Abstract
Introduction
Down syndrome (DS) patients have a 100-fold increase in the risk of Hirschsprung syndrome of the colon and rectum (HSCR), a lack of enteric neurons in the colon. The leading DS candidate gene is trisomy of the Down syndrome cell adhesion molecule (DSCAM).
Results
We find that Dscam1 protein is expressed in the Drosophila enteric/stomatogastric nervous system (SNS). Axonal Dscam1 phenotypes can be rescued equally by diverse isoforms. Overexpression of Dscam1 resulted in frontal and hindgut nerve overgrowth. Expression of dominant negative Dscam1-ΔC led to a truncated frontal nerve and increased branching of the hindgut nerve. Larval locomotion is influenced by feeding state, and we found that the average speed of larvae with Dscam1 SNS expression was reduced, whereas overexpression of Dscam1-ΔC significantly increased the speed. Dscam1 overexpression reduced the efficiency of food clearance from the larval gut.
Conclusion
Our work demonstrates that overexpression of Dscam1 can perturb gut function in a model system.
Graphical Abstract
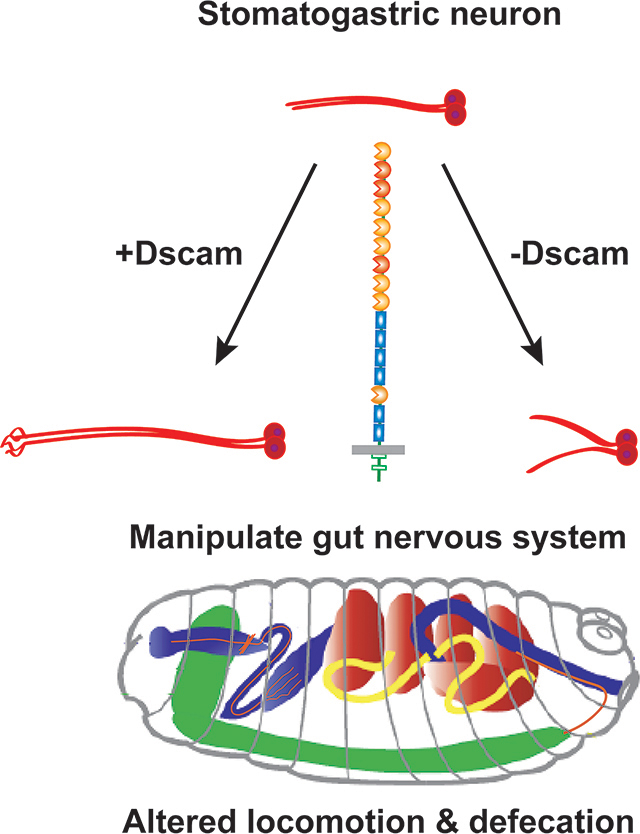
Introduction
Down syndrome (DS) is characterized by a wide range of symptoms, including variable levels of intellectual disability, congenital heart defects, and stereotypical facial features 1. DS patients additionally can have gastrointestinal abnormalities such as Hirschsprung disease of the colon and rectum (HSCR) 2. In HSCR, neural precursors derived from the neural crest either fail to migrate to the distal regions of the gut or have survival defects that lead to a lack of innervation 3. Trisomy for the DSCAM gene, located on chromosome 21, is the leading candidate for the increased occurrence of HSCR in DS 4,5. DSCAM is a large single-pass transmembrane protein with extensive roles in neural connectivity and other developmental processes 6,7. DSCAM is expressed in the migrating neural crest cells that give rise to the enteric nervous system (ENS) 8,9. Upon arriving at the correct rostro-caudal position, ENS neural precursors migrate orthogonally to form ganglia. It has been proposed that an extra copy of DSCAM disrupts this secondary migration to produce HSCR 5.
In Drosophila, disruption of Dscam1 function frequently leads to axon stalling 10–12, suggesting that Dscam1 normally promotes axon growth and expansion. Overexpression of Dscam1 increases axonal arbor size 13,14, produces synaptic targeting errors 13,15, alters dendritic patterning 16, and induces ectopic midline crossing of CNS axons 17. Functional studies reveal that Dscam1 overexpression can severely affect heart development and sensory perception 15,18.
Development of the Drosophila equivalent of the vertebrate ENS, the stomatogastric nervous system (SNS), resembles vertebrate placode-derived neurogenesis, rather than the neural crest. Nevertheless, there are strong genetic homologies between the SNS and ENS. Outside of DS, the principal gene underlying HSCR in humans is RET, which is inherited as an autosomal dominant mutation subject to modification by other loci 19. In the fly, dRet is expressed in the migrating neural precursors that give rise to the SNS, matching expression of the vertebrate RET gene 20,21. Mutation of dRet leads to axon branching and peristaltic defects 22, as well as disruption of sensory neuron dendrites outside the SNS 23. There are additional evolutionary homologies between the ENS and SNS. The neural crest placode is marked by Six1/2 expression even in primitive chordates and cells migrate to surround the future oral opening 24, and the fly paralogue, sine oculis, is required for SNS development 25. Intriguingly, the C. elegans homologue, ceh-34, is required for the initiation and maintenance of the entire enteric nervous system26. An additional set of motor neurons that innervate the hindgut, are responsible for defecation and are identified by Pigment-dispersing factor (Pdf) expression 27. PDF is functionally related to the vertebrate vasoactive intestinal polypeptide (VIP) and orexin/hypocretion 28,29, and both genes can be expressed in the same ENS neurons 30.
Here we investigate the contributions of Dscam1 to fly SNS innervation. We find that Dscam1 is expressed in the embryonic SNS suggesting an evolutionarily conserved role. Knock-out Dscam1 mutations resulted in reduced innervation of pharyngeal muscles. Based on prior over-expression studies 14,15,31, we predicted that Dscam1 over-expression in the SNS would lead to disruption of precursor cell migration and/or neural connectivity. We found that the overall structure of the SNS was unaffected, but axon connectivity defects were evident in the embryonic frontal nerve (innervates the muscles of the pharynx) and the hindgut nerves, with weaker effects in other parts of the SNS. These defects can be recapitulated by overexpressing the Dscam downstream signaling molecule PAK. Genetic manipulations of Dscam1 levels in the SNS led to impaired abilities to clear food from the gut as well as effects on larval locomotion. In summary, over-expression of Dscam1 has subtle positive effects on axon length or arbor size that nevertheless have functional consequences.
Results
Dscam1 is expressed in the Stomatogastric Nervous System
To assess whether the fly SNS is a suitable model system for modeling Dscam enteric function, we stained Drosophila embryos with anti-Dscam1 antibodies (Figure 1). Most of the staining was done with the “357” antibody directed against the cytoplasmic domain from D. Schmucker32, which gave the strongest signal, but has some background staining in the trachea. A second antibody from the Zipursky laboratory raised against the extracellular domain was used33. Finally, an antibody raised against exon 18 (ab18) from the Lee group was also used34. All three antibodies gave the same positive signal in the developing SNS. Dscam1 protein is found at the highest levels in CNS axons and can also be seen in muscle attachment sites and motor neurons10. The SNS gave a lower level of signal compared to other tissues. Dscam1 plays an important role in the peripheral nervous system and also heart development, but Dscam1 protein is difficult to observe in those tissues in the embryo. SNS staining was predominantly axonal, rather than in cell bodies. There may or may not be very low levels of Dscam1 protein in migrating neural precursors prior to axon outgrowth (Fig. 1A), which suggests that in flies Dscam1 is playing a role in axonal connectivity rather than precursor migration. We observed that Dscam1 protein is expressed in the dorsal pharyngeal muscles (Fig. 1B), prior to outgrowth of the frontal nerve which innervates the muscles (Fig. 1B’). Expression of Dscam1 is strongest in the frontal ganglion and recurrent nerve (Fig. 1B’, 1C). The Dscam1 positive neurons are responsible for controlling foregut movements 35,36, along with serotonergic axons from the CNS37. In older embryos, expression in the frontal nerve becomes evident (Fig. 1E), and the overall pattern is directly comparable to Fas2 staining in the SNS (Fig. 1D). In embryos about to hatch, for which staining is overdeveloped, expression of Dscam1 is observed throughout the entire SNS including the esophageal and ventricular ganglia, likely in the cell bodies (Fig. 1F). We attempted co-labeling of the SNS with anti-Dscam1 and anti-Fas2, but the weak signal of Dscam1 in the SNS makes co-localization challenging, even with computational clearing (Fig. 1G). We have tried to make the complex three-dimensional nature of the SNS clearly using a color-coded depth projection (Fig. 1H) and a cartoon of SNS organization in relation to other tissues (Fig. 1I). In summary, Dscam1 protein is observed in the axonal portions of the anterior SNS throughout embryogenesis, and appears at a low level in the more posterior ganglia shortly before hatching.
Figure 1. Dscam1 is expressed in the developing stomatogastric nervous system.
