Abstract
Detecting the presence of important parasites within a host and its environment is critical to understanding the dynamics that influence a pathogen's ability to persist, while accurate detection is also essential for the implementation of effective control strategies. Pseudoloma neurophilia is the most common pathogen reported in zebrafish (Danio rerio) research facilities. The only assays currently available for P. neurophilia are through lethal sampling, often requiring euthanasia of the entire population for accurate estimates of prevalence in small populations. We present a non-lethal screening method to detect P. neurophilia in tank water based on the detection of environmental DNA (eDNA) from this microsporidium, using a previously developed qPCR assay that was adapted to the digital PCR (dPCR) platform to complement current surveillance protocols. Using the generated dPCR data, a multi-state occupancy model was also implemented to predict the probability of detecting the microsporidium in tank water under different flow regimes and pathogen prevalence. The occupancy model revealed that samples collected in static conditions were more informative than samples collected from flow-through conditions, with a probability of detection at 80% and 47%, respectively. There was also a positive correlation between the frequency of detection in water and prevalence in fish based on qPCR.
Keywords: Occupancy modeling, Zebrafish, Microsporidia, eDNA, ddPCR, Sonication
The zebrafish as a biomedical model is widely used in many studies, ranging from immunological and infectious disease to developmental biology and neuro-behavioral studies. The ease of breeding and housing and the development of genetic tools has facilitated the expansion of this model into nearly every field of biology. Some genetic lines produced for specific experiments breed poorly and require labor-intensive husbandry conditions to maintain even a small population, creating several challenges to the production of embryos needed to fulfill experimental protocols (Avdesh et al., 2012). A common threat to laboratory zebrafish and overall zebrafish facility operations is the presence of infectious diseases. While many aspects of the husbandry conditions of this animal are known, many remain unknown or poorly understood, which may contribute to the negative impacts of pathogens. The presence of infectious diseases in laboratory zebrafish has significant impact on the maintenance of zebrafish populations and may be a confounding factor in research results, potentially biasing the conclusions of many studies (Kent et al., 2012).
A microsporidian parasite, Pseudoloma neurophilia, is an ongoing threat to the zebrafish model. This parasite continues to be prevalent in zebrafish research facilities that report to the diagnostic service of the Zebrafish International Resource Center (ZIRC) in Eugene, Oregon (Murray et al., 2011; Kent et al., 2020b). An obligate intercellular parasite, P. neurophilia, causes chronic infections in zebrafish and infects a broad range of fishes (Sanders et al., 2016). Infections by P. neurophilia are largely asymptomatic. However, a subset of infected populations may present general clinical signs, such as emaciation and skeletal deformities. Histologically, spores occur in the central nervous system and may cause associated gliosis (Spagnoli et al., 2015), whereas the parasite infects other organs (Sanders et al., 2014) causing other various forms of inflammation (myositis, menixitis, and encephalitis) that are also associated with the infection. Important to the integrity of the zebrafish model, P. neurophilia causes significant alterations in behavior and has been reported to alter transcripts in the brain, downregulating several genes involved in immune function (Midttun et al., 2020). Moreover, infections cause reduced fecundity and growth, while stress exacerbates P. neurophilia prevalence in large populations (Ramsay et al., 2009).
The biology of the parasite promotes its ability to survive in the environment, as it develops into a hearty resistant spore that is highly resistant to disruption and even regular disinfection protocols (Ferguson et al., 2007). The development of P. neurophilia has 3 major phases: an infectious phase, which is free spores found in the environment, and 2 intracellular phases, proliferative and sporogonic. During the intracellular growth phases, several developmental stages occur, but it is free spores in the environment that can be a useful target for non-lethal diagnostics. Additionally, transmission occurs both vertically and horizontally, which creates numerous opportunities for detection of the parasite, as spores are released in the environment during spawning, from decomposing carcasses, and through urine and feces (Sanders et al., 2013).
Environmental DNA (eDNA) assays are commonly used by ecologists to detect and quantify organisms in water, air, and soil (Barnes et al., 2014; Rees et al., 2014; Bass et al., 2015). In terrestrial systems, parasites are often detected in the soil or feces (Almazán et al., 2001; Mandarino-Pereira et al., 2010; Nagamori et al., 2018). Whereas analysis of feces is usually not practical for aquatic parasites, the water itself provides a useful medium for detecting parasites simply through filtration. Hence, surveillance in aquatic systems has been deployed for the detection of several human and wildlife pathogens, including parasites (Mocho et al., 2017; Berger and Aubin-Horth, 2018; Peters et al., 2018; Sieber et al., 2020; Amarasiri et al., 2021). Notably, these have been developed for detecting common water-borne human parasites, such as Cryptosporidium spp. and Giardia lamblia using filtration of water (Guy et al., 2003).
Water tests have also been developed for a variety of fish parasite taxa, e.g., myxozoans, helminths, and protozoa. These include Ceratonova shasta (Hallett et al., 2012), Nanophyetus salmincola (Purcell et al., 2017), Gyrodactylus salaris (Rusch et al., 2018), Dactylogyrus spp. (Trujillo-González et al., 2019), Neobenedenia girellae (Agawa et al., 2016), Chilodonella hexasticha (Bastos Gomes et al., 2017), and Ichthyophthirius multifiliis (Howell et al., 2019). Furthermore, Shea et al. (2020) used a multiplex PCR assay to screen for a panel of salmon parasites in seawater. In the case of zebrafish parasites, an eDNA assay has recently been developed for the capillarid nematode Pseudocapillaria tomentosa. The assay for P. tomentosa was shown to effectively detect the pathogens in feces, detritus, and water samples (Norris et al., 2020). In contrast, P. neurophilia is not easily detectable in water with current assays (Sanders et al., 2013; Crim et al., 2017; Miller et al., 2019).
Detection by PCR is commonly used in eDNA methods, but there are some general limitations, particularly when the target species is present in low abundance, which may be compounded by the fact PCR is unable to distinguish between the life stages of the parasite (Kralik and Ricchi, 2017). Often, quantitative PCR (qPCR) is utilized in the case of environmental testing, but due to advancements in technologies, such as the implementation of digital PCR (dPCR), the sensitivity of diagnostic assays has dramatically improved in recent years (Koepfli et al., 2016). Regarding pathogen detection in environmental samples, dPCR can be more sensitive than qPCR (Yang et al., 2014; Wilson et al., 2015; Norris et al., 2020) due to its inherent resistance to inhibition and ability to provide more precise quantification without the need for a standard curve (Quan et al., 2018). Quantification of target DNA by dPCR is achieved by partitioning each sample into thousands of reactions, in such a fashion that each reaction is analyzed for the amplification product in an endpoint PCR and measured.
While molecular methods provide an avenue for detection, occupancy models enable the estimation of the proportion of an area occupied by an organism, when the target species is not detected with certainty (MacKenzie et al., 2004). Thus, these models are used to account for imperfect detection of organisms in surveys and to determine the probability of the true presence or absence of a species in a specific area (Colvin et al., 2015). This is often done by calculating the detection probability of a species in a specified area (or tank) based on quantifiable data such as surveillance surveys, histological surveys, or PCR data (Schmidt et al., 2013; Hunter et al., 2017).
Occupancy models have been implemented to evaluate the performance characteristics of several methods for detecting parasites in wildlife, including serological assays for Toxoplasma gondii in arctic-nesting geese (Elmore et al., 2014), to assess test sensitivity and prevalence estimates of Plasmodium spp. (Lachish et al., 2012) in wild blue tits (Cyanistes caeruleus), and to estimate detection probabilities of Schistosoma mansoni in water (Sengupta et al., 2019). Additionally, our group recently used an occupancy model to evaluate the accuracy of detecting salmon pathogens in histologic sections, including metacercariae of Apophallus spp., Nanophyetus salmincola, and the myxozoan (Parvicapsula minibicornis) (Colvin et al., 2015). These models provide a powerful tool to quantify detection uncertainty across disparate test systems, allowing for greater accuracy in prevalence estimates.
Here we describe the detection and quantification of P. neurophilia DNA in zebrafish tank water using dPCR. The limit of detection was established by spiking water samples with known concentrations of spores, while specificity was verified by testing against Pleistophora hyphessobryconis, the only other microsporidium currently recognized to infect zebrafish in a research setting (Sanders et al., 2010). We also evaluated assay performance using various sample types from aquaria holding P. neurophilia–infected zebrafish, while implementing a novel multi-state occupancy model to evaluate relationships between habitat, sampling method, distribution, abundance, and detection of parasites in the environment. This newly developed application provides the zebrafish community with a non-lethal disease surveillance tool that is specific for P. neurophilia using tank water and could complement current detection efforts.
