Abstract
Mass spectrometry-based discovery glycoproteomics is highly dependent on the use of chromatography paradigms amenable to analyte retention and separation. When compared against established stationary phases such as reversed phase and hydrophilic interaction liquid chromatography, reports utilizing porous graphitic carbon (PGC) have detailed its numerous advantages. Recent efforts have detailed the utility in porous graphitic carbon in high throughput glycoproteomics, principally through enhanced profiling depth and liquid phase resolution at higher column temperatures. However, increasing column temperature has shown to impart disparaging effects in glycopeptide identification. Herein we further elucidate this trend, describing qualitative and semi-quantitative effects of increased column temperature on glycopeptide identification rates, signal intensity, resolution, and spectral count linear response. Through analysis of enriched bovine and human glycopeptides, species with high mannose and sialylated glycans were shown to most significantly benefit and suffer from high column temperatures, respectively. These results provide insight as to how porous graphitic carbon separations may be appropriately leveraged for glycopeptide identification while raising concerns over quantitative and semi-quantitative label free comparisons as temperature changes. RAW MS glycoproteomic data are available via ProteomeXchange with identifier PXD034354.
Keywords: Glycopeptide analysis, glycoproteomics, mass spectrometry, porous graphitic carbon chromatography, PGC separations
Graphical Abstract
Introduction
Glycosylation is one of the most prevalent and heterogenous post-translational modifications (PTMs) within the human proteome1–3. The inherent complexity of its study is not felt just in considering the vast expanse of known and theoretical modification sites2, but also through appreciation of the high degree of compositional and structural complexity4. Decades of targeted research have revealed the high degree of glycosylation diversity presents an equally diverse set of functions. Glycans and glycoproteins are known to participate in a litany of biological processes such as cell signaling5–7, host-pathogen interaction8–10, and protein folding11–13, and are ever increasingly implicated in health and human disease. Despite the importance of these PTMs and the extensive effort contributed to their study, analytical strategies that can keep pace with biological complexity remain limited.
In order to offset the overwhelming difficulties in glycopeptide analysis, numerous areas of analytical development have received significant attention in recent years. Glycopeptide enrichment strategies have been developed to compensate for low analyte abundance in proteolytic mixtures14–16, with some being tailored to unique glycan classes17, 18. As well, ubiquitous utilization of tandem-MS for glycopeptide identification has brought about validation and application of numerous unique dissociation modalities19–22. Even efforts for accurate quantitation of intact glycopeptides have seen a rise in popularity as isotopic and isobaric labeling strategies become more accessible23. These areas of development, however, largely ignore any potential benefits that may come through more effective chromatography.
Glycoproteomic analyses continue to utilize traditional reversed-phase liquid chromatography (RPLC) due to the effective retention mechanism and ease of implementation. This separation strategy provides meaningful access to the glycoproteome, but analyte retention is dependent on a dominant hydrophobic character of the peptide backbone – a characteristic not observed for all glycopeptides. Furthermore, the large, hydrophilic glycan moiety often reduces the efficacy of RPLC for glycopeptide retention and separation24, 25. Hydrophilic interaction chromatography (HILIC) and electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) are popular separation modalities that promote greater retention of glycopeptides26–28 but are less commonly implemented on-line due to the need for salt-containing buffers27, 29 and sample phase changes prior to MS analysis.
Offering reprieve from these shortcomings, porous graphitic carbon (PGC) has demonstrated great utility in the retention and separation of polar analytes. PGC has been extensively used in the analysis of released glycans30–35 and has even shown baseline resolution of glycan isomers when run at elevated temperatures36. Increasing column running temperatures promotes an expanded glycan morphology, increasing the partition coefficient and improving resolution. These improvements in glycan retention and separation have recently been validated for targeted37 and discovery-based glycoproteomics analyses where higher temperature PGC separations were shown to enhance glycoproteomic coverage and profiling depth38. However, these improvements were shown to come at a cost.
Whereas previous studies demonstrated that modest increases in column temperature result in higher peptide and glycopeptide identification rates, these improvements often diminished at higher column temperatures38. With specific attention drawn to high mannose and sialylated glycopeptides, these two glycopeptide classes were shown to yield the most significant changes in identification rates and signal response. As PGC separations are increasingly employed for glycan and glycopeptide analysis – and due to the biological significance of affected glycopeptide classes – these observations present substantial roadblocks in the pursuit of successful glycoproteomic analysis. Understanding the cause of altered identification rates and the effects higher temperatures impart on intact glycopeptides is imperative to promoting enhanced glycoproteomic coverage and providing guidance over experimental conditions that reduce analytical efficacy.
To survey the effects of higher column temperatures in PGC separations, we performed discovery-based glycoproteomic analysis on glycopeptides enriched from human prostate cancer cell lysate, supplemented with sialoglycopeptides from bovine standard proteins. Recreating previous experimental conditions38, we reaffirm profiling depth is enhanced with modest increases in column temperature (45°C). Increasing temperature to 60°C, however, results in significant disparities in glycopeptide detection. Confining much of our focus to high mannose and sialylated glycopeptides – those species most differentially affected – we demonstrate how elevated temperatures are responsible for altered reporting signal and peak shape, affecting detection and identification. Further knowing the prevalence of label-free and reaction monitoring quantitative approaches that rely on precursor peak intensity or area, we also analyzed serial dilutions of enriched glycopeptide mixtures to evaluate impact of column temperature on glycopeptide quantitation. Mirroring the observations seen in glycopeptide identification, glycopeptide spectral matches were shown to significantly deviate as temperature climbs, indicating label-free comparisons across temperatures are not viable without special consideration. Nevertheless, for almost all glycopeptide classes, our data demonstrate greater spectral-counting-based quantitative accuracy at the highest temperature, 60°C. The disparity in observations between discovery and quantitative analyses suggest that column temperature must be individually tailored to suit biological discovery or quantitative accuracy. Overall, the findings presented within serve to highlight notable limitations and topical concerns for future PGC-based glycoproteomics analyses.
Methods
Materials
Dithiothreitol (DTT, D9779), iodoacetamide (IAA), sodium dodecyl sulfate (SDS), trifluoroacetic acid (TFA), bovine ribonuclease B (R7884), bovine fetuin (F3004), bovine asialofetuin (A4781), and bovine α1-acid glycoprotein (G3643) were purchased from Millipore Sigma (Burlington, MA). Urea (U15), tris-base (BP152), hydrochloric acid (A144SI), formic acid (A117), water, acetonitrile, and PGC guard columns (35003–014001) were purchased from Fisher Scientific (Waltham, MA). Capillary tubing (1068150019) was purchased from PolyMicro. Trypsin (V5113) was purchased from Promega (Madison, WI). PolyHYDROXYETHYL-A packing material was purchased from PolyLC. Pencil column heater was purchased from Pheonix S&T. Sources of all other materials are noted.
Protein Digestion
300mg of each standard glycoprotein was aliquoted into separate reaction microtubes and dried under vacuum. Urea was dissolved in 50mM Tris-HCl to a concentration of 8M, which was then used to resuspend standard glycoproteins at a concentration of 2.0μg/μL. Disulfide bonds were reduced with 5mM DTT at room temperature for 1 hour, followed by alkylation with 15mM IAA at room temperature for 30 minutes in the dark. Alkylation was quenched with excess DTT prior to diluting the mixture 1:10 to reduce Urea concentration to <1M. Trypsin was added 1:100 for 4 hours at 37°C, followed by 1:50 and overnight incubation at 37°C.
Cell Preparation
Prostate cancer cell lines (BCaPMT10) were generated and described previously39. Cell lines were grown and maintained in phenol-free DMEM/Ham’s F12 (Gibco) supplemented with 5% fetal bovine serum (HyClone) and 1% penicillin-streptomycin solution (Gibco). T175 culture flasks were placed in an incubator at 5% CO2 and 98% humidity. Cells were grown to 90% confluency, washed with 1× phosphate-buffered saline (Cytiva), and harvested using a cell scraper. Approximately 1e6 cells were collected after culture. Cell pellets were washed twice using phosphate-buffered saline and stored at −80°C for subsequent processing.
Cell pellets were resuspended in 4 volumes of 4% SDS prior to lysis via ultrasonication. Protein concentration was estimated via bicinchoninic acid (BCA) assay (ThermoFisher Scientific). Disulfide bonds were reduced with 450nM DTT for 30 minutes at 55°C followed by alkylation with 10mM IAA at room temperature for 15 minutes. Protein was extracted through repeated additions of cold 80% acetone and incubation at −20°C. Protein was reconstituted in 8M Urea with 1x protease inhibitor cocktail. 300mg aliquots were taken, diluted, and digested similarly to the standard proteins.
