Abstract
Cytotoxic necrotizing factor type 1 (CNF1) of uropathogenic Escherichia coli belongs to a family of bacterial toxins that target the small GTP-binding Rho proteins that regulate the actin cytoskeleton. Members of this toxin family typically inactivate Rho; however, CNF1 and the highly related CNF2 activate Rho by deamidation. Other investigators have reported that the first 190 amino acids of CNF1 constitute the cellular binding domain and that the CNF1 enzymatic domain lies within a 300-amino-acid stretch in the C terminus of the toxin. Amino acids 53 to 75 appear to be critical for cell receptor recognition, while amino acids Cys866 and His881 are considered essential for deamidation activity. To delineate further the functional domains of CNF1, we generated 16 monoclonal antibodies (MAbs) against the toxin and used them for epitope mapping studies. Based on Western blot immunoreactivity patterns obtained from a series of truncated CNF1 proteins, this panel of MAbs mapped to epitopes located throughout the toxin, including the binding and enzymatic domains. All MAbs showed reactivity to CNF1 by Western and dot blot analyses. However, only 7 of the 16 MAbs exhibited cross-reactivity with CNF2. Furthermore, only three MAbs demonstrated the capacity to neutralize toxin in either HEp-2 cell assays (inhibition of multinucleation) or 5637 bladder cell assays (inhibition of cytotoxicity). Since CNF1 epitopes recognized by neutralizing MAbs are likely to represent domains or regions necessary for the biological activities of the toxin, the epitopes recognized by these three MAbs, designated JC4 (immunoglobulin G2a [IgG2a]), BF8 (IgA), and NG8 (IgG2a), were more precisely defined. MAbs JC4 and BF8 reacted with epitopes that were common to CNF1 and CNF2 and located within the putative CNF1 binding domain. MAb JC4 recognized an epitope spanning amino acids 169 to 191, whereas MAb BF8 mapped to an epitope between amino acids 135 and 164. Despite the capacity of both MAbs to recognize CNF2 in Western blot analyses, only MAb BF8 neutralized CNF2. MAb NG8 showed reactivity to a CNF1-specific epitope located between amino acids 683 and 730, a region that includes a very small portion of the putative enzymatic domain. Taken together, these findings identify three new regions of the toxin that appear to be critical for the biological activity of CNF1.
The cytotoxic necrotizing factors type 1 (CNF1) and type 2 (CNF2) of Escherichia coli belong to a new, expanding family of bacterial toxins and type III secretion effector molecules that modulate the activities of Rho GTPases or their associated regulatory proteins (1, 7, 25). Rho proteins belong to a subfamily of small GTP-binding molecules that regulate the actin cytoskeleton by cycling between GTP-bound active and GDP-bound inactive states (18). CNF1 was first identified in E. coli strains isolated from children with enteritis (8). However, recent epidemiological studies indicate that CNF1 is more frequently associated with E. coli strains that cause extraintestinal infections in humans, particularly those of the urinary tract such as cystitis, pyelonephritis, and prostatitis (2, 3, 9, 30). In CNF1-producing uropathogenic E. coli strains, cnf1 is chromosomally encoded and typically resides on a pathogenicity island that also contains hemolysin and P fimbria-related genes (6, 38). Both CNF1 and the highly related, plasmid-encoded CNF2 are monomeric, cytoplasmic toxins of approximately 115 kDa (14, 31).
In vivo, CNF1 is lethal when injected into mice and can produce necrotic lesions after intradermal injection into rabbits or guinea pigs (8, 10, 11). HeLa, CHO, Vero, and HEp-2 cell lines exhibit a series of striking features when intoxicated with CNF1. These CNF1-mediated responses include membrane ruffling, the formation of focal adhesions and actin stress fibers, and endomitosis without concomitant cytokinesis, a phenomenon that results in enlarged, multinucleated cells (12, 15). Cell lines derived from the human uroepithelium (J82, T24, SV-HUC-1, and 5637) also exhibit a multinucleation phenotype when intoxicated with CNF1. However, actin stress fiber formation is notably absent in all uroepithelial cell lines after toxin exposure, and CNF1 is frankly cytotoxic for the 5637 human bladder cell line (29).
Toxin-induced cellular manifestations are indicative of a dominant active Rho phenotype and result directly from the CNF1-catalyzed deamidation of Rho Gln63 and of Rac and Cdc42 Gln61 (17, 27, 34). The conversion of glutamine to glutamic acid occurs in the switch 2 domain of these small GTPases, a region that is critical for intrinsic GTP hydrolysis and is involved in the interactions with Rho GTPase-activating proteins (Rho-GAPs) (16, 26). CNF1-mediated deamidation of Rho thereby inhibits the intrinsic GTPase activity of Rho and abolishes Rho-GAP-stimulated GTP hydrolysis. These CNF1-induced events result in a constitutively active GTP-bound form of Rho that continually signals its cellular effector molecules.
Like other single polypeptide bacterial toxins, such as Pseudomonas aeruginosa exotoxin A and diphtheria toxin, CNF1 can be structurally organized into three functional domains (7, 24). The N-terminal portion of CNF1 shows amino acid similarity to the Pasteurella multocida toxin (PMT), a potent mitogen associated with progressive atrophic rhinitis in animals (14). This region of CNF1 is proposed by Fabbri et al. (13) to contain the cell receptor-binding domain. These investigators showed that the CNF1 cellular binding domain resides in the first 190 amino acids of the toxin and reported that hydrophilic amino acid residues 53 to 75 are critical for cell receptor recognition (13, 24). The central portion of the toxin contains two hydrophobic domains (amino acids 336 to 372 and amino acids 392 to 412) that are predicted by sequence analysis to be membrane-spanning regions that may be involved in toxin translocation. The C-terminal portion of CNF1 (amino acids 709 to 1014) exhibits the catalytic activity of the toxin, and two specific residues in this region, Cys866 and His881, have been reported to be essential for deamidation activity (24, 35). This C-terminal region also shares amino acid homology with the dermonecrotic toxin (DNT) of Bordetella pertussis, B. bronchiseptica, and B. parapertussis (39). DNT is the only other Rho-activating bacterial toxin currently identified. Like CNF1, DNT can deamidate Rho but instead preferentially uses a transglutaminase activity to add either primary amines or ubiquitous polyamines, such as putrescine, to Rho at Gln63. This DNT-mediated modification of Rho also results in the constitutive activation of the molecule (20, 28, 33).
In the present study, we further analyzed the relationship between the structure of CNF1 and its toxic activity. Our approach was to generate a panel of monoclonal antibodies (MAbs) that were used in conjunction with a series of CNF1 deletion constructs to more precisely delineate the functional regions of the toxin. Additionally, we were able to identify epitopes that were unique to CNF1 or shared with CNF2.
MATERIALS AND METHODS
Bacterial strains, plasmids, and recombinant DNA techniques.
