Abstract
Attachment of Mycobacterium avium subsp. paratuberculosis to host tissue and penetration of mucosal surfaces are pivotal events in the pathogenesis of Johne's disease. Fibronectin (FN) binding is required for attachment and internalization of several mycobacteria by epithelial cells in vitro. The objective of this study was to further characterize the FN binding activity of M. avium subsp. paratuberculosis. Although the bacteria bound FN poorly at pH above 7, brief acid pretreatment greatly enhanced FN binding within the pH range (3 to 10) studied. A 4.6-kbp fragment from an M. avium subsp. paratuberculosis genomic library was found to contain a 1,107-bp open reading frame that shows very high nucleotide sequence identity with that of the FN attachment protein (FAP) gene of M. avium subsp. avium. Pretreatment of FN with an FN-binding peptide from M. avium subsp. avium FAP abolished FN binding, indicating that M. avium subsp. paratuberculosis binds FN in a FAP-dependent manner. Pretreatment of M. avium subsp. paratuberculosis with anti-FAP immunoglobulin G did not abrogate FN binding; blocking occurred only when anti-FAP was added together with FN. FAP was detected by immunofluorescence only in lipid-extracted M. avium subsp. paratuberculosis. Western blotting and immunoelectron microscopy revealed that FAP is located near the interior of the cell envelope of M. avium subsp. paratuberculosis. The results indicate that a FAP homologue mediates the attachment of FN to M. avium subsp. paratuberculosis. Further, given the subcellular location of FAP, it is considered that this protein operates at the terminus of a coordinated FN binding system in the cell envelope of M. avium subsp. paratuberculosis.
Johne's disease, or paratuberculosis, is a chronic intestinal disease of cattle and ruminants that is caused by Mycobacterium avium subsp. paratuberculosis. This acid-fast bacillus invades macrophages in lymphoid tissue in the ileum, where it inhibits phagosome maturation and induces the recruitment of inflammatory cells, resulting in granulomatous enteritis. These granulomas cause the intestinal villi to become distorted, leading to the loss of absorptive surface area. Consequently, the animal's ability to absorb nutrients through the intestine is greatly impaired, leading ultimately to wasting and death. Although animals are usually infected before 6 months of age by ingesting contaminated feces, the profuse diarrhea and wasting are not observed until several years later, complicating the effort to control disease.
Johne's disease causes severe economic losses to the dairy cattle industry. A recent survey estimated that 21.6% of dairy herds in the United States are infected with M. avium subsp. paratuberculosis (21). Annual losses to the cattle industry due to Johne's disease have been estimated to be as high as $1 billion (29), principally due to lost milk production and the reduced value of clinically affected cull cows.
Studies with neonatal bovine calves demonstrated that M. avium subsp. paratuberculosis enters intestinal tissue through M cells, which are found in the dome epithelium covering Peyer's patches (17). Unlike villous enterocytes, which restrict integrin expression to their basolateral surfaces, M cells display integrins at high density on their apical surfaces (9). Many of these surface integrins contain binding sites for fibronectin (FN), a ubiquitous 450-kDa glycoprotein found in body fluids and extracellular matrix (ECM) of vertebrates.
Binding of FN has been shown to be important for attachment and internalization of many bacterial species by cultured cells. In addition, FN binding by Streptococcus pyogenes correlated with persistent carriage in humans (20), and disruption of FN binding in Staphylococcus aureus and Streptococcus sanguis led to attenuated virulence in animal models of infective endocarditis (7, 15). FN attachment proteins (FAPs) are a family of FN-binding proteins that present in several species of mycobacteria (25, 27, 28). Although these organisms may express several FN-binding proteins (32), FN-binding peptides from FAP abrogated in vitro FN binding by M. bovis BCG and M. avium subsp. avium (27, 37). Addition of recombinant FAP to human respiratory tract organ cultures led to dose-dependent inhibition of M. avium subsp. avium binding to ECM in areas of epithelial damage. In this experimental system, FAP-knockout mutants of M. smegmatis failed to bind to exposed ECM; binding was restored when the mutation was complemented by the M. avium subsp. avium FAP (FAP-A) gene on a multicopy plasmid (16). Further, internalization of mycobacteria by cultured epithelial cells and Schwann cells has been shown to occur in a FAP-dependent manner using polyclonal antibodies raised against M. vaccae FAP, and this process is mediated by β1 integrins on host cells (13, 28).
