PURPOSE
Multiple FGFR inhibitors are currently in clinical trials enrolling adults with different solid tumors, while very few enroll pediatric patients. We determined the types and frequency of FGFR alterations (FGFR1-4) in pediatric cancers to inform future clinical trial design.
METHODS
Tumors with FGFR alterations were identified from two large cohorts of pediatric solid tumors subjected to targeted DNA sequencing: The Dana-Farber/Boston Children's Profile Study (n = 888) and the multi-institution GAIN/iCAT2 (Genomic Assessment Improves Novel Therapy) Study (n = 571). Data from the combined patient population of 1,395 cases (64 patients were enrolled in both studies) were reviewed and cases in which an FGFR alteration was identified by OncoPanel sequencing were further assessed.
RESULTS
We identified 41 patients with tumors harboring an oncogenic FGFR alteration. Median age at diagnosis was 8 years (range, 6 months-26 years). Diagnoses included 11 rhabdomyosarcomas, nine low-grade gliomas, and 17 other tumor types. Alterations included gain-of-function sequence variants (n = 19), amplifications (n = 10), oncogenic fusions (FGFR3::TACC3 [n = 3], FGFR1::TACC1 [n = 1], FGFR1::EBF2 [n = 1], FGFR1::CLIP2 [n = 1], and FGFR2::CTNNA3 [n = 1]), pathogenic-leaning variants of uncertain significance (n = 4), and amplification in combination with a pathogenic-leaning variant of uncertain significance (n = 1). Two novel FGFR1 fusions in two different patients were identified in this cohort, one of whom showed a response to an FGFR inhibitor.
CONCLUSION
In summary, activating FGFR alterations were found in approximately 3% (41/1,395) of pediatric solid tumors, identifying a population of children with cancer who may be eligible and good candidates for trials evaluating FGFR-targeted therapy. Importantly, the genomic and clinical data from this study can help inform drug development in accordance with the Research to Accelerate Cures and Equity for Children Act.
INTRODUCTION
Oncogenic FGFR1-4 alterations occur at different frequencies in multiple types of solid tumors including head and neck cancer, lung cancer, breast cancer, and cholangiocarcinoma.1-6 FGFR1-4 oncogenic mutations, fusions, and amplifications can activate the downstream MAPK-ERK, PI3K/AKT, and/or JAK-STAT pathway.7-10 Pediatric cancers such as rhabdomyosarcoma (RMS) harbor recurrent FGFR4 sequence variants and low-grade gliomas (LGG) harbor recurrent FGFR1/2 variants and fusions, including intragenic duplications in FGFR1 affecting the tyrosine kinase domain.11-13
CONTEXT
Key Objective
FGFR alterations occur in multiple adult cancers and have previously been found in some pediatric tumors including rhabdomyosarcoma and low-grade glioma. To better define the relevance of FGFR as a molecular target in children with cancer, this study identified and characterized FGFR-activating alterations in a large cohort of sequenced pediatric cancers.
Knowledge Generated
Activating FGFR1-4 alterations were identified in 3% of pediatric solid tumors across 19 different histologies, thus highlighting a population of patients that may benefit from pediatric trials of FGFR-targeted therapies. The study also provides proof-of-concept evidence of clinical efficacy in an infant with a spindle-cell sarcoma with a novel FGFR1 fusion treated with erdafitinib.
Relevance
Although many FGFR-targeting compounds are under development for adults, the availability of these drugs for children with cancer is quite limited. The findings from this study significantly inform pediatric drug development in accordance with the Research to Accelerate Cures and Equity for Children Act.
FGFR-tyrosine kinase inhibitors (TKIs) are under clinical investigation, with early trials reporting response rates up to 20%-40% primarily in patients with well-characterized FGFR-activating alterations.14-16 These include specific FGFR4 inhibitors and pan-FGFR inhibitors (FGFR1-3 and FGFR1-4).14-16 Pemigatinib, infigratinib, and erdafitinib, three highly selective US Food and Drug Administration (FDA)–approved pan-FGFR inhibitors, have defined on-target toxicities including hyperphosphatemia, vision changes, and skin and nail changes.14 To decrease toxicity and address the development of resistance variants in FGFR1-4, additional compounds are under clinical investigation including derazantinib, rogaratinib, and futibatinib.14 A recent trial of futibatinib (TAS-120), the only irreversible pan-FGFR inhibitor, showed an objective response rate of 13.7% and notably included patients with previously uncharacterized FGFR1-3 alterations.17 Importantly, futibatinib can overcome resistance to reversible ATP-competitive FGFR inhibitors including infigratinib.17,18
Despite the numerous compounds under development targeting FGFR, the number of clinical trials and the availability of these drugs to children with different cancers are quite low. The Research to Accelerate Cures and Equity (RACE) for Children Act was enacted to remove clinical indication and orphan drug designation–based waivers that previously limited requirements for pediatric cancer drug development. This law authorizes the FDA to mandate pediatric studies of molecular targeted therapies if the target is relevant to the growth or progression of one or more pediatric cancers.19 Here, we aimed to determine the frequency and types of FGFR1-4 variants in pediatric solid tumors and the diagnoses in which they occur to better define the relevance of FGFR as a molecular target in pediatric cancer and facilitate the design of clinical trials of FGFR inhibitors in children.20 We integrated genomic data from two cohorts of patients with pediatric CNS and extracranial solid tumors subjected to targeted DNA sequencing to identify pediatric patients with oncogenic FGFR alterations and describe the genomic alterations and clinical features of these pediatric FGFR-altered malignancies.
METHODS
Patients and Samples
This study was approved by the Dana-Farber Cancer Institute (DFCI, Boston, MA) Institutional Review Board with a waiver of informed consent. The study included patients from two sequencing studies who had tumor samples sequenced with OncoPanel, a targeted DNA next-generation sequencing panel, between January 1, 2013, and July 1, 2021. The GAIN Consortium study (ClinicalTrials.gov identifier: NCT02520713) is a prospective cohort study enrolling patients age ≤ 30 years with relapsed/refractory and high-risk pediatric extracranial solid tumors at 12 institutions to determine the clinical impact of molecular tumor profiling.21 The Dana-Farber/Boston Children's Profile Cancer Research Study, offered to all pediatric oncology patients with a variety of solid malignancies and lymphomas since 2013, facilitates research on cancer biology and informs ongoing and potential trials assessing targeted therapies. Patients enrolled on either study were included in this analysis if they had at least one successful targeted DNA sequencing result and a CNS or extracranial solid malignancy. Patients with lymphoma were excluded.
