Abstract
Bats are increasingly studied as model systems for longevity and as natural hosts for some virulent viruses. Yet the ability to characterize immune mechanisms of viral tolerance and to quantify infection dynamics in wild bats is often limited by small sample volumes and few species-specific reagents. Here, we demonstrate how proteomics can overcome these limitations by using data-independent acquisition-based shotgun proteomics to survey the serum proteome of 17 vampire bats (Desmodus rotundus) from Belize. Using just 2 μL of sample and relatively short separations of undepleted serum digests, we identified 361 proteins across 5 orders of magnitude. Levels of immunological proteins in vampire bat serum were then compared to human plasma via published databases. Of particular interest were antiviral and antibacterial components, circulating 20S proteasome complex and proteins involved in redox activity. Lastly, we used known virus proteomes to putatively identify Rh186 from Macacine herpesvirus 3 and ORF1a from Middle East respiratory syndrome-related coronavirus, indicating that mass spectrometry-based techniques show promise for pathogen detection. Overall, these results can be used to design targeted mass-spectrometry assays to quantify immunological markers and detect pathogens. More broadly, our findings also highlight the application of proteomics in advancing wildlife immunology and pathogen surveillance.
Keywords: vampire bat, serum, data-independent acquisition, shotgun proteomics
Graphical Abstract
INTRODUCTION
Bats are a hyper-diverse and geographically widespread order (Chiroptera), accounting for over 20% of all mammal species.1 Owing to their capacity for flight and wide range of trophic habits (e.g., frugivory, nectarivory, insectivory, carnivory, sanguivory), bats provide critical ecosystem services that include seed dispersal, pollination, and predation of insects.2 Unique features of bats among mammals, such as the capacity for powered flight and long lifespans despite small body sizes, also make this order interesting for basic research in ecology and evolution.3,4 Recently, bats have also become model systems for studies of the microbiome and sociality.5,6
However, because some bats are common in anthropogenic landscapes, they are increasingly studied for their ability to harbor pathogens with high virulence in humans and domestic animals.7,8 In particular, bats carry more zoonotic viruses than most other mammalian orders,9 and they are the confirmed reservoir hosts for Hendra virus, Nipah virus, Marburg virus, an array of lyssaviruses (e.g., rabies virus), and SARS-like coronaviruses. 10–13 Spillovers of these sometimes lethal viruses, from bats to humans, are often driven by ecological changes that alter infectious disease dynamics in wild bat populations and change interactions between bats and recipient hosts.14,15 However, with some exceptions (e.g., lyssaviruses), these viruses may not typically cause clinical disease in bats.16 Although the high richness of zoonotic viruses in bats may simply be a function of their vast species diversity,17 tolerance to virulent viruses is likely driven by distinct aspects of immunity in these flying mammals.18 These include but are not limited to robust complement, constitutive expression of type I interferons (e.g., IFN-α), and high combinatorial diversity in immunoglobulin genes.4,19–22 Accordingly, contemporary studies of zoonotic pathogens in bats have focused on characterizing immunological factors that facilitate tolerance or shedding of viruses, quantifying viral diversity, and identifying spatial and temporal pulses of infection. 23–25
Traditionally, efforts to assess the immunological state of wild bats and detect viruses have relied on relatively simple tools. Most bat species are sufficiently small that only modest volumes of blood can be safely collected under nonlethal procedures;26 74% of bat species weigh less than 30 g as adults.27 Small blood volumes and lack of species-specific reagents for bats generally restrict the scope of possible immune assays.28,29 Remote field sites also pose significant challenges for sample storage and transport at ideal temperatures. Despite these obstacles, techniques such as in vitro microbial killing assays, enzyme-linked immunosorbent assays, and in vivo antigen challenge have helped to broadly characterize complement, antibody, and cellular immune response in wild bats and how these phenotypes vary with life history (e.g., reproduction) and environmental conditions.30–32 Similarly, polymerase chain reaction (PCR) has formed the primary basis of virus detection.33 However, because bat colonies can be large (e.g., hundreds to over 20 million individuals34) and prevalence of active viral infection is generally low, serology and the detection of virus-specific antibodies has also commonly been employed.35 Although serological surveys have been important for characterizing virus circulation in bats, antibody cross-reactivity and inconsistent cutoff thresholds can limit their interpretability.36
Given these restrictions, studies of bat immunology and virology have increasingly benefitted from modern bioanalytical approaches. Researchers employing global profiling techniques, such as RNA-Seq-based transcriptomics, have begun to contribute new resources for bat immunology and to illuminate the broad immune response of bats to viral and other infections.22,37–39 Similarly, metagenomic approaches have helped reduce bias in broad characterization of bat viral diversity.40,41 In addition to these approaches, proteomics can provide a complementary modality to define the molecular landscape. Proteomics is uniquely useful when applied to blood, because the blood proteome is mostly secreted from organs such as the liver,42 thereby permitting interrogation of the circulating protein phenotype beyond cellular transcript profiling. Using the relatively small sample volumes that typify bat field studies (e.g., <10 μL), proteomic analysis of blood can identify and relatively quantify hundreds of proteins, including but not limited to those involved in host response to infection and other useful immunological biomarkers.43 Prior applications of serum proteomics in bats have described proteins involved in shifts in innate immunity and blood coagulation between hibernating and active greater mouse-eared bats (Myotis myotis)44 as well as between North American and European Myotis species that vary in infection with Pseudogymnoascus destructans, which causes white-nose syndrome.45 Proteomic applications to other bat tissues, such as saliva, brain, and lung, have identified possible immunomodulatory properties in vampire bats (Desmodus rotundus),46 neuronal differences between active and torpid horseshoe bats (Rhinolophus ferrumequinum),47 and upregulation of cellmediated immunity in flying foxes (Pteropus alecto) compared with ferrets infected with Hendra virus.48 Given the general utility of proteomic analysis, we expected a survey of the vampire bat serum proteome should provide complementary information to existing approaches, while also providing a more complete survey of the underlying molecular landscape.
Here, we used data-independent acquisition-based shotgun proteomics (i.e., bottom-up proteomics) to profile the undepleted serum proteome of 17 bats from two locations. We focused this work on vampire bats, a species that has an obligate diet of blood and feeds on prey as diverse as sea lions, tapirs, livestock, and humans,49–51 providing vast opportunities for transmission of viruses (e.g., rabies virus, adenovirus, herpesvirus) to and from these prey.52–54 Our results demonstrate the feasibility and capabilities of serum proteomic analyses in wild bats, including possibilities to simultaneously detect immunological components and viruses as well as to establish preliminary ranges of vampire bat proteins for comparison with other mammals.
METHODS
Vampire Bat Sampling
As part of an ongoing longitudinal study, vampire bats were sampled in 2015 at two adjacent localities in the Orange Walk District of Belize: Lamanai Archeological Reserve (LAR, 450 ha) and Ka’Kabish (KK, 45 ha). These sites are located in a highly agricultural mosaic landscape where deforestation has been driven by cropland expansion in the 1990s followed by heightened demand for livestock pasture in the 2000s.55 These land-use changes have fragmented previously intact forest and have provided vampire bats with an abundant prey in the form of livestock.31 In recent years (2016 onward), rabies outbreaks in domestic animals have increased across Belize and in Orange Walk District, and virus isolates from livestock have been characterized as vampire bat-associated variants.56 Although we are unaware of other viral detection efforts in vampire bats in Belize, this species has tested positive elsewhere in Central and South America for adenoviruses, coronaviruses, flaviviruses, hantaviruses, herpesviruses, and paramyxoviruses, as well as for antibodies against henipavirus-like viruses.54,57–65
Vampire bats were captured with mist nets and harp traps set along trails and adjacent to known roost sites. All individuals were issued a unique Incoloy wing band (3.5 mm, Porzana Inc.) and identified by sex, age, and reproductive status.66 We collected blood by lancing the propatagial vein with a 23-gauge needle followed by collection with heparinized capillary tubes. Blood was allowed to sit in serum separator tubes (BD Microtainer) for approximately 10 to 20 min before centrifugation and drawing off the serum. Sera were stored short-term at −20 °C until long-term −80 °C storage. Bleeding was stopped with styptic gel, and all bats were released at their capture location. Field protocols followed guidelines for safe and humane handling of bats issued by the American Society of Mammalogists26 and were approved by the University of Georgia Animal Care and Use Committee (A2014 04–016-Y3-A5). Bat sampling was authorized by the Belize Forest Department under permit number CD/60/3/15(21). Specimen use for proteomic analysis was approved by the NIST Animal Care and Use Coordinator (NIST ACUC) under approval MML-AR19–0018. We included 17 serum samples for proteomic analysis, from 11 bats sampled at LAR (10 males, 1 female) and six bats sampled at KK (5 males, 1 female). This information and the experimental key can be found in Supplemental Table S1.
Sample Processing and Digestion
Prior to analysis, the sample set was randomized and assigned an experimental key to avoid bias and minimize batch effects. The 17 serum samples were thawed then centrifuged at 13 000gn for 8 min at 4 °C. The S-Trap method was used for digestion with the S-Trap micro column (ProtiFi; ≤100 μg binding capacity), and the specific method is described below. On the basis of prior work with other mammalian sera, we assumed a protein concentration of approximately 50 μg/μL (this value can vary but assuming at least 50 μg/μLwas used for estimating enzyme for the digest). Therefore, 2 μL (approximately 100 μg protein) of each sample was mixed with 48 μL 50 mmol/L ammonium bicarbonate and 50 μL of 2× lysis buffer [10% sodium dodecyl sulfate (SDS) volume fraction in 100 mmol/L triethylammonium bicarbonate (TEAB), pH 8.5]. Additionally, since the samples were digested in two sets, each set included a 2 μL aliquot of NIST SRM 909c Frozen Human Serum, which is technically a converted plasma pool. This sample was used to qualitatively confirm digestion, LC-MS/MS performance, and downstream search methods. All samples were reduced with 10 μL of 90 mmol/L DL-dithiothreitol (DTT; final concentration of 10 mmol/L) at 60 °C for 30 min, then cooled and alkylated with 10 μL of 200 mmol/L 2-chloroacetamide (CAA; final concentration of 20 mmol/L) at room temperature in the dark for 30 min. The sample was acidified with 12 μL of 12% phosphoric acid (volume fraction) bringing the final volumetric ratio to 1:10. Next, 700 μL binding buffer [ProtiFi; 5% TEAB volume fraction in methanol] was added (approximately 1:7 volumetric ratio)]. Using a vacuum manifold, the sample was washed across the S-Trap with six sequential washes of 400 μL binding buffer. Next, 3 μL of 1 μg/μL trypsin (Pierce) was mixed with 122 μL, 50 mmol/L ammonium bicarbonate, and this 125 μL solution was added to each S-Trap, yielding approximately a 1:30 mass ratio (trypsin:total protein). Samples were incubated at 47 °C for 1 h, after which they were sequentially washed into 1.5 mL Lo-Bind microcentrifuge tubes (Eppendorf) by centrifugation with the following wash steps: 80 μL 50 mmol/L ammonium bicarbonate, 80 μL 0.2% formic acid (volume fraction), 80 μL 0.2% formic acid in 50% acetonitrile (volume fractions) at 1000gn, 1000gn, and 4000gn, respectively at 4 °C for 1 min. The resulting peptide mixtures were reduced to dryness in a vacuum centrifuge at low heat and stored at −80 °C. Prior to analysis samples were reconstituted with 100 μL 0.1% formic acid (volume fraction) and briefly vortexed, then centrifuged 10 000gn for 10 min at 4 °C. The peptide concentration of each sample was determined using the Pierce quantitative fluorometric peptide assay with a BioTek Synergy HT plate reader.
