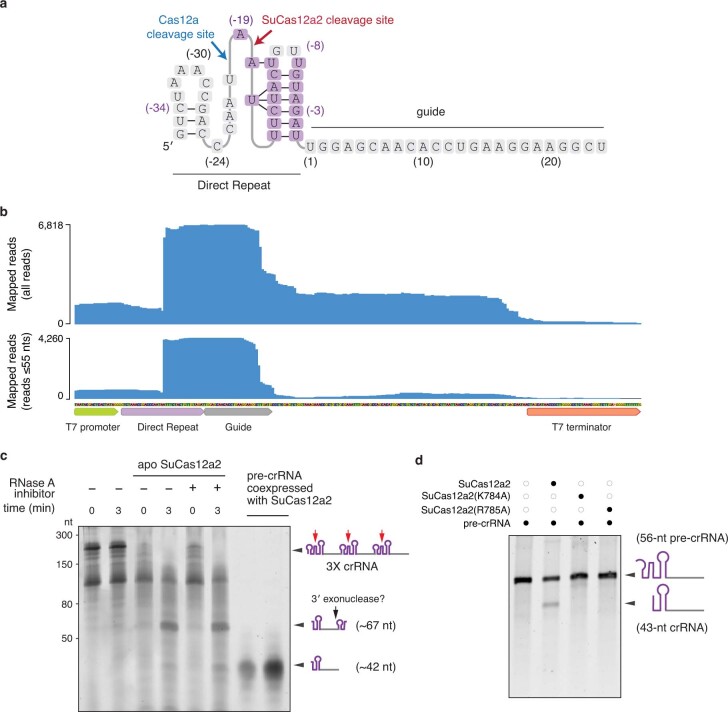
Extended Data Fig. 3. Pre-crRNA processing by SuCas12a2.
a, Diagram of the predicted secondary structure of the SuCas12a2 direct repeat. Cleavage sites of SuCas12a2 and Cas12a are indicated. b, Sequencing coverage of the cDNA mapped to the crRNA locus in E. coli BL21(AI) expressing SuCas12a2. Coverage for all of the quality-filtered reads above 10 nts as well as reads between 10 and 55 nts mapped to the plus strand are shown. c, In vitro processing of a 56-nt pre-crRNA incubated for 60 min in the presence of apo-SuCas12a2 or two mutants predicted to disrupt crRNA processing. d, In vitro SuCas12a2-mediated cleavage of a pre-crRNA containing three direct repeats and three spacers (3× crRNA). Reactions containing apo-SuCas12a2 are indicated. Time points after mixing the cleavage reaction with apo-SuCas12a2 are indicated. The estimated sizes of the pre-crRNA and crRNAs after cleavage are indicated on the left. The last two lanes contain RNA extracted from SuCas12a2 co-expressed with a CRISPR array. The difference in size between the major crRNA band in the in vitro assay (~ 67 nt) and the crRNA extracted from SuCas12a2 bound to E. coli-expressed crRNA (~42) may be due to further trimming by 3′ exonucleases in the cell. For gel source data, see Supplementary Fig. 1.