Patient Clinical Information
Medical records were reviewed to obtain demographic, clinical, and specimen variables including cancer diagnosis, age at diagnosis, sex, stage at diagnosis, tumor biopsy site, timing of tumor acquisition, and treatments. Timing of tumor acquisition was defined as at initial diagnosis (which included local control surgery during upfront treatment) or at relapse/progression. Sequencing data for all eligible patients were obtained from each study and used to identify patients with FGFR1-4 genomic alterations (single-nucleotide variants [SNVs], amplifications, or fusions) on OncoPanel sequencing.
Molecular Tumor Profiling
Patients had at least one tumor sample (fresh frozen or archival formalin-fixed, paraffin-embedded tissue) sequenced using OncoPanel (Data Supplement). OncoPanel analysis includes the detection of SNVs, insertions and deletions (indels), copy-number (CN) alterations, structural variants (SV), tumor mutational burden, and microsatellite instability, as previously described.22-24 FGFR2 introns are not covered for fusion detection by OncoPanelv1 (7% of cohort was tested with this version), and FGFR4 introns are not covered for fusion detection by any version as FGFR4 fusions are rarely reported in cancer. CN estimates for amplifications (defined as CN ≥ 7) were only available for tumors sequenced with OncoPanelv3. Of note, OncoPanel is not optimized to identify intragenic FGFR duplications but does detect extracellular in-frame deletions.25 A molecular pathology report was returned to treating providers at the time of sequencing, and data were made available for this analysis in August 2021.
Variant Interpretation
Variant curation was conducted using multiple databases, primary literature, and other criteria as detailed in the Data Supplement. The following were considered activating alterations of FGFR1-4: amplifications (CN ≥ 7) on the basis of the adult NCI-MATCH eligibility criteria for FGFR amplifications (ClinicalTrials.gov identifier: NCT02465060); in-frame oncogenic fusions with an intact protein tyrosine kinase domain (TKD); pathogenic SNVs and indels; and pathogenic-leaning variants of uncertain significant (PL-VUS). Other FGFR variants of uncertain significance (VUS) were identified and excluded from further assessment.
Clinical Trial Data Access
We identified clinical trials evaluating FGFR inhibitors using ClinicalTrials.gov. We collected recruitment status, phase, and age of eligibility for clinical trials of six FGFR inhibitors being investigated in the United States—derazantinib, rogaratinib, futibatinib, pemigatinib, infigratinib, and erdafitinib—as of January 20, 2022.
RESULTS
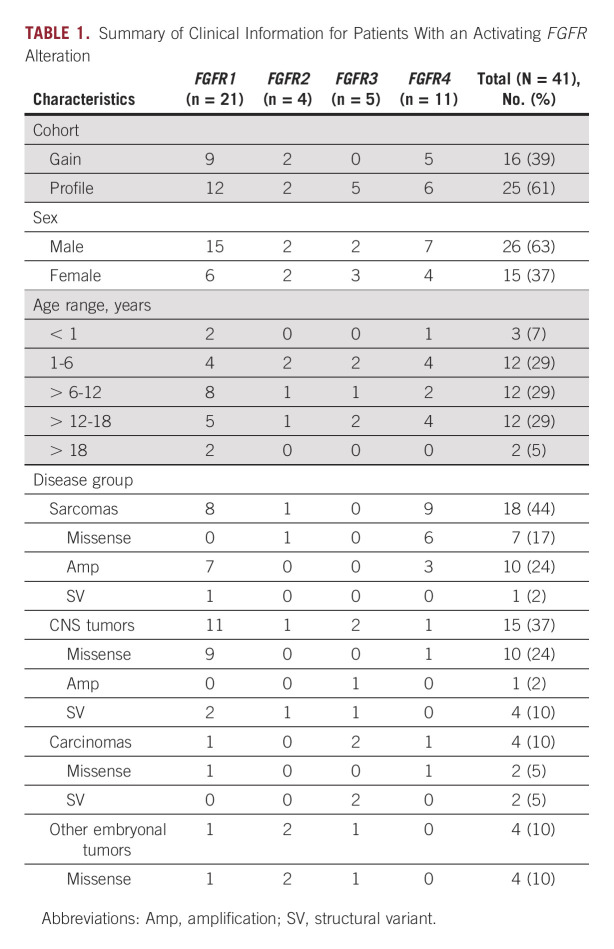
FGFR1-4 Activating Alteration Frequency and Case Characteristics
The study population included 888 patients enrolled on the Profile Cancer Research Study and 571 patients enrolled on the GAIN Consortium Study for a total of 1,395 patients (64 patients enrolled on both) with 93% sequenced on OncoPanel versions 2 or 3 (Data Supplement). Forty-one patients (3%) had tumors with activating FGFR1-4 alterations (Fig 1). Twenty-six patients (63%) were male. Median age at diagnosis was 8 years (range, 6 months-26 years; Data Supplement). Four disease groups were represented (sarcomas [n = 18], CNS tumors [n = 15], carcinomas [n = 4], other embryonal tumors [n = 4]; Table 1 and Data Supplement). Across these groups, there were 19 distinct diagnoses (Data Supplement). The most common diagnoses were RMS (n = 11) and LGG (n = 9), which together comprised 49% of the cohort. Other diagnosis included Ewing sarcoma, glioblastoma, osteosarcoma, and Wilms tumor. Of the 11 RMS, eight were fusion-negative and three were PAX3::FOXO1 fusion-positive (Fig 1). Sequenced samples were from the primary site at initial diagnosis for 68% of patients. One patient with a tumor harboring an activating FGFR alteration was treated with an FGFR inhibitor. An additional 95 patients had tumors with an FGFR VUS that were not considered activating (Data Supplement).
FIG 1.