The relevant characteristics of all bacterial strains and plasmids used in this study are described in Table 1. The pBluescript II SK(−) and pQE30 cloning vectors, in addition to all recombinant plasmids, were maintained and propagated in E. coli XL1-Blue. All histidine (His)-tagged toxin molecules were expressed in E. coli M15(pREP4), and E. coli NovaBlue (DE3) was used as the host strain for the CNF1 epitope library. All strains were grown in Luria broth or on Luria agar plates supplemented with the following concentrations of antibiotics when appropriate: ampicillin, 100 μg/ml; carbenicillin, 200 μg/ml; kanamycin, 25 μg/ml; and tetracycline, 12 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB+lacIqZΔM15::Tn10(Tetr)] | Stratagene |
| M15(pREP4) | NaIs Strs Rifs Δlac-ara-gal-mtl F−recA uvr (pREP4 lacI Kanr) | Qiagen |
| NovaBlue (DE3) | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB+ lacIqZΔM15::Tn10(Tetr)] (DE3) | Novagen |
| Cloning vectors | ||
| pBluescript II SK(−) | E. coli phagemid cloning vector (Ampr) | Stratagene |
| pQE30 | E. coli expression vector with His6 tag 5′ to the polylinker (Ampr) | Qiagen |
| pScreen T-Vector | E. coli expression vector with polylinker downstream of the T7 gene 10 260-amino-acid coding sequence (Ampr) | Novagen |
| Recombinant plasmids | ||
| pHLK102 | wt cnf1 gene (nt −31 to 3069) from UPEC strain J96 cloned into the SmaI site of pBluescript II SK(−) | Lockman and O'Brienb |
| pCNF24 | wt cnf1 gene (nt 1 to 3045) amplified from pHLK102 and cloned into BamHI/KpnI sites of pQE30 | This study |
| pΔC469 | 1.6-kb BamHI/PstI cnf1 fragment (nt 1 to 1637) cloned into pQE30 | This study |
| pΔN545 | 1.4-kb PstI cnf1 fragment (nt 1637 to 3045) cloned into pQE30 | This study |
| pΔ442 | pCNF24 with an internal in-frame BclI deletion (nt 1115 to 2348) | This study |
| pΔC146 | pCNF24 with a deletion of 442 nt from the 3′ end of the cnf1 gene | This study |
| pΔN63 | pCNF24 with a deletion of 187 nt from the 5′ end of the cnf1 gene | This study |
| pΔC25 | pCNF24 with a deletion of 75 nt from the 3′ end of the cnf1 gene | This study |
| pΔC230 | 2.3-kb BamHI/KpnI cnf1 fragment (nt 1 to 2355) cloned into pQE30 | This study |
| pΔC284 | 2.2-kb BamHI/KpnI cnf1 fragment (nt 1 to 2193) cloned into pQE30 | This study |
| pΔC331 | 2.0-kb BamHI/KpnI cnf1 fragment (nt 1 to 2052) cloned into pQE30 | This study |
| pΔC378 | 1.9-kb BamHI/KpnI cnf1 fragment (nt 1 to 1911) cloned into pQE30 | This study |
| pΔC433 | 1.7-kb BamHI/KpnI cnf1 fragment (nt 1 to 1746) cloned into pQE30 | This study |
| pΔN75 | 2.8-kb BamHI/KpnI cnf1 fragment (nt 226 to 3045) cloned into pQE30 | This study |
| pΔN134 | 2.6-kb BamHI/KpnI cnf1 fragment (nt 403 to 3045) cloned into pQE30 | This study |
| pΔN217 | 2.4-kb BamHI/KpnI cnf1 fragment (nt 652 to 3045) cloned into pQE30 | This study |
| pΔN272 | 2.2-kb BamHI/KpnI cnf1 fragment (nt 817 to 3045) cloned into pQE30 | This study |
| pEOSW30 | 3.3-kb BanI/HaeII fragment containing the cnf2 gene cloned into the SmaI site of pK184 | 31 |
Tetr, tetracycline resistance; Kanr, kanamycin resistance; Ampr, ampicillin resistance; nt, nucleotides; wt, wild type.
H. A. Lockman and A. D. O'Brien, Abstr. 97th Gen. Meet. Am. Soc. Microbiol. 1997, abstr. B-156, p. 55, 1997.
Standard techniques were used for the construction of all CNF1-encoding plasmids and deletion derivatives (4, 32). CNF1 deletion constructs were generated from wild-type toxin clones pCNF24 or pHLK102 either by subcloning restriction endonuclease-digested fragments or by PCR amplification using primers that incorporated sequences for restriction endonuclease sites at the 5′ end (Table 2).
TABLE 2.
Synthetic oligonucleotide primers
| Primer | Sequence (5′-3′)a | Purposeb |
|---|---|---|
| SCNF1 | TATTAATCTTCACAGAGGAC | Used with ASCNF1 to amplify wt cnf1 gene from UPEC strain J96 |
| ASCNF1 | GGCCAATAAATAATTTCCCGAATC | Used with SCNF1 to amplify wt cnf1 gene from UPEC strain J96 |
| 856 | GCGGATCCATGGGTAACCAATGGCAAC BamHI | Used with T7 primer to amplify wt cnf1 gene from pHLK102 and with primers KCM1 to -5 to generate C-terminal CNF1 deletion constructs |
| T7 | GTAATACGACTCACTATAGGGC | Used with 856 to amplify wt cnf1 gene from pHLK102 |
| 3195F | AAAGATCGCTTTGATAACCATGGC NcoI | Used with 3852R to amplify 657-bp cnf1 fragment for construction of pΔC25 |
| 3852R | ATGGTACCACCTTACGACAACATTGCC KpnI | Used with 3195F to amplify 657-bp cnf1 fragment for construction of pΔC25 |
| KCM1 | ATCGGTACCATAGCCATGGTTATCAAAGCG | Used with 856 to amplify 2.3-kb BamHI/KpnI cnf1 fragment |
| KCM2 | ATCGGTACCAAAGTTAGATTTGGAGGTGC | Used with 856 to amplify 2.2-kb BamHI/KpnI cnf1 fragment |
| KCM3 | ATCGGTACCGGCTTCATCAGGAATTGCC | Used with 856 to amplify 2.0-kb BamHI/KpnI cnf1 fragment |
| KCM4 | ATCGGTACCACCTAAGCTTGACTGTGGG | Used with 856 to amplify 1.9-kb BamHI/KpnI cnf1 fragment |
| KCM5 | ATCGGTACCAACAATATCCATTTCTGACG | Used with 856 to amplify 1.7-kb BamHI/KpnI cnf1 fragment |
| KCM6 | GCGGATCCACAGAAGTACTGACACTCAC | Used with 3900 to amplify 2.8-kb BamHI/KpnI cnf1 fragment |
| KCM7 | GCGGATCCGATTTTGCAGGTGTTTATGG | Used with 3900 to amplify 2.6-kb BamHI/KpnI cnf1 fragment |
| KCM8 | GCGGATCCGTAATGCTTTTCATTCCTGG | Used with 3900 to amplify 2.4-kb BamHI/KpnI cnf1 fragment |
| KCM9 | GCGGATCCGTAAATTCTGTTCTACATGC | Used with 3900 to amplify 2.2-kb BamHI/KpnI cnf1 fragment |
| 3900 | ACGGGTACCTCAAAATTTTTTTGAAAATACC | Used with primers KCM6 to -9 to generate N-terminal CNF1 deletion constructs |
Restriction sites are underlined.
wt, wild type.
Cell lines and media.
HEp-2 human laryngeal cells (ATCC CCL23) were maintained in Eagle's minimal essential medium with Earle's balanced salt solution supplemented with 10% fetal bovine serum (FBS). The 5637 human bladder cell line (ATCC HTB-9) was maintained in RPMI 1640 medium supplemented with 10% FBS. All tissue culture media contained 2 mM l-glutamine, 10 μg of gentamicin per ml, 10 U of penicillin G per ml, and 10 μg of streptomycin per ml. Cell lines were maintained at 37°C in a humidified atmosphere of 5% CO2.
Production of CNF1 MAbs.
