treatment status and also among combinations of affection and treatment status (normal untreated vs normal treated; PCOS untreated vs PCOS treated; normal untreated vs PCOS untreated; normal treated vs PCOS treated). The only gene identified in this study as associated with androgen levels that was also differentially expressed in any of the analyses was PA2G4. PA2G4 showed significantly decreased expression in normal (Wilcoxon rank sum test Bonferroni corrected padj = 8.02E-100) and PCOS (padj = 1.43E-48) cells after forskolin treatment. The fold change was greater after forskolin treatment for PCOS (average log2 fold change 0.26) than normal (average log2 fold change 0.18) cells. Expression levels were significantly (padj = 3.82E-30) less in PCOS cells (average expression 1.25) than normal cells (average expression 1.51) after forskolin treatment. We also compared PCOS samples heterozygous for rs773121 (n =2 ) to all samples, both normal and PCOS, without the SNV (n = 8). PA2G4 was not significantly (padj = 1) differentially expressed before forskolin treatment. After forskolin treatment, samples heterozygous for rs773121 showed significantly lower expression (padj = 3.82E-30; average log2 fold change 0.16; average expression 1.22) than samples without the rs773121 SNV (average expression 1.43). This is consistent with a functional effect of the PA2G4 promoter SNV.
3.3. Analysis of Whole Genome Sequences from a Cohort of European Ancestry Women.
Using the findings from our WES of theca cell DNA and variant associations with forskolin-stimulated theca cell DHEA production as a discovery phase, we next examined the chromosome 12 SNVs in whole genome sequencing data from a cohort of 318 individuals of European ancestry from 77 families with one or more daughters with PCOS (Dapas et al., 2019). This cohort includes 62 families that were previously published by Dapas, et al., 2019 (Dapas et al., 2019) and an additional 15 families enrolled using the same criteria. The chromosome 12 SNVs were not identified in Dapas, et al., 2019 (Dapas et al., 2019) which primarily examined rare SNVs. PLINK (Purcell et al., 2007) transmission disequilibrium tests (TDT) on the individual SNVs did not identify any of them as significantly associated with PCOS/HA (hyperandrogenemia) affection status (Urbanek et al., 1999) (Table S3). However, rs773123, rs812826, and rs773121, corresponding to one of the haplotype groups identified in the discovery phase, had p values ranging from 0.1011 to 0.1404 compared to p values ranging from 0.4652 to 0.8575 for the other SNVs. Based on the lower range of p values for rs773123, rs812826, and rs773121, we examined the possibility that they formed a haplotype with association to PCOS/HA using the Family-Based Association Tests (FBAT) (Horvath et al., 2001) HBAT test which is the haplotype version of the association test. Based on the HBAT test, the haplotype consisting of the minor alleles for rs773123, rs812826, and rs773121 and the major alleles for the remaining SNVs was found preferentially in women with PCOS and elevated androgen levels (p = 0.0583) (Table S4).
