Abstract
Meningococcal strains isolated during an outbreak were shown to belong to the ET-5 complex and to harbor a mutation in the VR2 region of the porA gene. They were less susceptible to the bactericidal effect of normal human serum than was the ET-5 wild-type strain. These results are of concern, as PorA is a potential target in vaccine design.
Neisseria meningitidis is an exclusive human respiratory tract bacterium causing either asymptomatic or invasive infections (bacteremia and/or meningitis). Meningococcal infections occur as sporadic or epidemic cases. Strains involved in epidemics are usually different from those isolated from sporadic cases and often belong to few clonal complexes (4). Herd immunity within a population is usually acquired by asymptomatic carriage of N. meningitidis or of closely related species, such as Neisseria lactamica (3). Lack of or an altered serum bactericidal effect against N. meningitidis is a major factor of host susceptibility, and serum resistance is a major virulence factor of strains causing invasive meningococcal infections (10).
From June 1995 to June 1998, 34 cases of meningococcal disease were recorded in the department of Maine et Loire, France (720,000 inhabitants). Seventeen cases were in the city of Saumur (40,000 inhabitants) in this department, and 13 of these occurred in the Chemin Vert (CV) district (3,000 inhabitants) of Saumur. During the peak of the outbreak in 1996, the national incidence of meningococcal disease in France was 0.69 per 100,000 inhabitants. However, it was 1.7, 20, and 200 in the department of Maine et Loire, the city of Saumur, and the district of CV, respectively.
Strains of N. meningitidis were obtained from 11 cases in the CV district and were studied at the National Reference Center for the meningococci. They were characterized by serological typing; multilocus DNA fingerprinting (MLDF) using pilA, pilD, and crgA restriction fragment length polymorphism; multilocus enzyme electrophoresis (MLEE) (6); and porA gene sequencing. The porA gene was amplified by PCR using oligonucleotides porA0 and porA100. The variable region VR1 of porA was sequenced using oligonucleotides porA1 and porA101 for coding and noncoding strands, respectively. The variable region VR2 of porA was sequenced using oligonucleotides porA4 and porA104 for coding and noncoding strands, respectively (Table 1). In one culture-negative case, MLDF typing and porA sequencing were performed directly on serum obtained from the patient as previously described (5).
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide (gene amplified) | Sequence | Relevant characteristic |
|---|---|---|
| porA0 (porA) | 5′-GATGTCAGCCTATACGGCGAAATCAAA-3′ | Coding strand |
| porA100 (porA) | 5′-GAATTTGTGGCGCAAACCGACGGAGGC-3′ | Noncoding strand |
| porA1 (porA) | 5′-GCCGGCGTGGAAGGCAGGAAC-3′ | Coding strand of VR1 |
| PorA101 (porA) | 5′-GCCGATAAACGAGCCGAAATC-3′ | Noncoding strand of VR1 |
| PorA4 (porA) | 5′-GGCAGCGTCCAATTCGTTCCG-3′ | Coding strand of VR2 |
| PorA104 (porA) | 5′-GCCATTTTTGTAATTCAGACCGGC-3′ | Noncoding strand of VR2 |
| A-412 (pilA) | 5′-CAATCCAGCAGTCGGTCCACA-3′ | Noncoding strand |
| B-127 (pilA) | 5′-GTTGTCGGTAACGACGGGCAG-3′ | Coding strand |
| PilD3 (pilD) | 5′-TGCCGCACAGATCCGGCGCGGAT-3′ | Coding strand |
| PilD4 (pilD) | 5′-TCTCACCGGATGGGTCAGCCA-3′ | Noncoding strand |
| 98-4 (crgA) | 5′-CGTTCAGCCGTGCGCGAGAGCTTGGCATGG-3′ | Coding strand |