The purification of native and electroeluted His-tagged CNF1 (His-CNF1) for use as immunogens has been previously described (29). All animal immunizations, enzyme-linked immunosorbent assay (ELISA) screenings of mouse sera for CNF1-reactive antibodies, and splenocyte-myeloma cell fusions were performed in conjunction with BioCon, Inc. (Rockville, Md.). Briefly, BALB/c mice were initially immunized subcutaneously with a 1:1 emulsion of 50 μg of electroeluted inactive His-CNF1 in complete Freund's adjuvant. At approximately 3-week intervals, mice were subcutaneously boosted with a 1:1 emulsion that contained incomplete Freund's adjuvant and either 25 μg of electroeluted inactive His-CNF1, 25 μg of formaldehyde-toxoided His-CNF1, or 10 to 20 ng of active His-CNF1. Throughout the immunization schedule, mice were bled and the sera were tested for reactivity with CNF1 by ELISA, Western blot analyses, and HEp-2 cell toxin neutralization assays. When sufficient serum neutralization titers were achieved (1:640 to 1:5,120), mice were given a final intraperitoneal (i.p.) and intravenous (i.v.) boost that consisted of 5 ng of active His-CNF1 per 100 μl mixed with either 250 μg (i.p. route) or 100 μg (i.v. route) of toxoided material per ml. Three days later, spleens from hyperimmunized mice were collected and fused to Sp2/O myeloma cells (19). Supernatants from the resulting fusion hybridomas were screened for the production of CNF1-specific MAbs by ELISA and toxin neutralization assays using HEp-2 cells (inhibition of multinucleation) and 5637 cells (inhibition of cytotoxicity). Hybridomas found to be positive by ELISA, toxin neutralization assays, or both were subsequently expanded and retested for anti-CNF1 activity. Selected hybridomas were cloned by two rounds of single-cell limiting dilutions (19). In total, 16 anti-CNF1 MAb-secreting hybridomas were established and used for the production of large-scale MAb tissue culture supernatants both in serum-free Hybridoma-SFM (Life Technologies) supplemented with antibiotics (10 U of penicillin G per ml, 10 μg of streptomycin per ml, and 10 μg of gentamicin per ml) and in RPMI 1640 supplemented with 10% FBS and antibiotics. The isotype of each MAb was determined with an ELISA-based mouse-hybridoma subtyping kit (Roche Molecular Biochemicals). The antibody concentration in hybridoma tissue culture supernatants was measured with a mouse immunoglobulin G (IgG) ELISA kit (Roche Molecular Biochemicals). To determine the concentration of MAb BF8 (IgA), the intensity of antibody light-chain bands in tissue culture supernatants was compared to those seen with known concentrations of purified mouse IgA (Sigma Aldrich) in colloidal Coomassie blue-stained sodium dodecyl sulfate (SDS)-polyacrylamide gels (serum-free samples) or by Western blot analysis using horseradish peroxidase (HRP)-conjugated goat anti-mouse IgA (Sigma Aldrich) (23).
CNF1-specific ELISA.
Each well of a Nunc Maxisorb 96-well plate was coated with 0.1 μg of electroeluted, inactive His-CNF1 per well in phosphate-buffered saline (PBS) for 2 h at room temperature or at 4°C overnight. Next, wells were washed four times with PBS that contained 0.05% Tween 20 (PBST) and then incubated for 1 h at room temperature with either 100 μl of mouse sera diluted in PBS or undiluted hybridoma supernatants. The wells were then washed with PBST and incubated for 1 h at room temperature with an HRP-conjugated goat anti-mouse IgG secondary antibody (Bio-Rad) diluted 1:2,000 in PBST. After four more washes with PBST, 100 μl of 2,2′-azinobis(3-ethylbenzthiazolinesulfonate) (ABTS) was added to each well. The plates were incubated at room temperature for 30 min to allow color development, and the A405 of samples was measured with a microplate reader.
Preparation of bacterial extracts.
For neutralization assays, clarified lysates containing full-length His-CNF1 were prepared as previously described (29). Briefly, overnight cultures of E. coli M15(pREP4) transformed with pCNF24 were diluted into Luria broth supplemented with kanamycin and ampicillin. Bacteria were grown for 1 h at 37°C and then induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and incubated for an additional 4 h at room temperature. These induction conditions were optimal for the expression of soluble toxin. Bacteria were harvested by centrifugation, concentrated 10-fold by resuspension in PBS supplemented with 100 μg of gentamicin per ml, and then disrupted by sonication. Extracts were clarified by centrifugation, and the supernatants were sterilized by filtration through a 0.2-μm (pore-size) syringe filter. The protein concentrations of clarified toxin extracts were determined using BCA protein assay reagents (Pierce), and each extract was adjusted to a concentration of 1 mg/ml. The titers of extracts that contained His-CNF1 were determined in both HEp-2 cell multinucleation assays and in 5637 cell cytotoxicity assays (29). The reciprocal of the dilution at which 50% of the monolayer was either multinucleated (HEp-2 cells) or destroyed (5637 cells) was considered to represent one 50% cytotoxic dose (CD50).
For Western and native dot blot analyses, whole-cell lysates of E. coli M15(pREP4) transformed with plasmids that encoded either wild-type or truncated His-CNF1 were prepared similarly to the extracts described above with the following two modifications. First, bacteria were grown at 37°C after induction with IPTG to permit maximal protein expression. Second, the sonically disrupted lysates were not clarified prior to use. To determine MAb cross-reactivity to CNF2, E. coli XL1-Blue transformed with pEOSW30 (31) was grown overnight in Luria broth supplemented with tetracycline and ampicillin and then directly used to produce a whole-cell lysate at a concentration of 1 mg/ml.
Toxin neutralization assays.
Twofold serial dilutions of either mouse sera or hybridoma tissue culture supernatants were incubated with four CD50s of a clarified toxin extract for 2 h at 37°C and then at 4°C overnight. On the following day, 96-well microtiter plates were seeded with either 4 × 103 HEp-2 or 8 × 103 5637 cells/well and incubated for 4 h at 37°C with 5% CO2. Next, toxin-antibody mixtures (100 μl) were added to the wells, and then the plates were incubated for an additional 72 h at 37°C with 5% CO2 prior to the fixing and staining of cells with Leukostat (Fisher Scientific). Neutralization assays using HEp-2 cells were assessed for the inhibition of multinucleation by light microscopy. For toxin neutralization assays using 5637 cells, the inhibition of cytotoxicity was determined by measuring the A600 of fixed and stained cells using a microplate reader (29).
Western and native dot blot analyses.
Whole-cell lysates of E. coli expressing wild-type His-CNF1, a series of truncated His-CNF1 molecules, or wild-type CNF2 were subjected to SDS-polyacrylamide gel electrophoresis (10% polyacrylamide) (23). Following electrophoretic separation, the proteins were transferred to Optitran nitrocellulose membranes (Schleicher & Schuell) with a Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell (Bio-Rad), and then the membranes were blocked overnight at 4°C in BLOTTO. Membranes were incubated with mouse polyclonal anti-CNF1 sera (1:500) or MAb hybridoma tissue culture supernatants (1:3 to 1:10), washed in TBST, and then incubated with either HRP-conjugated goat anti-mouse IgG (1:1,000) or HRP-conjugated goat anti-mouse IgA (1:500 to 1:1,000). Reactive proteins were detected with enhanced chemiluminescence (ECL; Amersham Pharmacia) or visualized with diaminobenzidine (DAB) substrate (Sigma-Aldrich).
For native dot blots, 15 μg of whole-cell E. coli lysates that expressed His-CNF1 or truncated His-CNF1 molecules was applied per well to an Optitran membrane with a microsample filtration manifold (Schleicher & Schuell). Wells were then washed twice with PBS, and the membrane was allowed to air dry. Dots blots were tested for CNF1 MAb reactivity as described above for Western blot analyses.
