Abstract
This study aims to investigate whether adding neoadjuvant radiotherapy (RT), anti-programmed cell death protein-1 (PD-1) antibody (anti-PD-1), or RT + anti-PD-1 to surgical resection improves disease-free survival for mice with soft tissue sarcomas (STS). We generated a high mutational load primary mouse model of STS by intramuscular injection of adenovirus expressing Cas9 and guide RNA targeting Trp53 and intramuscular injection of 3-methylcholanthrene (MCA) into the gastrocnemius muscle of wild-type mice (p53/MCA model). We randomized tumor-bearing mice to receive isotype control or anti-PD-1 antibody with or without RT (20 Gy), followed by hind limb amputation. We used micro-CT to detect lung metastases with high spatial resolution, which was confirmed by histology. We investigated if sarcoma metastasis was regulated by immunosurveillance by lymphocytes or tumor cell-intrinsic mechanisms. Compared to surgery with isotype control antibody, the combination of anti-PD-1, RT, and surgery improved local recurrence-free survival (p=0.035) and disease-free survival (p=0.005), but not metastasis-free survival. Mice treated with RT, but not anti-PD-1, showed significantly improved local recurrence-free survival and metastasis-free survival over surgery alone (p=0.043 and p=0.007, respectively). The overall metastasis rate was low (~12%) in the p53/MCA sarcoma model, which limited the power to detect further improvement in metastasis-free survival with addition of anti-PD-1 therapy. Tail vein injections of sarcoma cells into immunocompetent mice suggested that impaired metastasis was due to inability of sarcoma cells to grow in the lungs rather than a consequence of immunosurveillance. In conclusion, neoadjuvant RT improves metastasis-free survival after surgery in a primary model of STS.
Introduction:
Soft tissue sarcomas (STS) represent a diverse group of mesenchymal malignancies with over 70 different subtypes that account for approximately 1% of cancer in the United States (1). In patients with large and high-grade sarcomas, radiation therapy (RT) plays a critical role in improving local control (2,3), but approximately 50% of patients develop lung metastases and succumb to the disease within 18 months (4). Therefore, alternative therapeutic approaches are urgently needed to reduce STS metastases and thereby improve overall survival. Immune checkpoint inhibitors (ICIs), such as antibodies against cytotoxic T-lymphocyte antigen-4 (CTLA-4) and programmed cell death protein-1 (PD-1), reinvigorated the field of immuno-oncology and showed promising results in a subset of melanoma and non-small cell lung carcinoma patients (5–7). More than 3000 clinical trials are currently evaluating the effect of T-cell-targeted immunomodulators as first or second-line treatment for 50 cancer types (8). The recently concluded SARC028 trial (NCT02301039) showed activity of the anti-PD-1 (α-PD-1) antibody pembrolizumab in a subset of patients with metastatic undifferentiated pleomorphic sarcoma (UPS) or dedifferentiated/pleomorphic liposarcoma (LPS) (9). In addition, studies have suggested that RT can augment anti-tumor response by inducing the production of immunostimulatory cytokines and chemokines, increasing T-cell priming through the release of damage-associated molecular patterns, upregulating major histocompatibility complex-I, and exposing novel tumor antigens (10). Furthermore, preclinical and clinical studies suggest that RT has the potential to act synergistically with immune checkpoint blockade to enhance anti-tumor immune response outside of the radiation field, i.e., an abscopal effect (11–13). To determine whether the addition of α-PD-1 therapy to RT and surgery improves disease-free survival for high-risk STS, we are currently enrolling subjects in phase II clinical trial SU2C-SARC032 (NCT03092323), in which patients with stage III UPS or LPS are randomized to receive neoadjuvant RT and surgical resection, with or without neoadjuvant and adjuvant pembrolizumab (Supplementary figure 1).
SU2C-SARC032 presented the opportunity to implement a co-clinical trial that aims to: i) construct robust preclinical imaging protocols with MRI and micro-CT, and ii) employ these imaging modalities to assess the efficacy of immunotherapy combined with preoperative RT to reduce local recurrence and metastatic disease in a carcinogen-induced and genetically engineered autochthonous mouse model of STS that resembles human UPS (14,15). MRI is employed to evaluate the primary tumor site. The preclinical MRI imaging protocol and performance for the optimized extremity MRI in mice with sarcomas have been reported previously (16,17). Serial CT imaging of the lungs of mice (16,17) mimics a critical metric of therapeutic response in patients on SU2C-SARC032 because the lungs are the most common site of metastasis for patients with UPS or LPS. Here, we focus on the implementation and results of the preclinical arm of the co-clinical trial, which demonstrates that the combination of α-PD-1 treatment with RT and surgery improves disease-free survival compared to surgery alone by improving local control. Furthermore, neoadjuvant RT improves local recurrence-free and metastasis-free survival compared to surgery with or without α-PD-1 treatment.
Materials and Methods:
Animals
All animal procedures were approved by Duke University Institutional Care and Use Committee (IACUC) and adhere to the NIH Guide for the Care and Use of Laboratory Animals. Rag2tm1.1Flv (Rag2−/−) and 129/SvJae wild-type mice were obtained from The Jackson Laboratory and bred at Duke University. Trp53fl/fl (18) mice were obtained from Anton Berns at the Netherlands Cancer Institute and were bred at Duke University. All animals were kept in individually ventilated cages in a room with a 12:12h light:dark cycle and constant temperature (72 ± 2° F). All studies were performed using age-matched male and female littermate controls.
Sarcoma induction and treatment
Primary p53/MCA sarcomas were induced in mice with different genetic backgrounds to implement the preclinical arm of the co-clinical trial and to test hypotheses related to the control of metastases in the p53/MCA model. Primary p53/MCA sarcomas were induced in Rag2+/−, Rag2−/−, and 129/SvJae wild-type mice between the age of 6 and 10 weeks by intramuscular injection of adenovirus expressing Cas9 endonuclease and sgRNA targeting Trp53 (Adeno-sgp53-Cas9; Viraquest) (19) followed by intramuscular injection of 3-methylcholenthrene (MCA) into the gastrocnemius muscle as previously described (14). Rag2−/− and Rag2+/− mice were used to evaluate the role of lymphocytes in p53/MCA-induced sarcoma metastasis. Rag2−/− mice lack mature B- and T-cells, while immunocompetent Rag2+/− mice served as littermate controls for the experiment. After sarcomas reached 75–150 mm3, 129/SvJae mice were randomized into four treatment groups: isotype control antibody (ISO), anti-PD-1 antibody (α-PD-1), ISO + 20 Gy (ISO + RT), or α-PD-1 + 20 Gy (α-PD-1 + RT) (Figure 1A). α-PD-1 antibody (BioXCell, Cat # BE0146) and isotype control antibody (BioXCell, Cat # BE0086) were administered by intraperitoneal injection of 200 μl per dose at 1 mg/ml diluted in InVivoPure dilution buffer (BioXCell) on day 0 (pre-amputation) and day 7 (post-amputation). Sarcomas were irradiated with 20 Gy (single fraction) on day 0 with parallel-opposed anterior and posterior fields using the Precision Xrad 225Cx small animal image-guided irradiator with mice anesthetized with continuous 2% isoflurane at a rate of 2 L oxygen/minute (20). The irradiation field was centered on the tumor via fluoroscopy with 40 kilovolt peak (kVp), 2.5 mA X-rays using a 0.3 mm copper filter, followed by irradiation with an average dose rate of 300 cGy/min prescribed to midplane at treatment isocenter with 225 kVp, 13 mA X-rays using a 0.3 mm copper filter and a collimator with a 40 × 40 mm2 radiation field. Amputation of tumor-bearing hind limb was performed on day 3 as previously described (21–23). Mice were followed up to 7 months with micro-CT imaging performed at 3 months and 6–7 months post-amputation. For mice that developed local recurrence or otherwise met a humane endpoint before 6–7 months post-amputation, micro-CT was performed before euthanasia.
