Abstract
Purpose:
The gut microbiota is subject to multiple insults in allogeneic-hematopoietic cell transplantation (allo-HCT) recipients. We hypothesized that preparative conditioning regimens contribute to microbiota perturbation in allo-HCT.
Experimental design:
This was a retrospective study that evaluated the relationship between conditioning regimens exposure in 1,188 allo-HCT recipients and the gut microbiome. Stool samples collected from 20 days before transplantation up to 30 days after were profiled using 16S rRNA sequencing. Microbiota injury was quantified by changes in α-diversity.
Results:
We identified distinct patterns of microbiota injury that varied by conditioning regimen. Diversity loss was graded into three levels of conditioning-associated microbiota injury (CMBI) in a multivariable model that included antibiotic exposures. High-intensity regimens, such as total body irradiation (TBI)-thiotepa-cyclophosphamide, were associated with the greatest injury loss (CMBI III). In contrast, the non-myeloablative regimen fludarabine-cyclophosphamide with low-dose TBI (Flu/Cy/TBI200) had a low-grade injury (CMBI I). The risk of acute graft-versus-host disease correlated with CMBI degree. Pre-transplant microbial compositions were best preserved with Flu/Cy/TBI200, whereas other regimens were associated with loss of commensal bacteria and expansion of Enterococcus.
Conclusions:
Our findings support an interaction between conditioning at the regimen level and the extent of microbiota injury.
Keywords: Allogeneic hematopoietic cell transplantation, Conditioning, Microbiome, Microbiota injury, Graft-versus-host disease
TRANSLATIONAL RELEVANCE
Conditioning regimens have unique patterns of microbiota injury that correlate with GVHD. We propose a novel scale for conditioning-associated microbiota injury. It offers a new axis for assessing conditioning regimens distinct from typical conditioning intensity classification. Our work identifies conditioning strategies that could potentially benefit from microbiota-directed interventions.
INTRODUCTION
In allogeneic hematopoietic cell transplantation (allo-HCT) conditioning regimens facilitate engraftment and eradicate residual tumor cells. Conditioning regimens are classified into three intensity groups according to their ability to irreversibly ablate hematopoiesis: myeloablative (MAC), reduced intensity (RIC), and non-myeloablative (NMA).(1,2) Higher-intensity regimens are considered more toxic to the gastrointestinal tract and are associated with higher risk of graft-versus-host disease (GVHD).(3) While aggregation of conditioning regimens by intensity groups has proved clinically helpful,(1,2) conditioning drugs act through unique mechanisms and have distinct toxicity profiles. Furthermore, there is considerable intra-individual variability in toxicity patterns and clinical outcomes are incompletely accounted for by clinical features.(4)
Intestinal homeostasis is influenced by the intestinal microbiome and the neighboring mucosal tissue, as well as the interaction of microbe-derived factors with host cell populations.(5–8) In allo-HCT, the gut mucosa is inflamed and the microbiome markedly altered;(9–11) antibiotic exposures partially account for these microbial shifts.(12–16) Chemotherapy and radiation may also affect the microbiome,(17,18) but have not been extensively studied in allo-HCT. Furthermore, the microbiome may modulate the sensitivity and metabolism of these interventions.(19,20) In this observational study, we investigated the hypothesis that microbiota injury in allo-HCT is also dependent on conditioning regimens.
METHODS
Patients
Patients that received allo-HCT at Memorial Sloan Kettering Cancer Center between 2009-09-04 and 2019-12-26 were screened for inclusion. Patients were eligible if they had at least one stool sample collected between days −20 and +30. We excluded patients treated with uncommon conditioning regimens (n<40), those whose samples were collected at the time of a second or third allo-HCT for graft failure or a third allo-HCT, and those who received uncommon GVHD prophylaxis regimens (administered to <10 patients; Figure 1A). Conditioning regimens included were IV busulfan (pharmacokinetics-directed) 9.6 – 12.8 mg/kg, melphalan 140 mg/m2, and fludarabine 125 mg/m2 (Bu4/Mel/Flu); fludarabine 160 mg/m2 and IV busulfan (pharmacokinetics-directed) 9.6 – 12.8 mg/kg (Flu/Bu4); total body irradiation (TBI) 1375 cGY, thiotepa 10 mg/kg, and cyclophosphamide 120 mg/kg (TBI1375/Tt/Cy); clofarabine 100mg/m2, melphalan 140 mg/m2, and thiotepa 10 mg/kg (Clo/Tt/Mel); fludarabine 150 mg/m2, cyclophosphamide 50 mg/kg, thiotepa 10 mg/kg, and TBI 400 cGy (Flu/Cy/Tt/TBI400); melphalan 140 mg/m2, thiotepa 5 mg/kg, and fludarabine 160 mg/m2 (Mel/Tt/Flu); fludarabine 120 mg/m2 and nelphalan 140 mg/m2 (Flu/Mel); fludarabine 125 mg/m2, cyclophosphamide 50 mg/kg, and TBI 200 cGY (Flu/Cy/TBI200). Conditioning intensity was was classified according to standard criteria (Figure 1B).(1,2)
Figure 1. Fecal samples from 1,188 recipients of allo-HCT were analyzed.