Epitope mapping of CNF1 MAbs.
To localize the epitopes recognized by the CNF1 MAbs, the NovaTope Epitope Mapping System (Novagen) was used in accordance with the manufacturer's instructions. Briefly, the cnf1 gene was isolated from pHLK102 and digested with DNase I for 15 min at 37°C in the presence of 10 mM MnCl2, and then double-stranded cnf1 fragments of 50 to 150 bp were purified from 2.5% BioGels with a MERmaid kit (Bio 101). Purified cnf1 fragments were blunt ended with T4 DNA polymerase, and a single 3′ deoxyribosyladenine residue was added with Tth DNA polymerase. Fragments were then ligated into the pScreen T-Vector that had been linearized and single 3′ deoxyribosylthymine tailed at a unique EcoRV site. This vector is designed to express small inserts as a carboxy-terminal fusion to the T7 bacteriophage gene 10 capsid protein. Ligation mixes were transformed into NovaBlue (DE3) competent cells to generate a CNF1 epitope library that was subsequently screened by colony immunoblot as follows. Transformants that grew on Luria agar plates supplemented with tetracycline and carbenicillin were lifted onto nitrocellulose filters (BA85; Schleicher & Schuell). Bacteria were lysed in a chloroform vapor chamber, and bacterial proteins were then denatured with 20 mM Tris-HCl (pH 7.9), 6 M urea, and 0.5 M NaCl. Filters were washed in TBST to remove colony debris and blocked overnight at 4°C in BLOTTO. Next, filters were incubated consecutively in pools of five to six MAb hybridoma tissue culture supernatants (diluted 1:10 to 1:30 in BLOTTO), washed in TBST, and then incubated with an HRP-conjugated goat anti-mouse immunoglobulin secondary antibody (Roche Molecular Biochemicals) diluted 1:4,000 in BLOTTO. Immunoreactive colonies were detected with ECL or visualized with DAB substrate. Colonies that were reactive were isolated and rescreened with individual MAb tissue culture supernatants. Plasmids isolated from immunoreactive clones were sequenced using a T7 terminator primer (5′-GCTAGTTATTGCTCAGCGG) (Novagen).
RESULTS
Relevant characteristics of the anti-CNF1 MAbs.
From the initial screening of over 2,000 hybridoma fusion wells, a panel of 16 anti-CNF1-secreting hybridomas was established. The isotype of each MAb was determined by an ELISA-based assay and, with the exception of MAb BF8, which was identified as an IgA molecule, all MAbs were found to be IgG class antibodies (Table 3). While all MAbs showed reactivity to CNF1 in both Western (Fig. 1A) and native dot blot analyses of toxin-containing bacterial lysates (data not shown), only three MAbs significantly neutralized toxin activity in HEp-2 cell assays (inhibition of multinucleation) when hybridoma tissue culture supernatants at equivalent antibody concentrations (5 μg/ml) were mixed with four CD50s of toxin lysate (Table 3). Of the 16 MAbs tested, only MAbs NG8, JC4, and BF8 were found to have neutralization titers of 1:32 or greater (MAb NG8, 1:32; MAb JC4, 1:64; MAb BF8, 1:512). Similar toxin neutralization results and titers were also obtained with MAbs NG8, JC4, and BF8 in 5637 bladder cell assays (inhibition of cytotoxicity; data not shown).
TABLE 3.
Isotype and neutralization capacity of CNF1 MAbs
| MAb | Isotypea | Neutralizationb |
|---|---|---|
| JC4 | IgG2a | +++ |
| BF8 | IgA | ++++ |
| GC2 | IgG2a | − |
| KD3 | IgG1 | − |
| PE3 | IgG1 | − |
| ND4 | IgG1 | − |
| CA6 | IgG1 | + |
| PC10 | IgG2a | + |
| AE8 | IgG1 | ++ |
| NG8 | IgG2a | +++ |
| DD1 | IgG2a | − |
| HA5 | IgG1 | + |
| JC3 | IgG1 | + |
| DC7 | IgG1 | + |
| OB8 | IgG2a | − |
| AC11 | IgG2a | − |
All antibodies were of the kappa light-chain type.
Four CD50s of toxin contained in a bacterial lysate were incubated with 5 μg of MAb hybridoma supernatants per ml and evaluated for blocking of multinucleation on HEp-2 cells. −, No neutralization capacity; +, 1:2 to 1:4 neutralization titer; ++, 1:8 to 1:16 neutralization titer; +++, 1:32 to 1:64 neutralization titer; ++++, >1:128 neutralization titer.
FIG. 1.
Reactivity of MAbs with CNF1 and cross-reactivity with CNF2 as demonstrated by Western blot analyses. Toxin-containing bacterial lysates were separated on SDS-polyacrylamide gels, transferred to nitrocellulose, and probed with individual MAb hybridoma supernatants. Reactive proteins were visualized with appropriate HRP-conjugated secondary antibodies and either DAB (all IgG MAbs) or ECL substrates (MAb BF8). (A) CNF1 immunoblot. The position of CNF1 is indicated with an arrow. (B) CNF2 immunoblot.
CNF1 and CNF2 of pathogenic E. coli are closely related, both in primary sequence and in enzymatic activity. These toxins share 85% identity at the amino acid level and activate members of the Rho family of small GTPases through deamidation (31, 37). To identify CNF2 cross-reactive MAbs, bacterial extracts that contained CNF2 were used in Western blot analyses with individual MAbs (Fig. 1B). Seven of the sixteen MAbs (JC4, BF8, GC2, KD3, CA6, DD1, and OB8) demonstrated reactivity to epitopes common to the two toxins, and the remaining MAbs (PE3, ND4, PC10, AE8, NG8, HA5, JC3, DC7, and AC11) showed reactivity to CNF1-specific epitopes only. Because MAbs JC4 and BF8 neutralized CNF1 and cross-reacted with CNF2 in Western blot analyses, these MAbs were also tested for their potential to neutralize CNF2. Only MAb BF8 demonstrated neutralization capacity against four CD50s of a CNF2-containing lysate in HEp-2 cell assays (data not shown).
Epitope mapping of the anti-CNF1 MAbs.
Since the binding and enzymatic domains of CNF1 had been recently identified (13, 24, 35), the capacity of each of the CNF1 MAbs to recognize toxin in Western blot analyses provided us with an opportunity to examine the relationship between the functional domains of the toxin and the CNF1-reactive epitopes. Two approaches were taken to determine the specific epitopes recognized by the anti-CNF1 MAbs. First, truncated toxin molecules were used in Western blot analyses with the various MAbs to establish immunoreactivity patterns that indicated the relative position of each epitope. Second, a CNF1 epitope-expressing library was constructed and screened to ascertain the precise sequences of the reactive epitopes.
General localization of the CNF1-reactive epitopes.