Figure 1:
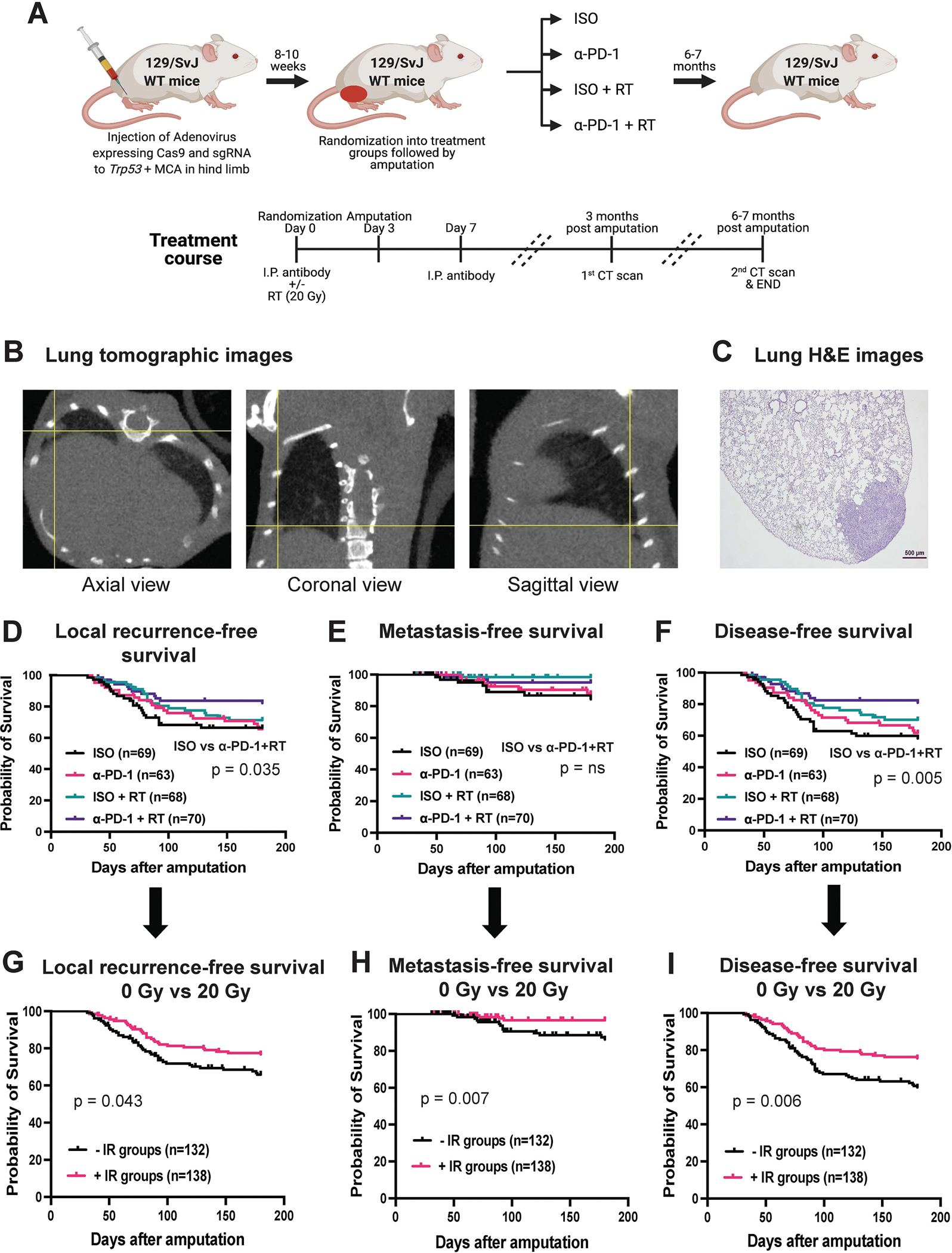
Long-term metastasis study performed using an autochthonous p53/MCA mouse model of soft-tissue sarcoma (STS) in immunocompetent 129/SvJae wild-type mice induced by CRISPR/Cas9 technology. (A) Schematic representation of experimental design showing the use of CRISPR/Cas9 technology to induce STS followed by randomization of mice into four treatment groups, i.e., isotype control antibody (ISO), anti-PD-1 antibody (α-PD-1), ISO + 20 Gy (ISO + RT), or α-PD-1 + 20 Gy (α-PD-1 + RT). (B) Representative tomographic images of lung with yellow cross-hairs centered on lung metastasis in axial, coronal, and sagittal views. (C) H&E-stained slide showing histological verification of STS metastasis corresponding with lung nodule on micro-CT in panel B. Kaplan-Meier graphs show (D) local recurrence-free survival, (E) metastasis-free survival, and (F) disease-free survival of mice after amputation of tumor-bearing hindlimb for the four treatment groups. Kaplan-Meier graphs represents (G) local recurrence-free survival, (H) metastasis-free survival, and (I) disease-free survival of mice of mice received 0 Gy RT with isotype or α-PD-1 antibody (-IR groups) vs. 20 Gy RT with isotype or α-PD-1 antibody (+IR groups). n = number of mice. p-values in Kaplan-Meier graphs were determined by Log-rank (Mantel-Cox) test. ns = non-significant.
Primary p53/MCA sarcomas were induced in Trp53fl/fl mice using intramuscular injection of adenovirus expressing Cre recombinase (Adeno-Cre; University of Iowa Viral Vector Core) followed by injection of MCA (24). Trp53fl/fl mice were randomized into two groups (ISO + RT or α-PD-1 + RT) when sarcomas reached 75–125 mm3. α-PD-1 antibody (BioXCell, BE0146) or isotype control antibody (BioXCell, BE0086) treatment was administered intraperitoneally as above on day 0 (pre-amputation), day 7 (day of amputation), and day 14 (post-amputation). Sarcomas were irradiated with 20 Gy (single fraction) on day 0 without image guidance. Mice anesthetized with continuous 2% isoflurane at a rate of 2 L oxygen/minute were positioned under lead shielding that exposed only the tumor-bearing hind limb. Irradiation was performed 50 cm from the radiation source with a dose rate of 204 cGy/min with 320 kVp X-rays, using 12.5mA and a filter consisting of 2.5mm Al and 0.1mm Cu (X-RAD 320 Biological Irradiator, Precision X-ray). Micro-CT scans were performed every three weeks post-amputation until the end of the study.
Micro-CT imaging
All micro-CT imaging was performed using a micro-CT system developed in-house (25). Animals were scanned while breathing freely under anesthesia using 2–3% isoflurane delivered by nose-cone. A pneumatic pillow was positioned on the animal’s thorax and connected to a pressure transducer to monitor respiratory motion and inform gating. Body temperature was maintained with heat lamps and a feedback controller. The X-ray source parameters were 80 kVp and 40 mA with 10 ms exposures. A total of 360 views were acquired over a full 360° rotation. The acquisition time was 3–5 min per set depending on respiratory rate and time needed for rotation to the next angle. We used prospective gating to synchronize our acquisition with respiration (16). With prospective gating, the X-ray exposure is triggered once each respiratory cycle in the desired respiratory phase, e.g., peak inspiration. The reconstruction resulted in a 63 μm isotropic voxel size using the Feldkamp algorithm (26) followed by bilateral filtration to reduce noise. The micro-CT images were converted to Hounsfield units (HU) and saved in DICOM format. The radiation dose for each micro-CT scan was ~0.017 Gy per mouse, which is ~294 to 411 times less than the LD50/30 lethal dose (5–7 Gy) in mice (27). We have previously demonstrated that our micro-CT with prospective respiratory gating facilitates sensitive detection of lung nodules down to 1.0 mm3 in size (16).
Histopathology
At the end of the study, mice were sacrificed using an IACUC-approved CO2 euthanasia technique. Gross necropsy was performed and any abnormal organ/mass, including local tumor recurrence at the amputation site, was collected and fixed in 10% neutral buffered formalin (Ricca, Cat # R319000–20F) overnight. Mouse lungs were inflated with 10% neutral buffered formalin before collection. Tissues were embedded in paraffin, sectioned, and stained with Hematoxylin and Eosin (H&E) for histological evaluation. H&E slides were blinded for the treatment group before histological analysis by a veterinary pathologist (J.E.).
Tumor initiating clone enumeration using Trp53 barcodes
Rag2+/− and Rag2−/− mice were euthanized when they developed sarcomas measuring 75–150mm3. Whole tumors were harvested and mechanically homogenized. Genomic DNA was extracted from 25 mg of the homogenized tumor using the DNeasy Blood and Tissue Kit (Qiagen, Cat # 69504). The Trp53 barcode region was amplified via PCR with high-fidelity Taq polymerase (ThermoFisher Scientific, Cat # 12337–016) and primers (ATGCTAAGCAAGTGTTGGGC, GCTGGGGAAGAAACAGGCTA). The PCR product was submitted to Genewiz for Amplicon EZ NGS sequencing with variant detection and enumeration of the barcode region. To avoid counting PCR error-related reads as Trp53 barcodes, unique sequences were defined as Trp53 barcodes if they contributed greater than 1% of the total reads. In all samples, wild-type Trp53 was identified due to the presence of stroma within the tumor; this was removed from the barcode enumeration but used to calculate total reads for each sample.