Glycopeptide Enrichment
Glycopeptides were enriched using a custom spin tip method. Briefly, 200μL pipette tips were packed with 3mg sterile cotton and loaded with polyHYDROXYETHYL-A packing resin at a 30:1 bead-to-peptide ratio. Beads were hydrated in 1% TFA prior to loading. Beads were flushed with 1% TFA and conditioned with 80%ACN+1%TFA. Samples were resuspended in 80% ACN+1% TFA and applied to the beads a total of six times, which was followed by six washes in the same buffer to remove non-glycosylated peptides. Glycopeptides were eluted in 10% ACN+5% FA prior to drying under vacuum. Samples were reconstituted in 0.1% FA and serially diluted 1x to 16x prior to LC-MS analysis (Figure 1).
Figure 1).
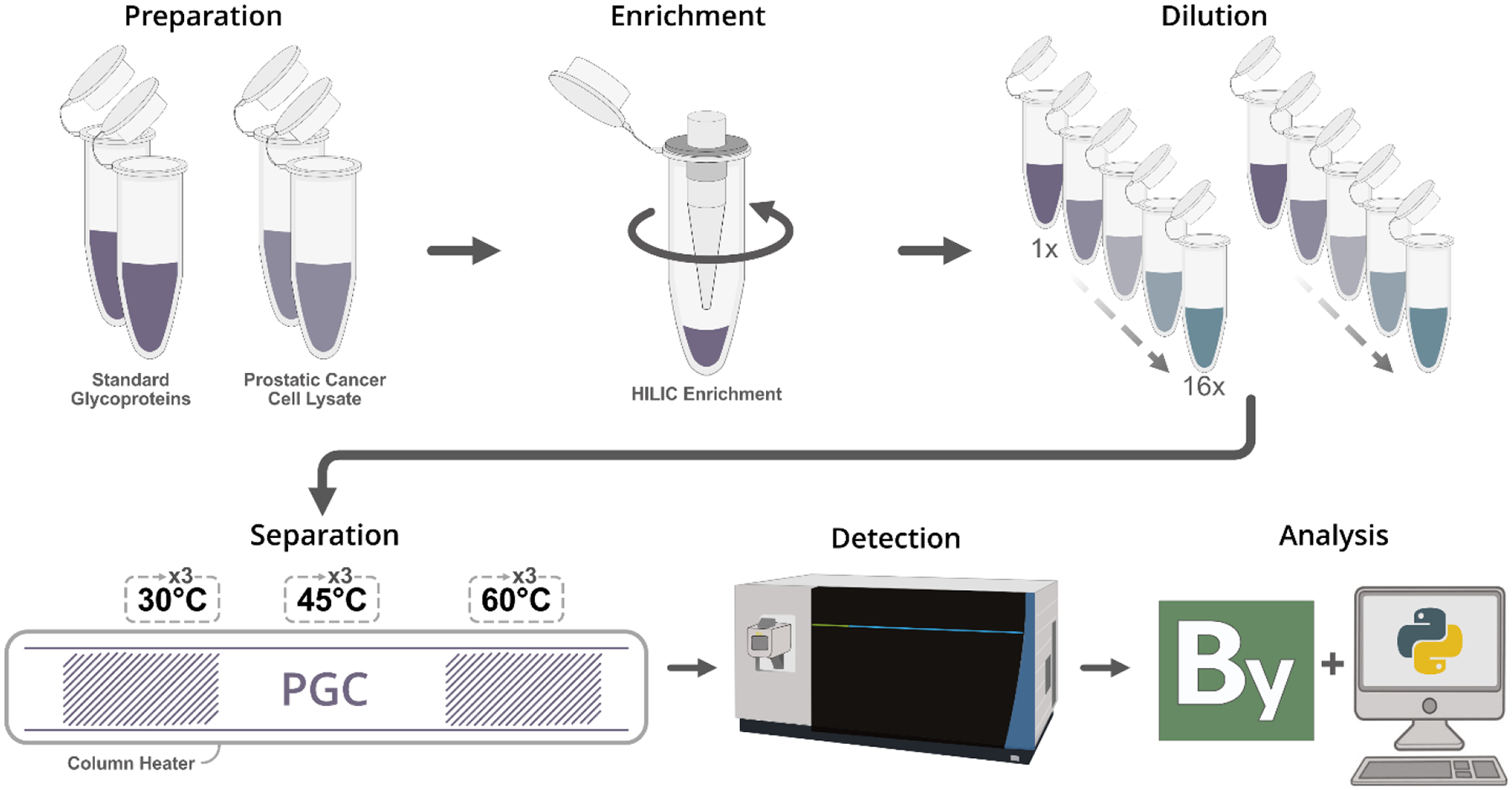
Outline of workflow. Glycoprotein mixtures or cell lysate were enzymatically digested prior to glycopeptide enrichment. Glycopeptides were reconstituted, serially diluted, and analyzed in technical triplicate on a custom PGC-packed capillary column. Data were collected on an Orbitrap Fusion Lumos with database searching performed in Proteome Discoverer with the Protein Metrics Byonic node.
Column Preparation
Capillary tubing (o.d.=360um, i.d.=75um) was trimmed to a length >30cm with a small portion of the polyimide coating removed. A Sutter P-2000 micropipette laser puller was used to stretch the glass capillary into the final emitter geometry. The closed, tapered tip was then opened via emersion in 48% hydrofluoric acid for 2.5 minutes. 3um PGC packing material was harvested from PGC guard columns (Thermo Fisher Scientific, 35003–014001) and resuspended in optima grade methanol in a 1.5mL glass vial. A micro flea stir bar was added to provide agitation, and the slurry and etched capillary were placed in a pressure injection cell (Next Advance, PC77-MAG). The capillary was packed with PGC material using ~1000psi industrial grade nitrogen until 30cm of the capillary was full. Pressure was released and the same packing conditions were used to flush the column with methanol and air, sequentially. Columns were stored at room temperature until use.
LC-MS Analysis
Enriched glycopeptide mixtures were analyzed in technical triplicate using a Nano Ultimate 3000 chromatography stack coupled to an Orbitrap Fusion Lumos mass spectrometer. Glycopeptides were trapped on the column at 3% B for 18 minutes, followed by a 52-minute gradient from 15% B to 40% B. Column was washed at 75% B for ten minutes followed by another ten-minute wash at 95%B. The column was then equilibrated to 3% for the final ten minutes. The Fusion Lumos was set to perform DDA analysis using a 3-second MS2 acquisition window rather than selecting top N precursors. MS1 settings were as follows: resolution, 120,000; m/z range, 400–2,000; RF lens, 30%; AGC target, 2.0e5; maximum injection time, 50ms; microscans, 1; polarity, positive. MS2 settings were as follows: resolution, 60,000; detector, orbitrap; isolation width, 1.6m/z; activation, HCD; collision energy mode, stepped HCD; collision energies, 20-30-40; first mass, 120m/z; AGC target, 5.0e4; maximum injection time, 118ms. Precursors were also required to meet an intensity threshold of 2.5e4 for selection. Allowed charges states were 2+ to 7+; precursors were excluded after 1 occurrence for 15 seconds.
Standard Peptide Analysis
A standard disialylated glycopeptide (KVANK[HexNAc4Hex5NeuAc2]T) was purchased from TCI America (S0523). Listed at >95% pure with uniformity confirmed through gel and capillary electrophoresis, this product enables analysis of a highly uniform glycopeptide not subject to biological variation or abundance constraints. Preparation and analysis of this standard is detailed in the Supplemental Information.
Data and Code Availability
The mass spectrometry glycoproteomics data have been deposited to the ProteomeXchange40 Consortium via the PRIDE41, 42 partner repository with the dataset identifier PXD034354 and 10.6019/PXD034354. Data were searched using Proteome Discoverer 2.5 with the Protein Metrics Byonic node; searching and filtering parameters are described in the Supplemental Information. Search results are available in the PRIDE repository. All code used for analysis is available at https://github.com/lingjunli-research/pgc-glycosylation-lfq.
