Abstract
Neurodevelopmental disorders arise due to various risk factors that can perturb different stages of brain development, and a combinatorial impact of these risk factors programs the phenotype in adulthood. While modeling the complete phenotype of a neurodevelopmental disorder is challenging, individual developmental perturbations can be successfully modeled in vivo in animals and in vitro in human cellular models. Nevertheless, our limited knowledge of human brain development restricts modeling strategies and has raised questions of how well a model corresponds to human in vivo brain development. Recent progress in high-resolution analysis of human tissue with single-cell and spatial omics techniques has enhanced our understanding of the complex events that govern the development of the human brain in health and disease. This new knowledge can be utilized to improve modeling of neurodevelopmental disorders and pave the way to more accurately portraying the relevant developmental perturbations in disease models.
Subject terms: Psychiatric disorders, Neuroscience, Biological techniques
Introduction
Brain development is a tremendously complex process in which a myriad of neuronal and non-neuronal cell types is generated and assembled into functional circuits in a highly organized manner. Neurodevelopmental disorders (NDDs) are common brain disorders, which usually have a major impact on the affected patients and their families and arise when developmental processes are perturbed by various genetic and environmental factors [1–7]. Given the complexity of human brain development and the variety of possible perturbations, the clinical presentations and suspected disease etiologies are very heterogenous within the NDD group. Historically, the NDD concept has been restricted to disorders in which perturbations are thought to occur during embryonic brain development, e.g., as for autism spectrum disorder, attention deficit hyperactivity disorder, and intellectual disability. However, with a better understanding of the maturational processes continuing up to early adulthood (Fig. 1A), disorders with a later age of onset, e.g., schizophrenia, are now also referred to as NDDs.
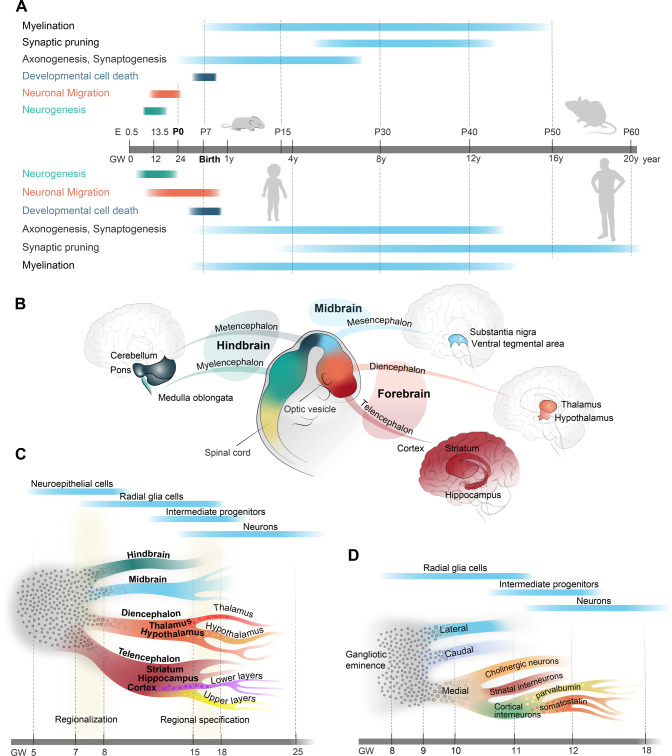
Fig. 1. Overview of brain development.
A The schedule of major developmental processes during brain development in mice (upper part) and humans (lower part). B Anatomical correspondence between developing brain regions in the early embryonic brain and the postnatal brain. C Schematic representation of differentiation strategy during brain development, starting from undifferentiated neuroepithelial cells (gray) to more differentiated radial glia cells, intermediate progenitors, and neurons. The timeline in gestational weeks (GW) is not-to-scale. Subtype differentiation within brain regions (such as upper/deeper layer principal neurons in the cortex or different neuropeptide-specific neurons in the hypothalamus) is organized rather orthogonally to areal signatures (such as rostro-caudal differentiation) and have strong temporal dependence. For instance, in the cortex, deeper layer principal neurons are produced as early as GW9, whereas switch to upper layer principal neurons happens at GW14-16. Colored cells that appear before the actual branching indicate that the cell fate is primed, mainly by epigenetic mechanisms. D Schematic representation of differentiation strategy for the ganglionic eminence that generates telencephalic inhibitory neurons. The medial ganglionic eminence is expanded to show an example for differentiation into classes, families, and subtypes of neurons.
In the last decade, our understanding of the brain development at the cellular level has been deepened by technological breakthroughs in single-cell and spatial omics. Combined with more precise methods for genetic engineering, improved disease models, and the growing number of genetic variants found to be associated with disease, we are now much better positioned to identify biological mechanisms causing NDDs. Nonetheless, great experimental and conceptual challenges lie ahead and will require careful considerations. In the current update, we summarize emerging new concepts in our understanding of the developing human brain and discuss how these advances could be implemented in NDD models.
The developing human brain in high resolution
Adult brain regions develop from embryonic brain vesicles, which are organized along an anterior-posterior axis of the neural tube (Fig. 1B). The telencephalon and diencephalon (forebrain regions) give rise to the cortex, striatum, and hippocampus, and the thalamus and hypothalamus, respectively. The mesencephalon (midbrain region) instead gives rise to the superior part of the brainstem, including structures such as the substantia nigra and ventral tegmental area; while the metencephalon and myelencephalon (hindbrain regions) give rise to the cerebellum and pons, and the medulla oblongata, respectively.
Recent developments in single-cell and spatial omics have revolutionized our understanding of the cellular diversity across regions and time periods in the developing human brain (Fig. 1C, D). Generally, early neural stem cells in embryonic brain, neuroepithelial (NE) cells, transform into more differentiated radial glia (RG) cells, which in turn differentiate into intermediate progenitor cells and neurons (Fig. 1C). Single-cell analysis of first-trimester human brain tissue have revealed that region-specific transcriptomic signatures first appear in NE and RG cells at gestational week (GW) 7–8, whereas prior progenitors are highly similar across developing brain regions [8] (Fig. 1C). Regional transcriptomic signatures expand over late first and second trimester, peaking into regional specification at GW15–18 [9, 10], when regions are being further divided into specific areas (Fig. 1C).
Cortex area-specific signatures can be detected at ~GW17–18 [9–11] (Fig. 1C), as a rostro-caudal gradient without clear borders [9, 10]. These areal transcriptomic signatures can already be identified in progenitors [9–11], which are likely primed by a few transcription factors and preassembled epigenetic mechanisms. Such priming in progenitors triggers large transcriptomic specification programs in early postmitotic neurons, with further differentiation taking place during migration and the early stages of neuronal maturation [12–14]. Importantly, specification of laminar position of cortical neurons (i.e., upper to lower layers) is organized orthogonally to areal signatures (i.e., anterior to posterior). Lamination specification also has a strong temporal dependence, where newborn, lower-layer neurons can be identified as early as GW9, switching to mainly upper layers between GW14 and 16 [12].
