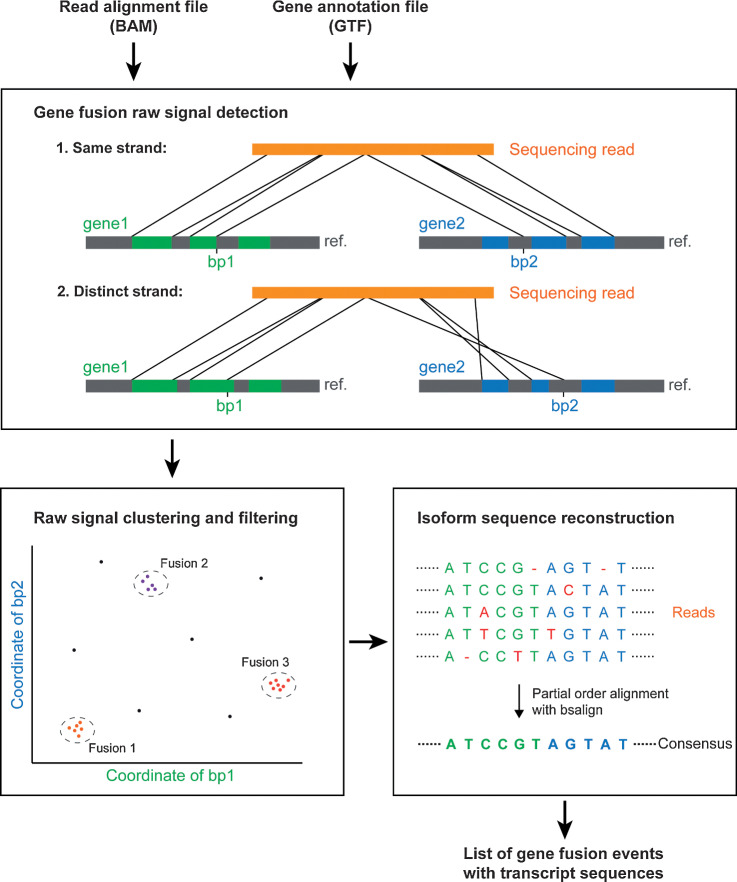
FusionSeeker is a new method to discover gene fusions and reconstruct fused transcript sequences in long-read cancer transcriptome sequencing data to help identify novel gene fusions important for tumorigenesis and progression.
Abstract
Gene fusions are prevalent in a wide array of cancer types with different frequencies. Long-read transcriptome sequencing technologies, such as PacBio, Iso-Seq, and Nanopore direct RNA sequencing, provide full-length transcript sequencing reads, which could facilitate detection of gene fusions. In this work, we developed a method, FusionSeeker, to comprehensively characterize gene fusions in long-read cancer transcriptome data and reconstruct accurate fused transcripts from raw reads. FusionSeeker identified gene fusions in both exonic and intronic regions, allowing comprehensive characterization of gene fusions in cancer transcriptomes. Fused transcript sequences were reconstructed with FusionSeeker by correcting sequencing errors in the raw reads through partial order alignment algorithm. Using these accurate transcript sequences, FusionSeeker refined gene fusion breakpoint positions and predicted breakpoints at single bp resolution. Overall, FusionSeeker will enable users to discover gene fusions accurately using long-read data, which can facilitate downstream functional analysis as well as improved cancer diagnosis and treatment.
Significance:
FusionSeeker is a new method to discover gene fusions and reconstruct fused transcript sequences in long-read cancer transcriptome sequencing data to help identify novel gene fusions important for tumorigenesis and progression.
Introduction
Gene fusions are recognized as important cancer-driving events for over 30 years (1). They often play critical roles in tumorigenesis and progression and sometimes serve as therapeutic targets (2). A large number of tools have been developed and applied to short-read cancer transcriptome sequencing data for gene fusion detection. However, it is always challenging to identify chimeric reads or discordant read pairs that represent gene fusions from short reads, especially given the innate splicing structures of isoforms. Recent development of long-read RNA sequencing technologies enables full-length transcript sequencing and may alleviate these issues, therefore showing great potential in gene fusion detection. However, only two tools, JAFFAL (3) and LongGF (4), are currently available for long-read gene fusion detection, and their performance is limited when detecting gene fusions occurred in intronic regions. Accurate sequences of the reported gene fusions also remain unknown, which limits further functional analysis of identified gene fusions.