| 98-11 (crgA) | 5′-AGAATTATCCACGAGAGATTGTTTCCC-3′ | Noncoding strand |
All strains were indistinguishable through MLEE and MLDF analysis and belonged to the ET-5 complex, a prominent clonal complex widely distributed in Europe (2). Serological typing showed that strains isolated in the CV district had the same antigenic formula, B:15:P1.7. However, four strains were also of subtype 16 (Table 2). Since subtype 16 was not detected in all strains from the CV district outbreak, we determined the DNA sequences encoding the variable regions (VR1 and VR2) of porA. PorA has eight surface-exposed loops, of which VR1 (loop 1) and VR2 (loop 4) are recognized by monoclonal antibodies used in N. meningitidis subtyping. DNA sequencing of VR1 indicated that all strains tested had the wild-type “classical” sequence of VR1 (subtype 7). However, DNA sequencing of VR2 showed the sequence of subtype 16, with the exception that all strains isolated from the CV district had a mutation in codon 184 (within VR2) of porA (AAC to AGC), which led to an Asn-to-Ser substitution in PorA (GenBank accession number AF287956). This mutation was not detected in strains isolated outside the CV district. Several strains of antigenic formula B:15:P1.7,16 were isolated elsewhere in France before, during, or after this period (1995 to 1998), and all shared a wild-type VR2 (Table 2 and data not shown). MLDF typing and porA sequencing gave identical results in all these strains. The VR2 mutation could not be detected by serosubtyping alone, therefore emphasizing the need for molecular methods for careful surveillance of N. meningitidis spread within populations.
TABLE 2.
Strains tested and their characteristics
| Strain | Origin (department) | Year | Antigenic formula | MLEE typing | MLDF typing for alleles of pilA, pilD, and crgA | DNA sequencing
|
|
|---|---|---|---|---|---|---|---|
| VR1 | VR2 | ||||||
| LNP13820 | Saumur-49 | 1995 | B:15:P1.7 | ET-5 | pilA5 / pilD5 / crgA1 | Wild type (7) | N→S mutation (16s)a |
| LNP14243 | Saumur-49 | 1996 | B:15:P1.7 | ET-5 | pilA5 / pilD5 / crgA1 | Wild type (7) | N→S mutation (16s) |
| LNP14522 | Saumur-49 | 1996 | B:15:P1.7 | ET-5 | pilA5 / pilD5 / crgA1 | Wild type (7) | N→S mutation (16s) |
| LNP14523 | Saumur-49 | 1996 | B:15:P1.7,16 | ET-5 | pilA5 / pilD5 / crgA1 | Wild type (7) | N→S mutation (16s) |
| LNP14714 | Saumur-49 | 1996 | B:15:P1.7,16 | ET-5 | pilA5 / pilD5 / crgA1 | Wild type (7) | N→S mutation (16s) |
| LNP14772 | Saumur-49 | 1996 | B:15:P1.7,16 | ET-5 | pilA5 / pilD5 / crgA1 | Wild type (7) | N→S mutation (16s) |
| LNP14839 | Saumur-49 | 1997 | B:15:P1.7 | ET-5 | pilA5 / pilD5 / crgA1 | Wild type (7) | N→S mutation (16s) |
| LNP14912 | Saumur-49 | 1997 | B:15:P1.7 | ET-5 | pilA5 / pilD5 / crgA1 | Wild type (7) | N→S mutation (16s) |
| LNP15367 | Saumur-49 | 1997 | B:15:P1.7 | ET-5 | pilA5 / pilD5 / crgA1 | Wild type (7) | N→S mutation (16s) |
| LNP15687 | Saumur-49 | 1998 | B:15:P1.7,16 | ET-5 | pilA5 / pilD5 / crgA1 | Wild type (7) | N→S mutation (16s) |
| LNP15709 | Saumur-49 | 1998 | B:15:P1.7 | ET-5 | pilA5 / pilD5 / crgA1 | Wild type (7) | N→S mutation (16s) |
| LNP10846 | LeMans-72b | 1992 | B:15:P1.7,16 | ET-5 | pilA5 / pilD5 / crgA1 | Wild type (7) | Wild type (16) |
GenBank accession number AF287956. The numbers in parentheses identify the subtypes of VR1 and VR2.
This strain was isolated in Department 72 (Sarthe), which neighbors Department 49.