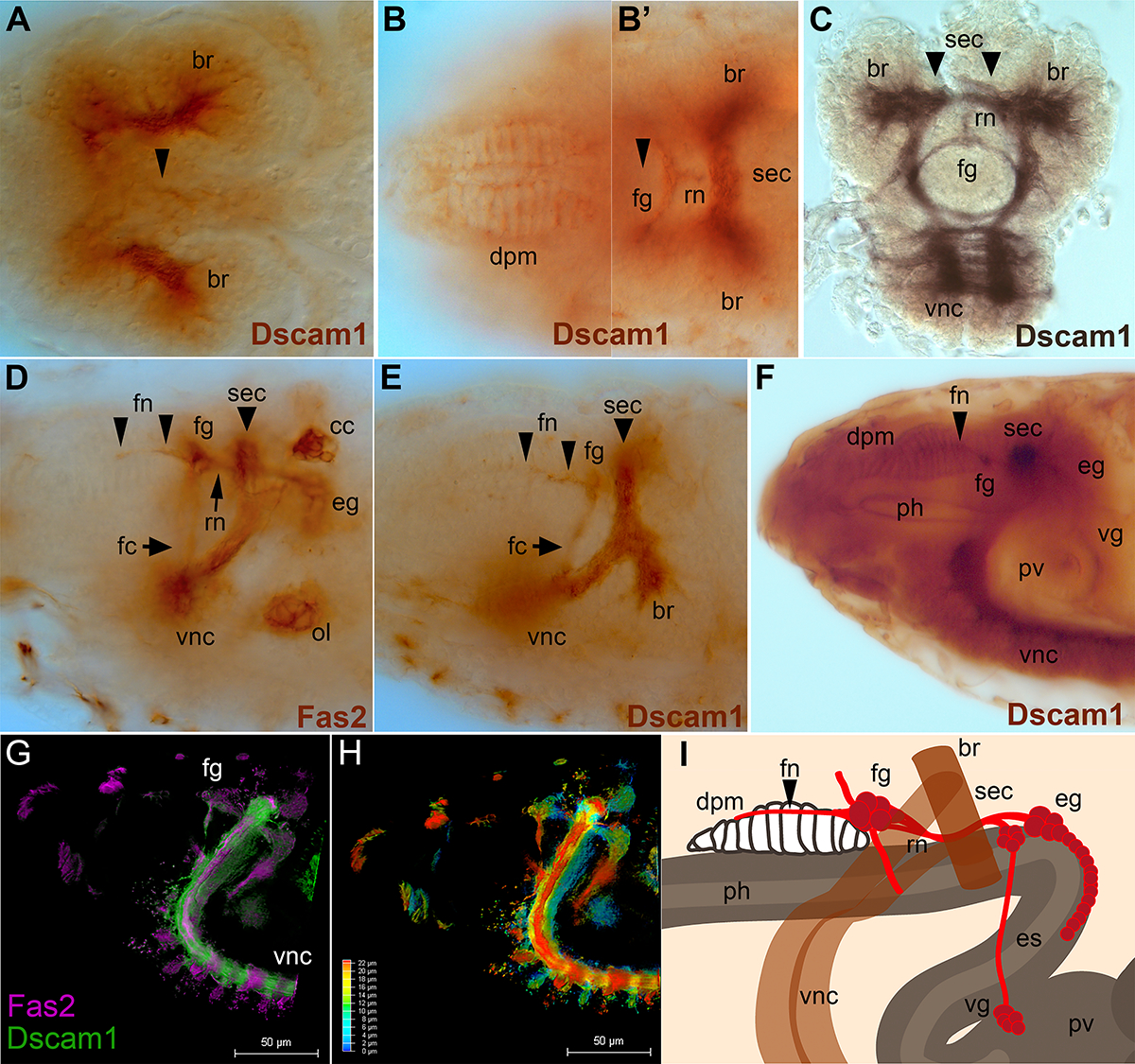
Embryos labeled with anti-Dscam1 “357” antibody, except (C) ab18, and (D) anti-Fas2 (1D4 monoclonal antibody). (G) and (H) are double labeled with anti-Dscam1 and 1D4. Anterior is to the left in all panels except (C). (A) Early stage 15 embryo in which the stomatogastric nervous system (SNS) precursor clusters begin to end their migration. Strong Dscam1 reactivity (brown) is seen in CNS axons within the brain lobes (br). The first hints of Dscam1 reactivity are visible in cells on top of the esophagus (arrowhead). The weakness of staining does not allow double labeling. (B, B’) Two focal planes of the same stage 16 embryo. (B). Dscam1 protein is visible in the dorsal pharyngeal muscles (dpm). (B’). The frontal ganglion (fg) and recurrent nerve (rn) of the SNS have formed. The frontal nerve has not yet appeared; its future path is marked with an arrowhead. Axons of the supraesophageal commissure (sec, also known as the brain commissure and part of the CNS) which links the brain lobes (br) are heavily labeled. (C). Frontal (coronal) view of a dissected stage 17 embryo labeled with anti-Dscam1 ab18 followed by Ni+ stain (black) to enhance visualization. Dscam1 expression is evident in the frontal ganglion (fg) and recurrent nerve (rn). The brain lobes, supraesophageal commissure, and the start of the ventral nerve cord are also labeled. (D). Stage 17 embryo labeled with anti-Fasciclin2 staining revealing the mature embryonic SNS. The frontal nerve (fn) projects anteriorly from the frontal ganglion (fg). The recurrent nerve (rn, arrow) runs from the esophageal ganglion (eg) along the esophagus underneath the supraesophageal commissure (sec) to the frontal ganglion (fg). The frontal connective (fc) projects ventrally from the frontal ganglion. Outside of the SNS, the anterior portion of the ventral nerve cord (vnc), the olfactory lobe (ol) and the corpora cardiaca (CC) are labeled. (E) Late stage 16 embryo reveals Dscam1 in the frontal nerve (fn, arrowheads), the frontal ganglion and brain commissure (sec) connecting the brain lobes (br). (F) Late stage 17 embryo with overdeveloped for DAB immunoreactivity. Dscam1 is visible in the dorsal pharyngeal muscles (dpm), which have been innvervated by the frontal nerve (fn; arrowhead). The frontal ganglion (fg) is visible in cross-section as is the brain commissure (sec). The posterior portion of the SNS, the esophageal ganglion (eg) displays light immunoreactivity, as do neurons of the ventricular ganglion (vg) which lie on top of the proventriculus (pv). Immunoreactivity is also seen in the lining of the pharynx (ph), pv, and heavily in the axons of the ventral nerve cord (vnc). (G). Fluorescent double labeling of a stage 17 embryo with anti-Fas2 (magenta) and anti-Dscam1 (green). To enhance the weak Dscam1 staining in the SNS, the embryo was imaged and computationally cleared on a Lecia Thunder microscope. The frontal ganglion is indicated (fg). (H). The same embryo as in (G), but color coded to indicate depth according to the inset scale. (I). Schematic of the embryonic stomatogastric nervous system (SNS; red). The frontal ganglion (fg) lies on top of the pharynx (ph), and the frontal nerve (fn) projects anteriorly to the pharyngeal muscles (dpm). The digestive tract (gray) and the recurrent nerve (rn) pass through the CNS (brown) underneath the brain commissure (sec). The esophageal ganglia (eg) lie on top of the esophagus (es), and the ventricular ganglion (vg) lies on top of the proventriculus (pv).
Phenotypic Analysis of Dscam1 in the Stomatogastric Nervous System
The spectacular isoform diversity of the Dscam1 gene plays well known roles in dendritic spacing and axon branching6. In contrast, Dscam1 defects in sensory axon arbors can be rescued by distinct Dscam1 isoforms13. These results also demonstrate that Dscam1 plays a role in axon growth. Dscam1 mutants have relatively mild embryonic CNS defects that are greatly enhanced by the loss of either the fra or robo1 receptors10,17. We wondered whether isoform diversity is required in CNS axons, and drove pan-neural expression of distinct isoforms, 7-27-25-1 (dendritic) and 1-30-30-2 (axonal), in Dscam1 fra double mutants (Fig. 2A–D). Both isoforms were able to rescue equally well as the Dscam1 fra double mutant is statistically different from the other three genotypes (p<0.01, one way ANOVA with Tukey HSD test on commissural defects; Table 1). Combined with our previous work on Dscam1 robo1 interactions10, we believe that Dscam1 can promote axonal growth in an isoform dependent fashion, with the caveat that the axonal isoform promotes growth to higher levels13. Having established that Dscam1 is expressed in the SNS, we examined Dscam1 mutants for alterations to SNS patterning using anti-Fas2. In control embryos, the frontal nerve projects to the dorsal pharyngeal muscles (Fig. 2E, G, G’). This phenotype is only 75% penetrant suggesting that innervation may complete after hatching. We use w1118 as a control strain as we find that body wall muscle innervation is most complete at stage 17 compared to wild type strains. Dscam1 mutants have a highly penetrant (0.89) reduced or absent frontal nerve phenotype (Fig 2F, H, H’), which is trending towards significance compared to the control strain (p=0.1, Fisher exact test). Future work may require scoring the frontal nerve projection in first instar larvae, rather than embryos. There appear to be no other defects compared to wild type. We also examined Dscam1 fra and Dscam1 robo1 double mutants (Fig. 2I, I’, J, J’), but other than the frontal nerve defect, the rest of the SNS appears normal. These results suggest that Dscam1 may play a role in innervation of the dorsal pharyngeal muscles by the frontal nerve, but has no other or only minor roles in patterning of the SNS.
Figure 2. Dscam1 function in the CNS and SNS.
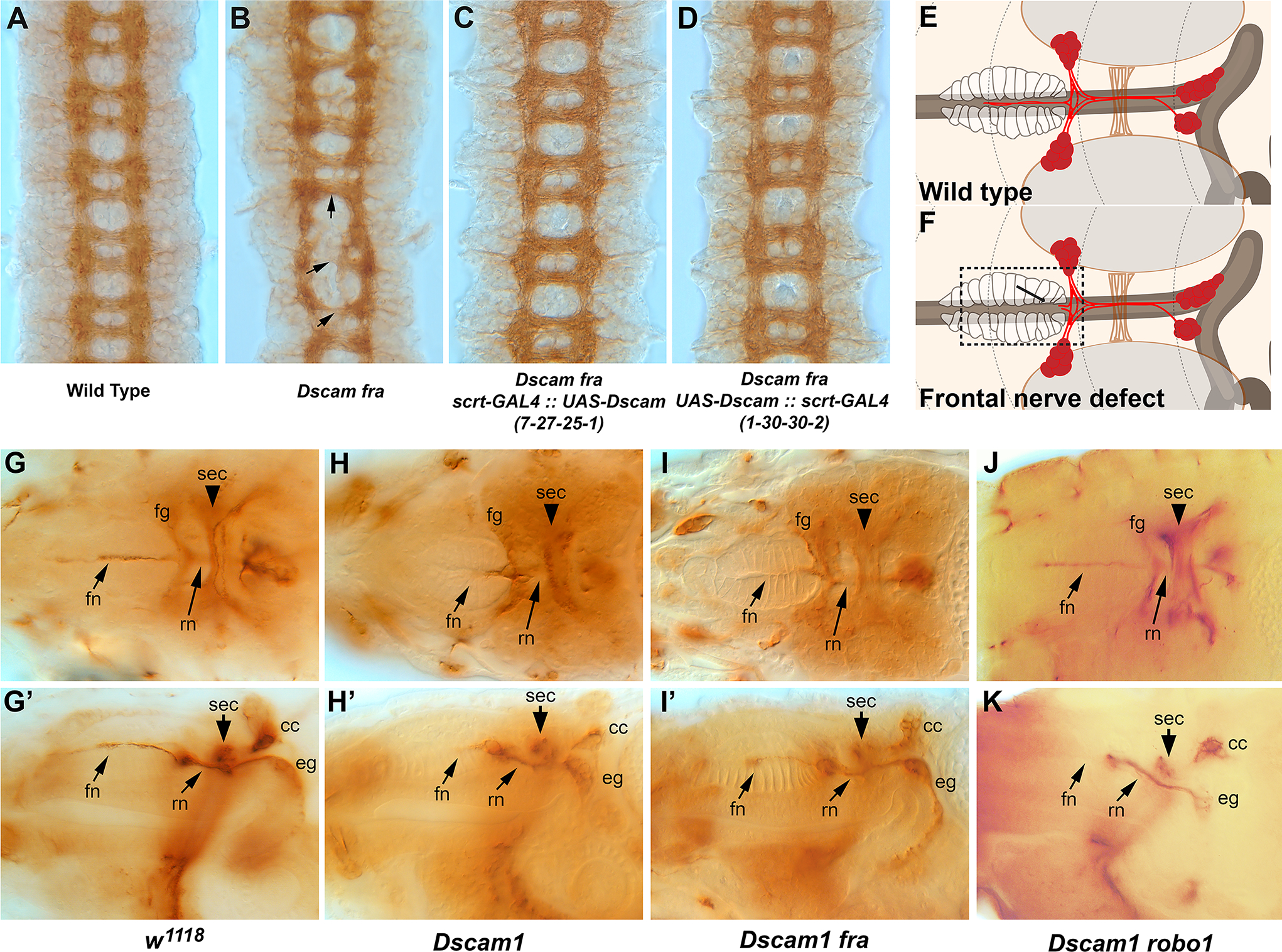
Stage 16 embryos labeled with anti-BP102 (A-D) or stage 17 embryos were labeled with anti-Fasciclin2 staining and photographed dorsally (G-J), or from the side (G’-J’). (A) Wild type (Oregon R) control embryos displaying the characteristic ladder-like pattern of the longitudinal axon tracts running vertically and connected by commissural axon tracts running horizonatally. (B). Dscam1 fra double mutant displaying absent, disorganized or thickened commissures (arrows). (C). Dscam1 fra double mutant expressing UAS-Dscam 7-27-25-1 under control of pan-neural scratch-GAL4. (D). Dscam1 fra double mutant expressing UAS-Dscam 1-30-30-2 driven by scratch-GAL4. Both (C) and (D) strongly resemble the wild type control. (E). Schematic of the SNS (red) viewed dorsally; the frontal ganglion (fg) and esophageal ganglia (eg) are labelled. The brain lobes (br), brain commissure (sec) and dorsal pharyngeal muscles (dpm) are indicated. (F). Schematic displaying failure of the frontal nerve to innervate the dorsal pharyngeal muscles (boxed area, arrow). (G). Control embryo (w1118) showing the frontal nerve (fn) extending over the pharynx towards the anterior of the embryo (left). The frontal ganglion (fg) and recurrent nerve (rn) are also visible. Part of the CNS, the supraesophageal commissure (sec) links the two brain lobes. (G’) Lateral view of a control embryo displaying the SNS components including the esophageal ganglion (eg) which extends along the foregut to the midgut. (H) Embryos lacking Dscam1 frequently display a thinned frontal nerve, and a slightly irregular frontal ganglia and frontal commissure. (H’) Lateral view of the same embryo in H. The frontal nerve is clearly thinner; the esophageal ganglion is out of the plane of focus and is unaffected. (I, I’) Dscam1 fra double mutants resemble Dscam1 single mutants with a thinner frontal nerve and slightly irregular frontal ganglion but no other defects. (J) Dscam1 robo double mutant with an intact frontal nerve and normal positioning of other SNS elements. (J’) Lateral view of a Dscam1 robo double mutant with an absent frontal nerve, but otherwise normal SNS.
