Abstract
CLASPs (cytoplasmic linker-associated proteins) are ubiquitous stabilizers of microtubule dynamics, but their molecular targets at the microtubule plus-end are not understood. Using DNA origami–based reconstructions, we show that clusters of human CLASP2 form a load-bearing bond with terminal non-GTP tubulins at the stabilized microtubule tip. This activity relies on the unconventional TOG2 domain of CLASP2, which releases its high-affinity bond with non-GTP dimers upon their conversion into polymerization-competent GTP-tubulins. The ability of CLASP2 to recognize nucleotide-specific tubulin conformation and stabilize the catastrophe-promoting non-GTP tubulins intertwines with the previously underappreciated exchange between GDP and GTP at terminal tubulins. We propose that TOG2-dependent stabilization of sporadically occurring non-GTP tubulins represents a distinct molecular mechanism to suppress catastrophe at the freely assembling microtubule ends and to promote persistent tubulin assembly at the load-bearing tethered ends, such as at the kinetochores in dividing cells.
Microtubule-binding by DNA origami-based clusters of CLASP2 reveals a distinct mechanism of plus-end stabilization.
INTRODUCTION
Proper regulation of microtubule dynamics and load bearing by microtubule tip–attached structures are essential for numerous cellular processes. Cytoplasmic linker-associated proteins (CLASPs) are an evolutionarily conserved family of ubiquitous factors that exert stabilizing effects on microtubules by suppressing catastrophes and promoting rescues (1–4). CLASPs’ importance has been recognized and extensively characterized in a variety of cellular and organismal systems, such as during cell migration, neuronal development, and cytoskeleton reorganization in interphase and during cell division [reviewed in (5, 6)]. CLASPs are highly potent regulators of the growing microtubule plus-ends, owing to their enrichment at these sites via the tip-binding proteins, such as end-binding protein (EB). Microtubule ends are also regulated by localized multimolecular ensembles of CLASPs. For example, membrane-associated clusters of CLASP molecules capture microtubule ends at the postsynaptic membrane, whereas cortical clusters of CLASPs tether and stabilize microtubule ends near focal adhesions in motile cells (7, 8) or at the cell cortex (9–11). These interactions are often modulated by cellular force, such as those produced during cell retraction (12), microtubule bundle protrusion into the lamella, or contact-mediated cell-cell repulsion (13).
Studies in mitotic cells have been particularly informative for understanding CLASP’s molecular mechanisms because, here, CLASP proteins are clustered at kinetochores to mediate their dynamic interactions with microtubule plus-ends (14–16). Although kinetochore-microtubule attachments are subjected to considerable forces, the tethered microtubule ends are not static and undergo dynamic transitions. During metaphase, tubulin dimers are added continuously to the kinetochore-embedded microtubule ends, contributing to their poleward flux (Fig. 1A). Human cells encode two CLASP proteins, CLASP1 and CLASP2 (each with several isoforms), which play redundant roles in mitosis. They are required for poleward flux by assisting the elongation of tethered microtubule ends under force (15–18).
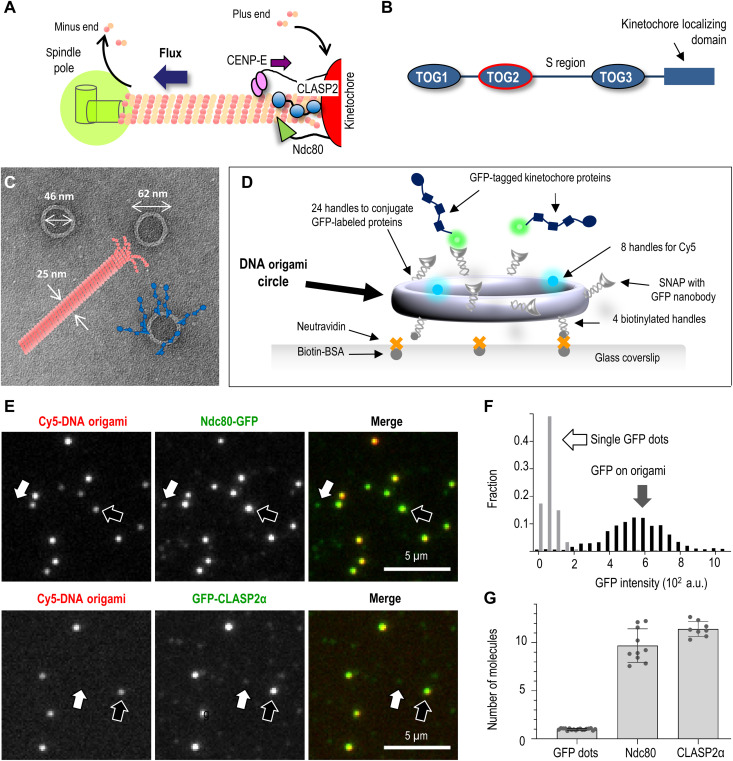
Fig. 1. Reconstruction of multivalent molecular clusters for kinetochore-microtubule interaction studies.
(A) Schematic of kinetochore microtubule plus-end and its coupling to the kinetochore, which clusters several microtubule-binding proteins, including CLASPs, the Ndc80 protein complex, and the plus-end–directed kinesin CENP-E. CLASP promotes the incorporation of tubulins at the kinetochore-bound microtubule plus-ends, sustaining poleward tubulin flux. (B) Schematic of functional domains in human CLASP2α (not to scale). (C) Electron microscopy image of the DNA origami circles with overlaid schematics of a microtubule (drawn to scale) and conjugated kinetochore proteins (blue). (D) The use of an immobilized DNA scaffold and conjugated GFP-tagged proteins to make a microtubule binding site. (E) Representative fluorescence images of Cy5-labeled DNA scaffolds (examples indicated by black arrows) with conjugated GFP-tagged proteins: human Ndc80 Broccoli and full-length CLASP2α. White arrows point to dim dots with GFP but not Cy5 fluorescence, representing single GFP-tagged protein molecules that attach nonspecifically. (F) Typical GFP intensity distribution of DNA scaffolds (n = 180) and dim dots (n = 527) based on one representative experiment with Ndc80-GFP, in which 13 image fields were analyzed. (G) The number of GFP-tagged molecules per DNA origami scaffold. Each dot represents the mean of the GFP intensity distributions from independent experiments plotted relative to the mean intensity of single GFP molecules. Error bars are SEMs. a.u., arbitrary units.
The model in which CLASPs promote tubulin assembly at the kinetochore-embedded microtubule plus-ends has been attractive in view of the prior proposal that CLASPs act as a tubulin polymerase (19, 20). Specifically, a linked array of tumor overexpression gene (TOG) domains (Fig. 1B), which are found in CLASPs and represent specialized tubulin-binding modules, has been thought to be instrumental in promoting the incorporation of tubulin dimers at microtubule ends (21–23). However, recent experiments in vitro have found that a single TOG2 domain, supplemented with the downstream flanking region of human CLASP2 (TOG2-S protein), is sufficient to provide anti-catastrophe and rescue activities characteristic of the full-length proteins (24, 25). Human TOG2 is unusual among the set of all TOG domains, as its highly bent shape is not compatible with binding to straight tubulins in the microtubule wall. The curvature of TOG2 also exceeds the curvature of the crystallized forms of soluble tubulin (26, 27). Accordingly, CLASPs have been proposed to bind to highly curved protofilaments at the microtubule tip (28). High-resolution fluorescence microscopy, however, localizes TOG2-S to a region trailing the growing microtubule tip by about 90 nm (10 to 12 tubulin layers), leading to a proposal that this is the actual site of its anti-catastrophe activity (24). Thus, the primary molecular target for TOG2 binding at the growing microtubule end and the mechanism of its stabilizing activity are ill understood. One underexplored model for CLASPs’ microtubule binding posits that it is regulated directly by guanosine triphosphate (GTP), based, in part, on the presence of molecular features of a GTP binding site in the CLASP ortholog in Drosophila (3). Whether this putative GTP binding site plays an important role in CLASP2 activity in vitro or in mitotic cells remains unknown.
Because of the pivotal role of CLASPs in numerous cellular functions, we have sought to gain insights into its role in regulating the dynamics of the tethered microtubule ends. With the DNA origami technique, we show that clusters of CLASP2 bind tightly to stabilized microtubule plus-ends, but these bonds dissociate quickly from the assembling microtubule end, arguing against the direct role of CLASP2 in promoting tubulin incorporation. We reconstructed high-affinity end association using only the TOG2 domain with flanking regions and found that the microtubule end is released by soluble GTP; however, this interaction does not require the putative GTP binding site in TOG2. Stoichiometry of CLASP2 binding to stabilized microtubule tip, as well as its marked GTP sensitivity, strongly suggests that TOG2 targets naturally occurring terminal non-GTP tubulins rather than curved tubulin protofilaments or wall tubulins at a tip-proximal location. We therefore propose a model in which CLASP2 promotes the elongation of kinetochore-tethered microtubules indirectly by stabilizing the catastrophe-promoting terminal non-GTP tubulin dimers, such as guanosine diphosphate (GDP)–tubulin or apotubulin subunits, which are generated stochastically during GTP-tubulin assembly.
RESULTS
DNA origami provides a multivalent molecular platform to reconstruct physiological clusters of kinetochore-associated proteins
To seek insight into the molecular mechanisms of CLASP2 activity, we used DNA origami technology (29, 30), which provides a versatile platform for organizing protein molecules with precisely controlled stoichiometry and spatial positioning (31, 32). Specifically, we modified a previously developed DNA origami nanocircle (33) to function as the structural scaffold for organizing kinetochore-associated proteins. Its outer diameter of 62 nm is a good match for the overall dimensions of one microtubule binding site at the kinetochore (Fig. 1C), which tethers flared tubulin protofilaments of the microtubule plus-end (34). This scaffold features three types of single-stranded DNA (ssDNA) handles (fig. S1; see Materials and Methods). First, there are eight identical handles originating from the inner surface of the circular core; these handles were hybridized with complementary ssDNA molecules (termed anti-handles) labeled with Cyanine 5 (Cy5) to assist scaffold visualization. The second type of handles at the “bottom” of the circles was labeled in a similar way with biotin and used to firmly flatten the circles in uniform orientation on NeutrAvidin-coated glass slides. Last, there are 24 ssDNA handles on the outer surface of the DNA origami scaffold. Their complementary anti-handles were functionalized with benzyl-guanine (BG) and covalently attached to a protein adaptor consisting of a soluble N-ethylmaleimide–sensitive factor attachment protein (SNAP)–tag fused to a green fluorescent protein (GFP)–binding protein (GBP), a GFP-binding nanobody (35). These SNAP-GBP–containing anti-handles enabled high-affinity recruitment of any soluble GFP-tagged protein that was brought into a microscopy chamber (Fig. 1D). After unbound protein was washed away, over 95% of the Cy5-labeled scaffolds colocalized with bright GFP dots, implying the formation of high-density molecular clusters (Fig. 1E).
Dim GFP dots were also seen nonspecifically attached to the coverslip. Their uniform brightness and single-step photobleaching kinetics indicated that they represented single molecules (fig. S2; see Materials and Methods). By dividing the average GFP intensity of the scaffold-associated protein clusters by the single-molecule brightness, we estimate that the clusters recruit, on average, 10 to 12 protein molecules (Fig. 1, F and G), which is a reasonable approximation of a microtubule binding site at a kinetochore (36). Thus, the generated molecular clusters include a sufficient number of each protein under study to engage in multivalent interactions with one microtubule end. Hence, they represent physiologically relevant assemblies whose study in vitro may elucidate kinetochore-microtubule interactions in cells.
Molecular clusters of CLASP2 form durable and specific attachment to stabilized microtubule plus-ends
Previous in vitro assays using soluble CLASP proteins revealed their diffusion on microtubule walls, as well as slight enrichment at the polymerizing microtubule plus-ends (20, 24, 37, 38). To investigate the microtubule-binding properties of the origami-clustered assemblies of CLASP molecules, we used full-length human CLASP2α, which is among the most studied CLASP isoforms. Taxol-stabilized microtubules labeled with rhodamine were flowed into the chambers with coverslip-immobilized clusters of CLASP2α and imaged using total internal reflection fluorescence (TIRF) microscopy (Fig. 2A; see Materials and Methods). Most of the microtubules associated with CLASP2α-containing clusters appeared as blurry spots, suggesting that tethered microtubules orient away from the coverslip surface (movie S1). In contrast, clusters containing a different kinetochore protein, the microtubule wall–diffusing Ndc80 complex (39), recruited microtubules by their walls and near the microtubule tip (Fig. 2B). These microtubules were often seen diffusing on the coverslip-bound Ndc80 clusters while remaining within the TIRF imaging plane (movie S2 and fig. S3).
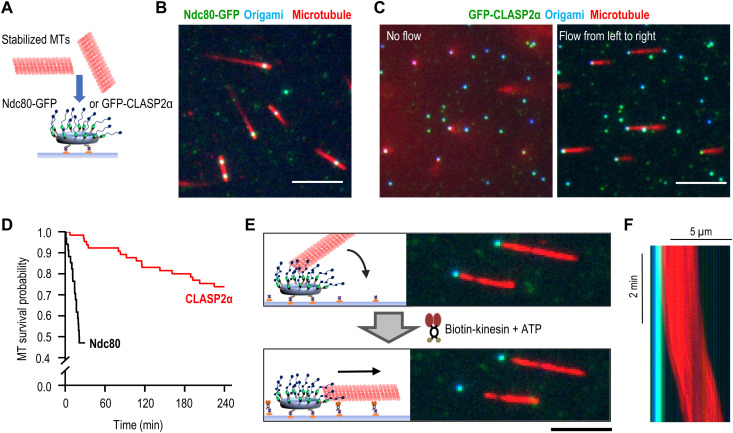
Fig. 2. End-on attachment of microtubules to molecular clusters of full-length CLASP2α protein.
(A) Schematic of experimental assay with a DNA origami circle attached to a coverslip and presenting clusters of microtubule-binding proteins to rhodamine-labeled taxol-stabilized microtubules (MTs). (B) Representative TIRF image of microtubules (red) bound to DNA origami scaffolds (blue) with clusters of GFP-tagged Ndc80 Broccoli (green). Scale bar, 5 μm. (C) Same as in (B) but with CLASP2α clusters and showing the same field with (right) or without (left) buffer flow. Scale bar, 5 μm. (D) Lifetime of microtubule association with the clusters of the indicated proteins. Kaplan-Meier survival plots were built on the basis of n microtubules observed in N independent experiments: for Ndc80, n = 34 and N = 2 and for CLASP2α, n = 65 and N = 3. (E) Schematic of the experiment and representative images of microtubules initially tethered at CLASP2α clusters but then dragged away by the coverslip-bound, plus-end–directed Kinesin-1 in the presence of 10 μM adenosine triphosphate (ATP). Scale bar, 5 μm. (F) Kymograph illustrating the lag period after addition of ATP and the persistence of the microtubule gliding away from the CLASP2α cluster [overlaid blue, green, and red images; see (B) for details]. Variations in red intensity are due to uneven illumination.
