Abstract
Inositol-requiring enzyme 1α (IRE1α) is the most conserved endoplasmic reticulum (ER) stress sensor with two catalytic domains, kinase and RNase, in its cytosolic portion. IRE1α inhibitors have been used to improve existing clinical treatments against various cancers. In this study we identified toxoflavin (TXF) as a new-type potent small molecule IRE1α inhibitor. We used luciferase reporter systems to screen compounds that inhibited the IRE1α-XBP1s signaling pathway. As a result, TXF was found to be the most potent IRE1α RNase inhibitor with an IC50 value of 0.226 μM. Its inhibitory potencies on IRE1α kinase and RNase were confirmed in a series of cellular and in vitro biochemical assays. Kinetic analysis showed that TXF caused time- and reducing reagent-dependent irreversible inhibition on IRE1α, implying that ROS might participate in the inhibition process. ROS scavengers decreased the inhibition of IRE1α by TXF, confirming that ROS mediated the inhibition process. Mass spectrometry analysis revealed that the thiol groups of four conserved cysteine residues (CYS-605, CYS-630, CYS-715 and CYS-951) in IRE1α were oxidized to sulfonic groups by ROS. In molecular docking experiments we affirmed the binding of TXF with IRE1α, and predicted its binding site, suggesting that the structure of TXF itself participates in the inhibition of IRE1α. Interestingly, CYS-951 was just near the docked site. In addition, the RNase IC50 and ROS production in vitro induced by TXF and its derivatives were negative correlated (r = −0.872). In conclusion, this study discovers a new type of IRE1α inhibitor that targets a predicted new alternative site located in the junction between RNase domain and kinase domain, and oxidizes conserved cysteine residues of IRE1α active sites to inhibit IRE1α. TXF could be used as a small molecule tool to study IRE1α’s role in ER stress.
Keywords: endoplasmic reticulum stress, toxoflavin, reactive oxygen species, IRE1α, XBP1
Introduction
Protein folding homeostasis in the endoplasmic reticulum (ER) is fundamentally important to match cellular demands and protect against stress conditions [1]. The accumulation of unfolded and misfolded proteins in the ER lumen, a condition called ER stress, affects various signaling processes, such as reduction-oxidation (redox) balance, energy production, inflammation, differentiation, and apoptosis [2]. The subsequent unfolded protein response (UPR) is engaged as an adaptive strategy for protein folding homeostasis under ER stress [3]. Mammalian cells have three major UPR signaling pathways: inositol-requiring enzyme 1α (IRE1α) - X-Box Binding Protein 1 (XBP1), PKR-like ER kinase – activating transcription factor 4 – C/EBP-Homologous Protein (CHOP), and activating transcription factor 6 [4].
IRE1α is the most conserved ER stress sensor with two catalytic regions, kinase and RNase domains, in its cytosolic portion. Unfolded or misfolded proteins in the ER lumen activate IRE1α by binding to its chaperone BiP (also known as GRP78) [4]. IRE1α mainly functions through its RNase activity, and kinase phosphorylation, mainly in the activation loop, is coupled to the activation state of RNase [5]. Activated IRE1α splices unspliced XBP1 (XBP1u) mRNA for translation of spliced XBP1 (XBP1s) as a transcription factor, which enhances the ability of the ER to cope with unfolded proteins and acts more broadly to upregulate secretory capacity [6, 7].
Currently, pharmacological inhibition of IRE1α has been the focus of several drug development strategies, and compounds such as IRE1α inhibitors have been disclosed and evaluated [8]. Combination with IRE1α inhibitors has been attempted to improve existing clinical treatments in many types of cancers, such as breast cancer [9], prostate cancer[10], multiple myeloma [11], leukemia [12], and glioblastoma [13]. Three types of IRE1α inhibitors were classified by their binding sites: (i) kinase pocket binders; (ii) RNase domain ligands; and (iii) binding-unconfirmed inhibitors [8]. Kinase pocket binders are a class of ligands that occupy the ATP-binding cleft of IRE1α kinase to inhibit RNase-mediated XBP1 mRNA splicing under endoplasmic reticulum stress [14]. The so-called RNase domain ligands represented by 4μ8C [15] share a common hydroxy-aryl-aldehyde moiety that reacts selectively with a specific lysine (LYS-907) in the RNase domain, thus forming a stable imine via Schiff base formation and effectively preventing XBP1 mRNA splicing. Other inhibitors, such as toyocamycin [16], doxorubicin [17], trierixin [18], and quinotrierixin [19], were shown to inhibit the IRE1α-XBP1 pathway in vitro and in vivo without clear demonstration of their mode of action. However, no IRE1α inhibitor has been approved for the market at present.
In this study, we identified toxoflavin (TXF) as a new-type potent small-molecule IRE1α inhibitor which targets a predicted new alternative site located in the junction between RNase domain and kinase domain, and oxidizes conserved cysteine residues to inhibit IRE1α. Four conserved cysteine sulfonations (CYS-605, CYS-630, CYS-715, and CYS-951) were observed using the compound, and the binding mode of the IRE1α–TXF complex was predicted, which will contribute to further study on clarifying IRE1α’s role in ER stress.
Materials and methods
Cell culture and chemicals
B16-F10, HeLa, and A549 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (#SH30022.01, HyClone, Logan, UT, USA) supplemented with 10% FBS (#FND500, ExCell Bio, Taicang, China). B16-F10-XBP1-luc cells were B16-F10 cells stably transfected with the pCAX-F-XBP1-ΔDBD-luciferase plasmid, a gift from Dr. Masayuki Miura (The University of Tokyo) [20], where XBP1 splicing activity could be indicated with luciferase activity. HEK293-CHOP-luc cells were constructed by our laboratory previously [21]. All cells were grown in the presence of 100 units/mL penicillin and 100 μg/mL streptomycin at 37 °C and 5% CO2. All cell lines were used within 15 passages and less than 6 months. Tunicamycin (TM) (#T101151, Aladdin, Shanghai, China), toxoflavin (TXF) (#HY-100760, MedChemExpress, Trenton, NJ, USA), 2-deoxy-D-glucose (2DG) (#25972-M, Sigma-Aldrich, St. Louis, MO, USA), disulfiram (DSF) (#T819460, Macklin, Shanghai, China), N-acetyl-L-cysteine (NAC) (#BD34319, Bidepharm, Shanghai, China), and Trolox (#HY-101445, MedChemExpress) were purchased from the manufacturers. The derivatives of TXF were obtained from the Chinese National Compound Library.
Luciferase assay
B16-F10-XBP1-luc cells were incubated with TM together with or without test compounds for 8 h. Then, the cells were lysed in Passive lysis buffer (Promega, Madison, WI, USA), and luciferase fluorescence value was measured using the luciferase assay system (Promega) and an EnVision® Multimode Microplate Reader (PerkinElmer, Waltham, MA, USA). Meanwhile, cell viability activities were assessed by the CellTiter 96® Aqueous Non-Radioactive Cell Proliferation Assay Reagent (Promega). The corrected fluorescence value was obtained from the ratio of the original fluorescence value to cell viability. Relative luciferase activity was represented by setting the result of TM treatment as 100% control.