MATERIALS AND METHODS
Test development and optimization
Microsporidian spore collection and purification: Pseudoloma neurophilia spores were obtained from the hind brain and anterior spinal cords as described by Sanders and Kent (2011). These tissues were mixed with 8 ml of deionized water (diH2O) and homogenized using successively smaller-gauged needles (18, 21, and 23 g). The homogenate was then allowed to sit in a refrigerator at 2 C for 48 hr while being rigorously vortexed every 24 hr. The homogenate was then centrifuged at max speed (1,600 g) for 25 min. Deionized water was used to hydrolyze host cells, as well as pre-sporogonic P. neurophilia stages. The spore suspension was then quantified using a hemocytometer and diluted in diH2O as needed to obtain the desired concentrations.
Assay design and optimization:
Taqman-based qPCR was performed using a forward and reverse primer and a probe specific to the ssuRNA gene of P. neurophilia, as previously described in Sanders and Kent (2011): Pn10F (5′ GTAATCGCGGGCTCACTAAG 3′), Pn10R (5′ GCTCGCTCAGCCAAATAAAC 3′), Pn10Probe (5′ 6-carboxyfluorescein (FAM)-ACACACCGCCCGTCGTTATCGAA 3′-Black Hole Quencher 1 (BHQ1)). Briefly, qPCR was performed in 20 μl reactions composed of 10 μM of forward and reverse primers, 10 μM of hydrolysis probe, 1× TaqMan Universal PCR Master Mix, and 2 μl of sample extract using the following reaction conditions: 50 C for 2 min, followed by 40 repetitions of 95 C for 15 sec and 60 C for 1 min using an Applied Biosystems 7500 Fast Real-Time PCR System and analyzed using the System Sequence Detection Software v1.4.1 (Applied Biosystems, Waltham, Massachusetts).
This assay was then adapted to the droplet digital (ddPCR) platform. ddPCR was performed using DNA extracted from P. neurophilia spores collected from zebrafish tank water using a commercial Qiagen DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) in combination with the forward and reverse P. neurophilia–specific primers. The reaction was composed of the following rations: 10 μl of supermix for probes—no dUTP (Bio-Rad), 1.8 μl of each primer, 0.5 μl probe (FAM), 1.9 μl water, and 4 μl extracted P. neurophilia DNA; total reaction volume = 20 μl. The reaction was run at the Oregon State University Center for Genome and Research and Biocomputing (CGRB) core facilities using a Bio-Rad Qx200 AutoDG Droplet Digital PCR system (Bio-Rad, Hercules, California) with the following conditions: 10 min at 95 C followed by 40 cycles of 30 sec at 94 C, 1 min at 60 C, followed by 98 C for 10 min and held at 4 C. Individual droplets were then classified as positive (fluorescence present) or negative (no fluorescence) for each reaction using the QX200 Droplet Reader. The number of copies of target DNA present per microliter in each reaction was determined using QuantaSoft Analysis Pro software (v1.0.596) (Biorad, 2016) by applying Poisson statistics to the ratio of positive droplets to total droplets (Hindson et al., 2013).
Specificity of the original qPCR assay for P. neurophilia was established in silico using the Primer-BLAST program (Sanders and Kent, 2011; Ye et al., 2012). This was further validated bioinformatically by performing BLAST searches across several common fish microsporidia, including Glugea anomala and Pl. hyphessobryconis. Specificity in the present study was further validated by testing cross-reaction with Pl. hyphessobryconis spores. Spores of Pl. hyphessobryconis were collected from zebrafish (casper strain) donated from a population with known Pl. hyphessobryconis infections and purified in diH2O and treated as described above to obtain an inoculum. Negative control system water was then inoculated with 10,000 and 50,000 spores/L concentrations, filtered, and evaluated with DNA extraction and subsequent ddPCR for absolute quantification as described below.
Optimizing the sonication protocol:
Earlier results indicated that sonication using a probe sonicator for 5 min at a voltage of 55 W and a frequency of 20 kHz was the most efficient mode of disrupting the spores to obtain quantifiable DNA from environmental samples (Sanders and Kent, 2011). To increase throughput and minimize the potential for cross-contamination, a sonicating water bath system, Bioruptor Pico (Diagenode, Denville, New Jersey), was used. To determine the optimal sonication time required to adequately disrupt spores of P. neurophilia, a time series was performed using water (negative control tank water) spiked with 8,500 spores/L and processed using the water filtration and DNA extraction protocol described above. The filters were dissolved, submerged in 100 μl PBS, and sonicated at 4, 5, 6, 7.5, 9, 10, and 12.5 min using a 30 sec on/off cycle to determine the optimal sonication time. Sonication for 9 min (18 cycles) using the Bioruptor Pico (Diagenode) resulted in minimal variability among PCR technical replicates with Ct values ranging from 35 to 37 (Suppl. Table S1a, b), ensuring consistent disruption of P. neurophilia spores in the water with minimal loss of signal. This sonication time was used for the processing of all subsequent samples.
Analytical sensitivity:
Incoming (parasite-free) supply water for the vivarium (dechlorinated city water) was inoculated with various spore concentrations for sensitivity testing. The limit of detection of the assay was determined using 2 separate 10-fold dilution series of P. neurophilia spores, each in 1 L of negative tank water. The first dilution series was generated using spores from 50 donor fish, resulting in a 105, 104, 103, and 102 spores/L series. The second dilution series was made from 40 fish, resulting in a 77.5 × 104, 7.75 × 103, 7.75 × 102, 7.75 × 101, and 7.75 spores/L series (Fig. 1; Table S2). Each sample was then filtered as described below. The filters were processed for DNA extraction and analyzed via ddPCR. A comparison of sensitivity between qPCR and ddPCR was also conducted using the first (105–102 spore/L) dilution series.
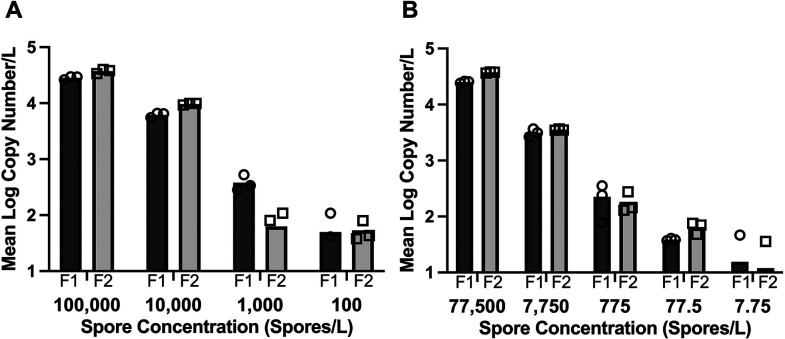
Figure 1.
Sensitivity of Pseudoloma neurophilia ddPCR by evaluation of inoculated filters in duplicate. Two separate experiments with starting concentrations of (A) 100,000 spores/L or (B) 77,500 spores/L. Each DNA assay was conducted in triplicate, with copy number reported for each PCR technical replicate at each concentration. Circles represent triplicate PCR reactions for filter 1 (F1), and squares represent the triplicate PCR reactions for filter 2 (F2).
Water filtration and DNA extraction:
Water samples for all experiments were processed as described in Norris et al. (2020). Briefly, 2 separate 1 L samples of water were collected from each tank and filtered through individual filters using a vacuum apparatus and a 0.45 μm nitrocellulose filter membrane (Nalgene, Rochester, New York). The filters were allowed to dry overnight in an open 15 ml conical tube. Once dry, 7 ml of acetone was added and vortexed until the filter was completely dissolved. The 15 ml conical tube was then centrifuged for 2 min at 3,000 g. The acetone was removed, and the pellet was resuspended in 1 ml of acetone. Five hundred microliters of the solution was then transferred to a 0.65 ml bioruptor tube and contents pelleted, which was completed twice to accommodate all 1 ml of acetone and set in a fume hood to dry. Once all of the acetone had evaporated, 100 μl of 1× PBS was added to the 0.65 ml bioruptor tube and was placed in a Bioruptor Pico, set at 4 C for 18 cycles (9 min) with 30 sec on and 30 sec off intervals. Once complete, DNA was extracted from the samples using the Qiagen Dneasy Blood and Tissue Kit (Qiagen) protocol for tissues and eluted in a total volume of 100 μl.