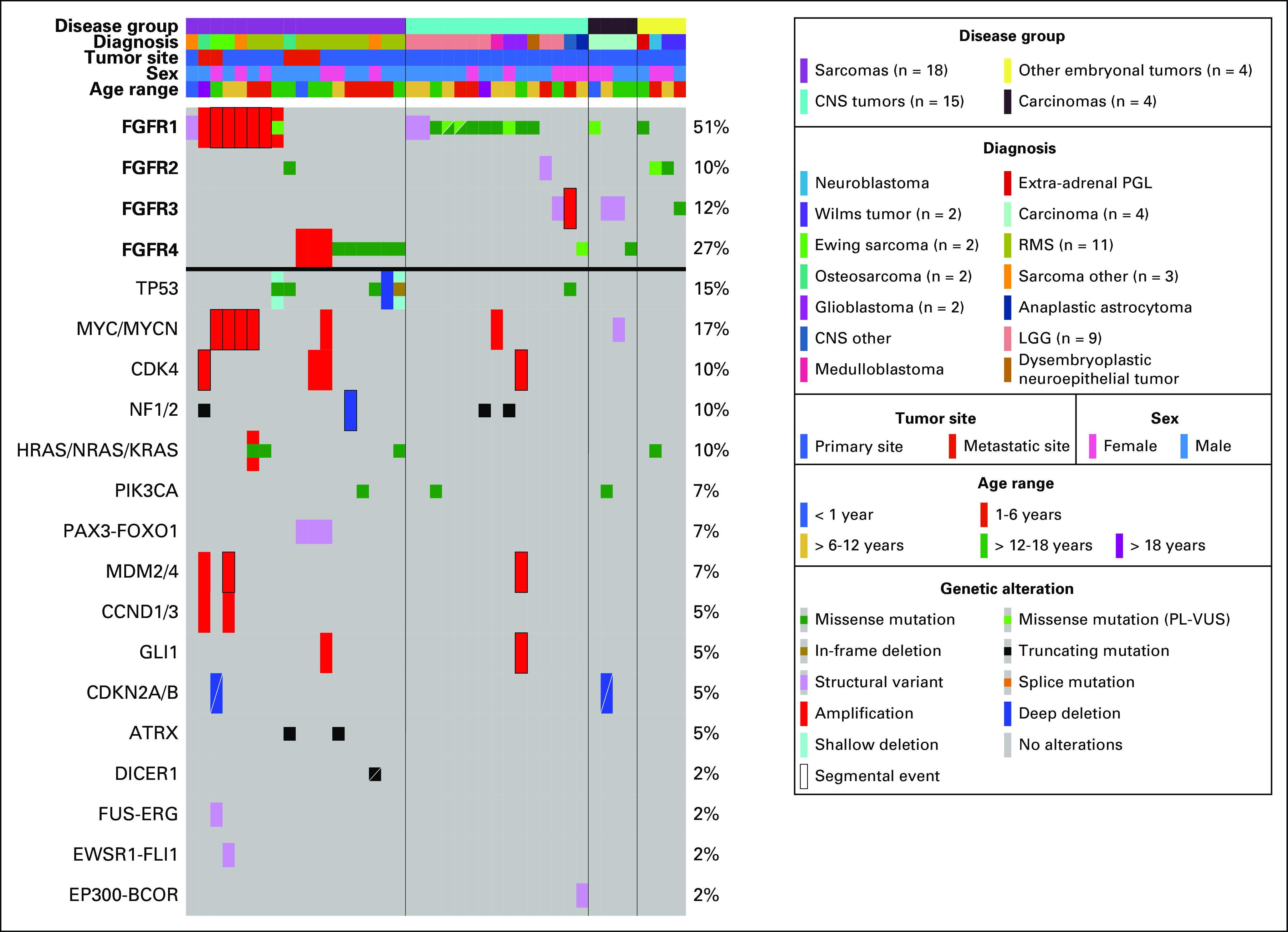
Genomic and clinical features of 41 pediatric patients with at least one activating FGFR alteration. OncoPrint shows clinical features including disease group, diagnosis, tumor site, sex, and age at diagnosis as well as oncogenic or likely oncogenic variants in genes altered in more than one patient or alterations with diagnostic implications. LGG, low-grade glioma; PGL, paraganglioma; PL-VUS, pathogenic-leaning variants of uncertain significance; RMS, rhabdomyosarcoma
TABLE 1.
Summary of Clinical Information for Patients With an Activating FGFR Alteration
Activating FGFR Alterations Identified
Pathogenic FGFR1-4 SNVs, identified in 19 tumors (46%), were the most frequent type of alteration in this cohort. Eleven tumors (27%) had an amplification (CN range, 7-30) and seven (17%) had a fusion. Four tumors (10%) had a PL-VUS, one of which also had an amplification. Activating alterations occurred most often in FGFR1 (n = 21) followed by FGFR4 (n = 11). Two tumors from the primary site harbored two activating FGFR1 alterations at initial diagnosis including a RMS (amplification [CN = 30], PL-VUS) and LGG (p.N546K; p.K656N; Fig 1). Tumors from patients with RMS harbored recurrent FGFR4 missense variants (5/11), FGFR1 amplifications (3/11), and FGFR4 amplifications (3/11; all in fusion-positive RMS). LGGs harbored FGFR1 variants (5/9) and FGFR1-3 fusions (4/9). The sarcomas harbored FGFR1 amplifications (4/7), FGFR1 fusion (1/7), and FGFR2/4 SNVs (2/7), while FGFR1 SNVs and FGFR3 alterations were found across other disease groups (Data Supplement). Extracellular FGFR2 in-frame deletions recently reported in cholangiocarcinoma (also via OncoPanel sequencing) were not seen in this cohort.25
Tumors with an FGFR missense variant were primarily those with an oncogenic hotspot variant in FGFR1 (N546K [n = 5]; K656E [n = 4]) and FGFR4 (N535K/D [n = 2]; V550L/E [n = 5]). Of note, 21 of 41 (51%) of the tumors had at least one FGFR missense variant located in the TKD (Fig 2A). The FGFR4 V550L/E gatekeeper mutation known to induce TKI resistance was identified in four of five patients with RMS, consistent with previous studies reporting this variant in embryonal RMS.26
FIG 2.