Here, we present FusionSeeker, a long-read gene fusion caller to accurately identify gene fusion events and reconstruct their transcript sequences. FusionSeeker takes read alignment file and gene annotation file as input and outputs a list of confident gene fusions and their transcript sequences (Fig. 1). It first scans the read alignments for candidate fusions when a single read is aligned to two or more genes. Candidate fusions are then grouped according to these genes and clustered with the density-based spatial clustering of applications with noise (DBSCAN) algorithm into gene fusion calls. The gene fusion calls are filtered on the basis of the number of supporting reads to remove noise signals caused by sequencing errors and incorrect read alignments. FusionSeeker then performs a partial order alignment (POA) using fusion-containing reads to generate a consensus transcript sequence for each confident gene fusion.
Figure 1.
Workflow of FusionSeeker. FusionSeeker scans the input file of the read alignments for split read alignments and records candidate fusions of gene fusions when two segments from one read are aligned to two distinct genes. It then clusters the candidate fusions into gene fusion calls and removes noise calls supported by only a few reads. For each fusion call, FusionSeeker generates a consensus transcript sequence by performing a POA with fusion-containing reads. The final output of FusionSeeker includes a list of confident gene fusion events and corresponding transcript sequences.
Materials and Methods
Gene fusion candidate detection
FusionSeeker first scans all read alignments for split-read patterns. To quickly annotate read alignments, FusionSeeker generates a list containing the coordinates of each gene and its exons on every chromosome based on the input genome annotation file (GTF). Input BAM file is then processed chromosome by chromosome. Reads with only one alignment are skipped to reduce computational burden. For reads with multiple alignments (with SA tags), FusionSeeker annotates each alignment and records essential information, including chromosome, alignment start and end positions, length of clipped sequences on both sides, read name, strand, mapping quality, simplified CIGAR tag, etc. As candidate fusion detection process is the most time-consuming step, FusionSeeker can process each chromosome in parallel to reduce the overall runtime. After all alignments are processed, FusionSeeker checks the alignment information from the same read and reports a candidate fusion when:
(i) two breakpoints from one read are annotated to two distinct genes (Gene A and Gene B),
(ii) length of alignment is longer than 100 bp on both genes,
(iii) length of overlap between two alignments (the part of read sequences present in both alignments) is shorter than 100 bp and 50% of the shorter alignment,
(iv) coordinates of Gene A and Gene B do not overlap in the GTF file,
(v) Gene A is not an antisense sequence of Gene B.
Gene fusion signal clustering and filtering
Candidate fusions are first grouped on the basis of gene names, for instance, Gene A and Gene B. The candidate fusions from the same pair of fused genes are then clustered on the basis of the breakpoint positions on the two genes. To achieve this, a density-based spatial clustering of applications with noise algorithm (DBSCAN) is adopted to cluster the candidate fusions, with a default of maximal distance of 20 bp for high accuracy reads and 40 bp for noisy reads. Next, the candidate fusions from each cluster are merged into a gene fusion call, with temporary breakpoint positions as the mean values from the candidate fusions. All gene fusion calls are then filtered on the basis of the number of fusion-supporting reads. By default, the cutoff of minimal supporting reads Nmin is set as Nmin = Ncan/50,000 + 3, where Ncan is the total number of the candidate fusions detected in the input dataset. Fusion calls supported by more than Nmin reads are reported as confident gene fusion calls.
Fused transcript reconstruction and breakpoint refinement
For each gene fusion event, FusionSeeker extracts the sequences of the fusion-supporting reads from the BAM file and writes into a new FASTQ file. It then performs POA for each call independently using bsalign (https://github.com/ruanjue/bsalign). All consensus sequences generated from POA are combined into a FASTA file and linked to each gene fusion call with its ID. When a reference genome is provided, FusionSeeker then aligns all the transcript sequences to the reference genome with minimap2 (5). The precise breakpoint positions of each gene fusion call are inferred from the transcript sequence alignment and used to replace the temporary positions inferred from the candidate fusions.