Cell-type specifications have also been studied in other brain regions, such as the hippocampus and hypothalamus (Fig. 1C). In the hippocampus, common progenitors of excitatory neurons are specified into dentate gyrus and nondentate gyrus neurons at GW16–18, followed by further specification associated with morphogenesis (GW20–22) and functional maturation (GW25–27) [15]. In the hypothalamus, specific transcriptomic signatures for hypothalamic nuclei can be distinguished at GW7–8, while nucleus diversification and onset of specific neuropeptide expression occurs at GW18–25 [16].
A number of studies have implicated inhibitory neurons in NDDs [2, 3, 17, 18]. Telencephalic inhibitory neurons are born in the transitory ganglionic eminences (GEs), which are subdivided into three regions—the medial, caudal, and lateral eminences (MGE, CGE, and LGE, respectively) (Fig. 1D). The MGE and CGE produce mainly cortical, striatal, and hippocampal interneurons, and LGE produces mainly striatal projection neurons and olfactory bulb interneurons. Similarly to principal neurons, telencephalic inhibitory neuron progenitors are noncommitted and primed to be specified during transition from precursors to neurons [19, 20]. However, it seems that stratification of neuronal cell fate is expedited for inhibitory neurons and individual subtypes can be distinguished in each GE already at GW11–18 [15, 19–21], for example, up to 11 subtypes of somatostatin neurons in MGE [20].
Developmental trajectories for non-neuronal cells have been also studied at high resolution. Oligodendrocyte precursor cells (OPCs) are generated via Pre-OPC states from common neural progenitors [22, 23] and the first wave of OPC generation is observed already at GW8–10 [23]. Microglia instead arises from erythromyeloid progenitor cells in the yolk sac and can be detected in the developing brain from at least GW9 [24]. Microglia are a highly heterogenous cell population during the fetal period and display temporal gene expression changes [24].
To date, the third trimester and the long postnatal period of the developing human brain have only been sparsely studied by single-cell analyses, mostly due to limitations in the availability of clinical samples. This means that while we have good resolution atlases for neurogenic, cell migration, early differentiation, and morphogenesis stages, data are sparse for the long and important maturational period of human brain development.
From developmental trajectories to modeling neurodevelopmental perturbations
When a risk factor for a NDD is first identified in epidemiologic or genetic association studies, the question arises as to where in the brain and during which developmental period(s) the relevant consequences for disease take place. Simply put, when, where, and how does risk transform into perturbation? Ideally, the initial modeling approach, before any certain mechanism is prioritized, should be broad with complex readouts for all brain regions, developmental stages, and cell types. For practical reasons, studies are, however, usually focused on a specific brain region, cell type, or developmental stage. Even if such decisions are based on prior knowledge, bias can easily be “inherited” as previous studies could have selected neuronal subtypes or developmental periods, for instance, according to available samples and tools. A good example is parvalbumin (PV) interneurons in the cortex: almost every study of a NDD evaluates an impact on PV interneurons, which might give the impression that PV interneurons are the major contributors to all NDDs. However, recent studies show that other types of interneurons, various types of excitatory neurons, and glia might be equally affected, and the unbiased view by single-cell analyses of postmortem human tissue reveals complex transcriptomic changes affecting multiple neuronal subtypes and glia [25, 26].
Fortunately, recent advances in our understanding of brain development by single-cell omics have improved our precision and could guide modeling strategies. By utilizing developmental trajectories, we can identify at single-cell level, when and where a gene is expressed (Fig. 2A). Hence, using reconstructed developmental trajectories, it can be determined for each gene in which brain regions, developmental periods, cell types, and differentiation stages this particular gene is expressed. For a given neurodevelopmental perturbation, we can then select a gene or a combination of genes that code for factors that are required to respond to a given perturbation, i.e., receiving gene sets [6] (Fig. 2B, C). For a genetic risk factor, a receiving gene set is then centralized around the primarily affected gene. For an environmental risk factor, complexity increases as an environmental factor can reach the cell via multiple routes. For instance, in case of maternal inflammation during pregnancy, the overall impact on a given cell in the embryonic brain is a combination of individual impacts of each pro-inflammatory molecule that reaches this brain cell [6]. The complexity is further augmented by secondary receiving gene sets, when microglia is directly activated by maternal inflammation and then in turn impacts surrounding neuronal cells. Using receiving gene sets, we can predict cells that should respond to the perturbation. As cell-state transcriptional signatures are dynamic and form a gradient throughout development, especially in the case of uncommitted progenitors [8–10], this suggests that there is a gradient sensitivity to risk factor exposure during development (Fig. 2D). Since gradient sharpness increases over the course of development from first commitment to regionalization and subtype differentiation, early developmental periods might have a greater regional sensitivity, which then changes during later developmental periods to areal- and cell type-specific sensitivity. Nonetheless, even in the adult stage, gradient gene expression between differentiated cell types and transitory cell states remains [27]. From the perspective of receiving gene sets with multiple regional and temporal gene functions, the plethora of clinical presentations for polygenic NDDs is then hardly surprising. This often requires additional phenotypic specification, preferably using quantitative traits, although the approach is still compatible to cross-disorder designs.
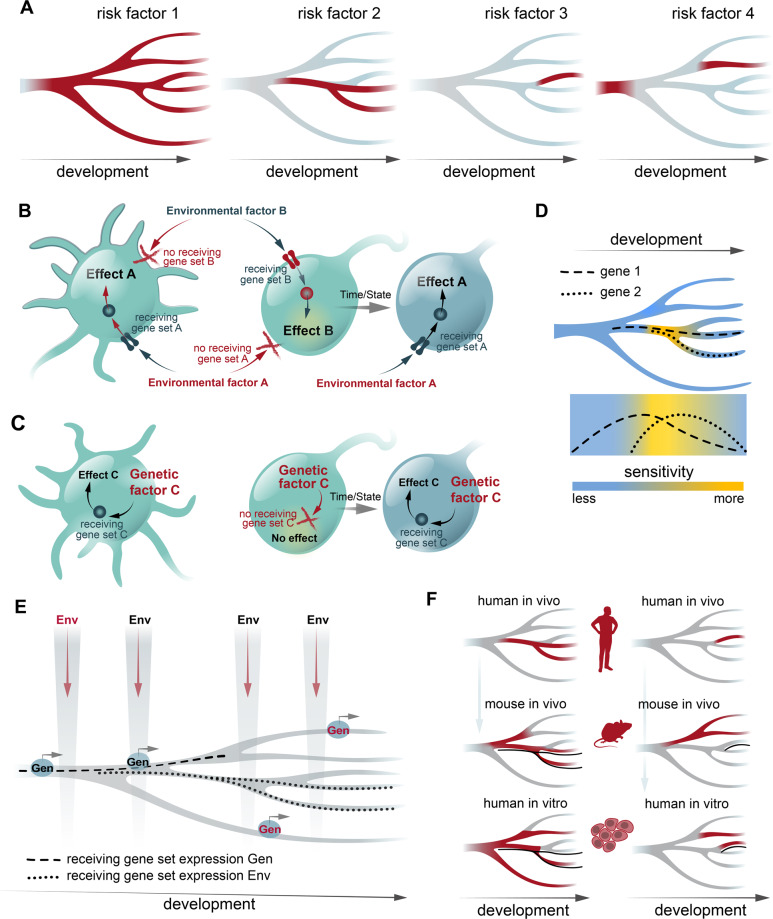
Fig. 2. Modeling neurodevelopmental perturbations.