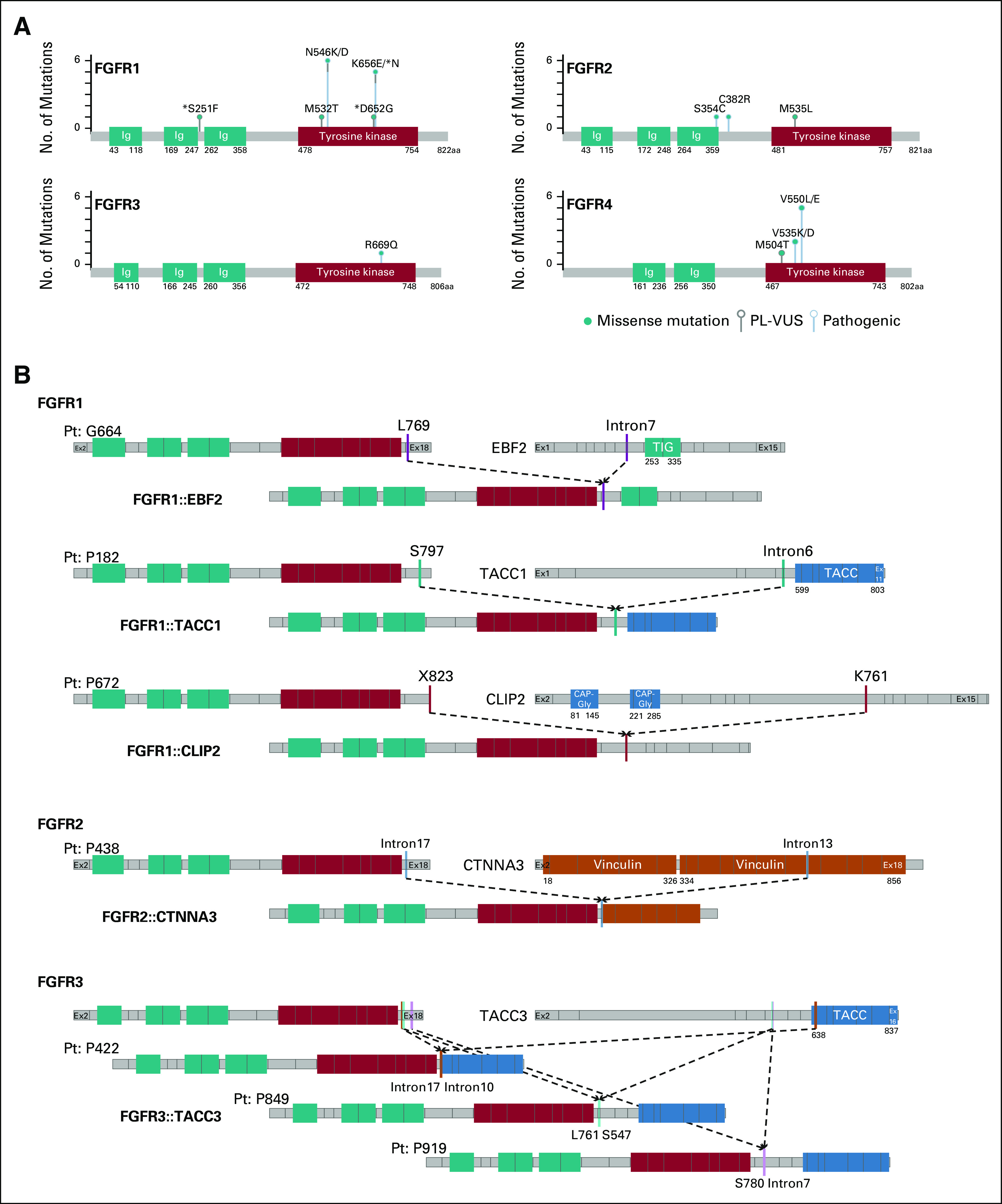
FGFR1-4 alterations identified in two large cohorts of pediatric patients. (A) Lollipop plot of FGFR1-4 activating variants identified. (B) Schematic representation of gene breakpoints and predicted fusion products of three FGFR1, one FGFR2, and three FGFR3 fusions in pediatric solid tumors. Light black lines represent start and end of exons. Solid lines of different colors depict breakpoints (CLIP2: NM_003388; EBF2: NM_022659; TACC1: NM_006283; TACC3: NM_006342; FGFR1: NM_023110; FGFR2: NM_000141; FGFR3: NM_000142; FGFR4 NM_0002011). *PL-VUS identified in patients who also have a pathogenic alteration. PL-VUS, pathogenic-leaning variants of uncertain significance.
FGFR fusions with an intact TKD accounted for 17% (7/41) of the activating FGFR alterations. An FGFR3::TACC3 fusion was identified in two carcinomas and one LGG. We also identified two previously reported fusions in LGGs including an FGFR1::TACC1 fusion and an FGFR2::CTNNA3 fusion.12,27 Two novel fusions were identified: FGFR1::CLIP2 fusion in a patient with LGG and FGFR1::EBF2 fusion in a patient with a spindle-cell sarcoma (Fig 2B).
Amplifications accounted for 27% (11/41) of the FGFR alterations (Data Supplement). Seven tumors harbored FGFR1 amplifications, four of which were further evaluated for FGFR1 gene expression using RNA sequencing and showed high expression levels (Data Supplement). One tumor harbored an FGFR3 amplification (CN = 7). Three tumors from patients with fusion-positive RMS harbored focal FGFR4 amplifications: one had an estimated CN = 7, and two had a high-level amplification (CN > 7; Fig 1).
Molecular Landscape of Pediatric Cancers Harboring FGFR Alterations
Co-occurring oncogenic alterations were commonly seen in the 41 tumors with activating FGFR alterations. MYC/MYCN amplification was the most recurrent alteration along with TP53-inactivating alterations identified in 17% and 15% of the FGFR-altered tumors, respectively. Of interest, the RAS pathway was activated through HRAS/NRAS/KRAS alterations, including a KRAS G12V variant, or NF1/2 inactivating alterations in 17% (7/41) of the FGFR-altered tumors. Activation of oncogenes (PIK3CA and MDM2/4), inactivation of tumor suppressors (ATRX), and cell cycle dysregulation (CDK4, CDKN2A/B, and CCND1/3) were also seen (Fig 1 and Data Supplement).
Additional alterations with diagnostic implications included three PAX3::FOXO1 fusions in RMS, a EWSR1::FLI1 fusion in a Ewing sarcoma, a EP300::BCOR fusion in a CNS tumor, and a DICER1-truncating alteration in a DICER1-associated sarcoma (Fig 1 and Data Supplement).
Eligibility for Clinical Trials
Review of clinical trials actively recruiting patients with FGFR alterations in ClinicalTrials.gov identified 36 clinical trials assessing six FGFR inhibitors. Children age < 18 years were only eligible for two phase II studies assessing efficacy of erdafitinib (Data Supplement). Many patients in this cohort would have been eligible for the NCI/COG Pediatric MATCH treatment trial (ClinicalTrials.gov identifier: NCT03210714) assessing erdafitinib since 39% of the patients (16/41) with eligible fusions and SNVs were between age 1 and 21 years at the time of sample acquisition. Three patients between age 1 and 4 years likely would not have been eligible for the Pediatric MATCH trial because of inability to swallow pills. The Janssen Research & Development clinical trial (ClinicalTrials.gov identifier: NCT04083976) assessing erdafitinib includes patients age ≥ 6 years. Nine patients (22%) were age ≥ 6 years at sample acquisition and harbored eligible FGFR variants and fusions (Data Supplement).
Overall, 20 patients (48% of those with FGFR-activating alterations) would not qualify for either trial because of the lack of eligible FGFR alterations including FGFR amplifications (n = 11), FGFR4 gatekeeper variant (n = 5), and PL-VUS missense variants (n = 4). Additionally, three patients with Janssen-eligible alterations did not meet the age cutoff at diagnosis: two with fusions and one with an FGFR3 R669Q SNV.