Evaluation of tanks with infected zebrafish
Tank assembly and population dynamics:
We further evaluated our assay by testing water from tanks with infected zebrafish. Five tanks of infected fish were tested at 3 monthly intervals with infected zebrafish held in 16 L tanks containing 30 adult fish/tank. The infected fish originated from 5 different populations housed at the Zebrafish International Resource Center, Eugene, Oregon, and were donated to us following the detection of P. neurophilia by their staff veterinarian, Dr. Katrina Murray. Populations varied in both sex composition and age. While they were separate populations, a few fish were mixed among some tanks so that the experiment would start with the same number of fish/tank. One tank of control (negative 5D line zebrafish fish) was included, originating from the Sinhubber Aquatic Resource Center (SARL), Oregon State University, which was established as a P. neurophilia–free facility in 2007 (Kent et al., 2011; Barton et al., 2016). The water system is a flow-through system with an inflow of 140 ml/min/tank. Water was maintained at 26–28 C, with conductivity maintained at around 115–130 micro-siemens. Fish were fed once daily with a 1:1 mix of Gemma 300 (Skretting, Westbrook, Maine) and Tetramin (Tetra, Melle, Germany). All live animal studies were conducted at Oregon State University, and all zebrafish researchers and platform staff were accredited as animal experimentation users according to the FELASA guidelines.
Flow/static/spawn water:
Experimental and control tanks were set up and water samples were taken during a flow-through period, a static period, and in a static group spawn setting with 3 monthly sample times. Before collecting water from each tank, tank water was mixed by using a sterile scrub brush, stirring tank bottom detritus, and brushing the tank internal wall surfaces. Two liters of tank water were then collected from each tank, filtered through a 0.45 μm filter, and placed in a −27 C freezer for future DNA extraction. Sampling methods were performed sequentially and applied to all tanks in the same order, with an 8 hr normalizing period in between treatment conditions. Flow sampling was measured first, since our facility is set up as a flow-through system, and sampled at 0900 hr. The flow was returned to the tanks and fish were fed, allowing tanks to normalize for 8 hr. At the end of the day (1700 hr) flow to each tank was then stopped and left static overnight. Two liters of tank water from the static tanks were then collected the following morning (0900 hr) and filtered. The flow was then resumed for the rest of the day (until 1700 hr).
At the end of the second day (1700 hr), group spawn tanks were then set up, and fish from each tank were moved from their regular housing tank into their designated 16 L spawning tanks with false bottoms and left static overnight to allow for a group spawning event the following morning. Two liters of water were then taken from each spawn tank and filtered through a 0.45 μm vacuum filter as described above. Each filter was saved back frozen at −27 C for later ddPCR analysis.
For each of the 2 water samples collected from a tank at a given time, ddPCR reactions were executed in triplicate (Table I). Copy number results for each reaction are provided in Table S3. A tank at a given time was scored as positive when either filter was positive. A filter was scored as positive when at least 2 of the 3 technical PCR replicates showed detection, regardless of the copy number.
Table I.
Summary of longitudinal evaluation of 5 tanks of zebrafish with Pseudoloma neurophilia infections. Each cell represents results, positive (+) or negative (−) following ddPCR testing of 2 (1 L) water samples taken from a tank with flowing water (flow), with water turned off for 8 hr (static) or water from a group spawn (spawn). DNA extracts from each filter sample were evaluated in triplicate. Number of positive replicates/tanks of the 6 replicates are in parentheses in each cell (No.). A positive score for a tank sample occurred when at least 1 of the 2 L samples showed detection of 2 or more replicates. Eggs from successful spawns were collected, counted, and equally divided into 10% pools and tested. Unless the spawns produced a small number of eggs, such as tank 1 in November and tank 2 in January, which has a total of 6 and 5 eggs (respectively), they were grouped into 1 pool. All egg samples were negative. Neural tissues from fish were evaluated by qPCR after the last water and spawn samples were collected.
| Tank No. |
Flow |
Static |
Spawn |
Total No. of eggs |
% Positive tank samples |
Tissue samples positive by QPCR (%) |
||||||||
| Nov |
Dec |
Jan |
Nov |
Dec |
Jan |
Nov |
Dec |
Jan |
Nov |
Dec |
Jan |
|||
| Tank 1 | − (1) | + (3) | − (0) | − (2) | + (4) | + (3) | + (3) | + (5) | + (5) | 6 | 0 | 0 | (67) | (54) |
| Tank 2 | − (1) | + (6) | + (3) | + (6) | + (6) | + (6) | + (4) | + (3) | + (5) | 159 | 83 | 5 | (89) | (83) |
| Tank 3 | − (1) | + (3) | − (1) | − (1) | + (5) | − (1) | + (4) | + (4) | + (4) | 0 | 0 | 90 | (56) | (42) |
| Tank 4 | + (3) | + (6) | − (2) | + (3) | + (5) | + (5) | − (2) | + (5) | − (1) | 0 | 30 | 360 | (67) | (68) |
| Tank 5 | + (3) | − (1) | − (1) | + (3) | + (6) | + (6) | − (0) | + (4) | + (5) | 0 | 80 | 72 | (67) | (63) |
| % positive by sampling method | (47) | (80) | (80) | |||||||||||
Tissue and egg qPCR:
Eggs from each tank were also collected from each spawn, counted, and divided into 10 pools/spawn, with a range of 3 to 58 eggs/pool, or if eggs count <10, the eggs were pooled in a single sample, as is the case of Tank 1 during the November timepoint (Table I). At the end of the experiment, each population was euthanized in an ice bath, and the anterior spinal cord and hindbrain tissue from each fish were extracted using fine forceps with the aid of a dissecting microscope. The tissues were then processed as outlined in Sanders and Kent (2011). Purified DNA was eluted in 100 μl of buffer AE, and extracted DNA from each fish was then quantified using the qPCR assay described above with the same running parameters.
Statistical analyses
Mixed effects ANOVA:
We initially fit a random effects analysis of variance (ANOVA) to partition variation in copy numbers among tanks, water samples within tanks, and technical PCR replicates within each water sample. We then evaluated differences in copy numbers between the 3 conditions (flow, static, and spawn) and sample month by adding covariates to the ANOVA. The baseline condition in the ANOVA was static, and the baseline month was December, so parameters should be interpreted relative to the baseline categories. Independence assumptions were evaluated by ordering residuals by tank. The models were fit using R package lme4 (Bates et al., 2015) implemented in R statistical software version 3.6.1 (R Core Team, 2019).
Multi-state occupancy modeling:
Preliminary results of the ANOVA indicated that the copy number counts were too variable among technical PCR replicates to be reliable indices. However, we believe that they would be useful for classifying counts as none (zero), present, and present and abundant. To evaluate trends and biases in parasite detection classes, we chose to use a multi-state occupancy model (MacKenzie et al., 2009). The multi-state occupancy model allows us to evaluate relationships between habitat, sampling method, distribution, abundance, and detection of parasites in the environment. Traditional occupancy models have focused on detection/non-detection data for detection probability estimation; However, in the multi-state occupancy model, detection/non-detection is estimated in consideration of 3 or more states. Thus, we estimated the probability of detecting P. neurophilia when it is present in a tank while accounting for treatment effects: flow, static, and spawn water and month. The underlying assumption in this model is that the population is closed, thus occupancy remains consistent during sampling. Occupancy states describe the true state, modeled with covariates using observations. The true state is a result of natural biological processes. Here, we considered 3 occupancy states: not detected, detected but not abundant, detected and abundant. Thereby, our model estimated the following parameters:
Ψ1i,j,k = Probability that the parasite is present regardless of abundance;
Ψ2i,j,k = Probability that the parasite is abundant given it is present;
p1i,j,k,s = Probability that the parasite is detected during sampling occasion, s, given that the true state is present, but not abundant;
p2i,j,k,s = Probability that the parasite is detected during sampling occasion, s, given that the true state is present and abundant; and
δi,j,k,s = Probability that evidence of the abundant state is detected during sampling occasion, s, given that true state is present and abundant,
where i, j, and k denote survey (water sample), tank, and sampling month, respectively. Because of the conditional nature of the probabilities, the probability that a tank contains a large number of parasites (the abundant state is present) is Ψ1 × Ψ2, and the probability of detecting the abundant state is p2 × δ. The abundant state was defined using the raw detection data for each survey from November to December. Based on recommendations from Peterson and Barajas (2018), we calculated the 80th percentile of the maximum copy number across tanks and through time and defined the abundant state as copy numbers that exceeded the 80th percentile (>123 copies/L).