Data availability
The source code of FusionSeeker is available at https://github.com/Maggi-Chen/FusionSeeker and https://codeocean.com/capsule/3525117/tree/v1 under MIT license, and the scripts used for benchmark in the article are available at https://github.com/Maggi-Chen/FS_code. The Nanopore direct RNA sequencing data of the MCF7 and HCT116 cell lines are available at https://github.com/GoekeLab/sg-nex-data/ (6). The PacBio Iso-Seq sequencing data of the MCF7 and the HCT116 cell lines are available at Sequence Read Archive (SRA) under the accessions SRP055913 (7) and SRP091981 (8). The PacBio Iso-Seq and continuous long read sequencing data of the SKBR3 cell line are downloaded from SRA under accession SRP150606 (9). PacBio Iso-Seq data of Human Genome Structural Variation Consortium (HGSVC) samples are available at HGSVC data portal (https://www.internationalgenome.org/data-portal/). Acute myeloid leukemia (AML) patient data are downloaded from SRA under SRR12048357 (4).
https://codeocean.com/widget.js?slug=3525117
Simulation and benchmark methods can be found in Supplementary Data S2.
Results
Benchmark gene fusion detection on the simulated datasets
We first benchmarked the accuracy of gene fusion detection of FusionSeeker on the simulated datasets. A total of 150 gene fusion transcripts (100 with breakpoints in exons, 50 in introns) were randomly generated and assigned to different expression levels (10×, 50×, and 100×). PacBio Iso-Seq–like and Nanopore-like reads were simulated with pbsim (10) and Badread (v0.2.0; ref. 11) and then aligned to the reference genome. FusionSeeker and another two long-read gene fusion callers, JAFFAL and LongGF, were used to detect gene fusions from the simulated reads. We repeated the simulation for three times, and FusionSeeker consistently achieved the highest F1 score among the three tools in both Iso-Seq and Nanopore datasets (Table 1). In all three simulated datasets, FusionSeeker identified more true-positive events than the other two tools, with slightly more false-positive calls than LongGF (Supplementary Fig. S1). The higher recall of FusionSeeker was mainly beneficial from its ability to detect gene fusions located in intronic regions, where FusionSeeker identified 94.67% of intronic events while JAFFAL and LongGF only reported 14.67% and 54.67%, respectively, using Iso-Seq data (Table 1; Supplementary Table S1). In general, all three fusion callers achieved higher recall in detecting fusions with high and medium expression levels than fusions with low expression level (Supplementary Table S2). Approximately 67% of the gene fusions missed by FusionSeeker were from the low-expression-level group, and the missing was caused by the low coverage of reads.
Table 1.
The accuracy of gene fusion detection on the simulated datasets.
| FusionSeeker | JAFFAL | LongGF | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Recall | Precision | F1 score | Recall | Precision | F1 score | Recall | Precision | F1 score | |
| Iso-Seq | |||||||||
| Exonic | 96.00 | 96.88 | 96.32 | 69.33 | 96.57 | 80.72 | 96.00 | 97.03 | 96.51 |
| Intronic | 94.67 | 90.00 | 92.28 | 14.67 | 66.67 | 24.04 | 54.67 | 94.87 | 69.36 |
| Total | 95.56 | 93.89 | 94.71 | 51.11 | 82.73 | 63.15 | 82.22 | 96.14 | 88.58 |
| Nanopore | |||||||||
| Exonic | 99.00 | 94.98 | 96.95 | 73.00 | 96.35 | 83.06 | 98.00 | 96.70 | 97.35 |
| Intronic | 99.33 | 78.72 | 87.84 | 18.00 | 53.81 | 26.98 | 56.67 | 91.80 | 70.08 |
| Total | 99.11 | 87.65 | 93.03 | 54.67 | 82.23 | 65.62 | 84.22 | 95.26 | 89.36 |
Note: Recall, precision, and F1 score in the table are the mean values of three replicate simulation datasets. Highest recall, precision, and F1 score among the three fusion callers are marked as bold.