A Examples of risk factor gene expression overlaid on developmental trajectories during cell differentiation and maturation, from neural stem cells on the left differentiating towards committed cell types on the right. Red denotes gene expression. B, C The concept of selective vulnerability to environmental and genetic risk factors, based on receiving gene sets, i.e., gene sets that define cellular components (as signaling network) that are necessary to respond to a particular risk factor, as first described in [6]. The response to genetic risk factors originates from within the cell, whereas for environmental risk factors the initial response is transmembrane. The susceptibility to a risk factor can change over the course of development due to changes in expression of receiving gene sets, illustrated by the cells on the right. D Sensitivity to risk factors over development is based on ability to respond to the risk factors. A combinations of two genes that constitute a receiving gene set is shown (dotted lines—gene expression). E Modeling strategies on what to model, and on what not to model. If a genetic (Gen) risk factor/receiving gene set is expressed only during certain period/cell lineage (dashed line), it makes sense to model within this period (black but not red Gen label). Similarly, for an environmental risk factor, only period when receiving gene set is present (dotted lines) is relevant to model (black but not red Env label). F Assessment of conservation of risk factor/receiving gene set expression across models. Left and right columns show examples when mouse in vivo or human in vitro models, respectively, are better in mimicking human in vivo.
Implementing developmental trajectories to assess the impact of developmental perturbations has an immense impact on a modeling strategy, i.e., what to model and what not to model (Fig. 2E). Thus, when a certain genetic factor has been proposed to be involved in a NDD and the gene is not expressed in principal neurons, but rather is expressed in GABAergic ones and maybe even in a certain subtype, modeling and analyses should initially be focused on this subtype. Similarly, for the developmental period, if the gene is expressed only in immature neurons and not in progenitor cells, then modeling should be initially focused on immature neurons. For environmental factors, modeling is more complex as there is typically a lack of prior knowledge for how an environmental factor reach the cell and triggers the response. Furthermore, there might be several routes for triggering the response. For instance, several individual soluble molecules that mediate environmental factor response might activate different transmembrane receptors in the cell. A combination of all receptors and their intracellular signaling pathways comprises a receiving gene set for the cell. Whereas it seems difficult to model an overall impact of an environmental factor, individual components can be readily modeled, e.g., such as IL-6 and IL-17 response in maternal inflammation [28, 29].
Such simple considerations could provide tremendous help and promote more efficient use of our resources. As an example, implementation of single-cell data to understand the mechanisms underlying the impact of the 15q13.3 microdeletion, a genetic risk factor for several psychiatric disorders [30], revealed a hidden impact on embryonic development, suggesting that a reduction in KLF13 expression in progenitors might be the major neurodevelopmental driver of the 15q13.3 microdeletion phenotype [3]. Importantly, although a considerable amount of work is still left to complete reconstruction of developmental trajectories, enough data have been generated to start predicting with good precision the impact of many neurodevelopmental perturbations.
Finally, conservation of developmental trajectories is also important to consider in regard to the model being used, typically in vivo animal models versus in vitro human models (Fig. 2F). For instance, if expression patterns along developmental trajectories for a risk factor are much different between humans and rodents, a mouse model of this risk factor might be a poor choice for reproducing the impact of such a perturbation in humans, and then a human cellular model or primate model should be considered instead. A concrete example is the component 4 (C4) genes. Here, detailed genotyping of the most strongly associated GWAS locus in schizophrenia has revealed that copy numbers of the isogene C4A explains most of the risk, while C4B copy numbers do not increase schizophrenia risk [31]. However, rodents lack specialized C4 genes, and it was first shown in human in vitro models that the unique link between C4A, excessive synaptic pruning, and schizophrenia could be established [4]. Another study showed similar data using a humanized mouse model, confirming in vivo that C4A binds synapses more efficiently than C4B, while overexpression of C4A leads to an increased microglial engulfment of synaptic structures [32].
Non-human primates (NHPs) offer an alternative strategy to model human NDD risk factors during brain development. Although there are a number of limitations in implementation of NHP models, such as long breeding periods, difficulties in genetic manipulations, and ethical considerations, advantages over rodent models are greater similarity in gestational time and physiology, brain architecture [33–35], cell type-specific signatures [36] and developmental trajectories [35, 37]. It is also worth noting that NHP, similarly to humans, exhibit a large outer subventricular zone—a region of developing brain that is rudimental in rodents and was proposed to have the major contribution to the large expansion of the cortex in primates [38]. Additionally, while genetic risk modeling is limited in NHPs, modeling environmental risk factors is rather feasible [39, 40] and allows to study developmental processes that are absent in rodents. Thus, NHP models help bridging rodent data with pathological processes in humans and enhance translatability of rodent results.
Overall, recent data from single-cell analyses show both considerable rodent-to-human conservation of cell-type specification as well as human-specific mechanisms, emphasizing that a dual approach using animal models as well as human cellular and NHP systems is required for modeling NDDs. Below, we discuss the latest progress in in vivo animal and in vitro human modeling of NDDs and propose how knowledge of developmental processes can be implemented in these strategies.
Improved modeling of NDDs: in vivo animal models
In recent years, the translational potential of animal modeling in NDDs, including psychiatric disorders, has been the subject of debate, and challenges as well as opportunities in this regard have been widely discussed elsewhere, see, for example [41]. We anticipate that while it might be difficult to model schizophrenia or autism in animals, it is feasible to model specific perturbations that are risk factors for NDDs, which will help us identify impaired developmental mechanisms that cause changes in neuronal network assembly and behavioral phenotypes relevant for NDDs. Thus, below we focus on how new knowledge of developmental processes can be implemented to design improved in vivo models of neurodevelopmental perturbations in rodents.