Pediatric Patient in the Cohort With Response to an FGFR Inhibitor
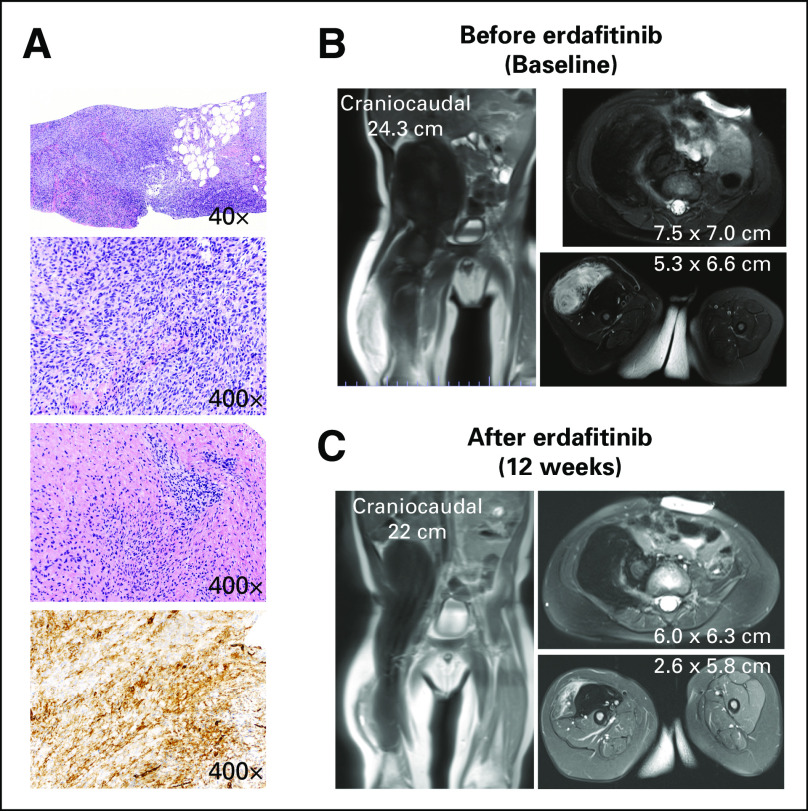
A previously healthy 9-month-old infant with a right thigh mass was the only patient with an FGFR-altered tumor who received targeted therapy with an FGFR inhibitor. Magnetic resonance imaging showed a large retroperitoneal heterogenous mass extending into the proximal right thigh. The predominantly T2 hypointense pelvic component measured 6.4 × 5.6 × 7.7 cm. The predominantly T2 hyperintense thigh component measured 4.1 × 3.5 × 7.5 cm. Both components were biopsied given differences in imaging characteristics. Pathology revealed a spindle and round cell neoplasm (Fig 3A). Clinical testing for germline cancer predisposition (156 gene panel) revealed no pathogenic variants. OncoPanel sequencing revealed an oncogenic FGFR1::EBF2 fusion.
FIG 3.
Infant with spindle-cell sarcoma who responded to erdafitinib treatment. (A) Histopathology at diagnosis showing the tumor is mostly hypercellular and infiltrating adipose tissue. Tumor cells are predominantly spindle shaped with foci showing round morphology and occasional intracytoplasmic vacuoles. The pelvic extension of the tumor is relatively less cellular with more collagenized stroma (top three panels; hematoxylin and eosin stain). By immunohistochemistry, tumor cells are diffusely positive for CD34 (bottom panel). MRI (B) at initiation of erdafitinib and (C) after 12 weeks. MRI, magnetic resonance imaging.
Lenvatinib, a multitargeted TKI, was selected as the initial therapy as dosing and administration had been studied in children age as young as 2 years in Children's Oncology Group (COG) protocol ADVL1711 (ClinicalTrials.gov identifier: NCT03245151).28 During cycle 1, he developed toxicities including nausea, vomiting, thrombocytopenia, and hypothyroidism requiring hormone replacement and hypertension, leading to a dose reduction of lenvatinib. Imaging after eight weeks of lenvatinib therapy showed progression. Lenvatinib was stopped, and therapy was initiated with vincristine and actinomycin on the basis of COG infantile fibrosarcoma protocol ARST03P1 (ClinicalTrials.gov identifier: NCT00072280). After two cycles, imaging showed progression of the thigh mass, and cyclophosphamide was added in accordance with the infantile fibrosarcoma protocol. Vincristine was omitted from subsequent cycles because of toxicities (grade 3 cranial nerve neuropathy causing aspiration of liquids and necessitating placement of a gastrostomy tube). He received five additional cycles of actinomycin and cyclophosphamide with stable disease on imaging. Given the lack of objective response after the first two cycles, the treating team worked to identify an FGFR inhibitor with pediatric dosing and administration data. Surgery was consulted and resection was felt to be high risk for morbidity, given the extensive nature of the tumor. After seven cycles of chemotherapy, insurance approval was obtained for erdafitinib, which has pediatric toxicity data and guidelines for administration as a suspension. Three weeks before planned resection, erdafitinib was started at a dose of 4.7 mg/m2 by G-tube once daily continuously. Cycle 1 of erdafitinib was complicated by hyperphosphatemia requiring a dose interruption, initiation of sevelamer, and a low-phosphorus diet. Physical examination after 5 days of erdafitinib was remarkable for a softer and smaller anterior thigh mass. After 12 weeks of erdafitinib (with two 7-day interruptions for hyperphosphatemia), the thigh mass was no longer tender to palpation, he started walking, and imaging showed reduction in size and enhancement of the thigh mass (Figs 3B and 3C). In the setting of clinical and radiographic response, erdafitinib treatment was continued with a 50% dose reduction (every other day dosing), given the prior toxicities and surgery was deferred. Treatment was ongoing at the time of this report.
DISCUSSION
In this study, we used the sequencing data from two large pediatric studies to describe the types and frequency of actionable FGFR alterations identified in pediatric solid tumors. Approximately 3% of pediatric patients across these two cohorts had tumors with an activating FGFR alteration and in some cases two alterations, suggesting possible selection for a second alteration for further pathway activation.
We also highlight a proof-of-principle clinical response to an FGFR inhibitor in a child with an FGFR1 fusion-positive soft tissue sarcoma who progressed on lenvatinib. Although two pediatric patients with gliomas have been previously reported in a case series of five patients to show a partial response to the FGFR inhibitor, Debio1347, these patients harbored well-characterized alterations including an oncogenic FGFR1::TACC1 fusion.29 Our study is, to our knowledge, the first to report a pediatric patient with a predicted oncogenic novel FGFR1 fusion to benefit from targeted therapy. All of these pediatric patients were treated under single patient protocols or with off-label use, highlighting the need for clinical trials investigating FGFR inhibitors allowing for a more uniform evaluation of these compounds in pediatric malignancies.