To implement a multi-state occupancy model, we utilized the program Mark (White and Burnham, 1999) and Rmark (Laake, 2013) interface package in R statistical software, which uses the conditional binomial version of multi-state occupancy to estimate detection probabilities in relation to the 3 occupancy states. Before fitting the model, we created binary (0, 1) covariates representing: the flow and spawning treatment with static treatment as the baseline category; the samples collected in the months of November and January with December as the statistical baseline; and tanks 2 through 5 with tank 1 as the baseline. Because the populations of tanks 1–5 were positively identified to be infected, but prevalence was unknown, we fixed Ψ1 to a value of 1 (present) and the control tank to a value of zero (absent). We tested for differences between treatments, months, and tanks by fitting a global model containing all of the covariates for the detection parameters (p1, p2, δ) holding the remaining parameters constant. After model fitting, we retained the statistically significant covariates and fit the abundant occupancy parameter (Ψ2) using all the covariates. A parameter was deemed statistically significant when 95% confidence intervals did not contain zero (i.e., α = 0.05). Only statistically significant covariates for the abundant occupancy parameter were retained in the final model. The occupancy detection parameters were also used to estimate the number of samples required to accurately determine detection of P. neurophilia in a tank under each treatment, placing the counts from each filter collected from a given tank into 3 categories: not detected, detected but not abundant, and detected and abundant.
RESULTS
Analytical sensitivity and assay specificity
The limit of detection for the first dilution series (105–102 spores/L) and second dilution series (77.5 × 104–7.75 spores/L) was 100 and 77 spores/L, respectively, with variable results at 7.75 spores/L (Fig. 1). Exponential regression values generated from both dilution series showed similar trends for detectable copies/spore (Fig. 1). For the first dilution series, the exponential line of best fit is y = 0.4284 × ln (x) − 0.3442, and for the section dilution series, y = 0.4188 × ln(x) − 0.2781 (omitting negative reactions). Details of copy numbers at each dilution are reported in Table S2. A comparison of analytical sensitivity between qPCR and ddPCR showed that the latter had an approximately 1 log lower limit of detection (Table S4).
No DNA detection was observed following testing of filters spiked with a large number of Pl. hyphessobryconis spores (50,000 or 10,000 spores/L) using the same test parameters as used with P. neurophilia samples.
Evaluation of infected tanks
Consistency in copy number between duplicate water samples:
Copy number/L among duplicate water samples from the same tank were consistent (Table S3), although disparate results between the 2 (1 L) samples from a tank were observed (i.e., detection in one 1 L sample and no detection in the second 1 L sample) when at the lower limit of detection. Data generated from the water test were analyzed using a random effects ANOVA, which found that the majority of variance (88%) occurred between technical PCR replicates, while the variance between 1 L samples from the same tank was much less substantial (<1%). The addition of covariates to the ANOVA indicated that the number of copies was significantly lower under the flow and spawning treatments relative to the static treatments (F = 47.6, 299 df, P < 0.001 and F = 23.6, 299 df, P < 0.001, respectively). Copy numbers were also significantly lower in November (F = 11.5, 302 df, P < 0.001) compared to December and January.
Flow/static/spawn water:
Evaluation of tank water and spawn water by ddPCR at the 3 monthly time points showed a range of prevalence amongst the 5 tanks, with static and spawn water showing a higher number of positive detections (Table I). The 5 tanks containing infected fish showed a range of prevalence of 56–89%, regardless of sampling method or sample period. Evaluation of data by sample type (flow, static, or spawn) showed that 47% of flow samples were positive, while 80% of the static or spawn water samples yielded were positive (Table I). These results were consistent with those from the multi-state occupancy model as discussed below. Based on these results, testing static water twice with independent samples spaced a few days apart increases the sensitivity from 80 to 96%, as calculated using the false negative rate (1 − 0.80 = 0.20 false negative rate). Repeating the test yields 0.20 × 0.20 = 0.04 false negative rate. Hence, 1.0 − 0.04 = a sensitivity of 96% sensitivity when a second set of samples is taken a few days later. Furthermore, the positive predictive value (PPV), which is the ratio of samples truly diagnosed as positive to all samples which truly had positive test results, was high (Table II).
Table II.
Positive (PPV) and negative (NPV) predictive values for each sampling regime by month.
|
|
Month |
Test Result |
Diseased |
Not diseased |
Total |
PPV or NPV |
| FLOW | NOV | Positive | 2 | 0 | 2 | 1 |
| Negative | 3 | 1 | 4 | 0.25 | ||
| DEC | Positive | 4 | 0 | 4 | 1 | |
| Negative | 1 | 1 | 2 | 0.5 | ||
| JAN | Positive | 1 | 0 | 1 | 1 | |
| Negative | 4 | 1 | 5 | 0.2 | ||
| STATIC | NOV | Positive | 3 | 0 | 3 | 1 |
| Negative | 2 | 1 | 3 | 0.33 | ||
| DEC | Positive | 5 | 0 | 5 | 1 | |
| Negative | 0 | 1 | 1 | 1 | ||
| JAN | Positive | 4 | 0 | 4 | 1 | |
| Negative | 1 | 1 | 2 | 0.5 | ||
| SPAWN | NOV | Positive | 4 | 0 | 4 | 1 |
| Negative | 1 | 1 | 2 | 0.5 | ||
| DEC | Positive | 5 | 0 | 5 | 1 | |
| Negative | 0 | 1 | 1 | 1 | ||
| JAN | Positive | 4 | 0 | 4 | 1 | |
| Negative | 1 | 1 | 2 | 0.5 |
Multi-state occupancy model:
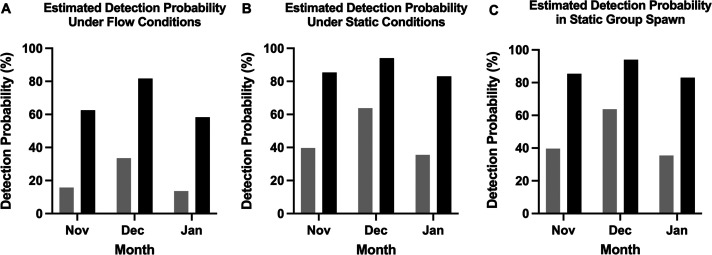
The model revealed that detection probabilities, regardless of abundance, were lower in flow samples (0.433) relative to static (0.668) and spawn samples (0.461). When taking abundance into account, detection probabilities for static and spawn increased to (0.85) and (0.90), respectively. Thus, detection probability estimates when the parasite is present but not abundant (Occupancy State 1), were collectively lower than when the parasite is present and abundant (Occupancy State 2). Also, when compared to static and spawn sampling methods, detection was predicted to be lowest when taking samples from tanks with a constant flow (Fig. 2). Overall, the detection estimates were much higher when the tanks were in an abundant state, however, the abundant state was less prevalent during the first month of the survey (November), compared to subsequent months (Table I). Detection was also lowest in November and January compared to samples collected in December. Furthermore, it was estimated that when the parasite is present, but not abundant, the required amount of water needed to detect the pathogen in static and spawn conditions was 3 and 5 L, respectively, while it would require 6 L under flow conditions.
Figure 2.
Estimated detection probabilities from the multi-state occupancy models by treatment and month for detection of the presence of Pseudoloma neurophilia. Probabilities are estimated for detecting the presence of the parasite when the true state is present but not abundant (gray) and present and abundant (black). The flow conditions are separated by panel: (A) flow conditions, (B) static conditions, (C) static group spawn conditions.
Fish prevalence compared to tank sample positivity:
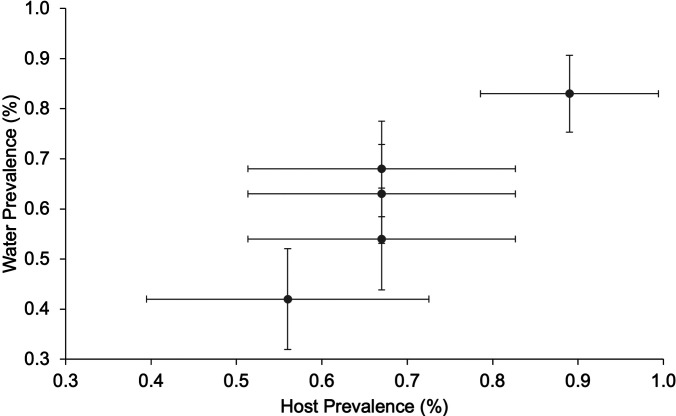
Prevalence based on PCR of host tissues at the end of the 3 mo experiment ranged from 42 to 83%, and the mean value correlated with the number of tanks deemed positive with our water test over the 3 monthly samples (r2 = 0.85) (Fig. 3). For example, in Tank 2, 89% of the tank samples were positive, and its fish showed 83% prevalence, whereas Tank 3 had 56% positivity with tank samples and only 42% of its fish were positive (Table I).