These lateral diffusive motions were notably absent when CLASP2α clusters were used. To reveal the preferred mode of microtubule binding by the CLASP2α clusters, we applied a gentle flow of imaging buffer (movie S3). Under the flow force, the scaffold-associated microtubules aligned with the flow, revealing one end of each microtubule tethered to a CLASP2α cluster (Fig. 2C). These end-on attachments were very durable, as half of microtubules maintained end association for 4 hours, substantially longer than the microtubule binding of Ndc80 clusters of similar size (Fig. 2D).
To determine whether the tethered microtubules had a specific polarity, we took advantage of the plus-end–directed Kinesin-1. After the microtubules formed end-on attachment to the CLASP2α clusters, we conjugated kinesin motors to the coverslip surface and introduced a flow of buffer supplemented with Mg–adenosine triphosphate (ATP; Fig. 2E). Microtubules pivoted at the CLASP2α-tethered ends to come in contact with the motor-coated surface and, after a brief delay, began to glide unidirectionally (Fig. 2F and movie S4). All microtubules examined (n = 124) glided away from the tethering sites, not toward them. Because kinesin propels microtubules with the minus-end leading, these findings reveal that clusters of CLASP2α form durable and highly specific attachment to the microtubule plus-ends, the same polarity of microtubule binding seen at kinetochores.
Microtubule ends tethered to the CLASP2α clusters can withstand substantial load
Microtubule ends embedded at kinetochores of dividing cells experience variable forces (40–43). To investigate the ability of CLASP2α clusters to maintain association with the stabilized microtubule plus-ends under load, we used a laser trapping approach (see Materials and Methods and fig. S4). Polystyrene beads coated with anti-tubulin antibodies were flowed into the chamber, and they attached spontaneously to the walls of microtubules whose ends were tethered to the coverslip-immobilized clusters of CLASP2α (Fig. 3, A and B). A microtubule-bound bead was then captured in a stationary optical trap, and the stage was moved at a constant speed to pull the cluster away from the laser-trapped bead. Because CLASP2α-tethered microtubules project into the chamber, the force acting on a CLASP2α cluster was calculated on the basis of the beads’ displacements in two directions: along the stage motion (y axis) and along the z axis, pointing away from the coverslip.
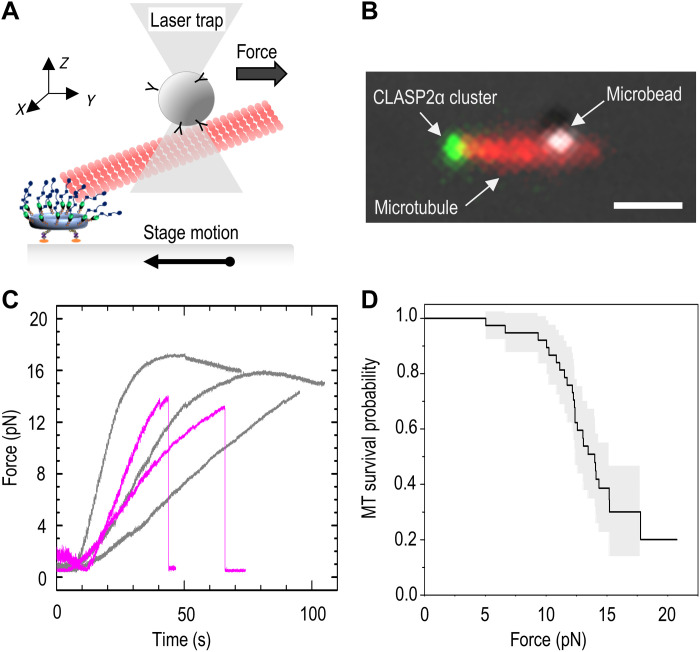
Fig. 3. Measuring the strength of a microtubule end-on attachment using laser trapping.
(A) Schematic of the experimental assay in which a pulling force is applied to the microtubule tethered at its plus-end to a coverslip-immobilized CLASP2α cluster. Microtubule is held in the laser trap via the trapped wall-bound microbead, while the stage is moved in the opposite direction until a bond is ruptured or the maximum force is reached with no rupture. (B) Representative fluorescence image of a taxol-stabilized rhodamine-labeled microtubule and the coverslip-bound CLASP2α cluster overlaid with the differential interference contrast image of the microtubule-bound bead. Because the bead is located several micrometers away from the coverslip (deep into the chamber), the microtubule image represents a projection, constructed by combining several images taken at different z planes. Scale bar, 1 μm. (C) Force application curves from individual experiments. Two typical outcomes are illustrated by these five curves. Gray curves show an increase in the pulling force followed by a gradual decrease, indicating that a maximal force was reached with no rupture. Curves in purple show similar increases in pulling force, followed by an abrupt drop, indicative of a rupture. (D) Kaplan-Meier survival plot based on 38 measurements from six independent experiments. The gray area corresponds to 95% confidence interval.
Using this approach, we detected a rupture in this mechanical chain in 26 of 39 experiments. Rupture was evident via an abrupt decrease in force sustained by the trapped bead (Fig. 3C, purple). In 25 of these events, the beads remained bound to the microtubule and GFP signal from CLASP2α persisted at the same location on the coverslip (fig. S4D), indicating that the rupture occurred specifically between the scaffold-associated CLASP2α and the microtubule plus-end. In only one case, the rupture took place between the origami scaffold and the coverslip. The remaining microtubules (n = 13) did not detach from the CLASP2α clusters even under the highest pulling force that our laser trap could apply: 15.2 ± 2.8 pN (Fig. 3C, gray). The Kaplan-Meier survival plot in Fig. 3D shows that the 50% survival probability for CLASP2α attachment to microtubule plus-end is 14 pN (95% confidence interval: 12.4 to 15.2 pN). These results strongly suggest that a cluster of 10 to 12 CLASP2α molecules is able to provide a sizeable contribution to microtubule attachment stability and to mediate force-dependent cellular functions, for example, at kinetochore-tethered microtubule ends during chromosome segregation.
Soluble GTP destabilizes the end-on microtubule attachment to CLASP2
Next, we investigated how microtubule plus-ends tethered to clusters of CLASP2α behave in the presence of soluble tubulin. Because disruption of CLASPs’ function in mitotic cells severely compromises soluble tubulin incorporation at the plus-ends of kinetochore-bound microtubules (14, 15, 44), we expected that CLASP2 clusters would promote the addition of soluble tubulins to tethered microtubule plus-ends in vitro (Fig. 4A). To enable free tubulin assembly, we used microtubule seeds polymerized with Guanosine-5'-[(α,β)-methyleno]triphosphate (GMPCPP). The seeds formed end-on attachment to the CLASP2α clusters similarly to the taxol-stabilized polymers. When soluble GTP-tubulin was added into the chamber, no tubulin addition was seen at the tethered ends. Instead, the end-bound microtubules detached immediately (Fig. 4B). We ruled out that our tubulin or buffer composition did not support microtubule elongation by observing in the same chambers that tubulin assembly was possible at the ends of rare microtubules that bound laterally to the CLASP2α clusters (movie S5). Thus, these results reveal unexpectedly that strong CLASP2α association with the microtubule end is incompatible with tubulin addition to that end.
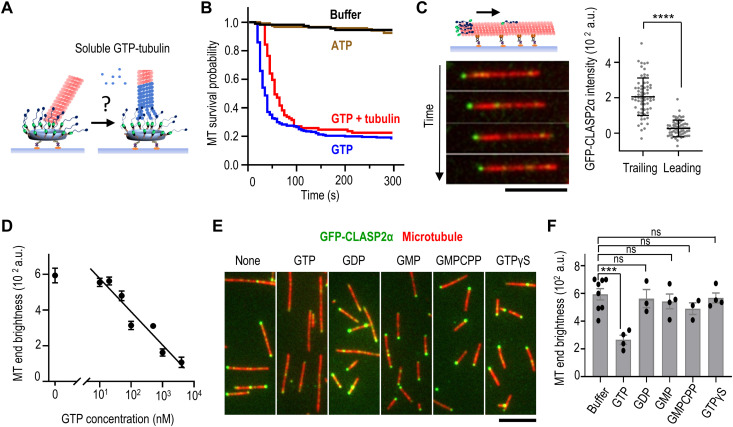
Fig. 4. Soluble GTP as a highly specific inhibitor of microtubule end-on attachment to CLASP2α.
(A) Schematic of the experiment to examine the ability of CLASP2α clusters to incorporate soluble tubulin (blue) at an associated microtubule plus-end (red). (B) Lifetime of microtubule end-on attachments to CLASP2α clusters. Kaplan-Meier plots show the probability of attachment survival after addition of imaging buffer, buffer with 1 mM Mg-ATP, 1 mM Mg-GTP, or soluble tubulin (8 μM tubulin with 1 mM Mg-GTP). Curves are based on n microtubules imaged in N independent experiments: buffer, n = 110 and N = 2; ATP, n = 120 and N = 2; GTP, n = 255 and N = 4; tubulin + GTP, n = 89 and N = 3. (C) Time-lapse images of a taxol-stabilized rhodamine-labeled microtubule gliding on a coverslip coated with Kinesin-1 in the presence of 1 nM soluble GFP-CLASP2α and 1 mM Mg-ATP. Scale bar, 5 μm. The graph on the right shows CLASP2α brightness at the trailing (minus) and leading (plus) microtubule ends: means ± SD, n = 66 microtubules, N = 2; ****P < 0.0001 (paired t test). (D) Inhibition curve for soluble GFP-CLASP2α (1 nM) binding to taxol-stabilized microtubule ends plotted on a semilog scale and with linear fitting. Dots correspond to the average brightness of the end-associated GFP-CLASP2α signal in the presence of the indicated concentration of Mg-GTP (>30 microtubules for each point). Error bars are SDs. (E) Representative images of taxol-stabilized microtubules incubated with 1 nM soluble GFP-CLASP2α in the presence of 1 μM indicated nucleotides. (F) Intensity of GFP-CLASP2α signal at the brightest microtubule end in the presence of 1 μM indicated nucleotides and 1 nM soluble GFP-CLASP2α. Each dot represents the average intensity of at least 50 microtubule ends collected in an independent experiment (see Data Source file for details). Error bars are SEMs, and significance was determined with Welch’s t test. ***P < 0.001; ns, not significant.
To tease apart this relationship, we varied experimental conditions and found that GTP alone, rather than GTP-tubulin, leads to a precipitous loss of end-on attachment by CLASP2α (Fig. 4B). The addition of ATP does not destabilize microtubule end attachment, suggesting that the effect is nucleotide specific. Because the close proximity of microtubule-binding proteins to the charged DNA nanocircle might affect interactions between the origami-conjugated proteins and the microtubule end (45), we next asked whether GTP-dependent CLASP2α dissociation was dependent on origami scaffolds. Taxol-stabilized microtubules were immobilized on the coverslip using tubulin antibodies and visualized via TIRF microscopy. When soluble GFP-tagged CLASP2α was added to this preparation in a GTP-free buffer, one end of most microtubules became decorated, with weaker spots seen along microtubule walls and at the opposite microtubule end. Using microtubule gliding assay with Kinesin-1 and soluble ATP (Fig. 4C), we observed that 95% of bright microtubule ends with associated GFP-CLASP2α were trailing (N = 66 microtubules; movie S6). Thus, the enhanced localization of CLASP2α at the plus-ends of stabilized microtubules does not depend on DNA origami scaffolds, consistent with (38).
Using this origami-free system, we investigated the impact of soluble GTP on stabilized microtubule end decoration by CLASP2α and found that it inhibits the reaction in a concentration-dependent manner with median inhibitory concentration (IC50) = 320 nM (Fig. 4D and fig. S5). This effect was specific to GTP, as other guanosine-containing nucleotides, such as GDP and guanosine monophosphate (GMP) and the nonhydrolyzable GTP analogs GMPCPP and GTPγS, had little effect on the interactions between CLASP2α and microtubule plus-ends (Fig. 4, E and F). Together, the results using soluble and clustered CLASP2α strongly suggest that GTP causes a rapid and highly specific dissociation of CLASP2α from the microtubule plus-end.
TOG2 domain of CLASP2 is sufficient for GTP-sensitive microtubule end binding
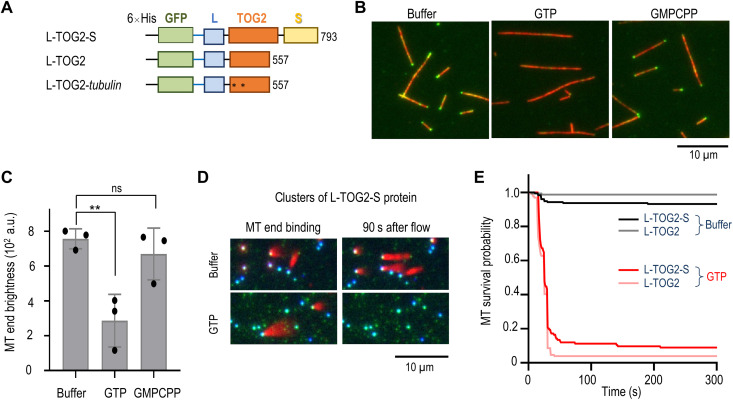
CLASP2 contains several TOG domains among which the TOG2 domain appears to play a primary role in mediating CLASP’s function as a regulator of microtubule dynamics. TOG2 with the flanking region S (TOG2-S protein) is sufficient to provide anti-catastrophe and rescue activities characteristic of the full-length proteins (24, 25). We investigated whether TOG2 domain is also involved in the GTP-sensitive microtubule end binding. We constructed L-TOG2-S protein in which the globular TOG2 domain is flanked by disordered L and S regions (Fig. 5A). Bacterially expressed and purified GFP-tagged L-TOG2-S readily decorated one end of taxol-stabilized microtubules [end brightness, 594 ± 41 arbitrary units (a.u.) for full-length protein and 757 ± 57 a.u. for L-TOG2-S protein], consistent with this domain’s importance for CLASP2α binding to microtubule plus-ends. Microtubule end decoration was also seen for GFP-L-TOG2, but not for the analogous protein with W340E and K425A substitutions (fig. S6E), which compromise microtubule binding by TOG2 in cells and in vitro (28). Thus, TOG2 domain binding to the microtubule tip is mediated by the same residues that are involved in microtubule wall recognition.
Fig. 5. Probing the role of TOG2 domain in GTP-sensitive microtubule end binding.
