PURPOSE
Understanding the differences in biomarker prevalence that may exist among diverse populations is invaluable to accurately forecast biomarker-driven clinical trial enrollment metrics and to advance inclusive research and health equity. This study evaluated the frequency and types of PIK3CA mutations (PIK3CAmut) detected in predicted genetic ancestry subgroups across breast cancer (BC) subtypes.
METHODS
Analyses were conducted using real-world genomic data from adult patients with BC treated in an academic or community setting in the United States and whose tumor tissue was submitted for comprehensive genomic profiling.
RESULTS
Of 36,151 patients with BC (median age, 58 years; 99% female), the breakdown by predicted genetic ancestry was 75% European, 14% African, 6% Central/South American, 3% East Asian, and 1% South Asian. We demonstrated that patients of African ancestry are less likely to have tumors that harbor PIK3CAmut compared with patients of European ancestry with estrogen receptor–positive/human epidermal growth factor receptor 2–negative (ER+/HER2–) BC (37% [949/2,593] v 44% [7,706/17,637]; q = 4.39E-11) and triple-negative breast cancer (8% [179/2,199] v 14% [991/7,072]; q = 6.07E-13). Moreover, we found that PIK3CAmut were predominantly composed of hotspot mutations, of which mutations at H1047 were the most prevalent across BC subtypes (35%-41% ER+/HER2– BC; 43%-61% HER2+ BC; 40%-59% triple-negative breast cancer).
CONCLUSION
This analysis established that tumor PIK3CAmut prevalence can differ among predicted genetic ancestries across BC subtypes on the basis of the largest comprehensive genomic profiling data set of patients with cancer treated in the United States. This study highlights the need for equitable representation in research studies, which is imperative to ensuring better health outcomes for all.
INTRODUCTION
Precision medicine clinical trials are increasingly common in drug development wherein the implementation of rational biomarker strategies to identify patients who are hypothesized as most likely to respond to treatment is a crucial component of these trials.1 This requires accurate estimations of the target biomarker prevalence in the indication(s) of interest. Such information affects study operations, including the projection of screen fail rates to inform the total number of potential study participants needed to ensure the complete enrollment of a clinical trial in a timely manner. Estimation of biomarker prevalence is primarily on the basis of publicly available and/or proprietary genomics databases. To date, the majority of genomics databases are largely composed of sequencing data from individuals of Western European descent.2 This lack of diversity translates into biomarker-driven patient selection strategies being designed without a clear understanding of whether these biomarkers may differ across diverse populations. This can lead to errors in study enrollment projections and, ultimately, may hinder the advancement of precision medicine for historically under-represented populations.
CONTEXT
Key Objective
As both biomarker-driven precision medicine trials and calls for diversity in clinical trials become increasingly common, accurate assessment of biomarker prevalence is critical for informing study enrollment metrics. In this study, we investigated the variation in the frequency and spectrum of PIK3CA mutations in breast cancer (BC) across predicted genetic ancestry subgroups.
Knowledge Generated
Patients of African ancestry are less likely to have tumors that harbor PIK3CA mutations compared with patients of European ancestry with estrogen receptor–positive/human epidermal growth factor receptor 2–negative BC and triple-negative BC. However, across predicted genetic ancestry groups, the most frequently observed PIK3CA mutations were generally similar and most, but not all, are able to be identified using commercially available polymerase chain reaction–based assays.
Relevance
This study highlights the need to systematically assess biomarker prevalence in historically under-represented populations to increase confidence in the generalizability and translatability of clinical trial outcomes to the population at large.
Although sampling biases are a topic of general concern when assessing biomarker prevalence, our analysis focuses on PIK3CA mutations (PIK3CAmut) as the biomarker of interest. The PIK3CA gene encodes for the p110α protein, the catalytic subunit of the phosphatidylinositol 3-kinase (PI3K) complex. p110α is a crucial cell-signaling component and is among the most frequently mutated genes in many solid tumor types, including endometrial cancer (approximately 53%), breast cancer (BC; approximately 36%), cervical cancer (approximately 26%), and head and neck squamous cell carcinoma (approximately 26%).3-6 Within BC, PIK3CA is mutated in approximately 40% of hormone receptor-positive/human epidermal growth factor receptor 2–negative (HR+/HER2−) BC, approximately 30% of HER2+ BC, and approximately 15% of triple-negative breast cancers (TNBCs). Furthermore, p110α is necessary for proper embryologic morphogenesis7,8 and PIK3CAmut drive noncancerous overgrowth syndromes including CLOVES (congenital lipomatous [fatty] overgrowth, vascular malformations, epidermal nevi and scoliosis/skeletal/spinal anomalies), PROS (PIK3CA-related overgrowth spectrum), and other vascular malformations.9 Mutations in PIK3CA lead to activation of the PI3K signaling pathway, a linchpin in the regulation of cell growth, proliferation, and survival, and its dysregulation has been characterized across a number of tumor types.10-12 Preclinical studies have demonstrated that hyperactivation of the PI3K signaling pathway is a resistance mechanism to endocrine therapy,13 and a possible resistance mechanism to HER2-targeted therapies14-17 and chemotherapy.18,19 To date, alpelisib (BYL719) is the only US Food and Drug Administration (FDA)–approved p110α inhibitor for use with fulvestrant in patients with HR+/HER2– PIK3CAmut advanced or metastatic BC.20 Both alpelisib and another p110α-targeting inhibitor, inavolisib (GDC-0077),21 are being evaluated in biomarker-driven phase III clinical trials in HR+/HER2– BC (ClinicalTrials.gov identifiers: NCT03439046, NCT05038735, NCT04191499), HER2+ BC (ClinicalTrials.gov identifiers: NCT04208178, NCT05063786), and TNBC (ClinicalTrials.gov identifier: NCT04251533). It is important to understand whether differences in biomarker prevalence exist across racial/ethnic populations and/or geographical regions to accurately forecast biomarker-driven clinical trial enrollment metrics, and to strive toward achieving diverse enrollment in clinical trials.
In this study, we used one of the largest real-world databases of patient tumors profiled with comprehensive genomic profiling (CGP) to evaluate whether the prevalence of PIK3CAmut in the major BC subtypes differs across predicted genetic ancestries. We subsequently delved into the types of PIK3CAmut detected and assessed how they may reflect underlying diversity among BC subtypes and predicted genetic ancestries. Finally, we contextualized our findings in terms of the impact to biomarker screening for clinical trials.