Semiquantitative RT-PCR
RNA was harvested with an RNA extraction kit (#BSC52M1, Bioer Technology, Hangzhou, China), and first-strand cDNA synthesis was performed with HiScript® III RT SuperMix for qPCR (#R323-01, Vazyme Biotech, Nanjing, China) according to the manufacturers’ specifications. The cDNA product was subjected to 35 cycles of PCR using the forward primer 5′-TTACGAGAGAAAACTCATGGCC-3′ and reverse primer 5′-GGGTCCAAGTTGTCCAGAATGC-3′ specific for XBP1 mRNA. PCR products (XBP1u = 289 bp, XBP1u = 263 bp) were separated by electrophoresis on a 3% agarose (#A500016-0250, Sangon Biotech, Shanghai, China) gel and visualized by GelRed (#A616697-0500, BBI Life Sciences, Shanghai, China) staining.
Western blotting
Cells (about 1 million per sample) were lysed in 100 μL of Laemmli sample buffer (#1610747, Bio-Rad Laboratories, Hercules, CA, USA) and boiled for 15 min. Proteins were then resolved via SDS-PAGE and transferred to nitrocellulose membranes (#10600002, GE Healthcare, Fairfield, CT, USA). After blocking with 5% nonfat milk, membranes were blotted with antibodies [primary antibodies: IRE1α rabbit mAb #3294 from Cell Signaling Technology (Danvers, MA, USA), phospho-IRE1α rabbit pAb #ab48187 from Abcam Company (Cambridge, MA, USA) and β-actin mouse mAb #AM1021B from Abgent (Suzhou, China); secondary antibodies: IRDye® 800CW anti-rabbit IgG secondary antibody #926-32211 and IRDye® 680RD anti-mouse IgG secondary antibody #926-68070 from LI-COR biosciences (Lincoln, NE, USA)]. Detection was achieved with an Odyssey® Imaging System (LI-COR Biosciences) and the band intensities were quantified by ImageJ software.
Biochemical assays for IRE1α
IRE1α RNase activity was measured by incubation of 6 nM purified recombinant human IRE1α protein with a cytoplasmic domain (Pro 465-Leu 977, #11905-H20B, Sino Biological Inc., Beijing, China) with 50 nM quenched RNA probe (FAM-5′-CAGUCCGCAGCACUG-3′-BHQ) for the substrate at room temperature in a Corning® 384-well low-volume round bottom plate (Corning, NY, USA). IRE1α reaction buffer (50 mM HEPES, 100 mM KOAc, 1 mM ADP, 0.005% Triton-X100, pH 7.2, and 1 mM DTT) was used for all RNase activity measurements. Fluorescence was measured on an EnVision® Multimode Microplate Reader using an excitation filter of 485 nm and an emission filter of 535 nm. IC50 values were determined from the dose-response curves of the inhibition of fluorescence intensity, setting the result of 1% DMSO treatment as 100% control.
IRE1α kinase activity was detected by the HTRF® KinEASE™ kit (# 62ST2PEB, Cisbio, Bedford, MA, USA). The incubation mixture consisted of the indicated chemicals and the abovementioned IRE1α protein (100 nM) with substrate S2 (300 nM) in reaction buffer (20 mM Tris base (pH 8.0), 10 mM MgCl2, 16 μM ATP, and 1 mM DTT) at room temperature in a ProxiplateTM-384 Plus plate (PerkinElmer, Waltham, MA, USA). Fluorescence was measured on an EnVision® Multimode Microplate Reader using an excitation filter of 340 nm and emission filters of 615 nm and 665 nm, and the activity was represented with the fluorescence ratio at 665 to 615 nm according to the protocol supplied by the manufacturer.
Reactive oxygen species (ROS) detection
Total ROS generation was detected using the ROS Assay Kit DCFH-DA (#HY-D0940, MedChemExpress, Trenton, NJ, USA). The incubation mixture consisted of the indicated chemicals and the enzymatic assay system containing 10 µM DCFH-DA. Fluorescence was measured on an EnVision® Multimode Microplate Reader using an excitation filter of 490 nm and an emission filter of 525 nm. For intracellular flow cytometry measurements, cells were treated with chemicals for 8 h, and the culture medium was replaced with fresh DMEM containing 10 μM DCFH-DA for another 30 min of incubation. Afterwards, the cells were washed three times with PBS, harvested, and resuspended in PBS for flow cytometry measurements.
Immunoprecipitation
Cells were lysed with RIPA lysis buffer (#PC101, Epizyme Biotech, Shanghai, China) containing protease and phosphatase inhibitor cocktail (#A32961, ThermoFisher, Carlsbad, CA, USA). Cell lysates (800 μg total protein) were incubated with the Anti-Cysteine sulfonate antibody (#ab176487, Abcam, Cambridge, MA, USA) overnight at 4 °C. Pre-wash Protein A/G magnetic beads (#HY-K0202, MedChemExpress) were added and incubated for 2 h. After immunoprecipitation, the samples were washed and then eluted with SDS sample buffer. The eluates were analyzed by Western blotting.
Identification of oxidized cysteine residues by mass spectrometry
The incubation mixture (25 μL) consisted of 400 nM recombinant human IRE1α protein, 20 mM Tris (pH 8.0), 5 mM MgCl2, and the indicated chemicals. After incubating for 1 h at room temperature, 5 μL catalase (final concentration: 6.25 μg/mL) was added. Another 1 h later, IRE1α protein was subjected to in-solution digestion and then desalted by a ZipTip C18 column (Merck KGaA, Darmstadt, Germany). The tryptic peptides were loaded into a homemade trapping column (75 µm × 2 cm, C18, Dikma, Beijing, China) and separated by a homemade analytical column (75 µm × 10 cm, C18, Dikma) with a 60 min gradient from 5% solvent B (2% H2O, 98% acetonitrile, 0.1% formic acid) to 30% solvent B in an EASY-nLC 1000 LC system (Thermo Fisher Scientific, Waltham, MA, USA). The eluted peptides were introduced into an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific) with a full MS scan range from 350 to 1800 m/z with a resolution of 240,000 at m/z 400. To fragment precursor ions, the twenty most intense precursor ions with intensities greater than 5000 and charge states of +2 or +3 in each full MS scan were sequentially fragmented in a linear ion trap by CID (collision-induced dissociation) with 35% NCE (normalized collision energy). The dynamic exclusion duration was set to 15 s, and the isolation window was 1.5 m/z.
The acquired MS raw files were transformed into MGF format by Proteome Discoverer 1.4 and then subjected to search with Mascot 2.3.01 against an in-house IER1α sequence database. Oxidation (-SOH), dioxidation (-SO2H), and trioxidation (-SO3H) modifications from the UniMod database (https://www.unimod.org/modifications_list.php) were set as variable modifications. For the full mass scan, the mass error was set as ±10 ppm, while fragment ions were set as 0.5 Da. Trypsin was set as the enzyme to digest proteins into peptides with a maximum of three missing cleavages. Before manually checking the matched spectra, spectra were filtered with a peptide ion score cutoff of 20. Spectral counting was applied to quantify the difference in modification among different conditions.