(A) Truncated GFP-CLASP2α constructs. Numbers correspond to full-length sequence; asterisks indicate locations of substitutions. GFP-L-TOG2-tubulin has W340E and K425A substitutions. (B) Representative TIRF images of taxol-stabilized microtubules (red) with 1 nM GFP-L-TOG2-S. Decoration of microtubule ends is visible in the buffer with or without 1 μM GMPCPP but not with 1 μM GTP. (C) Intensity of GFP-L-TOG2-S signal at the microtubule end under conditions as in (B). Each dot represents the average intensity from >50 microtubule ends observed in an independent experiment (see Data Source file for details). Error bars are SEMs; **P < 0.01, ns - not significant. (D) Still images from the video recording of experiments in which GFP-L-TOG2-S protein was conjugated via the N-terminal GFP tag to the coverslip-immobilized DNA scaffolds. Taxol-stabilized microtubules were added (left), revealing strong end-binding preference. Images on the right were taken while flowing in the buffer with or without 1 mM GTP. (E) Kaplan-Meier survival plots for microtubule association with different protein clusters in the buffers with or without 1 mM GTP (see Data Source file for details).
Microtubule end association of L-TOG2-S was reduced in the presence of 1 μM GTP but not GMPCPP (Fig. 5, B and C). We then conjugated L-TOG2-S and a shorter truncation, L-TOG2, to DNA scaffolds, with the unstructured L region serving as an extension for TOG2 to bridge the DNA circle and microtubule tip extremity. Both TOG2 proteins were able to tether the ends of stabilized microtubules, but these attachments dissociated rapidly upon addition of soluble GTP (Fig. 5, D and E). Thus, TOG2 domain is responsible for the GTP sensitivity of the CLASP2’s microtubule end binding.
CLASP2 binds specifically to terminal tubulins, which have exchangeable GDP-GTP binding pockets
The GTP sensitivity of binding between a stabilized microtubule end and CLASP2 is unexpected. We were intrigued by this finding in view of a prior work reporting that microtubule binding by the Drosophila CLASP ortholog is regulated by GTP via a putative GTP binding site in the TOG2 domain(3). However, when we used radical substitutions (46, 47) to disrupt the NFKD sequence within this site in human L-TOG2-S, mutant proteins had wild-type phenotypes in our in vitro assays and in mitotic cells (Supplementary Note and figs. S7 and S8), so we concluded that the NFKD sequence is dispensable for CLASP2’s role at the kinetochore-microtubule interface. We then considered an alternative hypothesis in which the GTP dependency is mediated by polymerized tubulins rather than CLASP2. Both α- and β-tubulins have GTP binding sites, but within the microtubule wall, GTP nucleotide is associated only with α-tubulin (N site), whereas β-tubulin’s “E-site” is occupied by GDP (48). Thus, soluble GTP might exert its effect on stabilized microtubules in our assays by interacting with the E-sites in β-tubulins (Fig. 6A). Although there is no evidence of a substantial GTP binding to these sites within the microtubule wall, the GTP binding sites at the most distal β-tubulins at the microtubule plus-end are exposed and permit nucleotide exchange (49, 50).
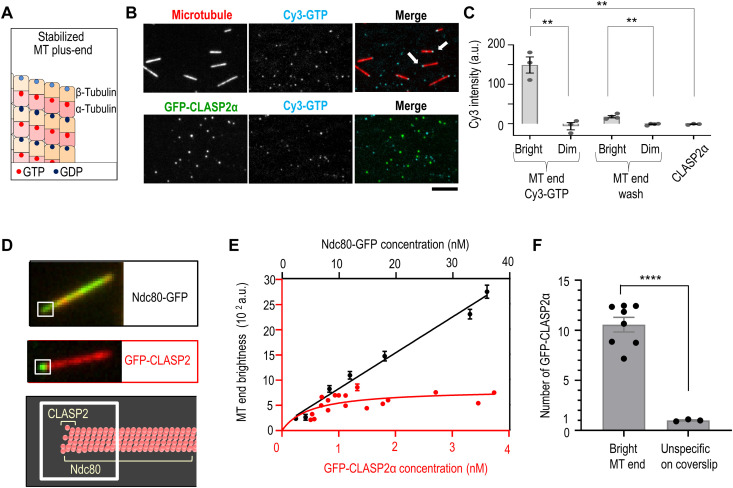
Fig. 6. GTP and CLASP2α binding to terminal tubulins at stabilized microtubule tips.
(A) Stabilized microtubule plus-ends showing GDP at β-tubulins and GTP at α-tubulins. Terminal E-sites are light blue. (B) Representative images of taxol-stabilized microtubules and origami-based clusters of full-length CLASP2α with 100 nM Cy3-GTP. Some Cy3-GTP molecules bind nonspecifically to the coverslips, but only microtubule ends show colocalization (arrows). Images in the Cy3 channel were taken and processed identically, allowing direct comparison of brightness at the microtubule tips and CLASP2α clusters. Scale bar, 5 μm. (C) Brightness of microtubule ends and CLASP2α clusters incubated with 100 nM Cy3-GTP, as in (B). “MT end wash” images were captured after Cy3-GTP was replaced with imaging buffer. Each dot represents the average brightness from an independent experiment. Error bars are SEMs; **P < 0.01, t test. (D) Example images of taxol-stabilized microtubules decorated with GFP-CLASP2α (1 nM) or Ndc80 Broccoli (12 nM). Microtubule end brightness was determined using the same size regions (squares) and settings, so maximal brightness is limited by the number of binding sites for these proteins (schematic). (E) GFP brightness for regions as in (D) as a function of soluble protein concentrations. After a similar initial rise in microtubule end brightness, the binding curves diverge, implying a different number of binding sites for these two proteins. Each dot represents the median intensity of >32 ends, and error bars are SEMs (see Data Source file for details). Lines are linear regression (black) or hyperbolic fit (red) with dissociation constant (Kd) = 0.5 ± 0.4 nM and plateau Imax = 800 ± 200 a.u. (F) Intensity of microtubule ends decorated with 1 nM GFP-CLASP2α was normalized to the intensity of coverslip-bound GFP-CLASP2α molecules imaged using the same settings. Each point represents average intensities for one experiment; total number of microtubule ends, 718 and non-specific dots, 305; ****P < 0.0001.
We investigated the role of soluble GTP in regulating CLASP2α association with stabilized microtubule end by using Cy3-labeled GTP (51). This fluorescent GTP analog reduced the intensity of CLASP2α-decorated microtubule end similarly to unlabeled GTP (fig. S9A), confirming that this modified nucleotide is functionally active. TIRF imaging revealed that Cy3-GTP highlighted the ends of taxol-stabilized microtubules, with one end showing much brighter intensity (Fig. 6B). In contrast, clusters of full-length CLASP2α protein incubated with Cy3-GTP and imaged under identical conditions showed no Cy3 signal (Fig. 6, B and C). Thus, microtubule ends but not CLASP2α are the primary receptor for GTP in our assays.
The Cy3-GTP signal at the microtubule ends decreased after washing with a nucleotide-free buffer (Fig. 6C), as expected for exchangeable E-sites. Microtubule end binding and unbinding by soluble Cy3-GTP is consistent with prior reports that the tips of stabilized microtubules in nucleotide-free buffer contain apotubulins, which can readily accept soluble GTP (49, 50). On the basis of these findings, we hypothesized that GTP binding to the terminal apotubulins is a direct cause of CLASP2α dissociation from stabilized microtubule tips. Consistent with this idea, CLASP2α end binding decreases with increasing brightness of the tip-associated Cy3-GTP (fig. S9, B and C). If the primary site for CLASP2α binding at the microtubule end is at the terminal tubulin dimers, then we also expect no more than 13 binding sites for CLASP2α at the microtubule tip. We tested this prediction by determining microtubule end brightness as a function of CLASP2α concentration in the absence of any soluble nucleotides (Fig. 6D). End association of CLASP2α increased gradually and plateaued, consistent with the saturation of its binding sites (Fig. 6E). To rule out that the plateau was caused by a technical limitation of our imaging procedure, we carried out an analogous experiment using GFP-tagged Ndc80. A higher soluble concentration of Ndc80-GFP is needed to achieve a similar initial increase in the intensity of the microtubule end than with CLASP2α because of their different microtubule binding affinities. For easier comparison, data for these two proteins were plotted using different concentration scales but with the same scale for microtubule end brightness (Fig. 6E). Brightness of the same region at the microtubule tips incubated with GFP-Ndc80 did not plateau but continued to increase above the maximal level seen for CLASP2α (fig. S10). This is consistent with a greater number of binding sites for Ndc80, which binds microtubule all along the wall, and this control verifies that the CLASP2α signal is not limited by our illumination or imaging settings. By measuring the brightness of single GFP-CLASP2α molecules in the same chambers (Fig. 6F), we estimate that end binding plateaued at 12 ± 2 molecules of CLASP2α. This number matches closely the number of microtubule protofilaments and, therefore, the number of terminal β-tubulins with exposed GTP binding pockets, leading us to conclude that these tubulins represent the major site for GTP-sensitive TOG2 binding at the stabilized microtubule tip. Similar molecular interactions and GTP-dependent regulation may take place at the tips of dynamic microtubules.
CLASP2α stabilizing activity correlates with reduced integrity of polymerizing microtubule tip
Our findings with stabilized and dynamic microtubules suggest that CLASP2α does not associate strongly with the tips of polymerizing microtubules because terminal GTP-tubulins have low affinity for TOG2. Since non-GTP tubulins can sometimes become exposed at the microtubule tip even during polymerization (50, 52), such rare subunits may represent a natural target for strong affinity binding by CLASP2α. Structural and physiological consequences of TOG2 binding to a terminal non-GTP subunit are not yet known, but it is possible that the affected protofilament may cease to grow while TOG2 is bound at its tip. This idea is supported, in part, by the prior finding that CLASP2α in combination with EB3 promotes the formation of incomplete tip structures, similar to those seen for a combination of EB3 and the protofilament-blocking agent eribulin (24, 53). Apparently, incomplete tips arise when one or several protofilaments are stalled (i.e., exhibit no assembly or disassembly), while the remaining protofilaments continue assembling, forming curly protrusions. A similar behavior reported for CLASP2α and eribulin suggests that incomplete tip structures may form occasionally at the tips of microtubules polymerizing in the presence of CLASP2α, owing to the TOG2 domain binding to an exposed non-GTP subunit. This prediction is difficult to test via direct visualization because small variability in the length of protofilaments at the microtubule tip cannot be resolved well. However, temporary reduction in tip integrity can be revealed using a sensitive mechanical test because individual protofilaments and their incomplete assemblies are much softer than a complete tubulin cylinder (54).
To test these ideas, we took advantage of our previously developed strategy to tether dynamic microtubule plus-ends at the surface of the coverslip-immobilized beads (55). Thermal pivoting of tethered microtubules around their attachment site can provide insights into the tip structure, serving as a proxy for the presence of stalled protofilaments at the tethered tip (Fig. 7A). We reasoned that if CLASP2α does not induce the formation of stalled protofilaments, then its activity should have no effect on the rigidity of the bead-tip coupling, and tethered microtubule pivoting should be similar for CLASP2α and other proteins that are not known to disrupt tip structure, such as Ndc80 and EB1. However, if the molecular target for CLASP2α is at the tip’s extremity and CLASP2α blocks tubulin addition to that site, then microtubule tip integrity may become compromised, leading to a temporary increase in pivoting.
Fig. 7. Dynamics of the microtubule end tethered near bead-immobilized CLASP2α versus other microtubule-binding proteins.
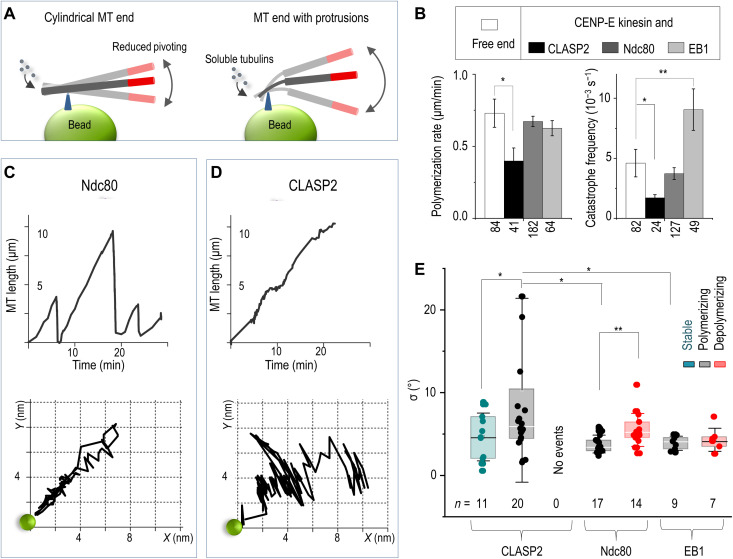
(A) Integrity of microtubule plus-end is examined by tethering it to a coverslip-immobilized bead. Thermal pivoting of tethered microtubules is a sensitive gauge of protruding protofilaments, which can arise if one or several protofilaments at the assembling end became stalled. Larger pivoting angles are expected when microtubule is tethered via several protofilaments than via the tube-like tip owing to low protofilament rigidity. Drawings are not to scale; triangles depict the bead attachment sites at assembling microtubule tips. Microtubule motions are monitored via the labeled segments (red). (B) Dynamic parameters of the plus-end of freely growing microtubules and of the plus-ends coupled to beads coated with the indicated proteins. Columns (means ± SEM) are based on N ≥ 3 independent experiments and n examined microtubules; *P < 0.05 and **P < 0.001(Mann-Whitney U test; see Data Source file for details). (C) Microtubule length versus time (top) and the corresponding trajectory (bottom) of the distal end of a HyLite647-labeled microtubule segment relative to the CENP-E/Ndc80–containing bead (green circle; not to scale). This CENP-E/Ndc80–coupled end underwent three cycles of microtubule growth and shortening while moving back and forth along a relatively straight path. (D) Same as in (C) but for a typical bead coated with CLASP2α, which couples growing but not shortening microtubule tips (fig. S12). Coupled microtubule end grows persistently with small pauses and pivots occasionally around the bead. (E) SD values of the pivoting angles (parameter σ) around the microtubule-bead attachment site for coatings with the indicated proteins and different microtubule ends: GMPCPP stabilized, polymerizing, and depolymerizing (color-coded). Box: first, second, and third quartiles; whiskers: SD; n: the total number of analyzed microtubule end tracings; *P < 0.05 and **P < 0.01, Welch’s t test.
We used coverslip-immobilized beads coated with different microtubule-binding proteins, each combined with the motor domains of the plus-end–directed CENP-E kinesin. As we have shown previously, bead-conjugated microtubule wall–diffusing proteins, including Ndc80 “Broccoli” and EB1, paired with CENP-E motors can maintain persistent association with the microtubule wall segment near the plus-end extremity, leaving the terminal subunits to exchange freely at the bead-tethered end (37). Full-length CLASP2α can also tether polymerizing microtubule ends owing to its ability to bind and diffuse on the microtubule wall via the charged residues in its S domain. To compare tip tethering mediated by these microtubule-binding proteins, coverslip-immobilized beads were coated with a mixture of CENP-E and a microtubule-binding protein of interest. Fluorescently labeled GMPCPP-microtubule seeds were introduced in the chambers, and microtubule seeds bound laterally to these beads. After the addition of ATP, the CENP-E motors drove the microtubule plus-ends closer to the bead’s surface, where the ends remained tethered with the help of microtubule-binding proteins (fig. S11A). Unlabeled GTP-tubulin was then added; as the bead-tethered plus-end incorporated new tubulins, the labeled seeds moved away from the beads at the rate of tubulin assembly.
