Abstract
Surfactant proteins A (SP-A) and D (SP-D) are members of the collectin family of calcium-dependent lectins and are important pulmonary host defense molecules. Human SP-A and SP-D and rat SP-D bind to Aspergillus fumigatus conidia, but the ligand remains unidentified. To identify a fungal ligand for SP-A and/or SP-D, we examined the interactions of the proteins with Saccharomyces cerevisiae. SP-D but not SP-A bound yeast cells, and EDTA inhibited the binding. SP-D also aggregated yeast cells and isolated yeast cell walls. Treating yeast cells to remove cell wall mannoprotein did not reduce SP-D binding, and SP-D failed to aggregate chitin. However, SP-D aggregated yeast glucan before and after treatment with a β(1→3)-glucanase, suggesting a specific interaction between the collectin and β(1→6)-glucan. In support of this idea, SP-D-induced yeast aggregation was strongly inhibited by pustulan [a β(1→6)-linked glucose homopolymer] but was not inhibited by laminarin [a β(1→3)-linked glucose homopolymer]. Additionally, pustulan but not laminarin strongly inhibited SP-D binding to A. fumigatus. The pustulan concentration for 50% inhibition of SP-D binding to A. fumigatus is 1.0 ± 0.3 μM glucose equivalents. Finally, SP-D showed reduced binding to the β(1→6)-glucan-deficient kre6 yeast mutant. Taken together, these observations demonstrate that β(1→6)-glucan is an important fungal ligand for SP-D and that glycosidic bond patterns alone can determine if an extended carbohydrate polymer is recognized by SP-D.
Pulmonary surfactant is a complex mixture of lipids and proteins. It is well known that surfactant lowers the surface tension at the air-liquid interface in the lung. Recent studies also support a host defense role for surfactant and particularly for pulmonary surfactant proteins A (SP-A) and D (SP-D).
SP-A and SP-D are produced by type II cells and Clara cells in the lung and are members of the C-type lectin protein superfamily. SP-A and SP-D share many structural features. Both proteins are composed of a short N-terminal region involved in covalent cross-linking, followed by a collagen-like domain, a neck region, and a C-terminal carbohydrate recognition domain (CRD) that binds carbohydrates in a calcium-dependent manner (9, 18, 29). Both proteins form higher-order structures but differ in the organization of these structures. SP-D predominantly forms a cruciform-like dodecamer, whereas SP-A forms a bouquet-like octadecamer (18). Although the proteins are very similar, important functional differences exist. For example, SP-A but not SP-D specifically binds phosphatidylcholine and dipalmitoylphosphatidylcholine, whereas SP-D but not SP-A binds phosphatidylinositol (17, 26).
SP-A and SP-D are thought to be important components of the innate immune system (5, 24, 30, 36), and recent animal studies have demonstrated host defense roles for these proteins. For example, SP-A-deficient mice are more susceptible to intratracheally instilled group B streptococci (20), Pseudomonas aeruginosa (22), and respiratory syncytial virus (21) than wild-type animals. Moreover, intranasally administered SP-D reduced respiratory syncytial virus replication in the lungs of infected mice (12). In many cases it is thought that SP-A and SP-D mediate their host defense roles by binding carbohydrates on the surface of pathogenic microorganisms, but the precise polysaccharide structures recognized by the proteins have not been determined. Additionally, although the monosaccharide specificity of SP-A and SP-D has been examined in detail (11, 29), very little is known about how the proteins interact with other carbohydrates such as long-chain polysaccharides present on the surface of many microorganisms.
Recent work has shown that human SP-A and SP-D and rat SP-D bind Aspergillus fumigatus conidia (1, 23). Inhibitor studies and use of mutant surfactant proteins led to the conclusion that the proteins bind to surface carbohydrate structures on the conidia, but the surfactant protein ligand(s) was not identified. The purpose of the present study was to identify SP-A and/or SP-D fungal ligands. This is important since elucidation of the ligand structures recognized by these proteins is critical to our understanding of their in vivo host defense functions. Additionally, these investigations will broaden our understanding of carbohydrate recognition by SP-A and SP-D.
Since A. fumigatus conidia are difficult to disrupt and the organism is not well defined genetically, we used Saccharomyces cerevisiae as a model fungus. This yeast is well characterized genetically and biochemically and is easily manipulated in the laboratory. Since cell wall composition is common in many fungi including S. cerevisiae (3, 6, 8, 15), we feel that the knowledge gained from this work will be directly applicable to other fungi, including important pulmonary pathogens.
MATERIALS AND METHODS
Materials.
Crab shell chitin, yeast glucan, and phenylmethylsulfonyl fluoride were purchased from Sigma (St. Louis, Mo.). Zymolyase 100T was purchased from U.S. Biological (Swampscott, Mass.), peptide N-glycosidase F (PNGase F) was purchased from New England Biolabs (Beverly, Mass.), pustulan was purchased from Calbiochem (San Diego, Calif.), and laminarin was purchased from Fluka (Buchs, Switzerland). The average molecular weight for pustulan was 20,000, and that for for laminarin was 8,600 (unpublished data). The S. cerevisiae kre6Δ::G418r mutant (Research Genetics strain 5574) and parental (Research Genetics strain Hansen BY 4741) strains were obtained from Research Genetics (Huntsville, Ala.).
Preparation of S. cerevisiae.
Unless otherwise indicated yeast strain SEY 6210 (MATα leu2 α ura3 his3 lys2 trp1 suc2) was used throughout this study. When other strains were used, the cells were prepared in the same manner as SEY 6210. The cells were grown in YEPD (1% yeast extract, 2% peptone, 2% glucose) at 30°C in a shaking incubator. The cells were harvested in log phase, washed three times with phosphate-buffered saline, pH 7.4 (PBS), and fixed with 2% paraformaldehyde in PBS for approximately 16 h at room temperature. Following fixation, the cells were again washed with PBS, counted with a hemacytometer, and stored at 4°C in PBS with 0.02% NaN3 until used. For the hydrofluoric acid (HF) treatment, the fixed cells were suspended in 48% HF and incubated on ice. After 48 h, the acid-treated cells were washed five times with PBS (sedimented at 100 × g) to remove the HF and liberated cellular material, counted with a hemacytometer, and stored at 4°C in PBS with 0.02% NaN3 until used. Protein deglycosylation with PNGase F was performed according to the manufacturer's instructions. An aliquot of 120 million cells was treated with 13 μl of PNGase F for 2 h at 37°C. The final reaction volume was 720 μl. Following deglycosylation, the cells were washed with PBS and stored at 4°C in PBS with 0.02% NaN3 until needed.