3.4. Functional Annotations of SNVs
To examine the potential functional impact of the chromosome 12 SNVs, we annotated them with CADD PHRED scores (Rentzsch et al., 2019). A CADD PHRED score of 10 predicts a SNV is among the 10% most functionally significant changes in the human genome while a CADD PHRED score of 20 indicates the change is among the 1% most functional changes. Three of the SNVs have a CADD PHRED score greater than 10 including two in the HBAT identified haplotype (Table S5). We also annotated the chromosome 12 SNVs using FATHMM-MKL (Shihab et al., 2015) and FATHMM-XF (Rogers et al., 2018) (Table S6) in which scores above 0.5 are predicted to be deleterious and scores close to 1 are high confidence predictions. rs773123 (CADD PHRED 24.6, FATHMM-MKL 0.9735, FATHMM-XF 0.855483) is a missense SNV in ERBB3 at amino acid position 1119 which is in the cytoplasmic topological domain (https://www.uniprot.org/uniprot/P21860#subcellular_location). PolyPhen-2 (Adzhubei et al., 2013) predicts rs773123 is probably damaging (HumDiv 1.0, HumVar 0.996 for canonical ERBB3 protein P21860). SIFT (Kumar et al., 2009; Ng and Henikoff, 2003) predicts whether an amino acid substitution affects protein function with scores ranging from 0.0 (deleterious) to 1.0 (tolerated) and scores less than 0.05 considered deleterious. SIFT predicts rs773123 is deleterious (0.023, 0.045, 0.049) or tolerated (0.096, 0.097, 0.12) depending on the ERBB3 protein isoform examined. rs773121 (CADD PHRED 13.2, FATHMM-MKL 0.72049, FATHMM-XF 0.100013) is in the promoter (Ensembl regulatory feature ENSR00000052477) for PA2G4 which is a transcriptional co-repressor of androgen receptor-regulated genes (Zhang et al., 2005). PA2G4 interacts with the cytoplasmic domain of ERBB3 (Yoo et al., 2000) which contains the rs773123 missense SNV. The PA2G4 Ensembl regulatory feature ENSR00000052477 is equivalent to the GeneHancer (Fishilevich et al., 2017) GH12J056102 regulatory element. This GeneHancer regulatory element is classified as having both promoter and enhancer functions and two of the genes for which it has a predicted enhancer function are ERBB3 and RAB5B. These interactions can be viewed in the UCSC Genome Browser “Interactions between GeneHancer regulatory elements and genes” track (https://genome.ucsc.edu/s/Rharris1/chr12.PCOS). Since these two SNVs occur in a haplotype with a p value approaching significance, there could be some interplay between SNV rs773121’s regulatory effect on the expression of PA2G4 and ERBB3 and SNV rs773123’s effect on the interaction between the ERBB3 cytoplasmic domain and PA2G4. Additionally, PA2G4 and ERBB3 are targets of transcription factor, ZNF217, another PCOS candidate gene identified by GWAS (https://maayanlab.cloud/Harmonizome/gene_set/ZNF217/ENCODE+Transcription+Factor+Targets) (Krig et al., 2010).
4. Discussion
We found evidence of an association between androgen (DHEA) production by cultures of human theca cells, a reflection of theca cell endocrine activity, and fifteen SNVs, including ten in a haplotype on chromosome 12. This haplotype was found preferentially in women with PCOS and elevated androgen levels (p = 0.0583) in an independent family cohort. Two minor alleles in the chromosome 12 haplotype were likely to have functional consequences based on CADD scores and genomic context, suggesting that they affect ERBB3 and PA2G4 interactions (rs773123) or PA2G4 expression (rs773121). PA2G4 is a transcriptional corepressor of androgen receptor and androgen receptor-regulated genes (Zhang et al., 2005). Interestingly, this gene has not been highlighted as a PCOS candidate in published GWAS, despite the fact that it resides in a locus containing ERBB3, which has been advanced as a PCOS candidate gene. Single cell RNA sequencing of the theca cells showed significantly (padj = 3.82E-30) less expression of PA2G4 in cells heterozygous for rs773121 compared to cells without the SNV. This is consistent with a functional effect of the PA2G4 promoter SNV. Moreover, a recent study (Censin et al., 2021) utilizing a different methodologic approach, colocalization analysis, identified the same region on chromosome 12 encompassing ERBB3, PA2G4, RAB5B and SUOX, as containing potential PCOS disease-mediating genes.
ERBB3 is a component of the EGF family of signal transduction factors. It forms heterodimers with other ERBB family members, including ERBB2, which is the receptor for neuregulins, which are produced by both theca cells and granulosa cells in response to LH (Chowdhury et al., 2017). Neuregulin-1 (NRG-1) also plays a role in the proliferation of Leydig cells, an androgen producing cell type in the male gonad that is functionally analogous to theca cells in the female gonad. Thus, the haplotype identified in this report encompasses genes that are implicated in the regulation/function of androgen producing cells. Moreover, both ERBB3 and RAB5B have been associated with metabolic phenotypes that are related to PCOS, including glucose metabolism/insulin resistance (Day et al., 2018, 2015; Jones et al., 2015).