First, we compared the character of these radial motions, reporting on the incorporation of tubulin dimers at the tethered plus-ends. Polymerization at the ends tethered by Ndc80 proceeded efficiently but unremarkably, similarly to the elongation of the bead-free microtubule ends (Fig. 7, B and C, and fig. S11) (37). Polymerization at the EB1-containing beads was also similar; these microtubule ends experienced frequent catastrophes, which is characteristic for EB proteins (56). Tubulin assembly at the ends tethered via CLASP2α proceeded with occasional pausing and at a slightly reduced rate, leading to longer polymerization-associated attachment time; tethering was lost during depolymerization (fig. S11, B and C) (37). The catastrophe frequency of the CLASP2α-tethered ends was noticeably lower, as reported previously for soluble CLASPs (5, 24, 37). Thus, tubulin assembly at the ends tethered by the bead-conjugated proteins follows the patterns described previously for their soluble counterparts. These findings confirm the functional activity of the bead-bound tethering proteins, which may be additionally assisted by their residual soluble fractions.
After establishing the microtubule-stabilizing activity of CLASP2α at the tethered microtubule ends, we next analyzed the pivoting behaviors of these microtubules and compared them with microtubules tethered via other proteins. End tethering via Ndc80 (movie S7) and EB1, which bind to lateral sites on the microtubule wall (57, 58), supported microtubule elongation along straight lines (Fig. 7C and fig. S12). This behavior implies a consistently rigid structure at the microtubule ends tethered by these proteins. CLASP2α-coupled polymerization was also directional; however, microtubules showed irregular pivoting (Fig. 7, D and E), suggesting occasional softening of the bead-microtubule connection. Large deviations had variable durations and were seen only sporadically (movie S8), so they were unlikely to result from the overall softer CLASP2α linkages compared to Ndc80 and EB1 proteins. We also observed significantly less pivoting for CLASP2α with stabilized microtubules, indicating that large angular deviations are caused at least in part by the changing rigidity of the dynamic microtubule end (Fig. 7E). Temporary loss of tip integrity observed with this assay is consistent with the idea that CLASP2α forms high-affinity bonds with the sporadically arising non-GTP subunits, stalling the assembly of corresponding tubulin protofilaments.
DISCUSSION
Understanding the molecular mechanisms that regulate microtubule dynamics is both important and challenging. Great progress has been achieved using in vitro assays that examine microtubule dynamics in the presence of soluble forms of microtubule regulators. CLASPs, however, often function in cells as multimolecular ensembles concentrated near the microtubule tips, such as those seen at mitotic kinetochores. By leveraging DNA origami technology and bead-tethering assay, we investigated the molecular activities of CLASP2α amplified by their clustering and without the interference from EB proteins, which are themselves potent regulators of microtubule structure and dynamics (59). Integrating these results with prior work by others [reviewed in (6, 60)] leads to a specific model for CLASP2-dependent stabilization of microtubule dynamics.
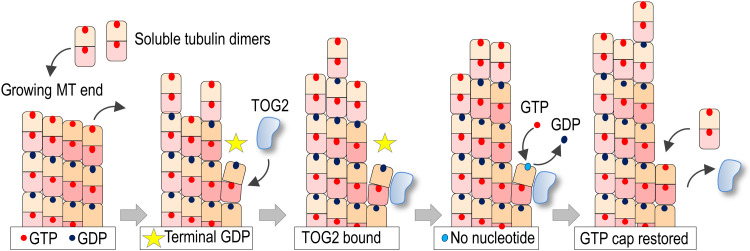
We propose that the TOG2 domain of CLASP2α has the ability to bind quickly and specifically to a non-GTP tubulin dimer in the most extreme, terminal position of a polymerizing microtubule tip. Although dynamic microtubule ends elongate by incorporating GTP-tubulins [reviewed in (48)], it is now well established that the structure and composition of the growing microtubule tip are highly complex and variable [reviewed in (61)]. Terminal non-GTP tubulins could arise at a growing microtubule tip via various mechanisms (48, 50, 52, 62), including an occasional dissociation of GTP-tubulin or accidental breakage of a vibrating protofilament extension, as seen in a computer model that accounts for protofilament mechanics under thermal forces (63). These pathways may generate terminal GDP-tubulins, but other non-GTP tubulin forms such as the GDP-Pi–containing subunits or apotubulins that have no associated nucleotides may also become exposed stochastically. Although the microtubule tip has considerable stability and can tolerate the brief presence of several terminal GDP-tubulins (63), non-GTP subunits have destabilizing and catastrophe-promoting effects because of their tendency to disassemble and also because they represent a poor site for incorporating new GTP-tubulins. It has been previously hypothesized that factors that influence the occurrence of the exposed non-GTP tubulins, e.g., via the rate of nucleotide exchange, may constitute a distinct mechanism for dynamic microtubule end regulation (50, 62); however, such regulators have not been identified. Here, we propose that CLASP2α exerts its anti-catastrophe activity at the assembling tip by binding to non-GTP terminal subunits and stabilizing them against dissociation while facilitating the acquisition of GTP from a soluble pool (Fig. 8). Justifications, possible implications of this model, and the arising questions are discussed below.
Fig. 8. Molecular model for CLASP2α function at polymerizing microtubule plus-end.
Two-dimensional schematic of a growing microtubule end illustrating the proposed cycle of TOG2 anti-catastrophe activity. TOG2 binds to terminal GDP-tubulin, which becomes exposed after a stochastic dissociation of a distal GTP-tubulin. As explained in the main text, other molecular pathways could generate exposed non-GTP subunits. Bound TOG2 stabilizes this polymerization-incompetent dimer against dissociation. As the neighboring protofilaments continue to elongate, a nanoscale indentation may form above the affected protofilament. Spontaneous GTP exchange at the exposed E-site releases TOG2, and the repaired protofilament resumes the assembly of GTP-tubulins. For simplicity, elongating protofilaments are depicted straight, and GTP hydrolysis is depicted to occur immediately after the addition of distal GTP-tubulins, leading to a single-layer GTP cap. This two-dimensional schematic also fails to show correctly the GDP-tubulin bending relative to the neighboring protofilaments, which is shown more accurately with a top view in fig. S13C. TOG2 domain is shown curved according to structural findings for human TOG2, but the exact shape of this domain is not a defining requirement in our model (see Discussion).
Structural work suggests that TOG2 binds along the tubulin dimer without hindering nucleotide exchange at the E-site of the associated β-tubulin (fig. S13A) (27). In our assay, the addition of soluble GTP caused CLASP2α dissociation from terminal tubulins with an IC50 value of 320 nM, which is well below cytoplasmic GTP concentration. Thus, soluble GTP concentration has no regulatory role for CLASP2α-tubulin interactions under the normal cellular environment. Instead, the proposed mechanism leverages the natural kinetics of GTP binding to terminal GDP-tubulin. For robust dynamic instability, the rate of GDP replacement within terminal tubulin dimers must not exceed the rate of GDP-tubulin dissociation. If the GDP-to-GTP exchange at the terminal GDP-tubulins was highly efficient, microtubules depolymerizing in the presence of GTP would quickly acquire terminal GTP-tubulins. The presence of polymerization-competent GTP-tubulins at the disassembling microtubule tip should lead to frequent pausing and possibly rescue, which is seen rarely with pure tubulin in vitro. This consideration about the relatively slow rate of soluble GTP binding to the exposed E-site suggests two possible mechanisms for TOG2-dependent microtubule stabilization. The parsimonious model that we currently favor is that TOG2 binding to terminal GDP-tubulin stalls this protofilament’s dis/assembly until GDP-to-GTP exchange takes place spontaneously. The alternative model is that TOG2 bound to terminal GDP-tubulin has a catalytic role and induces a polymerization-competent conformation of terminal GDP subunit. We considered the latter model in view of the prior finding of a putative GTP binding site within the CLASP ortholog in Drosophila (3); however, our mutational analysis of human CLASP2α in vitro and in mitotic cells has ruled out that this site is involved. It is interesting in this respect that the nonhydrolyzable GTP analogs, GMPCPP and GTPγS, have not been able to substitute for GTP in promoting TOG2 dissociation from microtubule tips in our assays. This could be explained by their lower affinity to the E-site relative to GTP. Alternatively, GTP hydrolysis might be required for TOG2 stabilizing activity, so these possibilities will require future investigation.
The model in which the TOG2 domain exerts its stabilizing activity without accelerating GTP exchange or modulating GTP hydrolysis is consistent with the prior finding that CLASPs do not affect the length of the GTP cap in microtubules growing in vitro (38). In addition, in the terminal-dimer binding mode to occasional rogue tubulins, CLASPs can exert stabilizing effects without significantly affecting the rates of microtubule assembly, a behavior reported by multiple studies (5). Strong TOG2 binding to terminal GDP-tubulins also explains CLASPs’ inability to affect shortening rates and track with depolymerizing ends because tip tracking by soluble proteins requires weak and diffusive interactions (64), which may be impeded by strong TOG2 binding. Our model is consistent with findings that in the absence of EB-dependent recruitment, CLASP2α is barely localizing to a dynamic microtubule tip (5), which is explained in the model by a relatively infrequent occurrence of non-GTP tubulins in terminal positions. In addition, prior mutational analysis demonstrated that conserved tubulin-binding residues in TOG2 domain abrogate both anti-catastrophe and rescue activities (25), suggesting that they are mediated by similar mechanisms. In the model, simultaneous binding of several TOG2 molecules to the ends of protofilaments during depolymerization could trigger microtubule rescue by the same mechanism as during CLASP’s anti-catastrophe activity at the polymerizing end. Consistent with this proposal, Aher and colleagues (24) observed brief CLASP2 binding events on shrinking microtubule ends immediately before rescue. Likewise, CLASP2 may become enriched at the damaged sites in the microtubule wall (65) by binding to GDP-tubulins that become exposed and acquire conformational features of terminal GDP subunits. In addition to the proposed binding mode for TOG2, CLASPs may recognize some yet-unidentified feature of the GTP cap or bind to curved protofilaments via other domains, such as TOG3 or S. These complex molecular interactions remain to be investigated.
The proposed molecular mechanism offers a plausible explanation for the apparent paradox that CLASP proteins are required for tubulin incorporation at the kinetochore-tethered ends, while at the same time, they show no strong effect on the rate of tubulin addition at the freely elongating microtubule ends. We explain this difference by the presence of tension at the kinetochore-tethered microtubule ends, which is likely to generate unstable protofilaments more frequently than at the freely polymerizing ends. However, tethering by various kinetochore-associated proteins ensures that microtubule ends remain bound regardless of their dynamic state. CLASPs may assist in capturing the ends of tethered protofilaments with terminal GDP-tubulins and in promoting their “conversion” into the polymerization-competent state. At the kinetochore, this activity would lead to persistent elongation of the tethered ends (fig. S13B), while the same molecular mechanism is manifested in reduced catastrophes at the microtubule ends that are not tethered. A cluster of about 10 CLASP2α molecules in our assays provides hours-long binding to a microtubule end. This collective, multivalent bond can withstand a pulling force up to 20 pN (Fig. 3); CLASP2α may also stabilize microtubule ends under compression (24). Thus, by forming molecular “nanoclasps” at the catastrophe-prone microtubule ends, clusters of CLASP2α that localized at the kinetochores or in other intracellular locations could promote persistent elongation of tubulin protofilaments under adverse force conditions.
Our work highlights several interesting and still unresolved questions about the mechanism of TOG2 activity. Notably, it is not clear how exactly TOG2 domain binds to terminal tubulin. Human TOG2 is highly bent, which has been interpreted to prevent it from forming an extended binding interface with tubulin dimers (26, 28, 60). However, molecular dynamic modeling suggests that tubulin dimers undergo significant bending and twisting under thermal forces (66). The intradimer bending occurs on a microsecond scale, with the maximum bending angle sufficient to provide contact with the convex surface of the TOG2 domain, so it is plausible that TOG2 binds to terminal tubulin quickly and with high affinity. We speculate that the highly curved surface of TOG2 domain serves to discriminate between terminal GDP- and GTP-tubulin dimers. Binding to the latter would lead to counterproductive interference with microtubule end dynamics, and it is likely avoided because GTP-tubulin is, on average, straighter than GDP dimers (67). Thus, although both forms of dimers at the protofilament tips are likely to undergo frequent outward bending, only GDP-tubulin may exhibit a sufficiently extreme conformation and become “captured” by TOG2.
It is also not known what happens at the site of TOG2 binding at a growing microtubule tip. We favor the idea that terminal non-GTP tubulin complexed with TOG2 is not competent to bond with incoming GTP-tubulin subunits, giving rise to a stalled protofilament. We base this proposal, in part, on findings by Aher and colleagues (24), who reported frequent occurrence of stalled protofilaments in the presence of EB3 and CLASP2α proteins. A similar behavior was seen when EB3 was combined with eribulin, a specific blocker of protofilament dynamics (53), suggesting that the tip-bound CLASP2α may also stall protofilament elongation. Long protofilament protrusions, which assemble adjacently to the stalled protofilaments, have not been observed with CLASP alone, implying that stalling events are likely to be on a nanoscale. We used microtubule pivoting as a proxy for CLASP2α-dependent formation of incomplete protofilament assemblies. Rare but large pivoting amplitudes were observed sporadically at the tethered polymerizing tips in the presence of CLASP2α but not with other microtubule-binding proteins (Fig. 7). Lesser pivoting was also observed when CLASP2α tethered the stabilized microtubule ends, consistent with their mechanically stable composition. Our observations provide indirect support for the idea that TOG2 binding to the stochastically arising non-GTP tubulins temporarily blocks tubulin assembly at that site. These events are transient because after the terminal E-site acquires GTP from a soluble pool, TOG2 dissociates (Fig. 8), and as reported in (53), the “repaired” protofilament can catch up quickly to restore typical microtubule end integrity. EB3 appears to have a synergistic effect with CLASP2α in the frequent formation of highly elongated protrusions (53). Perhaps, this synergism is explained by the EB3-induced maturation of GTP-tubulins into their catastrophe-prone forms (generating more frequent targets for TOG2 binding) in combination with the EB3-dependent acceleration of GTP-tubulin assembly (generating longer protrusions) (56, 68–70). Similar effects likely explain why, in the presence of EB3ΔTail (EB3 variant that does not bind CLASP2), TOG2-S protein localizes behind the microtubule tip extremity (24). This localization is not incompatible with our model because it suggests that TOG2 domain is actively engaged in “repairing” rogue GDP dimers, which may form frequently in the presence of catastrophe-promoting EB3ΔTail protein.