Results and Discussion
Changes in Glycopeptide Identification
Previous reports have signaled improved peptide and glycopeptide identification rates at elevated column temperatures38, indicating potential differences in analyte behavior or optimal separation and desolvation efficiency. This trend is rearticulated in this study where, especially at higher concentrations (1x, 2x, and 4x dilutions), 45°C provided the highest overall rates of glycopeptide identification (Figure 2A, Table S1). Though this behavior is not conserved at lower concentrations (8x and 16x dilutions), a consistent trend across all analyses is that 60°C provided the lowest rates of glycopeptide identification. This latter observation is in-line with previous reports38 and serves as an early marker for suboptimal chromatography conditions. Mapping glycopeptides back to their parent protein, our data demonstrate that the increase in glycopeptide identifications at 45°C stem from the detection of glycosites on previously undetected proteins, rather than new, additional glycosites or glycopeptides from proteins already identified (Figure 2A, right). Interestingly, while our data show that the majority of glycosites are identified across all temperatures (Figure 2B), each unique glycopeptide dilution contained proportional quantities of unique glycosites with statistically insignificant differences across all but the most concentrated sample (Figure S1). As our data showed high intra-sample reproducibility in the identification of glycosites at all temperatures (Figure 2B), we are confident this complementary detection is not due to analytical inaccuracies and serves to corroborate previous observations38 that column temperatures provide access to different portions of the glycoproteome.
Figure 2).
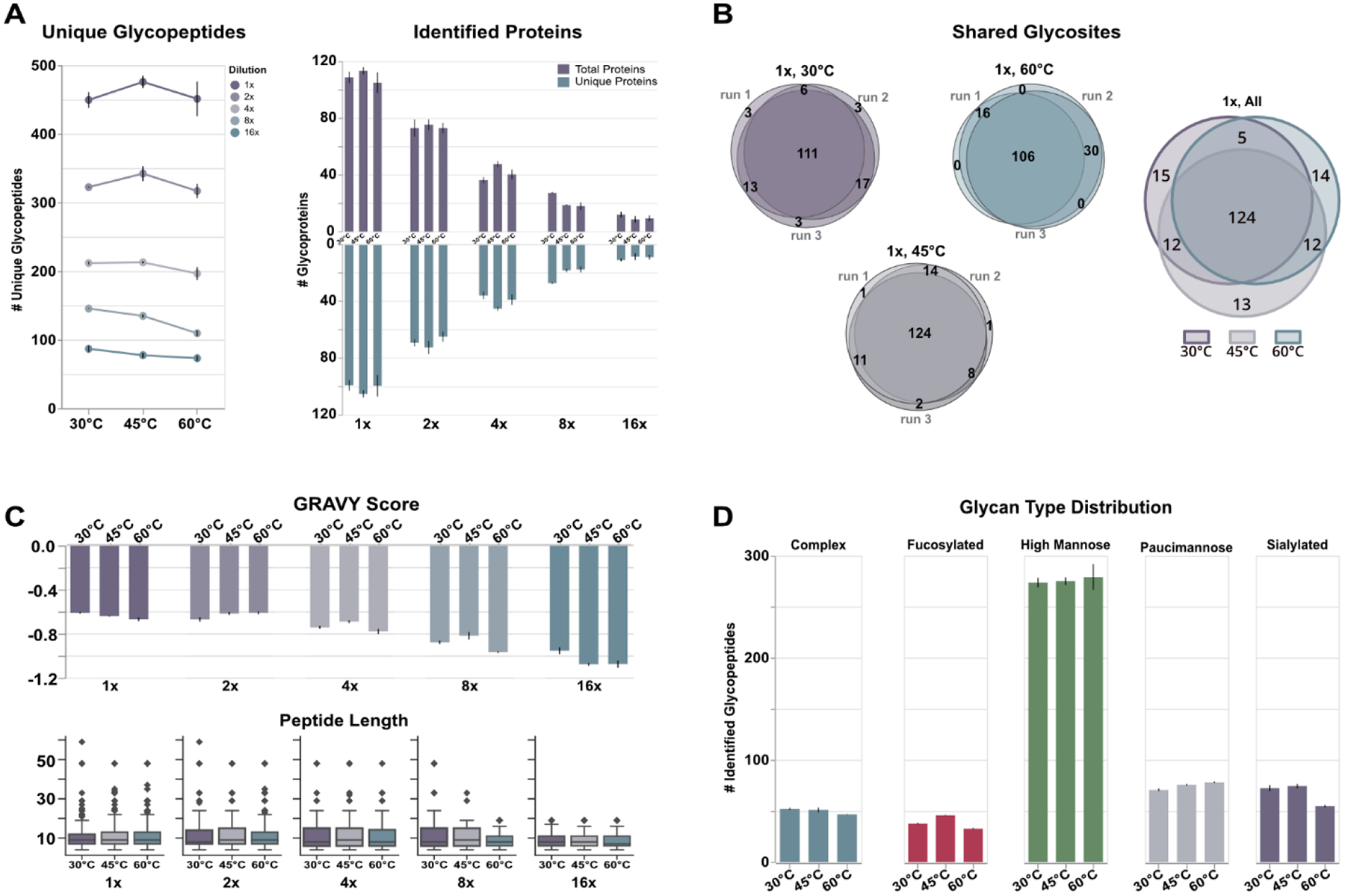
Results outlining temperature-based differences in glycopeptide identification. A) Higher concentration glycopeptide samples (i.e., the undiluted sample (1x), and first two serial dilutions (2x and 4x)) displayed the expected increase in identifications when separated at 45°C, stemming largely from accessing new glycoprotein constituents. Concentration and temperature are independent; all concentrations (i.e., dilutions) were analyzed at all temperatures. B) Our analyses showed high technical reproducibility indicating the changes in unique glycosites between temperature is unlikely due to analytical abnormalities. C) The peptide backbone information of glycopeptides was largely conserved across temperatures, suggesting major differences are not due to the backbone itself. D) Glycopeptides demonstrated a class-dependent response to temperature with mannose type glycans benefitting from higher temperatures; complex, fucosylated and sialylated types show decreased identifications at high temperatures.
We investigated whether the peptide backbone played a significant role in the retention of glycopeptides at different temperatures. As shown in Figure 2C, the relative hydrophilicity – presented here in the form of grand average of hydropathy (GRAVY) – and peptide length are well conserved across temperatures for each glycopeptide dilution. While the data show some slight preference for more hydrophilic analytes at higher temperatures, this observation is biased by fewer identified glycopeptides and should be further investigated in analyses of unmodified tryptic peptides where a hydrophilic glycan moiety plays no role. Interestingly, our data show a decrease in GRAVY score that mirrors glycopeptide concentration. This lower average value is mostly due to the lower number of identified glycopeptides, but it does speak to the power of PGC to selectively trap, retain, and elute these highly polar, hydrophilic analytes.
However, the differences in glycopeptide identification begin to take shape when examining the classes of glycans identified (Figure 2D). High mannose glycans were the most predominant modification identified across our analyses. These glycans, considered immature within the biosynthetic pathway4, may be seen in greater quantity due to the known relationship between cancer cell proliferation and glycan nascence43–45, though targeted investigation is needed for confirmation within this respective cell line model. Interestingly, while we anticipated complex glycans would be the second most prevalent glycan type, instead our data reveal paucimannose glycans as the next most common. Paucimannose glycans are relatively understudied in humans as it can be conjectured these glycans result from degradation or harsh preparation conditions46. However, recent studies have validated the occurrence of these glycan types in humans and have even been shown to play significant roles in human cancer47–49. Sialoglycopeptides, complex glycopeptides, and fucosylated glycopeptides follow in number of identifications. A clear disparity between mannose and the latter three glycan subtypes is the effect seen when elevating column temperature. While identifications of glycopeptides in all classes increased or were unaffected at 45°C, identifications of mannose subtypes continued to increase at 60°C (Figure 2D), albeit marginally. Conversely, the remaining glycan subtypes demonstrated overall worse performance at the highest temperature. This trend is less severe in complex glycopeptides but is immediately noticeable in fucosylated and sialylated glycopeptides.
Increasing glycopeptide identifications at 45°C are consistent with previous reports38; however, these data show more substantial increase of mannose subtypes at higher temperatures, which we attribute to different cell lysis methods and scale-up. Nevertheless, the reproducibility of declining identifications at high temperatures presented in our data indicates these observations are not due to experimental aberrations such as run-to-run variance or uncontrolled variables. Instead, we hypothesize that raising column temperatures induces some underlying chemical or chromatographic abnormalities that are responsible for affecting identification rates. Our results may be further dissected to provide qualitative and semi-quantitative investigation of this hypothesis.