METHODS
CGP of Patient BC Tissue Specimens
CGP of 39,572 formalin-fixed, paraffin-embedded breast tumor samples from patients with primary advanced or metastatic disease based in the United States was performed from 2013 to 2021 using the FoundationOne or FoundationOneCDx assay in a Clinical Laboratory Improvement Amendments–certified, College of American Pathologists (CAP)-accredited laboratory (Foundation Medicine Inc, Cambridge, MA). All sequenced samples featured ≥ 20% tumor content and yielded ≥ 50 ng extracted DNA. CGP was performed on hybrid-capture, adapter ligation–based libraries, to identify genomic alterations for ≥ 324 genes, as previously described.22,23 Sites of care included academic and community settings. Approval for this study, including waiver of informed consent and Health Insurance Portability and Accountability Act (HIPAA) waiver of authorization, was obtained from the Western IRB (Protocol No. 20152817).
Classification of PIK3CAmut Tissue Specimens
PIK3CAmut specimens were defined as those with ≥ 1 single-nucleotide variant in the PIK3CA gene that is predicted to be pathogenic, defined as known or likely oncogenic significance. Statistical testing used two-sided Fisher's exact tests unless otherwise noted. Statistical significance is defined as Benjamini-Hochberg–adjusted P value (q-value) < .05.
Estimation of Patient-Level Genetic Ancestry
Patient genetic ancestry was inferred using > 40,000 germline single-nucleotide polymorphisms that were covered as part of the CGP, as previously described.24 Patient-level self-reported ancestry and/or ethnicity data were unavailable. Individuals were classified into inferred population groups using a random forest classifier trained on phase III 1000 Genomes samples, wherein principal component analysis was run on alternate allele counts and a model was trained using the top 10 principal component analysis features. The classifier was then applied to the CGP patient samples. To quantitate the fraction of ancestral mixture in each patient, ADMIXTURE was run on the 1000 Genomes samples to define five population signatures, and then ADMIXTURE was applied to the CGP samples in projection mode using these five signatures.25,26
Estrogen Receptor Status for Tumor Tissue Specimens
Specimen estrogen receptor (ER) status (ER-positive or ER-negative) was derived from ER immunohistochemistry per local assessment in pathology reports provided to Foundation Medicine, Inc. It is possible that institutes did not score ER status according to ASCO/CAP guidelines, defined as ≥ 1% of tumor cells staining positive for ER. If ER status was unavailable, it was computationally imputed using a machine learning approach (Data Supplement).
Software
Statistics, computation, and plotting were carried out using R v3.6.1.
RESULTS
Demographics of the Study Cohort
The overall cohort consisted of samples from 39,572 individuals with BC. These samples were stratified by predicted ancestry and BC subtype, resulting in 36,151 samples that were further analyzed herein: 22,408 ER+/HER2– BC samples, 10,430 TNBC samples, and 3,313 HER2+ BC samples (Fig 1). A total of 3,421 samples were excluded from the analysis because of either low confidence of the ancestry classification for individuals of predicted Central/South American (AMR) ancestry with AMR fraction < 0.2 (n = 1,545) or indeterminate ER status for HER2– BC samples (n = 1,876). BC subtype was based on ER and HER2 status; progesterone receptor (PR) status was unavailable. As HER2-positivity is defined in this analysis by the presence of ERBB2 amplification, it may not encompass all HER2+ samples as defined per ASCO/CAP guidelines.27 Because the frequency of ER–/PR+ BC is rare,28,29 a small number of these samples may be misclassified as TNBC.
FIG 1.
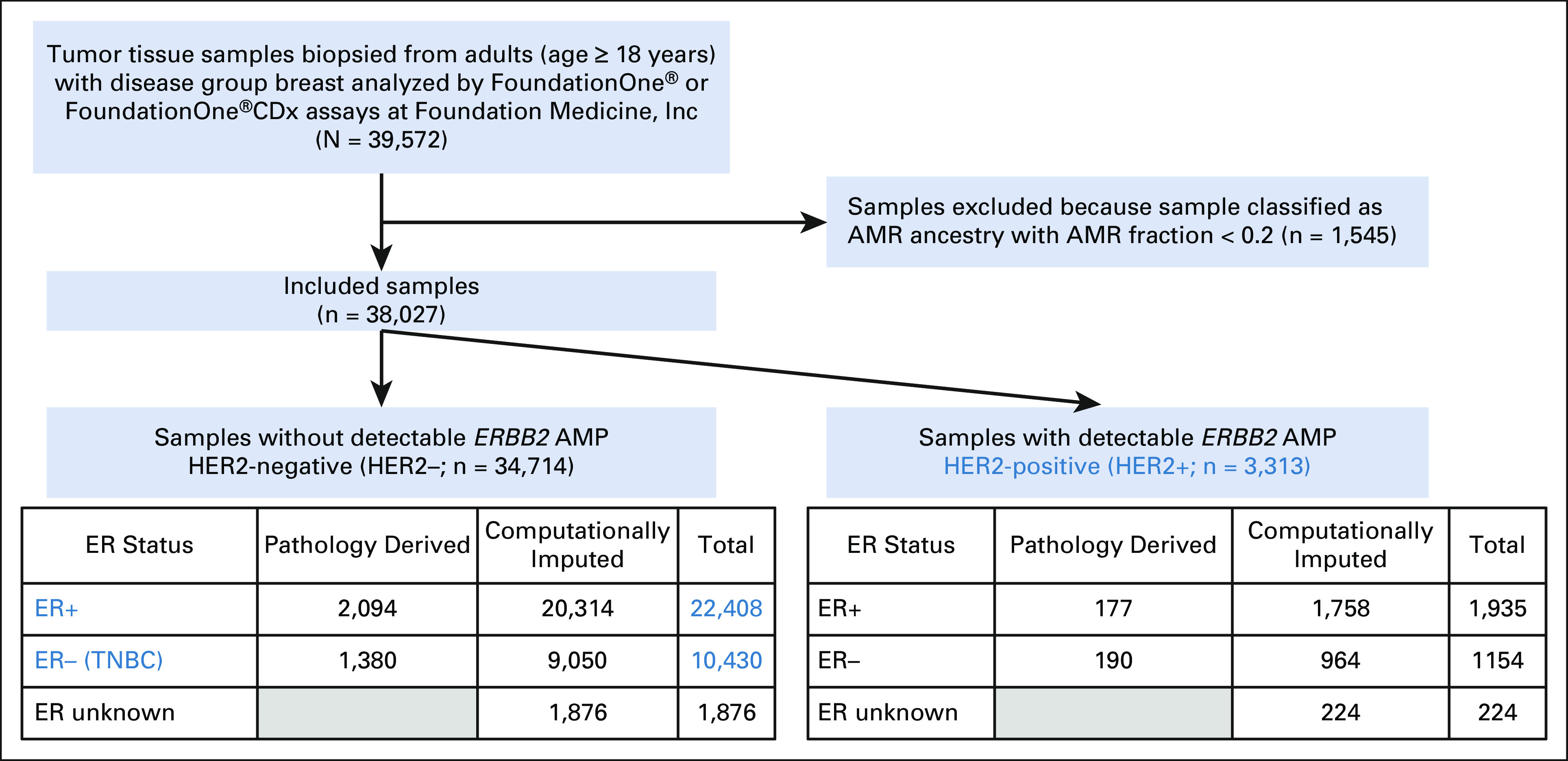
Flowchart of the analysis population. Blue-colored text denotes the breast cancer subtypes that are included in the analysis population. Details on the computational imputation of ER status are described in the Data Supplement. AFR, African; AMP, amplification; AMR, Central/South American; CDx, companion diagnostic; EAS, East Asian; ER, estrogen receptor; ERBB2, erb-b2 receptor tyrosine kinase 2; EUR, European; HER2, human epidermal growth factor receptor 2; n, sample size; SAS, South Asian; TNBC, triple-negative breast cancer.