We then evaluated the fused transcript sequences generated by FusionSeeker. To generate high-accuracy transcript sequences, FusionSeeker performs a POA using fusion-containing reads and calculates a consensus sequence for each gene fusion event. In the simulated datasets, FusionSeeker reconstructed full-length fused transcripts for more than 99.5% of events, with average sequence identities of 99.87% and 99.14% using Iso-Seq and Nanopore reads, respectively (Supplementary Table S3). When aligned to the reference genome, the FusionSeeker transcript sequences showed a better identity than raw reads (Supplementary Fig. S2). Taken together, we have demonstrated that FusionSeeker can accurately identify gene fusions and report full-length fused transcript sequences in the simulated datasets.
Gene fusion discovery in cancer transcriptomes
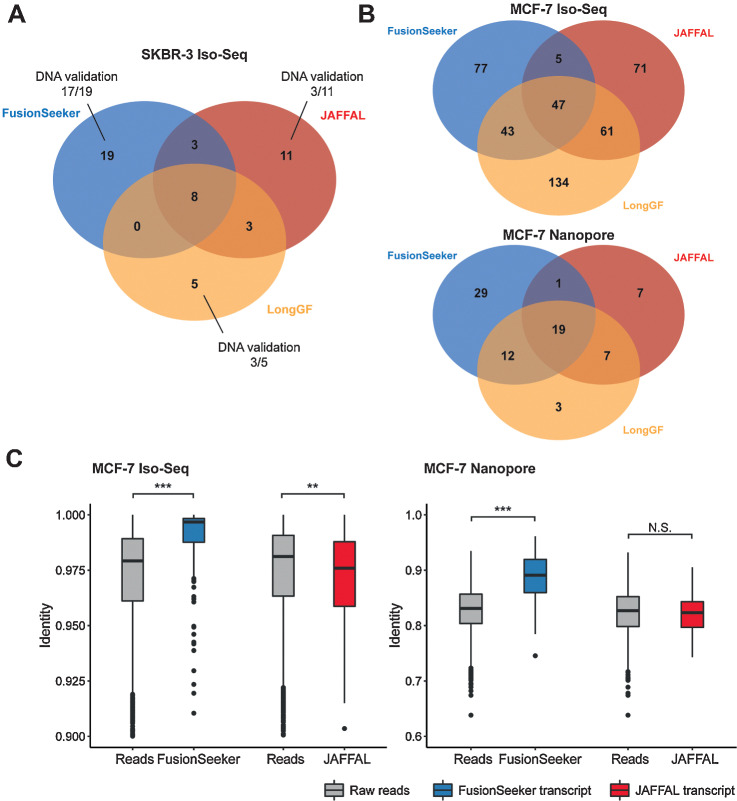
We then applied the three gene fusion callers on three cancer cell lines, SKBR3, MCF7, and HCT116. The PacBio Iso-Seq and Nanopore reads of each cell line were downloaded and aligned to the human reference genome (6–9). In the SKBR3 cell line, FusionSeeker identified 31 gene fusions, among which 15 events have been previously discovered and validated (Table 2; refs. 9, 12–14). Three of the previous studies for gene fusion detection in SKBR3 were based on short-read RNA sequencing data (12–14), except the Nattestad and colleagues (9) which used the PacBio Iso-Seq dataset. Tested on these Iso-seq data, FusionSeeker showed a better consistency with Nattestad and colleagues than the other short-read results (Supplementary Table S4). JAFFAL and LongGF identified 13 and 10 previously validated gene fusions, respectively. Comparing the gene fusion lists of three callers, eight gene fusions were reported by all the three tools, three gene fusions were reported by both FusionSeeker and JAFFAL, and three gene fusions were reported by both JAFFAL and LongGF (Fig. 2A). There were 19 FusionSeeker-unique, 11 JAFFAL-unique, and five LongGF-unique events. We cross-validated these unique gene fusion events with long-read DNA sequencing data and considered a gene fusion as validated when at least three DNA sequencing reads were aligned to both genes (Supplementary Fig. S3A and S3B). A total of 17 of 19 (89.47%) FusionSeeker-unique gene fusions were validated by DNA sequencing, which was higher than JAFFAL (3/11, 27.27%) and LongGF (3/5, 60.00%). In particular, with further investigation, we observed a 4-hop intronic gene fusion from FusionSeeker-unique calls, CSNK2A1:NCOA3:MMP24OS:TSHZ2, which was also supported by DNA sequencing data (Supplementary Fig. S4A and S4B).