LC-MS/MS
Peptide mixtures in 0.1% formic acid (volume fraction) were analyzed using an UltiMate 3000 Nano LC coupled to a Fusion Lumos Orbitrap mass spectrometer (Thermo Fisher Scientific). Using the original sample randomization yielded a randomized sample order and injection volumes were determined for 0.5 μg loading (between 0.37 and 0.95 μL sample). One sample from bat D141 (experimental key Bat_20) was used as a technical replicate to evaluate technical variability across the run (Supplemental Figure S1), while the two aforementioned human serum pools were analyzed in the same manner as the bat sera. Peptide mixtures were loaded onto a PepMap 100 C18 trap column (75 μm id × 2 cm length; Thermo Fisher Scientific) at 3 μL/min for 10 min with 2% acetonitrile (volume fraction) and 0.05% trifluoroacetic acid (volume fraction) followed by separation on an Acclaim PepMap RSLC 2 μm C18 column (75 μm id × 25 cm length; Thermo Fisher Scientific) at 40 °C. Peptides were separated along a 60 min two-step gradient of 5% to 30% mobile phase B (80% acetonitrile volume fraction, 0.08% formic acid volume fraction) over 50 min followed by a ramp to 45% mobile phase B over 10 min and last ramped to 95% mobile phase B over 5 min, and held at 95% mobile phase B for 5 min, all at a flow rate of 300 nL/min.
The Fusion Lumos was operated in positive polarity with a data-independent acquisition (DIA) method constructed using the targetedMS2 module (as opposed to the built-in DIA module). The full scan resolution using the Orbitrap was set at 120 000, the mass range was 399 to 1200 m/z (corresponding to the DIA windows used), 30% RF lens was set, the full scan ion target value was 4.0 × 105 allowing a maximum injection time of 20 ms. A default charge of 4 was set under MS Global Settings. As stated, DIA windows were constructed using the targetedMS2 module. Each window used higher-energy collisional dissociation (HCD) at a normalized collision energy of 32 with quadrupole isolation width at 21 m/z. The fragment scan resolution using the Orbitrap was set at 30 000, the scan range was specified as 200 to 2000 m/z, ion target value of 1.0 × 106 and 60 ms maximum injection time. Data were collected as profile data in both MS1 and MS2, though the authors wish to note that using centroid data is possible for most DIA software. The DIA window scheme was an overlapping static window strategy such that each of the 40 windows were 21 m/z wide, with 1 m/z overlap on each side covering the range of 399 to 1200 m/z. The window centers were specified in the mass list table (with z = 2), such that they were 409.5, 429.5, 449.5, …, 1189.5. The method file (85min_DIA_40×21mz.meth) is also included in the PRIDE submission. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE67 partner repository with the data set identifier PXD022885.
DIA Search Parameters
Analysis was performed using Spectronaut (v13.6.190905.43655). The following settings were used. Sequences: Trypsin selected, max pep length 52, min pep length 7, two missed cleavages, KR as special amino acids in decoy generation, toggle N-terminal M set to true. Labeling: no labeling settings were used. Applied modifications: maximum of five variable modifications using fixed carbamidomethyl (C), and variable acetyl (protein N-term) and oxidation (M). Identification: per run machine learning, Q-value cutoff of 0.01 for precursors and proteins, single hits defined by stripped sequence, and do not exclude single hit proteins, PTM localization set to true with a probability cutoff of 0.75, kernel density p-value estimator. Quantification: interference correction was used with excluding all multi-channel interferences with minimum of 2 and 3 for MS1 and MS2, respectively, proteotypicity filter set to none, major protein grouping by protein group ID, minor peptide grouping by stripped sequence, major group quantity set to mean peptide quantity, a Major Group Top N was used (min of 1, max of 3), minor group quantity set to mean precursor quantity, a Minor Group Top N was used (min of 1, max of 3), quantity MS-level used MS2 area, data filtering by q-value, cross run normalization was used with global median normalization and automatic row selection, no modifications or amino acids were specified, best N fragments per peptide was set to between 3 and 6, with ion charge and type not used. Workflow: no workflow was used. Post Analysis: no calculated explained TIC or sample correlation matrix, differential abundance grouping using major group (from quantification settings) and smallest quantitative unit defined by precursor ion (from quantification settings), differential abundance was not used for conclusions. The fasta file used for searching bat samples was the NCBI RefSeq Desmodus rotundus Release 100, GCF_002940915.1_ASM294091v2 (29 845 sequences), and for searching human samples was the 2020_01 release of the UniProtKB SwissProt with isoforms, using the taxonomy term 9606 (Homo sapiens; 42 385 sequences). For the host plus virus searching, these same fasta files were used along with a fasta of D. rotundus-associated virus sequences retrieved from the DBatVir database (156 sequences; downloaded 12 November 202065) and the following six fasta retrieved from the 2020_05 release of the UniProtKB SwissProt and TrEMBL: Adenoviridae (taxon ID: 10508; 30 051 sequences), Betacoronavirus (taxon ID: 694002; 21 892 sequences), Henipavirus (taxon ID: 260964; 462 sequences), Herpesvirales (taxon ID: 548681; 88 353 sequences), Morbillivirus (taxon ID: 11229; 23 142 sequences), and Rabies lyssavirus (taxon ID: 11292; 23 398 sequences). All fasta and Spectronaut.snes files (the human and bat data are searched with and without all virus databases) are included in PRIDE submission PXD022885. To facilitate reanalysis using other normalization methods or protein inference outside of Spectronaut, an expanded elution group, peptide group and protein group quantification table (Supplemental Table S7) was exported from “2020-4-16 bat directDIA tryp.sne” (available in PRIDE submission PXD022885).
Ortholog Mapping, Gene Ontology Term Sorting, and Rank Comparisons
Following DIA identification, there were 376 RefSeq vampire bat protein identifiers with MS2-based quantities across the experiment (Supplemental Table S2). These identifications were converted to human orthologs to aid in downstream analysis and for comparison to other studies. This was accomplished by using a series of python scripts (Anaconda v2019.07; conda v4.7.11; Python v3.6.8) from Github on the following two repositories: pwilmart/PAW_BLAST and pwilmart/annotations, retrieved November 18, 2019. Broadly, these tools take a list of identifiers to create a subset fasta (make_subset_DB_from_list_3.py), which is then searched against a human fasta (db_to_db_blaster.py) using a local installation of BLAST+ 2.9.0.68 The results were further annotated using add_uniprot_annotations.py. The 376 vampire bat proteins were assigned human orthologs, which were then manually inspected for incorrect assignments. Such assignments can happen when a protein is not present in humans (e.g., beta-lactoglobulin 1 or inhibitor of carbonic anhydrase), or if the blast hit disagrees with the original annotation, in which case the original annotation is preserved (e.g., apolipoprotein R was matched to C4b-binding protein alpha chain). Finally, duplicate entries were summed together into a single nonredundant entry. An intermediate table (Supplemental Table S2) is given that delineates which entries had manually assigned gene names, or were duplicates to be summed. The resulting table included mapped orthologs with UniProtKB links (Supplemental Table S3). The data in Supplemental Table S3 were evaluated using a Wilcoxon rank sum test (MATLAB R2015a; ranksum function) followed by Benjamini−Hochberg (BH) multiple hypothesis correction (mafdr function). There were no differentially abundant (BH-adjusted p-value <0.05) proteins detected between the two bat populations. Gene ontology (GO) term sorting was performed by using GO search terms in UniProtKB along with the human taxon identifier (9606), yielding a list of human gene symbols that was used to subset the vampire bat results. Specifically in Supplemental Table S3, the complement activation Gene Ontology (GO) term (GO:0006956) was used to identify 31 bat proteins. In addition to vampire bat proteome, identifications and relative abundances of the control pooled human serum can be found in Supplemental Table S4, but were not used for comparison.
The serum proteome of the vampire bat was compared to the plasma proteome of the human described on the Human Blood Atlas by using values from Table 1 “Protein detected in human plasma by mass spectrometry” from the Human Blood Atlas Web site (https://www.proteinatlas.org/humanproteome/blood/proteins+detected+in+ms; accessed 15 June 2020). This table was combined with the relative abundance and resulting ranks of the 361 proteins identified (Supplemental Table S5). The rank of the shared 323 proteins was compared by determining the absolute difference in rank divided by the minimum rank, referred to as “Rank Delta”. An arbitrary cutoff of 3 was chosen, but the complete list was still manually inspected for proteins of interest. Only proteins that were exceptionally elevated (or closely related to those that were elevated) in the vampire bat serum proteome were considered. Some proteins like PRDX1 (136th in bat and 432 in human) were less than 3 in their rank delta, but were still considered of interest. This final list of proteins of interest can be found in Supplemental Table S6, along with links to both UniProt and Human Blood Atlas entries.
RESULTS AND DISCUSSION
Studying the interactions between hosts and pathogens is important to understand infectious disease risks, yet such research is hampered by our poor knowledge of the underlying biochemical landscape for most host species. Bats offer a unique opportunity to investigate how immunological components may confer the ability to harbor virulent viruses, a topic of considerable interest particularly in light of the current COVID-19 pandemic. Because complement plays a key role in innate immunity and functions as a bridge between innate and adaptive responses,69–71 prior studies have mostly used microbicidal assays using pathogens with complement-dependent killing to profile levels of these proteins in bats, demonstrating individual and environmental sources of variation.30–32,72,73 We profiled the serum proteome of vampire bats as a first step at broadly describing the bat serum proteome. This inventory allowed investigating not only complement proteins, but also a global investigation of the bat serum proteome, including other components of the innate immune response, as well as detection of viral proteins, and can serve as a resource to develop targeted mass spectrometry-based assays.