Microscale thermophoresis (MST)
A microscale thermophoresis instrument (NanoTemper, Munich, Germany) was used to quantify biomolecular interactions. Recombinant human IRE1α protein was labeled using the Monolith NT Protein Labeling kit RED-NHS (NanoTemper). The incubation mixture consisted of labeled IRE1α (1 µM) and the indicated chemicals in 50 mM HEPES solution. After incubating for 30 min at room temperature, the incubation mixture was centrifuged at 14,000 × g for 10 min at 4 °C and loaded into a Monolith NT.115 MST Premium Capillaries. All the experimental parameters used by the MST instrument were fixed with an LED power of 20% and a laser power of 80%. The value of the dissociation constant (Kd) was calculated using NanoTemper Analysis software.
Cellular thermal shift assay (CETSA)
RPMI8226 cells were treated with 10 μM TXF or DMSO for 1 h, then collected and washed with PBS buffer three times to avoid excess compound residue. The cell suspensions were distributed into different 0.2 mL PCR tubes (1 million cells per tube), denatured at 45, 48, 51, 54 °C for 3 min on PCR instrument, and then freeze-thawed twice using liquid nitrogen. Samples were centrifuged and the supernatants were analyzed by Western blotting.
Docking simulation of binding mode
Docking simulation was performed using AutoDock 4.2.6 and MGLTools 1.5.7 against human IRE1α (PDB: 4YZD) [22]. 3D Conformers of TXF and its derivatives were produced using Chem 3D 20.0. When using a graphical user interface called AutoDockTools (https://ccsb.scripps.edu/mgltools/1-5-6/) to perform simulation, the grid box was set to contain the whole macromolecule, and the macromolecule was set as a rigid filename. Docking algorithm employed Lamarckian GA 4.2. Default settings were used for all other parameters. The docking result ranked with docked binding energy was wrote, and the first ranked docking conformation was analyzed and visualized by PyMOL 1.7.1.0.
Statistical analysis
Data are expressed as the mean ± SEM. Unpaired two-tailed Student’s t test was used for statistical analysis between two groups. P < 0.05 was considered statistically significant.
Results
Screening to identify compounds selectively inhibiting IRE1α-XBP1s
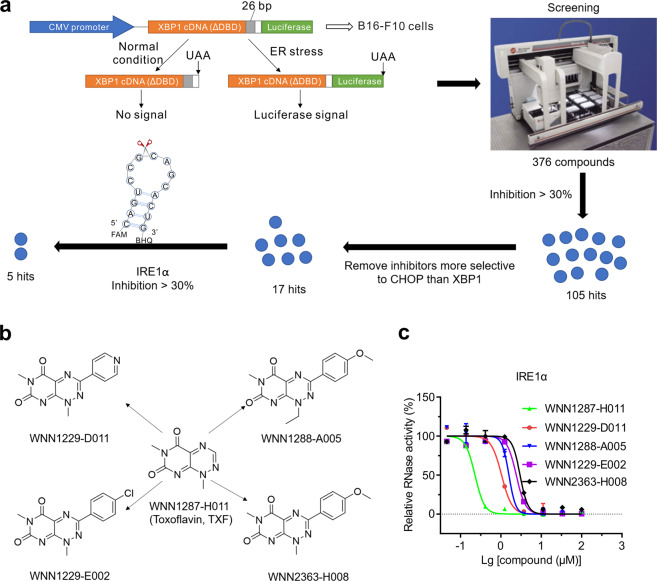
B16-F10 cells stably expressing an XBP1-ΔDBD luciferase splicing reporter (XBP1-luc) were used to identify compounds that inhibited the IRE1α-XBP1s signaling pathway (Fig. 1a). Of 376 compounds from the Chinese National Compound Library, our screening identified 105 hits (2 μg/mL) that inhibited XBP1-luc activity (inhibition >30%) induced by tunicamycin (TM, 1 μg/mL) (Fig. 1a and Supplementary Table S1). We then removed compounds that also selectively inhibit CHOP signaling arms using HEK293 cells stably expressing the CHOP-luciferase reporter (CHOP-luc). Thus, 17 hits were found to preferentially inhibit the XBP1-luc reporter (Fig. 1a and Supplementary Table S2). In addition, recombinant human IRE1α protein with a cytoplasmic domain with the quenched RNA probe for the substrate was used to determine whether compounds directly inhibited IRE1α. From the 17 hits, we identified that 5 hits potently inhibited IRE1α (Fig. 1a and Supplementary Table S3), which belongs to the same structural class (a triazine natural product toxoflavin and its derivatives) (Fig. 1b). Interestingly, TXF was the most potent IRE1α RNase inhibitor, with an IC50 value of 0.226 μM (Fig. 1c and Table 1).
Fig. 1. Screening to identify compounds selectively inhibiting IRE1α-XBP1s.
a Schematic showing the screening pipeline to identify prioritized compounds that selectively inhibit IRE1α. This pipeline includes identification of compounds that inhibit the XBP1-luc reporter, removal of compounds that inhibit the CHOP-luc reporter and assay of IRE1α RNase activity. b Chemical structures of five final prioritized IRE1α-XBP1s inhibitors identified. c RNase activities measured after indicated concentrations of the five final prioritized inhibitors were added to purified recombinant human IRE1α protein for 1 h.
Table 1.
RNase IC50, ROS production, and predicted binding energies of TXF and its derivatives for IRE1α.