GWAS conducted on Han Chinese identified RAB5B/SUOX as PCOS candidate loci (Shi et al., 2012). The minor alleles of the SNVs we identified have very different allele frequencies in non-Finnish European and East Asian populations with the exception of rs11550558, which is low frequency in East Asians (<0.05%) and 7–11% in Europeans (Table S7). A recent case-control study of SNVs encoding the 3’-UTR of the RAB5B gene conducted in Han Chinese revealed highly significant associations with PCOS phenotypes with large effect sizes (Yu et al., 2019). These findings support our conclusions that 12q13.2 contains important genetic determinants of PCOS, and that the specific SNVs that influence thecal androgen production are population-specific. The SNVs may impact the level of expression of the candidate genes, accounting for variation in cell/tissue function, including the endocrine functions of thecal cells and granulosa cells.
RAB5B is involved in intracellular trafficking of endosomes including those derived from the plasma membrane (Gulappa et al., 2011). We have previously shown that RAB5B is co-localized in compartments containing DENND1A.V2, another PCOS GWAS candidate gene associated with hyperandrogenemia (Kulkarni et al., 2019; McAllister et al., 2015). DENND1A.V2 has an N-terminal guanine nucleotide exchange function and a clathrin-binding domain, putting it at the nexus with plasma membrane signaling proteins like the ERBB family of proteins. DENND1A.V2 is translocated into the nucleus of theca cells along with RAB5B, suggesting a role in regulation of expression of genes involved in androgen synthesis (Kulkarni et al., 2019). SUOX, which overlaps the RAB5B gene, encodes a mitochondrial sulfite oxidase, which has not been linked to PCOS, thecal cell function or steroidogenesis. Thus, the SNVs in this local region are likely to affect PCOS through effects on RAB5B rather than through SUOX.
5. Conclusion
In summary, our findings provide support for functional contributions of genes on chromosome 12q13.2, including ERBB3, PA2G4, and RAB5B, to ovarian cell dysfunction in PCOS. Our findings align with in silico colocalization analyses (Censin et al., 2021) implicating these same genes in PCOS pathogenesis. The relatively small sample size of the theca cell lines and PCOS cohort precludes us from making any definitive statements. However, this chromosome 12 region warrants further study given that both datasets point to the same SNVs as being involved in PCOS and single cell RNA-seq suggests a functional role for the PA2G4 promoter SNV.
Supplementary Material
Table S1. WES results for selected PCOS candidate genes. The table presents the gene name, chromosome assignment, nucleotide position of the detected variant, rs number, major and minor alleles, location and/or variant effect on coding sequence, transcript, and distribution of minor alleles among the different theca cell preparations designated by their MC number (see below), with number of homozygous (left), heterozygous (middle) followed by the total number of minor alleles (right) detected.
Table S2. Number of variants analyzed by gene after filtering steps applied.
Table S3. Cohort PLINK transmission disequilibrium tests (TDT) based on affection status for the individual chromosome 12 SNVs.
Table S4. Cohort Family-Based Association Tests (FBAT) HBAT test based on affection status for chromosome 12 SNVs.
Table S5. CADD PHRED scores and a subset of annotation details for chromosome 12 SNVs.
Table S6. FATHMM-MKL and FATHMM-XF scores for chromosome 12 SNVs. Predictions are given as p-values in the range [0, 1]. Values above 0.5 are predicted to be deleterious, while those below 0.5 are predicted to be neutral or benign. P-values close to the extremes (0 or 1) are the highest-confidence predictions that yield the highest accuracy. Scores > 0.8 are highlighted in red.