Blood is a unique biofluid that is proximal to most organs, containing cells and secreted proteins. The considerable dynamic range of proteins in serum and plasma has been frequently described in humans, spanning at least 8 orders of magnitude, though approximately 10 proteins account for 90% of the plasma proteome.42,74 Similarly, we unsurprisingly found the vampire bat serum proteome encompassed a dynamic range of 5 orders of magnitude (Figure 1A). The top four proteins (albumin, immunoglobulin lambda-1 light chain, hemoglobin subunit beta, and alpha-1-antitrypsin) accounted for 50% of the total abundance estimated using DIA. Ranges for the top 20 proteins (based on average abundance; Figure 1B) were determined using protein abundance from the 17 individuals. Despite the difficulties in making direct comparisons between the vampire bat serum proteome and that of other mammals, it is possible to specifically evaluate complement components in bat serum. The complement activation Gene Ontology (GO) term (GO:0006956), which includes the lectin pathway, was used to highlight relative levels of 31 proteins in bat serum (Figure 1C) and indicate their rank and abundance in the serum proteome (Figure 1A). Apolipoprotein R and mannose-binding protein A were added since they are not present in humans but are likely involved in complement activation based on functional ontology. It would be interesting to infer that these complement protein levels are higher in bats than other mammals, but without adequate reference ranges, and given the technical artifacts of serum preparation across species, these comparisons are currently difficult at best and an avenue for future comparative study.
Figure 1.
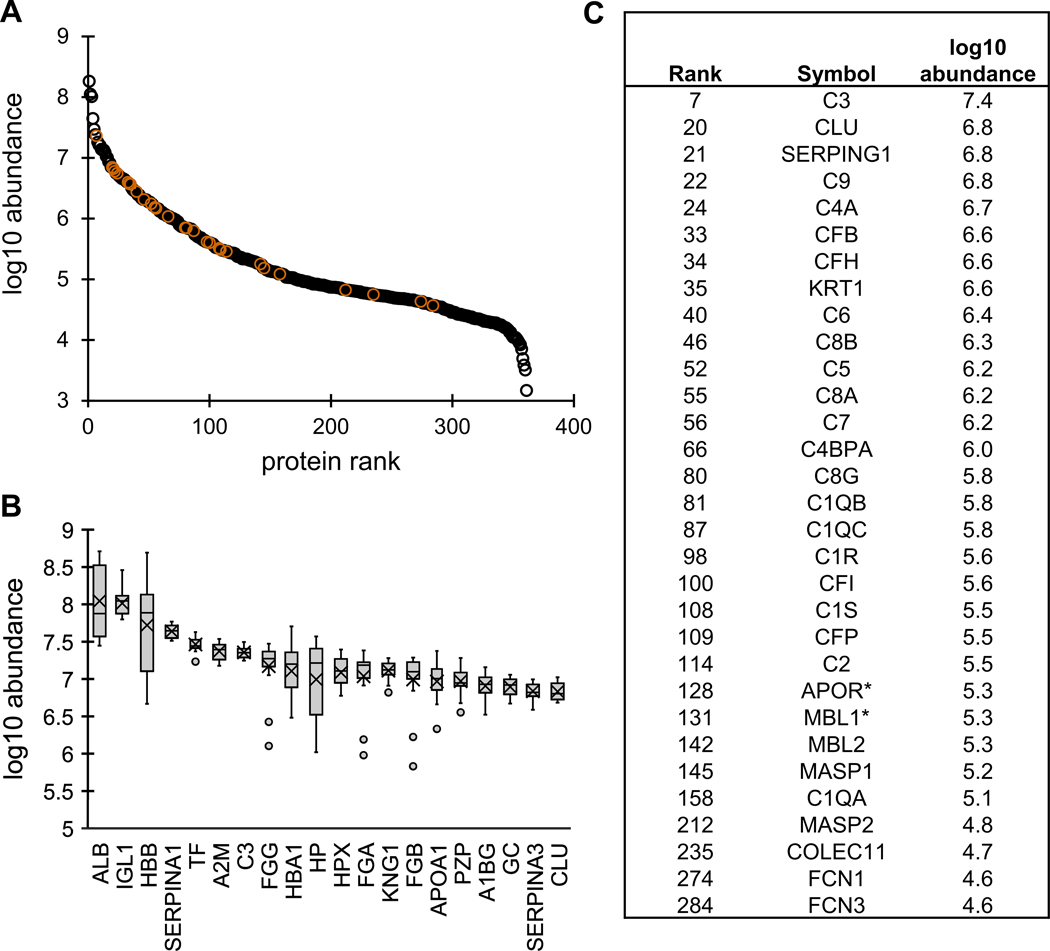
Average serum protein abundance. (A) The average intensity of the 361 identified proteins was plotted with rank to illustrate the dynamic range of the vampire bat serum proteome. Complement related proteins are in orange for reference. (B) Box and whisker plot of the top 20 proteins, listed as gene symbol. (C) 31 proteins involved in complement activation (GO:0006956). The rank and abundance are given for easy reference to the A panel. The * for MBL1 and APOR is to indicate that these are absent from GO:0006956 since they are not human proteins, but may be involved in complement activation based on functional ontology. Abbreviations: (B) ALB, serum albumin; IGL1, immunoglobulin lambda-1 light chain; HBB, hemoglobin subunit beta; SERPINA1, alpha-1-antitrypsin; TF, serotransferrin; A2M, alpha-2-macroglobulin; C3, complement C3; FGG, fibrinogen gamma chain; HBA1, hemoglobin subunit alpha; HP, haptoglobin; HPX, hemopexin; FGA, fibrinogen alpha chain; KNG1, kininogen-1; FGB, fibrinogen beta chain; APOA1, apolipoprotein A-I; PZP, pregnancy zone protein; A1BG, alpha-1B-glycoprotein; GC, vitamin D-binding protein; SERPINA3, alpha-1-antichymotrypsin. (C) C3, Complement C3; CLU, Clusterin; SERPING1, Plasma protease C1 inhibitor; C9, Complement component C9; C4A, Complement C4-A; CFB, Complement factor B; CFH, Complement factor H; KRT1, Keratin, type II cytoskeletal 1; C6, Complement component C6; C8B, Complement component C8 beta chain; C5, Complement C5; C8A, Complement component C8 alpha chain; C7, Complement component C7; C4BPA, C4b-binding protein alpha chain; C8G, Complement component C8 gamma chain; C1QB, Complement C1q subcomponent subunit B; C1QC, Complement C1q subcomponent subunit C; C1R, Complement C1r subcomponent; CFI, Complement factor I; C1S, Complement C 1s subcomponent; CFP, Properdin (Complement factor P); C2, Complement C2; APOR, Apolipoprotein R; MBL1, Mannose-binding protein A; MBL2, Mannose-binding protein C; MASP1, Mannan-binding lectin serine protease 1; C1QA, Complement C1q subcomponent subunit A; MASP2, Mannan-binding lectin serine protease 2; COLEC11, Collectin-11; FCN1, Ficolin-1; FCN3, Ficolin-3.
Although blood has been extensively studied in humans, it is surprisingly difficult to compare mass spectrometry-based analysis between human studies, much less between different species. The current study used a pooled human converted plasma sample (Supplemental Table S4) to enable this comparison. Additionally, bat serum protein identifications were converted to human orthologs (Supplemental Table S3) to aid in downstream analysis, including comparison to the Human Blood Atlas75 (Supplemental Table S5), with reported approximate concentrations of over 3000 proteins detected by mass spectrometry in human plasma. For an exploratory analysis, using these two human data sets is acceptable, but two key caveats should be acknowledged. First, ranges of human concentrations are needed, which is obscured in a pooled sample as well as in the current Human Blood Atlas. Second, the wild bat samples may have had technical artifacts (e.g., hemolysis), which is evident by the high levels of hemoglobin subunit beta (HBB) likely resulting from ruptured erythrocytes76 (Figure 1B). Ongoing efforts by our group and others will continue to define the serum and plasma proteome in humans and other species, and we expect more population-level data with protein abundance ranges to become available and more easily comparable across mammals.
Despite these caveats, we can highlight certain proteins that are higher in bat serum than in humans, without an obvious technical artifactual explanation. These were identified using the ranks of proteins in the parallel analyzed human pool and those available on the Human Blood Atlas versus the ranks of proteins in the vampire bat data set (Figure 2; Supplemental Table S5). Broadly, these proteins fall into four general categories: innate immunity, circulating proteasome, antioxidants, and “other”. We encourage readers to explore the full table for specific comparative proteins of interest (Supplemental Table S6).
Figure 2.