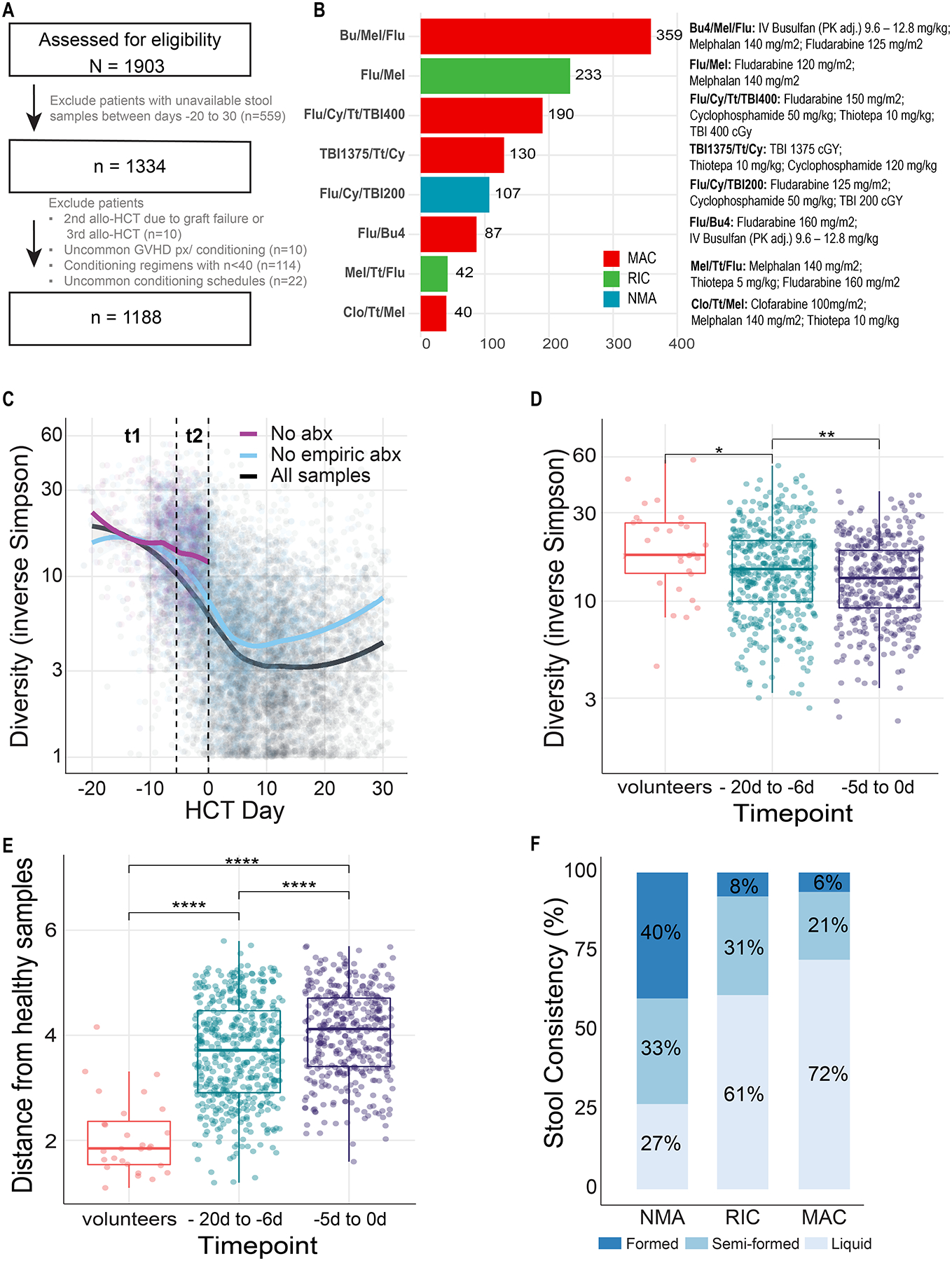
(A) CONSORT diagram for patient inclusion. The patients were conditioned with one of the eight regimens received by >40 patients with at least one stool sample collected between days −20 to 30. (B) Flu/Cy/TBI200 was the only nonmyeloablative (NMA) regimen. Flu/Mel and Mel/Tt/Flu were the reduced-intensity conditioning (RIC) regimens; the rest of the regimens were myeloablative (MAC). (C) Not all diversity loss can be attributed to antibiotic exposures. Each point is a stool sample whose α-diversity (as measured by 16S amplicon sequencing and the inverse Simpson index) is plotted over time relative to transplantation. Lines are smoothed average by the LOESS (locally estimated scatterplot smoothing) method, in which each data point (α-diversity measurement) is considered as an independent event. The black curve averages all samples, including those collected before or after any antibiotic exposure. The purple curve ignores any sample collected after exposure to any antibacterial antibiotic; as virtually all patients commenced prophylactic antibiotics by day −2 or with the onset of neutropenia, the purple curve does not extend beyond day 0. The blue curve ignores any samples collected after exposure to non-prophylactic antibiotics, i.e., samples exposed to the prophylactic antibiotics ciprofloxacin and intravenous vancomycin are included in the blue smoothed average. Time bin t1 (day −20 to −6) and t2 (day −5 to 0) are indicated with dashed vertical lines. (D) Among samples from n = 507 patients whose samples were not exposed to any antibacterial antibiotics, α-diversity declines significantly between t1 and t2, indicating that not all diversity loss can be attributed to antibiotic exposures. t1 α-diversity is also significantly lower than those of healthy volunteers (n = 30). Only one stool sample per patient was considered in each time window. (E) Among samples from n = 507 patients whose samples were not exposed to any antibacterial antibiotics, microbiota composition is significantly different between t1 and t2, as measured by Bray-Curtis distance to a centroid of healthy volunteers (n = 28) compositions, indicating that not all composition changes can be attributed to antibiotic exposures. (F) Liquid stool samples (n=957), collected between days 0 to 10, were more commonly collected from recipients of more intense conditioning regimens, as assessed by laboratory technicians at the time of sample aliquoting. X-squared for all samples= 118.44, df = 4, p-value < 2.2 × 10−16.
* p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001
Abbreviations: allo-HCT – allogeneic hematopoietic cell transplantation; MAC – myeloablative conditioning; RIC – reduced-intensity conditioning; NMA – nonmyeloablative conditioning; Abx – antibiotics.
To validate the association between conditioning intensity and diversity loss, we included an additional cohort of allo-HCT recipients from Duke University Medical Center in Durham, North Carolina; the University Medical Center, University Hospital Regensburg, in Regensburg, Germany; and Hokkaido University Hospital in Sapporo, Japan. The cohort features were previously reported by Peled et al.(11)
Patients provided written consent to an IRB-approved biospecimen-collection protocol.
Stool samples and sequencing
Stool samples were processed by disruption of bacterial cell walls with silica bead-beating, isolation of nucleic acids, and amplifying and sequencing the genomic 16S ribosomal-RNA gene V4–V5 variable region on the Illumina MiSeq platform, as previously described.(11) Amplicon sequence variants (ASVs) were called using the DADA2 pipeline and mapped to the NCBI 16S rRNA sequence database using BLAST.(21) α- and β-diversity were calculated with the Simpson reciprocal index and Bray-Curtis distances, respectively. Laboratory technicians ascertained stool consistency at the time of aliquoting.
The distances in Figure 2D were determined by applying multidimensional scaling (MDS) to the Bray-Curtis distance matrix. For each patient that had samples collected before conditioning, the earliest sample was selected for calculating per-regimen baseline centroids (based on the first two MDS components). Figure 1F shows the Euclidean distance between each patient’s latest sample and the regimen centroid.
Figure 2. Microbiota injury patterns in allo-HCT are conditioning regimen-specific.