Preparation of S. cerevisiae cell walls.
Yeast cell walls were prepared essentially as described elsewhere (4). Yeast cells were disrupted with a Bead Beater (10 pulses for 30 s each) using 0.5-mm-diameter glass beads at 4°C in 1 mM phenylmethylsulfonyl fluoride. Following disruption, the cell walls were collected and then washed twice with ethanol, three times with chloroform-methanol (1:1), three times with ethanol-ether (1:1), and finally three times with water. For all washes, the cell walls were collected by centrifugation at 3,000 × g at 4°C.
Preparation of SP-A and SP-D.
The purification of the surfactant proteins used in this study has been described previously (1). Briefly, recombinant human SP-D was purified from the culture medium of CHO-K1 cells expressing human SP-D, and AP human SP-D was purified from the bronchoalveolar lavage fluid of alveolar proteinosis patients by mannose-Sepharose affinity chromatography followed by elution with MnCl2 (33) and inositol. AP human SP-A was purified from the bronchoalveolar lavage fluid of alveolar proteinosis patients, and normal human SP-A was purified from the lavage fluid of a human lung not used for transplant as previously described (1). All proteins were judged pure by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (19), Coomassie blue staining, and Western blotting.
SP-A and SP-D binding to A. fumigatus and S. cerevisiae.
A. fumigatus binding by SP-D was performed exactly as described previously (1). Yeast binding was carried out in calcium binding buffer (CBB); 130 mM NaCl, 13 mM NaN3, 5 mM KCl, 3 mM sodium phosphate buffer, 10 mM HEPES, 2 mM CaCl2, 1 mM MgSO4 [pH 7.4] containing 1% heat-inactivated and dialyzed fetal bovine serum. For binding reactions, 4 × 106 yeast cells were suspended in CBB (or CBB containing 10 mM EDTA or carbohydrate inhibitor for inhibition experiments) and incubated with 20 μg of the appropriate surfactant protein per ml at 25°C for 1 h. The total volume was 100 μl. The cells were then washed three times with CBB and incubated with 10 μg rabbit polyclonal anti-human SP-A or anti-human SP-D immunoglobulin G (IgG) per ml at 25°C for 1 h. The cells were again washed three times with CBB and incubated with fluorescein isothiocyanate-conjugated F(ab′)2 fragment of donkey anti-rabbit IgG (10 μg/ml; Jackson ImmunoResearch Laboratories, West Grove, Pa.) at 25°C for 1 h. The cells were then washed twice with CBB. Control samples included (i) cells prepared as described above but without added SP-D (negative control) to determine background fluorescence and (ii) cells with SP-D but without inhibitor (positive control). Fluorescein isothiocyanate fluorescence was analyzed using a Becton Dickinson FACSCalibur flow cytometer and CELLQuest software. Binding was determined by subtracting the mean channel fluorescence of the negative control from the sample mean channel fluorescence. Binding is expressed as a percentage of the positive control binding.
Aggregation analysis.
For aggregation analysis, the yeast cells, cell walls, glucan, or zymolyase-treated glucan was suspended in CBB (or CBB containing the appropriate inhibitor) and diluted to the desired absorbance at 700 nm (A700). For a given experiment, the difference in starting A700 of all samples analyzed was generally less than 3%. SP-D or buffer was added to the appropriate samples after 5 min. The final volume for all samples tested was 800 μl. The A700 of the samples was measured every minute for 2 h after protein addition (except for glucan aggregation analysis, in which case the measurements were only recorded for 18 min after protein addition). Aggregation is indicated by a drop in A700 greater than the negative control (without added protein) as the aggregated material sediments to the bottom of the assay tube. Following analysis, the starting A700s were normalized to the negative control tube for ease of data interpretation.
Preparation of pustulan and laminarin.
Pustulan stock solutions were prepared by boiling the 20 mM glucose equivalents in water for 5 min. The stock solution was then diluted to the desired concentration in CBB. Laminarin solutions were prepared by directly dissolving solid laminarin in CBB at room temperature.
Zymolyase treatment of yeast glucan.
Yeast glucan (10 mg) was suspended in 10 mM Tris (pH 7.4) and digested with 8 mg of zymolyase [a β(1→3)-glucan digesting enzyme] for 16 h at 37°C. The total reaction volume was 3 ml. The digested material was dialyzed against H2O, washed with CBB, and subjected to aggregation analysis as described above.
Other methods.
All protein concentrations were determined by the bicinchoninic acid method (Pierce, Rockford, Ill.) with bovine serum albumin as the standard. Polyclonal anti-human SP-D was raised in rabbits against recombinant human SP-D produced by CHO-K1 cells. Polyclonal anti-human SP-A was raised in rabbits against human SP-A purified from the lavage fluid of alveolar proteinosis patients.
Statistical analysis.
Data are reported as means ± standard errors. Data were compared by Student's t test. P values of <0.05 were considered significant.