Figure 3.
Detection in water increases as prevalence in fish tissues increases. Horizontal error bars = standard error for prevalence amongst the population; Vertical error bars = standard error for water prevalence.
DISCUSSION
eDNA surveys of fish pathogens are particularly useful for fish kept in captivity, as they are often directed at the population level and defined by a population occupying the same space (e.g., tank). We developed a non-lethal assay that is specific and will complement current surveillance efforts being used in zebrafish facilities to diagnose disease. Effective biosecurity programs in modern zebrafish facilities require a combination of approaches, including daily health monitoring and utilization of quarantine rooms to segregate and screen incoming fish (Murray et al., 2016). The most common methods to survey for pathogens of zebrafish are lethal and require targeting of specific tissues. Additionally, these assays are usually directed on an individual fish basis, requiring sampling of a significant number of subjects to obtain a high confidence estimate of prevalence, abundance, and intensity (Kent et al., 2020a; Marques and Cabral, 2007). Ideally, an eDNA assay for P. neurophilia would be able to provide early detection of P. neurophilia in zebrafish populations at the tank or facility level, and it was a direct aim of this study to elucidate methods and conditions in which detection is strongest.
Advances in biotechnologies, specifically the development of ddPCR, have distinct advantages for environmental sampling compared to the qPCR method for screening water. Environmental samples often contain humic acid, fulvic acid, as well as debris, which have been shown to negatively impact the performance of DNA-based detection assays (Schrader et al., 2012; Guan et al., 2019). This inhibition can be variable as reflected in a study by Jane et al. (2015), where debris impacted high copy number samples, and in some cases, samples must be diluted to obtain positive results (Schrader et al., 2012; McKee et al., 2015). The effects of these inhibitory factors are mitigated using ddPCR, which divides the PCR into thousands of droplets and analyzes each droplet individually for the target marker. This minimizes the additive effects of inhibitory components in the sample and allows for greater precision when reporting on copy numbers, while also negating the need for a standard curve (Whale et al., 2012; Hayden et al., 2013).
Because the presence of environmental DNA is variable in a given system, the sensitivity of eDNA assays is very important, as target DNA in the environment will be dependent on many dynamics of the system (flow rates, stocking density, temperature, etc.) (Jane et al., 2015; Strickler et al., 2015; Troth et al., 2021). Recently, it was demonstrated that fly eDNA was only detectable using a ddPCR-based assay when comparing the qPCR and ddPCR assays used to detect Scarce Yellow Sally stonefly (Isogenus nubecula) eDNA in water samples (Mauvisseau et al., 2019). Particularly pertinent to the present study, our laboratory also found that a ddPCR test for the nematode P. tomentosa was much more sensitive than qPCR (Norris et al., 2020). Here we observed similar results for the detection of P. neurophilia in water (Table S4). Combined with pretreatment of samples by sonication, we found that the ddPCR test that we developed for P. neurophilia was sensitive and capable of consistently detecting down to 77.5 spores/L.
To our knowledge, this is the first implementation of a multi-state occupancy model using the zebrafish host to elucidate trends and biases in parasite detection. Occupancy modeling is typically used in wildlife ecology and has expanded into parasitology for the comparison of serological assays in the detection of important pathogens, estimating prevalence, and estimating the probability of detection of important parasites (Lachish et al., 2012; Elmore et al., 2014; Sengupta et al., 2019). Traditional occupancy models have focused solely on detection/non-detection data for detection probability estimations; however, in our multi-state occupancy model, probability estimates were determined using detection/non-detection data with consideration of 3 states. Thus, we were able to evaluate relationships that may affect the ability to effectively detect P. neurophilia in aquaria-holding populations of zebrafish (MacKenzie et al., 2009; Peterson and Barajas, 2018).
The occupancy model revealed that detection was lower when tanks were on a constant flow, relative to static and spawn, and that detection was much higher when parasites were predicted to be abundantly present (Fig. 2). In our study, we defined the abundant state as any detection that falls within the 80th percentile or higher, which corresponds to DNA concentrations greater than or equal to 123 copies/L. Detection was estimated to be lowest in November and January, relative to December, which highlights the variability of detection in the environment over time, as the microsporidian parasites replicate and move through their lifecycle. For this reason, static and December were set as baseline conditions in our model, as we wanted to evaluate the difference in detection under different conditions. Moreover, microsporidian spores are hydrophobic, which aids in their adherence to host cells (Hayman et al., 2005). This is likely the case with P. neurophilia, as we have observed that the spores often accumulate at the air/water interface of bubbles under the coverslip in wet mount preparation. Because of this phenomenon, in combination with the mixing of tank contents during sampling, spores are likely not evenly dispersed in the water column. Under flow conditions, it is less likely that spores settle and accumulate to the level they would under static conditions. This was certainly a dynamic contributing to the differences in the frequency of detection under static conditions relative to the flow.
Regarding the analysis of eDNA samples, PCR technical replicates for the same DNA extract frequently showed considerable variation. This phenomenon is common with PCR tests at the lower end of detection, and thus triplicate execution of the PCR was run for an accurate estimate of practical repeatability (Ahmed et al., 2009). The variability likely reflecting the variance of detection can be described by the uneven distribution of spores in the tank water, along with forms of parasite DNA (pre-sporgonic stages) being distributed differently between the 2 separate 1 L samples taken from each tank.
After optimizing the ddPCR assay to screen tank water, we evaluated 4 sample regimes on non-lethal samples from aquaria with infected zebrafish; water from tanks with flowing water (Flow), water from a tank in which the flow was stopped for 8 hr (Static), water from spawning events (Spawn), and eggs from the spawning events. Sensitivity of the assay with static or spawn water samples was similar and detected P. neurophilia DNA about 80% of the observed times. In contrast, flowing water samples were positive only 47% of the time (Table I). The results were similar to earlier studies using a qPCR-based assay (Sanders and Kent, 2011), where the parasite was not detected from tanks with flowing water but was detected in spawn water.
Pseudoloma neurophilia is commonly found in ovaries, and occasionally in the eggs; thus spores are likely regularly shed into the water during spawning events. In previous studies, P. neurophilia was detected in water from group spawning tanks, but detection significantly decreased in pair spawns with known infected females compared to when fish were spawned in groups (Sanders et al., 2013). In the present study, we consistently detected the parasite in tank water from group spawns, but only at the same rate as static water samples. Whereas P. neurophilia is capable of infecting developing eggs, our earlier study showed that the parasite was less frequently detected in embryos than spawn water from the same spawning event (Sanders et al., 2013). Likewise, while water from spawn tanks was frequently positive in the present study, none of the egg pools were positive. Evaluation of prevalence in populations at the end of the experiment revealed variable degrees of infection between the fish from different tanks, and the general trend was that tanks with the most infected populations exhibited the highest concentrations of the parasite in their respective tanks. The best example of this is tank 2, which had a prevalence of 83% in fish and had the most consistent water detection compared to the other tanks.
Previous studies have reported that environmental samples are not adequate for the detection of the microsporidium in zebrafish facilities (Crim et al., 2017; Miller et al., 2019). However, this conclusion was based on the sampling of flowing water from aquaria and a sample processing method that did not incorporate a sonication step to disrupt spores as previously recommended (Sanders and Kent, 2011). Sonication is a crucial step in the detection of spores of P. neurophilia, as sonicating adequately disrupts the resistant spores making DNA accessible, as seen with Ovipleistophora ovariae spores (Phelps, 2007). We also demonstrate that sonication improves detection for a better and more consistent result, particularly with tank water samples. We were limited to the use of a Diagenode Pico bioruptor, which had a volume limit of 100 μl. Despite the volumetric limitation, sonication was shown to effectively disrupt spores for environmental detection of P. neurophilia, resulting in the most consistent detection in water reported to date. Thus, sonication is a vital element that we recommend should be integrated at zebrafish facilities for accurate PCR diagnosis of P. neurophilia in tank water.