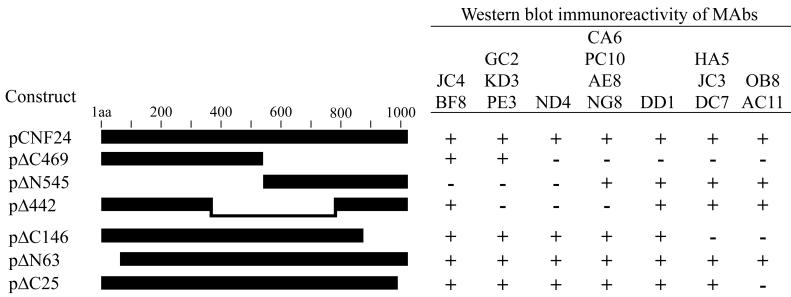
Epitopes recognized by the anti-CNF1 MAbs were localized by evaluating Western blot immunoreactivity patterns generated by each of the MAbs against a series of in-frame His-CNF1 deletion mutants (Fig. 2). This set of toxin deletion derivatives, comprised of toxin proteins with deletions of greater than a third of the toxin molecule, as well as relatively small amino- and carboxy-terminal truncations, allowed us to establish broad regions onto which the reactive epitopes mapped. Immunoblots were prepared with bacterial extracts containing each of the His-tagged toxin mutants and probed with each of the CNF1 MAbs (Fig. 2). Analysis of the reactivity patterns revealed that the CNF1 MAbs recognized epitopes located within seven distinct regions of the toxin. The epitopes detected by CNF1-neutralizing MAbs JC4 and BF8 lay within a 308-amino-acid N-terminal fragment that spanned amino acids 64 to 372. Neutralizing MAb NG8, as well as MAbs AE8, PC10, and CA6, reacted with epitopes in a region between amino acids 547 and 783. Based upon the immunoreactivity patterns produced by the remaining CNF1 MAbs, the positions of the other reactive epitopes were as follows: (i) MAbs GC2, KD3, and PE3 recognized epitopes contained between amino acids 373 and 546; (ii) MAbs HA5, JC3, and DC7 detected epitopes between amino acids 869 and 990; (iii) MAb DD1 reacted with an epitope between amino acids 784 and 868; and (iv) MAbs OB8 and AC11 recognized epitopes contained within the last 24 amino acids of the toxin. MAb ND4 recognized full-length toxin and the small deletion constructs ΔC25, ΔN63, and ΔC146 but could not detect Δ442 (amino acids 1 to 372 and amino acids 784 to 1014) or the N- and C-terminal halves of the toxin (ΔC469 and ΔN545). It should be noted that both pΔC469 and pΔN545 were constructed by the subcloning of PstI-digested cnf1 fragments. This suggests that the ND4 immunoreactive epitope may be encoded within this unique PstI site (amino acids 545 to 548). Another possibility may be that the epitope recognized by MAb ND4 lies proximal or distal to this site but that the conformation of this region is required for reactivity.
FIG. 2.
Schematic of the CNF1 toxin deletion mutants used to localize the epitopes recognized by the CNF1 MAbs (left) and Western blot immunoreactivity patterns obtained for each MAb (right). Amino acid residues deleted from each of the constructs were as follows: pΔC469, amino acids 547 to 1014; pΔN545, amino acids 1 to 546; pΔ442, amino acids 373 to 783; pΔC146, amino acids 869 to 1014; pΔN63, amino acids 1 to 62; and pΔC25, amino acids 990 to 1014. The pCNF24 construct expressed wild-type His-tagged toxin. Immunoreactive toxin proteins are indicated (+), as are those that were nonreactive (−).
Fine mapping of the epitopes recognized by CNF1 neutralizing MAbs.
Neutralization of CNF1 by MAbs JC4, BF8, and NG8 suggests that the epitopes recognized by these MAbs plays a functional role in at least one of the three probable stages of the CNF1 intoxication process, namely, toxin binding, translocation, and enzymatic activity. Western blot analyses showed that MAbs JC4 and BF8 recognized epitopes within a 308-amino-acid N-terminal domain and that MAb NG8 reacted with an epitope contained within a 236-amino-acid domain. Since the size of these regions provided only general localization information, more precise mapping of the epitopes that seemed to be critical for the biological activity of the toxin was undertaken. First, a commercially available epitope mapping kit was used to construct a plasmid-based CNF1 epitope library in which transformants expressed hybrid proteins containing small peptide fragments (16 to 50 amino acids) of CNF1. Immunological screening of the library thereby identifies reactive clones that can be sequenced to define the expressed CNF1 epitope.
With this system, immunoreactive clones were identified for only two CNF1 MAbs, even though more than 2,000 transformants were screened with the entire panel of anti-CNF1 MAbs. Five independent transformants demonstrated immunoreactivity with MAb DD1. Each of these five clones expressed CNF1 sequences within amino acids 750 to 825, and all shared a common epitope of 15 residues spanning amino acids 786 to 800 (YFYDNTVGLNGIPTL). MAb JC4 recognized three independent transformants that expressed CNF1 sequences from amino acids 165 to 215, and an epitope of 23 residues from amino acids 169 to 191 (YLYKNGELDEREYNFSMNALNRS) was common to all three clones. The remaining CNF1 MAbs failed to recognize any transformants. This lack of immunoreactivity, displayed by the majority of the MAbs to CNF1 epitope-expressing transformants, could reflect an unexpected bias in the cnf1 fragments that were cloned and expressed in the library. Alternatively, this might indicate the importance of adjacent sequences in the presentation and conformation of certain CNF1 epitopes.
While immunoreactive clones were not identified for CNF1-neutralizing MAbs NG8 or BF8, the results obtained from the DD1- and JC4-reactive transformants did provide additional information regarding the epitopes recognized by the two remaining neutralizing MAbs. One DD1-immunoreactive clone, which expressed a CNF1 peptide located between amino acids 755 and 825, did not cross-react with neutralizing MAb NG8 or MAbs AE8, PC10, or CA6 in either colony immunoscreens or in native dot blot analyses. In accordance with the results from previous Western blot analyses (Fig. 2), these findings suggested that the epitopes recognized by MAbs NG8, AE8, PC10, and CA6 were located between amino acids 547 and 754 but did not include amino acids 755 to 783. All three of the JC4-immunoreactive transformants which expressed CNF1 peptides between amino acids 165 to 215 failed to cross-react with MAb BF8 in Western and native dot blot analyses. These data, along with the previous immunoblot results, indicated that the epitope recognized by neutralizing MAb BF8 resides either between amino acids 64 and 164 or between amino acids 216 and 372.
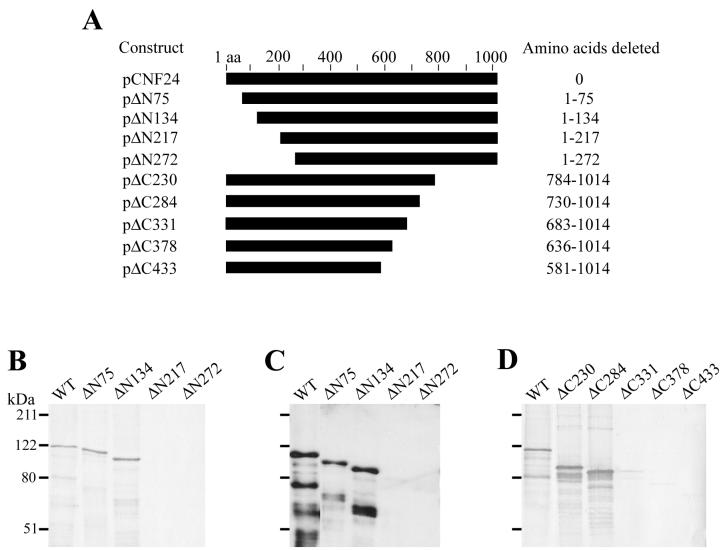
Immunoscreening of the CNF1 epitope library had identified a 23-amino-acid epitope recognized by MAb JC4; however, the location of the epitopes recognized by neutralizing MAbs BF8 and NG8 remained in large domains that spanned 100 to 200 amino acids. To more precisely define the reactive epitopes of MAbs BF8 and NG8, two additional sets of in-frame truncated toxin molecules were constructed for use in Western blot analyses. One set consisted of four CNF1 mutants that had progressively larger deletions from the N terminus of the toxin molecule, and these were used to ascertain the location of the epitope recognized by MAb BF8 (Fig. 3A). Another set of constructs was composed of five C-terminal CNF1 deletion derivatives that were used to map the NG8 reactive epitope (Fig. 3A). MAbs JC4 and BF8 produced identical immunoreactivity patterns in immunoblot analyses of the N-terminal truncated CNF1 molecules (Fig. 3B and C). Both of these neutralizing MAbs recognized wild-type CNF1 and toxins that had up to 134 amino acids removed from the N terminus (ΔN75 and ΔN134) but could not react with mutant toxins that had N-terminal deletions of 217 amino acids or greater (ΔN217 and ΔN272). These results imply that the epitopes recognized by MAbs JC4 and BF8 reside between amino acids 135 and 216. Taken together with the information obtained from the JC4 immunoreactive clones described previously, these findings strongly suggest that the BF8-reactive epitope lies between amino acids 135 and 164 and provides additional evidence in support of the location of the epitope recognized by MAb JC4 (amino acids 169 to 191) (Fig. 4).