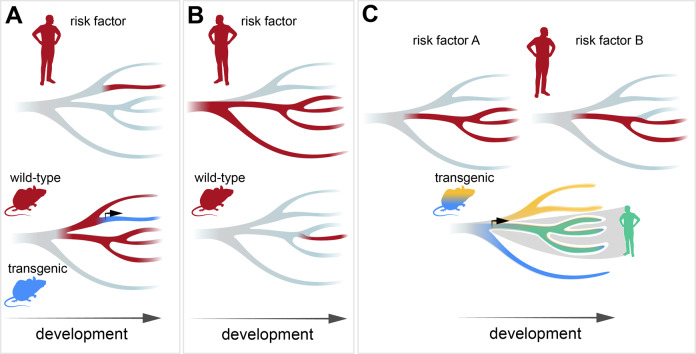
One of the key benefits of the available high-resolution data for brain development is that it allows for precision in modeling. In general, comparative studies report relatively good similarity in specification patterns during development of human and mouse brains [15, 20, 42]. Thus, the main features of developmental trajectories are conserved, such as prominent brain-region transcriptional signatures [43], largely orthogonal transcriptional signatures for spatial and temporal specification [44], noncommitted progenitors and first-commitment signatures for neuronal subtypes detected postmitotically [45–47], and priming progenitors via epigenetic mechanisms [42, 45, 46]. Nevertheless, notable differences between human and mouse developmental trajectories have been reported, such as human-enriched/-specific populations of progenitors [8] and neurons [48], or the ability of some progenitors to generate both excitatory and inhibitory neurons [49]. Therefore, we propose that when we aim to understand how a developmental perturbation affects “human” brain development, the credibility of a rodent model for this particular perturbation should be assessed first. If a risk factor (or receiving gene sets as responders to a risk factor) shows a very different developmental expression pattern between humans and an animal model of interest, the feasibility of using this type of animal model for modeling “human” developmental perturbation should be scrutinized. Nevertheless, even in those situations when there are significant differences in expression, advantages in recombinant technology in animal models enable us to “focus” an animal model on modeling the human situation. For instance, when a risk factor is present in a specific cell lineage in human brain, but its expression is broader in the animal model, then utilization of cell type-specific transgenics or turn on/off gene expression can restrict risk factor analysis to the cell lineage relevant for human brain (Fig. 3A). In contrast, when the expression of a risk factor is broader in human brain than in an animal model of interest (multiple examples of such cases have been identified [36, 50, 51]), the model can be utilized to target specific lineages or developmental periods (Fig. 3B). Importantly, rich resources in animal transgenics allow for multi-factor modeling by intersectional and/or substructional approaches [52, 53]. For instance, when two risk factors are present in overlapping, but different developmental period(s) or developmental cell lineage(s), the risk factor interplay can be modeled by intersection of specific promoters, including potential temporal dimension by turning on/off promoters (Fig. 3C). Finally, pooled CRISPR screens with single-cell RNA-sequencing readouts, such as Perturb-seq, can be applied to in vivo models [54] and include thousands of genetic perturbations across coding regions [55]. This allows for modeling interactions between a large number of genetic risk factors as well as interactions between genetic risk factors and environmental perturbations. Perturb-seq has already been implemented to study how multiple NDD genetic risk factors located in coding regions influence the developing mouse brain [54]; and with the possibility to target CRISPR screens at non-coding regions [56], Perturb-seq also allows for investigating NDD risk variants located in non-coding regions.
Fig. 3. Strategies for modeling neurodevelopmental perturbations in rodents.
A When a risk factor is present in a specific cell lineage in the human brain (in red, top), but the expression is broader in the rodent brain (in red, bottom), utilization of cell type-specific transgenics can restrict risk factor analysis to the cell lineage relevant for human brain (in blue). B When the expression of a risk factor is broader in the human brain (in red, top) than in an animal model of interest (in red, bottom), the model can be utilized to target specific cell lineages or developmental periods. C To model an interplay of two risk factors (in red, top), transgenic models with intersectional gene expression can be utilized (bottom, blue and yellow are individual transgenic lines, the intersection is in green).
Improved modeling of neurodevelopmental disorders: human in vitro models
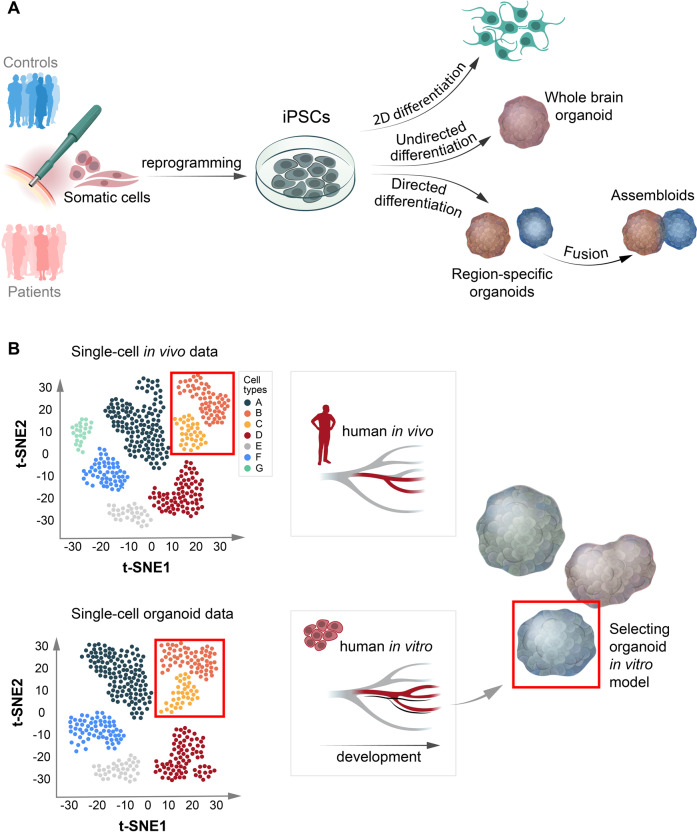
The ability to reprogram human somatic cells to induced pluripotent stem cells (iPSCs) has become a valuable tool in NDD modeling. Early efforts to differentiate patient-derived neural-like cells in monolayer have rapidly been complemented by models that use cellular 3D aggregates to recapitulate the tissue architecture of the developing human brain [57] (Fig. 4A). By supporting more complex cell-cell interactions and cell diversity while reducing artificial interactions with plastic surfaces, these approaches are likely to more adequately mimic human brain development, both in transcriptomic signatures and cell-type composition. Nonetheless, brain organoids are not “mini-brains” and can only approximate the developing brain in vivo. Data for developmental trajectories from single-cell omics can then help to evaluate each in vitro 3D modeling strategy for a given developmental perturbation. As in vitro 3D models display limitations in regard to developmental stages, cell types, and/or brain regions, the expression pattern of a risk factor (or receiving gene sets as responders to a risk factor) across cell types/states, brain regions, and developmental stages should be assessed in vivo to select those that are relevant for modeling (Fig. 4B).
Fig. 4. Strategies for modeling neurodevelopmental perturbations in human brain organoids.
A Overview of different strategies to model neurodevelopmental perturbations in vitro in human cellular systems, including brain organoids. B When an organoid model is deemed adequate for modeling a neurodevelopmental perturbation, careful consideration should be made concerning the protocol that is most suitable for the given research question instead of applying a “one size fits all” approach to organoid modeling. Single-cell and spatial omics datasets are then a crucial resource that can help us evaluate the protocol that is most adequate for modeling a given developmental perturbation.