A tank on a given sample day was determined to be positive if at least 1 of the 2 samples of 1 L of water resulted in consistent detection of the parasite, based on 2 of the 3 technical PCR replicates resulting in detection from an individual 1 L extract. Although we only evaluated 5 tanks, there was a correlation between the prevalence of infection in fish at the end of the study and the number of positive water samples over the 3 mo period, and this was also supported by a higher rate of detection as determined by the multi-state occupancy model. Positivity tended to fluctuate throughout the study, probably reflecting an increase of prevalence in fish over time (Ramsay et al., 2009). In the current study, the true prevalence before the start of the study was not known; however, the populations were positively identified as infected by microscopy. Future studies coupling current diagnostic methods (histology, PCR on fish tissues, etc.) and this non-lethal water assay would be useful for a longitudinal study to further characterize the progression of infection and transmission dynamics. Thus, targeting the early onset of infection would elucidate trends regarding the distribution of the parasite in an aquarium setting following initial infection and provide insights on the frequency of parasite concentration in the environment through the duration of infection. We are now using this test in a collaboration with the ZIRC, a large zebrafish facility with a history of P. neurophilia. With their staff veterinarian (Dr. K. Murray), we are moving forward with screening various populations of fish from their main facility and applying the static water approach to test water from fish following transport.
The occurrence of false negatives, in which a water sample was negative from a tank that contained infected fish, demonstrated that the sampling from flowing aquaria reduces the sensitivity of the test. Nevertheless, our eDNA test is still very useful for the non-lethal detection of the parasite in its present format with the following recommended applications. Static or spawn water had predicted an occupancy that was always greater than 82% when the parasite was abundant, which was consistently better than predictions under flowing water. Also, static and spawn water detection probability estimates were similar, but the latter would require more effort, as it would require setting up spawning tanks/chambers. We, therefore, recommend leaving tanks static for 8 hr with aeration before taking a water sample for screening, filtering 1 L water samples following the described extraction protocol.
The positive predictive value of this test is high, as this assay has been proven to be very specific for the microsporidium, thus indicating that a positive detection is likely correct (Table II). However, if the test is negative or inconclusive (failed PCR replicates), we recommend tagging the tank as suspect, waiting for 24–48 hr, and testing the tank again, as subsequent sampling will increase the probability of detection. This approach has commonly been used with other pathogens, in which the test is not optimally sensitive, but the populations to be tested are defined and can be easily retested. In such cases, serial testing has resulted in a nearly 10% increase in positivity (van Prehn et al., 2015; Larremore et al., 2021). eDNA survey sensitivity has also been demonstrated to be improved by simply increasing sample volume, sample number, and PCR replicates (Schultz and Lance, 2015). The occupancy model also supports this idea, as it was calculated that increasing the number of water samples when abundance is unknown, from 3 to 5 L would be required for detection in static and spawn conditions, whereas at least 6 L are required for detection in flow conditions.
Another strategy is to target water from older populations of zebrafish, as the infection naturally spreads within a population, resulting in a very high prevalence as fish approach 1 yr of age (Ramsay et al., 2009; Murray et al., 2011). Importantly, when our assay is used with static water, it falls within the new recommendations for assay sensitivity (>85%), according to the USDA NAHPS (USDA APHIS, 2021).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the Zebrafish International Resource Center (ZIRC) for providing us with a consistent supply of infected subject animals for investigation. We thank Christopher Lawrence, Children's Hospital Boston, Aquatic Resources Program for providing Pleistophora hyphessobryconis. We thank the Sinnhuber Aquatic Resource Center at Oregon State University for providing pathogen-free fish. The Oregon Cooperative Fish and Wildlife Research Unit is jointly sponsored by the U.S. Geological Survey, the U.S. Fish and Wildlife Service, the Oregon Department of Fish and Wildlife, Oregon State University, and the Wildlife Management Institute. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. government. All work was completed at Oregon State University, a land grant university, located on the traditional homelands of the Kalapuya people, who are now absorbed into the Grande Ronde and Siletz Nations.
Funding Statement
This study was supported by the National Institutes of Health under the Research Supplements to promote Diversity in health-related research (NIH ORIP 3 R24 OD010998).
LITERATURE CITED
- Agawa Y, Tani K, Yamamoto S, Hirano C, Shirakashi S. Development of a quantitative polymerase chain reaction assay for the detection of skin fluke Neobenedenia girellae larvae from environmental water. Fisheries Science. 2016;82:827–833. [Google Scholar]
- Ahmed A, Engelberts M. F. M, Boer K. R, Ahmed N, Hartskeerl R. A. Development and validation of a real-time PCR for detection of pathogenic Leptospira species in clinical materials. PLOS One. 2009;4:e7093. doi: 10.1371/journal.pone.0007093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazán C, Avila G, Quiroz H, Ibarra F, Ochoa P. Effect of parasite burden on the detection of Fasciola hepatica antigens in sera and feces of experimentally infected sheep. Veterinary Parasitology. 2001;97:101–112. doi: 10.1016/s0304-4017(01)00376-4. [DOI] [PubMed] [Google Scholar]
- Amarasiri M, Furukawa T, Nakajima F, Sei K. Pathogens and disease vectors/hosts monitoring in aquatic environments: Potential of using eDNA/eRNA based approach. Science of the Total Environment. 2021;796:148810. doi: 10.1016/j.scitotenv.2021.148810. [DOI] [PubMed] [Google Scholar]
- Avdesh A, Chen M, Martin-Iverson M. T, Mondal A, Ong D, Rainey-Smith S, Taddei K, Lardelli M, Groth D. M, Verdile G, et al. Regular care and maintenance of a zebrafish (Danio rerio) laboratory: An introduction. Journal of Visualized Experiments JoVE. 2012;69:e4196–e4196. doi: 10.3791/4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes M. A, Turner C. R, Jerde C. L, Renshaw M. A, Chadderton W. L, Lodge D. M. Environmental conditions influence eDNA persistence in aquatic systems. Environmental Science & Technology. 2014;48:1819–1827. doi: 10.1021/es404734p. [DOI] [PubMed] [Google Scholar]
- Barton C. L, Johnson E. W, Tanguay R. L. Facility design and health management program at the Sinnhuber Aquatic Research Laboratory. Zebrafish 13 S39–S43. 2016. [DOI] [PMC free article] [PubMed]
- Bass D, Stentiford G. D, Littlewood D. T. J, Hartikainen H. Diverse applications of environmental DNA methods in parasitology. Trends in Parasitology. 2015;31:499–513. doi: 10.1016/j.pt.2015.06.013. [DOI] [PubMed] [Google Scholar]
- Bastos Gomes G, Jerry D. R, Miller T. L, Hutson K. S. Current status of parasitic ciliates Chilodonella spp. (Phyllopharyngea: Chilodonellidae) in freshwater fish aquaculture. Journal of Fish Diseases. 2017;40:703–715. doi: 10.1111/jfd.12523. [DOI] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. Journal of Statistical Software. 2015;67:1–48. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- Berger C. S, Aubin-Horth N. An eDNA-qPCR assay to detect the presence of the parasite Schistocephalus solidus inside its threespine stickleback host. Journal of Experimental Biology 221 jeb178137. 2018. [DOI] [PubMed]
- Biorad. QuantaSoft Analysis Pro software (v1.0.596) 2016. Available at: www.biorad.com Accessed 12 January 2020.