Table 2.
Detection of previously validated gene fusions in cancer cell lines.
| FusionSeeker | JAFFAL | LongGF | |||||
|---|---|---|---|---|---|---|---|
| Cell line | Data type | Reported | Previously validate | Reported | Previously validate | Reported | Previously validated |
| SKBR3 | |||||||
| Iso-Seq | 30 | 15 | 25 | 13 | 16 | 10 | |
| MCF7 | |||||||
| Iso-Seq | 172 | 21 | 184 | 23 | 285 | 20 | |
| Nanopore | 61 | 20 | 34 | 18 | 41 | 20 | |
| HCT116 | |||||||
| Iso-Seq | 3 | 1 | 2 | 1 | 2 | 1 | |
| Nanopore | 17 | 1 | 12 | 1 | 10 | 1 | |
Note: Reported, number of gene fusions reported by each fusion caller. Previously validated, number of previously validated gene fusions detected by each fusion caller. Bold, the highest number of previously validated gene fusions reported among the three tools.
Figure 2.
Gene fusion discovery in cancer cell lines. A, Venn diagram of gene fusion calls by FusionSeeker, JAFFAL, and LongGF in SKBR3 cell line. B, Venn diagrams of gene fusion calls by the three fusion callers in MCF7 cell line using Iso-Seq (top) and Nanopore direct RNA-sequencing (bottom) data. C, The identity of raw reads and transcript sequences reported by FusionSeeker and JAFFAL in MCF7 Iso-Seq (left) and Nanopore (right) direct RNA-sequencing dataset. **, P < 0.01. ***, P < 0.001. N.S., not significant. The P values were calculated by Mann–Whitney U test.
In MCF7 cell line, FusionSeeker identified 172 gene fusions in Iso-Seq dataset and 61 gene fusions in Nanopore dataset (Table 2), with 21 and 20 previously validated gene fusions identified using Iso-Seq and Nanopore datasets, respectively (Supplementary Table S5). In HCT116 cell line, FusionSeeker reported three and 17 gene fusions in Iso-Seq and Nanopore dataset, respectively. In particular, a previously known gene fusion, TXLNG:SYAP1, in MCF7 cell line has two validated alternative breakpoint positions in TXLNG, with one located in the first exon and the other located in the first intron of TXLNG (14). FusionSeeker reported both exonic and intronic breakpoints for this fusion event, while JAFFAL and LongGF only reported the exonic breakpoint and missed the intronic breakpoint (Supplementary Fig. S5). The few previously validated events detected by JAFFAL but not by FusionSeeker were supported by ≤4 reads and therefore failed to pass the filter of FusionSeeker (Supplementary Table S6). When comparing gene fusion callsets of the three callers, 47 and 19 gene fusions were reported by all three callers in Iso-Seq and Nanopore dataset, respectively (Fig. 2B). Gene fusions reported by JAFFAL or LongGF but not by FusionSeeker were usually supported by fewer reads, with 88.35% of them supported by ≤3 reads in MCF7 Iso-Seq dataset (Supplementary Fig. S6). Within the 77 and 29 FusionSeeker-unique calls in MCF7 Iso-Seq and Nanopore dataset, we designed PCR primers for 10 most confident novel events and validated seven of them using RNA extracted from MCF7 cell line (Supplementary Table S7). All four events discovered in both Iso-Seq and Nanopore datasets were validated by PCR.