Proteins elevated in vampire bat serum versus human plasma. (A) Average protein abundance of 361 proteins in vampire bat serum. (B) Reported protein abundance of 3222 proteins reported on the Human Plasma Atlas. (C) Innate immunity proteins with bat and human ranks. The * for MBL1 and APOR is to indicate that these are not human proteins and not present in human plasma. (D) Proteasome related to proteins with bat and human ranks. (E) Redox related proteins with bat and human ranks. (F) Other uncategorized (“other”) proteins of interest with bat and human ranks. All protein ranks are given in Supplemental Table S5, while just select proteins here are described in Supplemental Table S6. Abbreviations: (C) ANG, angiogenin; APOR, apolipoprotein R; ARF1, ADP-ribosylation factor 1; BPIFA2, BPI fold-containing family A member 2; CAMP, cathelicidin antimicrobial peptide; CREG1, protein CREG1; DMBT1, deleted in malignant brain tumors 1 protein; EEF1A1, elongation factor 1-alpha 1; GBP2, guanylate-binding protein 2; GBP6, guanylate-binding protein 6; GBP7, guanylate-binding protein 7; IFI6, interferon alpha-inducible protein 6; MBL1, mannose-binding protein A; SFTPD, pulmonary surfactant-associated protein D. (D) PSMA1, proteasome subunit alpha type-1; PSMA2, proteasome subunit alpha type-2; PSMA3, proteasome subunit alpha type-3; PSMA4, proteasome subunit alpha type-4; PSMA5, proteasome subunit alpha type-5; PSMA6, proteasome subunit alpha type-6; PSMA7, proteasome subunit alpha type-7; PSMB1, proteasome subunit beta type-1; PSMB2, proteasome subunit beta type-2; PSMB3, proteasome subunit beta type-3; PSMB4, proteasome subunit beta type-4; PSMB5, proteasome subunit beta type-5; PSMB6, proteasome subunit beta type-6; PSMD2, 26S proteasome non-ATPase regulatory subunit 2; RAD23A, UV excision repair protein RAD23 homologue A; RPS27A, ubiquitin-40S ribosomal protein S27a; USP14, ubiquitin carboxyl-terminal hydrolase 14. (E) GPX1, glutathione peroxidase 1; GPX3, glutathione peroxidase 3; PRDX1, peroxiredoxin-1; PRDX2, peroxiredoxin-2; PRDX6, peroxiredoxin-6; RFESD, Rieske domain-containing protein; SOD1, superoxide dismutase [Cu–Zn]; TXN, thioredoxin; VNN2, vascular noninflammatory molecule 2; VNN3, vascular noninflammatory molecule 3. (F) ACE, angiotensin-converting enzyme; ADIPOQ, adiponectin; AFP, alpha-fetoprotein; AHCY, adenosylhomocysteinase; CD177, CD177 antigen; CD97, CD97 antigen; CLCA1, calcium-activated chloride channel regulator 1; DPEP1, dipeptidase 1; EIF5A2, eukaryotic translation initiation factor 5A-2; FCGBP, IgGFc-binding protein; GCLM, glutamate–cysteine ligase regulatory subunit; HIST2H2BE, histone H2B type 2-E; HSPA2, heat shock-related 70 kDa protein 2; HSPA4, heat shock 70 kDa protein 4; HYAL1, hyaluronidase-1; IMPDH2, inosine-5′-monophosphate dehydrogenase 2; ITIH2, interalpha-trypsin inhibitor heavy chain H2; ITIH3, interalpha-trypsin inhibitor heavy chain H3; ITIH4, interalpha-trypsin inhibitor heavy chain H4; KLK1, kallikrein-1; LIPG, endothelial lipase; PAFAH1B2, platelet-activating factor acetylhydrolase IB subunit beta; PCBP1, poly(rC)-binding protein 1; PF4, platelet factor 4; SERPINB10, serpin B10; SMPD1, sphingomyelin phosphodiesterase; SRI, sorcin; TFRC, transferrin receptor protein 1; TMPRSS9, transmembrane protease serine 9.
Innate Immunity
Investigation of apparent differences in proteins related to innate immunity is important because vampire bats harbor diverse pathogens24,54,57–59,77 and because bats more generally appear to tolerate some viral infections without showing disease.8,18,21,22 In addition to the complement proteins described above, we identified notable proteins related to the innate immune system that are highly ranked in bat serum (Figure 2C). Given that some bats can constitutively express interferons (e.g., IFN-α in Pteropus alecto),19 it was not surprising to detect interferon-inducible proteins (e.g., guanylate-binding proteins GBP2, GBP6, and GBP7). Of note is that IFN-α and IFN-γ induce many similar proteins in a highly dynamic manner.78 Specific to detected guanylate-binding proteins (GBPs), GBP2 was detected and shares antiviral properties with GBP5 (undetected), although circulating blood GBPs do not appear common in humans. 79–81 It would be interesting to determine if certain proteins are stratified into exosomes, similar to the IFN-induced antiviral proteome contained in macrophage-derived exosomes.82 There were also proteins identified related to antiviral and antibacterial activity, notably relatively high levels of CAMP (cathelicidin antimicrobial peptide). Finally, two scavenging proteins reported in human blood were ranked higher in bat serum: SFTPD (pulmonary surfactant-associated protein D) and DMBT1 (deleted in malignant brain tumors 1 protein). Though their functions are related, DMBT1 specifically binds viral and bacterial antigens83,84 and can reduce viral infectivity.85 The question remains whether these proteins confer a unique ability to keep pathogen virulence at bay or if these elevated protein levels instead indicate an active infection.
Circulating Proteasome
In addition to focusing on classically immunological proteins, we observed 14 proteasome subunits (Figure 2D). Specifically, 13 of the 14 known 20S subunits (though missing PSMB7) were observed along with one 26S component (PSMD2), indicating the presence of the circulating 20S proteasome.86,87 The lack of PSMB7 may be species-specific, similar to lower PSMB7 values reported across different mammal species.88 Although circulating 20S proteasome is lower ranked in the Human Blood Atlas, it has been detected in human plasma, possibly within exosomes.89 Given that our group has observed similarly abundant 20S proteasome subunits in other mammalian serum and plasma (e.g., California sea lion plasma proteome90), circulating 20S proteasome is likely present in vampire bats and not a technical artifact of hemolysis or serum preparation. The 20S proteasome mediates proteasomal cleavage, with numerous cellular roles including response to disease and oxidative stress.87 Interestingly, IFN-γ can induce formation of the immunoproteasome, which is the circulating 20S proteasome incorporated with β8, β9, and β10 subunits.86 The immunoproteasome has diverse functions, including cleaving peptides into MHC class I epitopes.86 The immunoproteasome is likely activated within hematopoietic cells, and may exist within extracellular vesicles though not necessarily circulating in the blood.87 Though these additional immunoproteasome subunits were not observed, it is interesting to hypothesize the role of such high levels of circulating 20S proteasome. These results demonstrate the potential of using mass spectrometry-based methods to study proteasomes, providing a basis for future immunological studies.
Redox-Related Proteins
Beyond the possible uniqueness of bat immune systems is their ability to minimize or respond to oxidative stress. The evolution of flight in the chiropteran lineage was accompanied by mechanisms to minimize or repair the negative effects of oxidative stress generated by this metabolically costly activity, which may explain both the particular longevity of bats and their apparent viral tolerance.8,91,92 For reference in Figure 2C, we have included glutathione peroxidase levels, which do not appear very different from levels in humans, but we note that there are higher levels of other redox-related proteins in vampire bats. Increased levels of peroxiredoxins (PRDX1, PRDX2, and PRDX6), superoxide dismutase (SOD1), and thioredoxin (TXN) indicate an enhanced capacity to scavenge or oppose free-radicals, though these levels may also be a result of hemolysis as evident by high HBB levels, and that, except for TXN, these proteins have been shown to be markers of erythrocyte lysis.76 High-levels of Rieske domain-containing protein (RFESD), which facilitates binding excess iron, may be related to the iron-rich, blood diet of vampire bats, although ferritin light chain (FTL) levels were found to be similar between bats and humans, ranked 154 and 336, respectively. Finally, vascular noninflammatory molecule proteins (VNN2 and VNN3) have been described as being involved in redox reactions via the production of cysteamine.93–95 Overall, these results highlight that there may be increased oxidative resistance in vampire bats relative to humans; this pattern is likely to be conserved across bat species given the ubiquity of flight and long lifespans in bats, but this should be confirmed with broad comparative analyses.
“Other” Interesting Proteins
In addition to proteins with obvious relation to known bat physiology, there are other interesting proteins that do not fit into a single category (Figure 2D). From this list, two sets of relationships are noteworthy: angiotensin-converting enzyme (ACE)/kallikrein 1 (KLK1) and hyaluronan-related proteins. Both ACE and KLK1 are higher in bats than humans, though their functions are antagonistic: kallikrein increases bradykinin, which vasodilates, while ACE cleaves bradykinin and thus may reduce vasodilation. In comparison, angiotensinogen (AGT), a source of the vasoconstrictor peptide angiotensin 2, is almost the same rank in bats as humans (ranked 29 and 25, respectively; Supplemental Table S5). Of note, given the entry of SARS-CoV-2 in humans, is the detection of TMPRSS9 as a midabundance protein in bat serum, though ACE2, which is detected in humans (rank 2517; Supplemental Table S5), was not present in bat serum. There are also increased hyaluronan-related proteins in vampire bat serum. The hyaluronan receptor CD44 is the same rank in bats and humans (Supplemental Table S5), though two hyaluronan carrier proteins (ITIH2 and ITIH3) and hyaluronidase 1 (HYAL1), which hydrolyzes hyaluronan, are higher in bats (Figure 2F). This last protein, HYAL1, is ranked 3024 in human plasma, versus 173 in vampire bat serum. It is unclear whether this indicates high levels of hyaluronan in bat serum, which in the naked-mole rat may confer anticancer properties,96 or that these proteins are involved with keeping serum hyaluronan levels low in response to infection.97 There are additional undiscussed proteins listed in this “other” category, as well as a full list of protein ranks compared to the Human Blood Atlas (Supplemental Table S5–S6), that researchers are encouraged to browse for insight beyond the survey nature of this manuscript. It is interesting to postulate that vampire bat blood may have an antipathogenic phenotype, but it is unclear if this is in response to infection or constitutive. Similarly, given that many antiviral proteins are often involved in anticancer roles,98 these two roles could plausibly be connected.
Pathogen Detection
Data-independent acquisition (DIA) is an emerging technique in mass spectrometry-based proteomics, which in theory should allow deeper coverage than typical data-dependent acquisition (DDA)-based analysis by overcoming the stochastic nature of DDA data. We did not set out to compare the two approaches, but found this to be an interesting use-case of DIA to analyze undepleted serum. Detecting 361 proteins in undepleted serum using a relatively short 60 min gradient demonstrated that DIA-based proteomics of undepleted serum can scale reasonably well. Moreover, we took advantage of this technique to evaluate the presence of viral proteins. Desmodus rotundus has tested positive elsewhere in its geographic range for adenoviruses, coronaviruses, flaviviruses, hantaviruses, herpesviruses, and paramyxoviruses, as well as for antibodies against henipavirus-like viruses.54,57–65 Searches were performed with Rabies lyssavirus, Adenoviridae, Betacoronavirus, Henipavirus, Herpesvirales, and Morbillivirus databases. Using this approach, we detected no viral proteins in the human pooled sample but putatively identified two viral peptides in all 17 bat samples (Supplemental Figures S2–S4): PRSGIPDR from the Rh186 protein in Macacine herpesvirus 3 (i.e., Rhesus cytomegalovirus) and LVTTEVK from the ORF1a protein in Middle East respiratory syndrome-related coronavirus (MERS-CoV). Detection of the MERS-CoV protein is of particular interest, as other betacoronaviruses have been detected in vampire bats and cell line studies suggest their receptors (i.e., DPP4) can support MERS-CoV replication.62,99 Although we are unaware of specific MERS-CoV detection in vampire bats, a betacoronavirus with high amino acid similarity to MERS-CoV has been identified in an unrelated but sympatric bat species in Mexico (Nyctinomops laticaudatus).100 More generally, these peptides were detected using a relatively short gradient separation of undepleted serum, suggesting that a method enriching for viral particles prior to analysis could easily detect more viral proteins if present. Importantly, although PCR-based techniques are frequently used for viral surveillance in bats, a mass spectrometry-based approach could query a larger search space for virus proteins while maintaining acceptable detection limits. In cases where a specific viral taxon was targeted, mass spectrometry could provide a more accurate method of virus protein detection (e.g., as evidenced by mass spectrometry-based SARS-CoV-2 detection in humans101–104).