Future studies should also address the exact molecular mechanism of TOG2-dependent stabilization of a growing microtubule tip. In our model, terminal non-GTP tubulin complexed with TOG2 is prevented from dissociating from the microtubule tip. Durable CLASP2α attachment was observed with terminal non-GTP tubulins at the ends of stabilized microtubules (Figs. 2 and 3), and the model assumes similarly strong binding between TOG2 and non-GTP tubulin at the extremity of the dynamic microtubule. The reasons preventing the dissociation of the highly curved TOG2-tubulin complex from the microtubule tip are not intuitive, as the outward bending of terminal non-GTP dimers is a hallmark feature of the unstable microtubule ends. One probable explanation is that the TOG2-tubulin complex is stabilized against dissociation by lateral interactions with adjacent protofilaments. This assisting bonding can be provided by the charged residues within the L or S domains or by the TOG2 domain itself. The latter possibility is suggested by a theoretical analysis of terminal subunits’ conformation, which reveals that GDP-tubulins bend outward with a pronounced twist (66). Twisting of terminal GDP-tubulin complexed with TOG2 may permit the establishment of a lateral bond between the tubulin-bound TOG2 and the neighboring protofilament (fig. S13C). This three-step conformational mechanism, bend-twist-bond, may explain the stabilizing activity of TOG2, but critical analysis of such complex dynamic interactions will require advanced modeling in combination with high-resolution structural analysis and targeted mutations. This consideration, however, highlights a current lack of knowledge about the complex relationship between the stabilizing and destabilizing molecular features of the dynamic tip. The outcome (stabilizing or not) for different perturbations at the microtubule tip appears to be sensitive to the exact molecular and conformational composition of the tip, a biochemical milieu and other parameters. Another TOG protein, XMAP215, under different circumstances can exhibit both activities, polymerization promoting and microtubule destabilizing, which likely represent different aspects of a common biochemical mechanism (71). Thus, strong TOG2 binding to terminal tubulins may have a stabilizing effect at the normally polymerizing ends, while this interaction may lead to destabilization of terminal tubulins under some other conditions. We hope that our work and these ideas will guide and promote future research of the molecular mechanisms of CLASP-dependent regulation of dynamic microtubule tips.
MATERIALS AND METHODS
DNA origami construction and labeling
The DNA origami ring structure was designed in caDNAno2 (caDNAno.org). The cylinder part was designed as in (33). ssDNA handles extended from the 3′ end of staple strands at positions indicated in fig. S1C and had the following sequences: 24 handles at the outer surface of the ring (AAATTATCTACCACAACTCAC ssDNA), 8 handles at the inner surface (CTTCACACCACACTCCATCTA ssDNA), and 4 handles at the bottom of the ring (CGGTTGTACTGTGACCGATTC ssDNA). The origami structures were assembled through an 85° to 25°C annealing protocol for 36 hours in TE buffer [10 mM tris-HCl (pH 8.0) and 1 mM EDTA] supplemented with 10 mM MgCl2. Molecular mass of assembled DNA origami was 5 MDa. The assembled sample was purified by rate-zonal centrifugation through a 15 to 45% glycerol gradient in 1× TE supplemented with 10 mM MgCl2 in an SW 55 rotor (Beckman Coulter) at 303,800g rotational centrifugal force at 4°C for 1 hour, and fractions were collected and stored at −20°C. To carry out negative-stain electron microscopy, purified DNA origami samples were deposited onto glow-discharged 400-mesh formvar/carbon-coated copper grids (Electron Microscopy Sciences). Grids were then stained with 2% uranyl formate. Imaging was performed on a JEOL JEM-1400 Plus microscope operated at 80 kV with a bottom-mount 4k × 3k charge-coupled device (CCD) camera (Advanced Microscopy Technologies) (33).
Protein-conjugating outer surface anti-handles were generated using 5′-amino–labeled oligonucleotides (oligos) purchased from Integrated DNA Technologies (IDT, IA, USA). These oligos were labeled with BG by mixing 4 μl of 5′-NH2-oligos at 2 mM, 8 μl of 200 mM Hepes buffer (pH 8.5), and 12 μl of BG-GLA-NHS (catalog no. S9151S, NEB, MA, USA) prepared in dimethyl sulfoxide at 20 mM. The mixture was incubated for 30 min at 25°C, and the DNA oligos were precipitated by addition of 226 μl of 100% ethanol and 50 μl of solution containing 500 mM NH4Ac, 10 mM Mg(Ac)2, and 2 mM EDTA. The mixture was kept at −20°C overnight and spun at 21,000g for 30 min at 4°C. Ethanol supernatant was aspirated; pellet was air-dried, then resuspended in 200 μl of Milli-Q H2O, and stored in aliquots at −20°C. Successful BG modification for each preparation was confirmed by running a sample on a 20% polyacrylamide gel (fig. S1D).
To label inner and bottom anti-handles, we used antisense 5′-Cy5 anti-handles and 5′-biotin anti-handle, respectively (IDT). To attach anti-handles, DNA origami stock diluted to 5 nM with 1× TE buffer supplemented with 10 mM MgCl2 was mixed with 400 nM Cy5-oligos and 200 nM biotin-oligos. The mixture was incubated for 2 hours at 37°C, diluted 10 times with TE buffer supplemented with 10 mM MgCl2, aliquoted, snap-frozen, and stored at −80°C. Concentrations of DNA origami and ssDNA oligos were determined by measuring absorbance at 260 nm with NanoDrop (Thermo Fisher Scientific, MA, USA).
Construction of CLASP2α variants for in vitro studies
Truncated CLASP2α protein L-TOG2-S is analogous to the protein with the same name described in (24), but it was designed using human CLASP2α sequences (GenBank; NM_015097.3). It was constructed using synthesized DNA fragment with L-TOG2-S (from K261 to S793 amino acids; Genewiz) with codon usage optimized for expression in Escherichia coli. Tags (6xHis and GFP) and a linker containing SGGGGSGGGGSGGGG sequence were added at the 5′ end of the L-TOG2-S open reading frame and subcloned between the Spe I and Hind III restriction sites of the expression vector pRSETa, generating GFP-L-TOG2-S fusion protein. GFP-L-TOG2, containing sequence from K261 to S557 of CLASP2α, was generated by truncating the S domain at the Hind III and Psp XI restriction sites, followed by blunting with Klenow fragment and religation. GFP-L-TOG2 tubulin construct containing substitutions W340E and K425A at the tubulin-binding TOG2 interface was generated by subcloning the synthetic DNA fragment with these mutations (IDT) between Bst API and Afl II of the GFP-L-TOG2 plasmid. To determine the role of putative GTP binding site, we used radical substitutions of essential residues N, K, and D, which contribute to guanidine recognition (47). Site-directed mutagenesis was performed at the Penn Genomics Analysis Core to generate GFP-L-TOG2-S-SKFS (containing N403S and D406S) and GFP-L-TOG2-S-NMFD with K404M.
Protein purification for in vitro studies
Tubulin was purified from cow brains by thermal cycling and phosphocellulose (Poly Sciences, 19792-100) chromatography as in (72). Tubulin was labeled with rhodamine or HiLyte647 as in (73). Human GFP-tagged Ndc80 Broccoli was purified as in (74), GFP-tagged CENP-E kinesin motor domains as in (75), human EB1-GFP-6x His as in (70), and SNAP-GBP as in (35). Drosophila Kinesin-1 (K560 construct) was purified and labeled with biotin (biotin-K560) as in (76). Purification of human full-length GFP-CLASP2α was performed as in (37). Truncated CLASP2α constructs were expressed in E. coli Arctic Express (DE3) cells (Agilent, CA, USA), grown in 2× Yeast Extract Tryptone (YT) broth (Thermo Fisher Scientific, MA, USA) to reach optical density of 0.9 to 1.0, and purified following the protocol in (24) with modifications. Briefly, cultures were snap-cooled on ice for 30 min, and protein expression was induced by the addition of 0.5 mM 1-thio-β-galactopyranoside (LabScientific, NJ, USA) for 22 to 24 hours at 10°C with shaking at 240 rpm. Cells were harvested by centrifugation at 5000g for 30 min at 4°C and then resuspended in lysis buffer: 50 mM Hepes, 500 mM NaCl, 10 mM imidazole, 5% glycerol, 2 mM β-mercaptoethanol, 0.5 mM 4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF) (GOLDBIO, MO, USA), and “cOmplete protease inhibitors cocktail” (Roche, Basel, Switzerland). Cells were lysed by sonication, and a crude lysate was centrifuged at 22,000 rpm for 30 min (Ti50.2 rotor and optima XPN-80 ultracentrifuge; Beckman Coulter, CA, USA). Supernatant was loaded onto the HisTrap HP 1-ml Ni2+column operated with ÄKTA pure chromatography system (GE Healthcare, IL, USA). The column was washed with 20 column volumes of the lysis buffer without protein inhibitors, and protein was eluted via 10 to 250 mM imidazole gradient. Affinity purification was followed by size exclusion chromatography using a Superdex 200-pg 16/600 column (GE Healthcare, IL, USA) equilibrated with size exclusion chromatography buffer (50 mM Hepes, 500 mM NaCl, 5% glycerol, and 2 mM β-mercaptoethanol). Fractions containing target protein were pooled and concentrated using 10k MWCO centrifugal filters (Amicon, MA, USA). Protein was aliquoted with final 25% glycerol, snap-frozen, and stored at −80°C.
Before each experiment, a frozen protein aliquot was diluted 2 to 10 times in phosphate-buffered saline (PBS) [137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate buffer, and 1.8 mM KH2PO4 (pH 7.4)] and clarified by ultracentrifugation (TLA100 rotor; Beckman Coulter) at 27,904g for 15 min at 4°C. Concentration of GFP-tagged protein in the supernatant was determined by measuring GFP intensity by fluorescence microscopy and comparing it to a “standard” GFP-labeled protein whose concentration was determined by spectrometry, as described in (55).
Fluorescence microscopy assays
In vitro assays using fluorescence imaging with no force application were carried out with an inverted Eclipse Ti-E Nikon microscope equipped with a 1.49 numerical aperture 100× oil objective, Perfect Focus system, and Andor iXon3 CCD camera (Cambridge Scientific, MA, USA), as in (75). Three diode lasers (488, 561, and 640 nm from Coherent, CA, USA) were used as a light source with a C-TIRF Quad cube (Chroma, VT, USA). The multicolor TIRF was implemented by rapidly switching between different laser wavelengths with an acousto-optical tunable filter and corresponding emission filter wheels. Under these conditions, the microscope produced 512 × 512 pixel images with a resolution of 0.14 μm/pixel in both the x and y directions. Illumination was carried out with a partially opened iris diaphragm to limit laser illumination only by the field of view during each exposure. Each imaging channel was recorded with a 300-ms exposure time unless otherwise specified. All experiments were carried out at 32°C by heating the objective with an objective heater (Bioptechs, PA, USA).
Preparation of flow chambers with the coverslip-immobilized DNA origami
Coverslips (22 mm by 22 mm) were plasma-cleaned and silanized with Repel-Silane ES (GE Healthcare Life Sciences, 17-1332-01), as in (77). To form a flow chamber, a silanized coverslip was attached to a regular glass slide using four strips of the double-sided tape (Scotch, MN, USA) as spacers, which formed three independent parallel flow channels with ~10-μl volume each. Body Double fast silicone rubber (catalog no. 83741, Smooth-On, PA, USA) was painted on a glass slide to create inlet and outlet grooves that prevented solution mixing between the channels. Solutions were exchanged by placing the assembled chambers on a gently sloped surface, adding 15 μl at the inlet of each channel one by one, and letting solutions flow by gravity. Each incubation was performed for 10 min followed by washing with 50 μl of PBS-BSA (bovine serum albumin) buffer [PBS supplemented with BSA (4 mg/ml) and 2 mM dithiothreitol (DTT)], unless specified otherwise. To immobilize DNA origami, we first incubated chambers with 22.5 μM biotinylated BSA (biotin-BSA; catalog no. A8549, Sigma-Aldrich, MO, USA) and then with 25 μM NeutrAvidin (catalog no. 31000, Thermo Fisher Scientific, MA, USA), followed by 5 pM biotin- and Cy5-labeled DNA origami in TE buffer supplemented with 10 mM MgCl2; the surfaces were then blocked with 1% Pluronic F-127 (catalog no. T2443, Sigma-Aldrich).
To conjugate GFP-tagged proteins to coverslip-immobilized DNA origami, SNAP-GBP (5 μM) was incubated with BG-oligos (1 μM) in an Eppendorf tube for 1 hour at 25°C and diluted to 200 nM in Mg-BRB80-casein buffer containing 80 mM Pipes (pH 6.9), 4 mM MgCl2, 1 mM EGTA, casein (0.5 mg/ml; catalog no. C5890, Sigma-Aldrich), BSA (4 mg/ml), and 2 mM DTT. The mixture was added to each channel, incubated for 10 min, and washed with Mg-BRB80-casein. GFP-tagged protein (10 nM) in Mg-BRB80-casein buffer was incubated for 10 min, and the chamber was washed with “imaging buffer” [Mg-BRB80-casein buffer supplemented with up to 10 mM DTT, glucose (6 mg/ml; catalog no. g8270, Sigma-Aldrich), catalase (20 μg/ml; catalog no. C40, Sigma-Aldrich), and glucose oxidase (0.1 mg/ml; catalog no. G2133, Sigma-Aldrich)]. The chamber was placed on the prewarmed microscope stage and imaged in GFP and Cy5 channels to collect images of 10 to 25 fields separated by at least 100 μm to avoid prebleaching. Typical settings for imaging of functionalized DNA origami in GFP and Cy5 channels were 10% intensity of 488-nm laser and 30% intensity of 640-nm laser. The settings for Andor iXon3 camera for both fluorophores were as follows: gain, 5.0; EM gain, 300; 1-MHz readout speed; and 300-ms exposure time.
Determining the number of GFP-containing protein molecules conjugated to DNA origami
Conjugation efficiency of GFP-tagged protein to DNA origami was calculated as a ratio of the total GFP intensity of DNA origami and the intensity of single GFP-tagged molecule. For semiautomatic analysis of total GFP intensity of DNA origami, we used a custom-written MATLAB program, which identified coordinates of local maximal pixel intensity in the Cy5 channel and then determined the integrated fluorescence intensity of the square regions (7 × 7 pixel) centered on these coordinates in the GFP channel. This procedure eliminated selection bias and avoided contaminating our datasets with intensities of occasional protein aggregates.