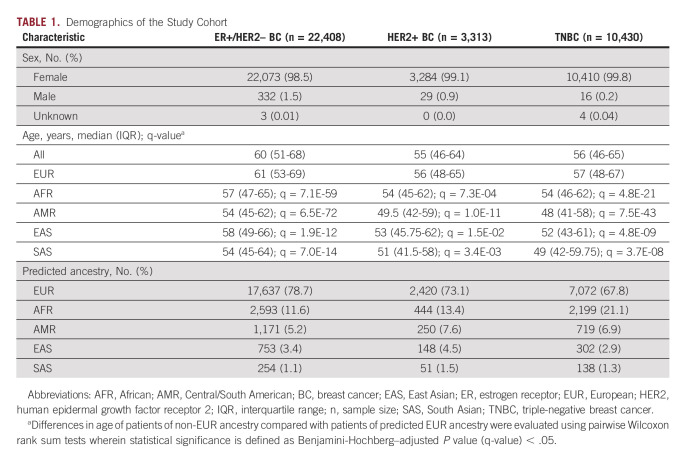
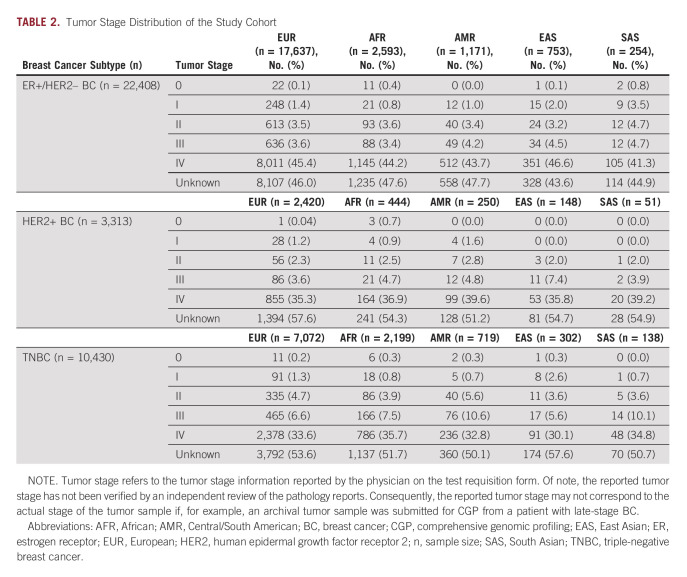
The patients were predominantly female (approximately 99%) with a median age at the time of biopsy of 60 years for ER+/HER2– BC, 55 years for HER2+ BC, and 56 years for TNBC. TNBC is not hormonally driven and is more frequently observed in a younger demographic group30 compared with HR+ BC, a disease largely diagnosed in postmenopausal women.31 We postulate the median age of the study cohort is younger than the reported median age of BC diagnosis32 because of a bias in the data wherein younger patients diagnosed with BC, who as a group tend to have worse outcomes,33,34 are more likely to have their tumors genomically profiled to guide treatment decisions. Across BC subtypes, patients of European (EUR) ancestry were significantly older than patients of other ancestries (Table 1, Data Supplement). Patients were predominantly of EUR ancestry (79% in ER+/HER2– BC; 73% in HER2+ BC; 68% in TNBC), followed by African (AFR) ancestry (12% in ER+/HER2– BC; 13% in HER2+ BC; 21% in TNBC); patients of South Asian (SAS) ancestry comprised the lowest percentage (1% in ER+/HER2– BC; 2% in HER2+ BC; 1% in TNBC; Table 1). The samples were largely from late-stage tumors per analysis of tumor stage and site of tumor biopsy (Table 2) and obtained from a mix of local tumor sites and metastatic lesions (Data Supplement). No other notable demographic differences were observed in the analysis population.
TABLE 1.
Demographics of the Study Cohort
TABLE 2.
Tumor Stage Distribution of the Study Cohort
PIK3CAmut Prevalence Across Predicted Ancestries
We next evaluated the prevalence of PIK3CAmut across the predicted ancestries within each BC subtype. As this study aimed to evaluate the impact of biomarker prevalence data on precision medicine clinical trial design, analysis of PIK3CAmut was limited to pathogenic single-nucleotide variants. This biomarker definition broadly encompasses the clinically relevant PIK3CAmut included in the biomarker eligibility criteria for clinical trials investigating PI3K inhibitors.20,35 PIK3CA indels, amplifications, and rearrangement events are incompletely characterized.
In ER+/HER2– BC, PIK3CAmut were identified in 44% (n = 7,706/17,637) of patients of EUR ancestry, 37% (n = 949/2,593) of patients of AFR ancestry, 40% (n = 472/1,171) of patients of AMR ancestry, 43% (n = 323/753) of patients of East Asian (EAS) ancestry, and 48% (n = 123/254) of patients of SAS ancestry. PIK3CAmut were detected significantly less frequently in patients of AFR (q = 4.39E-11) and AMR (q = 0.0417) ancestries compared with patients of EUR ancestry. No difference in PIK3CAmut prevalence was observed in patients of EAS (q = 0.680) and SAS (q = 0.171) ancestries compared with patients of EUR ancestry (Fig 2A).
FIG 2.