Primary mouse sarcoma cell line generation and transplantation
LSL-KrasG12D/+; Trp53fl/fl (KP) (24) and Trp53fl/fl (14) mice on a 129/SvJae background were used to generate primary sarcomas from which KP and p53/MCA cell lines were made. Sarcomas were induced by intramuscular injection of Adeno-Cre in LSL-KrasG12D/+; Trp53fl/fl mice (KP model) or injection of Adeno-Cre and MCA in Trp53fl/fl mice as above (p53/MCA model). Tumor tissue was excised and digested using a tumor dissociation kit according to the manufacturer’s protocol (Miltenyi Biotec, Cat # 130–096-730). Red blood cells were lysed using ACK lysis buffer (Lonza, Cat # BP10–548E), washed with PBS, and filtered using a 70 μM filter to get a single-cell suspension. Subsequently, cells were plated in complete DMEM media containing 4.5 mg/dL glucose, 110 mg/L pyruvate, 1% antibiotic/antimycotic, 10% fetal bovine serum, and 4 mM L-glutamine. Cells were passaged 5 times to deplete all stromal and immune cells before they were used in subsequent experiments. Cell lines were frequently tested for Mycoplasma using MycoAlert Mycoplasma Detection Kit (Lonza, Cat # LT07–318) as per the company’s protocol. However, mouse primary sarcoma cell lines were not authenticated using mouse cell-specific short tandem repeat (STR) profiling. For each experiment, cell lines were passaged 3–4 times before using them for the experiment and never used beyond passage 10 for any experiments. To generate transplant sarcomas, we injected 100,000 p53/MCA (402230 or 403865, generated from two independent primary tumors) or KP (403460 or 403492, generated from two separate tumors) mouse primary sarcoma cells resuspended in 50 μl of a 1:1 mixture of DMEM (complete media) and Matrigel (Corning, Cat # 354262) into the gastrocnemius muscle. We measured tumors three times a week with a caliper and sacrificed mice when tumors reached 1500 mm3.
Lung metastasis initiation by tail vein injection
To study the metastatic potential of sarcoma cell lines, we used primary mouse sarcomas cell lines generated in-house, as mentioned previously, and passaged 3–4 times before using them for the lung metastasis experiment. Briefly, we injected 10,000 p53/MCA (402230 or 403865) and KP (403460 or 403492) mouse primary sarcoma cells in 100 μl of PBS through an intravenous tail vein in 6–8 syngeneic 129/SvJae mice. Animals were sacrificed at 21 days post-tail vein injection using CO2 asphyxiation. Lungs were inflated with 10% neutral buffered formalin, and visible nodules were counted followed by overnight fixation. Lungs were embedded in paraffin, sectioned, and stained with H&E for histological verification of sarcoma metastases by an investigator blinded to tumor model/cell line.
Whole exome sequencing and analysis
A total of 24 sarcoma samples (two control liver samples, six amputated sarcomas with matched locally recurrent sarcomas from the ISO antibody treatment group, and five amputated sarcomas with matched locally recurrent sarcomas from α-PD-1 antibody + RT treatment group) were used for the whole exome sequencing (WES). DNA was extracted using a Quick-DNA Miniprep Plus kit (Zymo Research, cat # D4068) and sent to Novogene Co., Ltd for WES and data analysis. Initial sample purity, integrity, and quantity were measured by Agilent 5400, and samples with OD260/280 ≥ 1.8 were selected for the next step. Next, genomic DNA was randomly fragmented into small pieces (180–280 bp). DNA fragments were end-repaired, A-tailed, and ligated using Illumina adapters followed by DNA fragment PCR amplification, size selection, and purification. Finally, the prepped libraries were hybridized with biotin-labeled probes and the targeted exons were captured by streptavidin magnetic beads. After quality check, enriched captured libraries were pooled and sequenced on the Illumina platform using the PE150 strategy (6 Gb raw data per sample). Raw data quality control, sequence alignment, variant detection, and somatic mutation detection were performed by the bioinformatics group at Novogene Co., Ltd., while statistical analysis was performed by QuantBio LLC (Durham, NC).
Read pairs were excluded when one or both of the reads in the pair contained adapter contamination, >10% of the bases were ambiguous (N), or >50% of the bases had Phred score <5. BWA-MEM (v0.7.17) (28) was run with default parameters to align cleaned reads to the GRCm38 (mm10) genome (29). After sorting with samtools (v1.8) (30) PCR duplicates were marked with Picard tools MarkDuplicates (v2.18.9, http://sourceforge.net/projects/picard/). Somatic mutations (SNPs and indels) in tumor samples were called independently with each of the two normal liver samples. muTect (v1.1.4) (31) was used to call somatic single nucleotide variants (SNVs) from bam files utilizing tumor and normal samples. Somatic indels were identified with Strelka (v2.9.4) (32). SNVs and indels were annotated with ANNOVAR (v2015Mar22) (33) and with dbSNP v138 mm10 SNPs. SNPs were included in the analysis if they passed default filtering thresholds. Somatic copy number variants (CNV) were detected utilizing CNVkit (34) (v 0.9.6) with the batch command after constructing a copy number reference utilizing both normal samples. Gene level log2 copy-number ratios (log2 CNR) were calculated using the genemetrics command without thresholding or filtering the data. Log2 CNR normalized to copy-number reference was converted to absolute calls by exponentiating and assuming a normal diploid reference.
For each sample, variants were called independently with each of the separate liver samples. SNPs were retained for analysis if they met the following criteria: 1) identical SNP was identified when run with each of the normal samples, 2) had an allele frequency of >10%, and 3) was not annotated as intergenic, intronic, silent, found in RNA, or unknown, and 4) had a minimum of four reads for the alternate allele. For SNP and CNV analysis, paired t-tests were utilized to determine differences in the number of SNPs, copy number gains, and copy number losses between amputation (Amp) and local recurrence (LR) samples. For SNP overlap analysis, mutations were included if they were observed in at least three samples per tumor status/treatment group combination (e.g., Amp (ISO) vs Amp (LR)). For SNP pathway analysis, a hypergeometric test was utilized to test for overlap in shared mutations within each tumor status/treatment group in combination with MSigDB Hallmark pathways translated to mouse genes obtained from MSSigDB (35). All statistical analyses were conducted in the R statistical programming environment (v4.1.2). Amp_2221 was excluded from all WES analyses given its low-quality reads and outlier status on principal component analysis. In addition, LR_2221 was excluded from analyses where paired tests were conducted.
Statistical analysis
All amputated mice were followed for 6 to 7 months for signs of lung metastasis. Metastasis-free survival was defined as the time from amputation to euthanasia/death with histologic confirmation of distant metastases. Local recurrence-free survival was defined as the time from amputation to euthanasia/death with histologic confirmation of local recurrence at the amputation site. Disease-free survival was defined as the time from amputation to euthanasia/death with histologic confirmation of local recurrence and/or distant metastases. Survival curves were estimated after surgery using the Kaplan-Meier method, and pairwise significance was determined by log-rank test with the control arm depending on the experiment: (1) ISO group compared to α-PD-1, ISO + RT, or α-PD-1 + RT for 129/SvJae mice with p53/MCA tumors; (2) Rag2+/− mice with p53/MCA tumors compared to immunodeficient Rag2−/− mice; (3) ISO + RT group compared to α-PD-1 + RT for Trp53fl/fl mice with p53/MCA tumors. We performed Student’s t-test to compare time to tumor initiation and the number of tumor-initiating clones between Rag2+/− and Rag2−/− mice. We also performed Student’s t-test to compare the number of visible lung nodules found between KP and p53/MCA cell lines. A two-sided p < 0.05 was considered to be statistically significant. All statistical analyses were conducted in SAS 9.4 software.
Data availability:
Whole exome sequencing dataset is available for download from a publicly accessible database, Sequence Read Archive (SRA), using BioProject ID # PRJNA873947. Any other data will be available from the corresponding authors upon request.
Results:
Preoperative radiation therapy improves local control, metastasis-free survival, and disease-free survival
To mimic the phase II clinical trial SU2C-SARC032 (Supplementary figure 1), we generated autochthonous sarcomas with high tumor mutational burden (14,15) in immunocompetent 129/SvJae wild-type mice using CRISPR/Cas9 technology (Figure 1A). Micro-CT imaging was performed to detect lung metastasis using our previously published protocol (16), which provides reliable sensitivity to detect lesions ≥1.0 mm3 with respiratory gating. A representative micro-CT image of STS lung metastasis is shown in Figure 1B and was verified by lung histology (Figure 1C). In addition to lung metastases, we also detected other lung abnormalities such as inflammation (Supplementary figure 2A and 2B). Interestingly, a recent study showed that pneumonitis developed in roughly one-sixth of non-small cell lung cancer patients who received α-PD-1 treatment (36). However, we found that the frequency of lung inflammation was not associated with the treatment group (ISO vs α-PD-1) in this autochthonous STS mouse model (Supplementary figure 2B). Micro-CT images also identified lung infiltrates, nodules, or suspicious opacities that corresponded with abnormalities other than sarcoma metastases, such as lymphoma, pulmonary adenoma, or hemorrhage (Supplementary figure 2C).