RESULTS
S. cerevisiae binding and aggregation.
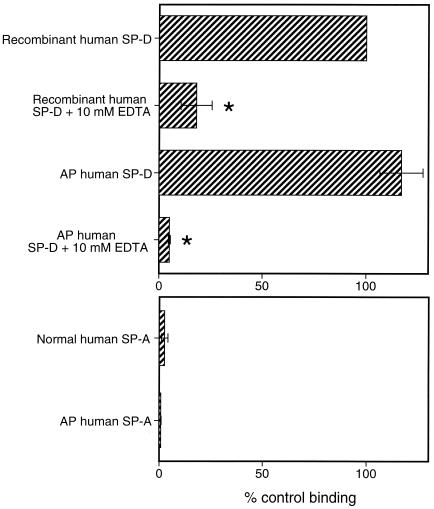
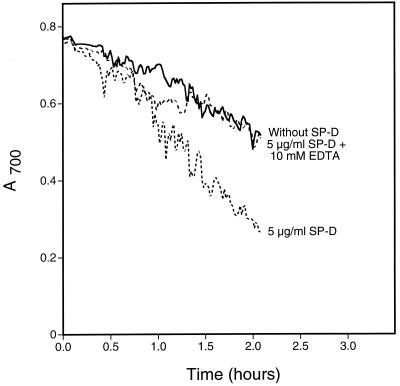
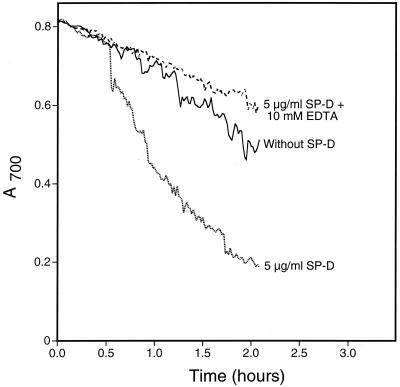
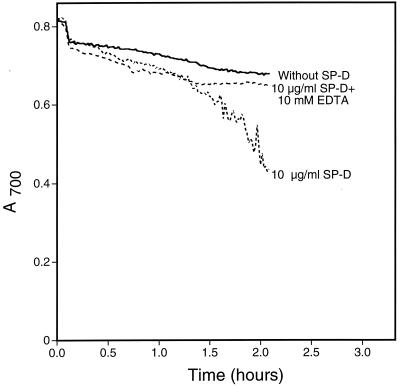
As an initial step toward identifying a fungal ligand for SP-A and/or SP-D, we first examined if either protein bound the well-characterized yeast S. cerevisiae. We performed binding experiments using proteins from various sources (Fig. 1). As can be seen, SP-D but not SP-A bound the cells and EDTA inhibited the binding. Since SP-D is known to aggregate microorganisms including Escherichia coli (16) and A. fumigatus (23), we also tested for SP-D-induced S. cerevisiae aggregation by measuring changes in A700 (Fig. 2). As shown, SP-D aggregated the cells and EDTA inhibited the aggregation. Similar results were seen when SP-D was tested for its ability to aggregate yeast cell walls (Fig. 3), suggesting that a SP-D ligand was cell wall associated. This provided a convenient method for rapidly evaluating the interaction of SP-D with the cells under a variety of conditions and to determine if the protein recognized insoluble cell wall material such as chitin or glucan (see below).
FIG. 1.
SP-A and SP-D binding to S. cerevisiae. Aliquots of 4 × 106 yeast cells were incubated with 20 μg of SP-A or SP-D per ml for 1 h at 25°C, followed by washing and similar incubations with primary and secondary antibodies. To determine background fluorescence, samples that contained cells and primary and secondary antibodies but not SP-A or SP-D were also included. For inhibition studies, the proteins were preincubated with EDTA for 15 min at 25°C, and the EDTA-surfactant protein mixture was then added to the cells. Binding was detected by flow cytometry and normalized to the mean fluorescence intensity of recombinant human SP-D (taken as 100% binding). Data represent the average ± standard error of three independent experiments (∗, P < 0.05 compared to binding without EDTA).
FIG. 2.
S. cerevisiae aggregation by SP-D. Yeast cells were suspended in calcium-containing buffer with or without 10 mM EDTA at room temperature. After 5 min, recombinant human SP-D was added to a final concentration of 5 μg/ml. Buffer was added to the negative control sample. The A700 of the suspensions was monitored every minute for 2 h after protein addition. For the graphs shown, the starting A700 for all samples was normalized to the buffer control for ease of interpretation. The graph shows representative data from duplicate experiments.
FIG. 3.
S. cerevisiae cell wall aggregation by SP-D. Yeast cell walls were suspended in calcium-containing buffer with or without 10 mM EDTA at room temperature. After 5 min, recombinant human SP-D was added to a final concentration of 5 μg/ml. Buffer was added to the negative control sample. The A700 of the suspensions was monitored every minute for 2 h after protein addition. For the graphs shown, the starting A700 for all samples was normalized to the buffer control for ease of interpretation. The graph shows representative data from duplicate experiments.
Mannan is a major constituent of the yeast cell wall and is a known inhibitor of SP-D binding to carbohydrate structures (12). We therefore considered it a likely target for SP-D binding to S. cerevisiae. To test this idea, we treated the cells with HF to remove the cell wall mannoprotein (15) and PNGase F to remove mannan attached to cell wall proteins. If SP-D bound only yeast mannan, we expected that these treatments would reduce or eliminate the observed binding. These treatments, however, did not reduce binding by SP-D (not shown). This finding demonstrates that SP-D ligands other than mannan are present on the yeast cell but does not exclude the possibility that SP-D recognizes mannan on the yeast cell surface.
Interactions with chitin and glucan.
In addition to mannoprotein, the yeast cell wall also contains chitin and glucan (3, 15). Chitin is a polymer of β(1→4)-linked N-acetylglucosamine subunits. When tested directly, SP-D failed to aggregate chitin (not shown). Therefore, this polymer is not likely to be a yeast ligand for SP-D. This was not unexpected since SP-D binds N-acetylglucosamine only very weakly (29), and as discussed in detail below, we feel that the nature of the linkages connecting the polymer subunits prohibits chitin recognition by SP-D.
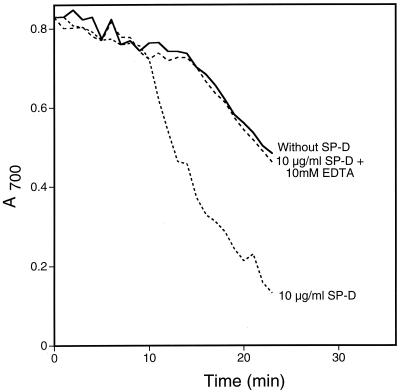
Next we tested yeast glucan in the aggregation assay (Fig. 4). Glucan is a glucose polymer with subunits connected by the indicated linkages. The material tested contained both β(1→3)- and β(1→6)-glucan. SP-D aggregated glucan, and EDTA inhibited the aggregation. The time required to demonstrate aggregation was significantly less than for yeast cells due to the rapid sedimentation of glucan even in the absence of SP-D. However, significant aggregation was apparent not only by spectrophotometric analysis but also by visual inspection (not shown). Finally, we tested both chitin and glucan for binding by SP-D using a depletion assay. Solutions containing chitin or glucan (50 mg/ml) and SP-D (5 μg/ml) were incubated for 2 h at 25°C. The bound SP-D was separated from unbound by centrifugation. No SP-D was lost from the centrifuged supernatant of the chitin solution, whereas approximately 80% of the added SP-D was lost from the supernatant of the glucan solution (not shown).
FIG. 4.