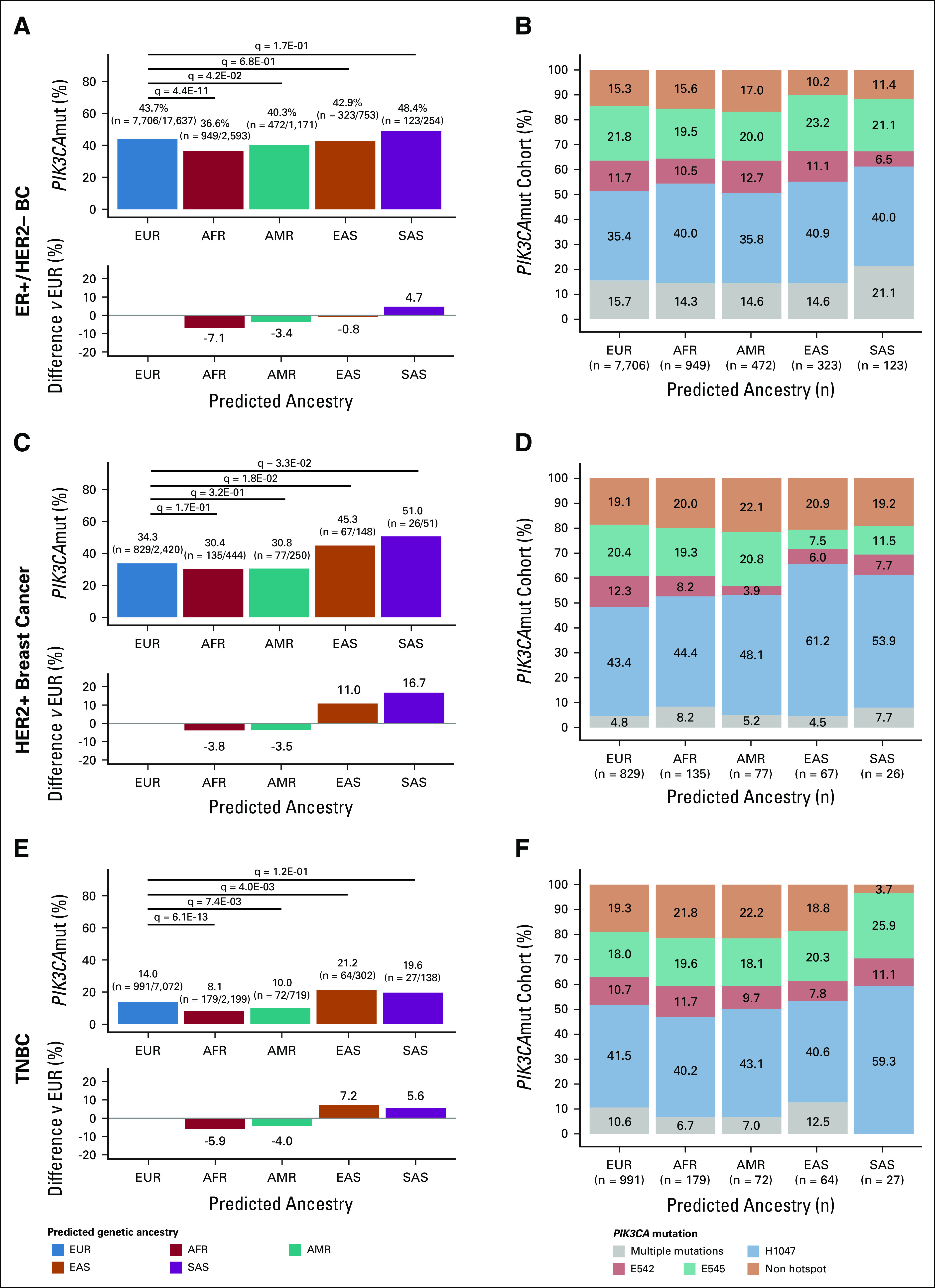
PIK3CAmut landscape across BC subtypes by predicted ancestries: (A) PIK3CAmut prevalence and (B) composition of PIK3CAmut tumors of ER+/HER2– BC; (C) PIK3CAmut prevalence and (D) composition of PIK3CAmut tumors of HER2+ BC; and (E) PIK3CAmut prevalence and (F) composition of PIK3CAmut tumors of TNBC. (A, C, E) Bar plots illustrate PIK3CAmut frequency within each predicted ancestry subgroup (top) and the difference in PIK3CAmut prevalence relative to patients of EUR ancestry (bottom). Differences in PIK3CAmut prevalence were evaluated using Fisher's exact test wherein statistical significance is defined as Benjamini-Hochberg–adjusted P value (q-value) < .05. (B, D, F) Percent stacked bar charts illustrate the relative frequency of the types of PIK3CAmut among PIK3CAmut samples within each predicted ancestry subgroup. Samples that harbor PIK3CAmut were classified as harboring either a single hotspot mutation (ie, occurring at H1047, E542, or E545), a single nonhotspot mutation, or multiple mutations (ie, tumor harbors ≥ 2 PIK3CAmut). BC, breast cancer; ER, estrogen receptor; TNBC, triple-negative breast cancer.
In HER2+ BC, PIK3CAmut were identified in 34% (n = 829/2,420) of patients of EUR ancestry, 30% (n = 135/444) of patients of AFR ancestry, 31% (n = 77/250) of patients of AMR ancestry, 45% (n = 67/148) of patients of EAS ancestry, and 51% (n = 26/51) of patients of SAS ancestry. PIK3CAmut were detected significantly more frequently in patients of EAS (q = 0.0182) and SAS (q = 0.0334) ancestries compared with patients of EUR ancestry. No difference in PIK3CAmut prevalence was observed in patients of AFR (q = 0.168) and AMR (q = 0.320) ancestries compared with patients of EUR ancestry (Fig 2C).
In TNBC, PIK3CAmut were identified in 14% (n = 991/7,702) of patients of EUR ancestry, 8% (n = 179/2,199) of patients of AFR ancestry, 10% (n = 72/719) of patients of AMR ancestry, 21% (n = 64/302) of patients of EAS ancestry, and 20% (n = 27/138) of patients of SAS ancestry. Compared with the patients of EUR ancestry, PIK3CAmut were detected significantly less frequently in patients of AFR (q = 6.07E-13) and AMR (q = 7.50E-03) ancestries, and more frequently in patients of EAS (q = 4.00E-03) ancestry. No difference in PIK3CAmut prevalence was observed in patients of SAS (q = 0.124) ancestry compared with patients of EUR ancestry (Fig 2E).
Our analysis demonstrates that tumor PIK3CAmut prevalence may differ among predicted ancestries across all BC subtypes evaluated. We observed similar findings when the analysis was limited to samples with pathology report–derived ER status. Specifically, PIK3CAmut were detected significantly less frequently in patients of AFR ancestry compared with patients of EUR ancestry in ER+/HER2– BC (29% [n = 71/247] v 39% [n = 630/1,619]; q = 2.37E-03) and TNBC (10% [n = 23/238] v 22% [n = 222/996]; q = 5.01E-06). Interestingly, PIK3CAmut were detected more frequently in patients of SAS ancestry compared with patients of EUR ancestry in ER+/HER2– BC (68% [n = 17/25] v 39% [n = 630/1,619]; q = 6.03E-03). However, this finding is based on a small sample size of ER+/HER2– patients of SAS ancestry (Data Supplement).