Table 1.
Quantification of commissural axon defects in Dscam1 fra mutants rescued by Dscam1 isoforms
| Anterior Commissures | Posterior Commissures | Both commissures | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Normal | Thin | Very thin | Absent | Normal | Thin | Very thin | Absent | Normal | Thin | Very thin | Absent | n |
| Oregon R | 99% | 1% | 0% | 0% | 99% | 1% | 0% | 0% | 99% | 1% | 0% | 0% | 110 |
| Dscam fra | 1% | 9% | 35% | 55% | 1% | 5% | 32% | 62% | 1% | 7% | 34% | 58% | 99 |
| Dscam fra, UAS-Dscam1-7-7-25-1, ScrtGAL4 | 93% | 7% | 0% | 0% | 93% | 7% | 0% | 0% | 93% | 7% | 0% | 0% | 105 |
| Dscam fra, UAS-Dscam1-1-30-30-2, ScrtGAL4 | 93% | 6% | 1% | 0% | 93% | 7% | 0% | 0% | 93% | 6% | 0% | 0% | 106 |
Characterization of larval expression of Ret-GAL lines
We previously reported on the generation of GAL4 lines driven by fragments of the Ret promoter38. The Ret-P2A and -P2B lines are independent isolates of identical transgenes that originally had slightly different expression patterns, presumably due to background mutations. Both express in the SNS precursor clusters prior to and during migration, and express in the frontal ganglion and ventricular ganglion during embryogenesis. Expression of Ret-P2A and -P2B in the ventricular ganglion continues during the first and second larval instars22,38. We confirmed this expression and observed that ventricular ganglion expression continued in the second and third instars (Fig. 3A, B). We also observed expression in a subset of CNS cells throughout larval development (Fig. 3C). A cluster along the midline may potentially be continuation of embryonic expression20. By increasing the dosage of the Ret-GAL4 lines and the UAS-CD8-GFP reporter, we were able to detect GFP expression in body wall sensory neurons (Fig. 3D). We assume this is a subset of the Ret positive sensory neurons previously reported23,39. We examined the anal sphincter for GFP expression in the defecation circuit, but could not detect any fluorescence above background (Fig. 3E). We compared expression to the pan-neuronal driver elav-GAL4 and found expression only in a very small subset of anterior SNS neurons, and never in the ventricular neurons in all larval stages (Fig. 3F). This is in contrast to adult expression of elav40.
Figure 3. Larval Expression of Ret-GAL4 lines.
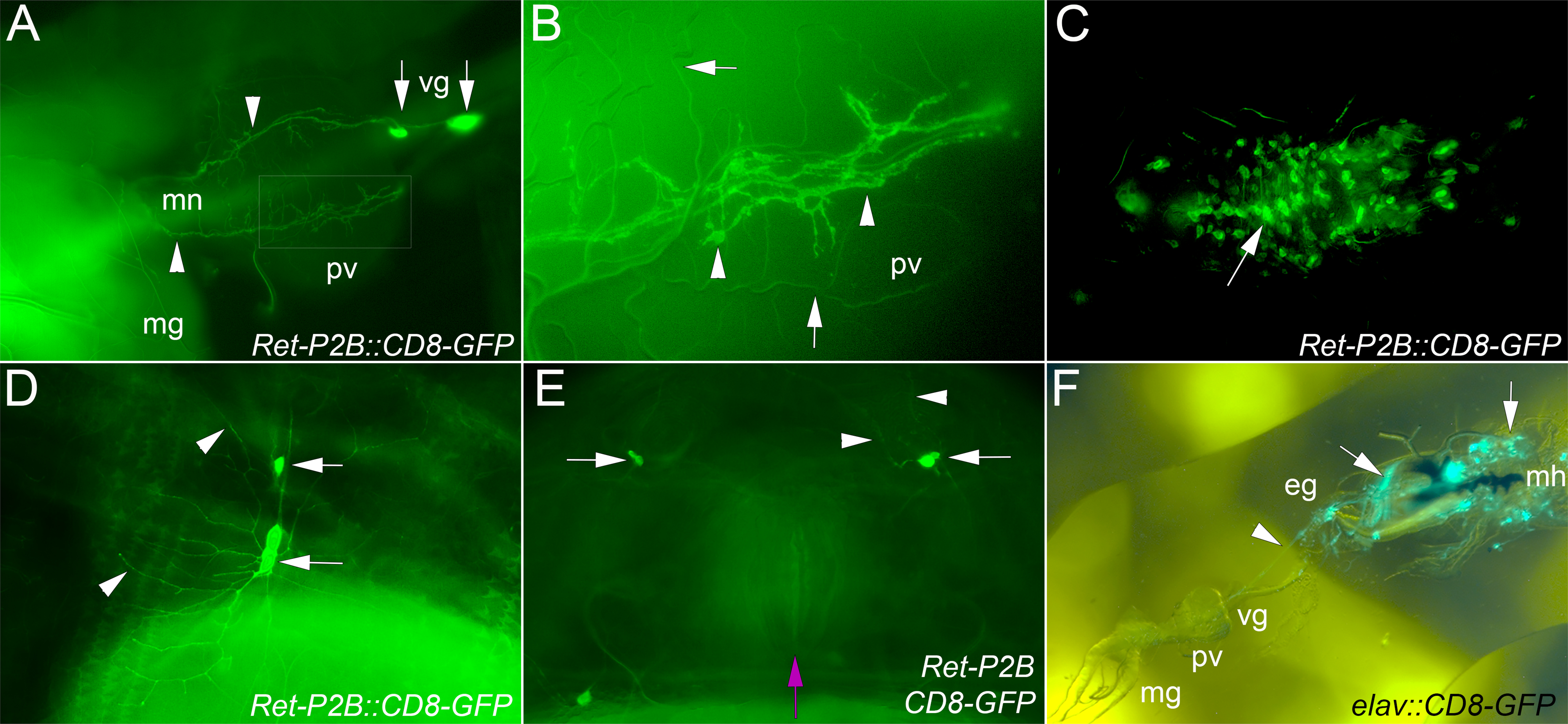
Ret-P2A-GAL4 and Ret-P2B-GAL4 were crossed to UAS-CD8-GFP and larvae were examined for GFP expression. Both lines displayed equivalent expression in the CNS and midgut neurons that persisted throughout all three larval stages. (A). Second instar larval displaying expression in the ventricular ganglion (vg; arrows) and axons of the midgut neurons (mn; arrows) projecting over the proventriculus (pv) to the midgut (mg). Gut autofluorescence prevents visualization of the mn axons as they continue towards the posterior. Anterior is to the right. Boxed area is enlarged in (B). Close up of the larva in (A) to show the broad nature of the mn axons as they project over the proventriculus (arrowheads). Trachea (arrowheads) are also visible due to reflection of light, but do not express GFP. (C). CNS expression in subset of neurons is rarely strong enough to visualize axons. A row of cells may correspond to the Ret positive midline neurons in the embryo (arrow). (D,E). When multiple copies of the Ret driver and GFP reporter are used, sensory axons in the body wall are visible (arrows). Expression in the dendrites (arrowheads) is also visible. (E). We examined the anal sphincter (magenta arrow) for expression in axons of the defecation circuit but could not detect GFP expression over endogenous autofluorescence. (F). Larval expression of elav-GAL4 driving UAS-CD8-GFP was examined. Neurons through the CNS and PNS were observed as expected. However, SNS expression was much more limited. Clear expression could be seen in the esophageal ganglion (eg) and axons projecting to the proventriculus (arrowhead, pv). The area around the mouth hooks (black structures, mh) was hard to dissect intact, but structures resembling small muscles and presumed sensory organs (arrows) were observed. Expression in the ventricular ganglion (vg) and axons projecting to the midgut (mg) was not observed.
Expression of Dscam1 transgenes in the developing SNS
To model trisomy for DSCAM in Down Syndrome, we overexpressed full-length Dscam1 (Dscam-FL) in the SNS. Transgenes were expressed by Ret-P2A and -P2B and Gsc-GAL438. Gsc-GAL4 shuts off expression at the end of embryogenesis. For all experiments reported here, we used UAS-Dscam1-1-30-30-2 (axonal). In parallel, we over-expressed a Dscam1-1-30-30-2 construct that lacks the cytoplasmic domain 16. We have found that this construct (Dscam1ΔC) is quite disruptive to CNS formation, with phenotypes exceeding those of fra or robo1 mutants alone, which are presumed or demonstrated co-receptors. These stronger defects suggest that Dscam1ΔC may be forming inactive complexes with other receptors 10. Expression of full-length Dscam1 altered morphology and distribution of Fas2 in the frontal nerve, with the tip of the frontal nerve expanded laterally and axons following the junctions between muscles (Figure 4B, C). Axons following intramuscular junctions is a phenotype that resembles the expansion of presynaptic arbors observed in C4da neurons when Dscam1 is overexpressed 13,14. The frontal nerve phenotype was visualized across the drivers used, along with marked phenotypes in the frontal ganglia and recurrent nerve (Fig. 4G). In contrast, expression of the dominant negative Dscam1ΔC frequently led to an opposite phenotype in which the frontal nerve was completely absent, and also caused defasciculation of the recurrent nerve (Figure 4D–F, G). Anti-Fas2 also stains the hindgut motor neurons that control defecation 27, and we found that overexpression of Dscam1 led to expansions at the tip of the hindgut nerves and increased numbers of side branches at the start of the axon trajectory (Fig. 5B, C). Dscam1ΔC instead induced splitting of the hindgut axon into two distinct branches (Fig. 5D–F). We confirmed these results using the hindgut neuron specific Pdf-GAL4 line (K.G. unpublished results). These results are quantified in Figure 4G and 5G. Overall, expression of Dscam1 appeared to act positively on axons increasing the area innervated, whereas Dscam1ΔC either inhibited growth or promoted branching.