Compared to mice treated with surgery and ISO, mice treated with surgery and α-PD-1 + RT showed significantly improved local recurrence-free survival (p=0.035), but not metastasis-free survival (Figure 1D and 1E). The improved local recurrence-free survival for mice treated with α-PD-1 + RT and surgery translated into significantly better disease-free survival compared to the surgery and ISO control group (p=0.005) (Figure 1F). However, we noted that α-PD-1 + RT and surgery was not significantly better in improving local recurrence-free, metastasis-free, and disease-free survival rate than ISO + RT and surgery, which represents the clinical standard of care for patients with large and/or high-grade sarcomas (Supplementary table 1). The rate of metastases in the ISO control group was unexpectedly low (12%, Figure 1E), as we had previously observed a rate of lung metastasis in the LSL-KrasG12D/+; Trp53fl/fl (KP) model (24) of approximately 40% (21–23). The unexpectedly low rate of metastases in the p53/MCA model limited the power to detect significant differences in metastases across the four treatment groups. To evaluate the effect of α-PD-1 and RT separately on outcomes after surgery, we combined the treatment groups as follows: ISO groups (ISO and ISO + RT), α-PD-1 groups (α-PD-1 and α-PD-1 + RT), - RT groups (ISO and α-PD-1), and + RT groups (ISO + RT and α-PD-1 + RT). Mice that received radiation treatment prior to surgery (ISO + RT and α-PD-1 + RT) showed significantly higher local recurrence-free survival (p = 0.043) and metastasis-free survival (p = 0.007) compared to unirradiated treatment groups (ISO and α-PD-1) (Figure 1G and 1H). Moreover, mice that received radiation therapy prior to surgery (ISO + RT and α-PD-1 + RT) showed significantly higher disease-free survival (p = 0.006) compared to mice in unirradiated treatment groups (ISO and α-PD-1) (Figure 1I). In contrast, the addition of α-PD-1 (α-PD-1 and α-PD-1 + RT) did not improve local-recurrence-free survival, metastasis-free survival, or disease-free survival over isotype control antibody (ISO and ISO+RT) (Supplementary figure 3A–C). These results indicate that in the p53/MCA model of STS, radiation therapy to the tumor-bearing hind limb may contribute to reducing metastases by decreasing local recurrence. Given the low rate of lung metastases in the p53/MCA autochthonous mouse model, we lack sufficient power to determine whether neoadjuvant and adjuvant α-PD-1 therapy with or without radiation therapy impacts the rate of metastases.
Comparing primary tumors to locally recurrent sarcomas
To identify possible genomic differences between primary and locally recurrent sarcomas, we performed whole exome sequencing (WES) of amputated (Amp) and matched locally recurrent (LR) tumors from mice treated with ISO antibody or α-PD-1 + RT. We analyzed WES of matched amputated and locally recurrent tumors to identify somatic mutations and copy number alterations within each tumor compared with two control liver samples. Interestingly, we found that LR tumors from ISO and α-PD-1 + RT treatment groups had significantly higher single nucleotide polymorphisms (SNPs) and copy number alterations (CNAs) compared to matched Amp tumors from ISO and α-PD-1 + RT treatment groups (Figure 2A and 2B). Since Amp and LR tumors from both treatment groups were sequenced at similar depth, it is possible that lower clonality in LR tumors was responsible for the detection of more mutational events compared to Amp tumors. Next, we performed a hypergeometric test to identify significant overlaps between genes mutated within treatment groups and evaluated frequently mutated hallmark pathways. We found that myogenesis genes were predominantly mutated in LR tumors (regardless of treatment), while Kras pathway genes were predominantly mutated in Amp and LR tumors from the ISO treatment group (Figure 2C). Interestingly, we noted that epithelial-mesenchymal transition (EMT) genes were frequently mutated in all tumors (Figure 2C), possibly contributing to the low rate of metastasis in mice with high-mutational burden p53/MCA-induced sarcomas.
Figure 2:
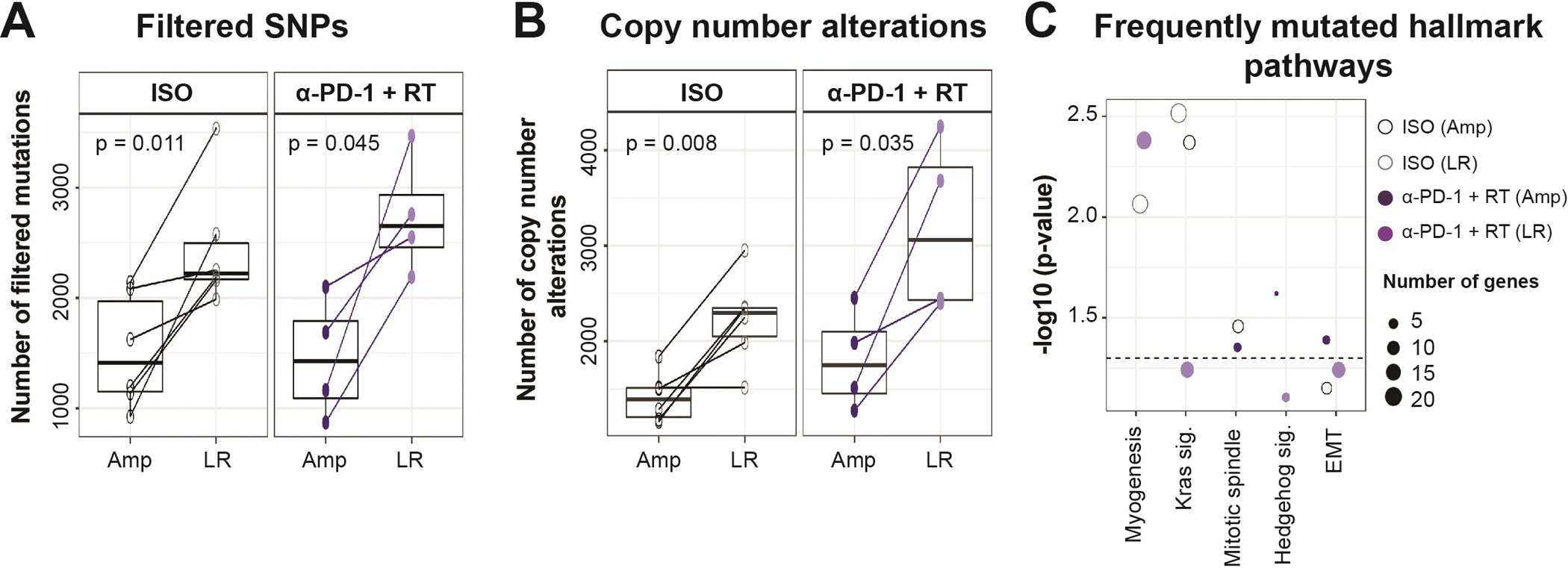
Whole exome sequencing (WES) analysis of amputated versus locally recurrent sarcomas. Boxplots represent (A) Filtered SNPs (excluding silent, RNA, intronic, and unknown mutations), and (B) Copy number alterations (including gains and losses) by treatment status and tumor type. (C) Top frequently mutated hallmark pathways determined by hypergeometric test. WES included 4–6 sarcoma samples for each tumor type and treatment group. Amp – amputated tumor, LR – locally recurrent tumor, EMT – epithelial-mesenchymal transition. p-values in boxplots were determined by paired t-tests.
Cas9 does not impact the rate of metastasis in the p53/MCA model
One difference between the KP sarcoma model with rates of lung metastasis of approximately 40% (21–23) and the p53/MCA sarcoma model with a much lower rate of metastasis (12%, Figure 1C) is that the p53/MCA sarcomas described above were generated with CRISPR/Cas9 technology, which could potentially activate the host immune system (37). To investigate if the low rate of lung metastasis was a consequence of initiating sarcomas with Cas9, we utilized Cre-LoxP technology to generate a high-mutational burden p53/MCA sarcomas in Trp53fl/fl mice. We induced sarcomas by injecting the gastrocnemius muscle with an adenovirus expressing Cre recombinase and then with MCA. We randomized tumor-bearing mice into two treatment groups prior to surgery: isotype control antibody + 20 Gy (ISO + RT) or anti-PD-1 antibody + 20 Gy (α-PD-1 + RT) (Supplementary figure 4A and 4B). As in the CRISPR/Cas9-induced p53/MCA model (Figure 1C), we observed a low rate of metastases in both treatment groups, suggesting that initiating tumors with CRISPR/Cas9 did not contribute to the low rate of lung metastasis (Supplementary figure 4C). Given the low rate of metastases in the Cre/loxP-induced p53/MCA model, we were underpowered to determine if metastasis-free survival was different between the α-PD-1 + RT and ISO + RT groups.