An FN-binding protein had been detected in M. avium subsp. paratuberculosis (31), but its identity was unknown. Moreover, for FN binding to play a role in the pathogenesis of M. avium subsp. paratuberculosis, this activity must be maintained as the organism passes through the host digestive tract. We sought to further characterize FN binding by M. avium subsp. paratuberculosis by examining the effect of pH on FN binding and to identify the FN-binding protein that mediates this activity. We observed that the efficiency of FN binding was pH dependent and could be enhanced by acid treatment. M. avium subsp. paratuberculosis was found to contain coding sequences for a FAP homologue and bound FN in a FAP-dependent manner. However, the FAP homologue was not expressed on the cell surface and was therefore unavailable for direct interaction with FN.
MATERIALS AND METHODS
Bacterial strains.
M. avium subsp. paratuberculosis strains 5781 and 6594 were recovered from feces of adult beef cattle with clinical paratuberculosis. Both were propagated in Middlebrook 7H9 broth (Becton Dickinson, Cockeysville, Md.) supplemented with 10% oleic acid-albumin-dextrose-catalase (Becton Dickinson), 0.05% Tween 80 (Sigma, St. Louis, Mo.), and 2 μg of mycobactin J (Allied Monitor, Fayetteville, Mo.) per ml in tissue culture flasks. Bacteria were grown to mid-logarithmic phase for all experiments.
Ligands, antisera, and peptides.
Bovine FN and bovine serum albumin (BSA) were obtained from Sigma and were biotinylated using a commercial kit (Pierce, Rockford, Ill.). Rabbit antiserum raised against M. vaccae FAP (anti-FAP antiserum) was a generous gift from J. S. Schorey (University of Notre Dame, South Bend, Ind.). Normal rabbit serum (NRS) was collected from female New Zealand White rabbits (Covance, Denver, Pa.). Immunoglobulin G (IgG) was purified from anti-FAP antiserum and NRS by protein A-agarose chromatography (Pierce). Rabbit anti-M. bovis BCG (anti-BCG) IgG was obtained from Dako (Carpinteria, Calif.). Fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG was purchased from Antibodies, Inc. (Davis, Calif.). Goat anti-rabbit IgG conjugated to horseradish peroxidase and gold-conjugated goat anti-rabbit IgG were acquired from Sigma. Peptides were synthesized using fluorenylmethoxycarbonyl chemistry and purified by high-pressure liquid chromatography (Gemini Biotech, Alachua, Fla.). FAPA 269-292 (GNRQRWFVVWLGTSSNDPVDKVAAK) corresponds to amino acid positions 269 to 292 of FAP-A (27). The scrambled peptide sequence WNQVTFAGPNWDVLKKVGRRVADS was synthesized as a control (27).
FN binding assay.
Bacterial cultures were centrifuged for 20 min at 1,000 × g, and the cell pellet was resuspended in phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBST) (Sigma). Vigorous pipetting and vortexing yielded a predominantly single-cell suspension, as judged by forward scatter in flow cytometry. Cell density was adjusted to 2 × 108 CFU/ml with PBST, and 100 μl of the cell suspension was added to each well of a 96-well V-bottom white microtiter plate (Whatman, Clifton, N.J.). Microplates containing bacteria were centrifuged for 15 min at 1,500 × g and then quickly inverted to decant the supernatant. Cell pellets were resuspended in 100 μl of either ANT buffer (10 mM ammonium acetate, 0.85% sodium chloride, 0.05% Tween 20) adjusted to pH 3, 4, or 6 or TBST (10 mM Tris-HCl, 0.85% sodium chloride, 0.05% Tween 20) adjusted to pH 7, 8, or 10. Biotinylated FN was diluted to 2 μg/ml in each of the above buffers, and 100 μl of each solution was added to the corresponding cell wells. Biotinylated BSA, prepared similarly, was added to companion cell wells as negative controls. Wells containing cells and buffer only were used as baseline controls. After FN binding was allowed to proceed for 90 min at room temperature, microplates were centrifuged as before, and cells were washed in PBST to remove unbound ligand. Cell pellets were then resuspended in 100 μl of Neutravidin-horseradish peroxidase conjugate (1 ng/ml in PBST; Pierce) and incubated for 30 min at room temperature. Following centrifugation and washing, cell pellets were resuspended in 100 μl of chemiluminescent peroxidase substrate solution (SuperSignal Pico; Pierce). Reactions were read in a TopCount luminometer (Packard, Meriden, Conn.).