Figure 4. Dscam1 overexpression causes SNS defects.
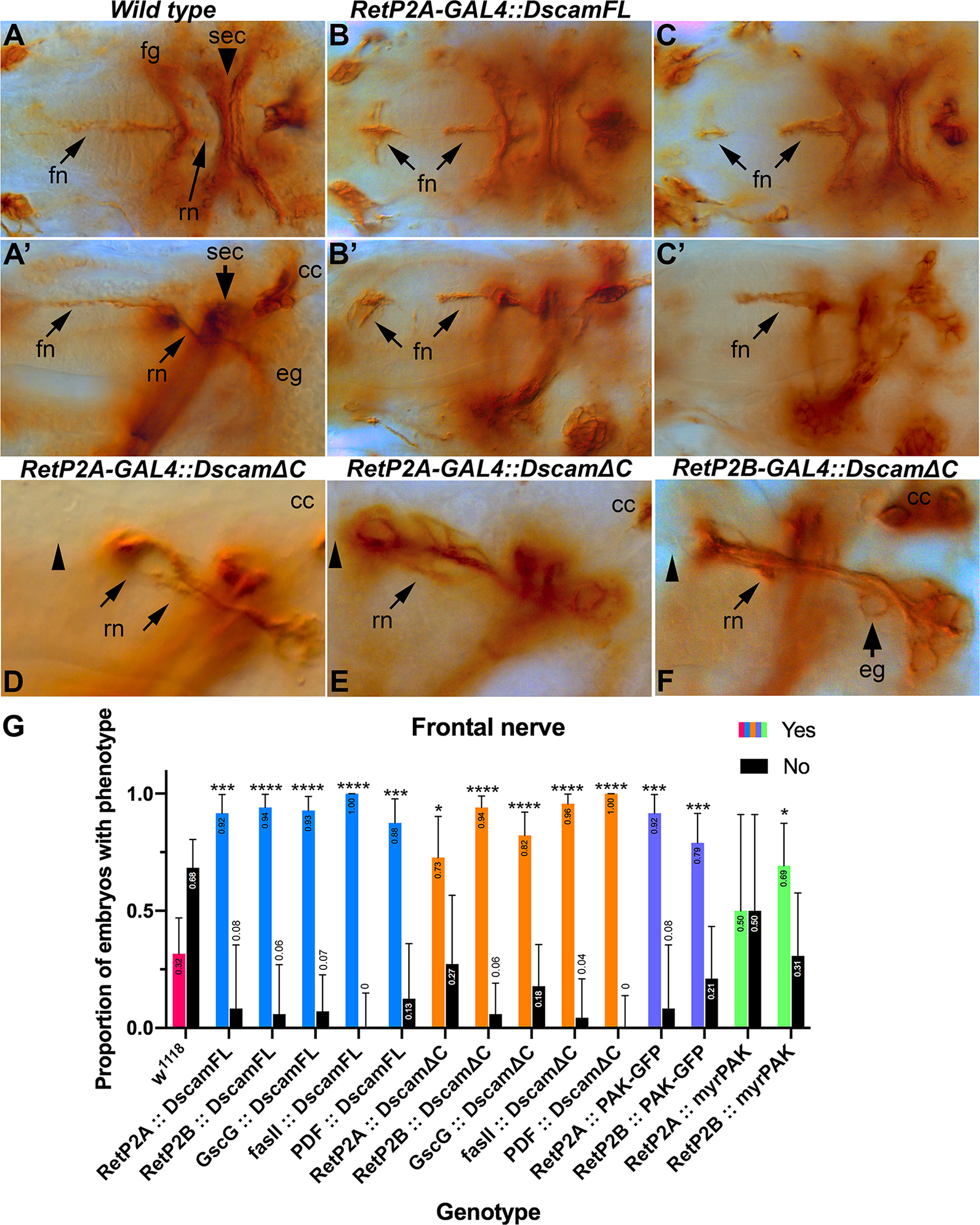
Stage 17 embryos stained with monoclonal 1D4 to visualize motor neurons comprising the characteristic anatomical landmarks of the mature SNS. (A) Dorsal view of stage 17 control embryo illustrating stereotypical/symmetrical SNS structures. (A’) Lateral view of the same embryo clearly showing the intact frontal nerve (fn), recurrent nerve (rn), brain commissure (sec), corpora cardiac (cc) and esophageal ganglia (eg). (B) Embryos carrying one copy of RetP2A-GAL4 and one copy of UAS-Dscam1-FL transgenes showing the frontal nerve extending anteriorly and ectopically expanding/innervating the pharyngeal muscles (arrows). Staining of the frontal nerve is absent in the middle section. (B’) Lateral view of the same embryo showing additional detail of the frontal nerve thickening/expansion phenotype. (C) Embryo expressing UAS-Dscam1-FL under control of the Gsc-GAL4 driver showing the same ectopic expansion of the frontal nerve (arrows). (C’) Lateral view of the same embryo. (D) Close-up of lateral view of stage 17 embryo carrying one copy of the RetP2A-GAL4 driver and one copy of the UAS-Dscam1ΔC transgene. The recurrent nerve displays strong defasciculation and separation (arrows). The frontal nerve is absent (arrowhead). (E) Embryo of the same genotype as D exemplifying additional recurrent nerve defasciculation and appreciable asymmetry in reference to the brain commissure (sec) (arrow). The frontal nerve is also absent (arrowhead). (F) Close-up of lateral view of embryo carrying RetP2B-GAL4 and UAS-Dscam1ΔC transgene. The frontal nerve is absent (arrowhead). The axons are defasciculated (arrow) and separated. Esophageal ganglia are also abnormal (arrow). (G) Quantification of embryonic phenotypic defects. Data from all three drivers (Ret-P2A, Ret-P2B and Gsc) was combined, scored blind, and statistically analyzed with Fisher exact tests conducted between all experimental genotypes and w1118 controls. The Odds ratio (Baptista-Pike) 95% confidence ratio was calculated and plotted as error bars. Key for p-value summary: * 0.01<p<0.05; ** 0.001<p<0.01; *** 0.0001<p<0.001; **** p<0.0001. Overexpression of Dscam1-FL disrupts frontal nerve and hindgut nerve formation, and to a lesser extent the frontal ganglion and recurrent nerve. Dscam1ΔC also disrupts the frontal and hindgut nerves and the recurrent nerve. Similar patterns are seen with over-expression PAK transgenes, including an activated PAK (myr-PAK).
Figure 5. Hindgut nerve phenotypes caused by Dscam1 or Dscam1-ΔC over-expression.
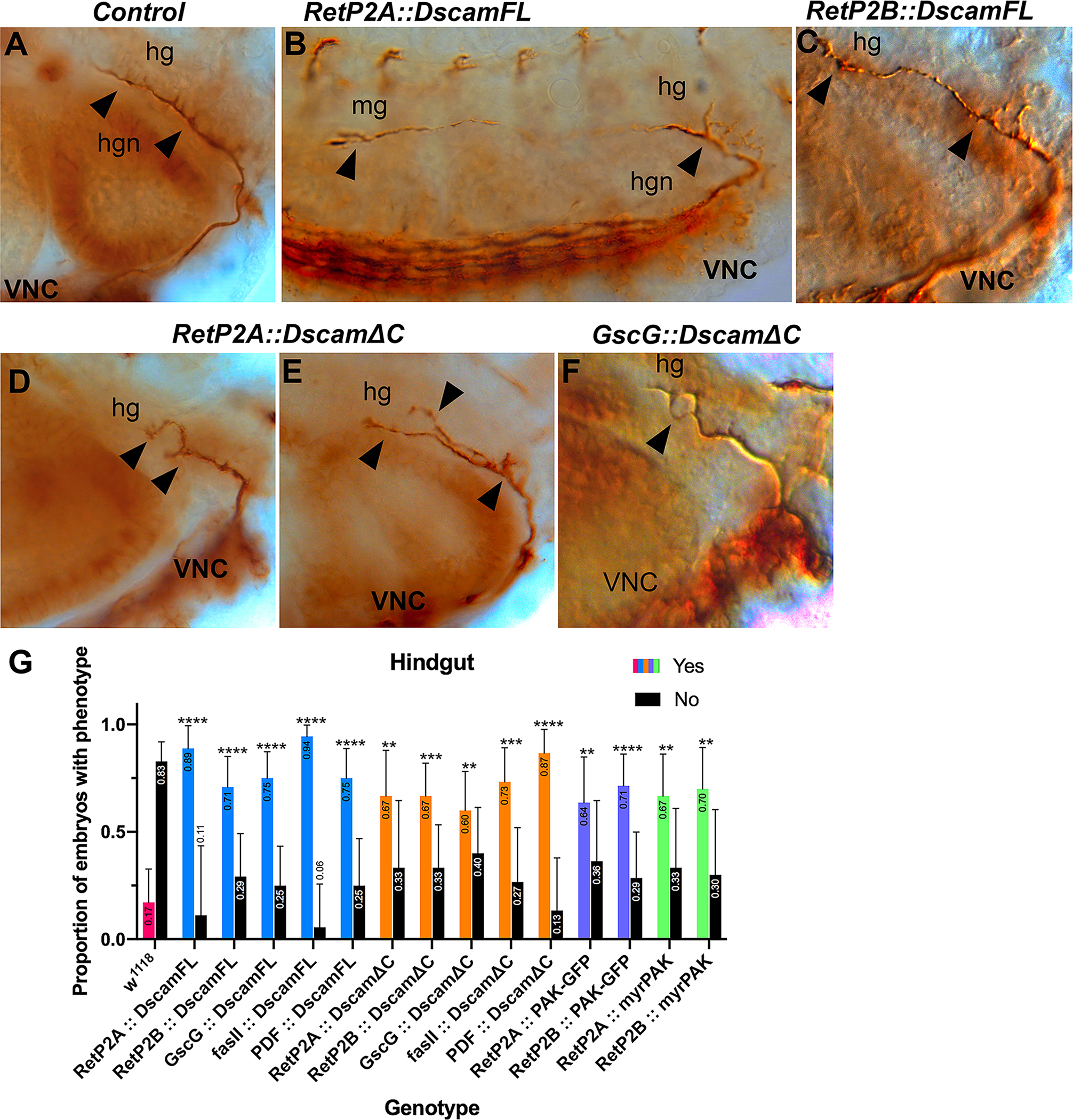
Stage 17 embryos stained with anti-Fas2 to visualize the hindgut nerve (hgn). (A). Control embryo showing the normal extent and branching of the hindgut nerve projecting from the ventral nerve cord (VNC). (B). Embryo over-expressing Dscam1 displaying a highly elongated hindgut nerve extending approximately six segments along the midgut (mg). Defasciculation is apparent at the end of the nerve, and larger than normal sidebranches are visible at the start (arrowheads). The longitudinal fascicles of the ventral nerve cord (VNC) can be seen running along the bottom of the panel. (C). Embryo over-expressing Dscam1 with a normal length hindgut nerve displaying subtle expansion of the nerve tip and more robust side branches (arrowheads). (D). Over-expression of Dscam1ΔC leads to a shorter, bifurcated hindgut nerve. (E). Embryo over-expressing Dscam1ΔC with distinct bifurcation and larger than normal side branch (arrowheads). (F). Short, defasciculated hindgut nerve (arrowhead) in an embryo over-expressing Dscam1ΔC. (G). Quantification of embryonic phenotypic defects. Deviations from control embryos, such as axon overgrowth or axon branching were combined and scored blind for the indicated genotypes. The data was statistically analyzed with Fisher exact tests conducted between all experimental genotypes and w1118 controls. The Odds ratio (Baptista-Pike) 95% confidence ratio was calculated and plotted as error bars. Key for p-value summary: * 0.01<p<0.05; ** 0.001<p<0.01; *** 0.0001<p<0.001; **** p<0.0001.