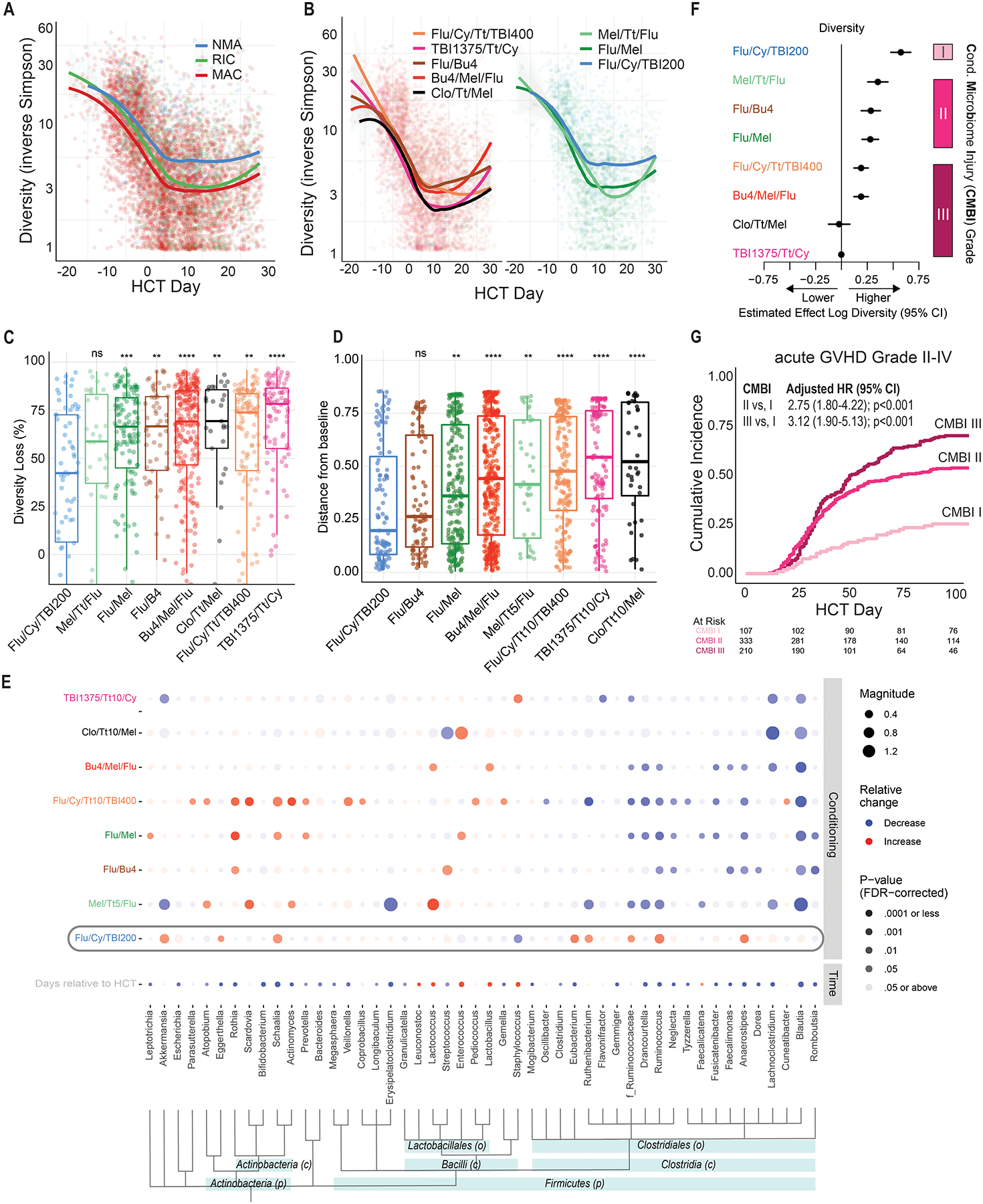
(A) Fecal microbiota α-diversity declines most in recipients of MAC and the least in recipients of NMA conditioning (Table S2). Each point is a stool sample whose α-diversity is plotted over time relative to transplantation. The smoothed curves plots are moving averages of recipients of different regimens. (B) Distinct patterns of α-diversity dynamics can be observed among recipients of specific conditioning regimens. Five MAC regimens are plotted in the left panel, two RIC and one NMA regimen are plotted in the right panel. (C) In a patient-level pair-matched analysis of the earliest sample collected before (days −20 to −1) and earliest sample after conditioning (days 0 to 10), the NMA regimen Flu/Cy/TBI200 was associated with the least reduction in diversity (42%). In contrast, the MAC regimens were associated with the greatest reduction (69%−78%). Regimens were compared to Flu/Cy/TBI200 with the Wilcoxon signed-rank test. (D) Microbiome dissimilarity, as determined by applying multidimensional scaling to the Bray-Curtis distance matrix, between samples collected at days 0 and 12 was significantly increased in all conditioning regimens except Flu/Cy/TBI200. The greatest distance was in the myeloablative regimen Clo/Tt/Mel. (E) A multivariable MaAsLin2 model(22) adjusting for time of sampling and conditioning regimens reveals a differential association between microbial taxa and conditioning regimens. Bacteria are ordered by taxonomical ranking. ASVs that could not be classified to the genus level were analyzed at the family level, as indicated by f_Ruminococcaceae. Flu/Cy/TBI200, highlighted with a gray rounded rectangle, was associated with the preservation of members of the Clostridia class. (F) A grading scheme classifying regimens into three categories of microbiota injury (low [I], intermediate [II], and high [III]) based diversity reduction between days −20 to 30 was introduced. The estimated effect on diversity of each regimen was derived from a generalized estimating equation (Table S4) regression model, adjusting for time, age, sex, exposure to GVHD prophylaxis, antibiotics, and conditioning. (G) Patients who received unmodified grafts had a higher incidence of acute GVHD Grade II-IV with regimens categorized as having greater microbiota toxicity. CMBI hazard ratios are derived from a multivariable Cox-regression adjusting for age, sex, comorbidity burden, donor and HLA matching, and GVHD prophylaxis.
* p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001
HCT – allogeneic hematopoietic cell transplantation; MAC – myeloablative conditioning; RIC – reduced-intensity conditioning; NMA – nonmyeloablative conditioning; CMBI – conditioning-associated microbiota injury; graft-versus-host disease (GVHD);
Statistical analysis
Descriptive statistics, including median and interquartile range [IQR] for continuous variables, and percentages for categorical variables, are provided. Acute GVHD was estimated using the cumulative incidence function, with death and relapse considered as competing events. A generalized estimating equation was constructed to assess correlations between clinical covariates, including the timing of sample collection and α diversity.