A growing number of protocols currently exist and can broadly be divided into undirected approaches, generating whole-brain organoids (relying more on the self-organizing capacity of pluripotent stem cells in 3D aggregates), and directed approaches to generate brain region-specific organoids (emphasizing more the external patterning factors to support intrinsic cues). Single-cell omics characterizations have been applied to many of the existing protocols and have revealed important information about how a protocol covers cellular diversity and differentiation states during development [58–63]. For most protocols, comparing organoid gene expression with in vivo references suggests that the maturation process typically reaches at least the second trimester. Nonetheless, directed protocols that culture cortical organoids over longer time periods suggest that early postnatal developmental stages can be reached at around 300 days in vitro [59], thus largely mimicking in vivo maturation. In contrast to a previous report [64], upregulation of cellular stress pathways in these organoids also remain relatively flat during differentiation and comparable to datasets generated from the fetal cortex [59]. In a recent systematic comparison of different organoid protocols using available single-cell transcriptomic datasets [58], genes related to endoplasmic reticulum were upregulated in both directed and undirected protocols. This suggests that despite using different technical approaches with orbital shaking or spinning bioreactors in vitro, the exchange of oxygen and nutrients into the core of the organoids is still compromised and needs to be taken into account. In support of this finding, transplanting human organoids into cortices of living mice markedly reduced stress-marker expression [64].
In comparison to primary tissue, organoids also show less defined subtypes in general and some cell types are represented in lower numbers (e.g., oligodendrocytes) or are absent (e.g., microglia) [65]. Especially the lack of microglia has obstructed organoid utility given their important regulatory roles in neurodevelopment [66]. By further reducing neuroectoderm stimulation, mesoderm-derived progenitors have, however, been shown to emerge spontaneously in undirected organoids and display microglia markers at later time points [67]. Single-cell transcriptomic profiling of such microglia-like cells at later stages has also shown a convincing clustering with fetal microglia [68]. Given the general limitations of less guided protocols, such as more pronounced organoid-to-organoid variability and relatively low and variable microglia counts, other approaches have also been proposed, such as adding mesodermal yolk sac progenitors to patterned brain region-specific organoids [69] or adding iPSC-derived microglia to undirected organoids [70]. Furthermore, as an alternative to undirected whole-brain organoids, fusing of brain region-specific organoids to form so-called assembloids has also been performed and applied to NDDs [71].
In summary, we propose that when an organoid model is deemed adequate for modeling a NDD, careful consideration should be made concerning the strategy that is most suitable for the given research question instead of applying a “one size fits all” approach to organoid modeling. The accumulated single-cell omics data will then be a useful resource to select the best strategy according to the most relevant cell types, brain region(s), and developmental stage(s) for a given developmental perturbation. Similarly, the experimental design needs to be carefully selected: is a case-control study more adequate or are isogenic lines preferable? In vivo validation should also be considered, either by using animal models or by observational clinical studies.
Limitations of the current state of single-cell analysis and future directions
Although the single-cell technology has progressed massively over the last few years and led to immense advance in understanding of human brain development, we are still far from complete multi-modal reconstruction of developmental trajectories. So far, only single-cell RNA-sequencing has reached a very high resolution, up to 10–12,000 genes per cell for technologies like Smart-seq [72], whereas other omics modalities at the moment are at lower resolution. For instance, current state-of-the-art for single-cell proteomics limits detectability to ~1000 of most abundant proteins per cell [73, 74], and there is a lack of methods to measure posttranslational modifications at single-cell level. Resolving these bottlenecks will be of major importance for the field as proteins regulate cellular functions, and mRNA and protein content for a given gene do not always match. Capturing of mRNA or protein context is another general limitation, as even for mRNA most protocols capture only 1–5% of total mRNA content. Additionally, single-cell analysis of brain tissue, whether at single-cell or single-nucleus level, involves rigorous tissue processing, which destroys neuronal/glial processes and the information about axonal and dendritic mRNAs is lost. Spatial transcriptomics has emerged as a method with subcellular resolution, e.g., MERFISH [75, 76], that allows for both detection of mRNA distribution in cellular compartments (including axons and dendrites) and large-scale brain tissue architecture reconstruction. Finally, neuronal connectivity can be also reconstructed at single-cell level by a combination of single-cell omics with cell-cell interaction [77].
In this review, we have updated the perspective of NDD modeling such that greater emphasis should be placed on the available single-cell omics data from primary tissue and disease models in selecting an appropriate modeling strategy. Combined approaches including human cells and in vivo conditions should be prioritized. A number of large-scale atlases are available for studying developmental trajectories at single-cell resolution (e.g., via Human Cell Atlas [78] or BRAIN Initiative—Cell Census Network [79]), and further initiatives have been established to study diseases at such resolution (e.g., LifeTime [80]). Nevertheless, classification of cell types still varies from dataset to dataset (although some initiatives try to deal with this problem [81]), number of individual samples is limited, interindividual variability is largely unstudied, and only some developmental periods are covered. Finally, while many studies have been conducted for embryonic development, postnatal development that include developmental cell death [82, 83] and maturational processes related to functional specification [84, 85] have not been studied as comprehensively in single-cell analyses, due to limitations in availability of material for this developmental period in tissue banks. Paradoxically, this is the key period for (1) understanding how the phenotype in many NDDs, including major psychiatric disorders, is built during maturation and network assembling, and (2) identifying drug targets and a therapeutic window. Studies targeting postnatal maturation would be tremendously important for the field and accelerate our understanding of the mechanisms driving NDDs.
Acknowledgements
The authors are grateful to Olga Kharchenko for work on figure illustrations. The work in Konstantin Khodosevich’s lab is supported by Novo Nordisk Foundation Hallas-Møller Investigator grants (NNF16OC0019920 and NNF21OC0067146) and Lundbeck Foundation Ascending Investigator grant (2020-1025). CMS is supported by grants from Erling Persson Foundation, Hjärnfonden, One Mind Foundation and Kaiser Permanente, and the Swedish Federal Government under the LUA/ALF agreement.
Author contributions
This Perspective was conceived by KK and CMS. KK and CMS jointly wrote the paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Konstantin Khodosevich, Email: konstantin.khodosevich@bric.ku.dk.