Aberrant Chromatographic Behavior
To survey any potential underlying abnormalities that may be responsible for the observations mentioned above, we compiled extracted ion chromatograms (XICs) for all identified glycopeptide masses in the highest concentration sample. Given the different quantities of glycopeptides as temperature rises, it would be reasonable to expect that glycopeptides not identified were truly absent or were seen at such low intensities they could be mistaken for noise. Neither of these suspicions were confirmed. Our analyses demonstrate that the vast majority of glycopeptide masses were both present and observed at appreciable intensity in every run – an observation that holds true regardless of glycan class, as evidenced below. This further supports our hypothesis that column temperatures are responsible for affecting identification rates.
To avoid misinterpretation of data, we first manually inspected the XIC images, removing any species that were poorly extracted or contained traces of insufficient quality (i.e., indistinguishable major peaks, co-extracted masses, etc.). Inspecting the remaining data, several trends became obvious. First, and most expectedly, increasing column temperature resulted in nearly unanimous increases in resolution, with XICs displaying narrower full width at half maximum (FWHM) and resolution of some putative glycopeptide isomers. More interestingly, however, is the disparity in overall signal intensity and peak height seen between glycopeptides of different classes. As temperature increased, the improved resolution for high mannose glycopeptides translated into higher signal intensity (i.e., narrower, taller peaks) (Figure 3A). This trend is largely conserved across high mannose glycopeptides to varying extents. At the same time, sialoglycopeptides show a similar improvement in resolution and signal intensity at 45°C but show a significant drop off at 60°C (Figure 3B). These observations rearticulate those seen elsewhere38 and provide direct evidence of temperature-correlated analyte response. If these temperature effects were directly related to glycopeptide detection and identification, we anticipate other glycopeptide subtypes to display similar correlations. Indeed, examining paucimannose and fucosylated glycopeptides reveal similar evidence. Paucimannose glycopeptides generally benefited from higher column temperatures while fucosylated glycopeptides showed higher intensity at 45°C that waned at higher temperatures (Figure S2). It should be noted that the associated trends are less significant for these latter two glycopeptide classes, but further study and broader collections of analytes may serve to definitively characterize their response to elevated temperatures.
Figure 3).
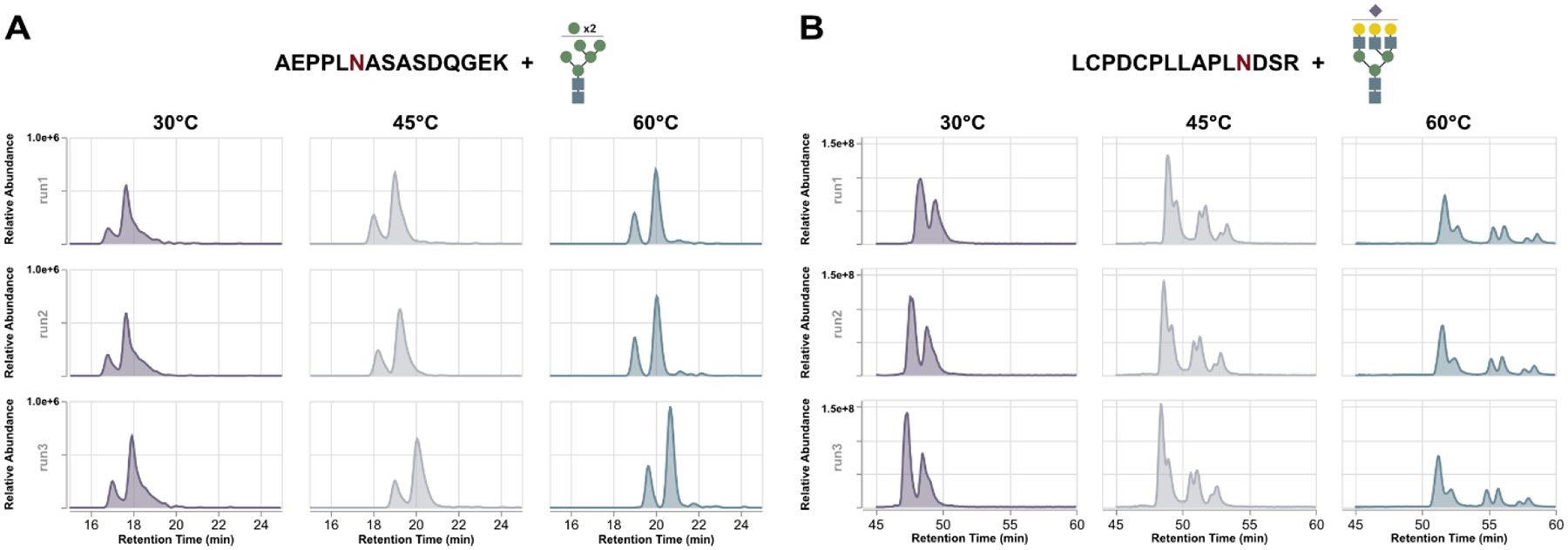
Representative extract ion chromatograms (XICs) of high mannose and sialylated glycopeptides. Both glycopeptides demonstrate improved resolution as temperature increases but high mannose types (A) displayed further increases in signal intensity as temperature climbs. Conversely, sialylated glycopeptides (B) increase in intensity at 45°C before showing significant attenuation at 60°C.
With evidence to support our hypothesis that temperature changes promote chromatographic behavior that impacts glycopeptide identification, we sought to provide further qualitative analysis to aid in characterizing these occurrences. For all glycopeptides identified in the highest concentration sample, we isolated the major peak and determined peak height, FWHM, and peak area via curve integration. We selected only the major peak for these analyses as the confident assignment of minor peaks must rely on exact match of isotopic envelopes to that of the major species that was selected for MS2 fragmentation; we found this to be untenable, given the quantity of unique glycopeptide identifications, replicates, and possible minor species.
Examining the data underpinning these glycopeptide identifications, all classes demonstrated the expected decrease in FWHM as temperature increases, reflecting the known improvements in liquid phase resolution achieved as glycan morphology expands (Figure 4A). As well, all glycopeptide classes demonstrated changes in peak intensity that directly reflect the observations made above, though these changes are most visible for sialylated glycopeptides. Similarly, when integrating area under the curve, high mannose and paucimannose glycopeptides demonstrate little discrepancy in peak area as temperature increases, contrasting that of complex and sialylated glycopeptides (Figure 4A). Taken together, we posit those improvements in liquid-phase resolution for mannose-type glycopeptides at high temperatures result in sharper, narrower elution peaks that conserve the overall peak area observed at lower temperatures. Because these observations directly correlate with identification rates, deviating peak intensities and areas are likely directly responsible for the incremental improvement in identifications, as greater intensity will raise precursor priority when performing DDA-MS/MS.
Figure 4).