Clinical trial opportunities for pediatric patients with FGFR-altered tumors are limited, with only two trials currently enrolling children age < 18 years. Overall, 44% of patients (18/41) in this study would be potentially eligible for at least one of these two trials, given age and alteration eligibility criteria; however, only 17% of patients (7/41) would have the option of either trial. The 9-month-old patient who responded to off-label erdafitinib did not meet the criteria for any clinical trial despite having an eligible fusion for the Janssen trial. By contrast, the 18-year-old patient with a hotspot FGFR1 variant would have been eligible for > 20 clinical trials actively enrolling adult patients with solid tumors. If the FGFR alterations identified in our cohort had been found in adults, at least 78% (32/41) of the patients would have met the alteration eligibility criteria for an FGFR inhibitor clinical trial.
Although three selective pan-FGFR inhibitors are FDA-approved, only erdafitinib is in clinical trials enrolling pediatric patients. Newer FGFR inhibitors, such as futibatinib, that are able to overcome gatekeeper mutations are now in clinical trials in adults (ClinicalTrials.gov identifier: NCT02052778).30 These FGFR inhibitors are important to study in children since the FGFR4 V550 gatekeeper mutation is frequently identified in pediatric patients with untreated RMS and patients with this mutation are not eligible for the Pediatric MATCH study as they are not expected to respond to erdafitinib. Clinical trials of FGFR inhibitors enrolling children are lacking both in number and diversity of FGFR inhibitors, highlighting the need to open new FGFR inhibitor trials or expand eligibility criteria for ongoing trials to allow enrollment of pediatric patients. The Pediatric Research Equity Act (PREA) was amended in August 2020 to include postmarketing requirements for pediatric studies of molecularly targeted oncology drugs and all New Drug Applications are required to have an Initial Pediatric Study Plan. Infigratinib, an FGFR inhibitor approved by the FDA for cholangiocarcinoma, met the postmarketing requirements with a planned pediatric study (ClinicalTrials.gov identifier: NCT05222165). There may be Initial Pediatric Study Plans that we have not reported on as these are not publicly available before FDA drug approval.
This study has several limitations. It is likely that we underestimate the frequency of FGFR alterations, especially fusions that can be missed when using a DNA assay. More specifically, the 7% of cases in the cohort (102/1,395) sequenced with OncoPanelv1 were not evaluated for FGFR2 fusions. In addition, we were stringent in assigning amplification status, which resulted in excluding cases with absolute CN < 7 and those sequenced with OncoPanelv1 and v2 for which absolute CN was not available. Additionally the VUSs, present in 95 patients, were not evaluated with functional studies and some may have been activating. An additional limitation is inclusion of FGFR amplification (CN ≥ 7) as an activating alteration as it is difficult to assess the pathogenicity of FGFR amplifications. That said, where assessed, FGFR expression was higher in FGFR-amplified compared with FGFR-nonamplified cases. To date, clinical trial results are unclear regarding whether FGFR amplifications are a biomarker for FGFR-TKI response, and whether responses differ between FGFR genes and inhibitor used.17,31-33 Finally, the infant with a FGFR1 fusion-positive soft tissue sarcoma who had a response to an FGFR inhibitor had a very short follow up time, so the durability of the response in this patient is yet unknown.
Despite these limitations, to our knowledge, this is the first and largest study of activating FGFR alterations in pediatric patients, which identifies an important patient population with overall limited treatment options. We describe the clinical characteristics of pediatric patients with targetable FGFR alterations, including response to an FGFR inhibitor in one patient with a novel fusion. Although FGFR alterations are rare, new molecular therapies in development that target FGFR should have early pediatric investigations in accordance with the RACE for Children Act. At a minimum, these trials should include assessment of dosing and liquid formulations. Approaches to address the infrequent occurrence of these alterations in pediatric cancer include international collaboration, prioritization of agents to be studied, and histology agnostic clinical trials.
ACKNOWLEDGMENT
The authors would like to thank the patients and families who participated in the Profile and GAIN studies. The authors would like to acknowledge the Profile study at Dana-Farber/Brigham and Women's Cancer Center and Dana-Farber/Boston Children's Cancer and Blood Disorders Center for generating the sequencing data for the Profile cohort. The authors would also like to acknowledge the DFCI Oncology Data Retrieval System (OncDRS) for the aggregation, management, and delivery of the clinical and operational research data used in this project for the Profile cohort.
Harrison K. Tsai
Consulting or Advisory Role: Vertex
Alanna J. Church
Honoraria: The Jackson Laboratory
Consulting or Advisory Role: AlphaSights, The Jackson Laboratory, Bayer
Travel, Accommodations, Expenses: Bayer
Navin R. Pinto
Patents, Royalties, Other Intellectual Property: I hold a patent 20210346431 that has been licensed to Umoja Biotherapeutics and may result in royalty payments. No payments have been received to date
(OPTIONAL) Uncompensated Relationships: Y-mAbs Therapeutics
Margaret E. Macy
Stock and Other Ownership Interests: Johnson & Johnson, GE Healthcare, Varian Medical Systems
Consulting or Advisory Role: Ymabs Therapeutics Inc
Research Funding: Bayer (Inst), Ignyta (Inst), Roche (Inst), Lilly (Inst), Merck (Inst), Oncternal Therapeutics, Inc (Inst), AbbVie (Inst), Jubilant DraxImage (Inst), Actuate Therapeutics (Inst)
Patents, Royalties, Other Intellectual Property: Patent for non-invasive methods of leukemia cell detection with MRI/MRS—Patent No. 8894975
Luke D. Maese
Honoraria: Jazz Pharmaceuticals
Consulting or Advisory Role: Jazz Pharmaceuticals
Speakers' Bureau: Jazz Pharmaceuticals
Amit J. Sabnis
Consulting or Advisory Role: Cardinal Health
Andrew D. Cherniack
Employment: LabCorp
Stock and Other Ownership Interests: Merck
Research Funding: Bayer
Steven G. DuBois
Consulting or Advisory Role: Bayer, Amgen, Jazz Pharmaceuticals
Research Funding: Merck (Inst), Roche/Genentech (Inst), Lilly (Inst), Curis (Inst), Loxo (Inst), BMS (Inst), Eisai (Inst), Pfizer (Inst), Turning Point Therapeutics (Inst), Bayer (Inst), Salarius Pharmaceuticals (Inst)
Travel, Accommodations, Expenses: Roche/Genentech, Salarius Pharmaceuticals
(OPTIONAL) Uncompensated Relationships: Ymabs Therapeutics Inc
Natalie B. Collins
Open Payments Link: https://openpaymentsdata.cms.gov/physician/977647
Bruce E. Johnson
Consulting or Advisory Role: Novartis, Foundation Medicine, Hengrui Therapeutics, Daiichi Sankyo, Lilly, Checkpoint Therapeutics, G1 Therapeutics, Jazz Pharmaceuticals, GlaxoSmithKline, Boston Pharmaceuticals, Janssen Scientific Affairs, Genentech
Research Funding: Novartis (Inst), Novartis
Patents, Royalties, Other Intellectual Property: Dana-Farber Cancer Institute
Katherine A. Janeway
This author is an Associate Editor for JCO Precision Oncology. Journal policy recused the author from having any role in the peer review of this manuscript.