Table S7. Minor Allele Frequencies for the 12q13.2 SNVs. Minor allele frequencies for non-Finnish Europeans, East Asians and African Americans were extracted from GnomAD (https://gnomad.broadinstitute.org). rs7297175 is independent of the other 9 SNVs and is not included in either haplotype block.
SNVs in genes relevant to PCOS show an association with DHEA levels in theca cells
SNVs in a chromosome 12 haplotype were found preferentially in women with PCOS
ERBB3 missense and PA2G4 promoter SNVs are predicted to have functional consequences
Single cell RNA-seq further suggests the PA2G4 promoter SNV has functional consequences
Acknowledgements
This research was funded in part by the National Institutes of Health grants R01HD083323 (to J.M.M and J.F.S.), R01 HD100812 (to A.D.), R01 HD108744 (to A.D.) and R21 HD102172 (to J.R.).
Abbreviations
- PCOS
Polycystic ovary syndrome
- SNV
single nucleotide variant
- GWAS
genome-wide association studies
- WES
whole exome sequencing
- LD
linkage disequilibrium
- CADD
Combined Annotation-Dependent Depletion
- HA
hyperandrogenemia
- FBAT
Family-Based Association Tests;
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Adzhubei I, Jordan DM, Sunyaev SR, 2013. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet Chapter 7. 10.1002/0471142905.HG0720S76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azziz R, Carmina E, Chen Z, Dunaif A, Laven JS, Legro RS, Lizneva D, Natterson-Horowtiz B, Teede HJ, Yildiz BO, 2016. Polycystic ovary syndrome. Nat Rev Dis Primers 2, 16057. 10.1038/nrdp.2016.57 [DOI] [PubMed] [Google Scholar]
- Censin JC, Bovijn J, Holmes M. v, Lindgren CM, 2021. Colocalization analysis of polycystic ovary syndrome to identify potential disease-mediating genes and proteins. Eur J Hum Genet 29, 1446–1454. 10.1038/s41431-021-00835-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Zhao H, He L, Shi Yuhua,Qin Y, Shi Yongyong,Li Z, You L, Zhao Junli, Liu J, Liang X, Zhao X, Zhao Junzhao, Sun Y, Zhang B, Jiang H, Zhao D, Bian Y, Gao X, Geng L, Li Yiran, Zhu D, Sun X, Xu JE, Hao C, Ren CE, Zhang Y, Chen S, Zhang W, Yang A, Yan J, Li Yuan, Ma J, Zhao Y, 2011. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet 43, 55–59. 10.1038/NG.732 [DOI] [PubMed] [Google Scholar]
- Chowdhury I, Branch A, Mehrabi S, Ford BD, Thompson WE, 2017. Gonadotropin-Dependent Neuregulin-1 Signaling Regulates Female Rat Ovarian Granulosa Cell Survival. Endocrinology 158, 3647–3660. 10.1210/EN.2017-00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapas M, Dunaif A, 2022. Deconstructing a Syndrome: Genomic Insights into PCOS Causal Mechanisms and Classification. Endocr Rev. 10.1210/ENDREV/BNAC001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapas M, Sisk R, Legro RS, Urbanek M, Dunaif A, Hayes MG, 2019. Family-based quantitative trait meta-analysis implicates rare noncoding variants in DENND1A in polycystic ovary syndrome. J Clin Endocrinol Metab 104, 3835–3850. 10.1210/JC.2018-02496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day F, Karaderi T, Jones MR, Meun C, He C, Drong A, Kraft P, Lin N, Huang H, Broer L, Magi R, Saxena R, Laisk T, Urbanek M, Hayes MG, Thorleifsson G, Fernandez-Tajes J, Mahajan A, Mullin BH, Stuckey BGA, Spector TD, Wilson SG, Goodarzi MO, Davis L, Obermayer-Pietsch B, Uitterlinden AG, Anttila V, Neale BM, Jarvelin MR, Fauser B, Kowalska I, Visser JA, Andersen M, Ong K, Stener-Victorin E, Ehrmann D, Legro RS, Salumets A, McCarthy MI, Morin-Papunen L, Thorsteinsdottir U, Stefansson K, Styrkarsdottir U, Perry JRB, Dunaif A, Laven J, Franks S, Lindgren CM, Welt CK, 2018. Large-scale genome-wide meta-analysis of polycystic ovary syndrome suggests shared genetic architecture for different diagnosis criteria. PLoS Genet 14. 