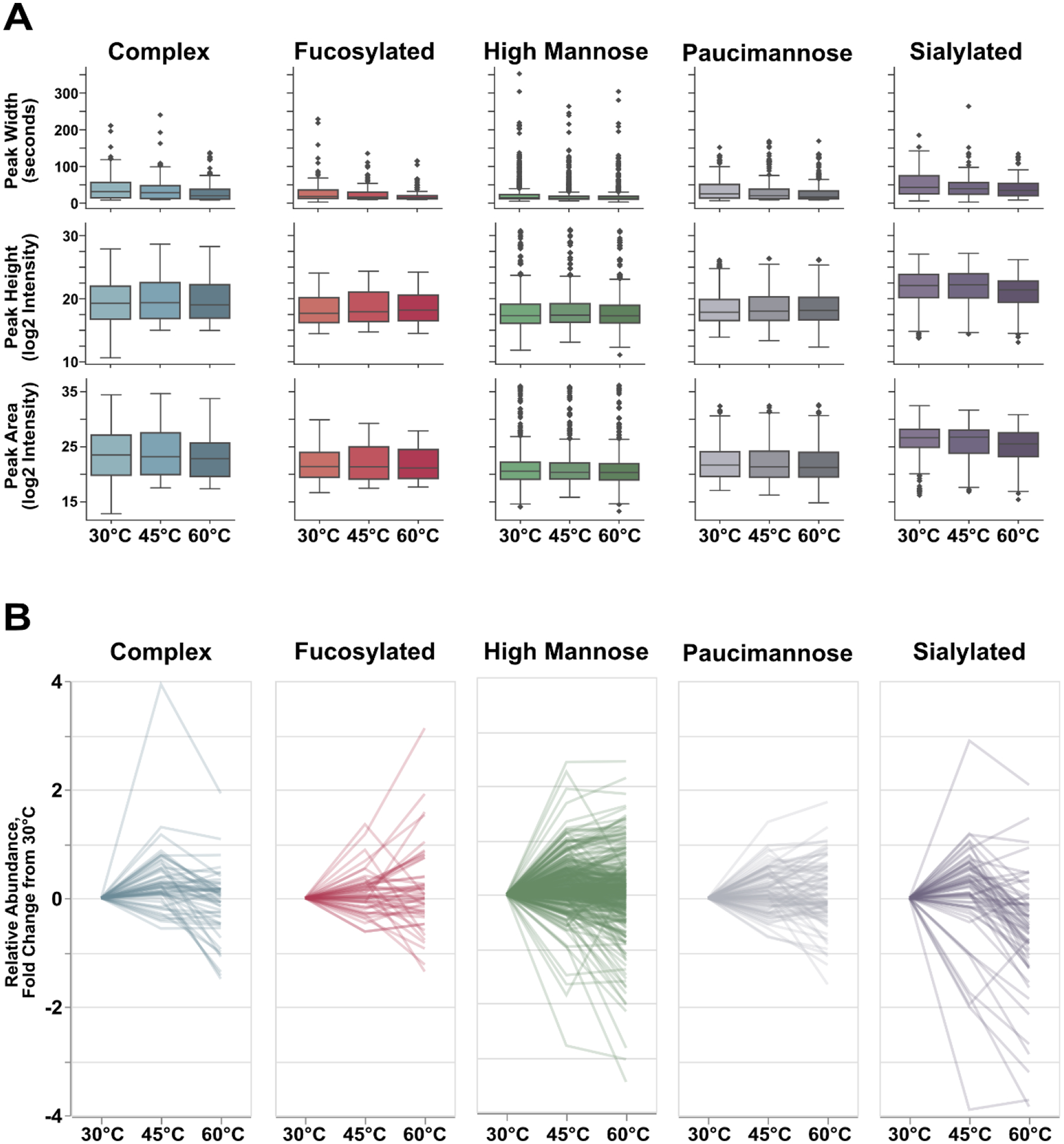
Qualitative metrics from identifiable glycopeptides. A) All glycopeptides demonstrate improved resolution at high temperatures, but most classes demonstrate little change in median peak height or peak area. Sialylated glycopeptides, however, do show disparities in these metrics. However, this change in median value is only reflective of the whole population. (B) When examining peak intensity for each individual glycopeptide, we observed that the majority of glycopeptides from each glycan class showed noticeable improvements in peak intensity at 45°C with a subset demonstrating statistical significance (P≤0.05, high mannose: 74 glycopeptides, 23.6% of high mannose population; paucimannose: 22 glycopeptides, 25.9% of population; complex: 16 glycopeptides, 30.8% of population; sialylated: 15 glycopeptides, 25.0% of population, fucosylated: 7 glycopeptides, 14.9% of population). Mannose type glycopeptide peak intensity is not largely impacted at 60°C, whereas complex and sialylated glycopeptides are – a finding that reflects their identification rates at high temperatures.
In order to validate this conjecture, we utilized the extracted information from each glycopeptide to represent the fold-change in relative abundance with respect to the base temperature, 30°C. Averaging across technical triplicates, the vast majority – though not all – glycopeptides exhibit higher relative abundance at 45°C (Figure 4B), mirroring the aforementioned XIC observations and identification rates. However, where these data begin to diverge is the relative abundance seen at the highest temperature, 60°C. For high mannose and paucimannose glycopeptides, relative abundances are often even higher than those observed at 45°C. While these glycopeptide classes do show some species to be lower in abundance at 60°C than at 45°C, the relative abundance at the highest temperature are often equivalent to or higher than the abundances seen at the base temperature of 30°C. Contrary to this observation, complex, fucosylated, and sialylated glycopeptides show significant drops in relative abundance at 60°C, with the majority of glycopeptides displaying lower or substantially lower abundances compared to that observed at either of the two lower temperatures.
These observations and conjectures notwithstanding, we acknowledge the presence of glycopeptides that deviate from these noted trends. However, these cases are the minority, giving way to the prevailing observations discussed here. This diversity in results should be expected for any biological population and therefore do not significantly hamper our interpretation of the overall trends seen across glycopeptide classes. Fucosylated glycopeptides, as well, show diversity in their relative abundances as temperature climbs. Given the lower number of these species and that fucosylation and sialylation often co-occur, these data may be further reorganized and investigated in later experiments to provide a more comprehensive dataset.
Overall, these data serve to confirm our hypothesis that increases in temperature induce chromatographic behavior that impacts glycopeptide identification. As shown, mannose type glycopeptides benefit from increased temperatures as their increased resolution results in greater peak intensity and therefore greater selection in DDA-MS/MS. Complex, fucosylated, and sialylated glycopeptides do benefit from improved resolution at higher temperatures but often yield lower peak intensities, affecting downstream identification. Beyond this, given the demonstrated abnormalities in glycopeptide detection and peak shape as temperature increases, we suspect common label-free quantitative methods may be unreliable under these chromatographic conditions.
Assessing Spectral Count-Based Quantitation
Strategies for glycopeptide quantitation have experienced a surge in innovation over the past decade23. While metabolic labeling, isotopic chemical tags, and isobaric labeling strategies have all been demonstrated as viable and effective, these approaches are often custom-tailored and require additional handling that can introduce sample loss. Label-free quantitation (LFQ) avoids these complications but is prone to missing values and run-to-run variance. Common LFQ approaches such as reaction monitoring rely on precursor area under the curve or intensity of transition ions50 as the quantitative marker, while others such as spectral counting assume relative quantity is proportional to frequency of MS selection. Given each of these methods relies on precursor and fragment intensity, severe limitations may be met when employing PGC separations at elevated temperatures.
Averaging the number of peptide spectral matches (PSMs) across technical replicates, a subset of glycopeptides was shown to be quantifiable (i.e., identified in ≥3 dilutions) with good linearity (Figure 5A). Notably, fucosylated glycopeptides demonstrated the worst linear response; this is due to the relatively low abundance of this class of glycopeptide – evidenced by the low number of identifications and PSMs. When comparing across the remaining glycopeptide subtypes, high mannose glycopeptides demonstrated the highest density of species with linear regression fit >0.9, though all classes demonstrated a meaningful distribution of high linearity species. More interestingly, quantifiable complex, fucosylated and sialylated glycopeptides showed greater PSM-based linearity as temperature climbed (Figure 5B). This observation presents a caveat to our existing discussion of how elevated temperatures bring limitations to glycopeptide identification. Though high temperatures (i.e., ≥60°C) reveal the fewest overall identifications, those that are identified tend to show excellent linear response and may be more easily relied upon for quantitation. On the other hand, mannose type glycopeptides generally showed a decrease in linear response at 45°C before becoming bimodally distributed at 60°C (Figure 5B). These data inform us that column temperatures may be tailored based on intended experimental outcome; 45°C typically provides best signal response but does not provide the best quantitation for all species.
Figure 5).