Carl M. Sellgren, Email: carl.sellgren@ki.se
References
- 1.Sanders B, D’Andrea D, Collins MO, Rees E, Steward TGJ, Zhu Y, et al. Transcriptional programs regulating neuronal differentiation are disrupted in DLG2 knockout human embryonic stem cells and enriched for schizophrenia and related disorders risk variants. Nat Commun. 2022;13:27. doi: 10.1038/s41467-021-27601-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vasistha NA, Pardo-Navarro M, Gasthaus J, Weijers D, Müller MK, García-González D, et al. Maternal inflammation has a profound effect on cortical interneuron development in a stage and subtype-specific manner. Mol Psychiatry. 2020;25:2313–29. doi: 10.1038/s41380-019-0539-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malwade S, Gasthaus J, Bellardita C, Andelic M, Moric B, Korshunova I, et al. Identification of vulnerable interneuron subtypes in 15q13.3 microdeletion syndrome using single-cell transcriptomics. Biol Psychiatry. 2022;91:727–39. doi: 10.1016/j.biopsych.2021.09.012. [DOI] [PubMed] [Google Scholar]
- 4.Sellgren CM, Gracias J, Watmuff B, Biag JD, Thanos JM, Whittredge PB, et al. Increased synapse elimination by microglia in schizophrenia patient-derived models of synaptic pruning. Nat Neurosci. 2019;22:374–85. doi: 10.1038/s41593-018-0334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosin JM, Sinha S, Biernaskie J, Kurrasch DM. A subpopulation of embryonic microglia respond to maternal stress and influence nearby neural progenitors. Dev Cell. 2021;56:1326–45.e6. doi: 10.1016/j.devcel.2021.03.018. [DOI] [PubMed] [Google Scholar]
- 6.Vasistha NA, Khodosevich K. The impact of (ab)normal maternal environment on cortical development. Prog Neurobiol. 2021;202:102054. doi: 10.1016/j.pneurobio.2021.102054. [DOI] [PubMed] [Google Scholar]
- 7.Mukai J, Tamura M, Fenelon K, Rosen AM, Spellman TJ, Kang R, et al. Molecular substrates of altered axonal growth and brain connectivity in a mouse model of schizophrenia. Neuron. 2015;86:680–95. doi: 10.1016/j.neuron.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eze UC, Bhaduri A, Haeussler M, Nowakowski TJ, Kriegstein AR. Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia. Nat Neurosci. 2021;24:584–94. doi: 10.1038/s41593-020-00794-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358:1318–23. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhaduri A, Sandoval-Espinosa C, Otero-Garcia M, Oh I, Yin R, Eze UC, et al. An atlas of cortical arealization identifies dynamic molecular signatures. Nature. 2021;598:200–4. doi: 10.1038/s41586-021-03910-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziffra RS, Kim CN, Ross JM, Wilfert A, Turner TN, Haeussler M, et al. Single-cell epigenomics reveals mechanisms of human cortical development. Nature. 2021;598:205–13. doi: 10.1038/s41586-021-03209-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan X, Fu Y, Zhou X, Sun L, Yang M, Wang M, et al. Single-cell transcriptome analysis reveals cell lineage specification in temporal-spatial patterns in human cortical development. Sci Adv. 2020;6:eaaz2978. doi: 10.1126/sciadv.aaz2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polioudakis D, de la Torre-Ubieta L, Langerman J, Elkins AG, Shi X, Stein JL, et al. A single-cell transcriptomic atlas of human neocortical development during mid-gestation. Neuron. 2019;103:785–801.e8. doi: 10.1016/j.neuron.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong S, Zhang S, Fan X, Wu Q, Yan L, Dong J, et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature. 2018;555:524–8. doi: 10.1038/nature25980. [DOI] [PubMed] [Google Scholar]
- 15.Zhong S, Ding W, Sun L, Lu Y, Dong H, Fan X, et al. Decoding the development of the human hippocampus. Nature. 2020;577:531–6. doi: 10.1038/s41586-019-1917-5. [DOI] [PubMed] [Google Scholar]
- 16.Herb BR, Glover HJ, Bhaduri A, Casella AM, Bale TL, Kriegstein AR, et al. Single-cell genomics reveals region-specific developmental trajectories underlying neuronal diversity in the prenatal human hypothalamus. BioRxiv. 2021. 10.1101/2021.07.20.453090. [DOI] [PMC free article] [PubMed]
- 17.Qvist P, Christensen JH, Vardya I, Rajkumar AP, Mørk A, Paternoster V, et al. The schizophrenia-associated BRD1 gene regulates behavior, neurotransmission, and expression of schizophrenia risk enriched gene sets in mice. Biol Psychiatry. 2017;82:62–76. doi: 10.1016/j.biopsych.2016.08.037. [DOI] [PubMed] [Google Scholar]
- 18.Shen M, Guo Y, Dong Q, Gao Y, Stockton ME, Li M, et al. FXR1 regulation of parvalbumin interneurons in the prefrontal cortex is critical for schizophrenia-like behaviors. Mol Psychiatry. 2021;26:6845–67. doi: 10.1038/s41380-021-01096-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Y, Zeng Z, Xie D, Chen R, Sha Y, Huang S, et al. Interneuron origin and molecular diversity in the human fetal brain. Nat Neurosci. 2021;24:1745–56. doi: 10.1038/s41593-021-00940-3. [DOI] [PubMed] [Google Scholar]
- 20.Shi Y, Wang M, Mi D, Lu T, Wang B, Dong H, et al. Mouse and human share conserved transcriptional programs for interneuron development. Science. 2021;374:eabj6641. doi: 10.1126/science.abj6641. [DOI] [PubMed] [Google Scholar]
- 21.Bocchi VD, Conforti P, Vezzoli E, Besusso D, Cappadona C, Lischetti T, et al. The coding and long noncoding single-cell atlas of the developing human fetal striatum. Science. 2021;372:eabf5759. doi: 10.1126/science.abf5759. [DOI] [PubMed] [Google Scholar]
- 22.Huang W, Bhaduri A, Velmeshev D, Wang S, Wang L, Rottkamp CA, et al. Origins and proliferative states of human oligodendrocyte precursor cells. Cell. 2020;182:594–608.e11. doi: 10.1016/j.cell.2020.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Bruggen D, Pohl F, Langseth CM, Kukanja P, Lee H, Albiach AM, et al. Developmental landscape of human forebrain at a single-cell level identifies early waves of oligodendrogenesis. Dev Cell. 2022;57:1421–36.e5. doi: 10.1016/j.devcel.2022.04.016. [DOI] [PubMed] [Google Scholar]
- 24.Kracht L, Borggrewe M, Eskandar S, Brouwer N, Chuva de Sousa Lopes SM, Laman JD, et al. Human fetal microglia acquire homeostatic immune-sensing properties early in development. Science. 2020;369:530–7. doi: 10.1126/science.aba5906. [DOI] [PubMed] [Google Scholar]