- Colvin M. E, Peterson J. T, Kent M. L, Schreck C. B. Occupancy modeling for improved accuracy and understanding of pathogen prevalence and dynamics. PLOS One. 2015;10:e0116605. doi: 10.1371/journal.pone.0116605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crim M. J, Lawrence C, Livingston R. S, Rakitin A, Hurley S. J, Riley L. K. Comparison of antemortem and environmental samples for zebrafish health monitoring and quarantine. Journal of the American Association for Laboratory Animal Science JAALAS. 2017;56:412–424. [PMC free article] [PubMed] [Google Scholar]
- Elmore S. A, Huyvaert K. P, Bailey L. L, Milhous J, Alisauskas R. T, Gajadhar A. A, Jenkins E. J. Toxoplasma gondii exposure in arctic-nesting geese: A multi-state occupancy framework and comparison of serological assays. International Journal for Parasitology Parasites and Wildlife. 2014;3:147–153. doi: 10.1016/j.ijppaw.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson J. A, Watral V, Schwindt A. R, Kent M. L. Spores of two fish microsporidia (Pseudoloma neurophilia and Glugea anomala) are highly resistant to chlorine. Diseases of Aquatic Organisms. 2007;76:205–214. doi: 10.3354/dao076205. [DOI] [PubMed] [Google Scholar]
- Guan X, Monroe E. M, Bockrath K. D, Mize E. L, Rees C. B, Lindsay D. L, Baerwaldt K. L, Nico L, Lance R. F. Environmental DNA (eDNA) assays for invasive populations of black carp in North America. Transactions of the American Fisheries Society. 2019;148:1043–1055. [Google Scholar]
- Guy R. A, Payment P, Krull U. J, Horgen P. A. Real-time PCR for quantification of Giardia and Cryptosporidium in environmental water samples and sewage. Applied and Environmental Microbiology. 2003;69:5178–5185. doi: 10.1128/AEM.69.9.5178-5185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallett S. L, Ray R. A, Hurst C. N, Holt R. A, Buckles G. R, Atkinson S. D, Bartholomew J. L. Density of the waterborne parasite Ceratomyxa shasta and its biological effects on salmon. Applied and Environmental Microbiology. 2012;78:3724–3731. doi: 10.1128/AEM.07801-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden R. T, Gu Z, Ingersoll J, Abdul-Ali D, Shi L, Pounds S, Caliendo A. M. Comparison of droplet digital PCR to real-time PCR for quantitative detection of cytomegalovirus. Journal of Clinical Microbiology. 2013;51:540–546. doi: 10.1128/JCM.02620-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman J. R, Southern T. R, Nash T. E. Role of sulfated glycans in adherence of the microsporidian Encephalitozoon intestinalis to host cells in vitro. Infection and Immunity. 2005;73:841–848. doi: 10.1128/IAI.73.2.841-848.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson C. M, Chevillet J. R, Briggs H. A, Gallichotte E. N, Ruf I. K, Hindson B. J, Vessella R. L, Tewari M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nature Methods. 2013;10:1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell C. K, Atkinson S. D, Bartholomew J. L, Hallett S. L. Development and application of a qPCR assay targeting Ichthyophthirius multifiliis in environmental water samples. Diseases of Aquatic Organisms. 2019;134:43–55. doi: 10.3354/dao03351. [DOI] [PubMed] [Google Scholar]
- Hunter M. E, Dorazio R. M, Butterfield J. S. S, Meigs-Friend G, Nico L. G, Ferrant J. A. Detection limits of quantitative and digital PCR assays and their influence in presence–absence surveys of environmental DNA. Molecular Ecology Resources. 2017;17:221–229. doi: 10.1111/1755-0998.12619. [DOI] [PubMed] [Google Scholar]
- Jane S. F, Wilcox T. M, McKelvey K. S, Young M. K, Schwartz M. K, Lowe W. H, Letcher B. H, Whiteley A. R. Distance, flow and PCR inhibition: eDNA dynamics in two headwater streams. Molecular Ecology Resources. 2015;15:216–227. doi: 10.1111/1755-0998.12285. [DOI] [PubMed] [Google Scholar]
- Kent M. L, Buchner C, Watral V. G, Sanders J. L, Ladu J, Peterson T. S, Tanguay R. L. Development and maintenance of a specific pathogen-free (SPF) zebrafish research facility for Pseudoloma neurophilia. Diseases of Aquatic Organisms. 2011;95:73–79. doi: 10.3354/dao02333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent M. L, Harper C, Wolf J. C. Documented and potential research impacts of subclinical diseases in zebrafish. ILAR Journal. 2012;53:126–134. doi: 10.1093/ilar.53.2.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent M. L, Murray K. N, Fisher K, Löhr C, Mulrooney D, Sanders J. L. Special procedures for zebrafish diagnostics. In: Cartner S. C, Eisen J. S, Farmer S. C, Guillemin K. J, Kent M. L, Sanders G. E, editors. The Zebrafish in Biomedical Research Biology Husbandry Diseases and Research Applications. American College of Laboratory Animal Medicine Series; Elsevier, Amsterdam: 2020. pp. 547–558. a. (eds.) p. [Google Scholar]
- Kent M. L, Sanders J. L, Spagnoli S, Al-Samarrie C. E, Murray K. N. Review of diseases and health management in zebrafish Danio rerio (Hamilton 1822) in research facilities. Journal of Fish Diseases. 2020;43:637–650. doi: 10.1111/jfd.13165. b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepfli C, Nguitragool W, Hofmann N. E, Robinson L. J, Ome-Kaius M, Sattabongkot J, Felger I, Mueller I. Sensitive and accurate quantification of human malaria parasites using droplet digital PCR (ddPCR) Scientific Reports. 2016;6:39183. doi: 10.1038/srep39183. https://doi.org/https://doi.org/10.1038/srep39183 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralik P, Ricchi M. A basic guide to real time PCR in microbial diagnostics: Definitions, parameters, and everything. Frontiers in Microbiology. 2017;8:108. doi: 10.3389/fmicb.2017.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laake J. L. An R interface for analysis of capture-recapture data with MARK. 2013. Available at: http://www.afsc.noaa.gov/Publications/ProcRpt/PR2013-01.pdf Accessed 26 June 2021.
- Lachish S, Gopalaswamy A. M, Knowles S. C. L, Sheldon B. C. Site-occupancy modelling as a novel framework for assessing test sensitivity and estimating wildlife disease prevalence from imperfect diagnostic tests. Methods in Ecology and Evolution. 2012;3:339–348. [Google Scholar]
- Larremore D. B, Wilder B, Lester E, Shehata S, Burke J. M, Hay J. A, Tambe M, Mina M. J, Parker R. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Science Advances 7 eabd5393. 2021. [DOI] [PMC free article] [PubMed]
- MacKenzie D. I, Nichols J. D, Seamans M. E, Gutiérrez R. J. Modeling species occurrence dynamics with multiple states and imperfect detection. Ecology. 2009;90:823–835. doi: 10.1890/08-0141.1. [DOI] [PubMed] [Google Scholar]
- MacKenzie D. I, Royle J. A, Brown J. A, Nichols J. D. Occupancy estimation and modeling for rare and elusive populations. In: Thompson W. L, editor. Sampling Rare or Elusive Species Concepts Designs and Techniques for Estimating Population Parameters. Island Press; Washington, D.C: 2004. pp. 149–172. (ed.) p. [Google Scholar]
- Mandarino-Pereira A, de Souza F. S, Lopes C. W. G, Pereira M. J. S. Prevalence of parasites in soil and dog feces according to diagnostic tests. Veterinary Parasitology. 2010;170:176–181. doi: 10.1016/j.vetpar.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Marques J. F, Cabral H. N. Effects of sample size on fish parasite prevalence, mean abundance, and mean intensity estimates. Journal of Applied Ichthyology. 2007;23:158–162. [Google Scholar]
- Mauvisseau Q, Davy-Bowker J, Bulling M, Brys R, Neyrinck S, Troth C, Sweet M. Combining ddPCR and environmental DNA to improve detection capabilities of a critically endangered freshwater invertebrate. Scientific Reports. 2019;9:14064. doi: 10.1038/s41598-019-50571-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee A. M, Spear S. F, Pierson T.W. The effect of dilution and the use of a post-extraction nucleic acid purification column on the accuracy, precision, and inhibition of environmental DNA samples. Biological Conservation. 2015;183:70–76. [Google Scholar]
- Midttun H. L. E, Vindas M. A, Whatmore P. J, Overli O, Johansen I. B. Effects of Pseudoloma neurophilia infection on the brain transcriptome in zebrafish (Danio rerio) Journal of Fish Disease. 2020;43:863–875. doi: 10.1111/jfd.13198. [DOI] [PubMed] [Google Scholar]
- Miller M, Sabrautzki S, Beyerlein A, Brielmeier M. Combining fish and environmental PCR for diagnostics of diseased laboratory zebrafish in recirculating systems. PloS One. 2019;14:e0222360. doi: 10.1371/journal.pone.0222360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocho J. P, Martin D. J, Millington M. E, Torres Y. Saavedra. Environmental screening of Aeromonas hydrophila Mycobacterium spp., and Pseudocapillaria tomentosa in zebrafish systems. Journal of Visualized Experiments. 2017;130:55306. doi: 10.3791/55306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K. N, Dreska M, Nasiadka A, Rinne M, Matthews J. L, Carmichael C, Bauer J, Varga Z, Westerfield M. Transmission, diagnosis, and recommendations for control of Pseudoloma neurophilia infections in laboratory zebrafish (Danio rerio) facilities. Comparative Medicine. 2011;61:322–329. [PMC free article] [PubMed] [Google Scholar]