Spectrum of PIK3CAmut in BC
We next evaluated the spectrum of PIK3CAmut detected. Activating mutations in PIK3CA occur largely in exons 9 and 20, which encode the helical and kinase domains, respectively, of p110α.12 Samples with PIK3CAmut were classified as harboring a single hotspot mutation (ie, occurring at H1047, E542, or E545), a single nonhotspot mutation (Data Supplement), or multiple mutations (ie, tumor harbors ≥ 2 PIK3CAmut; Data Supplement). Across all subgroups evaluated, hotspot PIK3CAmut were most predominant; mutations at H1047 (35%-41% ER+/HER2– BC; 43%-61% HER2+ BC; 40%-59% TNBC) were the most frequent and mutations at E542 (7%-13% ER+/HER2– BC; 4%-12% HER2+ BC; 8%-12% TNBC) were the least frequent (Figs 2B, 2D and 2F) among these hotspot mutations. Multiple PIK3CAmut were more frequently observed in ER+/HER2– BCs (14%-21%) compared with HER2+ BCs (4%-8%) and TNBCs (0%-13%) across all predicted ancestries. Within BC subtypes, no significant difference was observed in the proportional distribution of PIK3CAmut across predicted ancestries (q-value > 0.05; Figs 2B, 2D and 2F).
DISCUSSION
Our findings demonstrated that tumor PIK3CAmut prevalence differs among predicted ancestries in BC. We have shown that ER+/HER2– BC and TNBC tumors from patients of AFR and AMR ancestries are less likely to harbor PIK3CAmut compared with patients of EUR ancestry. Additionally, compared with patients of EUR ancestry, patients of EAS ancestry with HER2+ BC and TNBC, as well as patients of SAS ancestry with HER2+ BC, were more likely to have tumors that harbor PIK3CAmut (Figs 2A, 2C and 2E). The ancestry-specific tumor PIK3CAmut prevalences observed across BC subtypes is supported by the findings from studies that sampled substantially fewer participants in Africa,36-39 Latin America,40-44 China or Korea,45-52 and South Asian countries.53-57 Inconsistencies between PIK3CAmut prevalences identified in our study and those reported in the literature were observed when sample sizes were small and when differences existed in the clinical characteristics of the populations evaluated and/or assay methodologies. It is as yet unknown whether statistically significant differences in PIK3CAmut prevalence across the predicted ancestries translate into clinically meaningful differences in study enrollment metrics.
Furthermore, our analysis of the types of PIK3CAmut identified in BC revealed several similarities across all subgroups evaluated. Overall, within BC subtypes, the proportional distribution of PIK3CAmut did not differ among predicted ancestries (Figs 2B, 2D and 2F). The spectrum of PIK3CAmut was predominantly composed of hotspot mutations, of which mutations at H1047 were the most prevalent. These observations suggest that the overall types of PIK3CAmut that arise in BC tumors are not different between patients of different predicted ancestries. From a clinical trial perspective, these data suggest that no ancestry subgroup would be at an innate disadvantage toward meeting the biomarker definition for inclusion in a PI3K-inhibitor clinical trial. Single nonhotspot mutations comprised a small but sizable proportion of the identified mutations (10%-17% ER+/HER2– BC; 19%-22% HER2+ BC; 4%-22% TNBC; Figs 2B, 2D and 2F; Data Supplement). Although some nonhotspot mutations are included on commercial testing panels, the clinical utility of nonhotspot PIK3CAmut may vary depending on the drug and indication under investigation. Although resources58 exist to support clinical interpretation of somatic variants, studies evaluating their functional consequences may be lacking because of the rarity of some mutations. Therefore, it is unsurprising that commercial polymerase chain reaction–based PIK3CAmut assays were developed to detect PIK3CA hotspot mutations. Consequently, the most frequent PIK3CAmut detected across predicted ancestries can be identified in academic and community clinical settings. However, CGP offers the opportunity to identify a broader spectrum of mutations and more complex mutation patterns (eg, double PIK3CAmut), thus enabling the execution of clinical trials with compound or expanded biomarker inclusion criteria or even the opportunity for referral to other clinical trials on the basis of a single test result. Unfortunately, populations without access to high-quality health care may have limited or no access to CGP that would afford them greater opportunities to participate in such trials.
Finally, we hypothesize that the differences observed in the prevalence of tumors that harbor multiple PIK3CAmut across BC subtypes highlight the molecular diversity of BC. ER+/HER2– breast tumors are more likely to harbor multiple PIK3CAmut than other BC subtypes (14%-21% of ER+/HER2– BCs v 4%-8% of HER2+ BCs and 0%-13% of TNBCs; Figs 2B, 2D and 2F; Data Supplement). The prevalence of multiple PIK3CAmut across solid tumors is uncommon, occurring in < 1% of cancers.59 In preclinical studies, multiple PIK3CAmut in cis led to robust activation of the PI3K pathway, along with enhanced cell proliferation and tumor growth compared with single PIK3CAmut.60 The lower prevalence of multiple PIK3CAmut observed in HER2+ BCs and TNBCs relative to ER+/HER2– BCs may indicate decreased reliance on PI3K signaling mediated by PIK3CAmut, and dependence on alternative mechanisms to dysregulate PI3K signaling. For example, the HER2-PI3K axis is involved in regulation of cell proliferation, survival, and growth,61,62 and overexpressed HER2 (often because of amplified ERBB2) activates PI3K signaling.61,63,64 Likewise, genomic loss of PTEN, detected in 15% TNBCs,4 contributes to dysregulation of PI3K signaling.65,66 Nevertheless, preclinical studies are required to elucidate whether the breadth of genomic alterations capable of activating PI3K signaling are able to perturb downstream signaling effectors at comparable measures.
In summary, this study leverages one of the largest available genomic databases to enable detailed subanalyses of PIK3CAmut prevalence across predicted ancestries (ie, EUR, AFR, AMR, EAS, and SAS) and BC subtypes (ie, ER+/HER2–, HER2+, and TNBC); this information may inform study enrollment metrics for biomarker-based clinical trials and encourage similar analyses for biomarkers relevant to other cancer types. Nevertheless, our study has limitations. First, the data are confined to samples from patients in the United States, lack information about generational status of the patients (ie, length of familial residence in the United States), and represent a population with access to a level of health care that offered next-generation sequencing (NGS)-based genomic testing, all factors that may affect the generalizability of the findings. In the absence of publicly available somatic mutation data sets with a sizable number of patients from diverse backgrounds residing outside North America, it is challenging to know whether the genetic ancestry classifications are a suitable proxy for the world's diverse populations as historically stratified on the basis of geographical location in clinical trial analyses. Moreover, a subset of ER statuses was imputed (Data Supplement), with a classification accuracy of 83% (95% CI, 76 to 89) on an independent validation cohort (Data Supplement). While of consideration, similar findings were observed when the analysis was restricted to samples whose ER status was solely pathology report–derived (Data Supplement). Finally, the analysis may be underpowered because of the small sample size of some non-EUR ancestry cohorts.