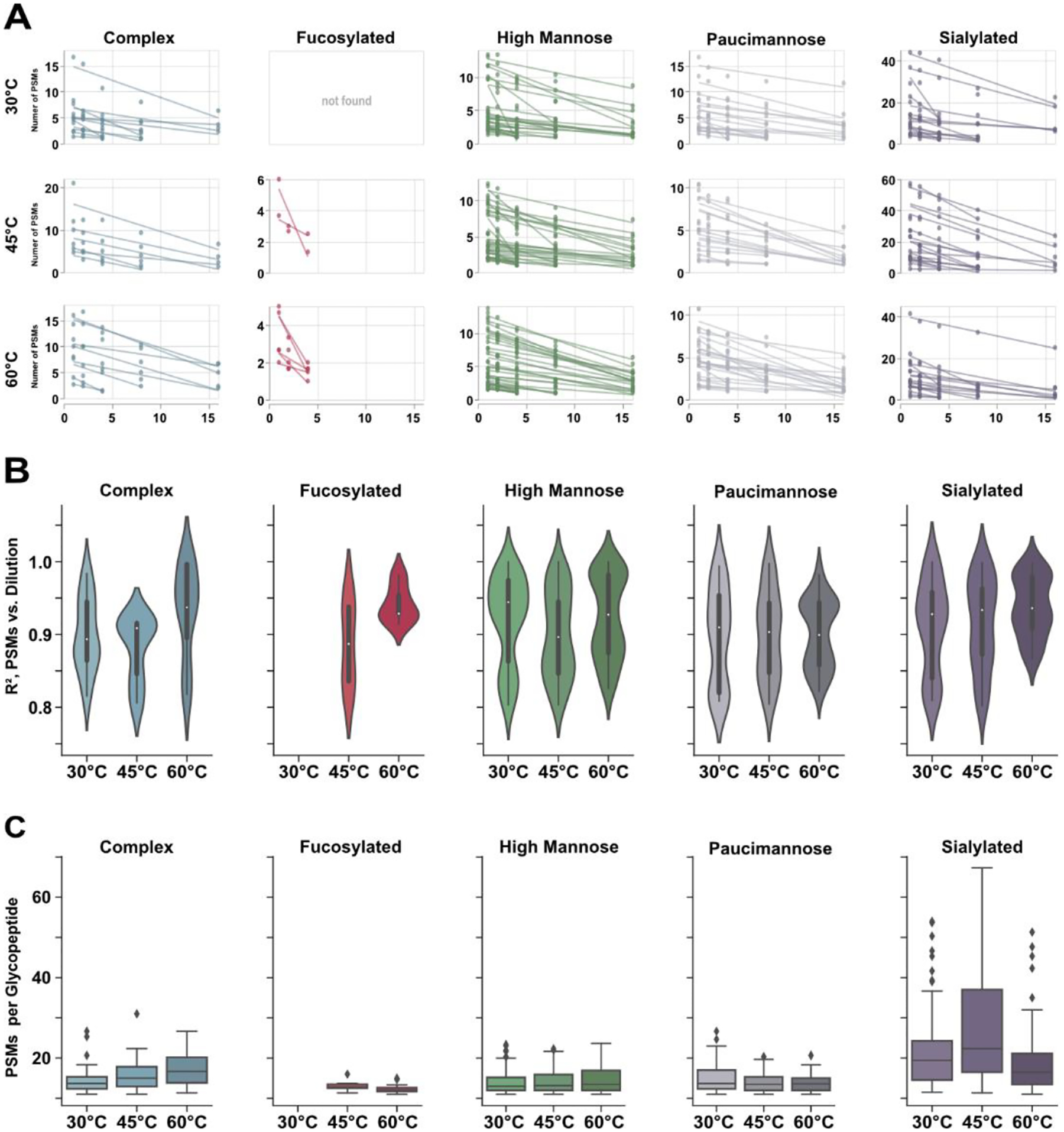
Semi-quantitative evaluations of glycopeptide dilution series across temperature. A) Plots demonstrating the linear response of glycopeptide identifications across dilutions. When temperature is held constant, spectral counting may be a viable option for LFQ. B) Distribution of linear regression for quantifiable glycopeptides as temperature increases. While 45°C is most beneficial for glycopeptide identifications, it is not the most optimal for LFQ. Conversely, identifiable glycopeptides may be best quantified at 60°C. C) Change in spectral count as temperature climbs. While PSM count for mannose type glycans is relatively conserved, complex and sialylated glycopeptides show significant differences across temperatures. This observation suggests that LFQ methods may be significantly impacted at different temperatures.
While these data suggest at which temperatures spectral counting-based quantitation may be achieved, we have not established whether individual glycopeptides may be accurately compared across temperatures. As shown in Figure 5C, all glycopeptide classes yield different quantities of PSMs at different temperatures. This observation may be obvious, given the discussion of chromatographic behavior. However, mannose and fucosylated subtypes generally showed a more conserved spectral count as temperatures increased compared to complex and sialylated glycopeptides. As seen, sialylated glycopeptides displayed a demonstrable increase in the number of PSMs at 45°C before decreasing again at 60°C, which reflects the deviation in signal intensity at these temperatures. Complex glycopeptides, on the other hand, exhibited steady increases in the number of PSMs as temperature climbs higher, being only one of two peptide classes that display such trend.
Taken together, these data are not intended to confer any quantitative information beyond the efficacy of using a label-free strategy for glycopeptide analysis when column temperature is an experimental variable. While our data demonstrate good linear response when PSMs are averaged as a function of sample dilution, this is only true when temperature is constant. In large part, glycopeptides cannot be directly compared across different temperatures in a label-free fashion. This conclusion is evidenced first by the significant changes in PSM count as temperature changes (Figure 5C) and when considering the noted differences in precursor peak intensity and area (Figure 3, S2). While quantitative accuracy remains to be evaluated when using area under the curve, our data suggest this approach may not be suitable; we did attempt to perform XIC area based LFQ in these analyses, but the breadth of identifications combined with technical replicates at three temperatures was untenable and computationally inefficient. Perhaps MS2-based transition monitoring is a more appropriate strategy for glycopeptide LFQ when using elevated column temperatures, but this claim should be investigated on its own.
Hypotheses and Future Directions
Given the nature of our data, it is imperative we consider the underlying phenomena that induce the observed chromatographic behavior shown to impact glycopeptide analysis. Focusing on sialylated glycopeptides, those species most adversely affected, two prevailing hypotheses exist that may explain the significant decrease in signal intensity at high temperatures. The first hypothesis is that sialoglycopeptides exhibit greater structural diversity in nature (i.e., antennae linkage, α/β orientation, etc.) and this structural diversity is readily resolved at high temperatures, as shown previously36, 37. This increased resolution distributes the density of glycopeptides eluting per unit time, lowering overall peak heights. The alternative explanation is that liquid phase separations at high temperatures impart greater energy into the system, provoking early dissociation of labile sialic acid linkages. As well, sialylated glycopeptides are known to be labile under acidic conditions17, 51, 52; our 0.1% FA additive during LC-MS, though common practice, likely exacerbates their lability and may contribute to early dissociation. We do not find strong evidence to support this hypothesis in our data as examining the identified sialoglycopeptides did not provide any such correlation between high sialylation states and lower sialylation states. To state tersely, we did not observe any disalylated glycopeptides converted to monosialylated glycopeptides, and so on. We did further investigate this claim through analysis of a commercially available sialoglycopeptide standard and no early dissociation could be definitively observed (Figure S3). In addition, we did not observe any relationship between the five high mannose glycans and identification rate, peak height, etc., further reinforcing the trends discussed in this report as global observations and not specific to a subset of modifications.
In all, the chemical or physical cause of altered glycopeptide identifications at elevated temperatures is still not clear. Anecdotal evidence supports the idea that the acidic conditions chosen for LC-MS analysis does cause dissociation of sialic acid. Considering the addition of higher temperatures during separation and desolvation, indiscriminate cleavage of glycosidic bonds is not outside the realm of possibilities. As our study provided some semi-quantitative investigations of this anomaly, we propose a more rigorous means of quantitation may be employed to fully elucidate this trend. Establishing specific transition ions for a broad array of glycopeptides and utilizing reaction monitoring (MRM or PRM) would eliminate any errors in precursor identification, extraction, and quantitation. Regardless of future approaches that provide a succinct connection between column temperature and glycopeptide identification, the data presented here provide heuristic guidance towards appropriate experimental design, depending on analytical objectives.
Conclusions
Porous graphitic carbon separations are a powerful addition to mass spectrometry-based glycoproteomics. Providing excellent retention of hydrophilic glycopeptides with a dominant glycan moiety, PGC provides facile access to regions of the glycoproteome that may be unobtainable through traditional separation modalities. As shown within, elevated column temperatures inspire significant improvements in liquid-phase resolution for all glycopeptide classes and yield greater reporting signal that aids in auto-MS identification. However, further increasing column temperature presents a tradeoff between liquid-phase resolution and glycopeptide identification. Our analyses show unique glycopeptide classes are differentially impacted – mannose type glycopeptides appear to benefit from high temperatures while complex and sialylated glycopeptides do not. Beyond this, we demonstrate the feasibility of performing label-free glycopeptide quantitation when temperature is held constant. However, glycopeptide species cannot be reliably compared from one temperature to the next as their spectral count and precursor area under curve are shown to deviate substantially according to their glycan composition. In summary, PGC-based glycopeptide separation and discovery is most effective at 45°C, providing excellent reporting signal and modest resolution. However, in chasing isomeric resolution at higher temperatures, special consideration must be taken to avoid misinterpretation of glycopeptide identifications – or lack thereof – and when drawing comparisons to analyses under different experimental conditions.
Supplementary Material
Supplemental Methods: Standard peptide analysis, database searching.
Figures S1-S3: Comparisons of glycosite overlap across dilutions, Representative XICs for paucimannose (top) and fucosylated (bottom) glycopeptides, and Extract ion chromatograms (XICs) of commercial sialoglycopeptide.