- Murray K. N, Varga Z. M, Kent M. L. Biosecurity and health monitoring at the Zebrafish International Resource Center. Zebrafish. 2016;13(Suppl. 1):S30–S38. doi: 10.1089/zeb.2015.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamori Y, Payton M. E, Duncan-Decocq R, Johnson E. M. Fecal survey of parasites in free-roaming cats in northcentral Oklahoma, United States. Veterinary Parasitology Regional Studies and Reports. 2018;14:50–53. doi: 10.1016/j.vprsr.2018.08.008. [DOI] [PubMed] [Google Scholar]
- Norris L, Lawler N, Hunkapiller A, Mulrooney D. M, Kent M. L, Sanders J. L. Detection of the parasitic nematode, Pseudocapillaria tomentosa in zebrafish tissues and environmental DNA in research aquaria. Journal of Fish Diseases. 2020;43:1087–1095. doi: 10.1111/jfd.13220. [DOI] [PubMed] [Google Scholar]
- Peters L, Spatharis S, Dario M. A, Dwyer T, Roca I. J. T, Kintner A, Kanstad-Hanssen O, Llewellyn M. S, Praebel K. Environmental DNA: A new low-cost monitoring tool for pathogens in salmonid aquaculture. Frontiers in Microbiology. 2018;9:3009. doi: 10.3389/fmicb.2018.03009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J. T, Barajas M. F. An evaluation of three fish surveys in the San Francisco Estuary, California, 1995–2015. San Francisco Estuary and Watershed Science. 2018;16:1995–2015. doi: 10.15447/sfews.2018v16iss4art2. [DOI] [Google Scholar]
- Phelps N. B. D. Validation of a quantitative PCR diagnostic method for detection of the microsporidian Ovipleistophora ovariae in the cyprinid fish Notemigonus crysoleucas. Diseases of Aquatic Organisms. 2007;76:215–221. doi: 10.3354/dao076215. [DOI] [PubMed] [Google Scholar]
- Purcell M. K, Powers R. L, Besijn B. L, Hershberger P. K. Detection of Nanophyetus salmincola in water, snails, and fish tissues by quantitative polymerase chain reaction. Journal of Aquatic Animal Health. 2017;29:189–198. doi: 10.1080/08997659.2017.1365780. [DOI] [PubMed] [Google Scholar]
- Quan P. L, Sauzade M, Brouzes E. dPCR: A technology review. Sensors. 2018;18:1271. doi: 10.3390/s18041271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2019. Available at: https://www.R-project.org Accessed 6 June 2020. [Google Scholar]
- Ramsay J. M, Watral V, Schreck C. B, Kent M. L. Pseudoloma neurophilia infections in zebrafish Danio rerio Effects of stress on survival, growth, and reproduction. Diseases of Aquatic Organisms. 2009;88:69–84. doi: 10.3354/dao02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees H. C, Maddison B. C, Middleditch D. J, Patmore J. R. M, Gough K. C. The detection of aquatic animal species using environmental DNA—A review of eDNA as a survey tool in ecology. Journal of Applied Ecology. 2014;51:1450–1459. [Google Scholar]
- Rusch J. C, Hansen H, Strand D. A, Markussen T, Hytterød S, Vrålstad T. Catching the fish with the worm: A case study on eDNA detection of the monogenean parasite Gyrodactylus salaris and two of its hosts, Atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss) Parasites & Vectors. 2018;11:333. doi: 10.1186/s13071-018-2916-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders J. L, Kent M. L. Development of a sensitive assay for the detection of Pseudoloma neurophilia in laboratory populations of the zebrafish Danio rerio. Diseases of Aquatic Organisms. 2011;96:145–156. doi: 10.3354/dao02375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders J. L, Lawrence C, Nichols D. K, Brubaker J. F, Peterson T. S, Murray K. N, Kent M. L. Pleistophora hyphessobryconis (Microsporidia) infecting zebrafish Danio rerio in research facilities. Diseases of Aquatic Organisms. 2010;91:47–56. doi: 10.3354/dao02245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders J. L, Peterson T. S, Kent M. L. Early development and tissue distribution of Pseudoloma neurophilia in the zebrafish, Danio rerio. Journal of Eukaryotic Microbiology. 2014;61:238–246. doi: 10.1111/jeu.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders J. L, Watral V, Clarkson K, Kent M. L. Verification of intraovum transmission of a microsporidium of vertebrates: Pseudoloma neurophilia infecting the Zebrafish, Danio rerio. PloS One. 2013;8:e76064. doi: 10.1371/journal.pone.0076064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders J. L, Watral V, Stidworthy M. F, Kent M. L. Expansion of the known host range of the microsporidium, Pseudoloma neurophilia. Zebrafish. 2016;13:S102–S106. doi: 10.1089/zeb.2015.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt B. R, Kéry M, Ursenbacher S, Hyman O. J, Collins J. P. Site occupancy models in the analysis of environmental DNA presence/absence surveys: A case study of an emerging amphibian pathogen. Methods in Ecology and Evolution. 2013;4:646–653. [Google Scholar]
- Schrader C, Schielke A, Ellerbroek L, Johne R. PCR inhibitors—Occurrence, properties, and removal. Journal of Applied Microbiology. 2012;11:1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- Schultz M. T, Lance R. F. Modeling the sensitivity of field surveys for detection of Environmental DNA (eDNA) PloS One. 2015;10:e0141503. doi: 10.1371/journal.pone.0141503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta M. E, Hellström M, Kariuki H. C, Olsen A, Thomsen P. F, Mejer H, Willerslev E, Mwanje M, Madsen H, Kristensen T. K, et al. Environmental DNA for improved detection and environmental surveillance of schistosomiasis. Proceedings of the National Academy of Sciences. 2019;116:8931–8940. doi: 10.1073/pnas.1815046116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea D, Bateman A, Li S, Tabata A, Schulze A, Mordecai G, Ogston L, Volpe J. P, Frazer N, Connors B, et al. Environmental DNA from multiple pathogens is elevated near active Atlantic salmon farms. Proceedings of the Royal Society B Biological Sciences. 2020;287:20202010. doi: 10.1098/rspb.2020.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber N, Hartikainen H, Vorburger C. Validation of an eDNA-based method for the detection of wildlife pathogens in water. Diseases of Aquatic Organisms. 2020;141:171–184. doi: 10.3354/dao03524. [DOI] [PubMed] [Google Scholar]
- Spagnoli S. T, Xue L, Murray K. N, Chow F, Kent M. L. Pseudoloma neurophilia A retrospective and descriptive study of nervous system and muscle infections, with new implications for pathogenesis and behavioral phenotypes. Zebrafish. 2015;12:189–201. doi: 10.1089/zeb.2014.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickler K. M, Fremier A. K, Goldberg C. S. Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biological Conservation. 2015;183:85–92. [Google Scholar]
- Troth C. R, Sweet M. J, Nightingale J, Burian A. Seasonality, DNA degradation and spatial heterogeneity as drivers of eDNA detection dynamics. Science of the Total Environment. 2021;768:144466. doi: 10.1016/j.scitotenv.2020.144466. [DOI] [PubMed] [Google Scholar]
- Trujillo-González A, Edmunds R. C, Becker J. A, Hutson K. S. Parasite detection in the ornamental fish trade using environmental DNA. Scientific Reports. 2019;9:5173. doi: 10.1038/s41598-019-41517-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA APHIS (U.S. Department of Agriculture: Animal and Plant Health Inspection Service) National Aquaculture Health Plan & Standards 2021–2023. U.S. Department of Agriculture; Animal and Plant Health Inspection Service, Riverdale, Maryland: 2021. 37 p [Google Scholar]
- van Prehn J, Vandenbroucke-Grauls C. M. J. E, van Beurden Y. H, van Houdt R, Vainio S, Ang C. W. Diagnostic yield of repeat sampling with immunoassay, real-time PCR, and toxigenic culture for the detection of toxigenic Clostridium difficile in an epidemic and a non-epidemic setting. European Journal of Clinical Microbiology & Infectious Diseases. 2015;34:2325–2330. doi: 10.1007/s10096-015-2484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whale A. S, Huggett J. F, Cowen S, Speirs V, Shaw J, Ellison S, Foy C, Scott D. J. Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation. Nucleic Acids Research. 2012;40:e82. doi: 10.1093/nar/gks203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White G. C, Burnham K. P. Program MARK: Survival estimation from populations of marked animals. Bird Study. 1999;46(Suppl. 1):S120–S139. doi: 10.1080/00063659909477239. [DOI] [Google Scholar]
- Wilson M, Glaser K. C, Adams-Fish D, Boley M, Mayda M, Molestina R. E. Development of droplet digital PCR for the detection of Babesia microti and Babesia duncani. Experimental Parasitology. 2015;149:24–31. doi: 10.1016/j.exppara.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Paparini A, Monis P, Ryan U. Comparison of next-generation droplet digital PCR (ddPCR) with quantitative PCR (qPCR) for enumeration of Cryptosporidium oocysts in faecal samples. International Journal for Parasitology. 2014;44:1105–1113. doi: 10.1016/j.ijpara.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden T. L. Primer-Blast: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.