Glucan aggregation by SP-D. Glucan powder was suspended in calcium-containing buffer with or without 10 mM EDTA at room temperature. After 5 min, recombinant human SP-D was added to a final concentration of 10 μg/ml. Buffer was added to the negative control sample. The A700 of the suspensions was monitored every minute for 18 min after protein addition. For the graphs shown, the starting A700 for all samples was normalized to the buffer control for ease of interpretation. The graph shows representative data from duplicate experiments.
After establishing that SP-D bound and aggregated glucan, we wanted to determine if a specific glucan component was recognized. Therefore, we treated glucan with zymolyase to remove β(1→3)-glucan and tested the remaining material for aggregation by SP-D. The results are shown in Fig. 5. SP-D aggregated zymolyase-treated yeast glucan, indicating that β(1→6)-glucan was recognized by SP-D. However, incomplete removal of β(1→3)-glucan or structures other than β(1→3)- and β(1→6)-glucan present in the starting material made it possible that ligands other than β(1→6)-glucan were being recognized by SP-D. It is also noteworthy that zymolyase treatment decreased the sedimentation rate for glucan, which accounts for the difference in aggregation rates in Fig. 4 and 5.
FIG. 5.
Aggregation of zymolyase-treated yeast glucan by SP-D. Insoluble material remaining after zymolyase digestion of yeast glucan was suspended in calcium-containing buffer with or without 10 mM EDTA at room temperature. After 5 min, recombinant human SP-D was added to a final concentrations of 10 μg/ml. Buffer was added to the negative control sample. The A700 of the suspensions was monitored every minute for 2 h after protein addition. For the graphs shown, the starting A700 for all samples was normalized to the buffer control for ease of interpretation. The graph shows representative data from duplicate experiments.
Inhibition of SP-D induced S. cerevisiae aggregation and A. fumigatus binding.
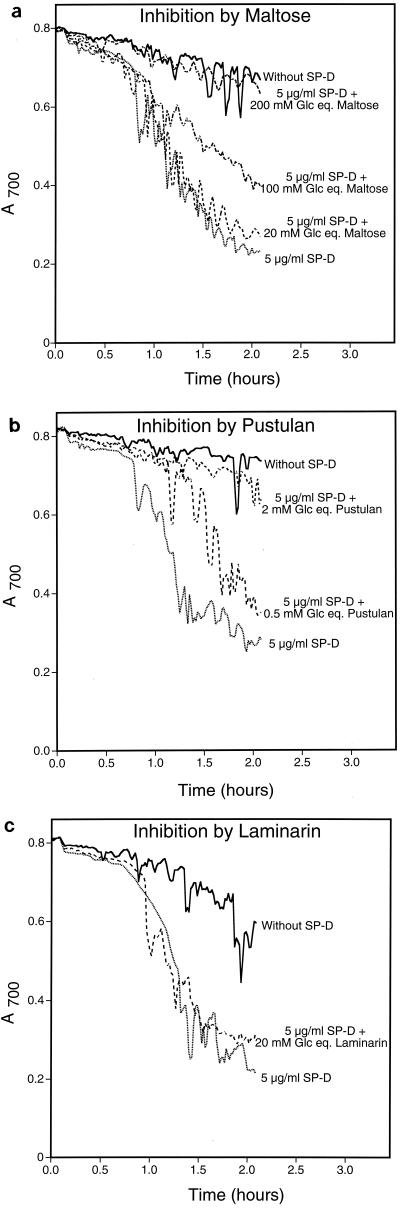
The results above demonstrate that SP-D recognizes components of the yeast cell wall. To test the hypothesis that SP-D specifically recognized β(1→6)-glucan, we examined pustulan and laminarin for the ability to inhibit SP-D-induced yeast aggregation (Fig. 6). Pustulan is a soluble glucose homopolymer linked via β(1→6) glycosidic bonds that should mimic yeast β(1→6)-glucan. Laminarin is a soluble glucose homopolymer linked via β(1→3) glycosidic bonds that should mimic yeast β(1→3)-glucan. For comparison we also tested maltose, a known carbohydrate-based SP-D inhibitor, for its ability to inhibit aggregation (Fig. 6). We have reported the inhibitor concentrations as glucose equivalents because the polymorphic nature of the long-chain carbohydrates makes direct molar comparisons less accurate. As expected, maltose inhibited SP-D induced yeast aggregation in a concentration-dependent manner (Fig. 6a). Essentially no inhibition was seen at 20 mM glucose equivalents, but complete inhibition was seen at 200 mM. By comparison, pustulan was a very strong inhibitor of the aggregation. Figure 6b shows that 0.5 mM glucose equivalents of pustulan partially inhibited yeast aggregation and 2 mM glucose equivalents almost completely inhibited the aggregation. Clearly, pustulan is a much more powerful inhibitor of SP-D-induced yeast aggregation than maltose. Finally, we tested laminarin for its ability to inhibit yeast aggregation. As shown in Fig. 6c, laminarin failed to inhibit aggregation at 20 mM glucose equivalents. Due to the experimental design and the limited solubility of laminarin, higher concentrations of this polysaccharide could not be tested in this assay. It is also noteworthy that in separate experiments we demonstrated that these carbohydrate inhibitors did not alter the sedimentation of yeast cells in the absence of SP-D (not shown).
FIG. 6.
Inhibition of SP-D-induced yeast aggregation. Yeast cells were suspended in calcium-containing buffer with or without carbohydrate inhibitor at room temperature. After 5 min, recombinant human SP-D was added to all samples except the negative control (final SP-D concentration was 5 μg/ml). Buffer was added to the negative control sample. The A700 of the suspensions was monitored every minute for 2 h after protein addition. For the graphs shown, the starting A700 for all samples was normalized to the buffer control for ease of interpretation. Maltose, pustulan, and laminarin concentrations are reported as glucose equivalents (Glc eq.). The graph shows representative data from duplicate experiments.