Intensity of single GFP-labeled molecules was determined in the same experimental chambers by quantifying the intensity of dim dots that did not colocalize with DNA origami. MATLAB program twobox based on DIPimage tool box (https://diplib.org/DIPimage.html; provided by P. Relich and M. Lakadamyali) selected local maximal pixel intensity in the GFP channel and collected integrated intensity I7 and I9 within two square regions centered on each selected dot: 7 × 7 pixel inner box and 9 × 9 pixel outer box. Data were collected in both GFP and Cy5 channels, and only GFP dots that lacked Cy5 brightness were used. Because of the selected regions’ geometry, background intensity in the GFP channel corresponds to the average pixel intensity of the space between inner and outer boxes multiplied by the number of pixels in the inner box. Thus, the integrated GFP intensity of each dot, Idot, can be calculated using the following equation
Intensity distribution for single molecules was then generated, and negative values (arising from image noise) were removed. Intensity of single GFP-labeled molecules was then calculated as a peak value of Gaussian fitting.
Because some proteins may cluster or aggregate, we additionally determined single-molecule intensity using photobleaching, as in (77). Briefly, dim GFP dots that attached nonspecifically in the DNA origami chambers were imaged for 2 min via stream acquisition at the rate of 300 ms per frame. Using MetaMorph software (Molecular Devices, CA, USA), each clearly discernible dot was surrounded with the circle 8 pixels in diameter while avoiding dots that were less than 4 pixels apart. After subtracting the background intensity, the intensity histogram for bleaching curves was fitted with an equidistant Gaussian function, leading to a highly similar estimate of single-molecule intensity as with the approach that used only the initial brightness of GFP dots rather than the entire photobleaching curve. The intensities of single molecules of Ndc80 Broccoli-GFP and GFP-CLASP2α determined with these methods were similar (fig. S2E), consistent with the predominantly monomeric form for these proteins at nanomolar concentrations in our assays (24, 37, 39). The achieved labeling efficiency (~44%) is typical of DNA origami–directed protein assemblies, considering the handle incorporation rate, SNAP-to-BG reaction yield, and GFP-GBP binding affinity.
Assays with stabilized microtubules and DNA origami–conjugated protein clusters
Taxol-stabilized microtubules were prepared as described in (55) using unlabeled tubulin (10 mg/ml) and HiLyte647-labeled tubulin (degree of labeling, 0.18; 8 mg/ml) in a 5:2 ratio, rhodamine-labeled tubulin (degree of labeling, 0.9; 6 mg/ml) in a 10:1 ratio, and 1 mM Mg-GTP (catalog no. G8877, Sigma-Aldrich). GMPCPP-stabilized microtubules were prepared analogously but with 1 mM GMPCPP (catalog no. NU405L, Jena Bioscience, Jena, Germany). After polymerization, microtubules were diluted 10 times with Mg-BRB80 buffer and spun at 16,000g at room temperature for 15 min. The pellet was resuspended, and the spinning procedure was repeated one more time to remove soluble tubulin. Microtubules were kept in Mg-BRB80 buffer supplemented with 10 μM taxol (catalog no. T7402, Sigma-Aldrich) or 1 mM GMPCPP at room temperature.
To assay microtubule binding to the origami-based protein clusters, taxol-stabilized microtubules were diluted 1:10 in imaging buffer supplemented with 10 μM taxol and flowed into the chamber with functionalized DNA origami and incubated for 30 min. Images in Cy5 and GFP channels were captured to define the position of origami-based CLASP2α clusters. Time-lapse video of microtubules was taken in rhodamine channel for 30 to 360 min, as indicated for each experiment, at 1 frame every 30 s (exposure, 300 ms). The following imaging conditions were used: 1% intensity of 561-nm laser and camera settings gain, 5.0×; EM gain, 300; 1-MHz readout speed; and 300-ms exposure time. The number of origami-bound microtubules was determined as a function of time, and results were plotted as survival probability using the Kaplan-Meier algorithm implemented with Origin software (OriginLab, MA, USA). Lifetime of the tethered microtubules in the presence of different nucleotides was determined in an analogous way.
To determine the polarity of microtubules bound to the CLASP2α clusters, experiment was carried out analogously but using custom-made flow chambers with a syringe pump, enabling solution exchange at well-controlled flow rates and without chamber drying. After microtubules formed end attachments to the origami-based CLASP2α clusters, biotinylated K560 kinesin (400 nM in PBS) was flowed into the chamber and incubated for 5 min to immobilize K560 on the NeutrAvidin-coated coverslip surface. Then, Mg-ATP (10 μM; catalog no. A9187, Sigma-Aldrich) in imaging buffer was flowed in, and microtubule gliding was recorded using a 561-nm laser. Kymographs of gliding microtubules were prepared with MetaMorph and overlaid with static DNA origami images taken in GFP channel.
To investigate whether soluble tubulin can incorporate at the microtubule plus-ends bound to clusters of CLASP2α, end-on attachments were formed between origami-bound CLASP2α and GMPCPP-containing rhodamine-labeled microtubule seeds via incubation for 5 to 10 min and a wash to remove unbound microtubules. Soluble HiLyte647-labeled tubulin (8 μM) was flowed at 25 μl/min in imaging buffer supplemented with 1 mM Mg-GTP. Images were taken alternating HiLyte647 (10% laser intensity) and rhodamine channels (1% laser intensity) for 300 s with 300-ms exposure time and 5-s intervals and the following camera settings: gain, 5.0×; EM gain, 300; and 1-MHz readout speed.
Rupture force measurements using optical trap
Our laser trap instrument has been described in (78, 79). Chambers with immobilized DNA origami were prepared and functionalized with full-length CLASP2α as described above. Taxol-stabilized rhodamine-labeled microtubules were flowed in and incubated for 30 min at 32°C to promote the formation of end-on attachments. After unbound microtubules were washed away with imaging buffer, beads coated with anti-tubulin antibodies were flowed in. Beads were prepared as in (79). Briefly, streptavidin-coated polystyrene beads (catalog no. SPV-05-10, Spherotech, IL, USA) were incubated for 30 min at room temperature with biotinylated anti-tubulin antibodies (10 μg/ml; catalog no. 801211, BioLegend, CA, USA) in PBS supplemented with BSA (4 mg/ml) and 2 mM DTT. Beads were washed by centrifuging at 4500g for 7 min at 4°C, incubated with 0.8 mM b-PEG (biotinylated polyethylene glycol; catalog no. 10776, Quanta BioDesign, OH, USA) to block bead surface, and stored at 4°C for no more than 2 months. Before each experiment, 10 μl of beads was washed and resuspended in 10 μl of imaging buffer before introducing into the chamber; the chamber was sealed with Kwik-Cast Sealant (World Precision Instruments, Kwik-Cast) and placed on a microscope stage prewarmed to 32°C.
Tethered bead was found using differential interference contrast imaging. To confirm that the bead is bound to a single microtubule, the bead was pulled gently with a low-power optical trap, and the bead-bound microtubule was imaged briefly in rhodamine channel. Next, the bead was released from the trap, and microtubule images were acquired in a Z-stack with a 200-nm step. This procedure was necessary because most origami-bound microtubules were oriented perpendicularly to the coverslip plane, so it was not possible to capture a microtubule image in one focal plane. Image of the coverslip surface was then acquired in GFP channel to confirm colocalization of the CLASP2α protein cluster and tethered microtubule end. Next, the tethered bead was recaptured in the optical trap at the appropriate depth to avoid exerting any forces on the bead. The piezo-stage (Physik Instrumente, Karlsruhe, Germany) was moved along the y axis at 20 nm/s for 1 to 2 min, and bead coordinates were recorded with a quadrant photodetector operated at 5 kHz. After the stage stopped moving, bead, microtubule, and GFP-CLASP2α images were acquired again to determine the outcome.
To take into account that the tethered bead moved in both y and z directions, the total force acting on the bead was calculated as a square root of the sum of squared forces along these axes. Calibration of the quadrant photodetector was carried out as in (78) using free-floating beads that moved in x and y directions with an acoustic-optical deflector and coverslip-immobilized beads that moved along the z axis with the piezo-stage. Trap stiffness along the y and z axes measured using power spectrum method was 0.078 ± 0.003 and 0.029 ± 0.003 pN/nm, correspondingly.
Assays with coverslip-bound stabilized microtubules
Anti-tubulin antibodies (catalog no. 801211, BioLegend, CA, USA) were diluted 1:20 in PBS and flowed into the microscopy chamber for 15 min at room temperature. The chamber was washed with Mg-BRB80, and the coverslip surface was blocked by the addition of 1% Pluronic F-127 in Mg-BRB80 for 10 min. The chamber was washed thoroughly, and taxol-stabilized microtubules labeled with HiLyte647 were added for 10 min, after which the unbound polymers and any residual nucleotides used to polymerize microtubules were removed by washing. Freshly prepared microtubules were used to minimize the appearance of CLASP2α-containing foci along microtubule walls, presumably at the damage sites (65). GFP-tagged protein of interest was flowed into the chamber and incubated for 10 min at 32°C. Chamber was sealed and moved to the microscope stage prewarmed to 32°C. Imaging of one microfluidic channel took 2 to 3 min, and all three channels were imaged one after another.
Microtubule end decoration by full-length GFP-CLASP2α and GFP-L-TOG2-S was examined at 1 nM soluble protein concentration; GFP-L-TOG2 and GFP-L-TOG2-tubulin were assayed at 10 nM. Typically, we collected images for GFP and HiLyte647 wavelengths for at least 25 fields of view. Images of CLASP2 decoration of stabilized microtubules were acquired using the same settings as for other assays involving different GFP-tagged CLASP proteins, enabling a direct comparison of CLASP2 intensities in different figures. Images of Ndc80 Broccoli-GFP were acquired analogously but at higher soluble concentration of Ndc80, and EM gain was reduced 30-fold to avoid saturation. Images of microtubules were taken at 300-ms exposure time with the following camera settings: gain, 5.0×; EM gain, 300; and 1-MHz readout speed.
To determine the identity of the microtubule end with bright GFP-CLASP2α signal, chambers prepared with silanized coverslip were incubated with biotin-BSA and then NeutrAvidin, followed by incubation for 30 min with biotin-K560 (400 nM). Taxol-stabilized microtubules labeled with HiLyte647 were added in imaging buffer supplemented with 1 nM GFP-CLASP2α and 1 mM Mg-ATP. Gliding microtubules were imaged by alternating HiLyte647 (3% laser intensity) and GFP (10% laser intensity) channels for 120 s with stream acquisition (300-ms exposure for each channel) and using the following camera settings: gain, 5.0×; EM gain, 300; and 1-MHz readout speed.
Assays using guanosine nucleotides
Guanosine-containing nucleotides were prepared as Mg-containing salts. Briefly, powder of GTP, GDP (catalog no. G7127, Sigma-Aldrich), GMPCPP (catalog no. NU-1028, Jena Bioscience), or GTPγS (catalog no. G8634, Sigma-Aldrich) was dissolved in 50 mM MgCl2 to prepare nucleotide stock solution at 50 mM (or 10 mM for GTPγS). Then, pH was adjusted to 7 with NaOH, as verified with a pH test paper. The prepared stock solutions, as well as the commercial stock of GMCPPP (10 mM), were diluted to 100 μM in Mg-BRB80, aliquoted, and stored at −80°C. On the day of experiment, nucleotide stock was thawed and further diluted to 1 μM with imaging buffer. This concentration was chosen to be only ~3-fold higher than the IC50 value for GTP to reduce the effect from possible GTP contamination in these reagents. In assays examining the effect of different GTP analogs on CLASP2α interactions with microtubules, nucleotide-containing buffer was added to the microscopy chamber with immobilized microtubules and incubated for 30 min to allow ample time for nucleotide binding to tubulin. Similar binding affinity of GTP and GDP to stabilized microtubule end was confirmed using a competition assay with Cy3-GTP (fig. S9D) (50).
GFP-tagged full-length CLASP2α protein was diluted to 1 nM with imaging buffer containing a specified nucleotide and incubated for 20 min to allow time for nucleotide binding before adding to the microscopy chamber with immobilized microtubules. Experiments with CLASP2 mutant proteins were carried out in an analogous way using concentrations indicated for each figure.
To investigate nucleotide exchange at the tips of taxol-stabilized microtubules labeled with HiLyte647, Cy3-GTP (catalog no. NU-820-CY3, Jena Bioscience) was diluted to 100 nM in imaging buffer and incubated with coverslip-immobilized taxol-stabilized microtubules for 10 min, followed by imaging in rhodamine (Cy3) and HiLyte647 channels. The following imaging conditions for Cy3-GTP signal were used: 1% intensity of 561-nm laser and camera settings gain, 5.0×; EM gain, 300; 1-MHz readout speed; and 300-ms exposure time. For nucleotide competition experiments, 1 μM indicated unlabeled nucleotides were mixed with 100 nM Cy3-GTP and imaged as above.
Analysis of microtubule end decoration
Images of coverslip-immobilized microtubules decorated with indicated GFP-tagged proteins were analyzed using general procedures described in (80) with modifications. Briefly, only the central quadrant of each image was analyzed to reduce any effects from uneven illumination. Images were examined visually to select nonoverlapping microtubules that were longer than 2 μm and with both ends clearly visible. To avoid any bias, both microtubule ends were selected in HiLyte647 channel by placing 7 × 7 pixel boxes at both ends. A custom-made MATLAB program was then used to measure the integrated fluorescence intensity within each box in the corresponding GFP (or Cy3) image. Background intensity in the same field of view was measured in three manually selected microtubule-free regions (7 × 7 pixel). Their average integrated intensity was subtracted from the intensity measured on microtubules in the same field. At least 40 microtubules were analyzed for each experiment, and median intensity of the brightest microtubule end was determined. Apparent dissociation constant Kd for GFP-CLASP2α was determined using one-site-binding model: I = Imax × c/(Kd + c), where I is the intensity of microtubule end, Imax is a plateau intensity, and c denotes soluble GFP-CLASP2α concentration. Microtubule end decoration by Cy3-GTP was measured and analyzed analogously. Average intensity of single Cy3 fluorophore was determined by measuring the brightness of dots formed by Cy3-labeled ssDNA handles (38 ± 9 a.u. based on six independent experiments in which 2112 dots were examined).
Establishment of stable cell lines and RNA interference treatment
The wild-type and SKFS (N403S and D406S) mRFP-CLASP2γ constructs were inserted between Nde I–Not I sites in a CSII-CMV-MCS vector (RIKEN BRC DNA BANK; RDB04377). The SKFS construct was generated from the wild-type construct by site-directed mutagenesis using two overlap primers bearing the desired mutations. Lentiviral supernatants were produced in human embryonic kidney 293T cells using pMD2.G (catalog no. 12259, Addgene, MA, USA), psPAX2 (catalog no. 12260, Addgene), and the CSII-CMV-MLS plasmids, in the presence of Lipofectamine 2000 (Invitrogen, CA, USA) and Opti-MEM (Gibco, MD, USA).