FIG. 3.
Epitope mapping of the CNF1-neutralizing MAbs JC4, BF8, and NG8 by Western blot analyses with N- and C-terminal CNF1 deletion proteins. (A) Schematic of the CNF1 deletion derivatives used as antigens. The designation of each construct is given on the left, and the deleted portion of the toxin is indicated on the right. (B to D) Immunoblots of E. coli lysates that contained wild-type toxin and either the N- or the C-terminal truncated toxin molecules probed with hybridoma supernatants of MAb JC4 (B), MAb BF8 (C), or MAb NG8 (D). Reactive proteins were detected with appropriate HRP-conjugated secondary antibodies and DAB (MAbs JC4 and NG8) or ECL substrates (MAb BF8).
FIG. 4.
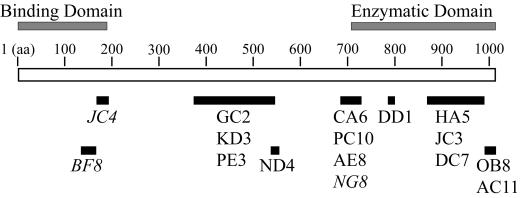
Proposed epitope map of the anti-CNF1 MAbs. The epitope of each MAb is denoted as a solid black bar below the linear sequence of CNF1. The putative CNF1 binding and enzymatic domains are indicated above the toxin molecule. The three neutralizing MAbs are shown in italics. Note that MAb BF8 also neutralized CNF2.
In Western blot analyses of the CNF1 C-terminal deletion mutants, MAb NG8 reacted with truncated proteins ΔC230 (amino acids 1 to 783) and ΔC284 (amino acids 1 to 729) but could not recognize mutants ΔC331 (amino acids 1 to 682), ΔC378 (amino acids 1 to 635), and ΔC433 (amino acids 1 to 580) (Fig. 3D). These findings suggest that the epitope recognized by MAb NG8 resides between amino acids 683 and 730 (Fig. 4). MAbs AE8, PC10, and CA6 had been shown previously to recognize epitopes between amino acids 547 and 754. Each of these three MAbs generated the same immunoreactivity patterns against the C-terminal deletion mutants as that seen with MAb NG8 (data not shown). These observations indicate that the reactive epitopes for MAbs AE8, PC10, and CA6 are also contained between amino acids 683 and 730 (Fig. 4).
DISCUSSION
From previous structure-function analyses, it appears that CNF1 can be organized into three functional domains which are reflective of the putative steps involved in the intoxication process, i.e., (i) binding of the toxin to a eukaryotic cell receptor, (ii) toxin internalization followed by translocation into the cytoplasm, and (iii) enzymatic activity on a specific cellular target. The region that contains the first 190 amino acids of CNF1 is considered to be the eukaryotic cell receptor binding domain, while a 300-amino-acid C-terminal portion of the toxin has demonstrated catalytic activity (13, 24, 35). Here, epitope mapping of 16 anti-CNF1 MAbs, including three with the capacity to neutralize toxin, has provided a way to delineate relatively small regions of CNF1 which appear to be critical for the biological function of the toxin (summarized in Fig. 4). The epitopes recognized by the three neutralizing MAbs are contained within the large domains that were originally established for the binding and catalytic activity of CNF1. However, these epitopes are independent of the smaller regions or residues that have been reported previously to be essential for function.
In separate reports, both Lemichez et al. (24) and Schmidt et al. (35) identified the C terminus of CNF1 (amino acids 720 to 1007 and amino acids 709 to 1014, respectively) as the catalytic domain through the use of glutathione S-transferase (GST)-CNF1 fusion proteins. Microinjection of this region alone into eukaryotic cells induces actin stress fiber formation, alters the electrophoretic mobility of the target Rho proteins, and abolishes the GTPase activity of these proteins in vitro. In an attempt to discern the precise region of enzymatic activity, Lemichez et al. (24) generated four smaller C-terminal GST-CNF1 fusion proteins that were based upon amino acid sequence similarities of CNF1 with DNT. However, none of these smaller fusion proteins could modify Rho in vitro, an observation that led the authors to suggest that amino acids essential for the deamidation activity lay throughout the C-terminal domain of the toxin. More recent findings with site-directed mutagenesis of the C-terminal half of CNF1 revealed that two residues are critical for the enzymatic activities of CNF1. Schmidt et al. (35) found that exchanging Cys866 with serine or exchanging His881 with alanine completely abolishes the deamidase and transglutaminase activities of CNF1 and therefore proposed that a cysteine-histidine catalytic dyad is contained within the C terminus of the toxin.
Similar types of structure-function analyses of DNT have demonstrated a C-terminal enzymatic domain with amino acids Cys1292, His1307, and Lys1310 essential for deamidation and transglutaminase activities (21, 33). Amino acid alignment of the C termini of CNF1 (amino acids 709 to 1014) and DNT (amino acids 1136 to 1451) indicates an overall identity of only 13%. However, a small region between amino acids 824 and 888 of CNF1 and amino acids 1250 and 1314 of DNT shares 45% identity (33, 39). The residues that have been reported to be essential for both CNF1- and DNT-mediated deamidation are contained within this region and appear to be functionally equivalent.
CNF1-neutralizing MAb NG8, as well as MAbs CA6, AE8, and PC10, recognizes epitopes located between amino acids 683 and 730. This 48-amino-acid region shares little homology with its aligned DNT counterpart and comprises only a small percentage of the putative enzymatic domain of CNF1. In contrast, this portion of CNF1 is 85% homologous with CNF2, and there is complete identity over the first 21 amino acids (14, 31). Since only one of the four CNF1 MAbs that maps to this region (MAb CA6) can recognize CNF2, the reactive epitope of CA6 probably lies between amino acids 683 and 703 (the region of shared CNF1 and CNF2 homology). Thus, the CNF1-specific MAbs AE8, PC10, and NG8 presumably recognize epitopes in the more disparate stretch between residues 704 and 730. The other MAbs that have been mapped to the catalytic domain of CNF1 (MAbs DD1, HA5, JC3, and DC7) do not exhibit significant CNF1 neutralization capacity, a finding that implies that these epitopes may not be essential for toxin function.