Number of tumor-initiating clones and metastasis-free survival not affected by the presence of mature lymphocytes
Clinical studies have shown that high tumor mutational load is associated with a better response to immune checkpoint blockade therapies and consequently improved overall survival (38,39). Previously, we have shown that the p53/MCA murine STS model exhibits a high mutational burden, leading to the expression of neoantigens (14,15). Novel neoantigen expression could make p53/MCA sarcoma cells highly immunogenic and thus susceptible to immune-mediated elimination, reducing the development of lung metastases. Therefore, we hypothesized that immunosurveillance by lymphocytes in immunocompetent 129/SvJae wild-type mice contributed to a low rate of lung metastases (12%) through the elimination of immunogenic clones with the potential to form metastases. To test this hypothesis, we used CRISPR/Cas9 technology (19) to induce p53/MCA sarcomas (15) in lymphocyte-deficient Rag2−/− mice and littermate control immunocompetent Rag2+/− mice (Figure 3A). We have previously shown that primary p53/MCA sarcomas initiated in Rag2−/− mice have approximately a 2-fold higher number of non-synonymous mutations and a higher level of neoantigen expression compared to p53/MCA sarcomas initiated in Rag2+/− mice (15). Time to tumor initiation did not differ between Rag2+/− versus Rag2−/− mice (Figure 3B). When sarcomas reached 75–125 mm3, we amputated the tumor-bearing hind limb and observed mice for up to 7 months for signs of lung metastases. CRISPR-barcoding was performed to determine the number of tumor-initiating clones from the resected tumor. The number of tumor-initiating clones did not differ between Rag2+/− vs. Rag2−/− mice, suggesting that mature B and T cells were unable to eliminate tumor-initiating clones from high-mutational burden p53/MCA sarcomas (Figure 3C). Next, we compared local recurrence-free and metastasis-free survival after surgery alone in Rag2+/− and Rag2−/− mice. We found that Rag2−/− mice had similar local recurrence-free survival (Figure 3D) and metastasis-free survival (Figure 3E) compared to Rag2+/− mice. These results indicate that the presence of mature lymphocytes does not affect local recurrence-free and metastasis-free survival of mice bearing high-mutational load p53/MCA sarcomas. Thus, the low rate of metastasis in the p53/MCA-induced STS mouse model is not secondary to lymphocyte-mediated elimination of tumor cells with metastatic potential.
Figure 3:
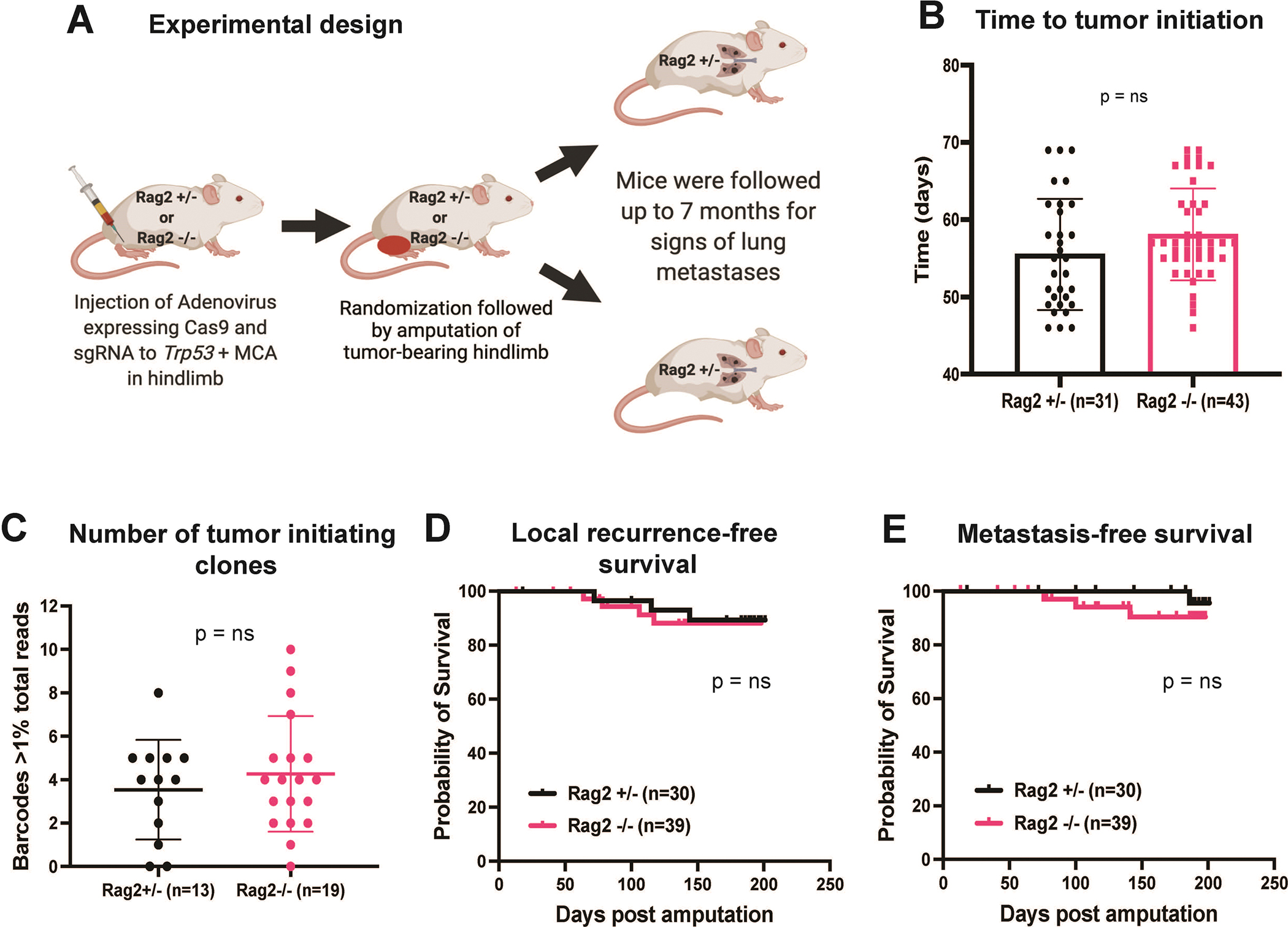
Long-term metastasis study performed using an autochthonous p53/MCA mouse model of STS in immuno-proficient (Rag2+/−) vs. immuno-deficient (Rag2−/−) mice. (A) Schematic representation of tumor induction in Rag2 mice using CRISPR/Cas9 technology followed by amputation of tumor-bearing hindlimb. (B) Bar graph represents a time to tumor initiation in Rag2+/− vs. Rag2−/− mice. (C) Bar graph shows the number of tumor-initiating clones determined by CRISPR barcoding in STS arising from Rag2+/− vs. Rag2−/− mice. Kaplan-Meier graphs represent (D) local recurrence-free survival and (E) metastasis-free survival of mice post amputation of the tumor-bearing hindlimb. n = number of mice; data plotted are means ± SD; p-values were determined by student’s t-test in bar graph and Log-rank (Mantel-Cox) test in Kaplan-Meier graph. ns = non-significant.
Low rate of sarcoma metastases in p53/MCA sarcomas due to tumor cell-intrinsic factors
Others have similarly observed the low metastatic potential for MCA-induced sarcomas in mice (40). To determine whether the low metastatic potential of p53/MCA sarcomas relative to KP sarcomas is due to tumor cell-intrinsic factors, we generated STS cell lines from p53/MCA (Trp53fl/fl + MCA) and KP (LSL-KrasG12D/+; Trp53fl/fl) mouse models using adenovirus expressing Cre recombinase. We transplanted MCA-induced (p53/MCA: 402230 and 403865) and non-MCA-induced (KP: 403460 and 403492) STS cell lines orthotopically into the hind limb of syngeneic mice. While transplantation of both MCA-induced and non-MCA-induced STS cell lines led to tumor formation, the p53/MCA cell lines had a slightly lower penetrance and tumor growth rate (Figure 4A and 4B) compared to KP cell lines (figure 4C and 4D). Next, we performed intravenous tail vein injection of KP or p53/MCA STS cells and sacrificed mice 21 days later to determine the metastatic potential of these cell lines. KP cell lines had a significantly higher potential to metastasize to the lung compared to p53/MCA cell lines, as evidenced by the quantification of visible lung nodules at the time of euthanasia (Figure 4E, p<0.0001). Histologic analysis of lungs from mice with tail vein injection of p53/MCA cell lines showed no metastases, whereas sarcoma metastases were confirmed in the lungs of all mice with tail vein injection of KP cell lines (Figure 4F). These results suggest that the low rate of metastases in the p53/MCA model is due to tumor cell-intrinsic processes, potentially related to mutations induced by MCA.