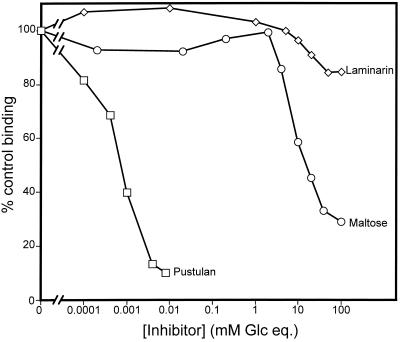
We next wanted to determine the 50% inhibitory concentrations (IC50s) for pustulan and laminarin. We first attempted to use these carbohydrates to inhibit SP-D binding to S. cerevisiae. We found that laminarin failed to inhibit SP-D binding to S. cerevisiae even when used at 100 mM glucose equivalents (not shown). However, although nearly 50% inhibition of SP-D binding to S. cerevisiae was seen at 10 μM glucose equivalents of pustulan, we were unable to determine the IC50 using our detection system. The most likely explanation for this observation lies in the fact that lectins are known to cause aggregation and precipitation of multivalent carbohydrate ligands (28). Thus, if SP-D bound and caused precipitation of pustulan, the aggregated material would be intensely stained for the lectin in our assay. These aggregates either could be mistaken for yeast cells when analyzed by flow cytometry or could adhere to the yeast cell surface. In either case, these stained aggregates could prevent accurate IC50 determinations. Indeed, small, intensely stained, amorphous aggregates were seen when yeast cells that had been subjected to pustulan IC50 determinations were examined microscopically (not shown). These aggregates were seen both free in solution and adhering to the yeast cell surface, and their numbers increased with increasing pustulan concentration. Thus, we conclude that SP-D aggregates pustulan. These aggregates prevented us from determining the IC50 for pustulan inhibition of yeast binding by SP-D.
We also tested pustulan and laminarin for the ability to inhibit SP-D binding to A. fumigatus. Although pustulan aggregates were also seen in these samples, they did not prevent IC50 determinations (Fig. 7) probably because the aggregates do not adhere as well to the A. fumigatus conidia as to the yeast surface. Pustulan was found to be an extremely powerful inhibitor of SP-D binding to A. fumigatus, with an IC50 of (1 ± 3) × 10−3 mM glucose equivalents. In contrast, maltose inhibited the binding weakly (IC50 of [1.9 ± 0.2] × 101 mM) and lamarin failed to inhibit binding (IC50 of >1.0 × 102 mM).
FIG. 7.
Inhibition of SP-D binding to A. fumigatus. Aliquots of 2 × 106 A. fumigatus conidia were incubated with 20 μg of recombinant human SP-D per ml for 1 h at 25°C, followed by washing and similar incubations with primary and secondary antibodies. To determine background fluorescence, samples that contained conidia and primary and secondary antibodies but not SP-D were also included. For inhibition studies, the proteins were preincubated with the indicated carbohydrate for 15 min at 25°C, and the carbohydrate-surfactant protein mixture was then added to the conidia. Binding was detected by flow cytometry and normalized to the mean fluorescence intensity of recombinant human SP-D without carbohydrate (taken as 100% binding). The graph shows representative data from three experiments for each inhibitor.
The inhibition of aggregation and binding data presented above clearly demonstrate that SP-D specifically recognized the glucose polymer linked via β(1→6) glycosidic bonds but failed to recognize the β(1→3)-linked polymer. This finding supports our hypothesis that SP-D binds β(1→6)-glucan on the yeast cell surface.
SP-D binding to β(1→6)-glucan-deficient yeast mutants.
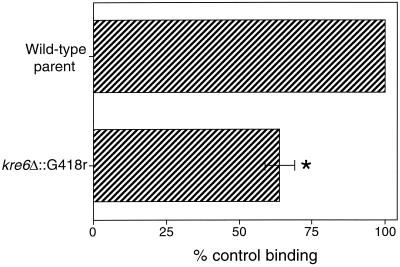
To further investigate our hypothesis that β(1→6)-glucan is a ligand for SP-D, we tested the kre6 (2, 31, 32) yeast mutant in our binding assay. Deletion of the KRE6 gene has been shown to cause a 50% reduction in the amount of β(1→6)-glucan in yeast cell walls without alterations in the size or structure of the polymer (31). Additional analysis has shown that deletion of the KRE6 gene does not alter the amount of cell wall mannoprotein (31), chitin (31), or β(1→3)-glucan (32), making it an ideal mutant to test in our binding assay. If SP-D bound β(1→6)-glucan on intact yeast cells, we expected to see reduced binding to this mutant. As shown in Fig. 8, SP-D showed reduced binding to the kre6 mutant yeast compared to the wild-type parent strain. The observed 35% reduction is consistent with the conclusion that SP-D binds yeast β(1→6)-glucan. We also tested kre6 skn1 and kre5 mutant yeast for SP-D binding. These mutants have essentially no β(1→6)-glucan in their cell walls (25, 32). SP-D bound these cells, but significantly less aggregation was seen for the mutants than the wild-type parents (not shown), suggesting a defective interaction between the lectin and the mutant yeast cells.
FIG. 8.
SP-D binding to kre6 mutant S. cerevisiae. Aliquots of 4 × 106 yeast cells were incubated with 20 μg of recombinant human SP-D per ml for 1 h at 25°C, followed by washing and similar incubations with primary and secondary antibodies. Binding was detected by flow cytometry and normalized to the mean fluorescence intensity of the untreated control (taken as 100% binding). Data represent the average ± standard error of three independent experiments (∗, P < 0.05 compared to wild-type parent cells).
DISCUSSION
SP-A and SP-D are important pulmonary host defense molecules. Human SP-A and SP-D and rat SP-D have been shown to bind the fungus A. fumigatus (1, 23). These studies indicated that the fungal ligand was likely carbohydrate based, but the specific target molecule was not identified. More recently, van Rozendaal et al. (34) have demonstrated that SP-D binds Candida albicans and inhibits cell growth by aggregating the organism. C. albicans binding by SP-D was inhibited by EDTA and competing sugars, suggesting that SP-D recognizes carbohydrate structures on these cells as well. To study fungal recognition by SP-A and SP-D further, we examined the interactions of the proteins with the well-characterized yeast S. cerevisiae. Human SP-D but not SP-A bound yeast cells in a manner that was inhibited by EDTA. In the present study, we used 20 μg of SP-A and SP-D per ml for our binding studies. Similar concentrations were used in a previous study examining binding to A. fumigatus (1). Estimates of the in vivo SP-D concentration in rats range from 36 to 216 μg/ml (36). SP-A concentrations are approximately 10 times that of SP-D, but most of the SP-A is lipid associated (36). We feel that the conditions tested are relevant to the physiologic surfactant protein concentrations that may be encountered in the lung.
SP-D also aggregated yeast cells. Since functional differences have been noted between human and rat SP-A (1), we also tested rat SP-A and SP-D for yeast binding (not shown). The results were similar to those shown for the human proteins; no functional differences were seen between human and rat SP-A or SP-D in interactions with S. cerevisiae.