Targeted mass spectrometry methods using stable isotope-labeled peptides allow for creation of sensitive, precise, and accurate assays that avoid interferences that plague antibody-based methods. The first step of developing these targeted assays is empirically confirming the presence of predicted peptides, which is possible even in nonmodel species. For example, shotgun proteomics were first used to identify proteotypic adiponectin peptides in dolphin plasma, followed by validation of a parallel-reaction monitoring (PRM) method to measure adiponectin at nmol/L levels using heavy isotope labeled peptides.105 Any of the proteins identified in the current study can be measured using this technique. For example, PRM assays could be constructed for each of the 31 complement activation proteins listed, and this could be performed in high-throughput manner. These assays would be free from measurement interference (i.e., cross-reactivity seen in antibody assays) with all the benefits of accuracy and precision found in PRM-based mass spectrometry assays.106,107
Limitations
There are inherent limitations when studying blood that are discussed above and briefly expanded here. First, because collection of blood from small animals requires small volumes to be drawn, blood was collected using a heparinized tube into a serum-separator tube. The residual heparin in the tube may have prevented complete clot formation. The main difference between plasma and serum proteomes is that clotting yields much lower fibrinogen levels.76 On the basis of the high levels of FGG, FGA, and FGB (ranked 8, 12, and 14, respectively), which closely correspond to ranks in human plasma (ranked 43, 14, and 21, respectively; Supplemental Table S5), it is likely that the bat blood sample did not fully clot. To avoid confusion, we refer to samples processed in the manner described above as serum, not plasma. Still, we encourage readers to not misinterpret high fibrinogen levels in bat serum. Second, the current study relied on the analyte-centric nature of DIA108 to detect target proteins and not experimental contaminant proteins, such as human keratin. Conceptually this is true, except the similarity between mammalian proteins means that protein identifications, such as bat keratins (e.g., KRT1) are ambiguous. In the case of KRT1, the observed abundance in bat serum aligns with values in the human blood atlas (Supplemental Table S5) and therefore experimental contamination is unlikely; however, contributions of human keratins cannot be completely excluded. Moreover, the levels of KRT1 and other keratins are not meant to be a major finding of this study. Finally, there is an opportunity to expand on work that can quantify technical artifacts due to clotting or hemolysis, as emphasized by Geyer et al.76 There is also an opportunity to develop new methods to compare blood proteomes within and between species. In the current study, we use protein ranks, which is the crudest metric available, although ranking allows comparisons between methods capable of different depth of coverage (e.g., we have compared levels of 361 bat serum proteins to 3222 human plasma proteins75). More sophisticated measures are possible that could use the relative quantification of DIA-based proteomics, such as spiking hundreds of stable isotope standard (SIS) peptides to quantify the complete blood proteome.109 Due to the high cost, this approach is currently limited to humans and widely used laboratory models. Alternatively, a set of mammalian blood markers could be used, similar to the endogenous common internal retention time standards (CiRT) concept.110 This would allow for more direct comparisons between proteomes. Despite these limitations, this study provides insight into the vampire bat blood proteome and is foundational to future and ongoing studies in comparative proteomics.
CONCLUSIONS
Our results highlight the power of modern proteomic techniques to provide new insights where other techniques might struggle. There is tremendous value in genomic and transcriptomic analyses; however, to understand the molecular landscape of blood, proteomic analysis is unique. This survey of the serum proteome of a small group of vampire bats provides a first look at the abundance of immunological proteins as well as proteins with unclear roles in the vampire bat phenotype. Moreover, we show that mass spectrometry-based viral protein detection in serum is possible and may be a viable tool for pathogen surveillance. Overall, these data can be used to develop targeted assays for future vampire bat research while also serving as a demonstration of the potential of proteomic studies for research on various topics in other bat species. Our understanding of the physiology of white-nose syndrome has already benefited from modern biomolecular analytical techniques,45,111 and proteomics could provide additional insights into the physiology of longevity and immunological tolerance of various bat species to zoonotic pathogens.18,112 Future work will continue to investigate if proteins elevated in vampire bats compared to humans are similarly elevated in other mammals, or whether they are more common in wild animals than expected and not specific to a vampire bat phenotype. There is an ongoing need for comparative physiological studies to move beyond two-species comparisons and broaden our horizons across multiple clades of the tree of life.
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to thank the anonymous peer-reviewers for critical insight and feedback. For assistance with bat sampling logistics and research permits, we thank Mark Howells, Neil Duncan, and the staff of the Lamanai Field Research Center. Identification of certain commercial equipment, instruments, software, or materials does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the products identified are necessarily the best available for the purpose.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00995.
Supplemental Figure S1: Relative standard deviation distribution of protein abundance in technical replicate; Supplemental Figure S2: Rh186 viral peptide identification; Supplemental Figure S3: ORF1a viral peptide identification; Supplemental Figure S4: Viral peptide quantities (PDF)
Supplemental Table S1: Experimental key with sample metadata; Supplemental Table S2: Intermediate data table of vampire bat serum protein ortholog mapping and quantities; Supplemental Table S3: Vampire bat serum protein orthologs and quantities; Supplemental Table S4: NIST 909c human serum protein relative quantities; Supplemental Table S5: Rank comparisons of the vampire bat proteome to the Human Blood Atlas; Supplemental Table S6: Proteins of interest with rank comparisons between vampire bat proteome and the Human Blood Atlas (XLSX)
Supplemental Table S7: Expanded elution group, peptide group, and protein group quantification and associated data (XLSX)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.jproteome.0c00995
The authors declare no competing financial interest.
Contributor Information
Benjamin A. Neely, Chemical Sciences Division, National, Institute of Standards and Technology, Charleston, South, Carolina 29412, United States
Daniel J. Becker, Department of Biology, University of, Oklahoma, Norman, Oklahoma 73019, United States
Michael G. Janech, Hollings Marine Laboratory, Charleston, South Carolina 29412, United States; Department of, Biology, College of Charleston, Charleston, South Carolina, 29424, United States
M. Brock Fenton, Department of Biology, Western University, London, Ontario N6A 3K7, Canada.
Nancy B. Simmons, Department of Mammalogy, Division of, Vertebrate Zoology, American Museum of Natural History, New York 10024, United States
Alison M. Bland, Hollings Marine Laboratory, Charleston, South Carolina 29412, United States; Department of, Biology, College of Charleston, Charleston, South Carolina, 29424, United States
REFERENCES
- (1).Wilson DE; Reeder DM Mammal Species of the World: A Taxonomic and Geographic Reference; JHU Press, 2005. [Google Scholar]
- (2).Kunz TH; Braun de Torrez E; Bauer D; Lobova T; Fleming TH Ecosystem Services Provided by Bats. Ann. N. Y. Acad. Sci 2011, 1223, 1–38. [DOI] [PubMed] [Google Scholar]
- (3).Wilkinson GS; South JM Life History, Ecology and Longevity in Bats. Aging Cell 2002, 1 (2), 124–131. [DOI] [PubMed] [Google Scholar]
- (4).Jebb D; Huang Z; Pippel M; Hughes GM; Lavrichenko K; Devanna P; Winkler S; Jermiin LS; Skirmuntt EC; Katzourakis A; Burkitt-Gray L; Ray DA; Sullivan KAM; Roscito JG; Kirilenko BM; Dávalos LM; Corthals AP; Power ML; Jones G; Ransome RD; Dechmann DKN; Locatelli AG; Puechmaille SJ; Fedrigo O; Jarvis ED; Hiller M; Vernes SC; Myers EW; Teeling EC Six Reference-Quality Genomes Reveal Evolution of Bat Adaptations. Nature 2020, 583 (7817), 578–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Ingala MR; Simmons NB; Perkins SL Bats Are an Untapped System for Understanding Microbiome Evolution in Mammals. mSphere 2018, DOI: 10.1128/mSphere.00397-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Ripperger SP; Carter GG; Duda N; Koelpin A; Cassens B; Kapitza R; Josic D; Berrío-Martínez J; Page RA; Mayer F. Vampire Bats That Cooperate in the Lab Maintain Their Social Networks in the Wild. Curr. Biol 2019, 29 (23), 4139–4144. [DOI] [PubMed] [Google Scholar]