Table S1. All glycopeptide identifications compiled from across all analyses.
Acknowledgments
Support for this research is provided in part by the National Institutes of Health (NIH) grants RF1 AG052324 (LL), R01 DK071801 (LL), and U54DK104310 (WAR, LL). The Orbitrap instruments were purchased through the support of an NIH shared instrument grant (NIH-NCRR S10RR029531) and Office of the Vice Chancellor for Research and Graduate Education at the University of Wisconsin-Madison. LL acknowledges the National Science Foundation funding support (CHE- 2108223 and IOS-2010789), NIH grant support R21AG065728, S10OD028473, and S10OD025084 as well as a Vilas Distinguished Achievement Professorship and Charles Melbourne Johnson Distinguished Chair Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
Footnotes
The Supporting Information is available free of charge on the ACS Publications website.
References
- 1.Abou-Abbass H; Abou-El-Hassan H; Bahmad H; Zibara K; Zebian A; Youssef R; Ismail J; Zhu R; Zhou S; Dong X; Nasser M; Bahmad M; Darwish H; Mechref Y; Kobeissy F, Glycosylation and other PTMs alterations in neurodegenerative diseases: Current status and future role in neurotrauma. Electrophoresis 2016, 37 (11), 1549–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khoury GA; Baliban RC; Floudas CA, Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Scientific Reports 2011, 1 (1), 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramazi S; Zahiri J, Post-translational modifications in proteins: resources, tools and prediction methods. Database 2021, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varki A; Cummings RD; Esko JD; Stanley P; Hart GW; Aebi M; Mohnen D; Kinoshita T; Packer NH; Prestegard JH; Schnaar RL; Seeberger PH, Essentials of glycobiology. Fourth edition. ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, 2022; p pages cm. [PubMed] [Google Scholar]
- 5.Patwardhan A; Cheng N; Trejo J, Post-Translational Modifications of G Protein–Coupled Receptors Control Cellular Signaling Dynamics in Space and Time. Pharmacological Reviews 2021, 73 (1), 120–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandler KB; Leon DR; Kuang J; Meyer RD; Rahimi N; Costello CE, N-Glycosylation regulates ligand-dependent activation and signaling of vascular endothelial growth factor receptor 2 (VEGFR2). Journal of Biological Chemistry 2019, 294 (35), 13117–13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freitas D; Campos D; Gomes J; Pinto F; Macedo JA; Matos R; Mereiter S; Pinto MT; Polónia A; Gartner F; Magalhães A; Reis CA, O-glycans truncation modulates gastric cancer cell signaling and transcription leading to a more aggressive phenotype. EBioMedicine 2019, 40, 349–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin B; Qing X; Liao J; Zhuo K, Role of Protein Glycosylation in Host-Pathogen Interaction. Cells 2020, 9 (4), 1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehaffy C; Belisle JT; Dobos KM, Mycobacteria and their sweet proteins: An overview of protein glycosylation and lipoglycosylation in M. tuberculosis. Tuberculosis 2019, 115, 1–13. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe Y; Bowden TA; Wilson IA; Crispin M, Exploitation of glycosylation in enveloped virus pathobiology. Biochimica et Biophysica Acta (BBA) - General Subjects 2019, 1863 (10), 1480–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H; Li S; Wang J; Chen S; Sun X-L; Wu Q, N-glycosylation in the protease domain of trypsin-like serine proteases mediates calnexin-assisted protein folding. eLife 2018, 7, e35672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagashima Y; von Schaewen A; Koiwa H, Function of N-glycosylation in plants. Plant Science 2018, 274, 70–79. [DOI] [PubMed] [Google Scholar]
- 13.Tannous A; Pisoni GB; Hebert DN; Molinari M, N-linked sugar-regulated protein folding and quality control in the ER. Seminars in Cell & Developmental Biology 2015, 41, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riley NM; Bertozzi CR; Pitteri SJ, A Pragmatic Guide to Enrichment Strategies for Mass Spectrometry-based Glycoproteomics. Molecular & Cellular Proteomics 2021, 100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue Y; Xie J; Fang P; Yao J; Yan G; Shen H; Yang P, Study on behaviors and performances of universal N-glycopeptide enrichment methods. Analyst 2018, 143 (8), 1870–1880. [DOI] [PubMed] [Google Scholar]
- 16.Chen Z; Wang D; Yu Q; Johnson J; Shipman R; Zhong X; Huang J; Yu Q; Zetterberg H; Asthana S; Carlsson C; Okonkwo O; Li L, In-Depth Site-Specific O-Glycosylation Analysis of Glycoproteins and Endogenous Peptides in Cerebrospinal Fluid (CSF) from Healthy Individuals, Mild Cognitive Impairment (MCI), and Alzheimer’s Disease (AD) Patients. ACS Chemical Biology 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M; Huang J; Ma M; Shi X; Li L, Selective Enrichment of Sialylglycopeptides Enabled by Click Chemistry and Dynamic Covalent Exchange. Analytical Chemistry 2022, 94 (18), 6681–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J; Liu X; Wang D; Cui Y; Shi X; Dong J; Ye M; Li L, Dual-Functional Ti(IV)-IMAC Material Enables Simultaneous Enrichment and Separation of Diverse Glycopeptides and Phosphopeptides. Analytical Chemistry 2021, 93 (24), 8568–8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riley NM; Malaker SA; Driessen MD; Bertozzi CR, Optimal Dissociation Methods Differ for N- and O-Glycopeptides. Journal of Proteome Research 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riley NM; Hebert AS; Westphall MS; Coon JJ, Capturing site-specific heterogeneity with large-scale N-glycoproteome analysis. Nature Communications 2019, 10 (1), 1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Escobar EE; King DT; Serrano-Negrón JE; Alteen MG; Vocadlo DJ; Brodbelt JS, Precision Mapping of O-Linked N-Acetylglucosamine Sites in Proteins Using Ultraviolet Photodissociation Mass Spectrometry. Journal of the American Chemical Society 2020, 142 (26), 11569–11577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Escobar EE; Wang S; Goswami R; Lanzillotti MB; Li L; McLellan JS; Brodbelt JS, Analysis of Viral Spike Protein N-Glycosylation Using Ultraviolet Photodissociation Mass Spectrometry. Analytical Chemistry 2022, 94 (15), 5776–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delafield DG; Li L, Recent Advances in Analytical Approaches for Glycan and Glycopeptide Quantitation. Molecular & Cellular Proteomics 2021, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furuki K; Toyo’oka T, Retention of glycopeptides analyzed using hydrophilic interaction chromatography is influenced by charge and carbon chain length of ion-pairing reagent for mobile phase. Biomedical Chromatography 2017, 31 (11), e3988. [DOI] [PubMed] [Google Scholar]
- 25.Frost DC; Li L, Chapter Three - Recent Advances in Mass Spectrometry-Based Glycoproteomics. In Advances in Protein Chemistry and Structural Biology, Donev R, Ed. Academic Press: 2014; Vol. 95, pp 71–123. [DOI] [PubMed] [Google Scholar]
- 26.Qing G; Yan J; He X; Li X; Liang X, Recent advances in hydrophilic interaction liquid interaction chromatography materials for glycopeptide enrichment and glycan separation. TrAC Trends in Analytical Chemistry 2020, 124, 115570. [Google Scholar]
- 27.Cui Y; Yang K; Tabang DN; Huang J; Tang W; Li L, Finding the Sweet Spot in ERLIC Mobile Phase for Simultaneous Enrichment of N-Glyco and Phosphopeptides. Journal of the American Society for Mass Spectrometry 2019, 30 (12), 2491–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui Y; Tabang DN; Zhang Z; Ma M; Alpert AJ; Li L, Counterion Optimization Dramatically Improves Selectivity for Phosphopeptides and Glycopeptides in Electrostatic Repulsion-Hydrophilic Interaction Chromatography. Analytical Chemistry 2021, 93 (22), 7908–7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alpert AJ, Effect of salts on retention in hydrophilic interaction chromatography. Journal of Chromatography A 2018, 1538, 45–53. [DOI] [PubMed] [Google Scholar]