- 25.Pfisterer U, Petukhov V, Demharter S, Meichsner J, Thompson JJ, Batiuk MY, et al. Identification of epilepsy-associated neuronal subtypes and gene expression underlying epileptogenesis. Nat Commun. 2020;11:1–19. doi: 10.1038/s41467-020-18752-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Batiuk MY, Tyler T, Dragicevic K, Mei S, Rydbirk R, Petukhov V, et al. Upper cortical layer–driven network impairment in schizophrenia. Sci Adv. 2022;8:eabn8367. doi: 10.1126/sciadv.abn8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao Z, van Velthoven CTJ, Nguyen TN, Goldy J, Sedeno-Cortes AE, Baftizadeh F, et al. A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell. 2021;184:3222–41.e26. doi: 10.1016/j.cell.2021.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garbett KA, Hsiao EY, Kálmán S, Patterson PH, Mirnics K. Effects of maternal immune activation on gene expression patterns in the fetal brain. Transl Psychiatry. 2012;2:e98. doi: 10.1038/tp.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirabella F, Desiato G, Mancinelli S, Fossati G, Rasile M, Morini R, et al. Prenatal interleukin 6 elevation increases glutamatergic synapse density and disrupts hippocampal connectivity in offspring. Immunity. 2021;54:2611–31.e8. doi: 10.1016/j.immuni.2021.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharp AJ, Mefford HC, Li K, Baker C, Skinner C, Stevenson RE, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–8. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sekar A, Bialas AR, De Rivera H, Davis A, Hammond TR, Kamitaki N, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–83. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yilmaz M, Yalcin E, Presumey J, Aw E, Ma M, Whelan CW, et al. Overexpression of schizophrenia susceptibility factor human complement C4A promotes excessive synaptic loss and behavioral changes in mice. Nat Neurosci. 2021;24:214–24. doi: 10.1038/s41593-020-00763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernard A, Lubbers LS, Tanis KQ, Luo R, Podtelezhnikov AA, Finney EM, et al. Transcriptional architecture of the primate neocortex. Neuron. 2012;73:1083–99. doi: 10.1016/j.neuron.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Y, Sousa AMM, Gao T, Skarica M, Li M, Santpere G, et al. Spatiotemporal transcriptomic divergence across human and macaque brain development. Science. 2018;362:eaat8077. doi: 10.1126/science.aat8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bakken TE, Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, et al. A comprehensive transcriptional map of primate brain development. Nature. 2016;535:367–75. doi: 10.1038/nature18637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakken TE, Jorstad NL, Hu Q, Lake BB, Tian W, Kalmbach BE, et al. Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature. 2021;598:111–9. doi: 10.1038/s41586-021-03465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitz MT, Sandoval K, Chen CP, Mostajo-Radji MA, Seeley WW, Nowakowski TJ, et al. The development and evolution of inhibitory neurons in primate cerebrum. Nature. 2022;603:871–7. doi: 10.1038/s41586-022-04510-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bauman MD, Iosif AM, Smith SE, Bregere C, Amaral DG, Patterson PH. Activation of the maternal immune system during pregnancy alters behavioral development of rhesus monkey offspring. Biol Psychiatry. 2014;75:332–41. doi: 10.1016/j.biopsych.2013.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boktor JC, Adame MD, Rose DR, Schumann CM, Murray KD, Bauman MD, et al. Global metabolic profiles in a non-human primate model of maternal immune activation: implications for neurodevelopmental disorders. Mol Psychiatry. 2022 doi: 10.1038/s41380-022-01752-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hyman SE. Use of mouse models to investigate the contributions of CNVs associated with schizophrenia and autism to disease mechanisms. Curr Opin Genet Dev. 2021;68:99–105. doi: 10.1016/j.gde.2021.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Li Z, Tyler WA, Zeldich E, Santpere Baró G, Okamoto M, Gao T, et al. Transcriptional priming as a conserved mechanism of lineage diversification in the developing mouse and human neocortex. Sci Adv. 2020;6:eabd2068. doi: 10.1126/sciadv.abd2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.La Manno G, Siletti K, Furlan A, Gyllborg D, Vinsland E, Mossi Albiach A, et al. Molecular architecture of the developing mouse brain. Nature. 2021;596:92–6. doi: 10.1038/s41586-021-03775-x. [DOI] [PubMed] [Google Scholar]
- 44.Moreau MX, Saillour Y, Cwetsch AW, Pierani A, Causeret F. Single-cell transcriptomics of the early developing mouse cerebral cortex disentangle the spatial and temporal components of neuronal fate acquisition. Development. 2021;148:dev197962. doi: 10.1242/dev.197962. [DOI] [PubMed] [Google Scholar]
- 45.Di Bella DJ, Habibi E, Stickels RR, Scalia G, Brown J, Yadollahpour P, et al. Molecular logic of cellular diversification in the mouse cerebral cortex. Nature. 2021;595:554–9. doi: 10.1038/s41586-021-03670-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allaway KC, Gabitto MI, Wapinski O, Saldi G, Wang CY, Bandler RC, et al. Genetic and epigenetic coordination of cortical interneuron development. Nature. 2021;597:693–7. doi: 10.1038/s41586-021-03933-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bandler RC, Vitali I, Delgado RN, Ho MC, Dvoretskova E, Ibarra Molinas JS, et al. Single-cell delineation of lineage and genetic identity in the mouse brain. Nature. 2022;601:404–9. doi: 10.1038/s41586-021-04237-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berg J, Sorensen SA, Ting JT, Miller JA, Chartrand T, Buchin A, et al. Human neocortical expansion involves glutamatergic neuron diversification. Nature. 2021;598:151–8. doi: 10.1038/s41586-021-03813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Delgado RN, Allen DE, Keefe MG, Mancia Leon WR, Ziffra RS, Crouch EE, et al. Individual human cortical progenitors can produce excitatory and inhibitory neurons. Nature. 2022;601:397–403. doi: 10.1038/s41586-021-04230-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonnefont J, Tiberi L, van den Ameele J, Potier D, Gaber ZB, Lin X, et al. Cortical neurogenesis requires Bcl6-mediated transcriptional repression of multiple self-renewal-promoting extrinsic pathways. Neuron. 2019;103:1096–108.e4. doi: 10.1016/j.neuron.2019.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanz-Morello B, Pfisterer U, Winther Hansen N, Demharter S, Thakur A, Fujii K, et al. Complex IV subunit isoform COX 6A2 protects fast-spiking interneurons from oxidative stress and supports their function. EMBO J. 2020;9:e105759. doi: 10.15252/embj.2020105759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matho KS, Huilgol D, Galbavy W, He M, Kim G, An X, et al. Genetic dissection of the glutamatergic neuron system in cerebral cortex. Nature. 2021;598:182–7. doi: 10.1038/s41586-021-03955-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He M, Tucciarone J, Lee S, Nigro MJ, Kim Y, Levine JM, et al. Strategies and tools for combinatorial targeting of GABAergic neurons in mouse cerebral cortex. Neuron. 2016;91:1228–43. doi: 10.1016/j.neuron.2016.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin X, Simmons SK, Guo A, Shetty AS, Ko M, Nguyen L, et al. In vivo Perturb-Seq reveals neuronal and glial abnormalities associated with autism risk genes. Science. 2020;370:eaaz6063. doi: 10.1126/science.aaz6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Replogle JM, Saunders RA, Pogson AN, Hussmann JA, Lenail A, Guna A, et al. Mapping information-rich genotype-phenotype landscapes with genome-scale Perturb-seq. Cell. 2022;185:2559–75.e28. doi: 10.1016/j.cell.2022.