- (7).Guth S; Visher E; Boots M; Brook CE Host Phylogenetic Distance Drives Trends in Virus Virulence and Transmissibility across the Animal–human. Philos. Trans. R. Soc., B 2019, 374 (1782), 20190296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Brook CE; Dobson AP Bats as “Special” Reservoirs for Emerging Zoonotic Pathogens. Trends Microbiol. 2015, 23 (3), 172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Olival KJ; Hosseini PR; Zambrana-Torrelio C; Ross N; Bogich TL; Daszak P. Host and Viral Traits Predict Zoonotic Spillover from Mammals. Nature 2017, 546 (7660), 646–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Halpin K; Hyatt AD; Fogarty R; Middleton D; Bingham J; Epstein JH; Rahman SA; Hughes T; Smith C; Field HE; Daszak P; Henipavirus Ecology Research Group. Pteropid Bats Are Confirmed as the Reservoir Hosts of Henipaviruses: A Comprehensive Experimental Study of Virus Transmission. Am. J. Trop. Med. Hyg 2011, 85 (5), 946–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Amman BR; Jones MEB; Sealy TK; Uebelhoer LS; Schuh AJ; Bird BH; Coleman-McCray JD; Martin BE; Nichol ST; Towner JS Oral Shedding of Marburg Virus in Experimentally Infected Egyptian Fruit Bats (Rousettus aegyptiacus). J. Wildl. Dis 2015, 51 (1), 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Banyard AC; Hayman D; Johnson N; McElhinney L; Fooks AR Bats and Lyssaviruses. Adv. Virus Res 2011, 79, 239–289. [DOI] [PubMed] [Google Scholar]
- (13).Li W; Shi Z; Yu M; Ren W; Smith C; Epstein JH; Wang H; Crameri G; Hu Z; Zhang H; Zhang J; McEachern J; Field H; Daszak P; Eaton BT; Zhang S; Wang L-F Bats Are Natural Reservoirs of SARS-like Coronaviruses. Science 2005, 310 (5748), 676–679. [DOI] [PubMed] [Google Scholar]
- (14).Plowright RK; Eby P; Hudson PJ; Smith IL; Westcott D; Bryden WL; Middleton D; Reid PA; McFarlane RA; Martin G; Tabor GM; Skerratt LF; Anderson DL; Crameri G; Quammen D; Jordan D; Freeman P; Wang L-F; Epstein JH; Marsh GA; Kung NY; McCallum H. Ecological Dynamics of Emerging Bat Virus Spillover. Proc. Biol. Sci 2015, 282 (1798), 20142124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kessler MK; Becker DJ; Peel AJ; Justice NV; Lunn T; Crowley DE; Jones DN; Eby P; Sanchez CA; Plowright RK Changing Resource Landscapes and Spillover of Henipaviruses. Ann. N. Y. Acad. Sci 2018, 1429, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Williamson MM; Hooper PT; Selleck PW; Westbury HA; Slocombe RF Experimental Hendra Virus Infection in Pregnant Guinea-Pigs and Fruit Bats (Pteropus poliocephalus). J. Comp. Pathol 2000, 122 (2–3), 201–207. [DOI] [PubMed] [Google Scholar]
- (17).Mollentze N; Streicker DG Viral Zoonotic Risk Is Homogenous among Taxonomic Orders of Mammalian and Avian Reservoir Hosts. Proc. Natl. Acad. Sci. U. S. A 2020, 117, 9423–9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Mandl JN; Schneider C; Schneider DS; Baker ML Going to Bat(s) for Studies of Disease Tolerance. Front. Immunol 2018, 9, 2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zhou P; Tachedjian M; Wynne JW; Boyd V; Cui J; Smith I; Cowled C; Ng JHJ; Mok L; Michalski WP; Mendenhall IH; Tachedjian G; Wang L-F; Baker ML Contraction of the Type I IFN Locus and Unusual Constitutive Expression of IFN-α in Bats. Proc. Natl. Acad. Sci. U. S. A 2016, 113 (10), 2696–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Becker DJ; Czirják GÁ; Rynda-Apple A; Plowright RK Handling Stress and Sample Storage Are Associated with Weaker Complement-Mediated Bactericidal Ability in Birds but Not Bats. Physiol. Biochem. Zool 2019, 92, 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Schountz T; Baker ML; Butler J; Munster V. Immunological Control of Viral Infections in Bats and the Emergence of Viruses Highly Pathogenic to Humans. Front. Immunol 2017, 8, 1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Banerjee A; Baker ML; Kulcsar K; Misra V; Plowright R; Mossman K. Novel Insights Into Immune Systems of Bats. Front. Immunol 2020, 11, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Becker DJ; Crowley DE; Washburne AD; Plowright RK Temporal and Spatial Limitations in Global Surveillance for Bat Filoviruses and Henipaviruses. Biol. Lett 2019, 15 (12), 20190423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Bergner LM; Orton RJ; Benavides JA; Becker DJ; Tello C; Biek R; Streicker DG Demographic and Environmental Drivers of Metagenomic Viral Diversity in Vampire Bats. Mol. Ecol 2020, 29 (1), 26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Brook CE; Boots M; Chandran K; Dobson AP; Drosten C; Graham AL; Grenfell BT; Müller MA; Ng M; Wang L-F; van Leeuwen A. Accelerated Viral Dynamics in Bat Cell Lines, with Implications for Zoonotic Emergence. eLife 2020, DOI: 10.7554/eLife.48401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Sikes RS; Gannon WL Guidelines of the American Society of Mammalogists for the Use of Wild Mammals in Research. J. Mammal 2011, 92 (1), 235–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Jones KE; Bielby J; Cardillo M; Fritz SA; O’Dell J; Orme CDL; Safi K; Sechrest W; Boakes EH; Carbone C; Connolly C; Cutts MJ; Foster JK; Grenyer R; Habib M; Plaster CA; Price SA; Rigby EA; Rist J; Teacher A; BinindaEmonds ORP; Gittleman JL; Mace GM; Purvis A. PanTHERIA: A Species-Level Database of Life History, Ecology, and Geography of Extant and Recently Extinct Mammals. Ecology 2009, 90, 2648–2648. [Google Scholar]
- (28).Baker ML; Schountz T; Wang L-F Antiviral Immune Responses of Bats: A Review. Zoonoses Public Health 2013, 60 (1), 104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Schountz T. Immunology of Bats and Their Viruses: Challenges and Opportunities. Viruses 2014, 6 (12), 4880–4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Allen LC; Turmelle AS; Mendonca MT; Navara KJ; Kunz TH; McCracken GF Roosting Ecology and Variation in Adaptive and Innate Immune System Function in the Brazilian Free-Tailed Bat (Tadarida brasiliensis). J. Comp. Physiol., B 2009, 179 (3), 315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Becker DJ; Czirjak GA; Volokhov DV; Bentz AB; Carrera JE; Camus MS; Navara KJ; Chizhikov VE; Fenton MB; Simmons NB; Recuenco SE; Gilbert AT; Altizer S; Streicker DG Livestock Abundance Predicts Vampire Bat Demography, Immune Profiles and Bacterial Infection Risk. Philos. Trans. R. Soc., B 2018, 373, 20170089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Ruoss S; Becker NI; Otto MS; Czirják GÁ; Encarnação JA Effect of Sex and Reproductive Status on the Immunity of the Temperate Bat Myotis daubentonii. Mamm. Biol 2019, 94 (1), 120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Young CCW; Olival KJ Optimizing Viral Discovery in Bats. PLoS One 2016, 11 (2), e0149237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Stepanian PM; Wainwright CE Ongoing Changes in Migration Phenology and Winter Residency at Bracken Bat Cave. Glob. Chang. Biol 2018, 24 (7), 3266–3275. [DOI] [PubMed] [Google Scholar]
- (35).Plowright RK; Peel AJ; Streicker DG; Gilbert AT; McCallum H; Wood J; Baker ML; Restif O. Transmission or Within-Host Dynamics Driving Pulses of Zoonotic Viruses in Reservoir-Host Populations. PLoS Neglected Trop. Dis. 2016, 10 (8), e0004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Gilbert AT; Fooks AR; Hayman DTS; Horton DL; Müller T; Plowright R; Peel AJ; Bowen R; Wood JLN; Mills J; Cunningham AA; Rupprecht CE Deciphering Serology to Understand the Ecology of Infectious Diseases in Wildlife. Ecohealth 2013, 10 (3), 298–313. [DOI] [PubMed] [Google Scholar]
- (37).Gerrard DL; Hawkinson A; Sherman T; Modahl CM; Hume G; Campbell CL; Schountz T; Frietze S. Transcriptomic Signatures of Tacaribe Virus-Infected Jamaican Fruit Bats. mSphere 2017, DOI: 10.1128/mSphere.00245-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Lee AK; Kulcsar KA; Elliott O; Khiabanian H; Nagle ER; Jones MEB; Amman BR; Sanchez-Lockhart M; Towner JS; Palacios G; Rabadan R. De Novo Transcriptome Reconstruction and Annotation of the Egyptian Rousette Bat. BMC Genomics 2015, 16, 1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Field KA; Johnson JS; Lilley TM; Reeder SM; Rogers EJ; Behr MJ; Reeder DM The White-Nose Syndrome Transcriptome: Activation of Anti-Fungal Host Responses in Wing Tissue of Hibernating Little Brown Myotis. PLoS Pathog. 2015, 11 (10), e1005168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Bergner LM; Orton RJ; da Silva Filipe A; Shaw AE; Becker DJ; Tello C; Biek R; Streicker DG Using Noninvasive Metagenomics to Characterize Viral Communities from Wildlife. Mol. Ecol. Resour. 2019, 19 (1), 128–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Donaldson EF; Haskew AN; Gates JE; Huynh J; Moore CJ; Frieman MB Metagenomic Analysis of the Viromes of Three North American Bat Species: Viral Diversity among Different Bat Species That Share a Common Habitat. J. Virol 2010, 84 (24), 13004–13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Uhlén M; Karlsson MJ; Hober A; Svensson A-S; Scheffel J; Kotol D; Zhong W; Tebani A; Strandberg L; Edfors F; Sjöstedt E; Mulder J; Mardinoglu A; Berling A; Ekblad S; Dannemeyer M; Kanje S; Rockberg J; Lundqvist M; Malm M; Volk A-L; Nilsson P; Månberg A; Dodig-Crnkovic T; Pin E, Zwahlen M; Oksvold P; von Feilitzen K; Häussler RS; Hong M-G; Lindskog C; Ponten F; Katona B; Vuu J; Lindström E; Nielsen J; Robinson J; Ayoglu B; Mahdessian D; Sullivan D; Thul P; Danielsson F; Stadler C; Lundberg E; Bergström G; Gummesson A; Voldborg BG; Tegel H; Hober S; Forsström B; Schwenk JM; Fagerberg L; Sivertsson Å The Human Secretome. Sci. Signaling 2019, 12, eaaz0274. [DOI] [PubMed] [Google Scholar]