Overall, this study sought to better understand potential differences in biomarker prevalence among diverse populations to improve the design of biomarker-driven clinical trials. Currently, enrollment forecasting for multicountry/multiregion clinical trials rarely account for variations in biomarker prevalence, which may directly affect study operations and lead to a lack of diversity in clinical trials. These issues are underscored by the > 400 FDA-approved drugs with biomarker(s) in their label.67 Such concerns may, in part, be mitigated by efforts such as the NIH Revitalization Act of 1993, the FDA's Drug Trials Snapshots Program, and the FDA's recent push for clinical trial sponsors to include diversity plans in clinical trial applications.68,69 We hope this study encourages the scientific community to improve efforts to collect specimens from diverse populations irrespective of race, ethnicity, and socioeconomic status, perform inclusivity analyses as part of routine demographics analyses, and consider incorporation of molecular diagnostic testing into routine care. Collectively, these efforts would increase representation of real-world patient populations in genomics databases, which may enable more equitable representation in research studies and advance personalized care for all patients with cancer.
ACKNOWLEDGMENT
The authors thank the patients and their families who contributed to this study, as well as the staff and investigators at the participating institutions. The authors would also like to thank Ching-Wei Chang for her guidance on the appropriate statistical analyses.
Jessica W. Chen
Employment: Genentech/Roche
Stock and Other Ownership Interests: Roche/Genentech
Karthikeyan Murugesan
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche Pharma AG
Patents, Royalties, Other Intellectual Property: Antibiotic resistance causation identification (US10629291B2) filed with Koninklijke Philips NV (Inst), Analytic prediction of antibiotic susceptibility (US20190279738A1) filed with Koninklijke Philips NV (Inst), Methods and devices for characterizing and treating combined hepatocellular cholangiocarcinoma, PCT/US2022/014148, filed with Foundation Medicine Inc (Inst), Methods of using somatic HLA-I loss of heterozygosity to predict response to immune checkpoint inhibitor-treated patients with lung cancer, PCT/US2022/073166, filed with Foundation Medicine Inc (Inst), Methodology for measuring the quality of phylogenetic and transmission trees and for merging trees (US20200357491A1) filed with Koninklijke Philips NV (Inst)
Justin Y. Newberg
Employment: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine
Patents, Royalties, Other Intellectual Property: I am on a patent stemming from research in my graduate school days related to identifying location biomarkers using imaging techniques. The patent is unrelated to the contents of this paper. https://patents.google.com/patent/US9092850B2/en?q=murphy&inventor=newberg&oq=newberg+murphy
Ethan S. Sokol
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Patents, Royalties, Other Intellectual Property: Submitted patent for HRD calling methodology (Inst)
Heidi M. Savage
Employment: Genentech/Roche
Stock and Other Ownership Interests: Bolt Biotherapeutics, Roche/Genentech
Thomas J. Stout
Employment: Genentech/Roche
Stock and Other Ownership Interests: Roche/Genentech
Sophia L. Maund
Employment: Genentech/Roche
Stock and Other Ownership Interests: Roche/Genentech
Katherine E. Hutchinson
Employment: Genentech/Roche
Stock and Other Ownership Interests: Roche/Genentech
No other potential conflicts of interest were reported.
SUPPORT
Supported by F. Hoffmann-La Roche Ltd, Basel, Switzerland (no grant number).
J.W.C. and K.M. contributed equally to this work.
DATA SHARING STATEMENT
Because of HIPAA requirements, Foundation Medicine, Inc (FMI) is not consented to share individualized patient genomic data, which contains potentially identifying or sensitive information. FMI is committed to collaborative data analysis and has well-established mechanisms by which investigators can query their core genomic database of > 400,000 deidentified sequenced cancer specimens to obtain aggregated data sets.
AUTHOR CONTRIBUTIONS
Conception and design: Jessica W. Chen, Karthikeyan Murugesan, Ethan S. Sokol, Katherine E. Hutchinson
Collection and assembly of data: Jessica W. Chen, Karthikeyan Murugesan, Ethan S. Sokol, Heidi M. Savage, Sophia L. Maund, Katherine E. Hutchinson
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Jessica W. Chen
Employment: Genentech/Roche
Stock and Other Ownership Interests: Roche/Genentech
Karthikeyan Murugesan
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche Pharma AG
Patents, Royalties, Other Intellectual Property: Antibiotic resistance causation identification (US10629291B2) filed with Koninklijke Philips NV (Inst), Analytic prediction of antibiotic susceptibility (US20190279738A1) filed with Koninklijke Philips NV (Inst), Methods and devices for characterizing and treating combined hepatocellular cholangiocarcinoma, PCT/US2022/014148, filed with Foundation Medicine Inc (Inst), Methods of using somatic HLA-I loss of heterozygosity to predict response to immune checkpoint inhibitor-treated patients with lung cancer, PCT/US2022/073166, filed with Foundation Medicine Inc (Inst), Methodology for measuring the quality of phylogenetic and transmission trees and for merging trees (US20200357491A1) filed with Koninklijke Philips NV (Inst)
Justin Y. Newberg
Employment: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine
Patents, Royalties, Other Intellectual Property: I am on a patent stemming from research in my graduate school days related to identifying location biomarkers using imaging techniques. The patent is unrelated to the contents of this paper. https://patents.google.com/patent/US9092850B2/en?q=murphy&inventor=newberg&oq=newberg+murphy
Ethan S. Sokol
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Patents, Royalties, Other Intellectual Property: Submitted patent for HRD calling methodology (Inst)
Heidi M. Savage
Employment: Genentech/Roche
Stock and Other Ownership Interests: Bolt Biotherapeutics, Roche/Genentech
Thomas J. Stout
Employment: Genentech/Roche
Stock and Other Ownership Interests: Roche/Genentech
Sophia L. Maund
Employment: Genentech/Roche
Stock and Other Ownership Interests: Roche/Genentech
Katherine E. Hutchinson
Employment: Genentech/Roche
Stock and Other Ownership Interests: Roche/Genentech
No other potential conflicts of interest were reported.