Modeling taxonomic abundance with multivariate models
In order to separate the association of the microbial community composition with conditioning regimens from the influence of the various patient variables, the data were modeled using Microbiome Multivariable Association with Linear Models (MaAsLin2).(22) In short, the dependency structure of the taxonomic composition within each patient was modeled by a patient-specific random effect. Covariates with fixed effects included conditioning regimens and time from HCT to sample collection. An alternative version of the model included additional covariates with fixed-effects – antibiotic exposures, diagnoses, GVHD-prophylaxis, previous allo-HCT, age, and graft type. Antibiotic exposure was encoded such that any sample collected the calendar day after the first dose of that antibiotic was considered exposed
MaAsLin2 was run with a minimum relative abundance threshold 2×10^−3 (corresponding to the limit of detection given the inclusion criteria of a minimum 1000 reads per sample), and a prevalence abundance of features being present in greater than or equal to 5% of the samples. Taxa indicated by a “f_” prefix indicate that no annotation was available at the Genus level. The base linear model was used on the log-scaled abundances; continuous metatada was left un-standardized. The data were analyzed in two windows: day −10 to 0, and day 0–12; these windows represent the conditioning period and the post-transplant/GVHD prophylaxis period, respectively. Hits were considered significant if they had a Benjamini-Hochberg corrected p-value less than 0.05.
Some of the various computational tools that have been developed for determining microbial differential abundance between groups have been observed to produce different sets of results under certain circumstances. To validate the bivariate results of MaAsLin2, we also used Corncob,(23) ANCOM2,(24) ANCOM-BC,(25) and metagenomeSeq (Bioconductor package)(26–28) to assess the compositional data from this study under various statistical approaches. Of these tools, only MaAsLin2 and ANCOM2 natively support the analysis of repeated measures from the same subjects.
Healthy volunteers
Healthy volunteers who provided stool samples provided written informed consent according to a biospecimen-collection protocol approved by the Memorial Sloan Kettering Cancer Center Institutional Review Board.
Data Availability Statement
Data sharing requests can be e-mailed to the corresponding author.
RESULTS
Patients
Of 1,903 allo-HCT recipients, 1,188 met the inclusion criteria for this observational cohort; key among these criteria were that the patient received one a relatively common conditioning regimen and had an donated an evaluable fecal specimen between day −20 and +30 relative to transplantation (Figure 1A, Table 1). CD34-selected peripheral blood stem cells (PBSC) were the most common graft source (498 [41.9%]), followed by unmodified PBSC (428 [36.0%]). Population and transplantation features differed between the conditioning regimens (Figure S1, Table S1). Most patients with lymphoma and chronic lymphocytic leukemia were conditioned with fludarabine, cyclophosphamide, and total body irradiation (TBI) 200 cGY (Flu/Cy/TBI200),(29) the only NMA regimen. Busulfan, melphalan, and fludarabine (Bu4/Mel/Flu), a myeloablative regimen frequently used before infusion of CD34-selected grafts, was commonly administered in those with myeloid malignancies and myeloma. Recipients of CD34-selected transplants did not receive any additional agents for GVHD prophylaxis. Unmodified PBSC grafts and methotrexate-based GVHD prophylaxis were most common after fludarabine and busulfan at a myeloablative dose (Flu/Bu4), fludarabine and melphalan (Flu/Mel), and Flu/Cy/TBI200 conditioning. Exposure to non-prophylactic antibiotics (i.e., antibiotics initiated empirically, as for fever or a documented infection) between days 0 to 21 was highest with TBI1375.
Table 1.
Population characteristics
| Overall | Missing | ||
|---|---|---|---|
| n | 1188 | ||
| Sex (%) | Male | 715 (60.2) | 0 |
| Female | 473 (39.8) | ||
| Age (median [IQR]) | 57 [47, 64] | 0 | |
| Disease (%) | Acute myeloid leukemia | 418 (35.2) | 0 |
| MDS or MPN | 286 (24.1) | ||
| Lymphoma/CLL | 231 (19.4) | ||
| ALL | 110 (9.3) | ||
| Plasma cell neoplasm | 111 (9.3) | ||
| Other leukemias | 22 (1.9) | ||
| Non-malignant disorders | 10 (0.8) | ||
| Previous Allo-HCT (%) | 1st allo-HCT | 1141 (96.0) | 0 |
| 2nd allo-HCT | 47 (4.0) | ||
| HCT-CI (median [IQR]) | 2 [1, 4] | 2.1 | |
| HCT year (median [IQR]) | 2015 [2013, 2017] | 0 | |
| Donor type (%) | Matched unrelated | 534 (44.9) | 0 |
| Matched related | 307 (25.8) | ||
| Cord blood | 174 (14.6) | ||
| Mismatched non-haploidentical | 128 (10.8) | ||
| Haploidentical | 45 (3.8) | ||
| Graft source (%) | PBSC T-cell depleted | 498 (41.9) | 0 |
| PBSC unmodified | 428 (36.0) | ||
| Cord blood | 174 (14.6) | ||
| BM unmodified | 88 (7.4) | ||
| Conditioning intensity (%) | Ablative | 806 (67.8) | 0 |
| Reduced Intensity | 275 (23.1) | ||
| Nonmyeloablative | 107 (9.0) | ||
| Conditioning regimens (%) | Bu4/Mel/Flu | 359 (30.2) | 0 |
| Flu/Mel | 233 (19.6) | ||
| Flu/Cy/Tt/TBI400 | 190 (16.0) | ||
| TBI1375/Tt/Cy | 130 (10.9) | ||
| Flu/Bu4 | 87 (7.3) | ||
| Flu/Cy/TBI200 | 107 (9.0) | ||
| Mel/Tt/Flu | 42 (3.5) | ||
| Clo/Tt/Mel | 40 (3.4) | ||
| GVHD prophylaxis (%) | none | 498 (41.9) | 0 |
| MTX-based | 426 (35.9) | ||
| MMF-based | 186 (15.7) | ||
| PTCy-based | 78 (6.6) | ||
| Exposure to antibiotics* up to day 21 (%) | Exposed | 804 (67.7) | 0 |
| Not exposed | 384 (32.3) | ||
The frequency of antibiotic exposure considered any exposure to drugs commonly used in this cohort of treatment of neutropenic fever or for C. difficile diarrhea (oral vancomycin, imipenem-cilastatin, meropenem, piperacillin-tazobactam, clindamycin, and metronidazole) between the first days of conditioning and days 21 post-allo-HCT.
