Abstract
Hereditary genetic diseases, cancer, and infectious diseases are affecting global health and become major health issues, but the treatment development remains challenging. Gene therapies using DNA plasmid, RNAi, miRNA, mRNA, and gene editing hold great promise. Lipid nanoparticle (LNP) delivery technology has been a revolutionary development, which has been granted for clinical applications, including mRNA vaccines against SARS-CoV-2 infections. Due to the success of LNP systems, understanding the structure, formulation, and function relationship of the lipid components in LNP systems is crucial for design more effective LNP. Here, we highlight the key considerations for developing an LNP system. The evolution of structure and function of lipids as well as their LNP formulation from the early-stage simple formulations to multi-components LNP and multifunctional ionizable lipids have been discussed. The flexibility and platform nature of LNP enable efficient intracellular delivery of a variety of therapeutic nucleic acids and provide many novel treatment options for the diseases that are previously untreatable.
Keywords: cationic lipid, ionizable lipid, lipid nucleic acid nanoparticles, nucleic acid delivery, structural effect
Introduction
Gene therapy corrects malfunctional genes and has brought hope to treat the diseases that are untreatable with conventional approaches [1]. Effective gene therapy requires correction of the mutated genes, replacement of the malfunctional genes with normal genes, or regulation of abnormal gene expression [1]. However, the limiting step for clinical development and application of gene therapy has been the safe and efficient delivery of genetic materials into target cells [2]. Despite of various challenges, significant progress has been made in the development of delivery systems for therapeutic genetic materials [3, 4]. Non-viral gene delivery platforms, which have demonstrated advantages of low immunogenicity, unlimited payload capacity, cost-friendly manufacturing, flexibility, and multi-dosing capability, have received substantial attention [5]. A variety of materials have been used for non-viral gene delivery, including lipid-based nanoparticles, cationic polymers, cell penetrating peptides, and inorganic nanoparticles [6–12].
Lipid nucleic acid nanoparticles (LNP) have emerged as the most promising delivery systems for clinical applications among the various non-viral delivery systems. Various lipids have been reported for intracellular delivery of nucleic acids in the past decades. The lipid structures play the most important role for safe and efficient intracellular delivery of nucleic acids. One key consideration in the design of lipids for nucleic acids delivery is to overcome endosomal barriers for efficient intracellular delivery. The pH-sensitive protonatable or ionizable lipids have emerged as the most promising class of lipids for efficient intracellular nucleic acid delivery. These lipids are neutral at physiological pH for safe systemic nucleic acid delivery. The protonation or ionization of the lipids in acidic endosome presents the lipids with amphiphilicity, which destabilizes endosome membrane for endosomal escape [13, 14]. The structures of the lipids can be fine-tuned to achieve safe and efficient intracellular delivery of nucleic acids.
LNP has been successfully used in the development of gene therapy using small interfering RNA (siRNA) for clinical applications [15] and mRNA vaccine to combat SARS-Cov-2 (Covid-19) [16]. Lipids with various structures have been reported in the literature to develop LNP for nucleic acids delivery. In this review, the evolution of lipid structures and functions as well as LNP development, especially the optimization of the essential lipid structure and formulations of LNP, has been discussed with a focus on the relationship of the structure, function, and formulation of cationic and ionizable lipids in LNP for nucleic acid delivery.
The Evolution of LNP for Gene Delivery
Liposomes
The development of LNP for nucleic acid delivery can be dated back to 1970s [17], when liposomes were explored for drug delivery, including enzyme replacement therapy, insulin delivery, cancer chemotherapy [18–20]. One subtype of the liposomes, large unilamellar vesicles (LUV), became the first generation of LNP technology for drug delivery [21]. An early attempt of using liposomes for intracellular DNA delivery showed reduced interaction of DNA containing liposomes with cells caused by those without DNA [22], suggesting that the DNA encapsulation efficiency in liposomes might play a role for gene transfection. In early 1980s, liposomes loaded with DNA were demonstrated with the ability to transfect both eukaryotic and prokaryotic cells [23, 24].
Continuous efforts have been made to optimize DNA encapsulation in formulating liposome based LNP to improve their physicochemical properties and gene transfection efficiency. Reverse-phase evaporation (REV), Ca2+-EDTA chelation, and detergent dialysis were some of the methods tested for DNA encapsulation in liposomes in 1980s. In the REV process, an emulsion containing DNA, lipid, a nonpolar solvent and a buffered aqueous solution was first prepared [25]. The nonpolar solvent was then removed under partial vacuum to form DNA encapsulated liposomes. Spermine or lysozyme was used in some cases to protect DNA from degradation caused by vortex or sonication and to improve the encapsulation efficiency of REV [26, 27]. Ca2+-EDTA could facilitate the loading of DNA in liposomes and was used to improve DNA encapsulation of liposome LNPs [28]. In detergent dialysis, a solution containing DNA, phospholipid, detergent, and aqueous buffer was dialyzed against an excess of buffer to remove the detergent. When the concentration of detergent is lower than critical micelle concentration (CMC), liposomes were formed spontaneously with DNA in the cavity [29]. These methods demonstrated promise in DNA encapsulation and LNP formulations. However, they were greatly limited by low encapsulation efficiency, narrow selection of phospholipids, and uncontrolled sizes [30, 31].
Cationic lipids were later introduced for efficient DNA encapsulation in liposomes with improved transfection efficiency. Quaternary ammonium lipids such as DOTMA (N-[1-(2,3-dioleyloxy)propyl]-N,N,N-trimethylammonium chloride) were used as the cationic lipids with the ability to mediate efficient intracellular transfection [32]. Cationic liposomes can condense and encapsulate DNA or other genetic materials in LNP formulations for intracellular delivery. Cationic liposome and DNA complexes have affinity to the negatively charged cell membrane and facilitate intracellular delivery of genetic materials.
Cationic Lipid/DNA Complex
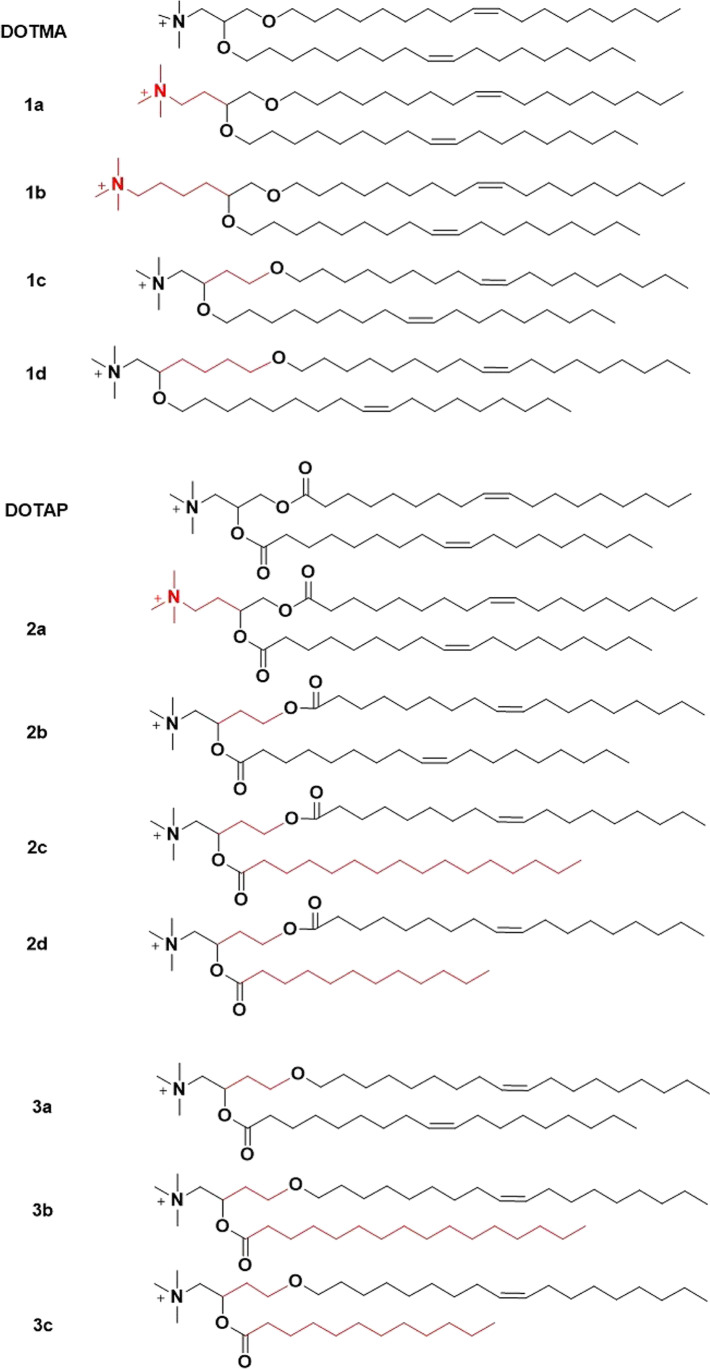
Cationic lipids are amphiphilic molecules, which consist of a hydrophilic and a hydrophobic region connected by a linker structure [33]. DOTMA and DOTAP (1,2-dioleoyl-3-trimethylammonium propane) are two most commonly investigated cationic lipids for transfection (Fig. 1). DOTMA directly interacts with plasmid DNA to form lipid-DNA complexes with 100% entrapment [32]. DOTMA facilitates fusion of the complexes with the membrane of cultured cells, resulting in intracellular uptake and expression of the DNA [32]. In order to reduce the cytotoxicity of DOTMA, a series of cationic lipids were synthesized and investigated [34]. DOTAP containing degradable ester bonds between the cationic head group and hydrophobic lipids showed higher transfection efficiency and reduced toxicity compared to DOTMA, when formulated with DOPE (dioleoylphosphatidylethanolamine) at a 1:1 ratio [34, 35].
Fig. 1.
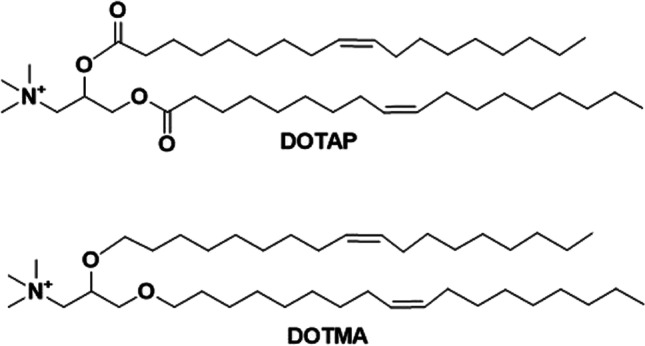
Chemical structures of DOTAP and DOTMA, which have different linkage bonds (two ester bonds in DOTAP and two ether bonds in DOTMA).
DOTMA and DOTAP differ only in the linkage bonds and share similar in vitro gene transfection efficiency in different cells. However, their efficiency in vivo was significantly different at the gene expression level in the lung after intravenous administration of DNA/DOTMA or DOTAP complexes, where DNA/DOTMA with stable ether linkage demonstrated better efficiency than DOTAP with the hydrolyzable ester linkage [36]. The structures of DOTMA and DOTAP were further altered in order to optimize the transfection efficiency of the cationic lipid and DNA complexes. Ren et al. synthesized a small library of cationic lipids to demonstrate the impact of each component of the structure of DOTMA and DOTAP on the transfection efficiency after intravenous administration (Fig. 2) [37]. A decrease in the efficiency in the lung was observed with an increase in the number of carbons between the cationic head group and the branch point of the lipid tails of DOTMA (DOTMA > 1a > 1b) [37]. Cationic lipids 1c and 1d showed similar transfection efficiency as DOTMA, indicating the branching position of the aliphatic chain with respect to the polar head group plays a role in transfection activity [37]. However, such phenomenon was not observed for the DOTAP analogues, which indicated that those effects might be exclusive for cationic lipids with two stable ether bonds [37]. Cationic lipids 2c, 2d with two different acyl chains showed a lower transfection activity than 2b, which has two identical unsaturated lipid chains. Cationic lipids 3b, 3c with different aliphatic lengths and ether/ester linkage, showed lower transfection than 3a, which has two identical hydrocarbon chains. These results demonstrated the importance of the composition of two identical unsaturated lipid chains for efficient transfection [37]. The variation of ester and ether linkages only showed difference under different transfection conditions, indicating other factors such as formulation and transfection conditions may be responsible for transfection efficiency of DOTMA, DOTAP, and their derivatives based DNA complexes in vivo.
Fig. 2.
Structures of the lipid analogues of DOTMA and DOTAP.