REFERENCES
- 1.Pritchard D, Wells CJ: Raising the bar: FDA accelerates the push toward personalized medicine. J Precis Med 5:36-39, 2018 [Google Scholar]
- 2.Popejoy AB, Fullerton SM: Genomics is failing on diversity. Nature 538:161-164, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Cancer Genome Atlas Research Network, Chen Z, Saller C, et al. : Integrated genomic and molecular characterization of cervical cancer. Nature 543:378-384, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The Cancer Genome Atlas Network, Fulton RS, McLellan MD, et al. : Comprehensive molecular portraits of human breast tumours. Nature 490:61-70, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Cancer Genome Atlas Network, Sougnez C, Lichtenstein L, et al. : Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517:576-582, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine DA: Integrated genomic characterization of endometrial carcinoma. Nature 497:67-73, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bi L, Okabe I, Bernard DJ, et al. : Proliferative defect and embryonic lethality in mice homozygous for a deletion in the p110α subunit of phosphoinositide 3-kinase. J Biol Chem 274:10963-10968, 1999 [DOI] [PubMed] [Google Scholar]
- 8.Hare LM, Schwarz Q, Wiszniak S, et al. : Heterozygous expression of the oncogenic Pik3caH1047R mutation during murine development results in fatal embryonic and extraembryonic defects. Dev Biol 404:14-26, 2015 [DOI] [PubMed] [Google Scholar]
- 9.Madsen RR, Vanhaesebroeck B, Semple RK: Cancer-associated PIK3CA mutations in overgrowth disorders. Trends Mol Med 24:856-870, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantrell DA: Phosphoinositide 3-kinase signalling pathways. J Cell Sci 114:1439-1445, 2001 [DOI] [PubMed] [Google Scholar]
- 11.Courtney KD, Corcoran RB, Engelman JA: The PI3K pathway as drug target in human cancer. J Clin Oncol 28:1075-1083, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samuels Y, Wang Z, Bardelli A, et al. : High frequency of mutations of the PIK3CA gene in human cancers. Science 304:554, 2004 [DOI] [PubMed] [Google Scholar]
- 13.Sabnis G, Goloubeva O, Jelovac D, et al. : Inhibition of the phosphatidylinositol 3-kinase/Akt pathway improves response of long-term estrogen-deprived breast cancer xenografts to antiestrogens. Clin Cancer Res 13:2751-2757, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Baselga J, Cortés J, Im S-A, et al. : Biomarker analyses in CLEOPATRA: A phase III, placebo-controlled study of pertuzumab in human epidermal growth factor receptor 2–positive, first-line metastatic breast cancer. J Clin Oncol 32:3753-3761, 2014 [DOI] [PubMed] [Google Scholar]
- 15.Hanker AB, Pfefferle AD, Balko JM, et al. : Mutant PIK3CA accelerates HER2-driven transgenic mammary tumors and induces resistance to combinations of anti-HER2 therapies. Proc Natl Acad Sci USA 110:14372-14377, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez EA, de Haas SL, Eiermann W, et al. : Relationship between tumor biomarkers and efficacy in MARIANNE, a phase III study of trastuzumab emtansine ± pertuzumab versus trastuzumab plus taxane in HER2-positive advanced breast cancer. BMC Cancer 19:517, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rexer BN, Chanthaphaychith S, Dahlman KB, et al. : Direct inhibition of PI3K in combination with dual HER2 inhibitors is required for optimal antitumor activity in HER2+ breast cancer cells. Breast Cancer Res 16:R9, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao C, Yuan X, Jiang Z, et al. : Regulation of AKT phosphorylation by GSK3β and PTEN to control chemoresistance in breast cancer. Breast Cancer Res Treat 176:291-301, 2019 [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Li Y, Wang Q, et al. : The PI3K subunits, P110α and P110β are potential targets for overcoming P-gp and BCRP-mediated MDR in cancer. Mol Cancer 1910:10, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.André F, Ciruelos E, Rubovszky G, et al. : Alpelisib for PIK3CA-mutated, hormone receptor–positive advanced breast cancer. N Engl J Med 380:1929-1940, 2019 [DOI] [PubMed] [Google Scholar]
- 21.Song KW, Edgar KA, Hanan EJ, et al. : RTK-dependent inducible degradation of mutant PI3Kα drives GDC-0077 (Inavolisib) efficacy. Cancer Discov 12:204-219, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frampton GM, Fichtenholtz A, Otto GA, et al. : Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 31:1023-1031, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milbury CA, Creeden J, Yip W-K, et al. : Clinical and analytical validation of FoundationOne®CDx, a comprehensive genomic profiling assay for solid tumors. PLoS One 17:e0264138, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Connelly CF, Carrot-Zhang J, Stephens PJ, et al. : Abstract 1227: Somatic genome alterations in cancer as compared to inferred patient ancestry. Epidemiology 78, 2018. (suppl 13; abstr 1227) [Google Scholar]
- 25.Alexander DH, Novembre J, Lange K: Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19:1655-1664, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newberg J, Connelly C, Frampton G: Determining patient ancestry based on targeted tumor comprehensive genomic profiling. Epidemiology 79, 2019. (suppl 13; abstr 1599) [Google Scholar]
- 27.Cho-Phan CD, Snider J, Zhang L, et al. : Concordance of HER2+ status by IHC/ISH and ERBB2 status by NGS in a real-world clinicogenomic database and analysis of outcomes in patients (pts) with metastatic breast cancer (mBC). J Clin Oncol 39, 2021. (suppl 15; abstr 1036) [Google Scholar]
- 28.Fan Y, Ding X, Xu B, et al. : Prognostic significance of single progesterone receptor positivity. Medicine (Baltimore) 94:e2066, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Yang D, Yin X, et al. : Clinicopathological characteristics and breast cancer–specific survival of patients with single hormone receptor–positive breast cancer. JAMA Netw Open 3:e1918160, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bauer KR, Brown M, Cress RD, et al. : Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype. Cancer 109:1721-1728, 2007 [DOI] [PubMed] [Google Scholar]
- 31.Goldschmidt D, Dalal AA, Romdhani H, et al. : Current treatment patterns among postmenopausal women with HR+/HER2− metastatic breast cancer in US community oncology practices: An observational study. Adv Ther 35:482-493, 2018 [DOI] [PubMed] [Google Scholar]
- 32.Howlader N, Noone AM, Krapcho M, et al. (eds): SEER Cancer Statistics Review, 1975-2018. Bethesda, MD, National Cancer Institute, 2021 [Google Scholar]
- 33.Lee HJ, Seo AN, Kim EJ, et al. : HER2 heterogeneity affects trastuzumab responses and survival in patients with HER2-positive metastatic breast cancer. Am J Clin Pathol 142:755-766, 2014 [DOI] [PubMed] [Google Scholar]
- 34.Maggard MA, O’Connell JB, Lane KE, et al. : Do young breast cancer patients have worse outcomes?. J Surg Res 113:109-113, 2003 [DOI] [PubMed] [Google Scholar]