IQR - interquartile range; AML – acute myeloid leukemia; MDS – myelodysplastic syndrome; MPN – myeloproliferative neoplasm; CLL – chronic lymphocytic leukemia; ALL – acute lymphoblastic leukemia; Allo-HCT – allogeneic hematopoietic cell transplantation; HCT-CI - hematopoietic cell transplantation-specific comorbidity index; MTX – Methotrexate; MMF – mycophenolate mofetil; PTCy – post-transplantation Cyclophosphamide.
Factors other than antibiotics contribute to changes in the microbiome of allo-HCT recipients
To characterize patterns of microbiota injury, we used 16S rRNA gene sequencing to analyze 7,682 stool samples (mean of 6.5 samples per patient) collected between days −20 to 30 (Figure S2). Alpha-diversity, a measure that reflects the number of unique bacteria present and their relative frequencies, was measured by the inverse Simpson index. Consistent with our previous studies,(11) α-diversity markedly decreased during allo-HCT; in particular the steepest decline was observed early in the treatment course, especially before day 0 (Figure 1C- black curve, Figure S2). The reduction was only partially explained by exposure to non-prophylactic antibiotics and any antibacterial antibiotics (Figure 1C, blue and purple curves). Among samples collected before graft infusion and not exposed to any type of bacterial antibiotics, diversity was lower in samples collected between days −5 to 0 (Figure 1C, purple line, period t2) than those collected between days −20 to −6 (Figure 1C, purple line, period t1, Figure 1D; p<0.01). Furthermore, in samples not exposed to antibacterial antibiotics, the similarity of the patient samples and healthy controls (measured as the Bray-Curtis distance to the average of the healthy-volunteer samples) still increased over time (Figure 1E, p<0.001). Therefore, additional factors besides antibiotics must be invoked to explain gut-microbiome disruption in allo-HCT recipients.
Conditioning regimen-associated microbiota injury
Stool consistency is a surrogate for microbiome composition.(30) The fraction of stool samples with liquid consistency (Figure 1F) correlated with conditioning intensity (MAC 72%, RIC 61%, NMA 27%, p<0.001), suggesting microbiome injury is highest with myeloablative conditioning. Diversity declined over time in recipients of all three conditioning intensities (Figure 2A). However, the decrease was greatest in recipients of MAC, followed by RIC and NMA (Table S2). Notably, NMA was also associated with preservation of diversity compared to MAC in an external, previously reported(11) multicenter cohort (n=291, Table S3). Therefore, conditioning intensity partially explains the variance in diversity trajectories among allo-HCT recipients.
We hypothesized that microbiota injury varies not only by conditioning intensity category but also between specific conditioning regimens. Patient samples followed a similar overall pattern of diversity reduction regardless of intensity (Figure 2B). However, diversity loss in samples collected shortly after graft infusion was most pronounced with TBI 1375 cGY, thiotepa, and cyclophosphamide (TBI1375/Tt/Cy) and fludarabine, cyclophosphamide, thiotepa, and TBI 400 cGy (Flu/Cy/Tt/TBI400) and lowest with Flu/Cy/TBI200 (Figure 2C).
We next asked whether conditioning regimens induce specific taxonomic changes in the fecal microbiome. Microbial composition shifted over time with all regimens, as measured by Bray-Curtis distance: compared to pre-conditioning samples, recipients of TBI135/Tt/Cy and clofarabine, melphalan, and thiotepa (Clo/Tt/Mel) exhibited the greatest magnitude of change, and recipients of Flu/Cy/TBI200 the least (Figure 2D). Nonetheless, the cumulative incidence of bacterial monodomination (defined as a relative-abundance threshold of any single ASV of ≥30%)(11) exceeded 80% by day 30, irrespective of the conditioning regimen (Figure S3). To explore the specific taxonomic changes that underpinned these shifts in global composition, we constructed a MaAslin2 multivariable linear model(22) in which the outcome variables were the changes in genus relative abundances and the exposure variables were time relative to transplantation and exposure to each of the conditioning regimens (Figure 2E). The red and blue circles in the figure indicate relative enrichment or depletion of taxa between day 0 and day 12, respectively; the size and intensity of the circles convey the magnitude of change and statistical significance, respectively. For example, a relative decline in Blautia abundance was associated with all the regimens except Flu/Cy/TBI200. Furthermore, Flu/Cy/TBI200 had a pattern distinct from the others with relative preservation of members of the Clostridia class, including Anaerostipes and Ruminococcus; an inverse relationship was seen with many of the other regimens and clostridia. To validate the bivariate results of MaAsLin2, we used additional modeling tools that rely on different statistical approaches (Figure S4).(23–28) Overall, there was strong agreement between the methods.
In a similar MaAsLin2 model also taking into account antibiotic exposures, underlying disease, GVHD prophylaxis, and other clinical features, a preponderance of the significant associations between taxa and clinical variables were observed for the antibiotic exposures. Oral vancomycin and piperacillin/tazobactam were associated with extensive taxonomic shifts, including a reduction in many of the commensal bacteria (Figure S5A). Nevertheless, conditioning-associated microbiome changes were still evident in the multivariable models, despite a correlation between conditioning regimens and antibiotic exposures (Figure S6). For instance, many taxonomic shifts were observed in the Flu/Cy/Tt/TBI400 regimen including the loss of Blautia, Ruminococcus, and other Ruminococcaea. GVHD prophylaxis was associated with a significant reduction of Clostridia, including Blautia, Anaerostipes, Dorea, and Drancourtella. We therefore constructed an additional multivariable model separating GVHD prophylaxis to individual elements (Figure S5B). Methotrexate and cyclophosphamide, which were used in mutually exclusive fashion for GVHD prophylaxis (Figure S6), were both associated with Clostridia loss, although numerically a greater change was observed with methotrexate. In contrast, cyclosporine and mycophenolate mofetil were associated with minimal microbial changes. Notably, in analysis of individual antibiotic or conditioning drug exposure in the pre-infusion time window, antibiotic exposures had many strong associations (Figure S5C). Oral vancomycin was associated with the greatest shifts. Collectively, our findings support a differential association between conditioning regimens and microbiome composition.