PAK expression reproduces Dscam1 phenotypes
In Drosophila, Dscam1 in axons signals downstream via PAK (p21 activated kinase) 33. We expressed PAK (PAK-GFP) and an activated PAK isoform (myr-PAK)41 in the SNS to determine whether the same phenotypes were observed. Overexpression of wild-type PAK reproduced the phenotypes observed when Dscam1 was over-expressed, specifically an expansion of the tip of the frontal nerve coupled with reduction of Fas2 staining in the frontal nerve itself (Figure 5B). Over-expression of activated PAK mimicked expression of Dscam1-ΔC causing the frontal nerve to be absent or highly reduced (Figure 6C). These results are included in the quantification in Fig. 4G. In the hindgut, PAK overexpression caused the hindgut motor neurons to grow much farther than usual, sometimes along the entire length of the hindgut as opposed to approximately halfway in wild type (Fig. 8B; quantification in Fig. 5G). We have observed this phenotype at a much lower frequency in Dscam1 gain-of-function embryos (Fig. 5B). Activated PAK expression induced the major branching characteristic of Dscam1-ΔC expression. These results indicate that Dscam1 may be signaling through PAK in the SNS. If Dscam1 was purely permissive for axon growth, then activated PAK should have the same effect as PAK over-expression. However, the similarity between activated PAK and Dscam1-ΔC strongly suggests that PAK activation of the entire growth cone is as detrimental to axon growth as inhibition of Dscam1 signaling. It seems likely that Dscam1 is responding to localized cues in both the frontal nerve and the hindgut motor neurons.
Figure 6. SNS and hindgut nerve phenotypes caused by PAK over-expression.
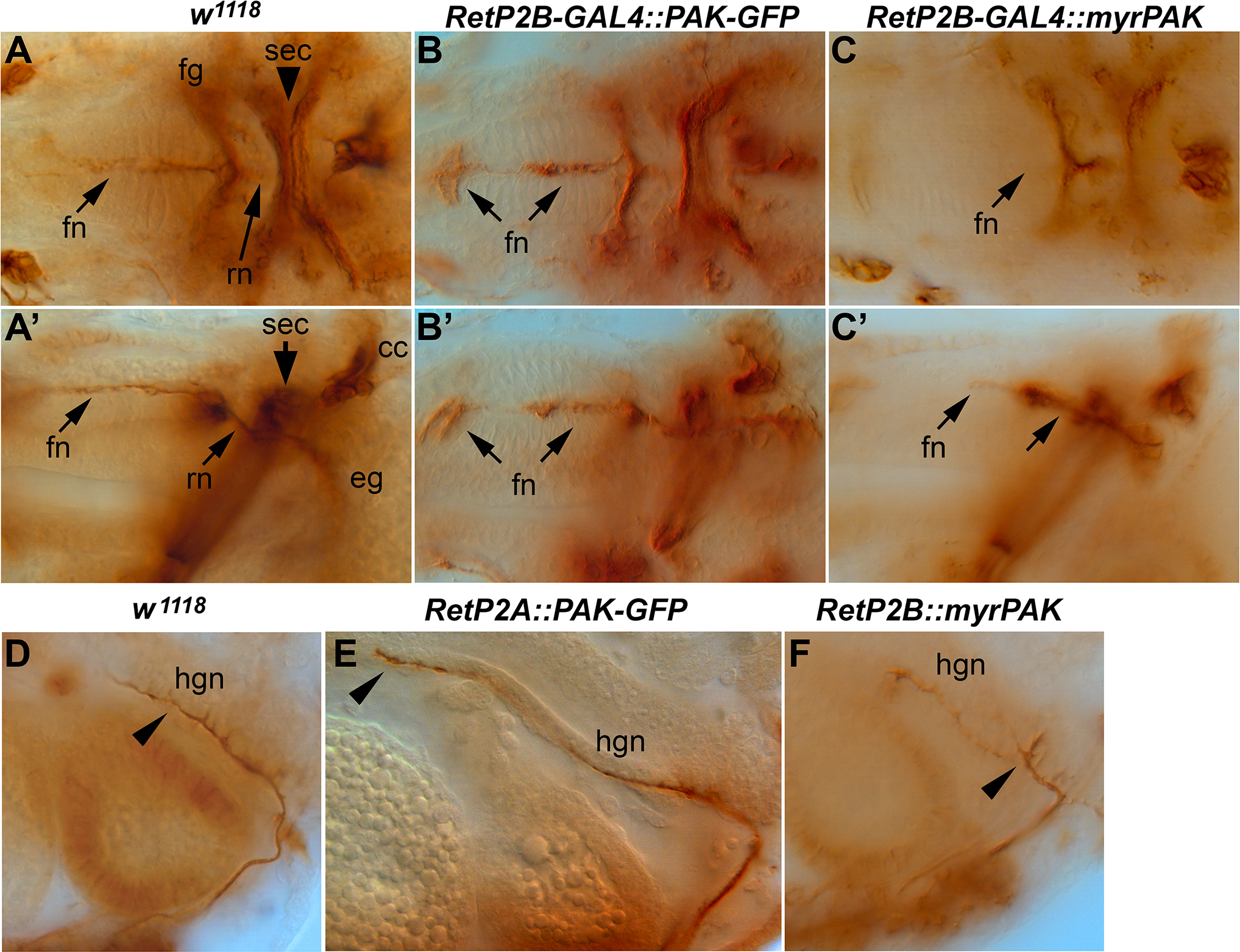
Stage 17 embryos stained with anti-Fas2 to visualize the stomatogastric nervous system and hindgut nerve (hgn). (A, A’) Control embryo with frontal nerve (fn), frontal ganglion (fg) and recurrent nerve (rn) labeled. (B, B’) Embryo over-expressing PAK-GFP. The frontal nerve is expanded both at the tip and further back (arrows). (C, C’) Embryo over-expressing myr-PAK. The frontal nerve is truncated (arrows). (D) Hindgut nerve (hgn) in a control embryo. (E) Over-expression of PAK-GFP increases the length of the hindgut nerve (arrowhead). (F) Over-expression of myr-PAK causes branching of the hindgut nerve (arrowhead).
Figure 8. Dscam1 overexpression affects food clearance from the digestive tract.
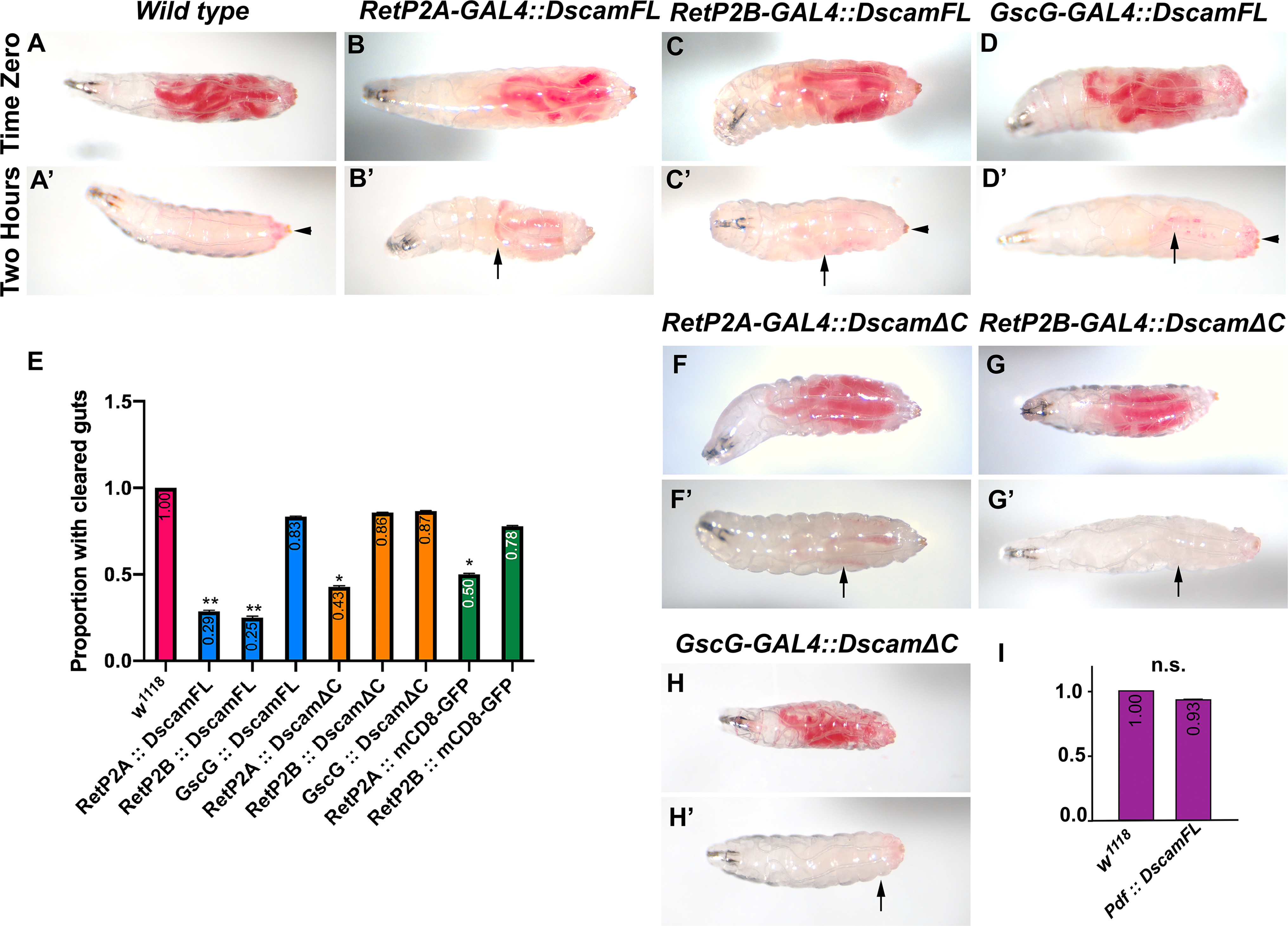
First instar larvae were fed yeast paste dyed red until the second instar. The larvae were washed and assayed for food intake. The larvae were allowed to feed on undyed food for two hours and re-examined for food clearance. (A) Control (w1118) larva displaying the presence of food in the intestines at time zero. (A’) Control embryo displaying complete clearance of food from the gut after two hours. Some red dye adheres to the cuticle but is not internal (arrowhead). (B-D) Second instar larvae over-expressing Dscam1-FL showing normal ingestion of food. (B’, C’) Reduced food clearance after two hours with expression driven by the RetP-GAL4 lines (arrows indicate red dye in the gut). (D’) Normal clearance of food after two hours when Dscam1-FL is driven by the GscG driver; the arrow indicates dye adhering to the cuticle. (E) Quantification of food clearance in indicated genotypes. A Fisher exact test was conducted between all experimental genotypes and w1118 controls with Odds ratio (Baptista-Pike) 95% confidence ratio calculated and plotted as error bars. P-value summary: * 0.01<p<0.05; ** 0.001<p<0.01. Note that the RetP2A-GAL4 driver gave a statistically significant result driving GFP localized to the cell membrane with a CD8 transmembrane domain. RetP2B-GAL4 did not. (F, F’) Overexpression of Dscam1-ΔC with the RetP2A driver impaired gut evacuation but at the same level as driving mCD8-GFP so is not significant. (G, G’, H, H’) Overexpression of Dscam1-ΔC with RetP2B and GscG drivers caused negligible effects on gut clearance. Arrowheads indicate external red food located on surface of the animals. (I) Food clearance was assessed in animals expressing Dscam1-FL driven by the Pdf-GAL4 driver and quantified. A Fisher exact test was conducted between the experimental genotype and w1118 control (n.s. is not significant).