Honoraria: Foundation Medicine, Takeda
Consulting or Advisory Role: Bayer, Ipsen, Ipsen
Travel, Accommodations, Expenses: Bayer
No other potential conflicts of interest were reported.
SUPPORT
Dana-Farber/Harvard Cancer Center is supported in part by an NCI Cancer Center Support Grant #NIH 5 P30 CA06516. Support for this study was provided by the Lamb Family Fund (K.A.J.), the Precision for Kids Pan Mass Challenge Team (K.A.J.), the 4C's Fund (K.A.J.), and NIH Career Development Award 1K08CA218691.
PRIOR PRESENTATION
Presented at the AACR Annual Meeting 2022, New Orleans, LA, April 13, 2022.
K.A.J. and S.J.F. contributed equally to this work.
DATA SHARING STATEMENT
Individual participant data that underlie the results reported in this article, after deidentification, are available in the article and in its online supplementary material.
AUTHOR CONTRIBUTIONS
Conception and design: Lorena Lazo De La Vega, Alanna J. Church, Neal I. Lindeman, Katherine A. Janeway, Suzanne J. Forrest
Financial support: Neal I. Lindeman, Bruce E. Johnson, Katherine A. Janeway
Administrative support: Abigail Ward, Neal I. Lindeman, Katherine A. Janeway
Provision of study materials or patients: AeRang Kim, Navin R. Pinto, Margaret E. Macy, Amit J. Sabnis, Megan E. Anderson, Steven G. DuBois, Bruce E. Johnson, Katherine A. Janeway
Collection and assembly of data: Lorena Lazo De La Vega, Hannah Comeau, Sarah Sallan, Harrison K Tsai, Wenjun Kang, Abigail Ward, Alanna J. Church, AeRang Kim, Margaret E. Macy, Neal I. Lindeman, Steven G. DuBois, Natalie B. Collins, Katherine A. Janeway, Suzanne J. Forrest
Data analysis and interpretation: Lorena Lazo De La Vega, Alyaa Al-Ibraheemi, Hersh Gupta, Yvonne Y. Li, Harrison K Tsai, Alanna J. Church, Navin R. Pinto, Margaret E. Macy, Luke D. Maese, Amit J. Sabnis, Andrew D. Cherniack, Neal I. Lindeman, Megan E. Anderson, Tabitha M. Cooney, Kee Kiat Yeo, Gregory H. Reaman, Steven G. DuBois, Bruce E. Johnson, Katherine A. Janeway, Suzanne J. Forrest
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Harrison K. Tsai
Consulting or Advisory Role: Vertex
Alanna J. Church
Honoraria: The Jackson Laboratory
Consulting or Advisory Role: AlphaSights, The Jackson Laboratory, Bayer
Travel, Accommodations, Expenses: Bayer
Navin R. Pinto
Patents, Royalties, Other Intellectual Property: I hold a patent 20210346431 that has been licensed to Umoja Biotherapeutics and may result in royalty payments. No payments have been received to date
(OPTIONAL) Uncompensated Relationships: Y-mAbs Therapeutics
Margaret E. Macy
Stock and Other Ownership Interests: Johnson & Johnson, GE Healthcare, Varian Medical Systems
Consulting or Advisory Role: Ymabs Therapeutics Inc
Research Funding: Bayer (Inst), Ignyta (Inst), Roche (Inst), Lilly (Inst), Merck (Inst), Oncternal Therapeutics, Inc (Inst), AbbVie (Inst), Jubilant DraxImage (Inst), Actuate Therapeutics (Inst)
Patents, Royalties, Other Intellectual Property: Patent for non-invasive methods of leukemia cell detection with MRI/MRS—Patent No. 8894975
Luke D. Maese
Honoraria: Jazz Pharmaceuticals
Consulting or Advisory Role: Jazz Pharmaceuticals
Speakers' Bureau: Jazz Pharmaceuticals
Amit J. Sabnis
Consulting or Advisory Role: Cardinal Health
Andrew D. Cherniack
Employment: LabCorp
Stock and Other Ownership Interests: Merck
Research Funding: Bayer
Steven G. DuBois
Consulting or Advisory Role: Bayer, Amgen, Jazz Pharmaceuticals
Research Funding: Merck (Inst), Roche/Genentech (Inst), Lilly (Inst), Curis (Inst), Loxo (Inst), BMS (Inst), Eisai (Inst), Pfizer (Inst), Turning Point Therapeutics (Inst), Bayer (Inst), Salarius Pharmaceuticals (Inst)
Travel, Accommodations, Expenses: Roche/Genentech, Salarius Pharmaceuticals
(OPTIONAL) Uncompensated Relationships: Ymabs Therapeutics Inc
Natalie B. Collins
Open Payments Link: https://openpaymentsdata.cms.gov/physician/977647
Bruce E. Johnson
Consulting or Advisory Role: Novartis, Foundation Medicine, Hengrui Therapeutics, Daiichi Sankyo, Lilly, Checkpoint Therapeutics, G1 Therapeutics, Jazz Pharmaceuticals, GlaxoSmithKline, Boston Pharmaceuticals, Janssen Scientific Affairs, Genentech
Research Funding: Novartis (Inst), Novartis
Patents, Royalties, Other Intellectual Property: Dana-Farber Cancer Institute
Katherine A. Janeway
This author is an Associate Editor for JCO Precision Oncology. Journal policy recused the author from having any role in the peer review of this manuscript.
Honoraria: Foundation Medicine, Takeda
Consulting or Advisory Role: Bayer, Ipsen, Ipsen
Travel, Accommodations, Expenses: Bayer
No other potential conflicts of interest were reported.