Helper Lipids for Cationic LNP of DNA
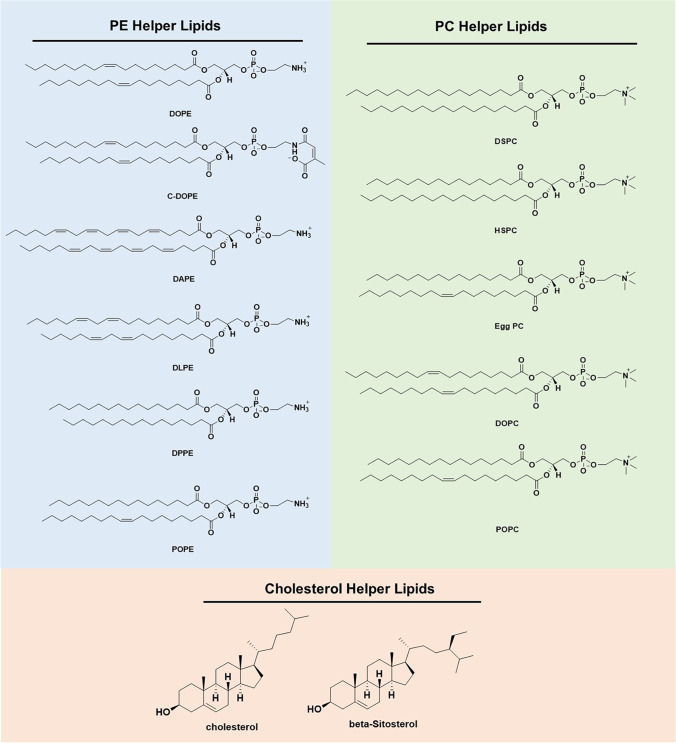
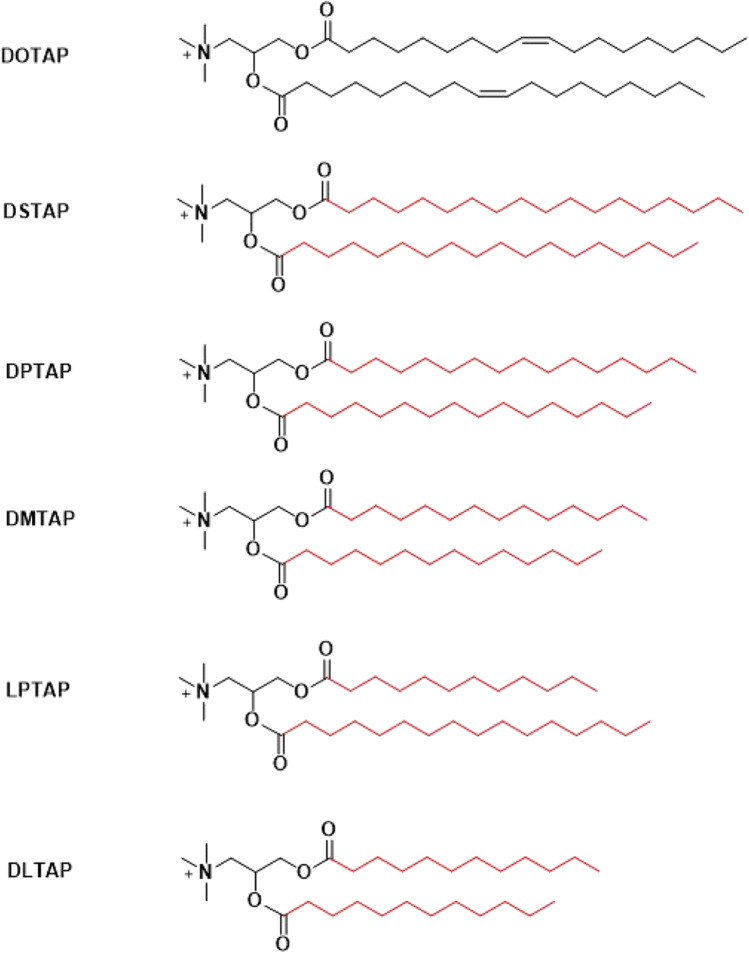
Since both DOTMA and DOTAP are cationic lipids containing a head group with a permanent positive charge [33], they don’t respond to pH changes during subcellular trafficking in endosomes. Their gene transfection efficiency could be enhanced by adding helper lipids, such as DOPE and other helper lipids (Fig. 3). DOPE has an amino head group, phosphoethanolamine, and two unsaturated oleoyl lipid chains, which have kinked structure and provide a cone geometry that is favorable for the non-bilayer hexagonal (HII) phase, which is believed crucial for membrane fusion and bilayer disruption during endosomal escape [38–40]. Formulating LNP of plasmid DNA, DOPE and DOTMA or DOTAP demonstrated enhanced efficiency compared with pDNA/DOTMA or pDNA/DOTAP complexes. The formulation processes involve in organic solvents such as chloroform and methanol, in which the lipids are dissolved. Lipid/DNA nanoparticles are formed after a process similar to reverse-phase evaporation [41]. Currently, ethanol is used to dissolve the lipids and to mix with an acidic aqueous solution of genetic materials using a microfluidic system [42, 43]. The cationic lipids and DOPE are often used at the molar ratio of 1:1 in the formulation of LNP [44]. Cholesterol has also been used as a helper lipid in DOTMA or DOTAP based LNP formulations for intravenous administration. It was demonstrated that the presence of at least 25 molar percent of cholesterol in LNP could significantly increase the stability and the retention within the circulation [45]. Massing et al. developed a small library of cationic lipids with various lipid chain lengths based on DOTAP (Fig. 4) [41, 46]. All the cationic lipids were formulated with DOPE (1:1 ratio) and cholesterol (2:1 ratio). The presence of DOPE or cholesterol could reduce the minimum temperature required for formulation of temporarily stable liposome formulations containing DOTAP and its analogs (Table I). The presence of DOPE and cholesterol could promote the transition of the LNP into the HII phase. Consequently, the LNP formulated with either DOPE and/or cholesterol could result in enhanced transfection efficiency due to their better performance in the membrane fusion and endosomal escape. No apparent efficacy was observed when cationic lipids directly formed lipid complexes with DNA [41, 46].
Fig. 3.
Structures of commonly used phosphatidylethanolamine (PE), phosphatidylcholine (PC) and cholesterol helper lipids.
Fig. 4.
Chemical structures of a lipid library based on DOTAP with various lipid chain lengths and combination.
Table I.
Minimum Temperature Required for Formation of Temporarily Stable Liposomes Containing DOTAP or DOTAP Analogs
| Lipid | Phase transition temperature (Tc) | Without helper lipid | With Dope (1:1) |
|---|---|---|---|
| DOTAP | < 5°C | RT | RT |
| DSTAP | 62.9°C | 55°C | 50°C |
| DPTAP | 52.8°C | 50°C | 35°C |
| DMTAP | 39.1°C | 40°C | RT |
| LPTAP | 43.1°C | 35°C | RT |
| DLTAP | 24.9°C | 25°C | RT |
The structure of cationic lipids was also modified using fluorinated lipid in order to enhance the efficiency of LNP formulations with helper lipids. For example, Vierling et al. synthesized a fluorinated version of DOTMA with one or both of the lipid chains fluorinated [47]. The fluorinated cationic lipids could condense DNA, with or without DOPE, to form fluorinated LNP, which demonstrated little cytotoxicity and high efficiency transfecting lung epithelial cells [47].
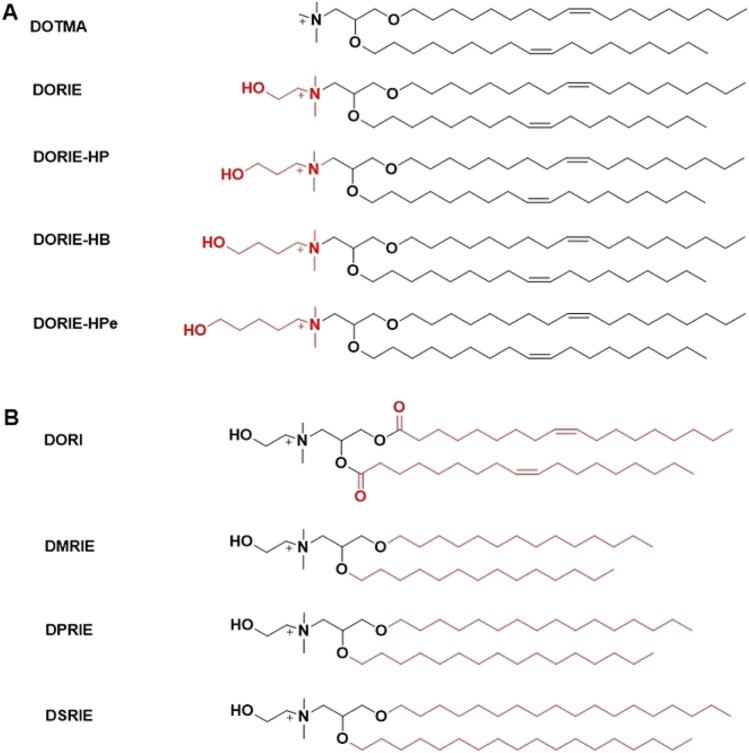
A homologous series of dioleyl (Cl8) compounds containing increasing hydroxyalkyl chain lengths on the quaternary amine were synthesized (Fig. 5A), formulated with 50 mol % DOPE [44]. The lipids showed structure-dependent transfection efficacy following an order of (DORIE) ethyl > (DORIE-HP) propyl > (DORIE-HB) butyl > (DORIE-HPe) pentyl > DOTMA [44]. These findings suggest an important role for the hydroxyl moiety in the activity of cationic lipid compounds, where the hydroxyl group may affect the interaction of the lipids with DNA or improve the interaction of the cationic LNP with cellular membranes leading to greater activity [44]. Based on the hydroxylalkyl quaternary amino head group structure, a series of cationic lipids of a hydroxyethyl quaternary ammonium with various alkyl chains were synthesized (Fig. 5B), and formulated with 50 mol % DOPE and plasmid DNA. The resulting LNP showed transfection efficacy in an order of dimyristyl (DMRIE) (di-Cl4) > dioleoyl (DORI) (di-Cl8, unsaturated) > dipalmityl (DPRIE) (di-Cl6) > disteryl (DSRIE) (di-C18). These results demonstrated shorter tails of the cationic lipid and unsaturated double bonds could facilitate better efficiency [44].
Fig. 5.
Structures of a lipid library containing hydroxyalkyl chain lengths on the quaternary amine head group (A) and different alkyl chains (B).