Based on amino acid sequence similarity, it has been suggested that CNF1 and PMT may bind to analogous cell receptors (24). CNF1 and PMT share 24% homology in the first 500 amino acids of their respective N termini and show the highest degree of similarity in regions that are proposed to encompass transmembrane domains in both toxin molecules (amino acids 199 to 420 of CNF1 and amino acids 245 to 467 of PMT) (14, 36, 40, 41). Despite this protein homology, recent work has indicated that the intracellular activity of PMT lies within the N terminus of the toxin. Using a voltage-clamped Xenopus oocyte assay system, Wilson et al. demonstrated that amino acids 1 to 568 of PMT could directly activate the inositol 1,4,5-trisphosphate (IP3) signaling pathway by targeting the Gq/11α-protein, whereas a C-terminal PMT molecule showed no activity in the oocyte assay (41). These findings, in addition to earlier studies that indicated that CNF1 cannot activate the IP3 pathway in Swiss 3T3 cells (22), suggest that CNF1 and PMT do not share either functional N-terminal binding domains or biological activities.
In recent work by Fabbri et al. (13), the authors proposed that the CNF1 receptor-binding domain lies within the first 190 amino acids of the toxin molecule. Fabbri et al. performed competitive binding assays with wild-type toxin and a series of GST-CNF1 N-terminal truncated fusion proteins and found that a fusion protein composed of amino acids 1 to 191 of CNF1 can inhibit the multinucleation activity of wild-type toxin when used at concentrations higher than 10−7 M. Furthermore, GST-CNF1 fusions that contain amino acids 54 to 191 or amino acids 76 to 191 exhibit only partial inhibition or completely fail, respectively, to protect cells from the effects of wild-type toxin. Although these results suggest that amino acids 53 to 75 are important in promoting the binding of CNF1 to its eukaryotic receptor, a synthetic peptide spanning this region did not inhibit toxin-mediated multinucleation. This latter finding led the authors to speculate that amino acids 53 to 75 may represent only a part of a receptor recognition site or that this predominantly hydrophilic region could instead be essential for the proper folding and exposure of a yet-unidentified binding domain.
Our findings suggest that a different region of the first 190 amino acids of CNF1 is critical for toxin function. Specifically, two of the CNF1-neutralizing MAbs identified here map to the toxin-binding domain contained between amino acids 1 and 190 but do not include the aforementioned hydrophilic block. The epitope recognized by MAb JC4 lies between amino acids 169 and 191, while the reactive epitope for MAb BF8 has been localized to amino acids 135 to 164. Both of these MAbs have demonstrated the capacity to recognize CNF2 in Western blot analyses. Nevertheless, only MAb BF8 appears to neutralize CNF2. The latter observation suggests that this 30-amino-acid stretch between amino acids 135 and 164 may contain the cell receptor recognition site for both CNF1 and CNF2.
The anti-CNF1 MAb mapping data described in the present study have identified three new regions of the toxin molecule that appear to be critical for function. Two of these regions are contained within the toxin binding domain, while the third resides at the edge of the catalytic domain. Based on these localization patterns, it is tempting to speculate that MAbs JC4 and BF8 block toxin binding, while MAb NG8 interferes with the deamidation of Rho. Studies are in progress to support or refute this hypothesis.
The MAbs generated and characterized here also may yield greater insight into the disparate nature of the biological activities attributed to CNF1 and CNF2. These two toxins share a high degree of amino acid identity and have similar catalytic activities (14, 17, 31, 34, 37), and yet CNF1 is a more potent inducer of multinucleation in vitro, while CNF2 appears to produce more necrosis in both mouse footpad assays and rabbit skin tests (5, 10). By exchanging the CNF1-specific or cross-reactive epitopes between the two toxins, new information may be revealed concerning the substrate and receptor specificities of CNF1 and CNF2.
ACKNOWLEDGMENTS
This work was supported by grant AI38281-05 from the National Institutes of Health.
We thank Edda Twiddy for assistance in the cloning and maintenance of the hybridoma cell lines.
REFERENCES
- 1.Aktories K. Rho proteins: targets for bacterial toxins. Trends Microbiol. 1997;5:282–288. doi: 10.1016/S0966-842X(97)01067-6. [DOI] [PubMed] [Google Scholar]
- 2.Alonso P, Blanco J, Blanco M, Gonzalez E A. Frequent production of toxins by Escherichia coli strains isolated from human urinary tract infections: relation with haemagglutination. FEMS Microbiol Lett. 1987;48:391–396. [Google Scholar]
- 3.Andreu A, Stapleton A E, Fennell C, Lockman H A, Xercavins M, Fernandez F, Stamm W E. Urovirulence determinants in Escherichia coli strains causing prostatitis. J Infect Dis. 1997;176:464–469. doi: 10.1086/514065. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1989. [Google Scholar]
- 5.Blanco J, Blanco M, González E A, Pilar Alonso M, Garabal J I. Comparative evaluation of three tests for the detection of Escherichia coli cytotoxic necrotizing factors (CNF1 and CNF2) using filtrates of cultures treated with mitomycin C. FEMS Microbiol Lett. 1990;69:311–316. doi: 10.1016/0378-1097(90)90086-6. [DOI] [PubMed] [Google Scholar]
- 6.Blum G, Falbo V, Caprioli A, Hacker J. Gene clusters encoding the cytotoxic necrotizing factor type 1, Prs-fimbriae and α-hemolysin form the pathogenicity island II of the uropathogenic Escherichia coli strain J96. FEMS Microbiol Lett. 1995;126:189–196. doi: 10.1111/j.1574-6968.1995.tb07415.x. [DOI] [PubMed] [Google Scholar]
- 7.Boquet P. Cytotoxic necrotizing factor 1 from Escherichia coli: a toxin with a new intracellular activity for eukaryotic cells. Folia Microbiol. 1998;43:285–289. doi: 10.1007/BF02818614. [DOI] [PubMed] [Google Scholar]
- 8.Caprioli A, Falbo V, Roda L G, Ruggeri F M, Zona C. Partial purification and characterization of an Escherichia coli toxic factor that induces morphological cell alterations. Infect Immun. 1983;39:1300–1306. doi: 10.1128/iai.39.3.1300-1306.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caprioli A, Falbo V, Ruggeri F M, Baldassarri L, Bisicchia R, Ippolito G, Romoli E, Donelli G. Cytotoxic necrotizing factor production by hemolytic strains of Escherichia coli causing extraintestinal infections. J Clin Microbiol. 1987;25:146–149. doi: 10.1128/jcm.25.1.146-149.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Rycke J, Gonzalez E A, Blanco J, Oswald E, Blanco M, Boivin R. Evidence for two types of cytotoxic necrotizing factor in human and animal clinical isolates of Escherichia coli. J Clin Microbiol. 1990;28:694–699. doi: 10.1128/jcm.28.4.694-699.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Rycke J, Phan-Thanh L, Bernard S. Immunochemical identification and biological characterization of cytotoxic necrotizing factor from Escherichia coli. J Clin Microbiol. 1989;27:983–988. doi: 10.1128/jcm.27.5.983-988.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donelli G, Fiorentini C. Cell injury and death caused by bacterial protein toxins. Toxicol Lett. 1992;64/65:695–699. doi: 10.1016/0378-4274(92)90249-j. [DOI] [PubMed] [Google Scholar]