- 30.Ashwood C; Lin C-H; Thaysen-Andersen M; Packer NH, Discrimination of Isomers of Released N- and O-Glycans Using Diagnostic Product Ions in Negative Ion PGC-LC-ESI-MS/MS. Journal of the American Society for Mass Spectrometry 2018, 29 (6), 1194–1209. [DOI] [PubMed] [Google Scholar]
- 31.Ashwood C; Waas M; Weerasekera R; Gundry RL, Reference glycan structure libraries of primary human cardiomyocytes and pluripotent stem cell-derived cardiomyocytes reveal cell-type and culture stage-specific glycan phenotypes. Journal of Molecular and Cellular Cardiology 2020, 139, 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashwood C; Pratt B; MacLean BX; Gundry RL; Packer NH, Standardization of PGC-LC-MS-based glycomics for sample specific glycotyping. The Analyst 2019, 144 (11), 3601–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei J; Tang Y; Bai Y; Zaia J; Costello CE; Hong P; Lin C, Toward Automatic and Comprehensive Glycan Characterization by Online PGC-LC-EED MS/MS. Analytical Chemistry 2020, 92 (1), 782–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong X; Peng W; Yu C-Y; Zhou S; Donohoo KB; Tang H; Mechref Y, 8-plex LC–MS/MS Analysis of Permethylated N-Glycans Achieved by Using Stable Isotopic Iodomethane. Analytical Chemistry 2019, 91 (18), 11794–11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gautam S; Banazadeh A; Cho BG; Goli M; Zhong J; Mechref Y, Mesoporous Graphitized Carbon Column for Efficient Isomeric Separation of Permethylated Glycans. Analytical Chemistry 2021, 93 (12), 5061–5070. [DOI] [PubMed] [Google Scholar]
- 36.Zhou S; Huang Y; Dong X; Peng W; Veillon L; Kitagawa DAS; Aquino AJA; Mechref Y, Isomeric Separation of Permethylated Glycans by Porous Graphitic Carbon (PGC)-LC-MS/MS at High Temperatures. Analytical Chemistry 2017, 89 (12), 6590–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu R; Huang Y; Zhao J; Zhong J; Mechref Y, Isomeric Separation of N-Glycopeptides Derived from Glycoproteins by Porous Graphitic Carbon (PGC) LC-MS/MS. Analytical Chemistry 2020, 92 (14), 9556–9565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delafield DG; Miles HN; Liu Y; Ricke WA; Li L, Complementary proteome and glycoproteome access revealed through comparative analysis of reversed phase and porous graphitic carbon chromatography. Analytical and Bioanalytical Chemistry 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu TT; Ewald JA; Ricke EA; Bell R; Collins C; Ricke WA, Modeling human prostate cancer progression in vitro. Carcinogenesis 2018, 40 (7), 893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deutsch EW; Bandeira N; Sharma V; Perez-Riverol Y; Carver JJ; Kundu DJ; García-Seisdedos D; Jarnuczak AF; Hewapathirana S; Pullman BS; Wertz J; Sun Z; Kawano S; Okuda S; Watanabe Y; Hermjakob H; MacLean B; MacCoss MJ; Zhu Y; Ishihama Y; Vizcaíno JA, The ProteomeXchange consortium in 2020: enabling ‘big data’ approaches in proteomics. Nucleic Acids Research 2019, 48 (D1), D1145–D1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez-Riverol Y; Csordas A; Bai J; Bernal-Llinares M; Hewapathirana S; Kundu DJ; Inuganti A; Griss J; Mayer G; Eisenacher M; Pérez E; Uszkoreit J; Pfeuffer J; Sachsenberg T; Yilmaz S; Tiwary S; Cox J; Audain E; Walzer M; Jarnuczak AF; Ternent T; Brazma A; Vizcaíno JA, The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 2019, 47 (D1), D442–d450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perez-Riverol Y; Bai J; Bandla C; García-Seisdedos D; Hewapathirana S; Kamatchinathan S; Kundu DJ; Prakash A; Frericks-Zipper A; Eisenacher M; Walzer M; Wang S; Brazma A; Vizcaíno JA, The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res 2022, 50 (D1), D543–d552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park DD; Phoomak C; Xu G; Olney LP; Tran KA; Park SS; Haigh NE; Luxardi G; Lert-itthiporn W; Shimoda M; Li Q; Matoba N; Fierro F; Wongkham S; Maverakis E; Lebrilla CB, Metastasis of cholangiocarcinoma is promoted by extended high-mannose glycans. Proceedings of the National Academy of Sciences 2020, 117 (14), 7633–7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radhakrishnan P; Dabelsteen S; Madsen FB; Francavilla C; Kopp KL; Steentoft C; Vakhrushev SY; Olsen JV; Hansen L; Bennett EP; Woetmann A; Yin G; Chen L; Song H; Bak M; Hlady RA; Peters SL; Opavsky R; Thode C; Qvortrup K; Schjoldager KT-BG; Clausen H; Hollingsworth MA; Wandall HH, Immature truncated O-glycophenotype of cancer directly induces oncogenic features. Proceedings of the National Academy of Sciences 2014, 111 (39), E4066–E4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boyaval F; Dalebout H; Van Zeijl R; Wang W; Fariña-Sarasqueta A; Lageveen-Kammeijer GSM; Boonstra JJ; McDonnell LA; Wuhrer M; Morreau H; Heijs B, High-Mannose N-Glycans as Malignant Progression Markers in Early-Stage Colorectal Cancer. Cancers (Basel) 2022, 14 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wuhrer M, Paucity of Paucimannosylation Revoked. PROTEOMICS 2019, 19 (21–22), 1900244. [DOI] [PubMed] [Google Scholar]
- 47.Thaysen-Andersen M; Venkatakrishnan V; Loke I; Laurini C; Diestel S; Parker BL; Packer NH, Human Neutrophils Secrete Bioactive Paucimannosidic Proteins from Azurophilic Granules into Pathogen-Infected Sputum *2. Journal of Biological Chemistry 2015, 290 (14), 8789–8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loke I; Østergaard O; Heegaard NHH; Packer NH; Thaysen-Andersen M, Paucimannose-Rich N-glycosylation of Spatiotemporally Regulated Human Neutrophil Elastase Modulates Its Immune Functions*. Molecular & Cellular Proteomics 2017, 16 (8), 1507–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chatterjee S; Lee LY; Kawahara R; Abrahams JL; Adamczyk B; Anugraham M; Ashwood C; Sumer-Bayraktar Z; Briggs MT; Chik JHL; Everest-Dass A; Förster S; Hinneburg H; Leite KRM; Loke I; Möginger U; Moh ESX; Nakano M; Recuero S; Sethi MK; Srougi M; Stavenhagen K; Venkatakrishnan V; Wongtrakul-Kish K; Diestel S; Hoffmann P; Karlsson NG; Kolarich D; Molloy MP; Muders MH; Oehler MK; Packer NH; Palmisano G; Thaysen-Andersen M, Protein Paucimannosylation Is an Enriched N-Glycosylation Signature of Human Cancers. PROTEOMICS 2019, 19 (21–22), 1900010. [DOI] [PubMed] [Google Scholar]
- 50.Miyamoto S; Stroble CD; Taylor S; Hong Q; Lebrilla CB; Leiserowitz GS; Kim K; Ruhaak LR, Multiple Reaction Monitoring for the Quantitation of Serum Protein Glycosylation Profiles: Application to Ovarian Cancer. Journal of Proteome Research 2018, 17 (1), 222–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ashwell M; Guo X; Sinnott ML, Pathways for the hydrolysis of glycosides of N-acetylneuraminic acid. Journal of the American Chemical Society 1992, 114 (26), 10158–10166. [Google Scholar]
- 52.Ito S; Asahina Y; Hojo H, Investigation of protecting group for sialic acid carboxy moiety toward sialylglycopeptide synthesis by the TFA-labile protection strategy. Tetrahedron 2021, 97, 132423. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Methods: Standard peptide analysis, database searching.
Figures S1-S3: Comparisons of glycosite overlap across dilutions, Representative XICs for paucimannose (top) and fucosylated (bottom) glycopeptides, and Extract ion chromatograms (XICs) of commercial sialoglycopeptide.
Table S1. All glycopeptide identifications compiled from across all analyses.
Data Availability Statement
The mass spectrometry glycoproteomics data have been deposited to the ProteomeXchange40 Consortium via the PRIDE41, 42 partner repository with the dataset identifier PXD034354 and 10.6019/PXD034354. Data were searched using Proteome Discoverer 2.5 with the Protein Metrics Byonic node; searching and filtering parameters are described in the Supplemental Information. Search results are available in the PRIDE repository. All code used for analysis is available at https://github.com/lingjunli-research/pgc-glycosylation-lfq.