We were originally surprised that SP-A failed to bind S. cerevisiae since the protein is known to bind yeast mannan (11), a component of the yeast cell wall. We speculate that charge repulsion may contribute to the failure of SP-A to bind the yeast cells. Yeast cells are known to carry a negative charge due to phosphate groups present in their cell wall mannan (13, 14), and SP-A would also carry a negative charge under test conditions (pH 7.4). Alternatively, the specific mannan conformation on the yeast cell surface may not allow SP-A binding, whereas different polysaccharide conformations present in the isolated mannan allow binding. No charge repulsion problem exists for SP-D. In fact, Håkansson et al. (10) have noted that SP-D carries a local positive charge in the cavity formed at the junction of its CRDs, and they have proposed that this may assist in recognition of negatively charged ligands.
After noting that SP-D also aggregated yeast cell walls (Fig. 3), we focused our ligand identification efforts on this cellular component. The cell wall is the outermost part of the cell and would therefore be more accessible to SP-D than underlying structures. The yeast cell wall is composed of mannoprotein, chitin, β(1→3)-glucan and β(1→6)-glucan (3, 15). Since yeast mannan is known to inhibit SP-D recognition of carbohydrate structures (12), it seemed likely that mannoproteins were ligands for SP-D. To test this idea, we treated the cells with HF to remove cell wall mannoproteins and PNGase F to remove cell wall mannan. Unexpectedly, these treatments did not reduce SP-D binding (not shown). While these data do not exclude the possibility that SP-D binds mannoprotein structures on the yeast cell wall, they do demonstrate that mannan is not the only SP-D ligand present.
Since SP-D failed to aggregate chitin, we next determined if SP-D interacted with glucan. Figure 4 shows that SP-D aggregated yeast glucan and EDTA inhibited the aggregation. Yeast glucan is a mixture of β(1→6)- and β(1→3)-linked glucose subunits. The fact that SP-D aggregated glucan demonstrated that the protein recognized this cell wall component. However, since both β(1→3)-and β(1→6)-linked polymers were present in the glucan preparation tested, we continued our investigations into SP-D ligand specificity. We next treated glucan with the β(1→3)-glucan-digesting enzyme zymolyase. SP-D aggregated zymolyase-treated glucan (Fig. 5), which suggested a direct interaction between the protein and β(1→6)-glucan.
To further confirm β(1→6)-glucan-specific recognition by SP-D, we tested pustulan and laminarin for the ability to inhibit SP-D-induced S. cerevisiae aggregation and A. fumigatus binding. If SP-D specifically recognized glucose polymers linked by β(1→6) glycosidic bonds, we expected that pustulan but not laminarin would strongly inhibit aggregation and binding. The data presented in Fig. 6 and 7 in Results confirm our expectations. Although it cannot be formally excluded that minor contaminants in the commercial pustulan preparation contribute to the observed inhibition, we consider this possibility highly unlikely. First, the pustulan conforms to the expected structure by nuclear magnetic resonance analysis (from the supplier), and second, dialyzed pustulan inhibited SP-D-induced yeast aggregation to the same extent as pustulan that was not dialyzed (not shown). For the experiment shown in Fig. 7, significant inhibition was found at concentrations as low as 0.1 μM glucose equivalents. Thus, any minor contaminant would have to be extraordinarily potent to be effective at such concentrations. We feel the most reasonable explanation for the collective data is that pustulan but not laminarin is a powerful SP-D inhibitor.
The fact that laminarin failed to inhibit SP-D-induced aggregation, while pustulan strongly inhibited the aggregation, is not unexpected considering existing structural knowledge: SP-D and mannose-binding protein A (MBP-A) are both C-type lectins and are highly homologous (the CRDs of human SP-D and rat MBP-A are 45% identical). The structures of rat MBP-A CRD in a complex with a carbohydrate (35) and SP-D (10) are known, and the structures of the CRDs of these two proteins are very similar. When the αC atoms of the SP-D CRD are superimposed with the MBP-A CRD, the root mean square deviation is 0.7 to 0.8 Å (10). Closer examination reveals that the region defining the carbohydrate-binding pocket in MBP-A is very similar to the corresponding region in SP-D (amino acids Glu 185 to Asp206 for MBP-A and amino acids Glu321 to Asp342 in SP-D). SP-D and MBP-A share 15 of 22 amino acids in this region, including all residues identified as critical for carbohydrate and calcium binding (Glu 185, Asn187, Glu 193, Asn205, and Asp206, using the MBP-A numbering scheme). The MBP-A and SP-D structures are also very similar in this region. The structure of MBP-A complexed with a carbohydrate revealed that the protein binds polysaccharides via interactions with hydroxyl groups at the 3 and 4 positions on the sugar ring and explained why the protein bound mannose and glucose but failed to bind galactose (35). A parallel mutagenesis study confirmed that by changing amino acids Glu 185 and Asn 187 to Gln and Asp respectively, the protein's monosaccharide affinity could be changed from mannose/glucose > galactose to galactose > mannose/glucose (7). Analogous mutagenesis work has been done with SP-D, which also shows higher affinity for glucose than for galactose (29), with similar results (27). Together, the carbohydrate recognition specificity, mutagenesis results, and structural similarity strongly suggests that SP-D binds carbohydrates by a mechanism similar to that used by MBP-A.
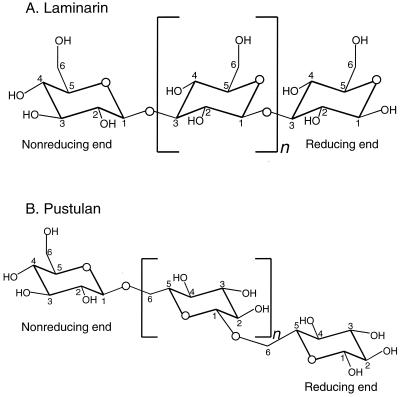
As stated above, MBP-A binds saccharides via interactions with the hydroxyl groups at positions 3 and 4 on the sugar. Assuming that SP-D interacts with the same positions on the sugar unit, the protein would be expected to only bind the nonreducing terminal glucose unit of a β(1→3)-linked glucosyl polysaccharide. This is because the 3 position on all other glucose units is involved in a glycosidic bond and is not available for interaction with the protein (Fig. 9 is presented for reference). This would be the case for laminarin and explains why this polysaccharide failed to inhibit SP-D-induced yeast aggregation or binding to A. fumigatus. However, in a β(1→6)-linked glucosyl polysaccharide, the hydroxyl groups at the 3 and 4 positions are available for interactions with the protein on every sugar unit (Fig. 9). This would be the case for pustulan and explains why this carbohydrate strongly inhibited SP-D-induced yeast aggregation and binding to A. fumigatus.