- (43).Geyer PE; Holdt LM; Teupser D; Mann M. Revisiting Biomarker Discovery by Plasma Proteomics. Mol. Syst. Biol 2017, 13 (9), 942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Hecht AM; Braun BC; Krause E; Voigt CC; Greenwood AD; Czirják GÁ Plasma Proteomic Analysis of Active and Torpid Greater Mouse-Eared Bats (Myotis myotis). Sci. Rep 2015, 5, 16604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Hecht-Höger AM; Braun BC; Krause E; Meschede A; Krahe R; Voigt CC; Greenwood AD; Czirják GÁ Plasma Proteomic Profiles Differ between European and North American Myotid Bats Colonized by Pseudogymnoascus destructans. Mol. Ecol 2020, 29 (9), 1745–1755. [DOI] [PubMed] [Google Scholar]
- (46).Francischetti IMB; Assumpção TCF; Ma D; Li Y; Vicente EC; Uieda W; Ribeiro JMC The “Vampirome”: Transcriptome and Proteome Analysis of the Principal and Accessory Submaxillary Glands of the Vampire Bat Desmodus rotundus, a Vector of Human Rabies. J. Proteomics 2013, 82, 288–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Yin Q; Zhang Y; Dong D; Lei M; Zhang S; Liao C-C; Pan Y-H Maintenance of Neural Activities in Torpid Rhinolophus ferrumequinum Bats Revealed by 2D Gel-Based Proteome Analysis. Biochim. Biophys. Acta, Proteins Proteomics 2017, 1865 (8), 1004–1019. [DOI] [PubMed] [Google Scholar]
- (48).Woon AP; Boyd V; Todd S; Smith I; Klein R; Woodhouse IB; Riddell S; Crameri G; Bingham J; Wang L-F; Purcell AW; Middleton D; Baker ML Acute Experimental Infection of Bats and Ferrets with Hendra Virus: Insights into the Early Host Response of the Reservoir Host and Susceptible Model Species. PLoS Pathog. 2020, 16 (3), e1008412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Greenhall AM Natural History of Vampire Bats; CRC Press, 2018. [Google Scholar]
- (50).Bobrowiec PED; Lemes MR; Gribel R. Prey Preference of the Common Vampire Bat (Desmodus rotundus, Chiroptera) Using Molecular Analysis. J. Mammal 2015, 96 (1), 54–63. [Google Scholar]
- (51).Streicker DG; Allgeier JE Foraging Choices of Vampire Bats in Diverse Landscapes: Potential Implications for Land-Use Change and Disease Transmission. J. Appl. Ecol 2016, 53 (4), 1280–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Schneider MC; Romijn PC; Uieda W; Tamayo H; da Silva DF; Belotto A; da Silva JB; Leanes LF Rabies Transmitted by Vampire Bats to Humans: An Emerging Zoonotic Disease in Latin America? Rev. Panam. Salud Publica 2009, 25 (3), 260–269. [DOI] [PubMed] [Google Scholar]
- (53).Condori-Condori RE; Streicker DG; Cabezas-Sanchez C; Velasco-Villa A. Enzootic and Epizootic Rabies Associated with Vampire Bats, Peru. Emerging Infect. Dis 2013, DOI: 10.3201/eid1909.130083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Wray AK; Olival KJ; Morán D; Lopez MR; Alvarez D; Navarrete-Macias I; Liang E; Simmons NB; Lipkin WI; Daszak P; Anthony SJ Viral Diversity, Prey Preference, and Bartonella Prevalence in Desmodus rotundus in Guatemala. Ecohealth 2016, 13 (4), 761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Patterson C. Deforestation, Agricultural Intensification, and Farm Resilience in Northern Belize: 1980–2010; University of Otago, 2016. [Google Scholar]
- (56).Becker DJ; Broos A; Bergner LM; Meza DK; Simmons NB; Fenton MB; Altizer S; Streicker DG Temporal Patterns of Vampire Bat Rabies and Host Connectivity in Belize. Transboundary Emerging Dis. 2020, DOI: 10.1111/tbed.13754. [DOI] [Google Scholar]
- (57).Drexler JF; Corman VM; Müller MA; Maganga GD; Vallo P; Binger T; Gloza-Rausch F; Cottontail VM; Rasche A; Yordanov S; Seebens A; Knörnschild M; Oppong S; Adu Sarkodie Y; Pongombo C; Lukashev AN; Schmidt-Chanasit J; Stöcker A; Carneiro AJB; Erbar S; Maisner A; Fronhoffs F; Buettner R; Kalko EKV; Kruppa T; Franke CR; Kallies R; Yandoko ERN; Herrler G; Reusken C; Hassanin A; Krüger, D. H.; Matthee, S.; Ulrich, R. G.; Leroy, E. M.; Drosten, C. Bats Host Major Mammalian Paramyxoviruses. Nat. Commun 2012, 3, 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Escalera-Zamudio M; Taboada B; Rojas-Anaya E; Löber U; Loza-Rubio E; Arias CF; Greenwood AD Viral Communities Among Sympatric Vampire Bats and Cattle. Ecohealth 2018, 15 (1), 132–142. [DOI] [PubMed] [Google Scholar]
- (59).de Araujo J; Lo MK; Tamin A; Ometto TL; Thomazelli LM; Nardi MS; Hurtado RF; Nava A; Spiropoulou CF; Rota PA; Durigon EL Antibodies Against Henipa-Like Viruses in Brazilian Bats. Vector Borne Zoonotic Dis. 2017, 17 (4), 271–274. [DOI] [PubMed] [Google Scholar]
- (60).Bergner LM; Orton RJ; Broos A; Tello C; Becker DJ; Carrera JE; Patel AH; Biek R; Streicker DG Diversification of Mammalian Deltaviruses by Host Shifting. Proc. Natl. Acad. Sci. U. S. A 2021, 118, e2019907118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Asano KM; Hora AS; Scheffer KC; Fahl WO; Iamamoto K; Mori E; Brandão PE Erratum to: Alphacoronavirus in Urban Molossidae and Phyllostomidae Bats, Brazil. Virol. J 2016, 13 (1), 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Brandao PE; Scheffer K; Villarreal LY; Achkar S; Oliveira R; de N; Fahl W; de O; Castilho JG; Kotait I; Richtzenhain LJ A Coronavirus Detected in the Vampire Bat Desmodus rotundus. Braz. J. Infect. Dis 2008, 12 (6), 466–468. [DOI] [PubMed] [Google Scholar]
- (63).Quan P-L; Firth C; Conte JM; Williams SH; Zambrana-Torrelio CM; Anthony SJ; Ellison JA; Gilbert AT; Kuzmin IV; Niezgoda M; Osinubi MOV; Recuenco S; Markotter W; Breiman RF; Kalemba L; Malekani J; Lindblade KA; Rostal MK; Ojeda-Flores R; Suzan G; Davis LB; Blau DM; Ogunkoya AB; Alvarez Castillo DA; Moran D; Ngam S; Akaibe D; Agwanda B; Briese T; Epstein JH; Daszak P; Rupprecht CE; Holmes EC; Lipkin WI Bats Are a Major Natural Reservoir for Hepaciviruses and Pegiviruses. Proc. Natl. Acad. Sci. U. S. A 2013, 110 (20), 8194–8199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).James S; Donato D; de Thoisy B; Lavergne A; Lacoste V. Novel Herpesviruses in Neotropical Bats and Their Relationship with Other Members of the Herpesviridae Family. Infect., Genet. Evol 2020, 84, 104367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Chen L; Liu B; Yang J; Jin Q. DBatVir: The Database of Bat-Associated Viruses. Database 2014, 2014, bau021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Delpietro HA; Russo RG Observations of the Common Vampire Bat (Desmodus rotundus) and the Hairy-Legged Vampire Bat (Diphylla ecaudata) in Captivity. Mamm. Biol 2002, 67 (2), 65–78. [Google Scholar]
- (67).Perez-Riverol Y; Csordas A; Bai J; Bernal-Llinares M; Hewapathirana S; Kundu DJ; Inuganti A; Griss J; Mayer G; Eisenacher M; Pérez E; Uszkoreit J; Pfeuffer J; Sachsenberg T; Yilmaz S; Tiwary S; Cox J; Audain E; Walzer M; Jarnuczak AF; Ternent T; Brazma A; Vizcaíno JA The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 2019, 47 (D1), D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Camacho C; Coulouris G; Avagyan V; Ma N; Papadopoulos J; Bealer K; Madden TL BLAST+: Architecture and Applications. BMC Bioinf. 2009, 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Dunkelberger JR; Song W-C Complement and Its Role in Innate and Adaptive Immune Responses. Cell Res. 2010, 20 (1), 34–50. [DOI] [PubMed] [Google Scholar]
- (70).Nordahl EA; Rydengård V; Nyberg P; Nitsche DP; Mörgelin M; Malmsten M; Björck L; Schmidtchen A. Activation of the Complement System Generates Antibacterial Peptides. Proc. Natl. Acad. Sci. U. S. A 2004, 101 (48), 16879–16884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Ricklin D; Hajishengallis G; Yang K; Lambris JD Complement: A Key System for Immune Surveillance and Homeostasis. Nat. Immunol 2010, 11 (9), 785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Moore MS; Reichard JD; Murtha TD; Zahedi B; Fallier RM; Kunz TH Specific Alterations in Complement Protein Activity of Little Brown Myotis (Myotis lucifugus) Hibernating in White-Nose Syndrome Affected Sites. PLoS One 2011, 6 (11), e27430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Pilosof S; Korine C; Moore MS; Krasnov BR Effects of Sewage-Water Contamination on the Immune Response of a Desert Bat. Mamm. Biol 2014, 79 (3), 183–188. [Google Scholar]
- (74).Anderson NL; Anderson NG The Human Plasma Proteome: History, Character, and Diagnostic Prospects. Mol. Cell. Proteomics 2002, 1 (11), 845–867. [DOI] [PubMed] [Google Scholar]
- (75).The Human Protein Atlas https://www.proteinatlas.org/humanproteome/blood/proteins+detected+in+ms (accessed Jul 19, 2020).