Larval locomotion is affected by SNS Dscam1 manipulations
Larval locomotor behavior can be influenced by feeding state, as larvae with eating impairments display wandering behavior42, and starved larvae displaying increased locomotion and altered foraging strategies 43–45. We tested the effect on distance travelled and the average speed of larvae over-expressing full-length Dscam1 and Dscam1-ΔC (Figure 7A)46,47. Dscam1 over-expressing larvae showed a decrease in average speed compared to control larvae when the Ret-P2A and -P2B drivers were used (Figure 7B). GscG-GAL4 did not display the same effect suggesting that persistent SNS expression of Dscam1-FL is required for this effect (GscG-GAL4 appears to turn off at the end of embryogenesis). These results suggest that larvae over-expressing Dscam1 are not starved, but which would result in increased foraging speed. Instead, decreased locomotive speed may indicate persistence of food in the gut, possibly through reduced defecation. In contrast, expression of Dscam1-ΔC produced an increase in larval speed when expressed by the Ret-P2A,B drivers (Figure 7B). Increased speed and foraging distance with expression of Dscam1-ΔC indicates either reduced food intake or increased passing of food. These results indicate that over-expression of Dscam1-FL and Dscam1-ΔC have opposite phenotypic effects, which we suspected based on the embryonic SNS neuroanatomy. We note that many alternative explanations are possible as Ret-P2-GAL4 lines express in the CNS and PNS. For example, alterations to CNS connectivity alter larval exploratory behavior44.
Figure 7. Over-expression of Dscam1 and Dscam1-ΔC have opposite effects on larval locomotor activity.
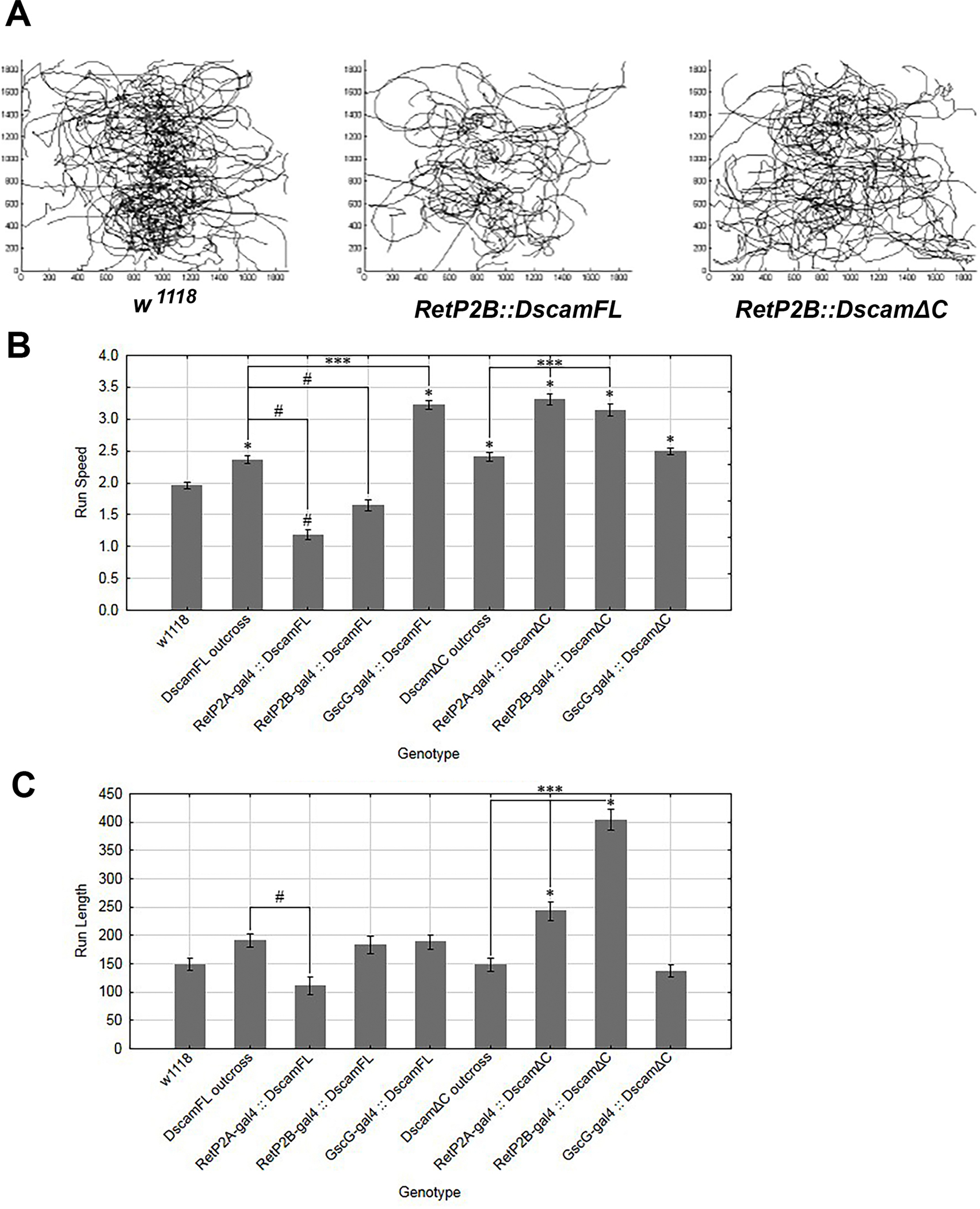
(A) Representative larval tracks recorded for the w1118 control and larvae overexpressing either Dscam1 or Dscam1-ΔC in the SNS. The Dscam1 over-expressing larvae appear to travel less than the controls or Dscam1-ΔC. (B) Analysis of run speeds in larvae overexpressing either Dscam1, which results in slower speeds, or Dscam1-ΔC which results in faster run speeds. (C) Analysis of run length in different genotypes overexpressing either Dscam1 or Dscam1-ΔC in the SNS. The Dscam1 and Dscam1-ΔC transgenes were outcrossed to w1118 to serve as a control. Statistics are one-way ANOVAs with Tukey’s multiple comparisons test. The mean is plotted with 95% confidence interval. Key for p-value summary: * 0.01<p<0.05; ** 0.001<p<0.01; *** 0.0001<p<0.001; **** p<0.0001.
Increased Dscam1 expression impairs food clearance but not feeding
In adult flies, foregut neurons are proposed to relay information from stretch receptors to the CNS, influencing feeding behavior 48. The reduced speed of larvae over-expressing Dscam1 suggested a food clearance defect could be responsible. To examine feeding behavior, first instar larvae were allowed to eat yeast paste mixed with carmine red dye 49. No apparent differences in the amount of food ingested or the distribution within the larvae were observed (Figure 8A–D, 8F–H). Larval development proceeded normally through the larval stages without any increases in mortality. As the hindgut motor neurons were affected by Dscam1 manipulation, we assayed larval defecation by feeding red yeast until the larvae reached second instar, washing them and transferring them to uncolored food for two hours. Wild type larvae (w1118) had no traces of red food internally but larvae expressing Dscam1 under control of Ret promoter fragments still had traces of food inside (Figure 8A’–C’, E). Expression under control of Gsc-GAL4, which switches off expression at the end of embryogenesis had no effect on food clearance (Fig. 8D’). This argues that continuous expression of Dscam1 is required to reduce food clearance. Expression of Dscam1ΔC under control of the Ret-P2A promoter reduced food clearance, but to the same level as a control experiment expressing UAS-mCD8-GFP (Fig. 8E, F’). Both genotypes had effects not as strong as Ret-P2A expressing Dscam-FL. The same control for RetP2B had no effect. We assume that background mutations either affect the phenotypes directly or through levels of GAL4 expression. Differences between P2A and P2B despite being supposedly identical constructs have been previously noted38. Differences between the drivers can be seen in the locomotor data as well (Fig. 7C), so the genetic background may be modulating distinct phenotypes differently. Expression of Dscam1ΔC by either RetP2B or GscG did not affect food clearance (Fig. 8G’, H’). The Abl kinase inhibitor Nilotinib has been used to counteract the effects of Dscam1 overexpression14, but when fed to larvae effects on food clearance did not reach significance (L.G. unpublished data). The continued CNS expression of the Ret-GAL4 drivers raised the possibility that the defecation circuit was being disrupted rather than the midgut neurons. We expressed UAS-Dscam1-FL under control of Pdf-GAL4, which expresses in neurons that regulate defection27, but saw no effect on food retention (Fig. 8I). Combined with them larval locomotion results, the reduced food clearance rate demonstrates that Dscam1 expression in the SNS affects gut function.
Discussion
In this study we establish that Dscam1 is likely playing a role in SNS development in Drosophila. Overexpression of full-length Dscam1 has correlated effects on SNS neuroanatomy, food clearance and locomotor activity. Driving Dscam1 expression continuously throughout larval development leads to behavioral phenotypes (Ret-P2A and 2B drivers), notably failing to clear food as efficiently as wild type. Expression solely in the embryo alters neuroanatomy but is not sufficient to disrupt larval behavior (GscG driver). Based on prior work and our observations, Dscam1 over-expression may increase local connections between SNS neurons and the gut musculature or other neurons, impairing digestive function. Dscam1 is known to affect virtually every aspect of neuronal development and function 50, so multiple explanations are possible.
Dscam1 signaling
Dscam1 acts as a homophilic cell adhesion molecule mediating repulsion between sister dendrites 51. Dscam1 also responds to Netrin and Slit ligands during axon guidance 10,17,32. In the embryonic CNS, Dscam1 mutants have mild defects in the longitudinal axons. However, Dscam1 mutants greatly enhance the phenotypes of the robo1 co-receptor and putative fra co-receptor, suggesting that axonal Dscam1 promotes axon growth in response to Slit or Netrin ligands. Expression of Dscam1-ΔC generates much stronger phenotypes, suggesting that Dscam1 can interact with unidentified ligands and co-receptors10. As we saw no enhancement of the Dscam1 SNS phenotype by robo1 or fra mutations, Dscam1 is either not responding to a ligand in the SNS, or responds to an unknown ligand using a different co-receptor. Expression of Dscam1-ΔC affected morphology of the recurrent nerve in a way not seen in Dscam1 loss-of-function mutations (Fig. 4D–F) supporting the idea of additional ligand(s). Dscam1 signals through the PAK kinase 33,52, and manipulation of PAK activity mimicked the Dscam1 phenotypes suggesting that downstream signaling in the SNS is similar to other systems.