10.1371/JOURNAL.PGEN.1007813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day FR, Hinds DA, Tung JY, Stolk L, Styrkarsdottir U, Saxena R, Bjonnes A, Broer L, Dunger DB, Halldorsson B. v., Lawlor DA, Laval G, Mathieson I, McCardle WL, Louwers Y, Meun C, Ring S, Scott RA, Sulem P, Uitterlinden AG, Wareham NJ, Thorsteinsdottir U, Welt C, Stefansson K, Laven JSE, Ong KK, Perry JRB, 2015. Causal mechanisms and balancing selection inferred from genetic associations with polycystic ovary syndrome. Nat Commun 6. 10.1038/NCOMMS9464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depristo MA, Banks E, Poplin R, Garimella K. v., Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ, 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43, 491–501. 10.1038/NG.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamanti-Kandarakis E, Dunaif A, 2012. Insulin resistance and the polycystic ovary syndrome revisited: an update on mechanisms and implications. Endocr Rev 33, 981–1030. 10.1210/ER.2011-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishilevich S, Nudel R, Rappaport N, Hadar R, Plaschkes I, Stein TI, Rosen N, Kohn A, Twik M, Safran M, Lancet D, Cohen D, 2017. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017. 10.1093/DATABASE/BAX028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulappa T, Clouser CL, Menon KM, 2011. The role of Rab5a GTPase in endocytosis and post-endocytic trafficking of the hCG-human luteinizing hormone receptor complex. Cell Mol Life Sci 68, 2785–2795. 10.1007/s00018-010-0594-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R, 2021. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. 10.1016/J.CELL.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes MG, Urbanek M, Ehrmann DA, Armstrong LL, Lee JY, Sisk R, Karaderi T, Barber TM, McCarthy MI, Franks S, Lindgren CM, Welt CK, Diamanti-Kandarakis E, Panidis D, Goodarzi MO, Azziz R, Zhang Y, James RG, Olivier M, Kissebah AH, Reproductive Medicine N, Stener-Victorin E, Legro RS, Dunaif A, 2015. Genome-wide association of polycystic ovary syndrome implicates alterations in gonadotropin secretion in European ancestry populations. Nat Commun 6, 7502. 10.1038/ncomms8502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Xu X, Laird NM, 2001. The family based association test method: strategies for studying general genotype--phenotype associations. Eur J Hum Genet 9, 301–306. 10.1038/SJ.EJHG.5200625 [DOI] [PubMed] [Google Scholar]
- Jones MR, Brower MA, Xu N, Cui J, Mengesha E, Chen YDI, Taylor KD, Azziz R, Goodarzi MO, 2015. Systems Genetics Reveals the Functional Context of PCOS Loci and Identifies Genetic and Molecular Mechanisms of Disease Heterogeneity. PLoS Genet 11. 10.1371/JOURNAL.PGEN.1005455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krig SR, Miller JK, Frietze S, Beckett LA, Neve RM, Farnham PJ, Yaswen PI, Sweeney CA, 2010. ZNF217, a candidate breast cancer oncogene amplified at 20q13, regulates expression of the ErbB3 receptor tyrosine kinase in breast cancer cells. Oncogene 29, 5500–5510. 10.1038/ONC.2010.289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni R, Teves ME, Han AX, McAllister JM, Strauss JF 3rd, 2019. Colocalization of Polycystic Ovary Syndrome Candidate Gene Products in Theca Cells Suggests Novel Signaling Pathways. J Endocr Soc 3, 2204–2223. 10.1210/js.2019-00169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC, 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4, 1073–81. 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- Legro RS, Arslanian SA, Ehrmann DA, Hoeger KM, Murad MH, Pasquali R, Welt CK, 2013. Diagnosis and treatment of polycystic ovary syndrome: an endocrine society clinical practice guideline. J Clin Endocrinol Metab 98, 4565–4592. 10.1210/jc.2013-2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legro RS, Driscoll D, Strauss JF, Fox J, Dunaif A, 1998. Evidence for a genetic basis for hyperandrogenemia in polycystic ovary syndrome. Proc Natl Acad Sci U S A 95, 14956–14960. 10.1073/PNAS.95.25.14956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM.