FIG. 9.
Glucosyl polysaccharides laminarin and pustulan. The carbon atom numbering schemes for each are indicated.
The preceding observations not only explain the inhibition by laminarin and pustulan but also support our conclusion that β(1→6)-glucan is a fungal ligand for SP-D. SP-D does not bind β(1→3)-glucan since the 3 position on all glucose units is unavailable for interactions with the protein. Similarly, SP-D does not bind chitin since the 4 position is unavailable for interactions with the protein in that polymer. However, both the 3 and 4 positions are available for interactions with SP-D on all glucose subunits on β(1→6)-glucan. Therefore, based on structural and experimental considerations, we feel that β(1→6)-glucan is an ideal ligand for SP-D. However, as stated previously, we cannot exclude the possibility that SP-D also binds mannoprotein structures on the yeast cell surface.
To provide additional support for our hypothesis that β(1→6)-glucan is a fungal ligand for SP-D, we tested the kre6 yeast mutant in our binding assay (Fig. 8). The cell wall of this mutant has approximately 50% less β(1→6)-glucan than the wild-type cell wall without alterations in mannoprotein or chitin (31). Additionally, no alteration in the amount of β(1→3)-glucan was found in the wall of this mutant (32). As expected, SP-D showed reduced binding to the kre6 mutant compared to the wild type, leading us to conclude that β(1→6)-glucan is a fungal ligand for SP-D. The fact that SP-D bound the kre6 skn1 and kre5 mutants (not shown) suggests that other SP-D ligands are present on the surface of these cells. These ligands may include yeast mannan. Additionally, it is possible that the mutants incorporate into their walls polymers not found in wild-type cells that also serve as ligands for SP-D.
In summary, we have shown that SP-D but not SP-A binds S. cerevisiae and that β(1→6)-glucan is a fungal ligand for SP-D. Since many fungi have similar cell wall compositions, we expect that these observations will be general to SP-D interactions with other fungi including Candida and Aspergillus. Additionally, our inhibitor data for pustulan and laminarin demonstrate that only certain carbohydrate conformers are recognized by SP-D and that bond patterns alone, regardless of subunit composition, can determine if a polysaccharide is recognized by SP-D. We are currently using computational modeling to explore this area further.
ACKNOWLEGMENTS
We thank Erika Crouch for providing CHO-K1 cells expressing human SP-D, Howard Bussey for providing kre6, skn1, and kre5 yeast mutants, Amanda Evans for excellent technical assistance, and Wen-I Wu and Mark Schumacher for helpful discussions.
This work was supported by grants from the National Institutes of Health (HL-29891, HL-45286) and Environmental Protection Agency (R825702). M.J.A. was funded by a Great West Life Assurance Fellowship at National Jewish. This work was performed in the Lord and Taylor Laboratory for Lung Biochemistry at National Jewish Medical and Research Center.
REFERENCES
- 1.Allen M J, Harbeck R, Smith B, Voelker D R, Mason R J. Binding of rat and human surfactant proteins A and D to Aspergillus fumigatus conidia. Infect Immun. 1999;67:4563–4569. doi: 10.1128/iai.67.9.4563-4569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boone C, Sommer S S, Hensel A, Bussey H. Yeast KRE genes provide evidence for a pathway of cell wall beta-glucan assembly. J Cell Biol. 1990;110:1833–1843. doi: 10.1083/jcb.110.5.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cabib E, Roberts R, Bowers B. Synthesis of the yeast cell wall and its regulation. Annu Rev Biochem. 1982;51:763–793. doi: 10.1146/annurev.bi.51.070182.003555. [DOI] [PubMed] [Google Scholar]
- 4.Cambell I, Duffus J H. Yeast. Oxford, United Kingdom: IRL Press Limited; 1988. [Google Scholar]
- 5.Crouch E C. Collectins and pulmonary host defense. Am J Respir Cell Mol Biol. 1998;19:177–201. doi: 10.1165/ajrcmb.19.2.140. [DOI] [PubMed] [Google Scholar]
- 6.Debono M, Gordee R S. Antibiotics that inhibit fungal cell wall development. Annu Rev Microbiol. 1994;48:471–497. doi: 10.1146/annurev.mi.48.100194.002351. [DOI] [PubMed] [Google Scholar]
- 7.Drickamer K. Engineering galactose-binding activity into a C-type mannose-binding protein. Nature. 1992;360:183–186. doi: 10.1038/360183a0. [DOI] [PubMed] [Google Scholar]
- 8.Fontaine T, Mouyna I, Hartland R P, Paris S, Latge J P. From the surface to the inner layer of the fungal cell wall. Biochem Soc Trans. 1997;25:194–199. doi: 10.1042/bst0250194. [DOI] [PubMed] [Google Scholar]
- 9.Haagsman H P, Hawgood S, Sargeant T, Buckley D, White R T, Drickamer K, Benson B J. The major lung surfactant protein, SP 28–36, is a calcium-dependent, carbohydrate-binding protein. J Biol Chem. 1987;262:13877–13880. [PubMed] [Google Scholar]
- 10.Håkansson K, Lim N K, Hoppe H J, Reid K B. Crystal structure of the trimeric alpha-helical coiled-coil and the three lectin domains of human lung surfactant protein D. Struct Fold Des. 1999;7:255–264. doi: 10.1016/s0969-2126(99)80036-7. [DOI] [PubMed] [Google Scholar]
- 11.Haurum J S, Thiel S, Haagsman H P, Laursen S B, Larsen B, Jensenius J C. Studies on the carbohydrate-binding characteristics of human pulmonary surfactant-associated protein A and comparison with two other collectins: mannan-binding protein and conglutinin. Biochem J. 1993;293:873–878. doi: 10.1042/bj2930873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hickling T P, Bright H, Wing K, Gower D, Martin S L, Sim R B, Malhotra R. A recombinant trimeric surfactant protein D carbohydrate recognition domain inhibits respiratory syncytial virus infection in vitro and in vivo. Eur J Immunol. 1999;29:3478–3484. doi: 10.1002/(SICI)1521-4141(199911)29:11<3478::AID-IMMU3478>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 13.Jigami Y, Odani T. Mannosylphosphate transfer to yeast mannan. Biochim Biophys Acta. 1999;1426:335–345. doi: 10.1016/s0304-4165(98)00134-2. [DOI] [PubMed] [Google Scholar]