| Compound | RNase IC50 (μM, 95% CI) | ROS ratio to DMSO (0.5 μM, average ± SD) | Binding energy (kcal/mol) |
|---|---|---|---|
| TXF | 0.226 (0.188–0.274) | 22.8 ± 0.950 | −5.05 |
| WNN1229-D011 | 1.04 (0.901–1.20) | 15.7 ± 1.38 | −5.17 |
| WNN1288-A005 | 1.58 (0.993–2.50) | 16. 6 ± 1.75 | −3.45 |
| WNN1229-E002 | 2.40 (2.01–2.86) | 11.9 ± 0.450 | −3.83 |
| WNN2363-H008 | 2.98 (2.27–3.92) | 13.6 ± 2.50 | −3.70 |
TXF potently inhibits IRE1α kinase and RNase
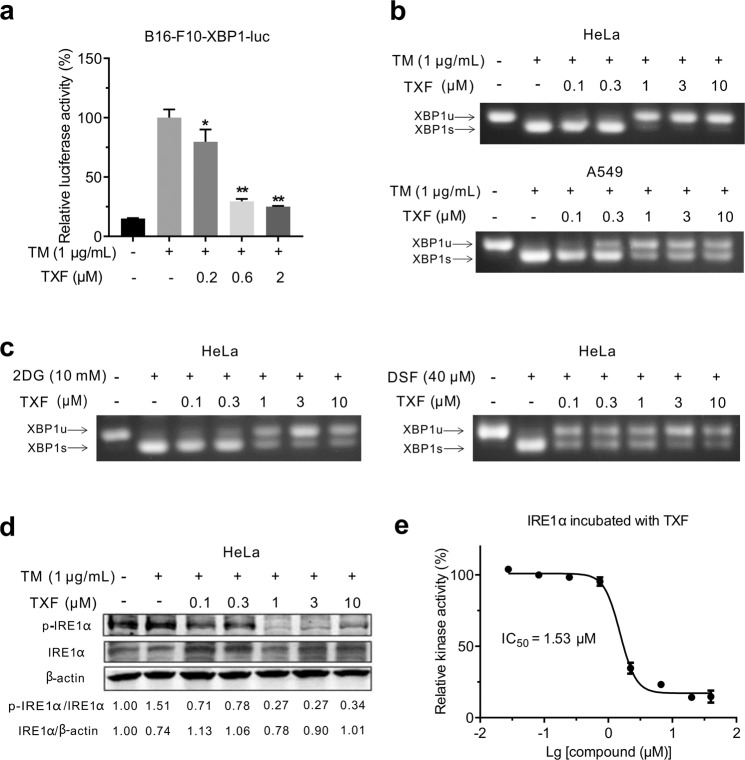
For B16-F10-XBP1-luc cells, TXF inhibited ER stress inducer TM-induced XBP1-luciferase activation in a dose-dependent manner (Fig. 2a). Next, by performing RT-PCR analysis of RNA isolated from TXF-treated or untreated HeLa or A549 cells, we confirmed that TXF inhibited TM-induced endogenous XBP1 mRNA splicing in a dose-dependent manner (Fig. 2b). Consistently, TXF inhibited ER stress inducers 2DG or DSF-induced endogenous XBP1 mRNA splicing in HeLa cells (Fig. 2c). Because kinase phosphorylation is coupled to the activation state of RNase, we performed Western blotting and found that TXF reduced the phosphorylation of IRE1α in HeLa cells (Fig. 2d). To further determine whether TXF inhibited IRE1α kinase, IRE1α protein was used to measure kinase activity. As shown in Fig. 2e, TXF directly inhibited IRE1α kinase activity, with an IC50 value of 1.53 μM (>0.226 μM for IRE1α RNase activity).
Fig. 2. TXF inhibits IRE1α kinase and RNase.
a B16-F10-XBP1-luc cells were treated with the indicated concentrations of TXF in the presence of TM (1 μg/mL). After 8 h, the cells were lysed and subjected to luciferase assay. *P < 0.05; **P < 0.01, versus vehicle group (n = 3), unpaired two-tailed Student’s t-test. b HeLa or A549 cells were treated with the indicated concentrations of TXF and TM (1 μg/mL), and c HeLa cells were treated with the indicated concentrations of TXF and 2DG or DSF for 8 h. XBP1 mRNA splicing was evaluated by semiquantitative RT-PCR. d HeLa cells were treated with the indicated concentrations of TXF and TM (1 μg/mL) for 8 h. The phosphorylation of IRE1α was evaluated by Western blotting. The band intensities were quantified by ImageJ software and the normalized ratio of p-IRE1α/IRE1α and IRE1α/β-actin were calculated. e Kinase activities measured after indicated concentrations of TXF were added to IRE1α for 1 h.
Time- and DTT-dependent irreversible inhibition of IRE1α by TXF
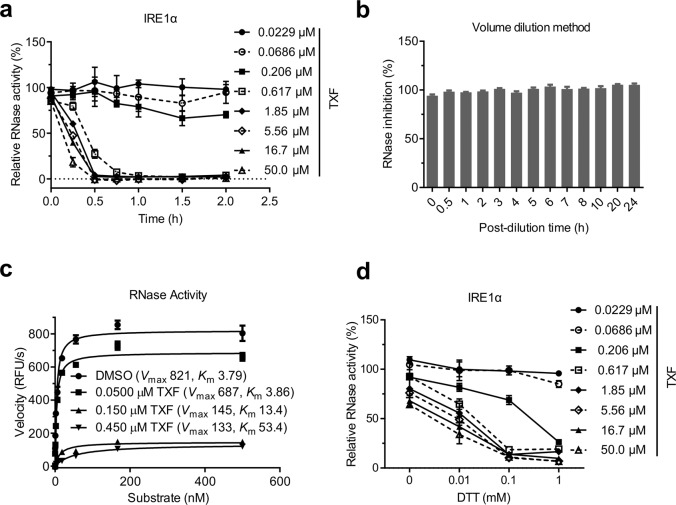
To clarify the mechanism of TXF inhibition, we performed kinetic analysis with an in vitro RNase assay. TXF displayed time-dependent inhibition by gradient concentrations of TXF when it was allowed to react with IRE1α before substrate addition (Fig. 3a). The volume dilution method indicated that the inhibition was irreversible after 24 h (Fig. 3b). In addition, TXF showed no classic competitive inhibition (Vmax reduction), which suggested a possible covalent or irreversible mode of action for the inhibitor (Fig. 3c). Interestingly, TXF affects the affinity between the enzyme and RNase substrate (Km increase, Fig. 3c). Because dithiothreitol (DTT) is an ingredient in general in vitro RNase assay of IRE1α [15, 23], we also studied DTT dependence in the inhibition of IRE1α mediated by TXF and observed that increased DTT concentrations significantly enhanced the inhibition to IRE1α RNase (Fig. 3d).
Fig. 3. Time- and DTT-dependent irreversible inhibition of IRE1α by TXF in vitro RNase assay.
a RNase activities measured after indicated concentrations of TXF were added to IRE1α for increasing time. b The inhibition of RNase over increasing time when the assay system was diluted 100 times after IRE1α was incubated with TXF (1 μM) for 1 h. c IRE1α RNase initial velocities over increasing RNA substrate in the presence of the indicated concentrations of TXF. Vmax and Km were calculated by GraphPad Prism 7. d IRE1α RNase activities over increasing DTT in the presence of the indicated concentrations of TXF.