Stable Nucleic Acid Lipid Nanoparticles (SNALP)
Stabilized Plasmid-lipid Particles (SPLP)
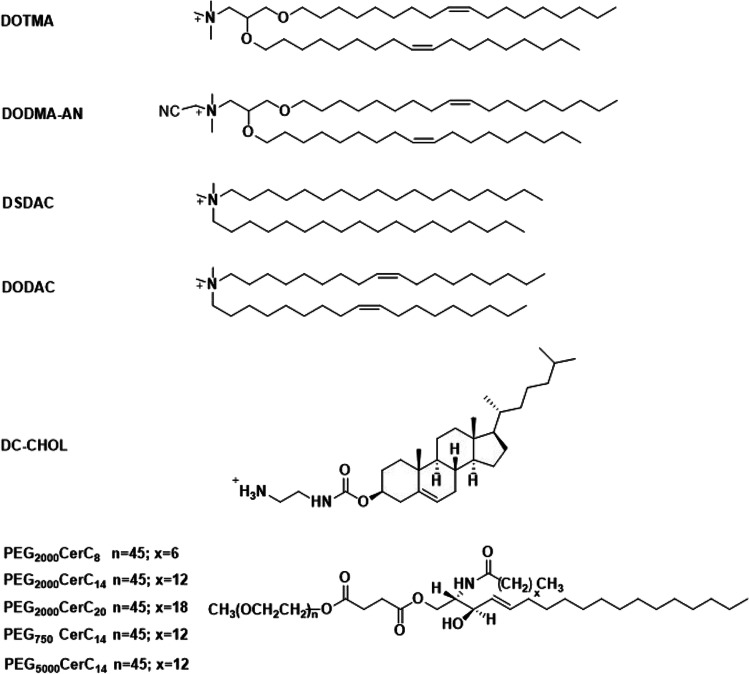
Modification of cationic lipid plasmid nanoparticles with PEG was shown to improve their stability. A stabilized plasmid-lipid particle (SPLP) formulation was established by incorporating poly(ethylene glycol)-ceramides (PEG-Cer, PEG2000 linked to ceramides) with cationic lipid dioleoyldimethylammonium chloride (DODAC), palmitoyloleoylphosphatidylcholine (POPC), or dioleoylphosphatidylethanolamine (DOPE) (Fig. 6) [48]. SPLP was formulated at a DOPE: DODAC: PEG-Cer molar ratio of 84:6:10. The plasmid DNA was first incubated with DODAC in 0.2 M octylglucoside, NaCl (5 mM) HEPES buffer (pH 7.4) and then the plasmid-DODAC mixture was added to DOPE and PEG-CerC14 or PEG-CerC20 dissolved in 0.2 M octylglucoside, NaCl (5 mM) HEPES buffer (pH 7.4) [48]. SPLP consisted of a unilamellar lipid bi-layer encapsulating a single copy of plasmid DNA with smaller size and better circulation stability. SPLP had an average size of 70 nm, with maximum entrapment observed at DODAC contents of 5 to 10 mol%. Helper lipid POPC demonstrated lower transfection efficiency than DOPE, which had better fusogenic properties. The in vitro transfection efficiency of this SPLP was also affected by the length of the acyl chain anchor of the PEG lipid, where shorter acyl chain lengths resulted in better ability of dissociation from the SPLP surface, thereby a better in vitro transfection efficiency [48]. A following study characterized the effects of the cationic lipid and PEG-Cer species on SPLP formation and in vitro transfection properties [49]. A series of cationic lipids and poly(ethylene glycol)-ceramides (PEG-Cer) were tested for SPLP formulation (Fig. 6). The in vitro transfection levels of SPLP were affected by the lipids following the order of DODMA-AN > DOTMA > DODAC > DSDAC > DC-CHOL. PEG molecular weight had modest effect on the transfection efficiency, which demonstrated slight increase in transfection efficiency with lower molecular weight PEG. It appears that the length of the acyl chain had the most dramatic effects, where PEG-Cer with short lipid anchor showed substantially better transfection [49]. SPLP composed of DOPE/DODAC/PEG-CerC20 (83:7:10; mol: mol: mol) were intravenously administered in mice and demonstrated a long circulation life, no evidence of systemic toxicities and significant levels of reporter gene expression were observed [50]. Taking this formulation strategy, lipid 1,2-dioleyloxy-3-dimethylaminopropane (DODMA) and 1,2-dioleoyl-3-dimethylaminopropane (DODAP) (Fig. 7) with ionizable head groups were also used to formulate SPLP using similar methods and formulation composition, and demonstrate excellent in vivo efficiency [51–55].
Fig. 6.
Structures of the cationic lipids and poly(ethylene glycol)-ceramides (PEG-Cer) for formulation of stabilized plasmid-lipid particles.
Fig. 7.
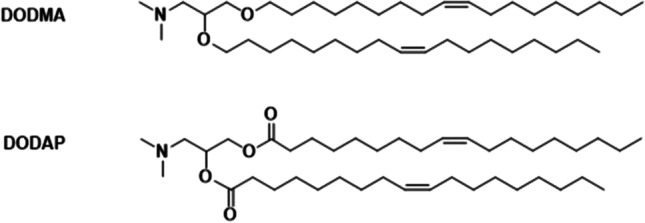
Chemical structures of 1,2-dioleyloxy-3-dimethylaminopropane (DODMA) and 1,2-dioleoyl-3-dimethylaminopropane (DODAP) lipids.
Stabilized Antisense-Lipid Particles (SALP)
PEGylation was also applied to develop stable lipid antisense nanoparticles or stabilized antisense-lipid particles (SALP) [56]. SALP was designed specifically for intravenous administration, which requires an ionizable lipid with pKa between pH 5.0 and 6.5, a PEGylated lipid, and neutral helper lipids [56]. The encapsulation and particle formulation were achieved through a spontaneous self-assembly process in an aqueous/ethanol solution with pH below the pKa of the ionizable lipid [56]. The protonated lipid complexed with an oligonucleotide by electrostatic interactions, and PEGylated lipid was used to reduce the particle aggregation. The helper lipids such as (phosphatidylcholine) PC and cholesterol assisted stable bilayer formation. SALP had diameters ranging from 150 to 200 nm. [56] An SALP with a composition of DODAP/DSPC/Cholesterol/PEG-C14 (25/20/45/10 mol%) demonstrated efficacy in the liver after systemic delivery [57], as well as other SALP nanoparticle formulations [58, 59].
Stable Nucleic Acid Lipid Nanoparticles (SNALP)
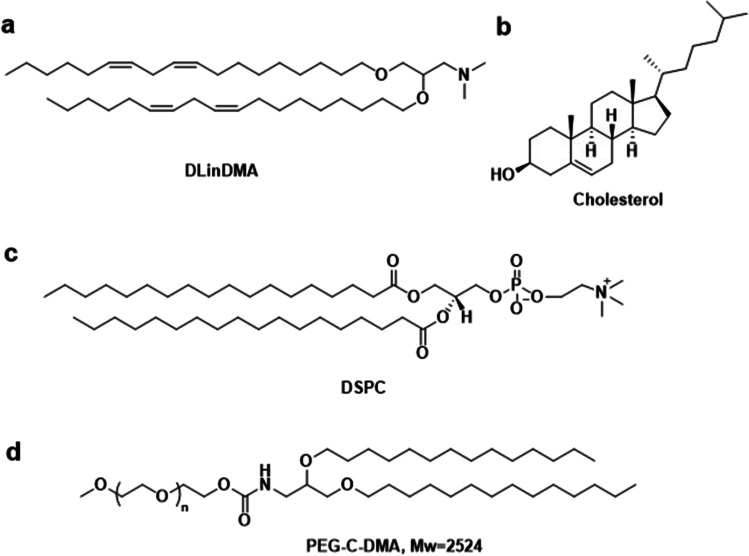
Stable nucleic acid lipid nanoparticles (SNALP) were established specifically for encapsulating siRNA into liposomes with a lipid composition of 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), cholesterol, 3-N-[(ω-methoxypoly(ethylene glycol)2000)carbamoyl]-1,2-dimyristyloxy-propylamine (PEG-C-DMA), and 1,2-dilinoleyloxy-3-(N,N-dimethyl)aminopropane (DLinDMA) (20:48:2:30 mol%), Fig. 8 [60]. SNALP was effective at mediating RNAi in vitro and in vivo, and effective for inhibiting viral replication in a murine model of hepatitis B and silencing gene expression in non-human primates [61–65].
Fig. 8.
SNALP Lipids. (a)1,2-dilinoleyloxy-N,N-dimethyl-3-aminopropane (DLinDMA), (b)cholesterol (Chol), (c)1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), (d)3-N-[(ω-methoxypoly(ethylene glycol)2000)carbamoyl]-1,2-dimyristyloxy-propylamine (PEG-C-DMA).
DlinDMA is a linolyl analogue of DODMA [66]. It was shown that the extra double bond in the lipid tails could increase propensity to form the non-bi-layer phase, thereby could increase the transfection efficiency of the LNP [67]. A series of DODMA analogues of 0, 1, 2 or 3 double bonds in the alkyl chains were synthesized and investigated in SNALP, Fig. 9 [66]. The effect of addition of double bonds on bi-layer transition temperature was studied using [31]P-NMR [66]. SNALP containing the saturated lipid DSDMA showed no sign of adopting the HII phase, even at relatively high temperatures. However, DODMA (1 double bond per alkyl chain) containing LNP exhibited a much lower phase transition temperature for HII phase transition. The presence of a 2nd double bond (DLinDMA) further reduced the phase transition temperature, while incorporation of a 3rd double bond (DLenDMA) has little additional effect [66]. Another study also demonstrated similar results, where lipid containing two double bonds in alkyl chains (DLinDMA) to be optimal [68].
Fig. 9.
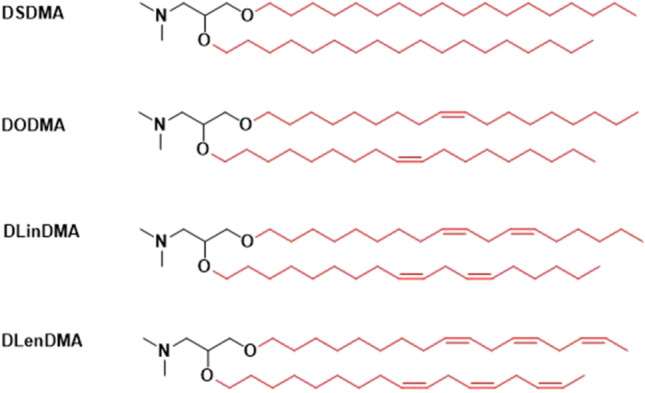
Structures of ionizable lipids DODMA analogues for SNALP.
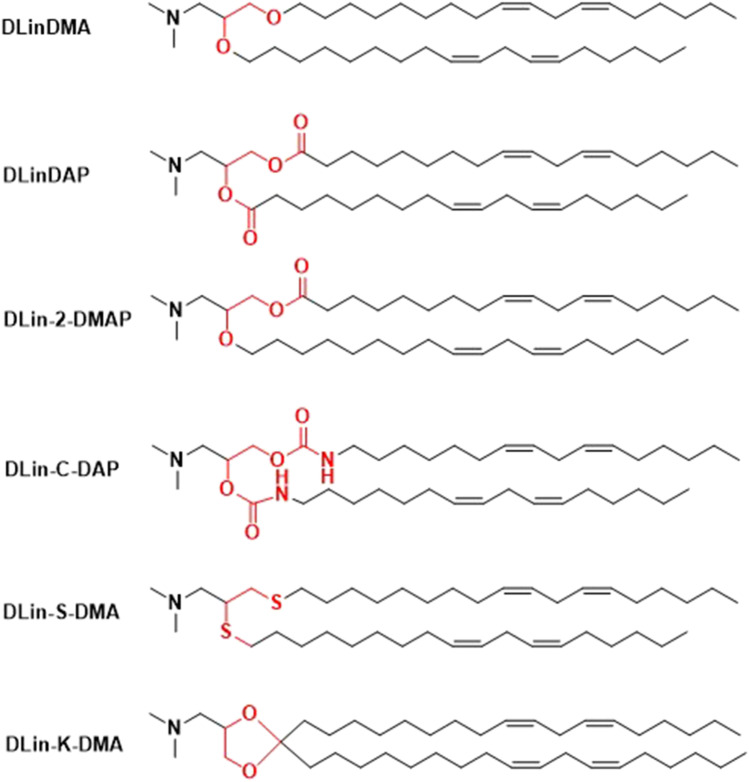
The ionizable lipid DLinDMA was further modified by varying the linker structure between the head group and lipid tails to introduce degradability, Fig. 10 [69]. Each of the ionizable lipids, distearoylphosphatidylcholine (DSPC), cholesterol, and PEG-lipid (40:10:40:10 mol/mol) were formulated to form SNALP for in vivo assessment of their efficacy in a mouse Factor VII model after a single intravenous injection [69]. The introduction of ester, carbamate, or thio-ether linkages to the ionizable lipids resulted in a substantial reduction in in vivo activity as compared to the LNP with DLinDMA [69]. DLin-2-DMA with one ether linkage and one ester linkage yielded intermediate activity [69]. Interestingly, the introduction of a ketal ring linker in DLin-K-DMA resulted in LNP with 2.5-fold potency in the same study [69].
Fig. 10.
Structures of DLinDMA analogues.
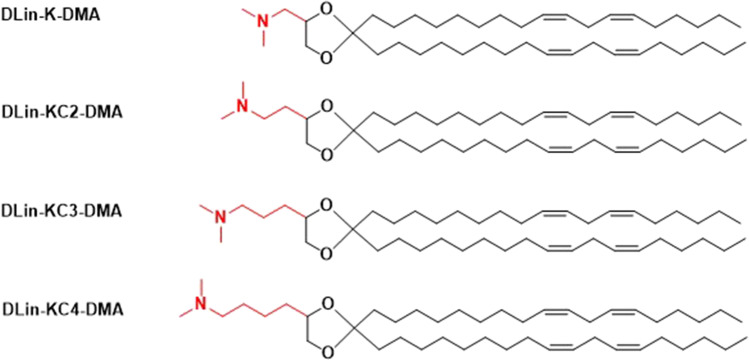
Further structural modification was made based on DLin-K-DMA by adding methylene units between the dimethylamino group and the dioxolane linker, Fig. 11 [69]. The modifications affected the pKa and flexibility of in the lipid bilayer interface [69]. Interestingly, the addition of a methylene unit in DLin-KC2-DMA produced significant increase in potency relative to DLin-K-DMA [69]. As a result, DLin-KC2-DMA became a better ionizable lipid for SNALP formulation than DLinDMA for systemic delivery to the liver.