- 14.Karson E M, Ballou C E. Biosynthesis of yeast mannan. Properties of a mannosylphosphate transferase in Saccharomyces cerevisiae. J Biol Chem. 1978;253:6484–6492. [PubMed] [Google Scholar]
- 15.Kollar R, Reinhold B B, Petrakova E, Yeh H J, Ashwell G, Drgonova J, Kapteyn J C, Klis F M, Cabib E. Architecture of the yeast cell wall. Beta(1→6)-glucan interconnects mannoprotein, beta(1→)3-glucan, and chitin. J Biol Chem. 1997;272:17762–17775. doi: 10.1074/jbc.272.28.17762. [DOI] [PubMed] [Google Scholar]
- 16.Kuan S F, Rust K, Crouch E. Interactions of surfactant protein D with bacterial lipopolysaccharides. Surfactant protein D is an Escherichia coli-binding protein in bronchoalveolar lavage. J Clin Investig. 1992;90:97–106. doi: 10.1172/JCI115861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuroki Y, Akino T. Pulmonary surfactant protein A (SP-A) specifically binds dipalmitoylphosphatidylcholine. J Biol Chem. 1991;266:3068–3073. [PubMed] [Google Scholar]
- 18.Kuroki Y, Voelker D R. Pulmonary surfactant proteins. J Biol Chem. 1994;269:25943–25946. [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.LeVine A M, Bruno M D, Huelsman K M, Ross G F, Whitsett J A, Korfhagen T R. Surfactant protein A-deficient mice are susceptible to group B streptococcal infection. J Immunol. 1997;158:4336–4340. [PubMed] [Google Scholar]
- 21.LeVine A M, Gwozdz J, Stark J, Bruno M, Whitsett J, Korfhagen T. Surfactant protein-A enhances respiratory syncytial virus clearance in vivo. J Clin Investig. 1999;103:1015–1021. doi: 10.1172/JCI5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeVine A M, Kurak K E, Bruno M D, Stark J M, Whitsett J A, Korfhagen T R. Surfactant protein-A-deficient mice are susceptible to Pseudomonas aeruginosa infection. Am J Respir Cell Mol Biol. 1998;19:700–708. doi: 10.1165/ajrcmb.19.4.3254. [DOI] [PubMed] [Google Scholar]
- 23.Madan T, Eggleton P, Kishore U, Strong P, Aggrawal S S, Sarma P U, Reid K B. Binding of pulmonary surfactant proteins A and D to Aspergillus fumigatus conidia enhances phagocytosis and killing by human neutrophils and alveolar macrophages. Infect Immun. 1997;65:3171–3179. doi: 10.1128/iai.65.8.3171-3179.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mason R J, Greene K, Voelker D R. Surfactant protein A and surfactant protein D in health and disease. Am J Physiol. 1998;275:L1–13. doi: 10.1152/ajplung.1998.275.1.L1. [DOI] [PubMed] [Google Scholar]
- 25.Meaden P, Hill K, Wagner J, Slipetz D, Sommer S S, Bussey H. The yeast KRE5 gene encodes a probable endoplasmic reticulum protein required for (1→6)-beta-d-glucan synthesis and normal cell growth. Mol Cell Biol. 1990;10:3013–3019. doi: 10.1128/mcb.10.6.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogasawara Y, Kuroki Y, Akino T. Pulmonary surfactant protein D specifically binds to phosphatidylinositol. J Biol Chem. 1992;267:21244–21249. [PubMed] [Google Scholar]
- 27.Ogasawara Y, Voelker D R. Altered carbohydrate recognition specificity engineered into surfactant protein D reveals different binding mechanisms for phosphatidylinositol and glucosylceramide. J Biol Chem. 1995;270:14725–14732. doi: 10.1074/jbc.270.24.14725. [DOI] [PubMed] [Google Scholar]
- 28.Olsen L R, Dessen A, Gupta D, Sabesan S, Sacchettini J C, Brewer C F. X-ray crystallographic studies of unique cross-linked lattices between four isomeric biantennary oligosaccharides and soybean agglutinin. Biochemistry. 1997;36:15073–15080. doi: 10.1021/bi971828+. [DOI] [PubMed] [Google Scholar]
- 29.Persson A, Chang D, Crouch E. Surfactant protein D is a divalent cation-dependent carbohydrate-binding protein. J Biol Chem. 1990;265:5755–5760. [PubMed] [Google Scholar]
- 30.Reid K B. Interactions of surfactant protein D with pathogens, allergens and phagocytes. Biochim Biophys Acta. 1998;1408:290–295. doi: 10.1016/s0925-4439(98)00074-x. [DOI] [PubMed] [Google Scholar]
- 31.Roemer T, Bussey H. Yeast beta-glucan synthesis: KRE6 encodes a predicted type II membrane protein required for glucan synthesis in vivo and for glucan synthase activity in vitro. Proc Natl Acad Sci USA. 1991;88:11295–11299. doi: 10.1073/pnas.88.24.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roemer T, Delaney S, Bussey H. SKN1 and KRE6 define a pair of functional homologs encoding putative membrane proteins involved in beta-glucan synthesis. Mol Cell Biol. 1993;13:4039–4048. doi: 10.1128/mcb.13.7.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strong P, Kishore U, Morgan C, Lopez Bernal A, Singh M, Reid K B. A novel method of purifying lung surfactant proteins A and D from the lung lavage of alveolar proteinosis patients and from pooled amniotic fluid. J Immunol Methods. 1998;220:139–149. doi: 10.1016/s0022-1759(98)00160-4. [DOI] [PubMed] [Google Scholar]
- 34.van Rozendaal B A, van Spriel A B, van De Winkel J G, Haagsman H P. Role of pulmonary surfactant protein D in innate defense against Candida albicans. J Infect Dis. 2000;182:917–922. doi: 10.1086/315799. [DOI] [PubMed] [Google Scholar]
- 35.Weis W I, Drickamer K, Hendrickson W A. Structure of a C-type mannose-binding protein complexed with an oligosaccharide. Nature. 1992;360:127–134. doi: 10.1038/360127a0. [DOI] [PubMed] [Google Scholar]
- 36.Wright J R. Immunomodulatory functions of surfactant. Physiol Rev. 1997;77:931–962. doi: 10.1152/physrev.1997.77.4.931. [DOI] [PubMed] [Google Scholar]