Figure 4:
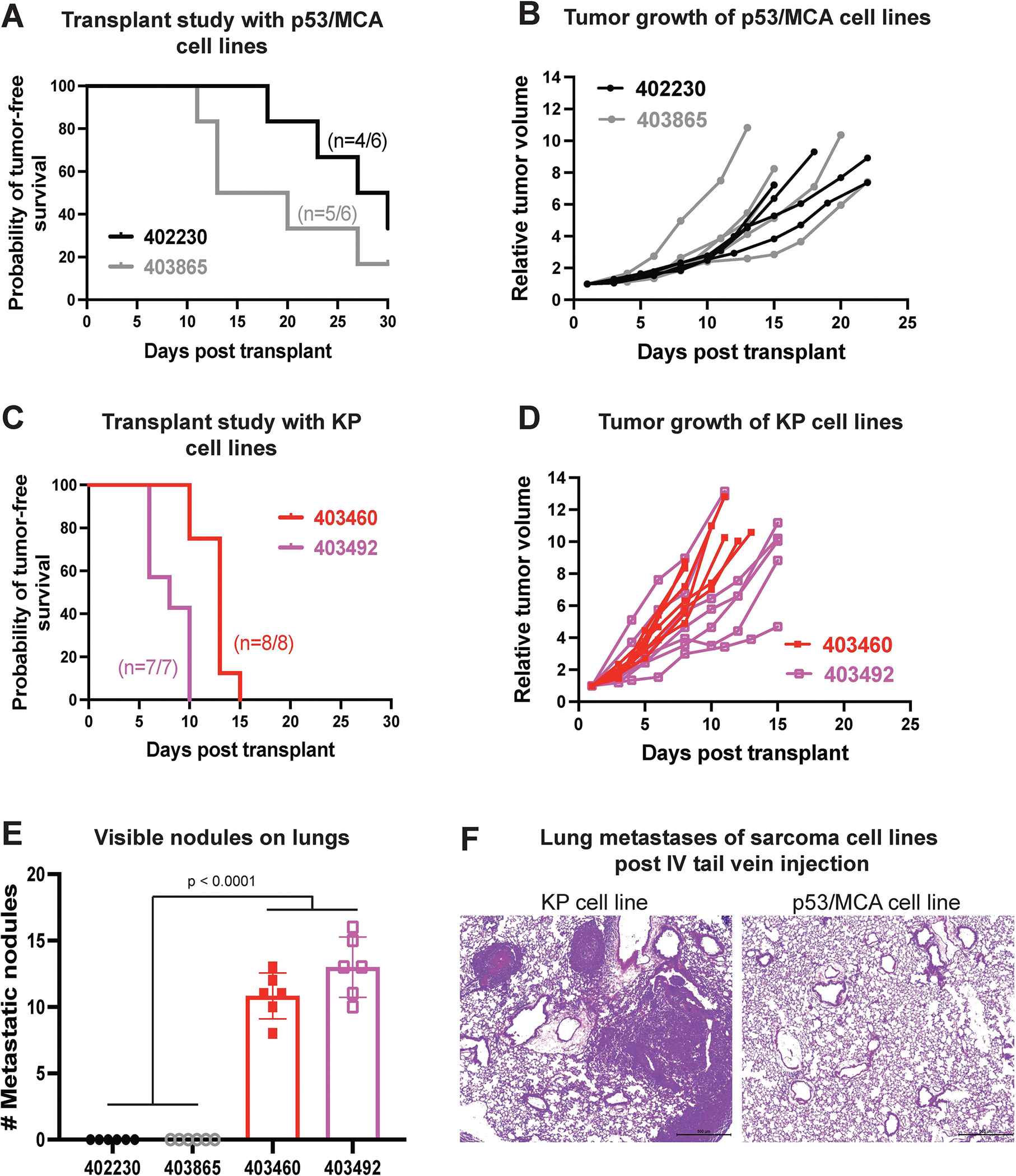
Metastatic potential of sarcoma cell lines measured by intravenous injection of sarcoma cells into immunocompetent 129/SvJae mice. In vivo transplant study shows (A) rate of tumor initiation and (B) rate of tumor growth post intramuscular injection of p53/MCA sarcoma cell lines generated using CRISPR/Cas9 technology. In vivo transplant study shows (C) rate of tumor initiation and (D) rate of tumor growth post intramuscular injection of KP sarcoma cell lines generated using Cre-LoxP technology. (E) Bar graph represents the number of visible nodules detected in the lungs of mice that received either p53/MCA or KP sarcoma cells intravenously via tail vein injection. (F) Representative histological images of lung metastases of sarcoma cell lines post intravenous tail vein injection of either p53/MCA or KP cell lines. n = number of mice; data plotted are means ± SD; p-values were determined by student’s t-test in bar graph.
Discussion:
The current ongoing phase II clinical trial SU2C-SARC032 (NCT03092323) provided a unique opportunity to perform a co-clinical trial using a genetically engineered mouse model of STS. The goal of the co-clinical trial was (1) to optimize and standardize preclinical imaging techniques, such as micro-MRI and micro-CT, and (2) to test the efficacy of neoadjuvant and adjuvant α-PD-1 treatment in combination with neoadjuvant RT and surgical resection. With prospective respiratory gating, metastatic lung nodules as small as 1.0 mm3 were detectable by micro-CT, but histological verification was required as imaging could not differentiate a lung nodule representing sarcoma metastasis from lung inflammation or lung adenoma. This is a clear limitation of anatomical CT imaging due to the low contrast capabilities of this X-ray-based modality. Nevertheless, in vivo micro-CT is still a useful screening method to survey for the development of lung metastases using low-dose scans because the same mice can be imaged at multiple time points rather than increasing the number of mice to perform histology at multiple time points. To address the challenge of detecting lung nodules in a large number of scans from mice, we have also recently implemented a deep learning method (41).
In this preclinical study, we found that treatment with neoadjuvant RT with or without α-PD-1 therapy significantly improved local recurrence-free survival and metastasis-free survival compared to surgery alone. Because the rate of metastasis was unexpectedly low, we were unable to determine the impact of adding neoadjuvant and adjuvant α-PD-1 treatment to neoadjuvant RT and surgery on metastasis-free survival. In the clinical management of STS, RT plays a crucial role to improve local control for patients with high-risk STS, and it also plays an increasing role in non-resectable and oligometastatic disease (42,43). Similarly, our preclinical study in mice revealed that RT decreased the rate of local recurrence (Figure 1E) and remarkably also decreased the rate of metastases (Figure 1F). While two randomized clinical trials (44,45) have not shown a statistically significant improvement in overall survival with the addition of RT to surgery for STS, they were relatively small studies that were not powered for an overall survival endpoint. Larger database studies (46,47) have suggested that neoadjuvant or adjuvant RT is associated with significantly improved overall survival. While these studies are limited by their retrospective nature and lack of data on cause-specific or metastasis-free survival, our preclinical results showing improved local control and reduced metastases in mice receiving radiation therapy support these clinical data suggesting a survival benefit with RT for patients with STS. Although limited by the low number of metastatic events (particularly in mice receiving RT), the available data from this study suggest that α-PD-1 treatment lacks an abscopal effect in this primary p53/MCA model of STS.
Our previously published data revealed that a single fraction of 20 Gy RT dramatically reshaped the distribution of myeloid cells compared to unirradiated p53/MCA-induced sarcomas. RT significantly increases macrophage infiltration in the TME, and these infiltrating macrophages express high levels of Ccl2 and Ly6C (consistent with active phagocytosis), Fcgr2B, Fcgr1, and Fcgr3 (consistent with antibody-dependent cell-mediated cytotoxicity), and B2m, Ctsl, Ctsd, Ctsb, and Ctss (consistent with antigen/protein processing) (15). In addition, RT affects the expression of transcription factors responsible for the increased expression of interferon response genes, such as Mx1, Ifit1, Ifit2, Ifit3, and Ifit3b. Thus, RT treatment repolarizes myeloid cells via activation of type I and II interferon response pathways. Studies have shown that highly inflamed TME caused by RT correlates with a better response to immune checkpoint blockade therapy (48). Furthermore, WES data from the p53/MCA model revealed frequent mutations in genes involved in the epithelial-mesenchymal transition (EMT) pathway, regardless of treatment status. Previous studies have shown that mutations in EMT genes reduce circulating cancer cells’ ability to extravasate from blood vessels to colonize distant sites, thus reducing their metastasizing potential (49). In future studies, we will utilize single cell clones generated from this model system to work towards identifying specific genes regulating sarcoma metastasis.