Origins of the food clearance defect
We observed expansions of the frontal and hindgut nerves when Dscam1 or PAK was over-expressed, consistent with previous observations of axonal arbors and synaptic targeting. The expansions could be due to general promotion of axon growth or errors in synaptic targeting. Promotion of axon growth by Dscam has been observed in vertebrates 53,54. A failure to form correct synaptic connections or the formation of too many connections would clearly disrupt function of the SNS. We began this work assuming that disruptions to the SNS would impair feeding ability and were surprised to see no change. The larval tracking data clearly indicated that our manipulations had an effect and led us to perform the food clearing assay. At this stage, we have three models for the reduced food clearance phenotype. The Drosophila SNS does not normally innervate the midgut, stopping shortly after the esophagus ends. The SNS may be responsible for initiating peristaltic contractions at the junction of the proventriculus and the midgut, eliminating the need for continuous innervation. The discovery of a set of enteroendocrine cells that are sufficient for larval gut motility argues against this model 55. A second model is that feedback from the midgut to the brain might require functional SNS connections 48. This mechanism seems more likely to regulate meal size and food search strategies rather than clearance of food that is already ingested. Previous data on disruption of signaling in the hindgut neurons has been shown to reduce the defecation rate, reducing food clearance 27. Expression of Dscam-FL in the anal sphincter circuit did not affect food clearance, suggesting that this circuit is not affected.
Implications for the vertebrate ENS
Analysis of dscamb mutants in zebrafish mutants showed only mild neuroanatomical changes with no alterations in enteric neuron number; problems ingesting food were observed 56. A complimentary study used morpholino knockdown of dscama and dscamb and observed reductions in the number of enteric neurons 57. dscamb mutations may be compensated for by dscama, whereas morpholino knockdown may have a stronger effect. Overexpression of human DSCAM had no effect on the number of enteric neurons in zebrafish 57. Recent data in mice failed to observe enteric neuron defects in Down syndrome models with trisomy for Dscam 58. However, this could be due to phenotypic variability, or differences between mouse and human. These studies suggest that a role for Dscam genes in enteric nervous system function may have stronger effects on neuronal connectivity rather than number.
Based on prior studies we expected Dscam1 overexpression to increase the size of axons or the number of connections made. DS patients frequently lack ENS neurons as seen for HSCR in non-DS patients. However, Jannot et al. suggested that trisomy for Dscam could disrupt local connections that form after neural precursors have finished migration 5. Mouse studies have found that ENS neurons can be present in animals with impaired gut function, and this may be due to incorrect specification of neuron types 59. Consistent with this, innervation defects have been detected in human patients 60. A key next step is to determine which vertebrate enteric neurons are Dscam positive.
Experimental Procedures
Drosophila Genetics
The Ret-P2A and -P2B lines and the Janelia Farm lines GscG-GAL4 (#40383) and fasII-GAL4 (#46123) were described in 38. Additional stocks, PDF-GAL4 (#6899), UAS-PAK-GFP (#52299) and UAS-PAKmyr (#8804) were obtained from Bloomington Drosophila Stock Center (BDSC). The UAS-Dscam1-1-30-30-2 and UAS-Dscam1-7-25-27-1 were obtained from L. Zipursky. Full-length UAS-Dscam1-GFP lines were obtained from P. Shen and Y. Pu, and are described in 61. Dscam lacking the cytoplasmic domain (UAS-Dscam1-ΔC) was obtained from L. Luo 16. UAS-CD8-GFP was obtained from BDSC. The most reliable stock for neuroanatomy was w1118 as it displays the wild type SNS neuroanatomy described in previous publications.
Immunohistochemistry
Antibody staining was performed as described in 62. Anti-Fas2 (1D4) was used in preference to 22c10 due to reliability in staining the frontal nerve. Anti-Dscam antibodies were generous gifts from D. Schmucker and S.L. Zipursky. Late stage 17 embryos were cleared in 70% glycerol in PBS and subsequently in RapiClear (Sunjin Labs; 63). For all genotypes 10+ embryos were randomly selected and analyzed. Fluorescence microscopy was performed using either a Lecia MZ FLIII, or a Leica Thunder model organism microscope with computational clearing.
Feeding and Defecation Assays
Feeding assays were performed as previous described in 42,49. Defecation assays were modified from 27. Adult flies were allowed to lay eggs in grape agar plates with yeast paste containing Carmine red, 0.02g per 10g of yeast paste (Sigma-Aldrich; St. Louis, MO, USA). Embryos were allowed to develop for 24 hours and counted to quantify mortality. For defecation assays, second instar larvae were imaged to document feeding state (time 0), then placed in a separate plate with regular yeast paste and monitored in 15-minute intervals. After two hours (time 2 hours), larvae were reimaged to assess gut clearance. Larval drug treatments were carried out as described in 14. Nilotinib dissolved in DMSO and added to yeast paste at a concentration of 380 μM (Abcam; Cambridge, MS, USA). For all genotypes, n > 20 second instar larvae.
Locomotion Assay
Larval tracking assays were executed as described elsewhere 46,47. In brief, larvae were collected from grape agar plates and gently washed in distilled water. Using a paintbrush, ~20 larvae were aligned along the y-axis of a petri plate (22×22 cm) atop a thin layer of 1.5% agarose. The plate was covered to contain animals and placed in an arena illuminated with infra-red LEDs (850nm, outside the range of larval phototaxis. Environmental lights Inc. San Diego, CA). Kinesis was recorded at 130 frames per minute, for five minutes with a monochrome USB 3.0 CCD camera (Basler Ace series, JH Technologies, San Jose CA) fitted with an IR long-pass 830-nm filter and an 8-mm F1.4 C-mount lens (JH Technologies, San Jose CA). Locomotion data was analyzed using MATLAB® software. For all genotypes, n > 60 second instar larvae.
Acknowledgements
We thank L. Luo, P. Shen and Y. Pu for Drosophila stocks, D. Schmucker, T.Lee and S.L. Zipursky for antibodies. Antibodies were obtained from the Developmental Studies Hybridoma Bank (DHSB) developed under the auspices of the NICHD and maintained by the University of Iowa. Drosophila stocks were obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537). We thank J.H. Kim, L. Myers, H. Perera, K. Sweeney, M. Contreras and members of the Kidd and Mathew laboratories for technical assistance. We thank L. Myers for use of his SNS schematics. We thank A. Yanaz for help with the Leica Thunder microscope. K.H. was supported by the Michael (Mick) J. M. Hitchcock, Ph.D. Graduate Student Research Fund. This project was supported by grants from the National Institutes of Health (R15 NS075918) and (R01 NS11421901) to T.K. This study also received funding from the NIH National Institute of General Medical Sciences grants P20 GM103650 and P20 GM103554.
Literature cited
- 1.Antonarakis SE, Skotko BG, Rafii MS, et al. Down syndrome. Nat Rev Dis Primers. 02 June 2020;6(1):9. 10.1038/s41572-019-0143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold S, Pelet A, Amiel J, et al. Interaction between a chromosome 10 RET enhancer and chromosome 21 in the Down syndrome-Hirschsprung disease association. Human mutation. May 2009;30(5):771–5. 10.1002/humu.20944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKeown SJ, Stamp L, Hao MM, Young HM. Hirschsprung disease: a developmental disorder of the enteric nervous system. Wiley interdisciplinary reviews Developmental biology. Jan-Feb 2013;2(1):113–29. 10.1002/wdev.57. [DOI] [PubMed] [Google Scholar]
- 4.Korbel JO, Tirosh-Wagner T, Urban AE, et al. The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proceedings of the National Academy of Sciences of the United States of America. Jul 21 2009;106(29):12031–6. 10.1073/pnas.0813248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jannot AS, Pelet A, Henrion-Caude A, et al. Chromosome 21 scan in Down syndrome reveals DSCAM as a predisposing locus in Hirschsprung disease. PloS one. 2013;8(5):e62519. 10.1371/journal.pone.0062519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmucker D, Chen B. Dscam and DSCAM: complex genes in simple animals, complex animals yet simple genes. Genes & development. Jan 15 2009;23(2):147–56. 10.1101/gad.1752909. [DOI] [PubMed] [Google Scholar]
- 7.Montesinos ML. Roles for DSCAM and DSCAML1 in central nervous system development and disease. Advances in neurobiology. 2014;8:249–70. [DOI] [PubMed] [Google Scholar]
- 8.Barlow GM, Lyons GE, Richardson JA, Sarnat HB, Korenberg JR. DSCAM: an endogenous promoter drives expression in the developing CNS and neural crest. Biochemical and biophysical research communications. Nov 22 2002;299(1):1–6. [DOI] [PubMed] [Google Scholar]
- 9.Yamakawa K, Huot YK, Haendelt MA, et al. DSCAM: a novel member of the immunoglobulin superfamily maps in a Down syndrome region and is involved in the development of the nervous system. Human molecular genetics. Feb 1998;7(2):227–37. [DOI] [PubMed] [Google Scholar]
- 10.Alavi M, Song M, King GL, et al. Dscam1 Forms a Complex with Robo1 and the N-Terminal Fragment of Slit to Promote the Growth of Longitudinal Axons. PLoS biology. Sep 2016;14(9):e1002560. 10.1371/journal.pbio.1002560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen BE, Kondo M, Garnier A, et al. The molecular diversity of Dscam is functionally required for neuronal wiring specificity in Drosophila. Cell. May 5 2006;125(3):607–20. 10.1016/j.cell.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 12.He H, Kise Y, Izadifar A, et al. Cell-intrinsic requirement of Dscam1 isoform diversity for axon collateral formation. Science (New York, NY). Jun 6 2014;344(6188):1182–6. 10.1126/science.1251852. [DOI] [PubMed] [Google Scholar]
- 13.Kim JH, Wang X, Coolon R, Ye B. Dscam expression levels determine presynaptic arbor sizes in Drosophila sensory neurons. Neuron. Jun 5 2013;78(5):827–38. 10.1016/j.neuron.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sterne GR, Kim JH, Ye B. Dysregulated Dscam levels act through Abelson tyrosine kinase to enlarge presynaptic arbors. Elife. 2015;4:e05196. 10.7554/eLife.05196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cvetkovska V, Hibbert AD, Emran F, Chen BE. Overexpression of Down syndrome cell adhesion molecule impairs precise synaptic targeting. Nature neuroscience. Jun 2013;16(6):677–82. 10.1038/nn.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu H, Hummel T, Clemens JC, Berdnik D, Zipursky SL, Luo L. Dendritic patterning by Dscam and synaptic partner matching in the Drosophila antennal lobe. Nature neuroscience. Mar 2006;9(3):349–55. 10.1038/nn1652. [DOI] [PubMed] [Google Scholar]