ROS production plays an important role in TXF-induced IRE1α inhibition
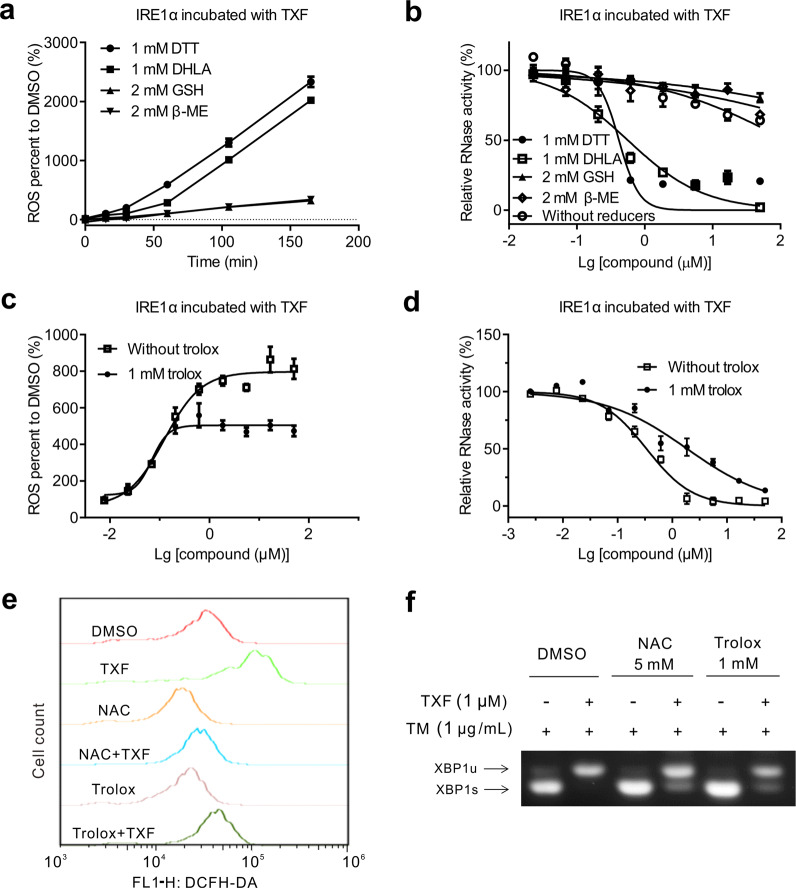
It has been generally shown that TXF acts as an electron carrier between some reducing reagents and oxygen and gives rise to ROS production in enzymatic assays or cells [24–26], which ultimately irreversibly inactivates enzymes [27]. This mode of action for TXF was suggested by our observations, as mentioned above (Fig. 3). We further detected ROS production with the addition of some reducers, DTT, dihydrolipoic acid (DHLA), GSH, or β-mercaptoethanol (β-ME), using the probe DCFH-DA. ROS production after TXF treatment increased in a time-dependent manner much more dramatically in the presence of DTT or DHLA than in the presence of GSH or β-ME (Fig. 4a). This result was similar to the ROS induction of isoquinoline-1,3,4-trione derivatives [27], speculating that the oxidation of DTT and DHLA to stable cyclic structures could accelerate electron transfer via TXF, which led to abundant ROS production. Accordingly, TXF inhibited RNase activity in the presence of DTT or DHLA and weakly inhibited RNase activity without reducers or in the presence of GSH or β-ME (Fig. 4b). These results indicated that ROS production may be the mechanism by which TXF inhibits IRE1α RNase.
Fig. 4. ROS production participates in the mechanism by which TXF inhibits IRE1α.
a ROS productions induced by TXF (500 nM) over increasing time and b IRE1α RNase activities over increasing TXF in the presence of reducing reagents DTT (1 mM), DHLA (1 mM), GSH (2 mM), and β-ME (2 mM). c ROS production and d IRE1α RNase activities over increasing TXF in the presence or absence of Trolox (1 mM). e Fluorescence intensity of ROS (FL1-H) probed by flow cytometry and f XBP1 splicing of HeLa cells treated with TXF (1 μM) and TM (1 μg/mL) in the presence or absence of NAC (5 mM) or Trolox (1 mM) for 8 h.
We next investigated the importance of ROS production in IRE1α RNase inhibition by TXF. Trolox was determined to be an ROS scavenger in the enzymatic reaction system because it did not affect IRE1α RNase activity (Supplementary Fig. S1). Trolox partially reduced ROS production (Fig. 4c) and RNase inhibition (Fig. 4d) induced by TXF. Furthermore, in HeLa cells, we observed that NAC and Trolox reversed ROS production (Fig. 4e) and partially reduced the inhibition of TXF to TM-induced endogenous XBP1 mRNA splicing (Fig. 4f). These results suggested that ROS production plays an important role in TXF-induced IRE1α RNase inhibition.
To further confirm the fact that ROS inactivates IRE1α, we firstly detected IRE1α RNase (Supplementary Fig. S2a) activity modulated by H2O2 and showed that H2O2 inhibited IRE1α RNase in the enzymatic reaction system. Furthermore, two other types of natural ROS inducers shikonin (SKN) [28] and cinnamaldehyde (CA) [29], which produce ROS in cells via mitochondrion, were used to detect XBP1 splicing in HeLa or A549 cells. SKN and CA were observed to inhibit TM-induced endogenous XBP1 mRNA splicing in a dose-dependent manner (Supplementary Fig. S3a and b). And NAC reversed the inhibition of SKN and CA to TM-induced endogenous XBP1 mRNA splicing in HeLa cells (Supplementary Fig. S3c). Besides, doxorubicin (DOX), a member of the anthracycline anticancer agents, has been identified as a ROS inducer [30] and recently an inhibitor of the IRE1α-XBP1 axis [17]. We found that DOX also harbored ROS-dependent inhibition to XBP1 mRNA splicing (Supplementary Fig. S3c). These results further confirmed that ROS inactivates IRE1α.
TXF oxidizes the thiol groups of IRE1α’s key catalytic cysteine to sulfonic groups
ROS has been reported to irreversibly inactivate enzymes by oxidizing the thiol group (-SH) of the catalytic cysteine residues to sulfonic groups (-SO3H) [27]. To determine whether the thiol group of IRE1α is oxidized to sulfonic acid by TXF, immunoprecipitation was performed with anti-sulfonate antibody to detect sulfonated form. The result showed that IRE1α was sulfonated (IRE1α: SO3H) in a dose-dependent manner in HeLa cells (Supplementary Fig. S4). Next, we performed an analysis of thiol modifications in IRE1α after treatment with TXF for 1 h using mass spectrometry (Table 2). Without DTT, we did not detect any modifications of the thiol groups in IRE1α. Nevertheless, we observed that thiol groups of four conserved IRE1α cysteine residues (CYS-605, CYS-630, CYS-715, and CYS-951) were oxidized to sulfonic groups by TXF in the presence of DTT. Furthermore, H2O2 was identified to oxidize thiol groups of three conserved IRE1α cysteine residues (CYS-605, CYS-715, and CYS-951). Among the four cysteine residues, there are three oxidized residues (CYS-605, CYS-630, and CYS-715) in the ATP-binding cleft of the kinase domain, and CYS-951 is proximal to the RNA cleavage site and the protein dimerization interface of the RNase domain (Fig. 5c).
Table 2.
Identified modification form and spectral counts of four oxidized conservative cysteine residues.
| Residues | Modification | Modified spectral counts | |||
|---|---|---|---|---|---|
| DMSO | 3 μM TXF | 3 μM TXF plus 2 mM DTT | 10 mM H2O2 | ||
| CYS-605 | -SO3H | 0 | 0 | 8 | 2 |
| CYS-630 | -SO3H | 0 | 0 | 2 | unidentify |
| CYS-715 | -SO3H | 0 | 0 | 5 | 2 |
| CYS-951 | -SO3H | 0 | 0 | 2 | 2 |
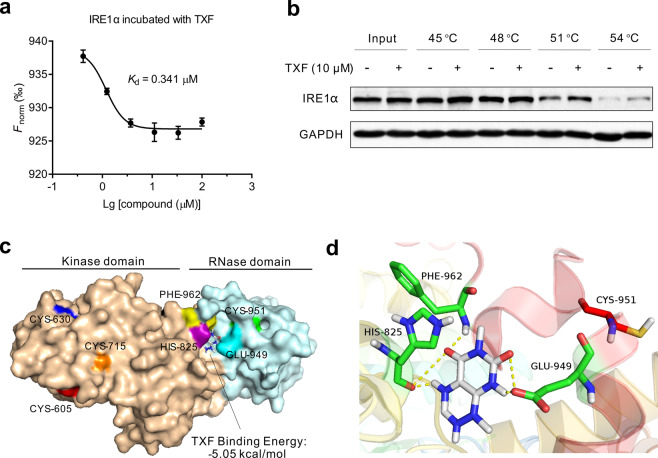
Fig. 5. The structure of TXF participates in the inhibition of IRE1α.