REFERENCES
- 1.Turner N, Pearson A, Sharpe R, et al. : FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res 70:2085-2094, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freier K, Schwaenen C, Sticht C, et al. : Recurrent FGFR1 amplification and high FGFR1 protein expression in oral squamous cell carcinoma (OSCC). Oral Oncol 43:60-66, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Weiss J, Sos ML, Seidel D, et al. : Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med 2:62ra93, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishizuka T, Tanabe C, Sakamoto H, et al. : Gene amplification profiling of esophageal squamous cell carcinomas by DNA array CGH. Biochem Biophys Res Commun 296:152-155, 2002 [DOI] [PubMed] [Google Scholar]
- 5.Arai Y, Totoki Y, Hosoda F, et al. : Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology 59:1427-1434, 2014 [DOI] [PubMed] [Google Scholar]
- 6.Wu YM, Su F, Kalyana-Sundaram S, et al. : Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 3:636-647, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ornitz DM, Itoh N: The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol 4:215-266, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner N, Grose R: Fibroblast growth factor signalling: From development to cancer. Nat Rev Cancer 10:116-129, 2010 [DOI] [PubMed] [Google Scholar]
- 9.Friesel RE, Maciag T: Molecular mechanisms of angiogenesis: Fibroblast growth factor signal transduction. FASEB J 9:919-925, 1995 [DOI] [PubMed] [Google Scholar]
- 10.Taylor JG VI, Cheuk AT, Tsang PS, et al. : Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J Clin Invest 119:3395-3407, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shern JF, Selfe J, Izquierdo E, et al. : Genomic classification and clinical outcome in rhabdomyosarcoma: A report from an international consortium. J Clin Oncol 39:2859-2871, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryall S, Tabori U, Hawkins C: Pediatric low-grade glioma in the era of molecular diagnostics. Acta Neuropathol Commun 8:30, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Wu G, Miller CP, et al. : Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet 45:602-612, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Facchinetti F, Hollebecque A, Bahleda R, et al. : Facts and new hopes on selective FGFR inhibitors in solid tumors. Clin Cancer Res 26:764-774, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krook MA, Reeser JW, Ernst G, et al. : Fibroblast growth factor receptors in cancer: Genetic alterations, diagnostics, therapeutic targets and mechanisms of resistance. Br J Cancer 124:880-892, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babina IS, Turner NC: Advances and challenges in targeting FGFR signalling in cancer. Nat Rev Cancer 17:318-332, 2017 [DOI] [PubMed] [Google Scholar]
- 17.Meric-Bernstam F, Bahleda R, Hierro C, et al. : Futibatinib, an irreversible FGFR1-4 inhibitor, in patients with advanced solid tumors harboring FGF/FGFR aberrations: A phase I dose-expansion study. Cancer Discov 12:402-415, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goyal L, Shi L, Liu LY, et al. : TAS-120 overcomes resistance to ATP-competitive FGFR inhibitors in patients with FGFR2 fusion-positive intrahepatic cholangiocarcinoma. Cancer Discov 9:1064-1079, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zettler ME: The RACE for children act at one year: Progress in pediatric development of molecularly targeted oncology drugs. Expert Rev Anticancer Ther 22:317-321, 2022 [DOI] [PubMed] [Google Scholar]
- 20.Chmielecki J, Bailey M, He J, et al. : Genomic profiling of a large set of diverse pediatric cancers identifies known and novel mutations across tumor spectra. Cancer Res 77:509-519, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Church AJ, Corson LB, Kao PC, et al. : Molecular profiling identifies targeted therapy opportunities in pediatric solid cancer. Nat Med 28:1581-1589, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abo RP, Ducar M, Garcia EP, et al. : BreaKmer: Detection of structural variation in targeted massively parallel sequencing data using kmers. Nucleic Acids Res 43:e19, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia EP, Minkovsky A, Jia Y, et al. : Validation of OncoPanel: A targeted next-generation sequencing assay for the detection of somatic variants in cancer. Arch Pathol Lab Med 141:751-758, 2017 [DOI] [PubMed] [Google Scholar]
- 24.Nowak JA, Yurgelun MB, Bruce JL, et al. : Detection of mismatch repair deficiency and microsatellite instability in colorectal adenocarcinoma by targeted next-generation sequencing. J Mol Diagn 19:84-91, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cleary JM, Raghavan S, Wu Q, et al. : FGFR2 extracellular domain in-frame deletions are therapeutically targetable genomic alterations that function as oncogenic drivers in cholangiocarcinoma. Cancer Discov 11:2488-2505, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shukla N, Ameur N, Yilmaz I, et al. : Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways. Clin Cancer Res 18:748-757, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huse JT, Snuderl M, Jones DTW, et al. : Polymorphous low-grade neuroepithelial tumor of the young (PLNTY): An epileptogenic neoplasm with oligodendroglioma-like components, aberrant CD34 expression, and genetic alterations involving the MAP kinase pathway. Acta Neuropathol 133:417-429, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsui J, Yamamoto Y, Funahashi Y, et al. : E7080, a novel inhibitor that targets multiple kinases, has potent antitumor activities against stem cell factor producing human small cell lung cancer H146, based on angiogenesis inhibition. Int J Cancer 122:664-671, 2008 [DOI] [PubMed] [Google Scholar]
- 29.Farouk Sait S, Gilheeney SW, Bale TA, et al. : Debio1347, an oral FGFR inhibitor: Results from a single-center study in pediatric patients with recurrent or refractory FGFR-altered gliomas. JCO Precis Oncol 5:876-883, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan L, Wang J, Tanizaki J, et al. : Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors. Proc Natl Acad Sci USA 111:E4869-E4877, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bahleda R, Italiano A, Hierro C, et al. : Multicenter phase I study of erdafitinib (JNJ-42756493), oral pan-fibroblast growth factor receptor inhibitor, in patients with advanced or refractory solid tumors. Clin Cancer Res 25:4888-4897, 2019 [DOI] [PubMed] [Google Scholar]
- 32.Nogova L, Sequist LV, Perez Garcia JM, et al. : Evaluation of BGJ398, a fibroblast growth factor receptor 1-3 kinase inhibitor, in patients with advanced solid tumors harboring genetic alterations in fibroblast growth factor receptors: Results of a global phase I, dose-escalation and dose-expansion study. J Clin Oncol 35:157-165, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearson A, Smyth E, Babina IS, et al. : High-level clonal FGFR amplification and response to FGFR inhibition in a translational clinical trial. Cancer Discov 6:838-851, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Individual participant data that underlie the results reported in this article, after deidentification, are available in the article and in its online supplementary material.