- McAllister JM, Legro RS, Modi BP, Strauss JF 3rd, 2015. Functional genomics of PCOS: from GWAS to molecular mechanisms. Trends Endocrinol Metab 26, 118–124. 10.1016/j.tem.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA, 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303. 10.1101/GR.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson VL, Legro RS, Strauss JF 3rd, McAllister JM, 1999. Augmented androgen production is a stable steroidogenic phenotype of propagated theca cells from polycystic ovaries. Mol Endocrinol 13, 946–957. [DOI] [PubMed] [Google Scholar]
- Nelson-DeGrave VL, Wickenheisser JK, Cockrell JE, Wood JR, Legro RS, Strauss JF 3rd, McAllister JM, 2004. Valproate potentiates androgen biosynthesis in human ovarian theca cells. Endocrinology 145, 799–808. [DOI] [PubMed] [Google Scholar]
- Nelson-Degrave VL, Wickenheisser JK, Hendricks KL, Asano T, Fujishiro M, Legro RS, Kimball SR, Strauss JF 3rd, McAllister JM, 2005. Alterations in mitogen-activated protein kinase kinase and extracellular regulated kinase signaling in theca cells contribute to excessive androgen production in polycystic ovary syndrome. Mol Endocrinol 19, 379–390. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S, 2003. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 31, 3812–3814. 10.1093/NAR/GKG509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC, 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M, 2019. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 47, D886–D894. 10.1093/NAR/GKY1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers MF, Shihab HA, Mort M, Cooper DN, Gaunt TR, Campbell C, 2018. FATHMM-XF: accurate prediction of pathogenic point mutations via extended features. Bioinformatics 34, 511–513. 10.1093/BIOINFORMATICS/BTX536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Yongyong, Zhao, Shi Yuhua, Cao Y, Yang D, Li Z, Zhang B, Liang X, Li T, Chen J, Shen J, Zhao Junzhao, You L, Gao X, Zhu D, Zhao X, Yan Y, Qin Yingying, Li Wenjin, Yan J, Wang Q, Zhao Junli, Geng L, Ma, Zhao, He, Zhang A, Zou S, Yang A, Liu J, Li Weidong, Li B, Wan C, Qin Ying, Shi J, Yang J, Jiang H, Xu JE, Qi X, Sun Y, Zhang Yajie, Hao C, Ju X, Zhao D, Ren CE, Li X, Zhang W, Zhang Yiwen, Zhang J, Wu D, Zhang C, He L, Chen ZJ, 2012. Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat Genet 44, 1020–1025. 10.1038/NG.2384 [DOI] [PubMed] [Google Scholar]
- Shihab HA, Rogers MF, Gough J, Mort M, Cooper DN, Day INM, Gaunt TR, Campbell C, 2015. An integrative approach to predicting the functional effects of noncoding and coding sequence variation. Bioinformatics 31, 1536–1543. 10.1093/BIOINFORMATICS/BTV009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanek M, Legro RS, Driscoll DA, Azziz R, Ehrmann DA, Norman RJ, Strauss JF, Spielman RS, Dunaif A, 1999. Thirty-seven candidate genes for polycystic ovary syndrome: strongest evidence for linkage is with follistatin. Proc Natl Acad Sci U S A 96, 8573–8578. 10.1073/PNAS.96.15.8573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickenheisser JK, Biegler JM, Nelson-Degrave VL, Legro RS, Strauss JF 3rd, McAllister JM, 2012. Cholesterol side-chain cleavage gene expression in theca cells: augmented transcriptional regulation and mRNA stability in polycystic ovary syndrome. PLoS One 7, e48963. 10.1371/journal.pone.0048963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickenheisser JK, Nelson-Degrave VL, McAllister JM, 2005. Dysregulation of cytochrome P450 17alpha-hydroxylase messenger ribonucleic acid stability in theca cells isolated from women with polycystic ovary syndrome. J Clin Endocrinol Metab 90, 1720–1727. [DOI] [PubMed] [Google Scholar]