- (76).Geyer PE; Voytik E; Treit PV; Doll S; Kleinhempel A; Niu L; Müller JB; Buchholtz M-L; Bader JM; Teupser D; Holdt LM; Mann M. Plasma Proteome Profiling to Detect and Avoid Sample-Related Biases in Biomarker Studies. EMBO Mol. Med 2019, 11, e10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Correa-Giron P; Calisher CH; Baer GM Epidemic Strain of Venezuelan Equine Encephalomyelitis Virus from a Vampire Bat Captured in Oaxaca, Mexico, 1970. Science 1972, 175 (4021), 546–547. [DOI] [PubMed] [Google Scholar]
- (78).Megger DA; Philipp J; Le-Trilling VTK; Sitek B; Trilling M. Deciphering of the Human Interferon-Regulated Proteome by Mass Spectrometry-Based Quantitative Analysis Reveals Extent and Dynamics of Protein Induction and Repression. Front. Immunol 2017, DOI: 10.3389/fimmu.2017.01139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Krapp C; Hotter D; Gawanbacht A; McLaren PJ; Kluge SF; Stürzel CM; Mack K; Reith E; Engelhart S; Ciuffi A; Hornung V; Sauter D; Telenti A; Kirchhoff F. Guanylate Binding Protein (GBP) 5 Is an Interferon-Inducible Inhibitor of HIV-1 Infectivity. Cell Host Microbe 2016, 19 (4), 504–514. [DOI] [PubMed] [Google Scholar]
- (80).Braun E; Hotter D; Koepke L; Zech F; Groß R; Sparrer KMJ; Müller JA; Pfaller CK; Heusinger E; Wombacher R; Sutter K; Dittmer U; Winkler M; Simmons G; Jakobsen MR; Conzelmann K-K; Pöhlmann S; Münch J; Fackler OT; Kirchhoff F; Sauter D. Guanylate-Binding Proteins 2 and 5 Exert Broad Antiviral Activity by Inhibiting Furin-Mediated Processing of Viral Envelope Proteins. Cell Rep. 2019, 27, 2092–2104. [DOI] [PubMed] [Google Scholar]
- (81).Tretina K; Park E-S; Maminska A; MacMicking JD Interferon-Induced Guanylate-Binding Proteins: Guardians of Host Defense in Health and Disease. J. Exp. Med 2019, 216 (3), 482–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Yao Z; Jia X; Megger DA; Chen J; Liu Y; Li J; Sitek B; Yuan Z. Label-Free Proteomic Analysis of Exosomes Secreted from THP-1-Derived Macrophages Treated with IFN-α Identifies Antiviral Proteins Enriched in Exosomes. J. Proteome Res 2019, 18 (3), 855–864. [DOI] [PubMed] [Google Scholar]
- (83).Ligtenberg AJM; Karlsson NG; Veerman ECI Deleted in Malignant Brain Tumors-1 Protein (DMBT1): A Pattern Recognition Receptor with Multiple Binding Sites. Int. J. Mol. Sci 2010, 11 (12), 5212–5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Bikker FJ; Ligtenberg AJM; Nazmi K; Veerman ECI; van’t Hof W; Bolscher JGM; Poustka A; Nieuw Amerongen AV; Mollenhauer J. Identification of the Bacteria-Binding Peptide Domain on Salivary Agglutinin (gp-340/DMBT1), a Member of the Scavenger Receptor Cysteine-Rich Superfamily. J. Biol. Chem 2002, 277 (35), 32109–32115. [DOI] [PubMed] [Google Scholar]
- (85).Hartshorn KL; Ligtenberg A; White MR; Van Eijk M; Hartshorn M; Pemberton L; Holmskov U; Crouch E. Salivary Agglutinin and Lung Scavenger Receptor Cysteine-Rich Glycoprotein 340 Have Broad Anti-Influenza Activities and Interactions with Surfactant Protein D That Vary according to Donor Source and Sialylation. Biochem. J 2006, 393 (2), 545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).McCarthy MK; Weinberg JB The Immunoproteasome and Viral Infection: A Complex Regulator of Inflammation. Front. Microbiol. 2015, 6, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Deshmukh FK; Yaffe D; Olshina MA; Ben-Nissan G; Sharon M. The Contribution of the 20S Proteasome to Proteostasis. Biomolecules 2019, 9, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Bayram HL; Claydon AJ; Brownridge PJ; Hurst JL; Mileham A; Stockley P; Beynon RJ; Hammond DE Cross-Species Proteomics in Analysis of Mammalian Sperm Proteins. J. Proteomics 2016, 135, 38–50. [DOI] [PubMed] [Google Scholar]
- (89).Lai RC; Tan SS; Teh BJ; Sze SK; Arslan F; de Kleijn DP; Choo A; Lim SK Proteolytic Potential of the MSC Exosome Proteome: Implications for an Exosome-Mediated Delivery of Therapeutic Proteasome. Int. J. Proteomics 2012, 971907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Neely BA; Ferrante JA; Mauro Chaves J; Soper JL; Almeida JS; Arthur JM; Gulland FMD; Janech MG Proteomic Analysis of Plasma from California Sea Lions (Zalophus californianus) Reveals Apolipoprotein E as a Candidate Biomarker of Chronic Domoic Acid Toxicosis. PLoS One 2015, 10, e0123295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Zhang G; Cowled C; Shi Z; Huang Z; Bishop-Lilly KA; Fang X; Wynne JW; Xiong Z; Baker ML; Zhao W; Tachedjian M; Zhu Y; Zhou P; Jiang X; Ng J; Yang L; Wu L; Xiao J; Feng Y; Chen Y; Sun X; Zhang Y; Marsh GA; Crameri G; Broder CC; Frey KG; Wang L-F; Wang J. Comparative Analysis of Bat Genomes Provides Insight into the Evolution of Flight and Immunity. Science 2013, 339 (6118), 456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Brunet-Rossinni AK Reduced Free-Radical Production and Extreme Longevity in the Little Brown Bat (Myotis lucifugus) versus Two Non-Flying Mammals. Mech. Ageing Dev 2004, 125 (1), 11–20. [DOI] [PubMed] [Google Scholar]
- (93).Min-Oo G; Ayi K; Bongfen SE; Tam M; Radovanovic I; Gauthier S; Santiago H; Rothfuchs AG; Roffe E; Sher A; Mullick A; Fortin A; Stevenson MM; Kain KC; Gros P. Cysteamine, the Natural Metabolite of Pantetheinase, Shows Specific Activity against Plasmodium. Exp. Parasitol 2010, 125 (4), 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Pitari G; Malergue F; Martin F; Philippe JM; Massucci MT; Chabret C; Maras B; Duprè S; Naquet P; Galland F. Pantetheinase Activity of Membrane-Bound Vanin-1: Lack of Free Cysteamine in Tissues of Vanin-1 Deficient Mice. FEBS Lett 2000, 483 (2–3), 149–154. [DOI] [PubMed] [Google Scholar]
- (95).Bartucci R; Salvati A; Olinga P; Boersma YL Vanin 1: Its Physiological Function and Role in Diseases. Int. J. Mol. Sci 2019, 20, 3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Tian X; Azpurua J; Hine C; Vaidya A; Myakishev-Rempel M; Ablaeva J; Mao Z; Nevo E; Gorbunova V; Seluanov A. High-Molecular-Mass Hyaluronan Mediates the Cancer Resistance of the Naked Mole Rat. Nature 2013, 499 (7458), 346–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Lin C-Y; Kolliopoulos C; Huang C-H; Tenhunen J; Heldin C-H; Chen Y-H; Heldin P. High Levels of Serum Hyaluronan Is an Early Predictor of Dengue Warning Signs and Perturbs Vascular Integrity. EBioMedicine 2019, 48, 425–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Alspach E; Lussier DM; Schreiber RD Interferon γ and Its Important Roles in Promoting and Inhibiting Spontaneous and Therapeutic Cancer Immunity. Cold Spring Harbor Perspect. Biol. 2019, 11, a028480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Letko M; Miazgowicz K; McMinn R; Seifert SN; Sola I; Enjuanes L; Carmody A; van Doremalen N; Munster V. Adaptive Evolution of MERS-CoV to Species Variation in DPP4. Cell Rep. 2018, 24 (7), 1730–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Anthony SJ; Ojeda-Flores R; Rico-Chávez O; Navarrete-Macias I; Zambrana-Torrelio CM; Rostal MK; Epstein JH; Tipps T; Liang E; Sanchez-Leon M; Sotomayor-Bonilla J; Aguirre AA; Ávila-Flores R; Medellín RA; Goldstein T; Suzán G; Daszak P; Lipkin WI Coronaviruses in Bats from Mexico. J. Gen. Virol 2013, 94 (5), 1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Orsburn BC; Jenkins C; Miller SM; Neely BA; Bumpus NN In Silico Approach toward the Identification of Unique Peptides from Viral Protein Infection: Application to COVID-19. bioRxiv, Apr 10, 2020. DOI: 10.1101/2020.03.08.980383. [DOI] [Google Scholar]
- (102).Bezstarosti K; Lamers MM; Haagmans BL; Demmers JA A. Targeted Proteomics for the Detection of SARS-CoV-2 Proteins. bioRxiv, Dec 18, 2020. DOI: 10.1101/2020.04.23.057810. [DOI] [Google Scholar]
- (103).Ihling C; Tänzler D; Hagemann S; Kehlen A; ttelmaier S; Arlt C; Sinz A. Mass Spectrometric Identification of SARS-CoV-2 Proteins from Gargle Solution Samples of COVID-19 Patients. J. Proteome Res 2020, 19, 4389. [DOI] [PubMed] [Google Scholar]
- (104).Gouveia D; Miotello G; Gallais F; Gaillard J-C; Debroas S; Bellanger L; Lavigne J-P; Sotto A; Grenga L; Pible O; Armengaud J. Proteotyping SARS-CoV-2 Virus from Nasopharyngeal Swabs: A Proof-of-Concept Focused on a 3 min Mass Spectrometry Window. J. Proteome Res 2020, 19, 4407. [DOI] [PubMed] [Google Scholar]
- (105).Neely BA; Carlin KP; Arthur JM; McFee WE; Janech MG Ratiometric Measurements of Adiponectin by Mass Spectrometry in Bottlenose Dolphins (Tursiops truncatus) with Iron Overload Reveal an Association with Insulin Resistance and Glucagon. Front. Endocrinol 2013, 4, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Shi T; Song E; Nie S; Rodland KD; Liu T; Qian W-J; Smith RD Advances in Targeted Proteomics and Applications to Biomedical Research. Proteomics 2016, 16 (15–16), 2160–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Meyer JG; Schilling B. Clinical Applications of Quantitative Proteomics Using Targeted and Untargeted Data-Independent Acquisition Techniques. Expert Rev. Proteomics 2017, 14 (5), 419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Ting YS; Egertson JD; Payne SH; Kim S; MacLean B; Käll L; Aebersold R; Smith RD; Noble WS; MacCoss MJ Peptide-Centric Proteome Analysis: An Alternative Strategy for the Analysis of Tandem Mass Spectrometry Data. Mol. Cell. Proteomics 2015, 14 (9), 2301–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Bruderer R; Muntel J; Mü S; Bernhardt OM; Gandhi T; Cominetti O; Macron C; Carayol J; Rinner O; Astrup A; Saris WHM; Hager J; Valsesia A; Dayon L; Reiter L. Analysis of 1508 Plasma Samples by Capillary-Flow Data-Independent Acquisition Profiles Proteomics of Weight Loss and Maintenance. Mol. Cell. Proteomics 2019, 18 (6), 1242–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Parker SJ; Rost H; Rosenberger G; Collins BC; Malmström L; Amodei D; Venkatraman V; Raedschelders K; Van Eyk JE; Aebersold R. Identification of a Set of Conserved Eukaryotic Internal Retention Time Standards for Data-Independent Acquisition Mass Spectrometry. Mol. Cell. Proteomics 2015, 14 (10), 2800–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Lilley TM; Prokkola JM; Blomberg AS; Paterson S; Johnson JS; Turner GG; Bartonicka T; Bachorec E; Reeder DM; Field KA Resistance Is Futile: RNA-Sequencing Reveals Differing Responses to Bat Fungal Pathogen in Nearctic Myotis lucifugus and Palearctic Myotis myotis. Oecologia 2019, 191 (2), 295–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Heck M; Neely BA Proteomics in Non-Model Organisms: A New Analytical Frontier. J. Proteome Res 2020, 19 (9), 3595–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.