We recently reported an in-depth characterization of the p53/MCA model (15), which included whole exome sequencing, bulk RNA sequencing (RNA-Seq), and single-cell analysis of the CD45+ hematopoietic cells using single-cell RNA-Seq and mass cytometry. CIBERSORTx analysis of the RNA-Seq data demonstrated that the immune infiltrates in the autochthonous p53/MCA sarcomas resemble most human STS. However, a subset of human sarcomas has an immune-enriched microenvironment (termed sarcoma immune class E), which may be a biomarker for response to α-PD-1 immunotherapy (50). We previously showed that injection of p53/MCA cell lines into the gastrocnemius muscle of syngeneic mice creates an inflamed tumor immune microenvironment that resembles human STS with sarcoma immune class E and these transplanted mouse p53/MCA sarcomas show the synergy between RT and α-PD-1 therapy (15). Therefore, it is conceivable that in the subset of human STS with an immune-enriched microenvironment, the combination of RT and α-PD-1 may lead to a systemic immune response that decreases the rate of lung metastases. We will investigate this possibility in planned correlative studies from SU2C-SARC032.
In the high tumor mutational burden p53/MCA primary sarcomas, mature lymphocytes did not play a role in regulating metastasis by immunosurveillance. These results are consistent with other studies in the MCA-induced sarcomas (51,52). Our results from tail vein injection experiments support the notion that MCA-driven high tumor mutation burden STS have an impaired ability to colonize the lung, possibly due to intrinsic rather than extrinsic factors. Many preclinical studies testing the impact of immune checkpoint inhibitors (ICIs) have been performed using transplant tumor models in syngeneic mice, which can create an inflamed tumor microenvironment (15,53) and may overestimate the response rate of ICIs in the clinic. Although these transplant tumor models may mimic a subset of immune-enriched human cancers (15), a primary mouse model may better reflect the response of most human cancers to ICIs because autochthonous tumors grow under constant immune surveillance and these primary tumors also model intra- and inter-tumor heterogeneity. Thus, we selected the p53/MCA autochthonous sarcoma model to study the impact of neoadjuvant and adjuvant α-PD-1 treatment on metastasis-free survival. Unfortunately, we were underpowered for this endpoint despite a robust sample size of an average of 70 mice per treatment group because of the low rate of metastasis in the p53/MCA sarcoma model (∼12%) compared to high-risk human STS patients (∼50%) (4). This low rate of metastasis underpowered the preclinical study to test whether or not adding α-PD-1 to RT and surgical resection provides systemic anti-tumor immune effects to improve metastasis-free survival. It is also possible that CD8+ T cells are not responsible for eliminating tumor clones from the circulation or lung microenvironment and thus have no impact on adding α-PD-1 to the metastasis rate. In fact, a previous study reported that Natural Killer T cells play a critical role in immunosurveillance of MCA-induced sarcomas through interleukin (IL)-12 or α-galactosylceramide (α-GalCer) (54). Thus, it is possible that Natural Killer T cells play a role in improving metastasis-free survival in MCA-induced sarcomas, which warrants further study.
In summary, we have successfully implemented the preclinical arm of a co-clinical trial of combined RT and immunotherapy in sarcoma. We find that RT improves the rate of local recurrence-free survival and metastasis-free survival. However, the addition of α-PD-1 to RT was unable to improve metastasis-free survival. Our work illustrates the strengths and weaknesses of the p53/MCA sarcoma model. Autochthonous mouse models can complement preclinical transplant models to study important clinical questions in RT and immunotherapy (55). This study also demonstrates the successful incorporation of optimized small animal imaging such as micro-CT into a co-clinical trial. By establishing clinically-driven preclinical imaging methods to serve in a co-clinical trial, we have created a pipeline that reduces the gap between preclinical and clinical studies of sarcoma therapy.
Supplementary Material
Acknowledgments:
We are grateful to all our funders who provided valuable resources to complete the study. This work was funded by U24 CA220245 (Y.M.M., C.T.B.), RF1AG070149 (C.T.B.), ASCO Young Investigator Award (Y.M.M.), RSNA Research Resident/Fellow Grant (Y.M.M.), Sarcoma Alliance for Research through Collaboration (SARC) SPORE 5U54CA168512 (D.G.K. and Y.M.M.), R35CA197616 (D.G.K), a grant from Emerson Collective (D.G.K.), and the Duke Cancer Center Support Grant (P30CA14236).
Footnotes
Conflict of Interest: D.G.K. and Y.M.M. are recipients of a Stand Up To Cancer (SU2C) MSD Catalyst Grant studying pembrolizumab and radiation therapy in sarcoma patients. D.G.K. has received research funding from XRAD Therapeutics, Eli Lilly & Co., Bristol Myers Squibb, Varian Medical Systems, and Merck. D.G.K. owns stock and is on the scientific advisory board of Lumicell, Inc., which is commercializing intraoperative imaging technology. D.G.K. is a cofounder of XRAD Therapeutics, which is developing radiosensitizers. The other authors declare no conflicts of interest.
References:
- 1.Jo VY, Doyle LA. Refinements in Sarcoma Classification in the Current 2013 World Health Organization Classification of Tumours of Soft Tissue and Bone. Surg Oncol Clin N Am 2016;25(4):621–43 doi 10.1016/j.soc.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Wang D, Zhang Q, Eisenberg BL, Kane JM, Li XA, Lucas D, et al. Significant Reduction of Late Toxicities in Patients With Extremity Sarcoma Treated With Image-Guided Radiation Therapy to a Reduced Target Volume: Results of Radiation Therapy Oncology Group RTOG-0630 Trial. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2015;33(20):2231–8 doi 10.1200/JCO.2014.58.5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Sullivan B, Davis AM, Turcotte R, Bell R, Catton C, Chabot P, et al. Preoperative versus postoperative radiotherapy in soft-tissue sarcoma of the limbs: a randomised trial. Lancet 2002;359(9325):2235–41 doi 10.1016/S0140-6736(02)09292-9. [DOI] [PubMed] [Google Scholar]
- 4.Borden EC, Baker LH, Bell RS, Bramwell V, Demetri GD, Eisenberg BL, et al. Soft tissue sarcomas of adults: state of the translational science. Clin Cancer Res 2003;9(6):1941–56. [PubMed] [Google Scholar]
- 5.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. The New England journal of medicine 2015;373(17):1627–39 doi 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. The New England journal of medicine 2015;372(21):2018–28 doi 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 7.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. The New England journal of medicine 2015;373(1):23–34 doi 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xin Yu J, Hubbard-Lucey VM, Tang J. Immuno-oncology drug development goes global. Nat Rev Drug Discov 2019;18(12):899–900 doi 10.1038/d41573-019-00167-9. [DOI] [PubMed] [Google Scholar]
- 9.Tawbi HA, Burgess M, Bolejack V, Van Tine BA, Schuetze SM, Hu J, et al. Pembrolizumab in advanced soft-tissue sarcoma and bone sarcoma (SARC028): a multicentre, two-cohort, single-arm, open-label, phase 2 trial. The Lancet Oncology 2017;18(11):1493–501 doi 10.1016/S1470-2045(17)30624-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaur P, Asea A. Radiation-induced effects and the immune system in cancer. Front Oncol 2012;2:191 doi 10.3389/fonc.2012.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalbasi A, June CH, Haas N, Vapiwala N. Radiation and immunotherapy: a synergistic combination. The Journal of clinical investigation 2013;123(7):2756–63 doi 10.1172/JCI69219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Twyman-Saint Victor C, Rech AJ, Maity A, Rengan R, Pauken KE, Stelekati E, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature 2015;520(7547):373–7 doi 10.1038/nature14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siva S, MacManus MP, Martin RF, Martin OA. Abscopal effects of radiation therapy: a clinical review for the radiobiologist. Cancer Lett 2015;356(1):82–90 doi 10.1016/j.canlet.