- Wickenheisser JK, Nelson-DeGrave VL, Quinn PG, McAllister JM, 2004. Increased cytochrome P450 17alpha-hydroxylase promoter function in theca cells isolated from patients with polycystic ovary syndrome involves nuclear factor-1. Mol Endocrinol 18, 588–605. [DOI] [PubMed] [Google Scholar]
- Wickenheisser JK, Quinn PG, Nelson VL, Legro RS, Strauss JF 3rd, McAllister JM, 2000. Differential activity of the cytochrome P450 17alpha-hydroxylase and steroidogenic acute regulatory protein gene promoters in normal and polycystic ovary syndrome theca cells. J Clin Endocrinol Metab 85, 2304–2311. [DOI] [PubMed] [Google Scholar]
- Yoo JY, Wang XW, Rishi AK, Lessor T, Xia XM, Gustafson TA, Hamburger AW, 2000. Interaction of the PA2G4 (EBP1) protein with ErbB-3 and regulation of this binding by heregulin. Br J Cancer 82, 683–690. 10.1054/BJOC.1999.0981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Ding C, Guan S, Wang C, 2019. Association of single nucleotide polymorphisms in the RAB5B gene 3’UTR region with polycystic ovary syndrome in Chinese Han women. Biosci Rep 39. 10.1042/BSR20190292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Akinmade D, Hamburger AW, 2005. The ErbB3 binding protein Ebp1 interacts with Sin3A to repress E2F1 and AR-mediated transcription. Nucleic Acids Res 33, 6024–6033. 10.1093/NAR/GKI903 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. WES results for selected PCOS candidate genes. The table presents the gene name, chromosome assignment, nucleotide position of the detected variant, rs number, major and minor alleles, location and/or variant effect on coding sequence, transcript, and distribution of minor alleles among the different theca cell preparations designated by their MC number (see below), with number of homozygous (left), heterozygous (middle) followed by the total number of minor alleles (right) detected.
Table S2. Number of variants analyzed by gene after filtering steps applied.
Table S3. Cohort PLINK transmission disequilibrium tests (TDT) based on affection status for the individual chromosome 12 SNVs.
Table S4. Cohort Family-Based Association Tests (FBAT) HBAT test based on affection status for chromosome 12 SNVs.
Table S5. CADD PHRED scores and a subset of annotation details for chromosome 12 SNVs.
Table S6. FATHMM-MKL and FATHMM-XF scores for chromosome 12 SNVs. Predictions are given as p-values in the range [0, 1]. Values above 0.5 are predicted to be deleterious, while those below 0.5 are predicted to be neutral or benign. P-values close to the extremes (0 or 1) are the highest-confidence predictions that yield the highest accuracy. Scores > 0.8 are highlighted in red.
Table S7. Minor Allele Frequencies for the 12q13.2 SNVs. Minor allele frequencies for non-Finnish Europeans, East Asians and African Americans were extracted from GnomAD (https://gnomad.broadinstitute.org). rs7297175 is independent of the other 9 SNVs and is not included in either haplotype block.