Conditioning-associated microbiota injury grading
To provide a scale quantifying the degree of microbiota injury by regimen, we constructed a generalized estimating equation multivariable model for α-diversity and then manually grouped the resulting diversity-reduction coefficients into three levels of Conditioning-associated Microbiome Injury (CMBI; Figure 2F, Table S4): low (CMBI I: Flu/Cy/TBI200), intermediate (CMBI II: Flu/Mel, Flu/Bu4, Mel/Tt/Flu [melphalan, thiotepa, and fludarabine]), and high (CMBI III: Flu/Cy/TBI400, Bu4/Mel/Flu, Clo/Tt/Mel, TBI1375/Tt/Cy). Notably, the myeloablative regimen Flu/Bu4(1,2) was associated with intermediate microbiota toxicity. Among recipients of unmanipulated grafts (i.e., excluding CD34-selected), regimens assigned to intermediate and high-grade CMBI had a higher risk of grade II-IV acute GVHD compared to low-grade MBI (Figure 2G). Altogether, our findings indicate that microbiota injury patterns are regimen-specific and can serve as a clinical biomarker.
DISCUSSION
This observational study is the first to show regimen-specific relationships between individual conditioning regimens and fecal microbiome composition. Conditioning-associated changes in the microbiome were observed with global metrics of microbiome composition, such as α- and β-diversity, as well as the taxonomic level. Multivariable modeling illustrated that antibiotic exposures and GVHD prophylaxis also contribute to microbiome perturbations after HCT. Our results in cancer patients extend prior reports of interactions between non-antibiotic drugs and gut microbiome in healthy volunteers, inflammatory bowel disease, and in vitro analyses.(17,31,32)
Conditioning regimes are the backbone of allogeneic HCT. Conditioning intensity categories (1,2) reflect the tradeoff between the degree of tumor eradication and toxicity, both being greatest with myeloablative regimens and reduced with nonmyeloablative regimens. We propose microbiota injury as a new axis for evaluating conditioning regimens and provide a classification of commonly used regimens that summarizes their degree of microbiota toxicity, which may be relevant to clinical outcomes such as GVHD and immune reconstitution.(9,11,33–35) In contrast to intensity classification, which was based on expert opinion,(1,2,36) we used a data-driven approach to quantitate microbiota changes and define injury levels. We defined injury based on α-diversity since it is a measure associated with survival after allo-HCT.(11,37) Given this association, there are ongoing efforts to increase diversity in allo-HCT recipients.(38,39) Our results suggest that conditioning regimens categorized with high-grade CMBI may serve as the optimal candidates for such interventions.
Non-antibiotic drugs can influence growth of bacterial population.(17,31,32) Our observation that changes in microbial patterns are conditioning-regimen dependent could be explained by several potential mechanisms. First, there could be direct effects of chemotherapy or radiation on bacteria. Indeed, radiation and cyclophosphamide, which are both commonly used as part of the conditioning backbone, have been shown to induce microbial changes in mice.(18,19,40) Conditioning may also lead to bacterial changes indirectly. Chemotherapy and radiation damage the mucosal membrane and elements of the intestinal epithelium, such as Paneth cells that secrete antibacterial peptides.(41–44)
Isolating the impact of conditioning regimens on the microbiota is challenging since confounders such as antibiotics, GVHD prophylaxis, and nutrition also affect bacterial populations. Nonetheless, we observed associations between conditioning regimens and microbiome perturbation, even after accounting for antibiotic and GVHD prophylaxis drug exposures. There are additional observations from the literature supporting conditioning as a driver of microbiome shifts. Maier and colleagues demonstrated that non-antibiotic drugs, including chemotherapeutic drugs, have an inhibiting effect on bacterial growth in-vitro.(17) Others have shown that radiation and cyclophosphamide (both commonly used for conditioning) have an impact on the gut microbiome.(18,19,40,45) The multivariable models illustrate the profound impact of antibiotics on the microbiome. Specifically, oral vancomycin and IV piperacillin-tazobactam were associated with dramatic changes in the microbiome, including butyrate-producing members of the Clostridiaceae family. Our findings raise the possibility that antibiotic stewardship and rational selection of antibiotics could substantially mitigate microbiota injury in allo-HCT recipients, a hypothesis we are testing in a randomized trial (NCT03078010).
Our findings indicate that conditioning regimens contribute to microbiota injury in allo-HCT recipients. Notably, the degree of injury is not directly correlated to classical conditioning intensity levels. For instance, conditioning with fludarabine and busulfan at a myeloablative dose (Flu/Bu4) was associated with intermediate microbiota injury (CMBI II). Since conditioning regimens are modifiable, there may be a rationale to select less microbiome-disruptive regimens in certain patients, perhaps those at higher risk of GVHD or those with already injured microbiome composition at baseline. Finally, the microbiome injury index could potentially be used for risk stratification in clinical trials studying microbiome-based interventions in HCT.
Supplementary Material
Financial Support
This research was supported by National Cancer Institute award numbers, R01-CA228358, R01-CA228308, P30 CA008748 MSK Cancer Center Support Grant/Core Grant and P01-CA023766; National Heart, Lung, and Blood Institute (NHLBI) award number R01-HL123340 and R01-HL147584; National Institute of Aging award number P01-AG052359; and Tri Institutional Stem Cell Initiative. Additional funding was received from The Lymphoma Foundation, The Susan and Peter Solomon Divisional Genomics Program, Cycle for Survival, and the Parker Institute for Cancer Immunotherapy.
TMH reports support from NIAID NIH Award R21AI156157.
JUP reports funding from NHLBI NIH Award K08HL143189.
RS was supported by the American Society of Transplantation and Cellular Therapy New Investigator Award, the American Society of Hematology Fellow Scholar Award, a grant from the Long Island Sound Chapter, Swim Across America, the Robert Hirschhorn Award, and the Memorial Sloan Kettering Steven Greenberg Lymphoma Research Award.
KAM would like to acknowledge funding from the DKMS and American Society for Hematology, the Royal Australasian College of Physicians, The Haematology Society of Australia and New Zealand, and the American Australian Association.
Disclosure of Conflicts of Interest
Roni Shouval served as a consultant for Medexus and MyBiotics.
Miguel Perales reports honoraria from Abbvie, Astellas, Bristol-Myers Squibb, Celgene, Equilium, Incyte, Karyopharm, Kite/Gilead, Merck, Miltenyi Biotec, MorphoSys, Novartis, Nektar Therapeutics, Omeros, Takeda, and VectivBio AG, Vor Biopharma. He serves on DSMBs for Cidara, Therapeutics, Medigene, Sellas Life Sciences, and Servier, and the scientific advisory board of NexImmune. He has ownership interests in NexImmune and Omeros. He has received research support for clinical trials from Incyte, Kite/Gilead, Miltenyi Biotec, and Novartis. He serves in a volunteer capacity as a member of the Board of Directors of the American Society for Transplantation and Cellular Therapy (ASTCT) and Be The Match (National Marrow Donor Program, NMDP), as well as on the CIBMTR Cellular Immunotherapy Data Resource (CIDR) Executive Committee.