Fig. 11.
Structures of DLin-K-DMA analogues.
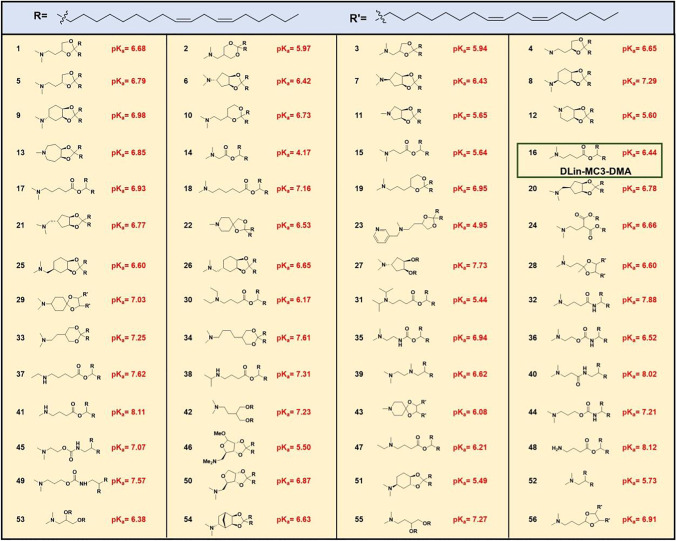
A library of 56 ionizable lipids were designed and tested by modifying the amino head groups of various pKas and linkage structures, Table II [70]. LNPs were formulated with the compositions of an ionizable lipid, DSPC, cholesterol and PEG-lipid in the molar ratio 40/10/40/10. (6Z,9Z,28Z,31Z)-Heptatriaconta-6,9,28,31-tetraen-19-yl-4-(dimethylamino)butanoate (DLin-MC3-DMA) (16 in Table II) demonstrated the best potency in mice [70]. The change of ratio of DLin-MC3-DMA, DSPC, cholesterol and PEG-lipid to 50/10/38.5/1.5 in the LNP composition resulted in a six-fold improvement relative to the 40/10/40/10 molar ratio composition in non-human primate [70]. DLin-MC3-DMA (MC3) is the ionizable lipid for the FDA approved siRNA-based drug ONPATTRO (Patisiran) for patients with hereditary transthyretin-mediated amyloidosis.
Table II.
Structures of the 56 Ionizable Lipids Tested for Gene Silencing Activity In Vivo
Multifunctional Envelope-type Nano Device (MEND)
The concept of multifunctional envelope-type nano device (MEND) was proposed to make a cleavable PEG ligand for LNP formulations to shed the PEG layer during the subcellular trafficking [71–73]. MEND was formulated using a cationic lipid, a phospholipid, cholesterol and a cleavable PEG-DMG or PEG-DSG. The cargo siRNA was complexed with protamine first before formulation [74]. A film of lipid mixture was formed by the evaporation of the solvent or solvents in a chloroform or chloroform/ethanol solution of the cationic lipid, DOPE, cholesterol and PEG lipids, and then mixed with the siRNA HEPES buffer solution [71–73]. A new ionizable lipid, YSK05, was later designed to improve the intracellular trafficking and gene silencing activity of MEND, Fig. 12 [74]. The initial MEND nanoparticles had a molar ratio of 30/40/30/3 for DOTAP, DODAP, or YSK05/DOPE/cholesterol/PEG-DMG, which was optimized to 50/25/25/3 for YSK05/POPE/cholesterol/PEG-DMG in YSK05-MEND [74]. YSK05-MEND could efficiently escape from endosomes and the addition of PEG-DSG (5 mol% of total lipid) in YSK05-MEND could facilitate excellent in vivo gene silencing activity [74]. YSK05-MEND was also effective to deliver nucleic acid to various organs in vivo [75–78].
Fig. 12.
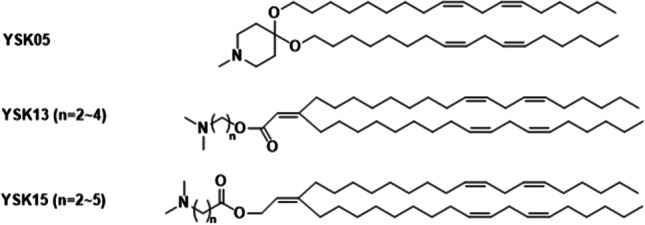
Structures of YSK13 and YSK15 lipids.
Two additional series of ionizable lipids, referred as YSK13 and YSK15, were developed for MEND formulations to enhance membrane destabilization, (Fig. 12) [79]. A carbon–carbon double bond was introduced in the acyl chains for enhanced fusogenic activity, and an ester bond for improved biodegradability. Both YSK13 and YSK15 series were formulated in MEND (YSK13 or YSK15/Cholesterol/PEG-DMG: 70/30/3) and the effect of the ester bond on siRNA delivery was examined [79]. The gene-silencing activity in hepatocytes dramatically reduced with the decrease in the pKa value of YSK13 and YSK15 lipids [79]. YSK15-C4-MEND (pKa = 7.10) demonstrated the strongest gene-silencing activity in the liver [79].
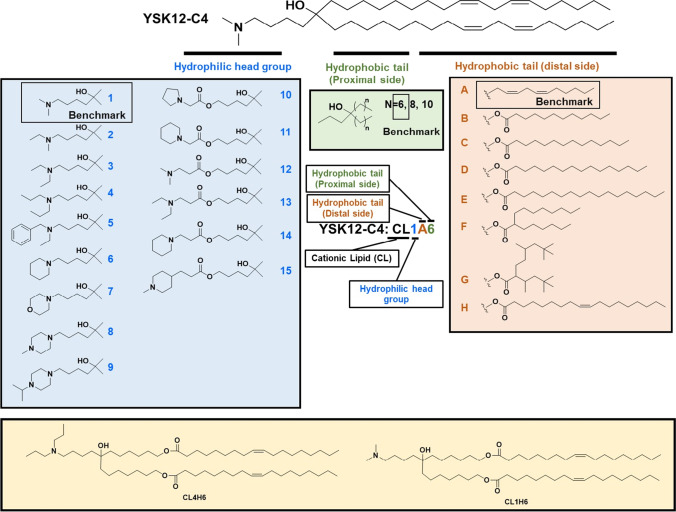
YSK12-MEND also demonstrated promising results as a delivery system of siRNA for dendritic cell-based therapy [80]. MEND formulated with YSK12-C4 loading siRNA (YSK12-MEND) (YSK12-C4/cholesterol/PEG-DMG: 85/15/1) was effective for endosomal escape in dendritic cells [80]. Based on the structure of YSK12-C4, a library of ionizable lipids have been established with various head groups, linkages and tail structures for liver siRNA delivery (Fig. 13) [81]. LNP formulated with a lipid CL4H6 (CL4H6-LNP) (Fig. 13) showed efficient gene silencing activity, biodegradability, and was tolerated. The in vivo experiments demonstrated that the CL4H6-LNP had a superior efficiency for endosomal escape, cytosolic release, and the RNA-induced silencing [81]. For human natural killer (NK) cells, LNP formulated with CL1H6 (CL1H6-LNP) (Fig. 13) demonstrated an increased gene silencing and cell viability as compared to YSK12-LNP due to the ability to avoid endosomal disruption, resulting in a decreased level of cytotoxicity [82].
Fig. 13.
Systematic derivatization of YSK12-C4. The YSK12-C4 is divided into 3 sections, an ionizable head group, distal, and proximal side of hydrophobic tails. Each part of the YSK12-C4 was derivatized to assess structure–activity relationship. The notation for the ionizable lipids used in this study are abbreviated as cationic lipid (CL), followed by the order of the number of carbons in the head group (1 to 15), distal (A to H) and proximal side (6 to 10) of the hydrophobic tails.
Multifunctional pH-Sensitive Amino Lipids
Multifunctional pH-sensitive amino lipids are a series of pH-sensitive protonatable or ionizable lipids that are designed to address the challenges of cytosolic delivery of nucleic acids [14, 83, 84]. Unlike the cationic or ionizable lipids discussed in the previous sections, these pH-sensitive amino lipids can form stable LNP by self-assembly with nucleic acids without helper lipids and mediate efficient cytosolic delivery of nucleic acids, including RNA and DNA of any sizes for various applications. The multifunctional pH-sensitive amino lipids were designed based on the concept of pH-sensitive amphiphilic endosomal escape and reductive cytosolic delivery, the PERC effect [14, 85]. Multiple functionalities were introduced in the structures of the pH-sensitive amino lipids to form stable LNP and to facilitate pH-sensitive amphiphilic endosomal escape and environment-sensitive release of nucleic acid cargo in cytosol.
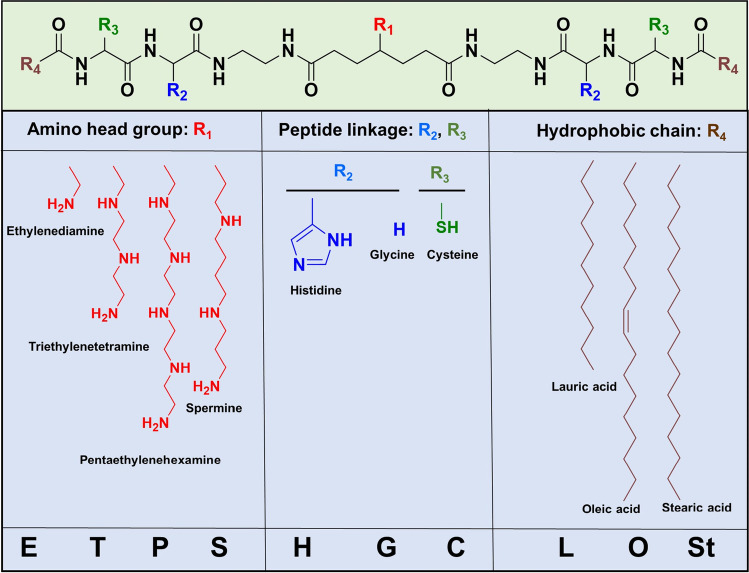
The core structure of the multifunctional amino lipids is shown in Fig. 14 [14, 84]. It is composed of three key components in its structure, including a protonatable or ionizable amino head group, dual cysteinyl residues, and dual lipid tails. The amino head group was designed to complex with the gene cargo and to enable pH-sensitive amphiphilicity of the lipids due to their ionization in acidic endosome during subcellular trafficking and to facilitate endosomal escape by amphiphilic interactions of the ionized lipids with endosomal membrane. Primary, secondary, tertiary, and aromatic amino groups have been tested as the protonatable amino head group. Lipophilic tails are designed to provide hydrophobic interactions to stabilize the LNP formulations and to induce lipophilicity to interact with cellular membrane during endosomal escape. Cysteinyl residues are introduced to further stabilize LNP by forming disulfide bonds, which are relatively stable in the plasma during the delivery process and can be reduced in the reductive cytosolic environment to facilitate the gene cargo release [13, 14]. The thiol groups can also be used for functionalization of the LNP with PEG or targeting agents. The pH-sensitive amphiphilicity can be tuned by varying the composition of amino head groups, including ethylenediamine, triethylenetetraamine, pentaethylenehexamine, spermine, and histidine, in combination with different lipid tails. Fatty acids of various chain lengths and saturation conditions were used, including lauric acid, stearic acid, and unsaturated oleic acid. The pH-sensitive amphiphilic cell membrane disruption, siRNA transfection, and silencing efficiency of the amino lipids were affected by the structures of both the head group and lipid tails. Among these amino lipids, EHCO with a combination of a ethylenediamine head group and unsaturated lipid tails exhibited no hemolytic activity at pH 7.4 and 6.5, and high hemolytic activity at the endosomal-lysosomal pH (5.4), indicating pH-sensitive membrane destabilization in acidic endosomes for efficient endosome escape [14]. The amino lipids with head group of more amino groups and/or saturated lipid did not show good pH sensitivity in this pH range. The siRNA delivery into U87 cells demonstrated that the lipid EHCO with an ethylenediamine head group, histidylcysteinyl linkers, and oleoyl tails had the best in vitro transfection and silencing efficiency [14]. More multifunctional amino lipids were also synthesized and investigated for plasmid DNA delivery [84]. The plasmid DNA transfection experiment demonstrated amino lipid SKACO showed high efficacy for LNP formulation with plasmid DNA [84].
Fig. 14.
The design of the multifunctional pH-sensitive amino lipids.