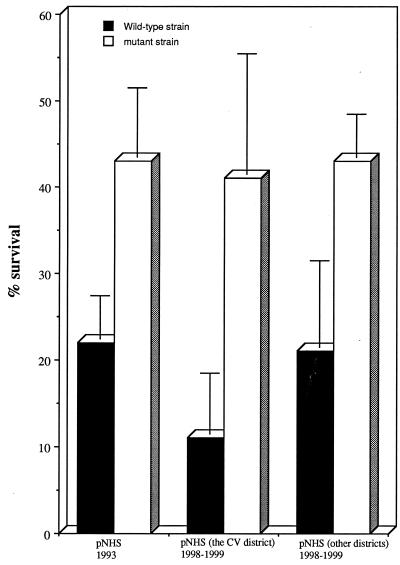
Antibodies directed against VR1 and VR2 are bactericidal (8). Lack of VR2 bactericidal antibodies could be an important factor explaining the behavior of the VR2 mutant among the population in the CV district. To test the susceptibility of the porA (VR2) mutant to the serum bactericidal effect, we compared the survival of the wild-type strain for VR2 to that of the porA mutant in 45% pooled normal human serum (NHS) using a whole serum assay (9). NHS (450 μl) was added to 550 μl of a bacterial suspension of 10,000 CFU/ml in Hanks' balanced salt solution (GIBCO BRL) containing 1 mM MgCl2 and 1.26 mM CaCl2. The mixture was incubated for 30 min at 37°C in the presence of 5% CO2. CFU were counted (at time zero and after 30 min) by plating the mixture on GCB medium (Difco) and incubating the plates for 24 h at 37°C in the presence of 5% CO2. Sera had been obtained before the outbreak (1993, n = 25) or after the outbreak (1998 to 1999). Sera obtained after the outbreak were from the CV district (n = 47) and from other districts in the city of Saumur (n = 36). The porA (VR2) mutant strain survived better than the wild-type strain when sera obtained before and after the outbreak were used (Fig. 1). Lipooligosaccharide is an important factor of serum resistance in pathogenic neisseriae (10). Lipooligosaccharide profiles of the porA mutant and the wild-type strain were analyzed as previously described (9) and were found identical (data not shown). These results suggest that natural bactericidal activity in NHS was more effective against the wild-type VR2 region than against the mutated VR2 region of PorA. No increase in bactericidal activity was observed in individual sera obtained from healthy subjects from the CV district obtained after the outbreak. These data suggest a low transmissibility of the mutant strain among the CV population during and after the outbreak, as also suggested by the prolonged duration of the outbreak (3 years).
FIG. 1.
Survival of N. meningitidis in pooled NHS. Bacterial strains were LNP10846 (wild type) and LNP13820 (mutant). Values represent the mean of three independent experiments ± standard deviation. The proportion of CFU surviving in sera after 30 min of exposure was significantly higher for the mutant than for the wild strain (P < 0.02, Student's t test). These findings were observed in three different settings: with sera obtained before the outbreak, with sera obtained after the outbreak from the CV inhabitants, and with sera obtained after the outbreak from Saumur inhabitants.
To demonstrate that antibodies in NHS reacted with PorA and that mutation in VR2 could affect this recognition, we performed competition enzyme-linked immunosorbent assays by modifying the classical subtyping method (1). Enzyme-linked immunosorbent assay plates were coated with heat-inactivated wild-type strain LNP10846 or with the mutant strain LNP14523 (Table 2). Serial dilutions of NHS were added and incubated for 2 h. After washing, specific monoclonal antibodies to subtype P1.7 or P1.16 were added to detect VR1 or VR2 regions on PorA, respectively, as in the classical subtyping method (1). Monoclonal antibody P1.7 (VR1) binding to both wild-type and mutant strains was inhibited by NHS at equivalent rates (30 and 28%, respectively). By contrast, when monoclonal antibody P1.16 (VR2) was used, NHS caused 37% binding inhibition to the wild-type strain, while no inhibition was detected for the mutant strain.