Michael Scordo served as a paid consultant for McKinsey & Company, Angiocrine Bioscience, Inc., and Omeros Corporation; received research funding from Angiocrine Bioscience, Inc. and Omeros Corporation; served on ad hoc advisory boards for Kite – A Gilead Company; and received a one-time speaker fee from i3Health for a CME speaking engagement.
Jonathan U. Peled reports research funding, intellectual property fees, and travel reimbursement from Seres Therapeutics, and consulting fees from DaVolterra, CSL Behring, and from MaaT Pharma. He serves on an advisory board of and holds equity in Postbiotics Plus Research. He has filed intellectual property applications related to the microbiome (reference numbers #62/843,849, #62/977,908, and #15/756,845). Memorial Sloan Kettering Cancer Center (MSK) has financial interests relative to Seres Therapeutics.
Marcel R.M. van den Brink has received research support from Seres Therapeutics; has consulted, received honorarium from or participated in advisory boards for Seres Therapeutics, WindMIL therapeutics, Rheos, Frazier Healthcare Partners, Nektar Therapeutics, Notch Therapeutics, Forty Seven inc., Priothera, Ceramedix, Lygenesis, Pluto Immunotherapeutics, Magenta Therapeutics, Merck & Co, Inc., and DKMS Medical Council (Board); has IP Licensing with Seres Therapeutics, Juno Therapeutics, and stock options from Seres and Notch Therapeutics.
Segio Giralt reports the following disclosures: AMGEN: Membership on an entity’s Board of Directors or advisory committees; BMS: Membership on an entity’s Board of Directors or advisory committees; SANOFI: Membership on an entity’s Board of Directors or advisory committees; PFIZER: Membership on an entity’s Board of Directors or advisory committees; JENSENN: Membership on an entity’s Board of Directors or advisory committees; GSK: Membership on an entity’s Board of Directors or advisory committees; JAZZ: Membership on an entity’s Board of Directors or advisory committees; CELGENE: Membership on an entity’s Board of Directors or advisory committees; Actinnum: Membership on an entity’s Board of Directors or advisory committees.
Kate A Markey serves on an advisory board of and holds equity in Postbiotics Plus Research.
Tobias M. Hohl has participated in a scientific advisory board for Boehringer-Ingolheim, Inc.
Ernst Holler served on the advisory board for Maat Pharma and Pharmabiom.
Takanori Teshima reports grants from Kyowa Kirin, Chugai, Sanofi, Astellas, Teijin Pharma, Fuji Pharma, Nippon Shinyaku, personal fees from Novartis, Merck, Kyowa Kirin, Takeda, Pfizer, Bristol-Myers Squibb, and non-financial support from Janssen, Novartis.
Daigo Hashimoto recieved honoraria from Novartis, Janssen, Eisai, Chugai, Ono, and Sumitomo Pharma, and MSD.
REFERENCES
- 1.Bacigalupo A, Ballen K, Rizzo D, Giralt S, Lazarus H, Ho V, et al. Defining the intensity of conditioning regimens: working definitions. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation 2009;15(12):1628–33 doi 10.1016/j.bbmt.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giralt S, Ballen K, Rizzo D, Bacigalupo A, Horowitz M, Pasquini M, et al. Reduced-intensity conditioning regimen workshop: defining the dose spectrum. Report of a workshop convened by the center for international blood and marrow transplant research. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation 2009;15(3):367–9 doi 10.1016/j.bbmt.2008.12.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooke KR, Luznik L, Sarantopoulos S, Hakim FT, Jagasia M, Fowler DH, et al. The biology of chronic graft-versus-host disease: a task force report from the National Institutes of Health Consensus Development Project on Criteria for Clinical Trials in Chronic Graft-versus-Host Disease. Biology of Blood and Marrow Transplantation 2017;23(2):211–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shouval R, Fein JA, Shouval A, Danylesko I, Shem-Tov N, Zlotnik M, et al. External validation and comparison of multiple prognostic scores in allogeneic hematopoietic stem cell transplantation. Blood Advances 2019;3(12):1881–90 doi 10.1182/bloodadvances.2019032268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Litvak Y, Byndloss MX, Baumler AJ. Colonocyte metabolism shapes the gut microbiota. Science 2018;362(6418) doi 10.1126/science.aat9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenq RR, Ubeda C, Taur Y, Menezes CC, Khanin R, Dudakov JA, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. The Journal of experimental medicine 2012;209(5):903–11 doi 10.1084/jem.20112408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansson GC, Johansson ME. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Gut Microbes 2010;1(1):51–4 doi 10.4161/gmic.1.1.10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy M, Thaiss CA, Zeevi D, Dohnalová L, Zilberman-Schapira G, Mahdi JA, et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell 2015;163(6):1428–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stein-Thoeringer CK, Nichols KB, Lazrak A, Docampo MD, Slingerland AE, Slingerland JB, et al. Lactose drives Enterococcus expansion to promote graft-versus-host disease. Science 2019;366(6469):1143–9 doi 10.1126/science.aax3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shono Y, van den Brink MR. Gut microbiota injury in allogeneic haematopoietic stem cell transplantation. Nature Reviews Cancer 2018;18(5):283–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peled JU, Gomes AL, Devlin SM, Littmann ER, Taur Y, Sung AD, et al. Microbiota as predictor of mortality in allogeneic hematopoietic-cell transplantation. New England Journal of Medicine 2020;382(9):822–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoma I, Littmann ER, Peled JU, Giralt S, van den Brink MR, Pamer EG, et al. Compositional flux within the intestinal microbiota and risk for bloodstream infection with gram-negative bacteria. Clinical Infectious Diseases 2021;73(11):e4627–e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morjaria S, Schluter J, Taylor BP, Littmann ER, Carter RA, Fontana E, et al. Antibiotic-Induced Shifts in Fecal Microbiota Density and Composition during Hematopoietic Stem Cell Transplantation. Infection and immunity 2019;87(9) doi 10.1128/IAI.00206-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rashidi A, Kaiser T, Holtan SG, Rehman TU, Weisdorf DJ, Khoruts A, et al. Levaquin Gets a Pass. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation 2020;26(4):778–81 doi 10.1016/j.bbmt.2019.12.722. [DOI] [PubMed] [Google Scholar]