- 17.Andrews GL, Tanglao S, Farmer WT, et al. Dscam guides embryonic axons by Netrin-dependent and -independent functions. Development (Cambridge, England). Dec 2008;135(23):3839–48. 10.1242/dev.023739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grossman TR, Gamliel A, Wessells RJ, et al. Over-expression of DSCAM and COL6A2 cooperatively generates congenital heart defects. PLoS genetics. Nov 2011;7(11):e1002344. 10.1371/journal.pgen.1002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallace AS, Anderson RB. Genetic interactions and modifier genes in Hirschsprung’s disease. World journal of gastroenterology : WJG. Dec 7 2011;17(45):4937–44. 10.3748/wjg.v17.i45.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn M, Bishop J. Expression pattern of Drosophila ret suggests a common ancestral origin between the metamorphosis precursors in insect endoderm and the vertebrate enteric neurons. Proceedings of the National Academy of Sciences of the United States of America. Jan 30 2001;98(3):1053–8. 10.1073/pnas.021558598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pachnis V, Mankoo B, Costantini F. Expression of the c-ret proto-oncogene during mouse embryogenesis. Development (Cambridge, England). Dec 1993;119(4):1005–17. [DOI] [PubMed] [Google Scholar]
- 22.Myers L, Perera H, Alvarado MG, Kidd T. The Drosophila Ret gene functions in the stomatogastric nervous system with the Maverick TGFbeta ligand and the Gfrl co-receptor. Development (Cambridge, England). Feb 2 2018;145(3). 10.1242/dev.157446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoyer N, Zielke P, Hu C, et al. Ret and Substrate-Derived TGF-β Maverick Regulate Space-Filling Dendrite Growth in Drosophila Sensory Neurons. Cell reports. August 2018;24(9):2261–2272.e5. 10.1016/j.celrep.2018.07.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abitua PB, Gainous TB, Kaczmarczyk AN, et al. The pre-vertebrate origins of neurogenic placodes. Nature. Aug 27 2015;524(7566):462–5. 10.1038/nature14657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Velasco B, Shen J, Go S, Hartenstein V. Embryonic development of the Drosophila corpus cardiacum, a neuroendocrine gland with similarity to the vertebrate pituitary, is controlled by sine oculis and glass. Developmental biology. Oct 15 2004;274(2):280–94. 10.1016/j.ydbio.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 26.Vidal B, Gulez B, Cao WX, et al. The enteric nervous system of the C. elegans pharynx is specified by the Sine oculis-like homeobox gene ceh-34. Elife. Mar 24 2022;11. 10.7554/eLife.76003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang W, Yan Z, Li B, Jan LY, Jan YN. Identification of motor neurons and a mechanosensitive sensory neuron in the defecation circuitry of Drosophila larvae. Elife. 2014;3. 10.7554/eLife.03293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vosko AM, Schroeder A, Loh DH, Colwell CS. Vasoactive intestinal peptide and the mammalian circadian system. General and comparative endocrinology. Jun-Jul 2007;152(2–3):165–75. 10.1016/j.ygcen.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crocker A, Sehgal A. Genetic analysis of sleep. Genes & development. Jun 15 2010;24(12):1220–35. 10.1101/gad.1913110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naslund E, Ehrstrom M, Ma J, Hellstrom PM, Kirchgessner AL. Localization and effects of orexin on fasting motility in the rat duodenum. American journal of physiology Gastrointestinal and liver physiology. Mar 2002;282(3):G470–9. 10.1152/ajpgi.00219.2001. [DOI] [PubMed] [Google Scholar]
- 31.Lowe SA, Hodge JJL, Usowicz MM. A third copy of the Down syndrome cell adhesion molecule (Dscam) causes synaptic and locomotor dysfunction in Drosophila. Neurobiology of disease. February 2018;110:93–101. 10.1016/j.nbd.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dascenco D, Erfurth M-L, Izadifar A, et al. Slit and Receptor Tyrosine Phosphatase 69D Confer Spatial Specificity to Axon Branching via Dscam1. Cell. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmucker D, Clemens JC, Shu H, et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. Jun 9 2000;101(6):671–84. [DOI] [PubMed] [Google Scholar]
- 34.Yu HH, Yang JS, Wang J, Huang Y, Lee T. Endodomain diversity in the Drosophila Dscam and its roles in neuronal morphogenesis. The Journal of neuroscience : the official journal of the Society for Neuroscience. Feb 11 2009;29(6):1904–14. 10.1523/jneurosci.5743-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ayali A The insect frontal ganglion and stomatogastric pattern generator networks. Neurosignals. Jan-Apr 2004;13(1–2):20–36. 10.1159/000076156. [DOI] [PubMed] [Google Scholar]
- 36.Hartenstein V Development of the insect stomatogastric nervous system. Trends in neurosciences. Sep 1997;20(9):421–7. [DOI] [PubMed] [Google Scholar]
- 37.Schoofs A, Huckesfeld S, Surendran S, Pankratz MJ. Serotonergic pathways in the Drosophila larval enteric nervous system. Journal of insect physiology. Oct 2014;69:118–25. 10.1016/j.jinsphys.2014.05.022. [DOI] [PubMed] [Google Scholar]
- 38.Hernandez K, Myers LG, Bowser M, Kidd T. Genetic Tools for the Analysis of Drosophila Stomatogastric Nervous System Development. PloS one. 2015;10(6):e0128290. 10.1371/journal.pone.0128290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soba P, Han C, Zheng Y, et al. The Ret receptor regulates sensory neuron dendrite growth and integrin mediated adhesion. Elife. 2015;4. 10.7554/eLife.05491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cognigni P, Bailey AP, Miguel-Aliaga I. Enteric neurons and systemic signals couple nutritional and reproductive status with intestinal homeostasis. Cell metabolism. Jan 5 2011;13(1):92–104. 10.1016/j.cmet.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hing H, Xiao J, Harden N, Lim L, Zipursky SL. Pak functions downstream of Dock to regulate photoreceptor axon guidance in Drosophila. Cell. Jun 25 1999;97(7):853–63. [DOI] [PubMed] [Google Scholar]
- 42.Melcher C, Pankratz MJ. Candidate gustatory interneurons modulating feeding behavior in the Drosophila brain. PLoS biology. Sep 2005;3(9):e305. 10.1371/journal.pbio.0030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Troncoso. The development of larval movement patterns in Drosophila. Heredity. 1987;58:321–329. [Google Scholar]
- 44.Berni J Genetic dissection of a regionally differentiated network for exploratory behavior in Drosophila larvae. Current biology : CB. May 18 2015;25(10):1319–26. 10.1016/j.cub.2015.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slankster E, Kollala S, Baria D, et al. Mechanism underlying starvation-dependent modulation of olfactory behavior in Drosophila larva. Scientific reports. Feb 20 2020;10(1):3119. 10.1038/s41598-020-60098-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gershow M, Berck M, Mathew D, et al. Controlling airborne cues to study small animal navigation. Nature methods. Jan 15 2012;9(3):290–6. 10.1038/nmeth.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mathew D, Martelli C, Kelley-Swift E, et al. Functional diversity among sensory receptors in a Drosophila olfactory circuit. Proceedings of the National Academy of Sciences of the United States of America. Jun 4 2013;110(23):E2134–43. 10.1073/pnas.1306976110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Al-Anzi B, Armand E, Nagamei P, et al. The leucokinin pathway and its neurons regulate meal size in Drosophila. Current biology : CB. Jun 8 2010;20(11):969–78. 10.1016/j.cub.2010.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zinke I, Kirchner C, Chao LC, Tetzlaff MT, Pankratz MJ. Suppression of food intake and growth by amino acids in Drosophila: the role of pumpless, a fat body expressed gene with homology to vertebrate glycine cleavage system. Development (Cambridge, England). Dec 1999;126(23):5275–84. [DOI] [PubMed] [Google Scholar]
- 50.Lemieux M, Thiry L, Laflamme OD, Bretzner F. Role of DSCAM in the Development of Neural Control of Movement and Locomotion. Int J Mol Sci. Aug 7 2021;22(16). 10.3390/ijms22168511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi L, Lee T. Molecular diversity of Dscam and self-recognition. Adv Exp Med Biol. 2012;739:262–75. 10.1007/978-1-4614-1704-0_17. [DOI] [PubMed] [Google Scholar]
- 52.Kamiyama D, McGorty R, Kamiyama R, Kim MD, Chiba A, Huang B. Specification of Dendritogenesis Site in Drosophila aCC Motoneuron by Membrane Enrichment of Pak1 through Dscam1. Developmental cell. Oct 12 2015;35(1):93–106. 10.1016/j.devcel.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bruce FM, Brown S, Smith JN, Fuerst PG, Erskine L. DSCAM promotes axon fasciculation and growth in the developing optic pathway. Proceedings of the National Academy of Sciences of the United States of America. Feb 14 2017;114(7):1702–1707. 10.1073/pnas.1618606114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simmons AB, Bloomsburg SJ, Sukeena JM, et al. DSCAM-mediated control of dendritic and axonal arbor outgrowth enforces tiling and inhibits synaptic plasticity. Proceedings of the National Academy of Sciences of the United States of America. Nov 21 2017;114(47):E10224–E10233. 10.1073/pnas.1713548114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.LaJeunesse DR, Johnson B, Presnell JS, Catignas KK, Zapotoczny G. Peristalsis in the junction region of the Drosophila larval midgut is modulated by DH31 expressing enteroendocrine cells. BMC physiology. 2010;10:14. 10.1186/1472-6793-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Julien DP, Chan AW, Barrios J, et al. Zebrafish expression reporters and mutants reveal that the IgSF cell adhesion molecule Dscamb is required for feeding and survival. Journal of neurogenetics. Dec 2018;32(4):336–352. 10.1080/01677063.2018.1493479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu YJ, Yu WW, Cui MM, et al. Association Analysis of Variants of DSCAM and BACE2 With Hirschsprung Disease Susceptibility in Han Chinese and Functional Evaluation in Zebrafish. Front Cell Dev Biol. 2021;9:641152. 10.3389/fcell.2021.641152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schill EM, Wright CM, Jamil A, LaCombe JM, Roper RJ, Heuckeroth RO. Down syndrome mouse models have an abnormal enteric nervous system. JCI Insight. Apr 18 2019;5. 10.1172/jci.insight.124510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Musser MA, Correa H, Southard-Smith EM. Enteric neuron imbalance and proximal dysmotility in ganglionated intestine of the Hirschsprung mouse model. Cellular and molecular gastroenterology and hepatology. Jan 1 2015;1(1):87–101. 10.1016/j.jcmgh.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Warren M, Kaul A, Bove K. Calretinin-immunoreactive hypoinnervation in Down syndrome (DS): report of an infant with very short-segment Hirschsprung disease (vsHD) and comparison to biopsy findings in 20 normal infants and 11 infants with DS and chronic constipation. Pediatric and developmental pathology : the official journal of the Society for Pediatric Pathology and the Paediatric Pathology Society. Jul 31 2015. 10.2350/15-01-1602-oa.1. [DOI] [PubMed] [Google Scholar]
- 61.Wang J, Ma X, Yang JS, et al. Transmembrane/juxtamembrane domain-dependent Dscam distribution and function during mushroom body neuronal morphogenesis. Neuron. Sep 2 2004;43(5):663–72. 10.1016/j.neuron.2004.06.033. [DOI] [PubMed] [Google Scholar]
- 62.Patel NH. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. Methods in cell biology. 1994;44:445–87. [DOI] [PubMed] [Google Scholar]
- 63.Liu YC, Chiang AS. High-resolution confocal imaging and three-dimensional rendering. Methods (San Diego, Calif). May 2003;30(1):86–93. [DOI] [PubMed] [Google Scholar]