Mutations in this immunodominant antigen may have contributed to the CV district outbreak by allowing the mutant strain to circumvent herd immunity and to colonize and invade immunologically naive hosts. Mutations in the VR2 region have been reported to be associated with an increase in endemic disease in several areas in England and Wales (7). Our report directly correlates such a mutation with a geographically restricted outbreak of N. meningitidis. Both wild-type and mutant strains in this study belong to the ET-5 clonal complex. However, we cannot exclude that other mutations in other chromosomal loci could be responsible for enhanced virulence of the isolates involved in the CV district outbreak.
PorA and, in particular, the VR1 and VR2 regions are targets for the development of meningococcal vaccines. Our study underlines the ability of N. meningitidis to escape target-specific bactericidal antibodies. Escape strategies seem to be a general virulence mechanism in N. meningitidis. We have recently reported a capsule switch (escape switch from serogroup B to serogroups C and W135) after a vaccination campaign during a clonal outbreak in the Czech Republic (6). Vaccines directed against meningococcal-protective antigens should be carefully evaluated. A “cocktail” of several different proteins may be a reliable choice for future meningococcal vaccines, due to the extreme genetic plasticity of N. meningitidis.
Acknowledgments
We are grateful to Magaly Ducos, Martine Guibourdenche, Dario Giorgini, and René Pirés for technical help.
REFERENCES
- 1.Abdillahi H, Poolman J T. Neisseria meningitidis group B serosubtyping using monoclonal antibodies in whole-cell ELISA. Microb Pathog. 1988;4:27–32. doi: 10.1016/0882-4010(88)90045-9. [DOI] [PubMed] [Google Scholar]
- 2.Achtman M. Global epidemiology of meningococcal disease. In: Cartwright K, editor. Meningococcal disease. Chichester, United Kingdom: John Wiley & Sons; 1995. pp. 159–175. [Google Scholar]
- 3.Cartwright K. Meningococcal carriage and disease. In: Cartwright K, editor. Meningococcal disease. Chichester, United Kingdom: John Wiley & Sons; 1995. pp. 115–146. [Google Scholar]
- 4.Caugant D A, Kristiansen B E, Frøholm L O, Bøvre K, Selander P K. Clonal diversity of Neisseria meningitidis from a population of asymptomatic carriers. Infect Immun. 1988;56:2060–2068. doi: 10.1128/iai.56.8.2060-2068.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giorgini D, Nassif X, Taha M-K. Rapid epidemiological characterization of Neisseria meningitidis using polymerase chain reaction from biological samplings. Presse Med. 1997;26:1516–1519. [PubMed] [Google Scholar]
- 6.Kriz P, Giorgini D, Musilek M, Larribe M, Taha M-K. Microevolution through DNA exchange among strains of Neisseria meningitidis isolated during an outbreak in the Czech Republic. Res Microbiol. 1999;150:273–280. doi: 10.1016/s0923-2508(99)80052-7. [DOI] [PubMed] [Google Scholar]
- 7.McGuinness B T, Clarke I N, Lambden P R, Barlow A K, Poolman J T, Jones D M, Heckels J E. Point mutation in meningococcal porA gene associated with increased endemic disease. Lancet. 1991;337:514–517. doi: 10.1016/0140-6736(91)91297-8. [DOI] [PubMed] [Google Scholar]
- 8.Saukkonen K, Abdillahi H, Poolman J T, Leinonen M. Protective efficacy of monoclonal antibodies to class 1 and class 3 outer membrane proteins of Neisseria meningitidis B:15:P1.16 in infant rat infection model: new prospects for vaccine development. Microb Pathog. 1987;3:261–267. doi: 10.1016/0882-4010(87)90059-3. [DOI] [PubMed] [Google Scholar]
- 9.Taha M-K. Increased sensitivity of gonococcal pilA mutants to bactericidal activity of normal human serum. Infect Immun. 1993;61:4662–4668. doi: 10.1128/iai.61.11.4662-4668.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogel U, Frosch M. Mechanisms of neisserial serum resistance. Mol Microbiol. 1999;32:1133–1139. doi: 10.1046/j.1365-2958.1999.01469.x. [DOI] [PubMed] [Google Scholar]