Unlike other LNP formulations where a lipid anchor is required for PEGylation, the multifunctional amino lipid-based LNP can be readily modified with PEG by conjugating a maleimido-PEG to a small portion (ca. 2.5 mol-%) of the free thiol groups of the amino lipids [86, 87]. A targeting ligand could be used at the other end of PEG. Bombesin peptide was used in EHCO/siRNA nanoparticles to target CHO-d1EGFP cells in vitro, which had specific overexpression of bombesin receptors, resulting in specific targeting and enhanced delivery and gene silencing efficiency [86]. RGD peptide on PEG was also used for EHCO based LNP formulation, which resulted in receptor-mediated cellular uptake and high gene silencing efficiency in U87 cells in vitro and in vivo [87].
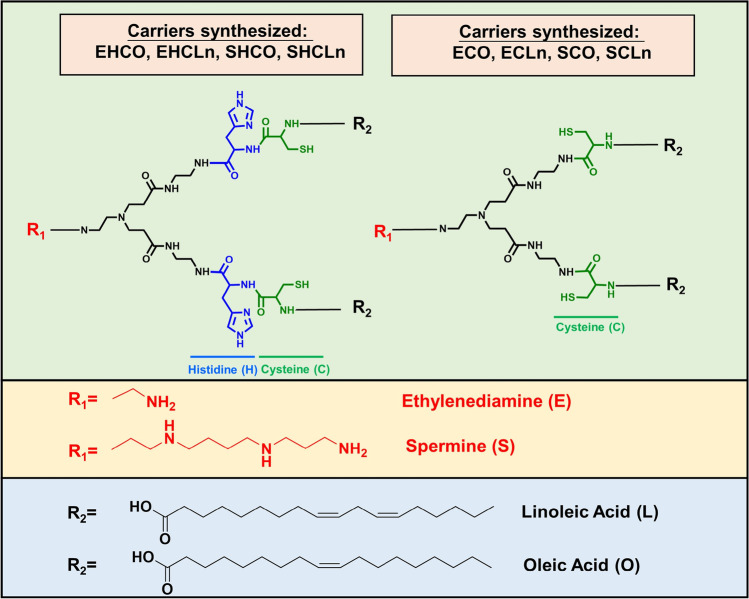
The structure of multifunctional amino lipids was modified by altering the head groups and lipid tails based on the core structure of EHCO, Fig. 15 [83]. The impact of the unsaturation degree in oleoyl or linoleoyl tails and the role of the protonatable amino groups and histidine residues on siRNA LNP formulation, pH sensitivity, gene silencing was further investigated. The presence of additional protonatable amines in the head group helped the encapsulation of siRNA at low N/P ratios, but show less sensitivity to pH changes. ECO and ECLn without the histidyl residues mediated the best gene silencing efficiency among these new amino lipids. The number of double bonds in the lipid tails did not affect gene silencing efficiency of the lipids with the same head group [83].
Fig. 15.
Structural modification of the multifunctional pH-sensitive amino lipids based on EHCO.
ECO with an ethylenediamine head group, two cysteine-based linker groups, and two oleoyl tails has been extensively explored for in vivo delivery of nucleic acids for various applications, including siRNA gene regulation for cancer therapy, CRISPR/Cas9 for gene editing, and gene replacement therapy with plasmid DNA for eye genetic disorders [88–101]. ECO readily self-assembles with nucleic acids of any sizes to form stable LNPs without helper lipids. The cysteinyl residues stabilize the LNP formulation by disulfide bonds formation, and can also be used for PEGylation with targeting functions. ECO is not amphiphilic and does not induce cell membrane destabilization at physiological pH 7.4. It can be protonated or ionized at the endosomal pH (5.4–6.5) to become amphiphilic to facilitate endosomal membrane destabilization and to promote efficient endosomal escape [85, 90]. In the reductive cytoplasm, the disulfide crosslinks in ECO LNP can undergo reductive cleavage, further aid on the release of the gene cargo [97], which is defined as the PERC effect [85].
In the formulation of multifunctional amino lipid LNP, the amino lipids are dissolved in ethanol, which accounts for 5% of total final volume, and mixed with nuclease free aqueous solution of nucleic acids to form LNP by self-assembly with uniformed distribution and size in the range of 80–200 nm with simple agitation. The presence of 5% ethanol in the final volume does not affect stability and transfection efficiency of the LNP. No dialysis process is needed after the formulations. Addition of sucrose in a range of 5 -10% significantly improve the stability of the LNP formulation. The frozen formulation of RGD-PEG-ECO/siRNA LNP with as low as 5% sucrose retained nanoparticle integrity (90% siRNA encapsulation), size distribution (polydispersity index [PDI] 20%), and functionality (at least 75% silencing efficiency) at -80 ºC for at least 1 year [97]. The frozen LNP formulation also exhibited excellent biocompatibility, with no adverse effects on hemocompatibility and minimal immunogenicity [97].
RGD peptide with a PEG spacer was used to modify siRNA ECO LNP for targeted delivery in cancer via systemic injection, which has demonstrated excellent in vivo efficacy in mouse tumor models [91, 96, 97, 101]. Retinoid analogues with a PEG spacer were also used to target the retinal pigmented epithelium for retinal gene therapy using ECO plasmid LNP [94, 99]. ECO was also able to deliver a large plasmid DNA encoding ABCA4 gene for treating Stargardt disease. ECO is a versatile and highly efficient multifunctional amino lipid for delivery of nucleic acid for treating disease with abnormal genetic functions [94, 98, 99, 102, 103]. A pH-sensitive hydrazone linker was also incorporated in the PEG spacer to develop dual pH-sensitive ECO LNP, enabling the shed of PEG layer at acidic endosomal pH, resulting in increased endosomal escape properties than previous targeted ECO LNP both for cancer and retinal gene therapies [89, 99].
Ionizable Lipids for mRNA LNP
Amino lipids have also been used to form mRNA LNP as vaccines to combat viral infections. The FDA approved two LNP based Covid-19 vaccines that contain mRNA encoding the spike protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [104, 105], which are the first mRNA vaccines approved for human use. The mRNA is encapsulated in LNPs containing an ionizable lipid, PEG-lipid, the phospholipid 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), and cholesterol. Ionizable lipid ALC-0315 ((4-hydroxybutyl)azanediyl)bis(hexane-6,1-diyl)bis(2-hexyldecanoate) is used in the mRNA vaccine developed by Pfizer, and SM-102 (heptadecan-9-yl 8-(2-hydroxyethyl)[6-oxo-6-(undecyloxy)hexyl]amino octanoate) is used in Moderna’s vaccine, Fig. 14. It was shown that SM-102 in Moderna’s mRNA vaccine out-performed MC3 [106, 107]. Interestingly, both ACL-315 and SM-102 share some common structure features, including a tertiary amine with a hydroxyalkyl group, branched tails, and ester linkers. ALC-0315 has four very short tails, while SM-102 has only three and one of them is along lipid [108]. As compared MC3, the unique structural feature of ALC-0315 and SM-102 is the branched lipid tails, which has also been seen from YSK based ionizable lipids development [81]. Ionizable lipid CL4F6 has demonstrated as excellent properties as CL4H6 (Fig. 13) [81]. It seems that multiple branched short tails could achieve similar property as unsaturated long tails, which largely affect the lipid phase transition and thereby endosomal escape properties. Molecular dynamics simulations demonstrated differences in the stability of SM-102 and ALC-0315 due to the tail structure difference, which SM-102 demonstrated better stability using a three-tailed design whereas ALC-0315 used a four-short-tail design [108]. The mRNA-loaded LNPs for Covid-19 vaccine are formulated through microfluidic mixing, where the pH of the solution is kept low to protonate the ionizable lipid's amino groups for electrostatic complexation of negatively charged mRNA. Dialysis or ultrafiltration is used to neutralize the pH, resulting in uncharged, solid-core LNPs that densely package the mRNA. PEGylation stabilizes the mRNA LNP by preventing aggregations during manufacturing and storage. Due to a short anchor and the biodegradability of the PEG lipid, it can quickly dissociate from the LNP following injection and allow efficient intracellular delivery (Fig. 16.).
Fig. 16.
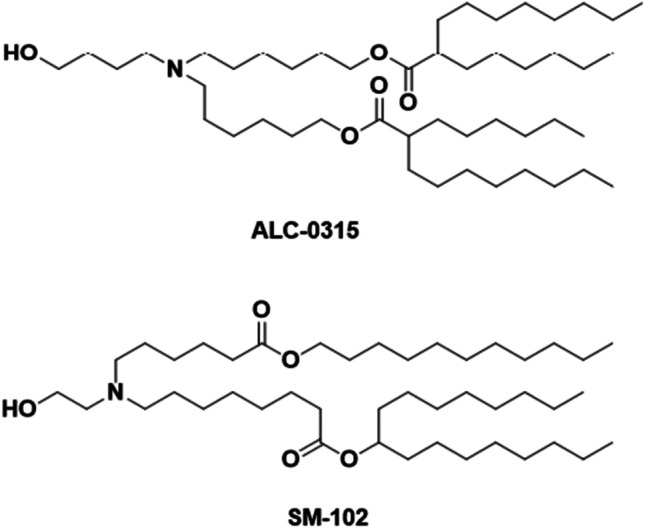
Chemical structures of ionizable lipids in mRNA LNP.
The Effects of Lipid Structure on Formulation and Intracellular Nucleic Acid Delivery Of LNP
Safe and efficient delivery of nucleic acids into cytoplasm of target cells is the key consideration in the design and development of LNP for biomedical applications. The structure of lipids plays the essential role in cytosolic delivery of nucleic acids with LNP. The main route of entry of LNP into cells is by endocytosis, including clathrin-mediated, caveolae-mediated endocytosis, receptor-mediated endocytosis, macropinocytosis, or phagocytosis, based on its size and properties. Once LNP is internalized by cells, it is transported through endosomes and lysosomes, where the internalized materials are digested or exocytosed. In order to allow nucleic acid to perform the intended function, the gene cargo has to escape the endocytic pathway and redistributes in intact form in cytoplasm or nucleus of target cells. Thus, endosomal escape is a crucial step for effective gene delivery with LNP. The ability of the lipids to promote endosomal escape is a critical factor in the design and development of LNP for nucleic acid delivery.
Cationic lipids, which have permanent positive charges, can facilitate better electrostatic interactions with negatively charged cell membrane, but are cytotoxic due to their permanent positive charges and amphiphilicity, which causes cytotoxicity by dissolving lipid bilayer cell membrane [109]. Neutral amino lipids such as DODMA and DODAP were then developed to reduce the cytotoxicity of the cationic lipids with permanent changes. The concept of pH-sensitive protonatable or ionizable amino lipids for pH-sensitive amphiphilic endosomal escape was first introduced in the design of the multifunctional pH-sensitive protonatable or ionizable lipids [14, 90] and systemically demonstrated with these lipids. The pH-sensitive protonatable or ionizable lipids are neural at physiological pH and are protonated or ionized at acidic endosomal pH to become amphiphilic to destabilize the endosomal membrane for endosome escape. It is shown that the structures of both amino head group and lipid tails are essential to control the pH sensitivity and pH-dependent cell membrane destabilization [14]. The amino lipids EHCO and ECO with a small amino head group, ethylenediamine, and unsaturated oleoyl tails exhibited the best pH sensitivity in the pH range of 5.4–7.4 [14, 83]. ECO shows effective pH-sensitive cell membrane destabilization and mediates efficient endosomal escape for cytosolic delivery of nucleic acids [90]. The concept has now been broadly adopted in the design and development of ionizable lipids to promote membrane destabilization and facilitate endosomal escape of LNP [70, 107, 108, 110–115].
Since the pH-sensitivity of ionizable lipid plays a crucial role in gene delivery using LNPs, the pKa of the head group has significant impact on the pH-sensitivity [111]. It has been shown that the most effective amino lipids have a pKa around 6.5 [111] and the lipids with pKa values less than or equal to 5.4 demonstrates significantly lower efficiency [113]. For systemic delivery for the liver targeting, it was reported that the pKa between 6.2 and 6.5 of amino lipids are optimal for systemic siRNA delivery to the liver [70]. If the criterion for a proper pKa value is not met, LNP may have with low efficiency. Proper pKa values can facilitate a non-bilayer (hexagonal HII) phase structure when mixed with anionic lipids, which serves as a measure of their bilayer-destabilizing capacity and relative endosomolytic potential [ 110, 115–117].