When comparing two lists of gene fusion calls from Iso-Seq and Nanopore datasets for each caller, we observed slightly higher overlapping ratio in FusionSeeker callsets than JAFFAL and LongGF, with Jaccard index of 0.1208 for FusionSeeker, 0.0741 for JAFFAL, and 0.0584 for LongGF in MCF7 cell line, respectively (Supplementary Fig. S7). This overall low overlapping rate was probably caused by the evolution of the cell line or inconsistent sequencing depth on each gene in the two datasets during sequencing (Supplementary Fig. S8). This systemic difference may need further investigation.
We then applied three fusion callers on noncancer datasets from HGSVC to assess the FDR of three tools. In all the 12 noncancer samples, FusionSeeker reported the fewest number of gene fusions, suggesting that FusionSeeker had lowest FDRs among the three tested fusion callers (Supplementary Table S8). We have also applied FusionSeeker on a patient sample with AML to demonstrate its clinical utility (4). FusionSeeker identified a prevalidated gene fusion between RUNX1 and RUNX1T1 and reported another seven confident gene fusion events in the patient sample (Supplementary Table S9).
Isoform sequence reconstruction with de novo assembly
We next evaluated the transcript sequences generated by FusionSeeker. Compared with the raw reads, FusionSeeker transcript sequences showed significant higher identity with reference gene sequences in both Iso-Seq and Nanopore datasets of MCF7 cell line (Fig. 2C).
JAFFAL also reported one of the fusion-containing reads as the transcript sequence, which showed no significant difference in identity comparing with the raw reads. In the Iso-Seq dataset of the SKBR3 and the Nanopore dataset of the HCT116 cell lines, FusionSeeker reported more accurate transcript sequences than the raw reads, while transcript sequences reported by JAFFAL showed no significant differences (Supplementary Fig. S9A–S9C). There was no significant difference between FusionSeeker transcript sequences and raw reads in HCT116 Iso-Seq dataset, likely due to only three gene fusions were reported. Note that the identity calculated by comparing with the reference is an underestimation of transcript sequence accuracy, owing to the presence of genetic variants in these cell lines. These genetic variants can often be maintained in the transcript sequences (Supplementary Fig. S10A and S10B).
Discussion
In this work, we presented FusionSeeker for gene fusion detection in long-read cancer transcriptome sequencing data. FusionSeeker can detect gene fusions in both exonic and intronic regions. On the basis of simulation and three cancer cell line data, we have demonstrated that FusionSeeker outperformed existing methods in characterizing gene fusion events. Besides, we have both orthogonally and experimentally validated many gene fusion events only detected by FusionSeeker. These novel gene fusions may be important for tumorigenesis and progression, which deserves further investigation. Because the long-read sequencing platform can almost generate full-length transcripts, FusionSeeker provides accurate full-length fusion transcripts based on an assembly approach. The full-length fusion transcripts may facilitate downstream functional and clinical research.
After candidate fusion detection, FusionSeeker used DBSCAN to cluster candidate fusions that share the same breakpoints. DBSCAN was implanted as it does not require predetermined number of clusters, which allows FusionSeeker to report gene fusions with one or multiple breakpoints in the same gene pair. DBSCAN can also robustly exclude outliers while clustering, which is necessary in this case as there are often abundant noise signals in long-read RNA-sequencing read alignments.
Supplementary Material
Supplementary figure S1-S10, Supplementary table S1-S6 and S8-S9, Supplementary note 1-2.
Supplementary Table S7
Acknowledgments
The authors acknowledge Dr. Anna Sorace for providing MCF7 cell line. This work was supported by a grant from National Institute of General Medical Sciences (1R35GM138212), the BioData Catalyst Fellowship from National Heart, Lung, and Blood Institute (a subaward from 1OT3HL147154) to Z. Chong, and funding from Robert Reed Foundation to H. Chen.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
A list of members of Human Genome Structural Variation Consortium is in Supplementary Data S1.