A U2OS cell line stably expressing photoactivatable GFP-α-tubulin (81) was transduced with the viral supernatants, in the presence of Polybrene (6 μg/ml; Sigma-Aldrich). The cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS), supplemented with antibiotic-antimycotic mixture (10 μg/μl; Gibco, MD, USA), and selected with Zeocin (1 μg/μl). Cell lines were maintained at 37°C in a 5% CO2, humidified atmosphere. Cells expressing the different mRFP-CLASP2γ constructs were sorted using a FACSAria II cell sorter (Becton Dickinson, NJ, USA). Briefly, cells were resuspended in basic sorting buffer (1× PBS, Ca2+- and Mg2+-free; 5 mM EDTA; 25 mM Hepes; and 2% FBS). Cells expressing red fluorescence were sorted into a tube containing DMEM with 10% FBS and then transferred to an appropriate growth flask. RNA interference experiments were performed using scrambled small interfering RNA (siRNA) CUUCCUCUCUUUCUCUCCCUUGUGA, CLASP1α siRNA GCCAUUAUGCCAACUAUCU, or CLASP2α siRNA GUUCAGAAAGCCCUUGAUG with Lipofectamine RNAiMAX (Invitrogen), as described in (18).
Cell imaging and spindle measurements
All experiments were performed in the presence of N-carbobenzyloxy-l-leucyl-l-leucyl-l-leucinal (5 μM) added 30 min before imaging to ensure that cells were in metaphase and to prevent mitotic exit. To measure spindle length, cells were fixed, and microtubules were stained using mouse anti–α-tubulin clone B-512 at 1:1500 (Sigma-Aldrich) and secondary antibodies Alexa Fluor 488 anti-mouse, diluted in PBS–Triton X-100 (0.1%) with 10% FBS. DNA staining was achieved by the addition of 4′,6-diamidino-2-phenylindole (1 μg/ml; Sigma-Aldrich).
Live-cell imaging was used to measure microtubule turnover rate. A thin strip of spindle microtubules was locally photoactivated in one half-spindle by pulsed near-ultraviolet irradiation (405-nm laser; 500-μs exposure time). Fluorescence images (seven 1-μm–separated z planes centered at the middle of the mitotic spindle) were captured every 15 s for 4.5 min with a 100× oil-immersion 1.4 numerical aperture plan-apochromatic objective. Under these conditions, the photoactivated region dissipated, yet the photoactivated molecules were retained within the cellular boundaries defining the cytoplasm. To calculate microtubule turnover, the sum intensity at each time point was fit to a double exponential curve, A1 × exp(−k1 × t) + A2 × exp(−k2 × t) using MATLAB (MathWorks), in which t is time. A1 represents the less stable population (typically interpreted as the fraction corresponding to nonkinetochore microtubules), and A2 represents the more stable population (typically interpreted as the fraction corresponding to kinetochore microtubules), with decay rates of k1 and k2, respectively. The half-life for each process was calculated as ln2/k for each microtubule population. Spindle poleward flux rates were determined in the same cells used for quantification of spindle microtubule half-life by determining the slope of the fluorescence signal over time (4.5 min), using a MATLAB custom-written routine in LAPSO software (82) and using the slope of the movement of the metaphase plate as reference. Other details were as described in (18).
Tethered microtubule end assay with protein-coated beads
Experiments were carried out as in (37, 55). Briefly, streptavidin-coated polystyrene beads (catalog no. SVP-08-10, Spherotech) were immobilized on the coverslips and functionalized with biotinylated anti-GFP antibodies (20 μg/ml; catalog no. ab6658, Abcam). Chambers were washed with Mg-BRB80 supplemented with BSA (4 mg/ml), 0.1 mM Mg-ATP, and 2 mM DTT; unreacted streptavidins were blocked with 100 μM biotinylated PEG (catalog no.10776, Quanta BioDesign). The beads were then coated with a mixture of GFP-tagged proteins: kinesin CENP-E-GFP (0.3 μM) and an indicated microtubule-associated protein (0.4 μM). Chambers were washed thoroughly to remove soluble proteins, but their residual soluble fractions are likely to persist owing to nonspecific protein interactions with the chamber walls despite blocking and repeated washes. GMPCPP-stabilized HiLyte647-labeled microtubules were allowed to bind and glide on beads in the ATP-containing imaging buffer [Mg-BRB80 supplemented with BSA (4 mg/ml), 10 mM DTT, 2 mM Mg-ATP, glucose (6 mg/ml), catalase (80 μg/ml), and glucose oxidase (0.1 mg/ml)]. Microtubule gliding and subsequent pivoting at the tethered ends were visualized and recorded as in (37). In experiments with dynamic tethered ends, after the stabilized microtubule plus-end reached the bead and became tethered, unlabeled tubulin (6 μM) was introduced in imaging buffer supplemented with 1 mM Mg-GTP. The fraction of dynamic microtubule attachments was calculated as the ratio of the number of microtubules that showed at least one away motion to the total number of microtubules bound to beads within a particular field. Velocities of the away and backward motions, their respective duration, and the maximum lengths of polymerized microtubules for each dynamic cycle were estimated from kymographs. The total dynamic microtubule attachment time was calculated as the time between the start and end of all dynamic motions, the detachment of the seed from the bead, or the end of imaging. Microtubule catastrophe frequency was calculated by counting the observed number of catastrophes and dividing it by the sum of the elongation time for all polymerizing microtubules. Data on microtubule dynamics in Fig. 7B and fig. S11B for the freely polymerizing end and for bead-coupled Ndc80 Broccoli reproduce the results reported in figure 4d in (37) and are plotted for a direct comparison with CLASP2α and EB1. To quantify the extent of tethered microtubule pivoting, two-dimensional trajectories for motions of the leading microtubule ends were obtained using a point tracking tool in MetaMorph or FIJI. Positions (x,y coordinates) of the bead-distant end of the labeled seed relative to the attachment site on the bead were used to calculate pivoting angles θ = arc tan (y/x) for each point, and the resulting histograms were approximated with Gaussian function. Whenever possible, data are presented as an average of N independent trials, i.e., experiments carried out on different days or on the same day in different microscopy chambers. Small n denotes the total number of observed events in all trials for each experimental condition.
Acknowledgments
Biotinylated Kinesin-1 protein, expression plasmid, and purification protocol were provided by D. Arginteanu and W. Hancock (Penn State University). We thank P. Relich and M. Lakadamyali (University of Pennsylvania) for providing a program for fluorescence intensity analysis; K. Miletic for assistance with image analysis; E. Mattes, P.-T. Chen, and E. Tarasovetc for help with protein purification; and Grishchuk laboratory members for discussions. We are also grateful to J. R. McIntosh (University of Colorado) for critical reading of the manuscript and R. F. Hariadi (Arizona State University) for introducing us to DNA origami technology and providing help and reagents during the initial stages of this project. We are grateful to B. Black (University of Pennsylvania) for suggesting the use of circular origami for kinetochore reconstruction in vitro and R. Dominguez (University of Pennsylvania) for fruitful discussions.
Funding: Research reported in this publication was supported by the National Institute of General Medical Sciences of the NIH under award number R35-GM141747 to E.L.G., R01 AI162260 and P50 AI150481 (via a collaboration development program of Pittsburgh Center for HIV Protein Interactions) to C.L., and R01GM098389 to E.L.G.; by the American Cancer Society grant RSG-14-018-01-CCG to E.L.G.; by the Pittsburgh Foundation under C. Kaufman Foundation grant KA2017-91783 to E.L.G. and B. B. Black; by the European Research Council (ERC) consolidator grant CODECHECK, under the European Union’s Horizon 2020 research and innovation programme (grant agreement 681443); by Fundação para a Ciência e a Tecnologia of Portugal (PTDC/MED-ONC/3479/2020); and by a La Caixa Health Research Grant (LCF/PR/HR21/52410025) to H.M. F.I.A. acknowledges support from the Russian Science Foundation #21-45-00012. H.G. is a recipient of a PhD studentship from Fundação para a Ciência e a Tecnologia of Portugal (SFRH/BD/141066/2018).
Author contributions: W.L. performed experiments using DNA origami technology. M.C. performed microtubule end-conversion assays. V.D. and F.I.A. designed and performed experiments using optical trap. H.G. and H.M. designed and performed experiments using U2OS cells. Q.S. and C.L. designed and synthesized DNA origami scaffolds. A.M. constructed and purified CLASP2α truncated proteins and carried out sequence analyses. E.L.G. designed the research. E.L.G. and W.L. analyzed data and wrote the paper with input from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper, the Supplementary Materials, and Data Source file.
Supplementary Materials
This PDF file includes:
Supplementary Note
Figs. S1 to S13
Other Supplementary Material for this : manuscript includes the following:
Movies S1 to S8
Data file S1
REFERENCES AND NOTES
- 1.A. Akhmanova, C. C. Hoogenraad, K. Drabek, T. Stepanova, B. Dortland, T. Verkerk, W. Vermeulen, B. M. Burgering, C. I. De Zeeuw, F. Grosveld, N. Galjart, Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell 104, 923–935 (2001). [DOI] [PubMed] [Google Scholar]
- 2.C. L. Lemos, P. Sampaio, H. Maiato, M. Costa, L. V. Omel’yanchuk, V. Liberal, C. E. Sunkel, Mast, a conserved microtubule-associated protein required for bipolar mitotic spindle organization. EMBO J. 19, 3668–3682 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Y. H. Inoue, M. do Carmo Avides, M. Shiraki, P. Deak, M. Yamaguchi, Y. Nishimoto, A. Matsukage, D. M. Glover, Orbit, a novel microtubule-associated protein essential for mitosis in Drosophila melanogaster. J. Cell Biol. 149, 153–166 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Y. Mimori-Kiyosue, I. Grigoriev, G. Lansbergen, H. Sasaki, C. Matsui, F. Severin, N. Galjart, F. Grosveld, I. Vorobjev, S. Tsukita, A. Akhmanova, CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 168, 141–153 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.E. Lawrence, M. Zanic, Rescuing microtubules from the brink of catastrophe: CLASPs lead the way. Curr. Opin. Cell Biol. 56, 94–101 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.E. J. Lawrence, M. Zanic, L. M. Rice, CLASPs at a glance. J. Cell Sci. 133, jcs243097 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.N. Schmidt, S. Basu, S. Sladecek, S. Gatti, J. van Haren, S. Treves, J. Pielage, N. Galjart, H. R. Brenner, Agrin regulates CLASP2-mediated capture of microtubules at the neuromuscular junction synaptic membrane. J. Cell Biol. 198, 421–437 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.S. J. Stehbens, M. Paszek, H. Pemble, A. Ettinger, S. Gierke, T. Wittmann, CLASPs link focal-adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nat. Cell Biol. 16, 561–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.J. C. Ambrose, G. O. Wasteneys, CLASP modulates microtubule-cortex interaction during self-organization of acentrosomal microtubules. Mol. Biol. Cell 19, 4730–4737 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.C. Ambrose, J. F. Allard, E. N. Cytrynbaum, G. O. Wasteneys, A CLASP-modulated cell edge barrier mechanism drives cell-wide cortical microtubule organization in Arabidopsis. Nat. Commun. 2, 430 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.G. Lansbergen, A. Akhmanova, Microtubule plus end: A hub of cellular activities. Traffic 7, 499–507 (2006). [DOI] [PubMed] [Google Scholar]
- 12.B. P. Bouchet, I. Noordstra, M. van Amersfoort, E. A. Katrukha, Y.-C. Ammon, N. D. Ter Hoeve, L. Hodgson, M. Dogterom, P. W. Derksen, A. Akhmanova, Mesenchymal cell invasion requires cooperative regulation of persistent microtubule growth by SLAIN2 and CLASP1. Dev. Cell 39, 708–723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.B. Stramer, S. Moreira, T. Millard, I. Evans, C.-Y. Huang, O. Sabet, M. Milner, G. Dunn, P. Martin, W. Wood, Clasp-mediated microtubule bundling regulates persistent motility and contact repulsion in Drosophila macrophages in vivo. J. Cell Biol. 189, 681–689 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.H. Maiato, A. Khodjakov, C. L. Rieder, Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat. Cell Biol. 7, 42–47 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.S. Maffini, A. R. Maia, A. L. Manning, Z. Maliga, A. L. Pereira, M. Junqueira, A. Shevchenko, A. Hyman, J. R. Yates III, N. Galjart, D. A. Compton, H. Maiato, Motor-independent targeting of CLASPs to kinetochores by CENP-E promotes microtubule turnover and poleward flux. Curr. Biol. 19, 1566–1572 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A. L. Pereira, A. J. Pereira, A. R. Maia, K. Drabek, C. L. Sayas, P. J. Hergert, M. Lince-Faria, I. Matos, C. Duque, T. Stepanova, C. L. Rieder, W. C. Earnshaw, N. Galjart, H. Maiato, Mammalian CLASP1 and CLASP2 cooperate to ensure mitotic fidelity by regulating spindle and kinetochore function. Mol. Biol. Cell 17, 4526–4542 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Y. Mimori-Kiyosue, I. Grigoriev, H. Sasaki, C. Matsui, A. Akhmanova, S. Tsukita, I. Vorobjev, Mammalian CLASPs are required for mitotic spindle organization and kinetochore alignment. Genes Cells 11, 845–857 (2006). [DOI] [PubMed] [Google Scholar]
- 18.H. Girão, N. Okada, T. A. Rodrigues, A. O. Silva, A. C. Figueiredo, Z. Garcia, T. Moutinho-Santos, I. Hayashi, J. E. Azevedo, S. Macedo-Ribeiro, H. Maiato, CLASP2 binding to curved microtubule tips promotes flux and stabilizes kinetochore attachments. J. Cell Biol. 219, e201905080 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.N. Yu, L. Signorile, S. Basu, S. Ottema, J. H. Lebbink, K. Leslie, I. Smal, D. Dekkers, J. Demmers, N. Galjart, Isolation of functional tubulin dimers and of tubulin-associated proteins from mammalian cells. Curr. Biol. 26, 1728–1736 (2016). [DOI] [PubMed] [Google Scholar]
- 20.J. Al-Bassam, H. Kim, G. Brouhard, A. van Oijen, S. C. Harrison, F. Chang, CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev. Cell 19, 245–258 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.J. Al-Bassam, M. Van Breugel, S. C. Harrison, A. Hyman, Stu2p binds tubulin and undergoes an open-to-closed conformational change. J. Cell Biol. 172, 1009–1022 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.J. Al-Bassam, N. A. Larsen, A. A. Hyman, S. C. Harrison, Crystal structure of a TOG domain: Conserved features of XMAP215/Dis1-family TOG domains and implications for tubulin binding. Structure 15, 355–362 (2007). [DOI] [PubMed] [Google Scholar]