Saturation status of the lipid tails also affects the overall property of the cationic or ionizable lipids that, in turn, determines the endosomal escape properties [118]. Cationic/ionizable lipids can have various dynamic structural phases, including the micellar, lamellar, cubic and inverted hexagonal phase [119]. The type of structure can be predicted by the packing parameter (P), which represents the ratio of the area occupied by the hydrophobic tails versus the hydrophilic head [120–122]. When P value exceeds 1, the area occupied by the hydrocarbon lipid tails is much larger than the head group, the lipid tends to adopt the inverted hexagonal phase, which is in favor of endosomal membrane destabilization, which is crucial for LNP gene delivery [112, 123]. The study utilized lipids of the same alkyl chain length (C18) modified by a systematic addition of double bonds and the effect of double bonds addition against bi-layer transition temperature demonstrated that the lipid tails containing 1 or 2 double bonds had much lower phase transition temperature for HII phase transition to get the ‘cone’ shape character [66]. Lower phase transition temperature and the ‘cone’ shape structure can facilitate efficient membrane fusion and thereby endosomal escape.
Helper lipid structure can also affect the endosomal escape properties of LNP formulations [124–127]. As one of the most commonly used helper lipid used for LNP formulations, DOPE has a small head group, phosphoethanolamine, and two unsaturated oleoyl lipid chains, providing kinked structure and creating a cone geometry that is favorable for the non-bilayer hexagonal (HII) phase crucial for membrane fusion and bilayer disruption process during endosomal escape [38–40]. Therefore, DOPE aids endosomal escape of some of the cationic and ionizable lipids, especially when they have low pH-sensitivity. For example, combination of DOPE with a cationic lipid that originally did not have effective transfection resulted in efficient gene transfection [114]. On the other hand, saturated phosphatidylcholines, such as distearoylphosphatidylcholine (DSPC) and hydrogenated soybean phosphatidylcholines (HSPC), have high phase transition temperatures [128–131]. They have been used to formulate LNP such as stabilized nucleic acid lipid particles (SNALP) to improve its stability [117, 132]. However, LNPs formulated with saturated phosphatidylcholine helper lipids potentially have lower endosomal release properties compared with unsaturated helper lipids, which may affect the gene delivery efficiency. Cholesterol is another commonly used component in LNP formulations, which stabilizes lipid bilayers by filling in the gaps between phospholipids and give better circulation stability to LNPs [133]. It can facilitate membrane fusion, thereby promoting the endosomal release [134]. The lower solubility of cholesterol in the LNP core may lead to its enrichment on the LNP surface, promoting its crystallization and potentially contributing to endosomal fusion [135]. As a result, cholesterol can enhance the activity of LNP especially at a high concentration [136]. Derivatives of CHOL has also demonstrated ability enhancing the efficiency of LNP, such as phytosterols and the derivatives of β-sitosterol [137, 138]. These derivatives with longer alkyl chains can improve the fusogenic properties and endosomal escape property [135].
Cationic or ionizable lipid can mediate electrostatic interaction between LNPs and the cellular or endosomal membranes, and facilitate cellular uptake and endosomal release of gene cargos [139]. At the same time, the interactions of LNP may cause cytotoxicity and related toxic side effects in vivo if the amphiphilic properties of lipids and surface property of the LNP are not precisely controlled in the range physiological pH (7.4) and endosomal pH. The pH sensitive protonation or ionization of amino lipids in this pH range is essential in the design of safe and effective LNP for clinical translation, especially for systemic administration. The pH sensitive ionizable lipids with little to no amphiphilicity at physiological pH and high amphiphilicity at endosomal pH can have better control of cytotoxicity and side effects of LNP for in vivo use. In addition, pH-sensitive amphiphilic cell membrane destabilization of LNP also depends on the concentrations of the lipids [83]. The pH-sensitive ionizable lipids with the ability to form stable LNP and mediate efficient cytosolic delivery of nucleic acids at low N/P ratios are preferred to minimize the potential toxic side effects for in vivo applications. Surface modification of LNP with biocompatible polymers or biopolymers could also minimize the toxic side effects of LNP [140, 141]. Nevertheless, the breakthrough of LNP based mRNA vaccines and other therapies has demonstrated the potential of LNP systems, especially the pH-sensitive ionizable LNP systems, for the delivery of therapeutic nucleic acids, including DNA, mRNA, miRNA, and siRNA. As such, it has ignited a new wave of the development of ionizable lipid based LNP systems for delivering various therapeutic nucleic acids for preventing and treating human diseases that are not treatable with conventional therapeutic regimens. It is expected that more LNP based gene therapies or vaccines will be approved for clinical use.
Summary
Lipid nanoparticle systems hold great promise in treating untreatable diseases including inherited genetic diseases, cancer, and infectious diseases. Through the modifications in lipid structures, formulations with helper components, and preparation processes, LNP systems have shown significantly improved delivery efficiency and therapeutic efficacy in the targeted tissue, from simple lipid/DNA complexes to the mRNA vaccine to combat SARS-CoV-2. Understanding the structure/formulation/function relationship allows tunable designs of LNP systems addressing different applications, which will facilitate more innovative design and development of LNP systems for broader applications in future clinical practice.
Funding
This research was funded by National Cancer Institute of the National Institutes of Health, grant numbers R01 CA235152. ZRL is M. Frank Rudy and Margaret Domiter Rudy Professor of Biomedical Engineering.
Declarations
Conflicts of Interests/Competing Interests
There is no conflict of interests/competing interests for the authors in this work.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Collins M, Thrasher A. Gene therapy: progress and predictions. Proceedings of the Royal Society B: Biological Sciences. 2015;282:20143003. doi: 10.1098/rspb.2014.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donkuru M, et al. Advancing nonviral gene delivery: lipid-and surfactant-based nanoparticle design strategies. Nanomedicine. 2010;5:1103–1127. doi: 10.2217/nnm.10.80. [DOI] [PubMed] [Google Scholar]
- 3.Young LS, Searle PF, Onion D, Mautner V. Viral gene therapy strategies: from basic science to clinical application. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland. 2006;208:299–318. doi: 10.1002/path.1896. [DOI] [PubMed] [Google Scholar]
- 4.Zu H, Gao D. Non-viral vectors in gene therapy: recent development, challenges, and prospects. AAPS J. 2021;23:1–12. doi: 10.1208/s12248-021-00608-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, et al. Recent development of gene therapy for pancreatic cancer using non-viral nanovectors. Biomaterials Science. 2021;9:6673–6690. doi: 10.1039/D1BM00748C. [DOI] [PubMed] [Google Scholar]
- 6.Li W, Szoka FC. Lipid-based nanoparticles for nucleic acid delivery. Pharm Res. 2007;24:438–449. doi: 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- 7.Pissuwan D, Niidome T, Cortie MB. The forthcoming applications of gold nanoparticles in drug and gene delivery systems. J Control Release. 2011;149:65–71. doi: 10.1016/j.jconrel.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Y, et al. Mesoporous silica nanoparticles for drug and gene delivery. Acta pharmaceutica sinica B. 2018;8:165–177. doi: 10.1016/j.apsb.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zare H, et al. Carbon nanotubes: Smart drug/gene delivery carriers. Int J Nanomed. 2021;16:1681. doi: 10.2147/IJN.S299448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discovery. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 11.Fernández-Carneado J, Kogan MJ, Pujals S, Giralt E. Amphipathic peptides and drug delivery. Pept Sci. 2004;76:196–203. doi: 10.1002/bip.10585. [DOI] [PubMed] [Google Scholar]
- 12.Carballo-Pedrares N, Fuentes-Boquete I, Díaz-Prado S, Rey-Rico A. Hydrogel-based localized nonviral gene delivery in regenerative medicine approaches—an overview. Pharmaceutics. 2020;12:752. doi: 10.3390/pharmaceutics12080752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gujrati M, Vaidya A, Lu Z-R. Multifunctional pH-sensitive amino lipids for siRNA delivery. Bioconjug Chem. 2016;27:19–35. doi: 10.1021/acs.bioconjchem.5b00538. [DOI] [PubMed] [Google Scholar]
- 14.Wang X-L, Ramusovic S, Nguyen T, Lu Z-R. Novel polymerizable surfactants with pH-sensitive amphiphilicity and cell membrane disruption for efficient siRNA delivery. Bioconjug Chem. 2007;18:2169–2177. doi: 10.1021/bc700285q. [DOI] [PubMed] [Google Scholar]
- 15.Rossi JJ, Rossi DJ. siRNA drugs: Here to stay. Mol Ther. 2021;29:431–432. doi: 10.1016/j.ymthe.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thi TTH, et al. Lipid-based nanoparticles in the clinic and clinical trials: From cancer nanomedicine to COVID-19 vaccines. Vaccines. 2021;9:359. doi: 10.3390/vaccines9040359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juliano RL, Stamp D. The effect of particle size and charge on the clearance rates of liposomes and liposome encapsulated drugs. Biochem Biophys Res Commun. 1975;63:651–658. doi: 10.1016/S0006-291X(75)80433-5. [DOI] [PubMed] [Google Scholar]
- 18.Enzyme-Carrier Potential of Liposomes in Enzyme Replacement Therapy N Engl J Med. 1975;292:215–215. doi: 10.1056/nejm197501232920423. [DOI] [PubMed] [Google Scholar]
- 19.Magee WE, Goff CW, Schoknecht J, Smith MD, Cherian K. The interaction of cationic liposomes containing entrapped horseradish peroxidase with cells in culture. J Cell Biol. 1974;63:492–504. doi: 10.1083/jcb.63.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rahman Y-E, et al. Liposome-encapsulated actinomycin D: potential in cancer chemotherapy. Proc Soc Exp Biol Med. 1974;146:1173–1176. doi: 10.3181/00379727-146-38268. [DOI] [PubMed] [Google Scholar]
- 21.Cullis PR, Mayer LD, Bally MB, Madden TD, Hope MJ. Generating and loading of liposomal systems for drug-delivery applications. Adv Drug Deliv Rev. 1989;3:267–282. doi: 10.1016/0169-409X(89)90024-0. [DOI] [Google Scholar]
- 22.Blumenthal R, Weinstein JN, Sharrow SO, Henkart P. Liposome–lymphocyte interaction: saturable sites for transfer and intracellular release of liposome contents. Proc Natl Acad Sci. 1977;74:5603–5607. doi: 10.1073/pnas.74.12.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deshayes A, Herrera-Estrella L, Caboche M. Liposome-mediated transformation of tobacco mesophyll protoplasts by an Escherichia coli plasmid. EMBO J. 1985;4:2731–2737. doi: 10.1002/j.1460-2075.1985.tb03996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tai-Kin W, Nicolau C, Hofschneider PH. Appearance of β-lactamase activity in animal cells upon liposome-mediated gene transfer. Gene. 1980;10:87–94. doi: 10.1016/0378-1119(80)90126-2. [DOI] [PubMed] [Google Scholar]
- 25.Fraley R, Subramani S, Berg P, Papahadjopoulos D. Introduction of liposome-encapsulated SV40 DNA into cells. J Biol Chem. 1980;255:10431–10435. doi: 10.1016/S0021-9258(19)70482-7. [DOI] [PubMed] [Google Scholar]
- 26.Tikchonenko TI, et al. Transfer of condensed viral DNA into eukaryotic cells using proteoliposomes. Gene. 1988;63:321–330. doi: 10.1016/0378-1119(88)90535-5. [DOI] [PubMed] [Google Scholar]
- 27.Jay DG, Gilbert W. Basic protein enhances the incorporation of DNA into lipid vesicles: model for the formation of primordial cells. Proc Natl Acad Sci. 1987;84:1978–1980. doi: 10.1073/pnas.84.7.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gould-Fogerite S, et al. Chimerasome-mediated gene transfer in vitro and in vivo. Gene. 1989;84:429–438. doi: 10.1016/0378-1119(89)90517-9. [DOI] [PubMed] [Google Scholar]