- 13.Fabbri A, Gauthier M, Boquet P. The 5′ region of cnf1 harbours a translational regulatory mechanism for CNF1 synthesis and encodes the cell-binding domain of the toxin. Mol Microbiol. 1999;33:108–118. doi: 10.1046/j.1365-2958.1999.01453.x. [DOI] [PubMed] [Google Scholar]
- 14.Falbo V, Pace T, Picci L, Pizzi E, Caprioli A. Isolation and nucleotide sequence of the gene encoding cytotoxic necrotizing factor 1 of Escherichia coli. Infect Immun. 1993;61:4909–4914. doi: 10.1128/iai.61.11.4909-4914.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiorentini C, Arancia G, Caprioli A, Falbo V, Ruggeri F M, Donelli G. Cytoskeletal changes induced in HEp-2 cells by the cytotoxic necrotizing factor of Escherichia coli. Toxicon. 1988;26:1047–1056. doi: 10.1016/0041-0101(88)90203-6. [DOI] [PubMed] [Google Scholar]
- 16.Flatau G, Landraud L, Boquet P, Bruzzone M, Munro P. Deamidation of RhoA glutamine 63 by the Escherichia coli CNF1 toxin requires a short sequence of the GTPase switch 2 domain. Biochem Biophys Res Commun. 2000;267:588–592. doi: 10.1006/bbrc.1999.1904. [DOI] [PubMed] [Google Scholar]
- 17.Flatau G, Lemichez E, Gauthier M, Chardin P, Paris S, Fiorentini C, Boquet P. Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature. 1997;387:729–733. doi: 10.1038/42743. [DOI] [PubMed] [Google Scholar]
- 18.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 19.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 20.Horiguchi Y, Inoue N, Masuda M, Kashimoto T, Katahira J, Sugimoto N, Matsuda M. Bordetella bronchiseptica dermonecrotizing toxin induces reorganization of actin stress fibers through deamidation of Gln-63 of the GTP-binding protein Rho. Proc Natl Acad Sci USA. 1997;94:11623–11626. doi: 10.1073/pnas.94.21.11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kashimoto T, Katahira J, Cornejo W R, Masuda M, Fukuoh A, Matsuzawa T, Ohnishi T, Horiguchi Y. Identification of functional domains of Bordetella dermonecrotizing toxin. Infect Immun. 1999;67:3727–3732. doi: 10.1128/iai.67.8.3727-3732.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lacerda H M, Pullinger G D, Lax A J, Rozengurt E. Cytotoxic necrotizing factor 1 from Escherichia coli and dermonecrotic toxin from Bordetella bronchiseptica induce p21rho-dependent tyrosine phosphorylation of focal adhesion kinase and paxillin in Swiss 3T3 cells. J Biol Chem. 1997;272:9587–9596. doi: 10.1074/jbc.272.14.9587. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Lemichez E, Flatau G, Bruzzone M, Boquet P, Gauthier M. Molecular localization of the Escherichia coli cytotoxic necrotizing factor CNF1 cell-binding and catalytic domains. Mol Microbiol. 1997;24:1061–1070. doi: 10.1046/j.1365-2958.1997.4151781.x. [DOI] [PubMed] [Google Scholar]
- 25.Lerm M, Schmidt G, Aktories K. Bacterial protein toxins targeting Rho GTPases. FEMS Microbiol Lett. 2000;188:1–6. doi: 10.1111/j.1574-6968.2000.tb09159.x. [DOI] [PubMed] [Google Scholar]
- 26.Lerm M, Schmidt G, Goehring U-M, Schirmer J, Aktories K. Identification of the region of Rho involved in substrate recognition by Escherichia coli cytotoxic necrotizing factor 1 (CNF1) J Biol Chem. 1999;274:28999–29004. doi: 10.1074/jbc.274.41.28999. [DOI] [PubMed] [Google Scholar]
- 27.Lerm M, Selzer J, Hoffmeyer A, Rapp U R, Aktories K, Schmidt G. Deamidation of Cdc42 and Rac by Escherichia coli cytotoxic necrotizing factor 1: activation of c-Jun N-terminal kinase in HeLa cells. Infect Immun. 1999;67:496–503. doi: 10.1128/iai.67.2.496-503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masuda M, Betancourt L, Matsuzawa T, Kashimoto T, Takao T, Shimonishi Y, Horiguchi Y. Activation of Rho through a cross-link with polyamines catalyzed by Bordetella dermonecrotizing toxin. EMBO J. 2000;19:521–530. doi: 10.1093/emboj/19.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mills M, Meysick K C, O'Brien A D. Cytotoxic necrotizing factor type 1 of uropathogenic Escherichia coli kills cultured human uroepithelial 5637 cells by an apoptotic mechanism. Infect Immun. 2000;68:5869–5880. doi: 10.1128/iai.68.10.5869-5880.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitsumori K, Terai A, Yamamoto S, Ishitoya S, Yoshida O. Virulence characteristics of Escherichia coli in acute bacterial prostatitis. J Infect Dis. 1999;180:1378–1381. doi: 10.1086/314976. [DOI] [PubMed] [Google Scholar]
- 31.Oswald E, Sugai M, Labigne A, Wu H C, Fiorentini C, Boquet P, O'Brien A D. Cytotoxic necrotizing factor type 2 produced by virulent Escherichia coli modifies the small GTP-binding proteins Rho involved in assembly of actin stress fibers. Proc Natl Acad Sci USA. 1994;91:3814–3818. doi: 10.1073/pnas.91.9.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Schmidt G, Goehring U-M, Schirmer J, Lerm M, Aktories K. Identification of the C-terminal part of Bordetella dermonecrotic toxin as a transglutaminase for Rho GTPases. J Biol Chem. 1999;274:31875–31881. doi: 10.1074/jbc.274.45.31875. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt G, Sehr P, Wilm M, Selzer J, Mann M, Aktories K. Gln63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature. 1997;387:725–729. doi: 10.1038/42735. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt G, Selzer J, Lerm M, Aktories K. The Rho-deamidating cytotoxic necrotizing factor 1 from Escherichia coli possesses transglutaminase activity: cysteine 866 and histidine 881 are essential for enzyme activity. J Biol Chem. 1998;273:13669–13674. doi: 10.1074/jbc.273.22.13669. [DOI] [PubMed] [Google Scholar]
- 36.Smyth M G, Sumner I G, Lax A J. Reduced pH causes structural changes in the potent mitogenic toxin of Pasteurella multocida. FEMS Microbiol Lett. 1999;180:15–20. doi: 10.1111/j.1574-6968.1999.tb08772.x. [DOI] [PubMed] [Google Scholar]
- 37.Sugai M, Hatazaki K, Mogami A, Ohta H, Pérès S Y, Hérault F, Horiguchi Y, Masuda M, Ueno Y, Komatsuzawa H, Suginaka H, Oswald E. Cytotoxic necrotizing factor type 2 produced by pathogenic Escherichia coli deamidates a Gln residue in the conserved G-3 domain of the Rho family and preferentially inhibits the GTPase activity of RhoA and Rac1. Infect Immun. 1999;67:6550–6557. doi: 10.1128/iai.67.12.6550-6557.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swenson D L, Bukanov N O, Berg D E, Welch R A. Two pathogenicity islands in uropathogenic Escherichia coli J96: cosmid cloning and sample sequencing. Infect Immun. 1996;64:3736–3743. doi: 10.1128/iai.64.9.3736-3743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker K E, Weiss A A. Characterization of the dermonecrotic toxin in members of the genus Bordetella. Infect Immun. 1994;62:3817–3828. doi: 10.1128/iai.62.9.3817-3828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward P N, Miles A J, Sumner I G, Thomas L H, Lax A J. Activity of the mitogenic Pasteurella multocida toxin requires an essential C-terminal residue. Infect Immun. 1998;66:5636–5642. doi: 10.1128/iai.66.12.5636-5642.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson B A, Ponferrada V G, Vallance J E, Ho M. Localization of the intracellular activity domain of Pasteurella multocida toxin to the N terminus. Infect Immun. 1999;67:80–87. doi: 10.1128/iai.67.1.80-87.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]