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
Y. Chen: Software, formal analysis, validation, methodology, writing–original draft, writing–review and editing. Y. Wang: Software, methodology. W. Chen: Validation. Z. Tan: Data curation. Y. Song: Data curation, writing–original draft. N. Human Genome Structural Variation Consortium: Resources. H. Chen: Funding acquisition, validation. Z. Chong: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Edwards PA. Fusion genes and chromosome translocations in the common epithelial cancers. J Pathol 2010;220:244–54. [DOI] [PubMed] [Google Scholar]
- 2. Forsythe A, Zhang W, Phillip Strauss U, Fellous M, Korei M, Keating K. A systematic review and meta-analysis of neurotrophic tyrosine receptor kinase gene fusion frequencies in solid tumors. Ther Adv Med Oncol 2020;12:1758835920975613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davidson NM, Chen Y, Sadras T, Ryland GL, Blombery P, Ekert PG, et al. JAFFAL: detecting fusion genes with long-read transcriptome sequencing. Genome Biol 2022;23:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu Q, Hu Y, Stucky A, Fang L, Zhong JF, Wang K. LongGF: computational algorithm and software tool for fast and accurate detection of gene fusions by long-read transcriptome sequencing. BMC Genomics 2020;21:793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 2018;34:3094–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen Y, Davidson NM, Wan YK, Patel H, Yao F, Low HM, et al. A systematic benchmark of nanopore long read RNA sequencing for transcript level analysis in human cell lines. bioRxiv; 2021. doi: 10.1101/2021.04.21.440736. [DOI]
- 7. University of Iowa. Full-length transcripts of the MCF-7 breast cancer cell line by PacBio SMRT sequencing. SRP055913 [Internet]. Gene Expression Omnibus. 2015. Available from: https:/www.ncbi.nlm.nih.gov/sra/?term=SRP055913.
- 8. BC Cancer Research Centre. Transcriptome dynamics of CLK dependent exon recognition and conjoined gene formation revealed with a novel small molecule inhibitor; 2017 [DOI] [PMC free article] [PubMed]
- 9. Nattestad M, Goodwin S, Ng K, Baslan T, Sedlazeck FJ, Rescheneder P, et al. Complex rearrangements and oncogene amplifications revealed by long-read DNA and RNA sequencing of a breast cancer cell line. Genome Res 2018;28:1126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ono Y, Asai K, Hamada M. PBSIM: PacBio reads simulator–toward accurate genome assembly. Bioinformatics 2013;29:119–21. [DOI] [PubMed] [Google Scholar]
- 11. Wick RR. Badread: simulation of error-prone long reads. J Open Source Software 2019;4:1316. [Google Scholar]
- 12. Chen K, Navin NE, Wang Y, Schmidt HK, Wallis JW, Niu B, et al. BreakTrans: uncovering the genomic architecture of gene fusions. Genome Biol 2013;14:R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edgren H, Murumagi A, Kangaspeska S, Nicorici D, Hongisto V, Kleivi K, et al. Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol 2011;12:R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Inaki K, Hillmer AM, Ukil L, Yao F, Woo XY, Vardy LA, et al. Transcriptional consequences of genomic structural aberrations in breast cancer. Genome Res 2011;21:676–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure S1-S10, Supplementary table S1-S6 and S8-S9, Supplementary note 1-2.
Supplementary Table S7
Data Availability Statement
The source code of FusionSeeker is available at https://github.com/Maggi-Chen/FusionSeeker and https://codeocean.com/capsule/3525117/tree/v1 under MIT license, and the scripts used for benchmark in the article are available at https://github.com/Maggi-Chen/FS_code. The Nanopore direct RNA sequencing data of the MCF7 and HCT116 cell lines are available at https://github.com/GoekeLab/sg-nex-data/ (6). The PacBio Iso-Seq sequencing data of the MCF7 and the HCT116 cell lines are available at Sequence Read Archive (SRA) under the accessions SRP055913 (7) and SRP091981 (8). The PacBio Iso-Seq and continuous long read sequencing data of the SKBR3 cell line are downloaded from SRA under accession SRP150606 (9). PacBio Iso-Seq data of Human Genome Structural Variation Consortium (HGSVC) samples are available at HGSVC data portal (https://www.internationalgenome.org/data-portal/). Acute myeloid leukemia (AML) patient data are downloaded from SRA under SRR12048357 (4).
https://codeocean.com/widget.js?slug=3525117
Simulation and benchmark methods can be found in Supplementary Data S2.