- 15.Shono Y, Docampo MD, Peled JU, Perobelli SM, Velardi E, Tsai JJ, et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Science translational medicine 2016;8(339):339ra71 doi 10.1126/scitranslmed.aaf2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holler E, Butzhammer P, Schmid K, Hundsrucker C, Koestler J, Peter K, et al. Metagenomic Analysis of the Stool Microbiome in Patients Receiving Allogeneic Stem Cell Transplantation: Loss of Diversity Is Associated with Use of Systemic Antibiotics and More Pronounced in Gastrointestinal Graft-versus-Host Disease. Biology of Blood and Marrow Transplantation 2014;20(5):640–5 doi 10.1016/j.bbmt.2014.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 2018;555(7698):623–8 doi 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo H, Chou WC, Lai Y, Liang K, Tam JW, Brickey WJ, et al. Multi-omics analyses of radiation survivors identify radioprotective microbes and metabolites. Science 2020;370(6516) doi 10.1126/science.aay9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crawford PA, Gordon JI. Microbial regulation of intestinal radiosensitivity. Proc Natl Acad Sci U S A 2005;102(37):13254–9 doi 10.1073/pnas.0504830102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Javdan B, Lopez JG, Chankhamjon P, Lee YJ, Hull R, Wu Q, et al. Personalized Mapping of Drug Metabolism by the Human Gut Microbiome. Cell 2020;181(7):1661–79 e22 doi 10.1016/j.cell.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 2016;13(7):581–3 doi 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mallick H, Rahnavard A, McIver LJ, Ma S, Zhang Y, Nguyen LH, et al. Multivariable association discovery in population-scale meta-omics studies. PLOS Computational Biology 2021;17(11):e1009442 doi 10.1371/journal.pcbi.1009442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin BD, Witten D, Willis AD. Modeling microbial abundances and dysbiosis with beta-binomial regression. The annals of applied statistics 2020;14(1):94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaul A, Mandal S, Davidov O, Peddada SD. Analysis of microbiome data in the presence of excess zeros. Frontiers in microbiology 2017;8:2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin H, Peddada SD. Analysis of compositions of microbiomes with bias correction. Nature communications 2020;11(1):3514 doi 10.1038/s41467-020-17041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nature methods 2013;10(12):1200–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paulson JN, Talukder H, Bravo HC. Longitudinal differential abundance analysis of microbial marker-gene surveys using smoothing splines. BioRxiv 2017:099457. [Google Scholar]
- 28.Paulson JN, Pop M, Bravo HC. metagenomeSeq: Statistical analysis for sparse high-throughput sequencing. Bioconductor package 2013(0):191. [Google Scholar]
- 29.Sauter CS, Barker JN, Lechner L, Zheng J, Devlin SM, Papadopoulos EB, et al. A phase II study of a nonmyeloablative allogeneic stem cell transplant with peritransplant rituximab in patients with B cell lymphoid malignancies: favorably durable event-free survival in chemosensitive patients. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation 2014;20(3):354–60 doi 10.1016/j.bbmt.2013.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vandeputte D, Falony G, Vieira-Silva S, Tito RY, Joossens M, Raes J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 2016;65(1):57–62 doi 10.1136/gutjnl-2015-309618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vich Vila A, Collij V, Sanna S, Sinha T, Imhann F, Bourgonje AR, et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nature communications 2020;11(1):362 doi 10.1038/s41467-019-14177-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson MA, Verdi S, Maxan M-E, Shin CM, Zierer J, Bowyer RCE, et al. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nature communications 2018;9(1):2655 doi 10.1038/s41467-018-05184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miltiadous O, Waters NR, Andrlova H, Dai A, Nguyen CL, Burgos da Silva M, et al. Early intestinal microbial features are associated with CD4 T cell recovery after allogeneic hematopoietic transplant. Blood 2022. doi 10.1182/blood.2021014255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Markey KA, Schluter J, Gomes ALC, Littmann ER, Pickard AJ, Taylor BP, et al. The microbe-derived short-chain fatty acids butyrate and propionate are associated with protection from chronic GVHD. Blood 2020;136(1):130–6 doi 10.1182/blood.2019003369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schluter J, Peled JU, Taylor BP, Markey KA, Smith M, Taur Y, et al. The gut microbiota is associated with immune cell dynamics in humans. Nature 2020;588(7837):303–7 doi 10.1038/s41586-020-2971-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spyridonidis A, Labopin M, Savani BN, Niittyvuopio R, Blaise D, Craddock C, et al. Redefining and measuring transplant conditioning intensity in current era: a study in acute myeloid leukemia patients. Bone Marrow Transplant 2020;55(6):1114–25 doi 10.1038/s41409-020-0803-y. [DOI] [PubMed] [Google Scholar]
- 37.Taur Y, Jenq RR, Perales MA, Littmann ER, Morjaria S, Ling L, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 2014;124(7):1174–82 doi 10.1182/blood-2014-02-554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Lier YF, Van den Brink MR, Hazenberg MD, Markey KA. The post-hematopoietic cell transplantation microbiome: relationships with transplant outcome and potential therapeutic targets. haematologica 2021;106(8):2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeFilipp Z, Peled JU, Li S, Mahabamunuge J, Dagher Z, Slingerland AE, et al. Third-party fecal microbiota transplantation following allo-HCT reconstitutes microbiome diversity. Blood Adv 2018;2(7):745–53 doi 10.1182/bloodadvances.2018017731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. science 2013;342(6161):971–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjöberg J, Amir E, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol 2010;11(1):76–83 doi 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eriguchi Y, Takashima S, Oka H, Shimoji S, Nakamura K, Uryu H, et al. Graft-versus-host disease disrupts intestinal microbial ecology by inhibiting Paneth cell production of α-defensins. Blood 2012;120(1):223–31 doi 10.1182/blood-2011-12-401166. [DOI] [PubMed] [Google Scholar]
- 43.Weber D, Frauenschläger K, Ghimire S, Peter K, Panzer I, Hiergeist A, et al. The association between acute graft-versus-host disease and antimicrobial peptide expression in the gastrointestinal tract after allogeneic stem cell transplantation. PloS one 2017;12(9):e0185265 doi 10.1371/journal.pone.0185265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levine JE, Huber E, Hammer ST, Harris AC, Greenson JK, Braun TM, et al. Low Paneth cell numbers at onset of gastrointestinal graft-versus-host disease identify patients at high risk for nonrelapse mortality. Blood 2013;122(8):1505–9 doi 10.1182/blood-2013-02-485813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jenq RR, Ubeda C, Taur Y, Menezes CC, Khanin R, Dudakov JA, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. Journal of Experimental Medicine 2012;209(5):903–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sharing requests can be e-mailed to the corresponding author.