- 23.K. C. Slep, R. D. Vale, Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol. Cell 27, 976–991 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.A. Aher, M. Kok, A. Sharma, A. Rai, N. Olieric, R. Rodriguez-Garcia, E. A. Katrukha, T. Weinert, V. Olieric, L. C. Kapitein, M. O. Steinmetz, M. Dogterom, A. Akhmanova, CLASP suppresses microtubule catastrophes through a single TOG domain. Dev. Cell 46, 40–58.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.S. Majumdar, T. Kim, Z. Chen, S. Munyoki, S.-C. Tso, C. A. Brautigam, L. M. Rice, An isolated CLASP TOG domain suppresses microtubule catastrophe and promotes rescue. Mol. Biol. Cell 29, 1359–1375 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.J. B. Leano, S. L. Rogers, K. C. Slep, A cryptic TOG domain with a distinct architecture underlies CLASP-dependent bipolar spindle formation. Structure 21, 939–950 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.J. B. Leano, K. C. Slep, Structures of TOG1 and TOG2 from the human microtubule dynamics regulator CLASP1. PLOS ONE 14, e0219823 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.T. Maki, A. D. Grimaldi, S. Fuchigami, I. Kaverina, I. Hayashi, CLASP2 has two distinct TOG domains that contribute differently to microtubule dynamics. J. Mol. Biol. 427, 2379–2395 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.P. W. Rothemund, Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006). [DOI] [PubMed] [Google Scholar]
- 30.H. Dietz, S. M. Douglas, W. M. Shih, Folding DNA into twisted and curved nanoscale shapes. Science 325, 725–730 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.G. Kong, M. Xiong, L. Liu, L. Hu, H.-M. Meng, G. Ke, X.-B. Zhang, W. Tan, DNA origami-based protein networks: From basic construction to emerging applications. Chem. Soc. Rev. 50, 1846–1873 (2021). [DOI] [PubMed] [Google Scholar]
- 32.S. Dey, C. Fan, K. V. Gothelf, J. Li, C. Lin, L. Liu, N. Liu, M. A. D. Nijenhuis, B. Saccà, F. C. Simmel, H. Yan, P. Zhan, DNA origami. Nat. Rev. Methods Primers 1, 13 (2021). [Google Scholar]
- 33.P. D. E. Fisher, Q. Shen, B. Akpinar, L. K. Davis, K. K. H. Chun, D. Baddeley, A. Saric, T. J. Melia, B. W. Hoogenboom, C. X. Lin, C. P. Lusk, A programmable DNA origami platform for organizing intrinsically disordered nucleoporins within nanopore confinement. ACS Nano 12, 1508–1518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.J. R. McIntosh, E. O’Toole, K. Zhudenkov, M. Morphew, C. Schwartz, F. I. Ataullakhanov, E. L. Grishchuk, Conserved and divergent features of kinetochores and spindle microtubule ends from five species. J. Cell Biol. 200, 459–474 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.N. Gudimchuk, E. V. Tarasovetc, V. Mustyatsa, A. L. Drobyshev, B. Vitre, D. W. Cleveland, F. I. Ataullakhanov, E. L. Grishchuk, Probing mitotic CENP-E kinesin with the tethered cargo motion assay and laser tweezers. Biophys. J. 114, 2640–2652 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.K. Johnston, A. Joglekar, T. Hori, A. Suzuki, T. Fukagawa, E. Salmon, Vertebrate kinetochore protein architecture: Protein copy number. J. Cell Biol. 189, 937–943 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.M. Chakraborty, E. V. Tarasovetc, A. V. Zaytsev, M. Godzi, A. C. Figueiredo, F. I. Ataullakhanov, E. L. Grishchuk, Microtubule end conversion mediated by motors and diffusing proteins with no intrinsic microtubule end-binding activity. Nat. Commun. 10, 1673 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.E. J. Lawrence, G. Arpag, S. R. Norris, M. Zanic, Human CLASP2 specifically regulates microtubule catastrophe and rescue. Mol. Biol. Cell 29, 1168–1177 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.A. V. Zaytsev, J. E. Mick, E. Maslennikov, B. Nikashin, J. G. DeLuca, E. L. Grishchuk, Multisite phosphorylation of the NDC80 complex gradually tunes its microtubule-binding affinity. Mol. Biol. Cell 26, 1829–1844 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.B. Akiyoshi, K. K. Sarangapani, A. F. Powers, C. R. Nelson, S. L. Reichow, H. Arellano-Santoyo, T. Gonen, J. A. Ranish, C. L. Asbury, S. Biggins, Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature 468, 576–579 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.S. Cane, A. A. Ye, S. J. Luks-Morgan, T. J. Maresca, Elevated polar ejection forces stabilize kinetochore–microtubule attachments. J. Cell Biol. 200, 203–218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R. B. Nicklas, Measurements of the force produced by the mitotic spindle in anaphase. J. Cell Biol. 97, 542–548 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.A. A. Ye, S. Cane, T. J. Maresca, Chromosome biorientation produces hundreds of piconewtons at a metazoan kinetochore. Nat. Commun. 7, 13221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.H. Maiato, E. A. Fairley, C. L. Rieder, J. R. Swedlow, C. E. Sunkel, W. C. Earnshaw, Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell 113, 891–904 (2003). [DOI] [PubMed] [Google Scholar]
- 45.V. Verma, L. Mallik, R. F. Hariadi, S. Sivaramakrishnan, G. Skiniotis, A. P. Joglekar, Using protein dimers to maximize the protein hybridization efficiency with multisite DNA origami scaffolds. PLOS ONE 10, e0137125 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.H. R. Bourne, D. A. Sanders, F. McCormick, The GTPase superfamily: Conserved structure and molecular mechanism. Nature 349, 117–127 (1991). [DOI] [PubMed] [Google Scholar]
- 47.A. Wittinghofer, I. R. Vetter, Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem. 80, 943–971 (2011). [DOI] [PubMed] [Google Scholar]
- 48.A. Desai, T. J. Mitchison, Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83–117 (1997). [DOI] [PubMed] [Google Scholar]
- 49.T. Mitchison, Localization of an exchangeable GTP binding site at the plus end of microtubules. Science 261, 1044–1047 (1993). [DOI] [PubMed] [Google Scholar]
- 50.M. Caplow, J. Shanks, Induction of microtubule catastrophe by formation of tubulin-GDP and apotubulin subunits at microtubule ends. Biochemistry 34, 15732–15741 (1995). [DOI] [PubMed] [Google Scholar]
- 51.E. K. Anderson, D. S. Martin, A fluorescent GTP analog as a specific, high-precision label of microtubules. Biotechniques 51, 43–48 (2011). [DOI] [PubMed] [Google Scholar]
- 52.F. A. Piedra, T. Kim, E. S. Garza, E. A. Geyer, A. Burns, X. Ye, L. M. Rice, GDP-to-GTP exchange on the microtubule end can contribute to the frequency of catastrophe. Mol. Biol. Cell 27, 3515–3525 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.H. Doodhi, A. E. Prota, R. Rodriguez-Garcia, H. Xiao, D. W. Custar, K. Bargsten, E. A. Katrukha, M. Hilbert, S. S. Hua, K. Jiang, I. Grigoriev, C.-P. H. Yang, D. Cox, S. B. Horwitz, L. C. Kapitein, A. Akhmanova, M. O. Steinmetz, Termination of protofilament elongation by eribulin induces lattice defects that promote microtubule catastrophes. Curr. Biol. 26, 1713–1721 (2016). [DOI] [PubMed] [Google Scholar]
- 54.O. Kononova, Y. Kholodov, K. E. Theisen, K. A. Marx, R. I. Dima, F. I. Ataullakhanov, E. L. Grishchuk, V. Barsegov, Tubulin bond energies and microtubule biomechanics determined from nanoindentation in silico. J. Am. Chem. Soc. 136, 17036–17045 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.M. Chakraborty, E. V. Tarasovetc, E. L. Grishchuk, In vitro reconstitution of lateral to end-on conversion of kinetochore–microtubule attachments. Methods Cell Biol. 144, 307–327 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.P. Bieling, L. Laan, H. Schek, E. L. Munteanu, L. Sandblad, M. Dogterom, D. Brunner, T. Surrey, Reconstitution of a microtubule plus-end tracking system in vitro. Nature 450, 1100–1105 (2007). [DOI] [PubMed] [Google Scholar]
- 57.G. M. Alushin, V. H. Ramey, S. Pasqualato, D. A. Ball, N. Grigorieff, A. Musacchio, E. Nogales, The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature 467, 805–810 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.S. P. Maurer, F. J. Fourniol, G. Bohner, C. A. Moores, T. Surrey, EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell 149, 371–382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.A. Aher, A. Akhmanova, Tipping microtubule dynamics, one protofilament at a time. Curr. Opin. Cell Biol. 50, 86–93 (2018). [DOI] [PubMed] [Google Scholar]
- 60.V. J. Farmer, M. Zanic, TOG-domain proteins. Curr. Biol. 31, R499–R501 (2021). [DOI] [PubMed] [Google Scholar]
- 61.N. B. Gudimchuk, J. R. McIntosh, Regulation of microtubule dynamics, mechanics and function through the growing tip. Nat. Rev. Mol. Cell Biol. 22, 777–795 (2021). [DOI] [PubMed] [Google Scholar]
- 62.J. M. Cleary, T. Kim, A. S. I. Cook, L. A. McCormick, W. O. Hancock, L. M. Rice, Measurements and simulations of microtubule growth imply strong longitudinal interactions and reveal a role for GDP on the elongating end. eLife 11, e75931 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.P. Zakharov, N. Gudimchuk, V. Voevodin, A. Tikhonravov, F. I. Ataullakhanov, E. L. Grishchuk, Molecular and mechanical causes of microtubule catastrophe and aging. Biophys. J. 109, 2574–2591 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.E. L. Grishchuk, Biophysics of microtubule end coupling at the kinetochore, in Centromeres and Kinetochores: Discovering the Molecular Mechanisms Underlying Chromosome Inheritance, B. E. Black, Ed. (Springer International Publishing, 2017), vol. 56, pp. 397–428. [DOI] [PubMed] [Google Scholar]
- 65.A. Aher, D. Rai, L. Schaedel, J. Gaillard, K. John, Q. Liu, M. Altelaar, L. Blanchoin, M. Thery, A. Akhmanova, CLASP mediates microtubule repair by restricting lattice damage and regulating tubulin incorporation. Curr. Biol. 30, 2175–2183.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.V. A. Fedorov, P. S. Orekhov, E. G. Kholina, A. A. Zhmurov, F. I. Ataullakhanov, I. B. Kovalenko, N. B. Gudimchuk, Mechanical properties of tubulin intra- and inter-dimer interfaces and their implications for microtubule dynamic instability. PLOS Comput. Biol. 15, e1007327 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.R. Ayukawa, S. Iwata, H. Imai, S. Kamimura, M. Hayashi, K. X. Ngo, I. Minoura, S. Uchimura, T. Makino, M. Shirouzu, H. Shigematsu, K. Sekimoto, B. Gigant, E. Muto, GTP-dependent formation of straight tubulin oligomers leads to microtubule nucleation. J. Cell Biol. 220, e202007033 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.B. Vitre, F. M. Coquelle, C. Heichette, C. Garnier, D. Chrétien, I. Arnal, EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat. Cell Biol. 10, 415–421 (2008). [DOI] [PubMed] [Google Scholar]
- 69.R. Zhang, G. M. Alushin, A. Brown, E. Nogales, Mechanistic origin of microtubule dynamic instability and its modulation by EB proteins. Cell 162, 849–859 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.S. P. Maurer, N. I. Cade, G. Bohner, N. Gustafsson, E. Boutant, T. Surrey, EB1 accelerates two conformational transitions important for microtubule maturation and dynamics. Curr. Biol. 24, 372–384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.M. Shirasu-Hiza, P. Coughlin, T. Mitchison, Identification of XMAP215 as a microtubule-destabilizing factor in Xenopus egg extract by biochemical purification. J. Cell Biol. 161, 349–358 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.H. P. Miller, L. Wilson, Preparation of microtubule protein and purified tubulin from bovine brain by cycles of assembly and disassembly and phosphocellulose chromatography. Methods Cell Biol. 95, 3–15 (2010). [DOI] [PubMed] [Google Scholar]
- 73.A. Hyman, D. Drechsel, D. Kellogg, S. Salser, K. Sawin, P. Steffen, L. Wordeman, T. Mitchison, Preparation of modified tubulins. Methods Enzymol. 196, 478–485 (1991). [DOI] [PubMed] [Google Scholar]
- 74.J. C. Schmidt, H. Arthanari, A. Boeszoermenyi, N. M. Dashkevich, E. M. Wilson-Kubalek, N. Monnier, M. Markus, M. Oberer, R. A. Milligan, M. Bathe, G. Wagner, E. L. Grishchuk, I. M. Cheeseman, The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev. Cell 23, 968–980 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.N. Gudimchuk, B. Vitre, Y. Kim, A. Kiyatkin, D. W. Cleveland, F. I. Ataullakhanov, E. L. Grishchuk, Kinetochore kinesin CENP-E is a processive bi-directional tracker of dynamic microtubule tips. Nat. Cell Biol. 15, 1079–1088 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.K. J. Mickolajczyk, N. C. Deffenbaugh, J. O. Arroyo, J. Andrecka, P. Kukura, W. O. Hancock, Kinetics of nucleotide-dependent structural transitions in the kinesin-1 hydrolysis cycle. Proc. Natl. Acad. Sci. U.S.A. 112, E7186–E7193 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.V. A. Volkov, A. V. Zaytsev, E. L. Grishchuk, Preparation of segmented microtubules to study motions driven by the disassembling microtubule ends. J Vis. Exp. , 51150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.M. Barisic, R. Silva e Sousa, S. K. Tripathy, M. M. Magiera, A. V. Zaytsev, A. L. Pereira, C. Janke, E. L. Grishchuk, H. Maiato, Microtubule detyrosination guides chromosomes during mitosis. Science 348, 799–803 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.S. K. Tripathy, V. M. Demidov, I. V. Gonchar, S. Wu, F. I. Ataullakhanov, E. L. Grishchuk, Ultrafast force-clamp spectroscopy of microtubule-binding proteins, in Optical Tweezers: Methods and Protocols, Methods in Molecular Biology, A. Gennerich, Ed. (Springer, 2022), vol. 2478, pp. 609–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.E. V. Tarasovetc, P. K. Allu, R. T. Wimbish, J. G. DeLuca, I. M. Cheeseman, B. E. Black, E. L. Grishchuk, Permitted and restricted steps of human kinetochore assembly in mitotic cell extracts. Mol. Biol. Cell 32, 1241–1255 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.N. J. Ganem, K. Upton, D. A. Compton, Efficient mitosis in human cells lacking poleward microtubule flux. Curr. Biol. 15, 1827–1832 (2005). [DOI] [PubMed] [Google Scholar]
- 82.O. Afonso, C. M. Castellani, L. P. Cheeseman, J. G. Ferreira, B. Orr, L. T. Ferreira, J. J. Chambers, E. Morais-de-Sa, T. J. Maresca, H. Maiato, Spatiotemporal control of mitotic exit during anaphase by an aurora B-Cdk1 crosstalk. eLife 8, e47646 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Note
Figs. S1 to S13
Movies S1 to S8
Data file S1