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bock C, Datlinger P, Chardon F, Coelho MA, Dong MB, Lawson KA, et al. High-content CRISPR screening. Nat Rev Methods Prim. 2022;2:1–23. doi: 10.1038/s43586-022-00098-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Youhanna S, Kemas AM, Preiss L, Zhou Y, Shen JX, Caka SD, et al. Organotypic and microphysiological human tissue models for drug discovery and development—current state-of-the-art and future perspectives. Pharm Rev. 2022;74:141–206. doi: 10.1124/pharmrev.120.000238. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka Y, Cakir B, Xiang Y, Sullivan GJ, Park I-H. Synthetic analyses of single-cell transcriptomes from multiple brain organoids and fetal brain. Cell Rep. 2020;30:1682–9.e3. doi: 10.1016/j.celrep.2020.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gordon A, Yoon SJ, Tran SS, Makinson CD, Park JY, Andersen J, et al. Long-term maturation of human cortical organoids matches key early postnatal transitions. Nat Neurosci. 2021;24:331–42. doi: 10.1038/s41593-021-00802-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–7. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trevino AE, Sinnott-Armstrong N, Andersen J, Yoon SJ, Huber N, Pritchard JK, et al. Chromatin accessibility dynamics in a model of human forebrain development. Science. 2020;367:eaay1645. doi: 10.1126/science.aay1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci USA. 2015;112:15672–7. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kanton S, Boyle MJ, He Z, Santel M, Weigert A, Sanchís-Calleja F, et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature. 2019;574:418–22. doi: 10.1038/s41586-019-1654-9. [DOI] [PubMed] [Google Scholar]
- 64.Bhaduri A, Andrews MG, Mancia Leon W, Jung D, Shin D, Allen D, et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature. 2020;578:142–8. doi: 10.1038/s41586-020-1962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pas SP. The rise of three-dimensional human brain cultures. Nature. 2018;553:437–45. doi: 10.1038/nature25032. [DOI] [PubMed] [Google Scholar]
- 66.Hammond TR, Robinton D, Stevens B. Microglia and the brain: complementary partners in development and disease. Annu Rev Cell Dev Biol. 2018;34:523–44. doi: 10.1146/annurev-cellbio-100616-060509. [DOI] [PubMed] [Google Scholar]
- 67.Ormel PR, Vieira de Sá R, van Bodegraven EJ, Karst H, Harschnitz O, Sneeboer MAM, et al. Microglia innately develop within cerebral organoids. Nat Commun. 2018;9:1–14. [DOI] [PMC free article] [PubMed]
- 68.Samudyata, Oliveira AO, Malwade S, Rufino de Sousa N, Goparaju SK, Gracias J, et al. SARS-CoV-2 promotes microglial synapse elimination in human brain organoids. Mol Psychiatry. 2022;2022:1–12. doi: 10.1038/s41380-022-01786-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu R, Boreland AJ, Li X, Erickson C, Jin M, Atkins C, et al. Developing human pluripotent stem cell-based cerebral organoids with a controllable microglia ratio for modeling brain development and pathology. Stem Cell Rep. 2021;16:1923–37. doi: 10.1016/j.stemcr.2021.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Popova G, Soliman SS, Kim CN, Keefe MG, Hennick KM, Jain S, et al. Human microglia states are conserved across experimental models and regulate neural stem cell responses in chimeric organoids. Cell Stem Cell. 2021;28:2153–66.e6. doi: 10.1016/j.stem.2021.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Birey F, Li MY, Gordon A, Thete MV, Valencia AM, Revah O, et al. Dissecting the molecular basis of human interneuron migration in forebrain assembloids from Timothy syndrome. Cell Stem Cell. 2022;29:248–64.e7. doi: 10.1016/j.stem.2021.11.011. [DOI] [PubMed] [Google Scholar]
- 72.Hagemann-Jensen M, Ziegenhain C, Chen P, Ramsköld D, Hendriks GJ, Larsson AJM, et al. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat Biotechnol. 2020;38:708–14. doi: 10.1038/s41587-020-0497-0. [DOI] [PubMed] [Google Scholar]
- 73.Specht H, Emmott E, Petelski AA, Huffman RG, Perlman DH, Serra M, et al. Single-cell proteomic and transcriptomic analysis of macrophage heterogeneity using SCoPE2. Genome Biol. 2021;22:50. doi: 10.1186/s13059-021-02267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schoof EM, Furtwängler B, Üresin N, Rapin N, Savickas S, Gentil C, et al. Quantitative single-cell proteomics as a tool to characterize cellular hierarchies. Nat Commun. 2021;12:1–15. doi: 10.1038/s41467-021-23667-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science. 2018;362:eaau5324. doi: 10.1126/science.aau5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Petukhov V, Xu RJ, Soldatov RA, Cadinu P, Khodosevich K, Moffitt JR, et al. Cell segmentation in imaging-based spatial transcriptomics. Nat Biotechnol. 2022;40:345–54. doi: 10.1038/s41587-021-01044-w. [DOI] [PubMed] [Google Scholar]
- 77.Clark IC, Gutiérrez-Vázquez C, Wheeler MA, Li Z, Rothhammer V, Linnerbauer M, et al. Barcoded viral tracing of single-cell interactions in central nervous system inflammation. Science. 2021;372:eabf1230. doi: 10.1126/science.abf1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, et al. The human cell atlas. Elife. 2017;6:e27041. doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Muñoz-Castañeda R, Zingg B, Matho KS, Chen X, Wang Q, Foster NN, et al. Cellular anatomy of the mouse primary motor cortex. Nature. 2021;598:159–66. doi: 10.1038/s41586-021-03970-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rajewsky N, Almouzni G, Gorski SA, Aerts S, Amit I, Bertero MG, et al. LifeTime and improving European healthcare through cell-based interceptive medicine. Nature. 2020;587:377–86. doi: 10.1038/s41586-020-2715-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yuste R, Hawrylycz M, Aalling N, Aguilar-Valles A, Arendt D, Arnedillo RA, et al. A community-based transcriptomics classification and nomenclature of neocortical cell types. Nat Neurosci. 2020;23:1456–68. doi: 10.1038/s41593-020-0685-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pfisterer U, Khodosevich K. Neuronal survival in the brain: neuron type-specific mechanisms. Cell Death Dis. 2017;8:e2643. doi: 10.1038/cddis.2017.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Korshunova I, Rhein S, García-González D, Stölting I, Pfisterer U, Barta A, et al. Genetic modification increases the survival and the neuroregenerative properties of transplanted neural stem cells. JCI Insight. 2020;5:e126268. doi: 10.1172/jci.insight.126268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chini M, Hanganu-Opatz IL. Prefrontal cortex development in health and disease: lessons from rodents and humans. Trends Neurosci. 2021;44:227–40. doi: 10.1016/j.tins.2020.10.017. [DOI] [PubMed] [Google Scholar]
- 85.Semple BD, Blomgren K, Gimlin K, Ferriero DM, Noble-Haeusslein LJ. Brain development in rodents and humans: identifying benchmarks of maturation and vulnerability to injury across species. Prog Neurobiol. 2013;106–107:1–16. doi: 10.1016/j.pneurobio.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]