- 35.Dent S, Cortés J, Im Y-H, et al. : Phase III randomized study of taselisib or placebo with fulvestrant in estrogen receptor-positive, PIK3CA-mutant, HER2-negative, advanced breast cancer: The SANDPIPER trial. Ann Oncol 32:197-207, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elwy F, Helwa R, El Leithy AA, et al. : PIK3CA mutations in HER2-positive breast cancer patients; frequency and clinicopathological perspective in Egyptian patients. Asian Pac J Cancer Prev 18:57-64, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jouali F, Marchoudi N, Talbi S, et al. : Detection of PIK3/AKT pathway in Moroccan population with triple negative breast cancer. BMC Cancer 18:900, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Omilian AR, Wei L, Hong C-C, et al. : Somatic mutations of triple-negative breast cancer: A comparison between Black and White women. Breast Cancer Res Treat 182:503-509, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pitt JJ, Riester M, Zheng Y, et al. : Characterization of Nigerian breast cancer reveals prevalent homologous recombination deficiency and aggressive molecular features. Nat Commun 9:4181, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castaneda CA, Lopez-Ilasaca M, Pinto JA, et al. : PIK3CA mutations in Peruvian patients with HER2-amplified and triple negative non-metastatic breast cancers. Hematol Oncol Stem Cell Ther 7:142-148, 2014 [DOI] [PubMed] [Google Scholar]
- 41.Olivier M, Bouaoun L, Villar S, et al. : Molecular features of premenopausal breast cancers in Latin American women: Pilot results from the PRECAMA study. PLoS One 14:e0210372, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Philipovskiy A, Dwivedi AK, Gamez R, et al. : Association between tumor mutation profile and clinical outcomes among Hispanic Latina women with triple-negative breast cancer. PLoS One 15:e0238262, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rojas-Jiménez E, Mejía-Gómez JC, Díaz-Velásquez C, et al. : Comprehensive genomic profile of heterogeneous long follow-up triple-negative breast cancer and its clinical characteristics shows DNA repair deficiency has better prognostic. Genes (Basel) 11:1367, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaca-Paniagua F, Alvarez-Gomez RM, Maldonado-Martínez HA, et al. : Revealing the molecular portrait of triple negative breast tumors in an understudied population through omics analysis of formalin-fixed and paraffin-embedded tissues. PLoS One 10:e0126762, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng L, Zhu X, Sun Y, et al. : Prevalence and prognostic role of PIK3CA/AKT1 mutations in Chinese breast cancer patients. Cancer Res Treat 51:128-140, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen L, Yang L, Yao L, et al. : Characterization of PIK3CA and PIK3R1 somatic mutations in Chinese breast cancer patients. Nat Commun 9:1357, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L, Zhai Q, Lu Q, et al. : Clinical genomic profiling to identify actionable alterations for very early relapsed triple-negative breast cancer patients in the Chinese population. Ann Med 53:1358-1369, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lian J, Xu E-W, Xi Y-F, et al. : Clinical-pathologic analysis of breast cancer with PIK3CA mutations in Chinese women. Technol Cancer Res Treat 19:153303382095083, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan C, Liu N, Fan S, et al. : Comprehensive multigene mutation spectra of breast cancer patients from Northeast China obtained using the Ion Torrent sequencing platform. Oncol Rep 42:1580-1588, 2019 [DOI] [PubMed] [Google Scholar]
- 50.Jia M, Liao N, Chen B, et al. : PIK3CA somatic alterations in invasive breast cancers: Different spectrum from Caucasians to Chinese detected by next generation sequencing. Breast Cancer 28:644-652, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee S, Park K, Kim GM, et al. : Exploratory analysis of biomarkers associated with clinical outcomes from the study of palbociclib plus endocrine therapy in premenopausal women with hormone receptor-positive, HER2-negative metastatic breast cancer. Breast 62:52-60, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li G, Guo X, Chen M, et al. : Prevalence and spectrum of AKT1, PIK3CA, PTEN and TP53 somatic mutations in Chinese breast cancer patients. PLoS One 13:e0203495, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahmad F, Badwe A, Verma G, et al. : Molecular evaluation of PIK3CA gene mutation in breast cancer: Determination of frequency, distribution pattern and its association with clinicopathological findings in Indian patients. Med Oncol 33:74, 2016 [DOI] [PubMed] [Google Scholar]
- 54.Azizi Tabesh G, Izadi P, Fereidooni F, et al. : The high frequency of PIK3CA mutations in Iranian breast cancer patients. Cancer Invest 35:36-42, 2017 [DOI] [PubMed] [Google Scholar]
- 55.Kim S-B, Do I-G, Tsang J, et al. : BioPATH: A biomarker study in asian patients with HER2+ advanced breast cancer treated with lapatinib and other anti-HER2 therapy. Cancer Res Treat 51:1527-1539, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niyomnaitham S, Parinyanitikul N, Roothumnong E, et al. : Tumor mutational profile of triple negative breast cancer patients in Thailand revealed distinctive genetic alteration in chromatin remodeling gene. PeerJ 7:e6501, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saikia KK, Panigrahi MK, Mehta A, et al. : Clinico-pathological features of PIK3CA mutation in HER2-positive breast cancer of Indian population. Indian J Surg Oncol 9:381-386, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsang H, Addepalli K, Davis SR: Resources for interpreting variants in precision genomic oncology applications. Front Oncol 7:214, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vasan N, Sivakumar S, Jin D, et al. : 1954P A pan-cancer analysis of double PIK3CA mutations. Ann Oncol 31:S1101, 2020 [Google Scholar]
- 60.Vasan N, Razavi P, Johnson JL, et al. : Double PIK3CAmutations in cis increase oncogenicity and sensitivity to PI3Kα inhibitors. Science 366:714-723, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruiz-Saenz A, Dreyer C, Campbell MR, et al. : HER2 amplification in tumors activates PI3K/Akt signaling independent of HER3. Cancer Res 78:3645-3658, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan TL, Cantley LC: PI3K pathway alterations in cancer: Variations on a theme. Oncogene 27:5497-5510, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ross DS, Zehir A, Cheng DT, et al. : Next-generation assessment of human epidermal growth factor receptor 2 (ERBB2) amplification status. J Mol Diagn 19:244-254, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.She QB, Chandarlapaty S, Ye Q, et al. : Breast tumor cells with PI3K mutation or HER2 amplification are selectively addicted to Akt signaling. PLoS One 3:e3065, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jia S, Liu Z, Zhang S, et al. : Essential roles of PI(3)K-p110β in cell growth, metabolism and tumorigenesis. Nature 454:776-779, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pascual J, Turner NC: Targeting the PI3-kinase pathway in triple-negative breast cancer. Ann Oncol 30:1051-1060, 2019 [DOI] [PubMed] [Google Scholar]
- 67.US. Food and Drug Administration : Table of Pharmacogenomic Biomarkers in Drug Labeling, 2021. https://www.fda.gov/media/124784/download [Google Scholar]
- 68.Kelsey MD, Patrick-Lake B, Abdulai R, et al. : Inclusion and diversity in clinical trials: Actionable steps to drive lasting change. Contemp Clin Trials 116:106740, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.US Food and Drug Administration : Diversity Plans to Improve Enrollment of Participants From Underrepresented Racial and Ethnic Populations in Clinical Trials (FDA-2021-D-0789), 2022. https://www.regulations.gov/document/FDA-2021-D-0789-0001 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Because of HIPAA requirements, Foundation Medicine, Inc (FMI) is not consented to share individualized patient genomic data, which contains potentially identifying or sensitive information. FMI is committed to collaborative data analysis and has well-established mechanisms by which investigators can query their core genomic database of > 400,000 deidentified sequenced cancer specimens to obtain aggregated data sets.