a Normalized fluorescence (Fnorm) using NANOTEMPER analysis software over increasing TXF by microscale thermophoresis. b CETSA was assayed in RPMI8226 cells in the presence of TXF (10 μM). c Diagram indicating four oxidized conserved cysteine residues of IRE1α by TXF and docking result of the IRE1α-TXF complex (IRE1α PDB entry: 4YZD [22]). The residues of amino acids are represented in a different color (red: CYS-605, blue: CYS-630, orange: CYS-715, green: CYS-951, purple: HIS-825, cyan: GLU-949, yellow: PHE-962). d Detailed presentation of the simulated interaction between TXF and IRE1α. The H-bonding interactions are displayed as dashed yellow lines.
The structure of TXF itself participates in the inhibition of IRE1α
Although the results above suggested that ROS plays an important role in TXF-induced IRE1α RNase inhibition, the structure of TXF itself may participate in the inhibition. This mode of action for TXF was suggested by our observations: (i) TXF affected the affinity between enzyme and RNase substrate (Km increase, Fig. 3c); (ii) Reversion of ROS production partially reduced the inhibition of TXF to TM-induced endogenous XBP1 mRNA splicing; (iii) The oxidative activity of H2O2 (Supplementary Fig. S2c) was higher than TXF incubation (Fig. 4c), but it modestly inhibited IRE1α kinase and RNase activity and especially IC50 to RNase activity achieved to 202.5 mM (Supplementary Fig. S2a and b); (iv) H2O2 had selectivity on IRE1α kinase compared with RNase (Supplementary Fig. S2a and b), while TXF had selectivity on IRE1α RNase compared with kinase (Figs. 1c and 2e); (v) NAC considerably reversed the inhibition of SKN and CA to TM-induced endogenous XBP1 mRNA splicing in HeLa cells (Supplementary Fig. S3c) but partially reduced the inhibition of TXF to TM-induced endogenous XBP1 mRNA splicing (Fig. 4f).
To confirm the interaction of IRE1α-TXF, we performed microscale thermophoresis. As shown in Fig. 5a, TXF showed good affinity for IRE1α, with a dissociation constant (Kd) value of 0.341 μM. Furthermore, we performed a cellular thermal shift assay that enables us to assay for affinity, cellular uptake, and target engagement. TXF was analyzed for thermal stabilization of IRE1α in RPMI8226 cells which express higher IRE1α than other cell lines we tested to clearly detect the soluble IRE1α protein (Supplementary Fig. S5). The detected soluble IRE1α protein level with treatment of TXF clearly was more than without TXF at indicated denaturation temperatures (Fig. 5b), indicating that TXF was able to enter cells and directly bind to the IRE1α protein. Next, a docking experiment was performed using the AutoDock suites to obtain insight into the binding mode of the IRE1α-TXF complex. The first-ranked results of the docking studies suggested that TXF would have a high predicted affinity with IRE1α, with a binding energy of −5.05 kcal/mol (Fig. 5c). The strong interaction of TXF with IRE1α is attributed to the H-bonding interaction of HIS-825, GLU-949, and PHE-962 residues located in the junction between RNase domain and kinase domain (Fig. 5d). Excitingly, we observed that CYS-951 was just near the docked site. These results suggested that the structure of TXF participates in the inhibition of IRE1α.
The inhibition of IRE1α by TXF and its derivatives is positively correlated with ROS production and binding affinity
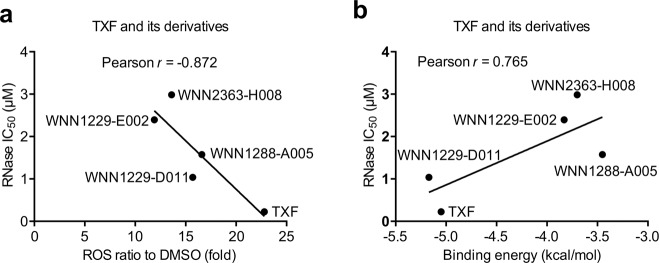
To further confirm the correlation between RNase inhibition and ROS production or the structure type of TXF, its derivatives (Fig. 1b) were also utilized. Among the compounds, TXF promoted the most ROS production after incubation for 1 h (Table 1), and a negative correlation (Pearson r = −0.872) was found between IRE1α RNase IC50 and ROS production (Fig. 6a). Next, the derivatives from TXF were used to perform docking experiments, and the binding energies of the first ranked results each were further analyzed together with TXF. TXF and WNN1229-D011 had higher binding energy values than the other compounds (Table 1). The results of correlation analysis showed that the binding energies were positively correlated (Pearson r = 0.765) with IRE1α RNase IC50 (Fig. 6b). These results suggested that both ROS production and binding affinity participate in TXF-induced IRE1α inhibition.
Fig. 6. The inhibition of IRE1α by TXF and its derivatives is positively correlated with ROS production or binding affinity.
a A plot of IRE1α RNase IC50 as a function of ROS production induced by TXF and its derivatives (0.5 μM) or b the binding energies docking TXF and its derivatives to IRE1α and a, b their correlation analysis.
Discussion
The structure of IRE1α suggests three alternative sites that could be targeted by small molecules: the ATP-binding pocket, the dimer-interface hydrophobic pocket, and the RNase active site. Interestingly, our study identified a potent small molecule (TXF) that targets a predicted new alternative site located in the junction between RNase domain and kinase domain, and oxidizes conserved cysteine residues of IRE1α active sites to inhibit IRE1α.
The clue of oxidative regulation is first from the mode of action for TXF with IRE1α. TXF displayed time- and DTT-dependent irreversible inhibition of IRE1α (Fig. 3). It has been generally shown that TXF gives rise to ROS production in enzymatic assays or cells [24–26], which ultimately irreversibly inactivates enzymes [27]. Therefore, we hypothesized that ROS production may participate in the mechanism by which TXF inhibits IRE1α. We further observed that ROS production after TXF treatment increased much more dramatically in the presence of DTT or DHLA than GSH or β-ME (Fig. 4a). Accordingly, TXF inhibited RNase activity in the presence of DTT or DHLA and weakly inhibited RNase activity without reducers or in the presence of GSH or β-ME (Fig. 4b). We next found that the ROS scavenger partially reduced RNase inhibition (Fig. 4d) induced by TXF. These results suggest that ROS participates in TXF-induced IRE1α RNase inhibition.