- 29.Zumbuehl O, Weder HG. Liposomes of controllable size in the range of 40 to 180 nm by defined dialysis of lipid/detergent mixed micelles. Biochimica et Biophysica Acta (BBA)-Biomembranes 640, 252–262 (1981). [DOI] [PubMed]
- 30.Gould-Fogerite S, Mannino RJ. Rotary dialysis: its application to the preparation of large liposomes and large proteoliposomes (protein-lipid vesicles) with high encapsulation efficiency and efficient reconstitution of membrane proteins. Anal Biochem. 1985;148:15–25. doi: 10.1016/0003-2697(85)90622-0. [DOI] [PubMed] [Google Scholar]
- 31.Szoka F, Jr, Papahadjopoulos D. Comparative properties and methods of preparation of lipid vesicles (liposomes) Annu Rev Biophys Bioeng. 1980;9:467–508. doi: 10.1146/annurev.bb.09.060180.002343. [DOI] [PubMed] [Google Scholar]
- 32.Felgner PL, et al. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rietwyk S, Peer D. Next-generation lipids in RNA interference therapeutics. ACS Nano. 2017;11:7572–7586. doi: 10.1021/acsnano.7b04734. [DOI] [PubMed] [Google Scholar]
- 34.Leventis R, Silvius JR. Interactions of mammalian cells with lipid dispersions containing novel metabolizable cationic amphiphiles. Biochimica et Biophysica Acta (BBA)-Biomembranes 1023, 124–132 (1990). [DOI] [PubMed]
- 35.Lee Y, et al. New cationic lipids for gene transfer with high efficiency and low toxicity: T-shape cholesterol ester derivatives. Bioorg Med Chem Lett. 2004;14:2637–2641. doi: 10.1016/j.bmcl.2004.02.061. [DOI] [PubMed] [Google Scholar]
- 36.Song YK, Liu F, Chu S, Liu D. Characterization of cationic liposome-mediated gene transfer in vivo by intravenous administration. Hum Gene Ther. 1997;8:1585–1594. doi: 10.1089/hum.1997.8.13-1585. [DOI] [PubMed] [Google Scholar]
- 37.Ren T, Song Y, Zhang G, Liu D. Structural basis of DOTMA for its high intravenous transfection activity in mouse. Gene Ther. 2000;7:764–768. doi: 10.1038/sj.gt.3301153. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Fan H, Levorse DA, Crocker LS. Interaction of cholesterol-conjugated ionizable amino lipids with biomembranes: lipid polymorphism, structure–activity relationship, and implications for siRNA delivery. Langmuir. 2011;27:9473–9483. doi: 10.1021/la201464k. [DOI] [PubMed] [Google Scholar]
- 39.Hou X, Zaks T, Langer R, Dong Y. Lipid nanoparticles for mRNA delivery. Nat Rev Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao, Y. et al. Emerging mRNA technologies: delivery strategies and biomedical applications. Chemical Society Reviews (2022). [DOI] [PubMed]
- 41.Regelin AE, et al.. Biophysical and lipofection studies of DOTAP analogs. Biochimica et Biophysica Acta (BBA)-Biomembranes 1464, 151–164 (2000). [DOI] [PubMed]
- 42.Kulkarni JA, Cullis PR, Van Der Meel R. Lipid nanoparticles enabling gene therapies: from concepts to clinical utility. Nucleic Acid Ther. 2018;28:146–157. doi: 10.1089/nat.2018.0721. [DOI] [PubMed] [Google Scholar]
- 43.Semple SC, et al.. Efficient encapsulation of antisense oligonucleotides in lipid vesicles using ionizable aminolipids: formation of novel small multilamellar vesicle structures. Biochimica et Biophysica Acta (BBA)-Biomembranes 1510, 152–166 (2001). [DOI] [PubMed]
- 44.Felgner JH, et al. Enhanced gene delivery and mechanism studies with a novel series of cationic lipid formulations. J Biol Chem. 1994;269:2550–2561. doi: 10.1016/S0021-9258(17)41980-6. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, et al. Factors influencing the efficiency of cationic liposome-mediated intravenous gene delivery. Nat Biotechnol. 1997;15:167–173. doi: 10.1038/nbt0297-167. [DOI] [PubMed] [Google Scholar]
- 46.Massing U, Kley JT, Gürtesch L, Fankhaenel S. A simple approach to DOTAP and its analogs bearing different fatty acids. Chem Phys Lipid. 2000;105:189–191. doi: 10.1016/S0009-3084(00)00121-3. [DOI] [PubMed] [Google Scholar]
- 47.Gaucheron J, Santaella C, Vierling P. Transfection with fluorinated lipoplexes based on fluorinated analogues of DOTMA, DMRIE and DPPES. Biochimica et Biophysica Acta (BBA)-Biomembranes 1564, 349–358 (2002). [DOI] [PubMed]
- 48.Wheeler J, et al. Stabilized plasmid-lipid particles: construction and characterization. Gene Ther. 1999;6:271–281. doi: 10.1038/sj.gt.3300821. [DOI] [PubMed] [Google Scholar]
- 49.Mok KW, Lam AM, Cullis PR. Stabilized plasmid-lipid particles: factors influencing plasmid entrapment and transfection properties. Biochimica et Biophysica Acta (BBA)-Biomembranes 1419, 137–150 (1999). [DOI] [PubMed]
- 50.Tam P, et al. Stabilized plasmid-lipid particles for systemic gene therapy. Gene Ther. 2000;7:1867–1874. doi: 10.1038/sj.gt.3301308. [DOI] [PubMed] [Google Scholar]
- 51.Chen Z, et al. Application of DODMA and derivatives in cationic nanocarriers for gene delivery. Curr Org Chem. 2016;20:1813–1819. doi: 10.2174/1385272820666160202004348. [DOI] [Google Scholar]
- 52.Heyes J, et al. Lipid encapsulation enables the effective systemic delivery of polyplex plasmid DNA. Mol Ther. 2007;15:713–720. doi: 10.1038/sj.mt.6300101. [DOI] [PubMed] [Google Scholar]
- 53.Jeffs LB, et al. A scalable, extrusion-free method for efficient liposomal encapsulation of plasmid DNA. Pharm Res. 2005;22:362–372. doi: 10.1007/s11095-004-1873-z. [DOI] [PubMed] [Google Scholar]
- 54.Gregory G. In Liposome Technology 503–520 (CRC Press, 2018).
- 55.Wang T, Upponi JR, Torchilin VP. Design of multifunctional non-viral gene vectors to overcome physiological barriers: dilemmas and strategies. Int J Pharm. 2012;427:3–20. doi: 10.1016/j.ijpharm.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 56.Webb MS, Klimuk SK, Semple SC, Hope MJ. in Manual of Antisense Methodology 167–190 (Springer, 1999).
- 57.Bramson JL, Bodner CA, Johnson J, Semple S, Hope MJ. Intravenous administration of stabilized antisense lipid particles (SALP) leads to activation and expansion of liver natural killer cells. Antisense Nucleic Acid Drug Dev. 2000;10:217–224. doi: 10.1089/oli.1.2000.10.217. [DOI] [PubMed] [Google Scholar]
- 58.Bartsch M, Weeke-Klimp AH, Meijer DK, Scherphof GL, Kamps JA. Cell-specific targeting of lipid-based carriers for ODN and DNA. J Liposome Res. 2005;15:59–92. doi: 10.1081/LPR-64961. [DOI] [PubMed] [Google Scholar]
- 59.Dass CR, Su T. Particle-mediated intravascular delivery of oligonucleotides to tumors: associated biology and lessons from genotherapy. Drug Delivery. 2001;8:191–213. doi: 10.1080/107175401317245886. [DOI] [PubMed] [Google Scholar]
- 60.Morrissey DV, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 61.Judge AD, et al. Confirming the RNAi-mediated mechanism of action of siRNA-based cancer therapeutics in mice. J Clin Investig. 2009;119:661–673. doi: 10.1172/JCI37515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Judge AD, Bola G, Lee AC, MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 63.Judge AD, et al. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 64.Zimmermann TS, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 65.Li L, et al. Developing lipid nanoparticle-based siRNA therapeutics for hepatocellular carcinoma using an integrated approachdeveloping siRNA therapy for hepatocellular carcinoma. Mol Cancer Ther. 2013;12:2308–2318. doi: 10.1158/1535-7163.MCT-12-0983-T. [DOI] [PubMed] [Google Scholar]
- 66.Heyes J, Palmer L, Bremner K, MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J Control Release. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 67.Sankaram M, Powell GL, Marsh D. Effect of acyl chain composition on salt-induced lamellar to inverted hexagonal phase transitions in cardiolipin. Biochimica et Biophysica Acta (BBA)-Biomembranes 980, 389–392 (1989). [DOI] [PubMed]
- 68.Liu Y, Huang L. Designer lipids advance systemic siRNA delivery. Mol Ther. 2010;18:669–670. doi: 10.1038/mt.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Semple SC, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 70.Jayaraman M, et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew Chem. 2012;124:8657–8661. doi: 10.1002/ange.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hatakeyama H, et al. Development of a novel systemic gene delivery system for cancer therapy with a tumor-specific cleavable PEG-lipid. Gene Ther. 2007;14:68–77. doi: 10.1038/sj.gt.3302843. [DOI] [PubMed] [Google Scholar]
- 72.Hatakeyama H, et al. Systemic delivery of siRNA to tumors using a lipid nanoparticle containing a tumor-specific cleavable PEG-lipid. Biomaterials. 2011;32:4306–4316. doi: 10.1016/j.biomaterials.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 73.Sakurai Y, et al. Endosomal escape and the knockdown efficiency of liposomal-siRNA by the fusogenic peptide shGALA. Biomaterials. 2011;32:5733–5742. doi: 10.1016/j.biomaterials.2011.04.047. [DOI] [PubMed] [Google Scholar]
- 74.Sato Y, et al. A pH-sensitive cationic lipid facilitates the delivery of liposomal siRNA and gene silencing activity in vitro and in vivo. J Control Release. 2012;163:267–276. doi: 10.1016/j.jconrel.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 75.Hatakeyama H, et al. The systemic administration of an anti-miRNA oligonucleotide encapsulated pH-sensitive liposome results in reduced level of hepatic microRNA-122 in mice. J Control Release. 2014;173:43–50. doi: 10.1016/j.jconrel.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 76.Sakurai Y, et al. Gene silencing via RNAi and siRNA quantification in tumor tissue using MEND, a liposomal siRNA delivery system. Mol Ther. 2013;21:1195–1203. doi: 10.1038/mt.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tamaru M, et al. Application of apolipoprotein E-modified liposomal nanoparticles as a carrier for delivering DNA and nucleic acid in the brain. Int J Nanomed. 2014;9:4267. doi: 10.2147/IJN.S65402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Watanabe T, et al. In vivo therapeutic potential of Dicer-hunting siRNAs targeting infectious hepatitis C virus. Sci Rep. 2014;4:1–11. doi: 10.1038/srep04750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sato Y, Hatakeyama H, Hyodo M, Harashima H. Relationship between the physicochemical properties of lipid nanoparticles and the quality of siRNA delivery to liver cells. Mol Ther. 2016;24:788–795. doi: 10.1038/mt.2015.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Warashina S, et al. A lipid nanoparticle for the efficient delivery of siRNA to dendritic cells. J Control Release. 2016;225:183–191. doi: 10.1016/j.jconrel.2016.01.042. [DOI] [PubMed] [Google Scholar]
- 81.Sato Y, et al. Understanding structure-activity relationships of pH-sensitive cationic lipids facilitates the rational identification of promising lipid nanoparticles for delivering siRNAs in vivo. J Control Release. 2019;295:140–152. doi: 10.1016/j.jconrel.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 82.Nakamura T, Nakade T, Yamada K, Sato Y, Harashima H. The hydrophobic tail of a pH-sensitive cationic lipid influences siRNA transfection activity and toxicity in human NK cell lines. Int J Pharm. 2021;609:121140. doi: 10.1016/j.ijpharm.2021.121140. [DOI] [PubMed] [Google Scholar]
- 83.Malamas AS, Gujrati M, Kummitha CM, Xu R, Lu Z-R. Design and evaluation of new pH-sensitive amphiphilic cationic lipids for siRNA delivery. J Control Release. 2013;171:296–307. doi: 10.1016/j.jconrel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu R, Lu Z-R. Design, synthesis and evaluation of spermine-based pH-sensitive amphiphilic gene delivery systems: Multifunctional non-viral gene carriers. SCIENCE CHINA Chem. 2011;54:359–368. doi: 10.1007/s11426-010-4198-2. [DOI] [Google Scholar]
- 85.Lu ZR, Laney VE, Hall R, Ayat N. Environment-Responsive Lipid/siRNA Nanoparticles for Cancer Therapy. Adv Healthcare Mater. 2021;10:2001294. doi: 10.1002/adhm.202001294. [DOI] [PubMed] [Google Scholar]
- 86.Wang X-L, Xu R, Lu Z-R. A peptide-targeted delivery system with pH-sensitive amphiphilic cell membrane disruption for efficient receptor-mediated siRNA delivery. J Control Release. 2009;134:207–213. doi: 10.1016/j.jconrel.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 87.Wang X-L, et al. Targeted systemic delivery of a therapeutic siRNA with a multifunctional carrier controls tumor proliferation in mice. Mol Pharm. 2009;6:738–746. doi: 10.1021/mp800192d. [DOI] [PubMed] [Google Scholar]
- 88.Sun D, Schur RM, Lu Z-R. Vol. 8 823–826 (Future Science, 2017).