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 14.Lee CL, Mowery YM, Daniel AR, Zhang D, Sibley AB, Delaney JR, et al. Mutational landscape in genetically engineered, carcinogen-induced, and radiation-induced mouse sarcoma. JCI Insight 2019;4(13) doi 10.1172/jci.insight.128698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wisdom AJ, Mowery YM, Hong CS, Himes JE, Nabet BY, Qin X, et al. Single cell analysis reveals distinct immune landscapes in transplant and primary sarcomas that determine response or resistance to immunotherapy. Nature communications 2020;11(1):6410 doi 10.1038/s41467-020-19917-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blocker SJ, Holbrook MD, Mowery YM, Sullivan DC, Badea CT. The impact of respiratory gating on improving volume measurement of murine lung tumors in micro-CT imaging. PLoS One 2020;15(2):e0225019 doi 10.1371/journal.pone.0225019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blocker SJ, Mowery YM, Holbrook MD, Qi Y, Kirsch DG, Johnson GA, et al. Bridging the translational gap: Implementation of multimodal small animal imaging strategies for tumor burden assessment in a co-clinical trial. PloS one 2019;14(4):e0207555 doi 10.1371/journal.pone.0207555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nature genetics 2001;29(4):418–25 doi 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 19.Huang J, Chen M, Whitley MJ, Kuo HC, Xu ES, Walens A, et al. Generation and comparison of CRISPR-Cas9 and Cre-mediated genetically engineered mouse models of sarcoma. Nature communications 2017;8:15999 doi 10.1038/ncomms15999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newton J, Oldham M, Thomas A, Li Y, Adamovics J, Kirsch DG, et al. Commissioning a small-field biological irradiator using point, 2D, and 3D dosimetry techniques. Med Phys 2011;38(12):6754–62 doi 10.1118/1.3663675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisinger-Mathason TS, Zhang M, Qiu Q, Skuli N, Nakazawa MS, Karakasheva T, et al. Hypoxia-dependent modification of collagen networks promotes sarcoma metastasis. Cancer discovery 2013;3(10):1190–205 doi 10.1158/2159-8290.CD-13-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sachdeva M, Mito JK, Lee CL, Zhang M, Li Z, Dodd RD, et al. MicroRNA-182 drives metastasis of primary sarcomas by targeting multiple genes. The Journal of clinical investigation 2014;124(10):4305–19 doi 10.1172/JCI77116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang YJ, Huang J, Tsushima H, Ban GI, Zhang H, Oristian KM, et al. Tracing Tumor Evolution in Sarcoma Reveals Clonal Origin of Advanced Metastasis. Cell reports 2019;28(11):2837–50 e5 doi 10.1016/j.celrep.2019.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirsch DG, Dinulescu DM, Miller JB, Grimm J, Santiago PM, Young NP, et al. A spatially and temporally restricted mouse model of soft tissue sarcoma. Nature medicine 2007;13(8):992–7 doi 10.1038/nm1602. [DOI] [PubMed] [Google Scholar]
- 25.Holbrook M, Clark D, Badea C. Low-dose 4D cardiac imaging in small animals using dual source micro-CT. Physics in Medicine & Biology 2018;63(2):025009 doi ARTN 025009 10.1088/1361-6560/aa9b45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feldkamp LA, Davis LC, Kress JW. Practical cone-beam algorithm. J Opt Soc Am 1984;1(6):612–19. [Google Scholar]
- 27.Badea CT, Hedlund LW, Johnson GA. Micro-CT with respiratory and cardiac gating. Medical Physics 2004;31(12):3324–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25(14):1754–60 doi 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome research 2002;12(6):996–1006 doi 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25(16):2078–9 doi 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature Biotechnology 2013;31(3):213–9 doi 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012;28(14):1811–7 doi 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 33.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research 2010;38(16):e164 doi 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput Biol 2016;12(4):e1004873 doi 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1(6):417–25 doi 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koyauchi T, Inui N, Karayama M, Kitahara Y, Takuma S, Amano Y, et al. Clinical Outcomes of Anti-programmed Death-1 Antibody Related Pneumonitis in Patients with Non-Small Cell Lung Cancer. SN Comprehensive Clinical Medicine 2020;2:570–8. [Google Scholar]
- 37.Ajina R, Zamalin D, Zuo A, Moussa M, Catalfamo M, Jablonski SA, et al. SpCas9-expression by tumor cells can cause T cell-dependent tumor rejection in immunocompetent mice. Oncoimmunology 2019;8(5):e1577127 doi 10.1080/2162402X.2019.1577127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348(6230):124–8 doi 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan TA, Wolchok JD, Snyder A. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. The New England journal of medicine 2015;373(20):1984 doi 10.1056/NEJMc1508163. [DOI] [PubMed] [Google Scholar]
- 40.Bottazzi B, Mantovani A, Taraboletti G, Giavazzi R. Characterization of spontaneous metastases from autochthonous 3-methylcholanthrene-induced tumors. Invasion Metastasis 1986;6(1):44–57. [PubMed] [Google Scholar]
- 41.Holbrook MD, Clark DP, Patel R, Qi Y, Bassil AM, Mowery YM, et al. Detection of Lung Nodules in Micro-CT Imaging Using Deep Learning. Tomography 2021;7(3):358–72 doi 10.3390/tomography7030032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roeder F Radiation Therapy in Adult Soft Tissue Sarcoma-Current Knowledge and Future Directions: A Review and Expert Opinion. Cancers (Basel) 2020;12(11) doi 10.3390/cancers12113242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beane JD, Yang JC, White D, Steinberg SM, Rosenberg SA, Rudloff U. Efficacy of adjuvant radiation therapy in the treatment of soft tissue sarcoma of the extremity: 20-year follow-up of a randomized prospective trial. Ann Surg Oncol 2014;21(8):2484–9 doi 10.1245/s10434-014-3732-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang JC, Chang AE, Baker AR, Sindelar WF, Danforth DN, Topalian SL, et al. Randomized prospective study of the benefit of adjuvant radiation therapy in the treatment of soft tissue sarcomas of the extremity. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 1998;16(1):197–203 doi 10.1200/jco.1998.16.1.197. [DOI] [PubMed] [Google Scholar]
- 45.Pisters PW, Harrison LB, Leung DH, Woodruff JM, Casper ES, Brennan MF. Long-term results of a prospective randomized trial of adjuvant brachytherapy in soft tissue sarcoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 1996;14(3):859–68 doi 10.1200/JCO.1996.14.3.859. [DOI] [PubMed] [Google Scholar]
- 46.Ramey SJ, Yechieli R, Zhao W, Kodiyan J, Asher D, Chinea FM, et al. Limb-sparing surgery plus radiotherapy results in superior survival: an analysis of patients with high-grade, extremity soft-tissue sarcoma from the NCDB and SEER. Cancer Med 2018;7(9):4228–39 doi 10.1002/cam4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koshy M, Rich SE, Mohiuddin MM. Improved survival with radiation therapy in high-grade soft tissue sarcomas of the extremities: a SEER analysis. Int J Radiat Oncol Biol Phys 2010;77(1):203–9 doi 10.1016/j.ijrobp.2009.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKelvey KJ, Hudson AL, Back M, Eade T, Diakos CI. Radiation, inflammation and the immune response in cancer. Mamm Genome 2018;29(11–12):843–65 doi 10.1007/s00335-018-9777-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yadavalli S, Jayaram S, Manda SS, Madugundu AK, Nayakanti DS, Tan TZ, et al. Data-Driven Discovery of Extravasation Pathway in Circulating Tumor Cells. Scientific reports 2017;7:43710 doi 10.1038/srep43710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petitprez F, de Reynies A, Keung EZ, Chen TW, Sun CM, Calderaro J, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 2020;577(7791):556–60 doi 10.1038/s41586-019-1906-8. [DOI] [PubMed] [Google Scholar]
- 51.Qin Z, Blankenstein T. A cancer immunosurveillance controversy. Nat Immunol 2004;5(1):3–4; author reply −5 doi 10.1038/ni0104-3. [DOI] [PubMed] [Google Scholar]
- 52.Stutman O Tumor development after 3-methylcholanthrene in immunologically deficient athymic-nude mice. Science 1974;183(4124):534–6 doi 10.1126/science.183.4124.534. [DOI] [PubMed] [Google Scholar]
- 53.Crittenden MR, Zebertavage L, Kramer G, Bambina S, Friedman D, Troesch V, et al. Tumor cure by radiation therapy and checkpoint inhibitors depends on pre-existing immunity. Scientific reports 2018;8(1):7012 doi 10.1038/s41598-018-25482-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crowe NY, Smyth MJ, Godfrey DI. A critical role for natural killer T cells in immunosurveillance of methylcholanthrene-induced sarcomas. The Journal of experimental medicine 2002;196(1):119–27 doi 10.1084/jem.20020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coleman CN, Higgins GS, Brown JM, Baumann M, Kirsch DG, Willers H, et al. Improving the Predictive Value of Preclinical Studies in Support of Radiotherapy Clinical Trials. Clin Cancer Res 2016;22(13):3138–47 doi 10.1158/1078-0432.CCR-16-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole exome sequencing dataset is available for download from a publicly accessible database, Sequence Read Archive (SRA), using BioProject ID # PRJNA873947. Any other data will be available from the corresponding authors upon request.