Next, we observed that thiol groups of four conserved IRE1α cysteines were oxidized to sulfonic groups (Table 2). There are three oxidized residues in the ATP-binding cleft of kinase domain, and one is located in the RNase domains. Consistently, TXF was identified to directly inhibit IRE1α RNase and kinase activities (Figs. 1c and 2e). Among the cysteine residues, CYS-605 and CYS-630 have not been reported to have any functions. CYS-715 is located in the activation loop of ATP-binding pocket and stabilizes the DFG-in active conformation of IRE1α [14], and CYS-951 is a target for S-nitrosylation, which occurs via oxidative reaction between NO and cysteine thiol and inhibits its RNase activity [31]. These results imply the mechanism of ROS-induced IRE1α inhibition. In future experiments, we hope to determine whether CYS-605 and CYS-630 are key catalytic cysteines modulating IRE1α.
In addition, we showed that the structure of TXF participated in the inhibition of IRE1α. It was found that TXF showed good affinity for IRE1α by microscale thermophoresis (Fig. 5a) and cellular thermal shift assay (Fig. 5b). Next, via a docking experiment, we obtained insight into the predicted binding mode of the IRE1α-TXF complex (Fig. 5c). The strong interaction of TXF with IRE1α could be attributed to the H-bonding interaction of HIS-825, GLU-949 and PHE-962 residues near CYS-951 (Fig. 5d). These results accounted for the selectivity of TXF to IRE1α RNase. Furthermore, we confirmed the positive correlation between RNase inhibition and ROS production of TXF using TXF and its derivatives and observed that the predicted binding affinity had a positive correlation with RNase inhibition (Fig. 6).
TXF, originally known as a toxin produced from bacteria with antibiotic function [32], was previously reported to inhibit Polo-like kinase 1 [33], tyrosyl DNA phosphodiesterase 2 [25], SIRT1/2 [34], and the Tcf4/β-catenin complex [35], but further exploration was precluded because ROS production by TXF misled them [36]. In fact, the strong antibiotic activity and toxicity of TXF in humans and animals have been attributed to ROS production [24]. In contrast to the above reports, we proved that TXF inhibits IRE1α structure- and ROS-dependently and found a potential binding site for TXF with IRE1α. Thus, TXF could not only induce oxidative stress via ROS production but also bring severe ER stress by inhibiting IRE1α, which led to its cytotoxicity.
Recent research shows that IRE1α is also a component coping with oxidative stress. Hourihan et al. showed that the thiol group (-SH) of an IRE-1 cysteine (CYS-663 of IRE-1 for C. elegans, equivalently CYS-715 of human IRE1α) in the kinase domain was oxidized to a sulfinyl group (-SO2H) by localized ROS, directing IRE-1 to activate the p38/SKN-1/Nrf2 antioxidant response [37]. In our study, we found that thiol groups of four IRE1α cysteines (CYS-605, CYS-630, CYS-715, and CYS-951) were oxidized to sulfonic groups (-SO3H) by TXF and inhibited both IRE1α kinase and RNase. The results together with the binding affinity and predicted binding mode of the IRE1α-TXF complex imply that TXF could induce persistent or excess ROS. Thus, TXF will act as a useful tool to perform further studies to clarify IRE1α’s role in ER stress in a situation with persistent or excess ROS.
Conclusion
In summary, we identified TXF as a novel potent IRE1α inhibitor. Furthermore, the inhibition of IRE1α by TXF is positively correlated with ROS production and binding affinity. Our findings suggest that TXF could act as a potent small molecule tool to study IRE1α’s role in ER stress.
Supplementary information
Acknowledgements
This work was supported by grants from the Guangdong High-level new R&D institute (2019B090904008), the Guangdong High-level Innovative Research Institute (2021B0909050003), the Shenzhen Basic Research Special (Natural Science Fund) Basic Research Surface Project (JCYJ20210324113006017), the Guangdong Basic and Applied Basic Research Fund Regional Joint Fund Project (Key Project) (2020B1515120091), the National Natural Science Foundation of China (81821005, 81473244, 81270942) and the National Science and Technology Major Project (2018ZX09711002).
Author contributions
KLJ, CML, QQZ, YBZ, and JL conceived and designed the study. KLJ, CML, LTN, KZZ, LXF, AHG, LX, and YBZ developed the methodology. KLJ, CML, LTN, HNJ, LX, and LLL performed the experiments and analyzed the data. KLJ and YBZ wrote, reviewed, and/or revised the manuscript. XYW, MJT, YBZ, QQZ, and JL contributed to the study supervision.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this article was revised: In this article the name of the 9th and 11th author have been misspelled.
Change history
8/4/2023
A Correction to this paper has been published: 10.1038/s41401-023-01132-4
Contributor Information
Qi-qing Zhang, Email: zhang.qiqing@szhospital.com.
Yu-bo Zhou, Email: ybzhou@simm.ac.cn.
Jia Li, Email: jli@simm.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41401-022-00949-9.
References
- 1.Lemberg MK, Strisovsky K. Maintenance of organellar protein homeostasis by ER-associated degradation and related mechanisms. Mol Cell. 2021;81:2507–19. doi: 10.1016/j.molcel.2021.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Kraskiewicz H, FitzGerald U. InterfERing with endoplasmic reticulum stress. Trends Pharmacol Sci. 2012;33:53–63. doi: 10.1016/j.tips.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 4.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–6. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 5.Prischi F, Nowak PR, Carrara M, Ali MM. Phosphoregulation of Ire1 RNase splicing activity. Nat Commun. 2014;5:3554. doi: 10.1038/ncomms4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjelkmar P, Larsson P, Cuendet MA, Hess B, Lindahl E. Implementation of the CHARMM force field in GROMACS: analysis of protein stability effects from correction maps, virtual interaction sites, and water models. J Chem Theory Comput. 2010;6:459–66. doi: 10.1021/ct900549r. [DOI] [PubMed] [Google Scholar]