- 89.Gujrati M, et al. Targeted Dual pH-Sensitive Lipid ECO/siRNA Self-Assembly Nanoparticles Facilitate In Vivo Cytosolic sieIF4E Delivery and Overcome Paclitaxel Resistance in Breast Cancer Therapy. Adv Healthcare Mater. 2016;5:2882–2895. doi: 10.1002/adhm.201600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gujrati M, et al. Multifunctional cationic lipid-based nanoparticles facilitate endosomal escape and reduction-triggered cytosolic siRNA release. Mol Pharm. 2014;11:2734–2744. doi: 10.1021/mp400787s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Parvani JG, Gujrati MD, Mack MA, Schiemann WP, Lu Z-R. Silencing β3 integrin by targeted ECO/siRNA nanoparticles inhibits EMT and metastasis of triple-negative breast cancer. Can Res. 2015;75:2316–2325. doi: 10.1158/0008-5472.CAN-14-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun D, et al. Self-assembly of a multifunctional lipid with core-shell dendrimer DNA nanoparticles enhanced efficient gene delivery at low charge ratios into RPE cells. Macromol Biosci. 2015;15:1663–1672. doi: 10.1002/mabi.201500192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Malamas AS, Jin E, Gujrati M, Lu Z-R. Dynamic contrast enhanced MRI assessing the antiangiogenic effect of silencing HIF-1α with targeted multifunctional ECO/siRNA nanoparticles. Mol Pharm. 2016;13:2497–2506. doi: 10.1021/acs.molpharmaceut.6b00227. [DOI] [PubMed] [Google Scholar]
- 94.Sun D, et al. Targeted multifunctional lipid ECO plasmid DNA nanoparticles as efficient non-viral gene therapy for Leber’s congenital amaurosis. Molecular Therapy-Nucleic Acids. 2017;7:42–52. doi: 10.1016/j.omtn.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sun D, et al. Synthesis and evaluation of pH-sensitive multifunctional lipids for efficient delivery of CRISPR/Cas9 in gene editing. Bioconjug Chem. 2018;30:667–678. doi: 10.1021/acs.bioconjchem.8b00856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vaidya AM, et al. Systemic delivery of tumor-targeting siRNA nanoparticles against an oncogenic LncRNA facilitates effective triple-negative breast cancer therapy. Bioconjug Chem. 2019;30:907–919. doi: 10.1021/acs.bioconjchem.9b00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ayat, N. R. et al. Formulation of biocompatible targeted ECO/siRNA nanoparticles with long-term stability for clinical translation of RNAi. nucleic acid therapeutics 29, 195–207 (2019). [DOI] [PMC free article] [PubMed]
- 98.Sun D, et al. Non-viral gene therapy for Stargardt disease with ECO/pRHO-ABCA4 self-assembled nanoparticles. Mol Ther. 2020;28:293–303. doi: 10.1016/j.ymthe.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sun D, et al. Stable retinoid analogue targeted dual pH-sensitive smart lipid ECO/pDNA nanoparticles for specific gene delivery in the retinal pigment epithelium. ACS Appl Bio Mater. 2020;3:3078–3086. doi: 10.1021/acsabm.0c00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schilb AL, et al. Efficacy of targeted ECO/miR-200c nanoparticles for modulating tumor microenvironment and treating triple negative breast cancer as non-invasively monitored by MR molecular imaging. Pharm Res. 2021;38:1405–1418. doi: 10.1007/s11095-021-03083-z. [DOI] [PubMed] [Google Scholar]
- 101.Nicolescu C, Vaidya A, Schilb A, Lu Z-R. Regulating oncogenic LncRNA DANCR with targeted ECO/siRNA nanoparticles for non-small cell lung cancer therapy. ACS Omega. 2022;7:22743–22753. doi: 10.1021/acsomega.2c02260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sun D, et al. Formulation and efficacy of ECO/pRHO-ABCA4-SV40 nanoparticles for nonviral gene therapy of Stargardt disease in a mouse model. J Control Release. 2021;330:329–340. doi: 10.1016/j.jconrel.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sun D, et al. Effective gene therapy of Stargardt disease with PEG-ECO/pGRK1-ABCA4-S/MAR nanoparticles. Molecular Therapy-Nucleic Acids. 2022;29:823–835. doi: 10.1016/j.omtn.2022.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oliver SE, et al. The advisory committee on immunization practices’ interim recommendation for use of Pfizer-BioNTech COVID-19 vaccine—United States, December 2020. Morb Mortal Wkly Rep. 2020;69:1922. doi: 10.15585/mmwr.mm6950e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Oliver SE, et al. The advisory committee on immunization practices’ interim recommendation for use of moderna COVID-19 vaccine—United States, December 2020. Morb Mortal Wkly Rep. 2021;69:1653. doi: 10.15585/mmwr.mm695152e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hassett KJ, et al. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Molecular Therapy-Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sabnis S, et al. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Paloncýová M, Čechová P, Šrejber M, Kührová P, Otyepka M. Role of ionizable lipids in SARS-CoV-2 vaccines as revealed by molecular dynamics simulations: from membrane structure to interaction with mRNA fragments. The journal of physical chemistry letters. 2021;12:11199–11205. doi: 10.1021/acs.jpclett.1c03109. [DOI] [PubMed] [Google Scholar]
- 109.Terada T, Kulkarni JA, Huynh A, Tam YYC, Cullis P. Protective effect of edaravone against cationic lipid-mediated oxidative stress and apoptosis. Biol Pharm Bull. 2021;44:144–149. doi: 10.1248/bpb.b20-00679. [DOI] [PubMed] [Google Scholar]
- 110.Liu S, et al. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR–Cas gene editing. Nat Mater. 2021;20:701–710. doi: 10.1038/s41563-020-00886-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Patel P, Ibrahim NM, Cheng K. The importance of apparent pKa in the development of nanoparticles encapsulating siRNA and mRNA. Trends Pharmacol Sci. 2021;42:448–460. doi: 10.1016/j.tips.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schlich M, et al. Cytosolic delivery of nucleic acids: The case of ionizable lipid nanoparticles. Bioengineering & Translational Medicine. 2021;6:e10213. doi: 10.1002/btm2.10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Whitehead KA, et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat Commun. 2014;5:1–10. doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Habrant D, et al. Design of ionizable lipids to overcome the limiting step of endosomal escape: application in the intracellular delivery of mRNA, DNA, and siRNA. J Med Chem. 2016;59:3046–3062. doi: 10.1021/acs.jmedchem.5b01679. [DOI] [PubMed] [Google Scholar]
- 115.Han X, et al. An ionizable lipid toolbox for RNA delivery. Nat Commun. 2021;12:1–6. doi: 10.1038/s41467-021-27493-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ramezanpour M, Tieleman DP. Computational insights into the role of cholesterol in inverted hexagonal phase stabilization and endosomal drug release. Langmuir (2022). [DOI] [PMC free article] [PubMed]
- 117.Cheng X, Lee RJ. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv Drug Deliv Rev. 2016;99:129–137. doi: 10.1016/j.addr.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 118.Epand RM, Epand RF, Ahmed N, Chen R. Promotion of hexagonal phase formation and lipid mixing by fatty acids with varying degrees of unsaturation. Chem Phys Lipid. 1991;57:75–80. doi: 10.1016/0009-3084(91)90051-C. [DOI] [PubMed] [Google Scholar]
- 119.Siegel DP. Inverted micellar intermediates and the transitions between lamellar, cubic, and inverted hexagonal lipid phases. II. Implications for membrane-membrane interactions and membrane fusion. Biophysical journal 49, 1171–1183 (1986). [DOI] [PMC free article] [PubMed]
- 120.Wasungu L, Hoekstra D. Cationic lipids, lipoplexes and intracellular delivery of genes. J Control Release. 2006;116:255–264. doi: 10.1016/j.jconrel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 121.Kumar V. Complementary molecular shapes and additivity of the packing parameter of lipids. Proc Natl Acad Sci. 1991;88:444–448. doi: 10.1073/pnas.88.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cavagnetto F, et al. Molecular packing parameters of bipolar lipids. Biochimica et Biophysica Acta (BBA)-Biomembranes 1106, 273–281 (1992). [DOI] [PubMed]
- 123.Hafez I, Maurer N, Cullis P. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 2001;8:1188–1196. doi: 10.1038/sj.gt.3301506. [DOI] [PubMed] [Google Scholar]
- 124.Kiaie SH, et al. Recent advances in mRNA-LNP therapeutics: immunological and pharmacological aspects. Journal of Nanobiotechnology. 2022;20:1–20. doi: 10.1186/s12951-022-01478-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Álvarez-Benedicto E, et al. Optimization of phospholipid chemistry for improved lipid nanoparticle (LNP) delivery of messenger RNA (mRNA) Biomaterials science. 2022;10:549–559. doi: 10.1039/D1BM01454D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kulkarni JA, et al. Design of lipid nanoparticles for in vitro and in vivo delivery of plasmid DNA. Nanomedicine: Nanotechnology, Biology and Medicine 13, 1377–1387 (2017). [DOI] [PubMed]
- 127.Akita H, et al. A neutral lipid envelope-type nanoparticle composed of a pH-activated and vitamin E-scaffold lipid-like material as a platform for a gene carrier targeting renal cell carcinoma. J Control Release. 2015;200:97–105. doi: 10.1016/j.jconrel.2014.12.029. [DOI] [PubMed] [Google Scholar]
- 128.Chen J, et al. Thermosensitive liposomes with higher phase transition temperature for targeted drug delivery to tumor. Int J Pharm. 2014;475:408–415. doi: 10.1016/j.ijpharm.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 129.Chen J, et al. Influence of lipid composition on the phase transition temperature of liposomes composed of both DPPC and HSPC. Drug Dev Ind Pharm. 2013;39:197–204. doi: 10.3109/03639045.2012.668912. [DOI] [PubMed] [Google Scholar]
- 130.Daniel I, Oger P, Winter R. Origins of life and biochemistry under high-pressure conditions. Chem Soc Rev. 2006;35:858–875. doi: 10.1039/b517766a. [DOI] [PubMed] [Google Scholar]
- 131.Gabizon A, Papahadjopoulos D. The role of surface charge and hydrophilic groups on liposome clearance in vivo. Biochimica Et Biophysica Acta (BBA)-Biomembranes 1103, 94–100 (1992). [DOI] [PubMed]
- 132.Di Martino MT, et al. In vivo activity of miR-34a mimics delivered by stable nucleic acid lipid particles (SNALPs) against multiple myeloma. PLoS ONE. 2014;9:e90005. doi: 10.1371/journal.pone.0090005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Andreoli TE. On the anatomy of amphotericin B-cholesterol pores in lipid bilayer membranes. Kidney Int. 1973;4:337–345. doi: 10.1038/ki.1973.126. [DOI] [PubMed] [Google Scholar]
- 134.Gindy ME, et al. Stabilization of Ostwald ripening in low molecular weight amino lipid nanoparticles for systemic delivery of siRNA therapeutics. Mol Pharm. 2014;11:4143–4153. doi: 10.1021/mp500367k. [DOI] [PubMed] [Google Scholar]
- 135.Patel S, et al. Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat Commun. 2020;11:1–13. doi: 10.1038/s41467-020-14527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Park S, Choi YK, Kim S, Lee J, Im W. CHARMM-GUI membrane builder for lipid nanoparticles with ionizable cationic lipids and PEGylated lipids. J Chem Inf Model. 2021;61:5192–5202. doi: 10.1021/acs.jcim.1c00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Eygeris Y, Patel S, Jozic A, Sahay G. Deconvoluting lipid nanoparticle structure for messenger RNA delivery. Nano Lett. 2020;20:4543–4549. doi: 10.1021/acs.nanolett.0c01386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pilkington EH, et al. From influenza to COVID-19: Lipid nanoparticle mRNA vaccines at the frontiers of infectious diseases. Acta Biomater. 2021;131:16–40. doi: 10.1016/j.actbio.2021.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.ur Rehman Z, Zuhorn IS, Hoekstra D. How cationic lipids transfer nucleic acids into cells and across cellular membranes: recent advances. Journal of Controlled Release 166, 46–56 (2013). [DOI] [PubMed]
- 140.Doktorovova, S., Shegokar, R. & Souto, E. B. in Nanostructures for novel therapy 811–843 (Elsevier, 2017).
- 141.Ganesan P, Ramalingam P, Karthivashan G, Ko YT, Choi D-K. Recent developments in solid lipid nanoparticle and surface-modified solid lipid nanoparticle delivery systems for oral delivery of phyto-bioactive compounds in various chronic diseases. Int J Nanomed. 2018;13:1569. doi: 10.2147/IJN.S155593. [DOI] [PMC free article] [PubMed] [Google Scholar]