- 7.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–47. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 8.Raymundo DP, Doultsinos D, Guillory X, Carlesso A, Eriksson LA, Chevet E. Pharmacological targeting of IRE1 in cancer. Trends Cancer. 2020;6:1018–30. doi: 10.1016/j.trecan.2020.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Logue SE, McGrath EP, Cleary P, Greene S, Mnich K, Almanza A, et al. Inhibition of IRE1 RNase activity modulates the tumor cell secretome and enhances response to chemotherapy. Nat Commun. 2018;9:3267. doi: 10.1038/s41467-018-05763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheng X, Arnoldussen YJ, Storm M, Tesikova M, Nenseth HZ, Zhao S, et al. Divergent androgen regulation of unfolded protein response pathways drives prostate cancer. EMBO Mol Med. 2015;7:788–801. doi: 10.15252/emmm.201404509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mimura N, Fulciniti M, Gorgun G, Tai YT, Cirstea D, Santo L, et al. Blockade of XBP1 splicing by inhibition of IRE1alpha is a promising therapeutic option in multiple myeloma. Blood. 2012;119:5772–81. doi: 10.1182/blood-2011-07-366633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun H, Lin DC, Guo X, Kharabi Masouleh B, Gery S, Cao Q, et al. Inhibition of IRE1alpha-driven pro-survival pathways is a promising therapeutic application in acute myeloid leukemia. Oncotarget. 2016;7:18736–49. doi: 10.18632/oncotarget.7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Reste PJ, Pineau R, Voutetakis K, Samal J, Jegou G, Lhomond S, et al. Local intracerebral inhibition of IRE1 by MKC8866 sensitizes glioblastoma to irradiation/chemotherapy in vivo. Cancer Lett. 2020;494:73–83. doi: 10.1016/j.canlet.2020.08.028. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Perera BG, Hari SB, Bhhatarai B, Backes BJ, Seeliger MA, et al. Divergent allosteric control of the IRE1alpha endoribonuclease using kinase inhibitors. Nat Chem Biol. 2012;8:982–9. doi: 10.1038/nchembio.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cross BC, Bond PJ, Sadowski PG, Jha BK, Zak J, Goodman JM, et al. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc Natl Acad Sci USA. 2012;109:E869–78. doi: 10.1073/pnas.1115623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ri M, Tashiro E, Oikawa D, Shinjo S, Tokuda M, Yokouchi Y, et al. Identification of Toyocamycin, an agent cytotoxic for multiple myeloma cells, as a potent inhibitor of ER stress-induced XBP1 mRNA splicing. Blood Cancer J. 2012;2:e79. doi: 10.1038/bcj.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang D, Lynch C, Medeiros BC, Liedtke M, Bam R, Tam AB, et al. Identification of Doxorubicin as an inhibitor of the IRE1alpha-XBP1 axis of the unfolded protein response. Sci Rep. 2016;6:33353. doi: 10.1038/srep33353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tashiro E, Hironiwa N, Kitagawa M, Futamura Y, Suzuki S, Nishio M, et al. Trierixin, a novel Inhibitor of ER stress-induced XBP1 activation from Streptomyces sp. 1. Taxonomy, fermentation, isolation and biological activities. J Antibiot. 2007;60:547–53. doi: 10.1038/ja.2007.69. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto K, Tashiro E, Imoto M. Quinotrierixin inhibited ER stress-induced XBP1 mRNA splicing through inhibition of protein synthesis. Biosci Biotechnol Biochem. 2011;75:284–8. doi: 10.1271/bbb.100622. [DOI] [PubMed] [Google Scholar]
- 20.Iwawaki T, Akai R, Kohno K, Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat Med. 2004;10:98–102. doi: 10.1038/nm970. [DOI] [PubMed] [Google Scholar]
- 21.Fan LX, Liu CM, Gao AH, Zhou YB, Li J. Berberine combined with 2-deoxy-d-glucose synergistically enhances cancer cell proliferation inhibition via energy depletion and unfolded protein response disruption. BBA-Gen Subj. 2013;1830:5175–83. doi: 10.1016/j.bbagen.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Concha NO, Smallwood A, Bonnette W, Totoritis R, Zhang G, Federowicz K, et al. Long-range inhibitor-induced conformational regulation of human IRE1alpha endoribonuclease activity. Mol Pharmacol. 2015;88:1011–23. doi: 10.1124/mol.115.100917. [DOI] [PubMed] [Google Scholar]
- 23.Wiseman RL, Zhang Y, Lee KP, Harding HP, Haynes CM, Price J, et al. Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1. Mol Cell. 2010;38:291–304. doi: 10.1016/j.molcel.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Latuasan HE, Berends W. On the origin of the toxicity of toxoflavin. Biochim Biophys Acta. 1961;52:502–8. doi: 10.1016/0006-3002(61)90408-5. [DOI] [PubMed] [Google Scholar]
- 25.Raoof A, Depledge P, Hamilton NM, Hamilton NS, Hitchin JR, Hopkins GV, et al. Toxoflavins and deazaflavins as the first reported selective small molecule inhibitors of tyrosyl-DNA phosphodiesterase II. J Med Chem. 2013;56:6352–70. doi: 10.1021/jm400568p. [DOI] [PubMed] [Google Scholar]
- 26.Gencheva R, Cheng Q, Arner ESJ. Efficient selenocysteine-dependent reduction of toxoflavin by mammalian thioredoxin reductase. BBA-Gen Subj. 2018;1862:2511–7. doi: 10.1016/j.bbagen.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 27.Du JQ, Wu J, Zhang HJ, Zhang YH, Qiu BY, Wu F, et al. Isoquinoline-1,3,4-trione derivatives inactivate caspase-3 by generation of reactive oxygen species. J Biol Chem. 2008;283:30205–15. doi: 10.1074/jbc.M803347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duan D, Zhang B, Yao J, Liu Y, Fang J. Shikonin targets cytosolic thioredoxin reductase to induce ROS-mediated apoptosis in human promyelocytic leukemia HL-60 cells. Free Radic Biol Med. 2014;70:182–93. doi: 10.1016/j.freeradbiomed.2014.02.016. [DOI] [PubMed] [Google Scholar]
- 29.Ka H, Park H-J, Jung H-J, Choi J-W, Cho K-S, Ha J, et al. Cinnamaldehyde induces apoptosis by ROS-mediated mitochondrial permeability transition in human promyelocytic leukemia HL-60 cells. Cancer Lett. 2003;196:143–52. doi: 10.1016/S0304-3835(03)00238-6. [DOI] [PubMed] [Google Scholar]
- 30.Zhu H, Sarkar S, Scott L, Danelisen I, Trush MA, Jia Z, et al. Doxorubicin redox biology: redox cycling, topoisomerase inhibition, and oxidative stress. React Oxy Species. 2016;1:189–98. doi: 10.20455/ros.2016.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L, Calay ES, Fan J, Arduini A, Kunz RC, Gygi SP, et al. S-Nitrosylation links obesity-associated inflammation to endoplasmic reticulum dysfunction. Science. 2015;349:500–6. doi: 10.1126/science.aaa0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Machlowitz RA, Fisher WP, Mc KB, Tytell AA, Charney J. Xanthothricin, a new antibiotic. Antibiot Chemother. 1954;4:259–61. [PubMed] [Google Scholar]
- 33.Goh KC, Wang H, Yu N, Zhou Y, Zheng Y, Lim Z, et al. PLK1 as a potential drug target in cancer therapy. Drug Dev Res. 2004;62:349–61. doi: 10.1002/ddr.10392. [DOI] [Google Scholar]
- 34.Choi G, Lee J, Ji JY, Woo J, Kang NS, Cho SY, et al. Discovery of a potent small molecule SIRT1/2 inhibitor with anticancer effects. Int J Oncol. 2013;43:1205–11. doi: 10.3892/ijo.2013.2035. [DOI] [PubMed] [Google Scholar]
- 35.Mao Y, Tian W, Huang Z, An J. Convenient synthesis of toxoflavin that targets β-catenin/Tcf4 signaling activities. J Heterocycl Chem. 2014;51:594–7. doi: 10.1002/jhet.1111. [DOI] [Google Scholar]
- 36.Baell J, Walters MA. Chemistry: chemical con artists foil drug discovery. Nature. 2014;513:481–3. doi: 10.1038/513481a. [DOI] [PubMed] [Google Scholar]
- 37.Hourihan JM, Moronetti Mazzeo LE, Fernandez-Cardenas LP, Blackwell TK. Cysteine sulfenylation directs IRE-1 to activate the SKN-1/Nrf2 antioxidant response. Mol Cell. 2016;63:553–66. doi: 10.1016/j.molcel.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.