This protocol was modified as follows for acid pretreatment experiments. After addition of bacteria to microplate wells (2 × 107 CFU/well), the microplate was centrifuged, and the supernatant was poured off. Cell pellets were resuspended in 100 μl of ANT (pH 3) and incubated for 5 min at room temperature. Cells were then pelleted and resuspended in the appropriate buffer followed by the addition of biotinylated ligand, and the assay proceeded as described above. For neutralization experiments, cells were resuspended in 100 μl of PBST (pH 7) following a 5-min acid pretreatment and incubated for 15 min at room temperature. The microplates were then centrifuged, and cell pellets were resuspended in the appropriate buffer. The assay continued as outlined above. Statistical evaluation was conducted by one-way analysis of variance and Tukey's multiple comparison test.
PCR amplification, library cloning, and DNA sequencing.
Primers FAPA-1 (CCCGCCGGCTGGGTGGAGTC) and FAPA-2 (CCACGCCTGGATCGACTCGGC) were designed to amplify a highly conserved region of FAP from M. tuberculosis, M. bovis, M. leprae, and M. avium subsp. avium (GenBank accession no. X80268, AF013569, L01095, and U53585, respectively). M. avium subsp. paratuberculosis strain 5781 genomic DNA used for PCR amplification was prepared by glass bead lysis (12). The PCR product was cloned into pcDNA3.1/NT-GFP (Invitrogen, Carlsbad, Calif.). The resulting clone, designated pTS011, was sequenced using dye terminators on an ABI Prism 377 DNA sequencer (Applied Biosystems, Foster, Calif.; ACGT, Northbrook, Ill.).
Intact genomic DNA was extracted from strain 5781 by the enzymatic lysis method (6) and digested overnight with a 20-fold excess of EcoRI (Promega, Madison, Wis.). DNA fragments were ligated into λ ZAP II (Stratagene, La Jolla, Calif.) and packaged in vitro using the Gigapack II Gold packaging extract as instructed by the manufacturer (Stratagene). The resulting primary library had a titer of 2 × 105 PFU/ml and contained >80% recombinant phage. Probe DNA was synthesized by PCR amplification of a 415-bp fragment of the insert contained in pTS011 with primers FAPA-1B (CGACGCGTCTCACCTCGGCTACGGCTCGGCGC) and FAPA-2B (CTTGTCCACCGGGTCGTTCGAGGTGCCCAGCCA) in the presence of digoxigenin-11-dUTP, as specified by the manufacturer (Roche Molecular Biochemicals, Indianapolis, Ind.), and was used to probe nylon membranes containing approximately 2 × 104 plaques. Hybridization and stringency washes were carried out at 72°C. Digoxigenin-labeled hybrids were detected in accordance with the manufacturer's recommendations. Reactive plaques were plaque purified and reprobed twice. Recombinant pBluescript SK− phagemids were excised from purified λ phage using ExAssist helper phage (Stratagene). One phagemid, designated pTS036, contained a 4.6-kb insert and was selected for sequencing as described above.
Southern hybridization.
Genomic DNA was extracted from M. avium subsp. paratuberculosis 5781 and 6594 by the enzymatic lysis method (6) and digested overnight with EcoRI (Promega). Following agarose gel electrophoresis, DNA fragments were transferred to a nylon membrane. Probe DNA was prepared as above. Following hybridization and stringency washes at 72°C, digoxigenin-labeled hybrids were detected as specified by the manufacturer (Roche Molecular Biochemicals).
Peptide blocking assay.
Biotinylated FN was added to a final concentration of 2 μg/ml to solutions containing either FAPA 269-292 or scrambled peptide (2.5 μg/ml in ANT [pH 6]) and allowed to react at room temperature for 1 h. One hundred microliters of either FN peptide solution was added to microplate wells containing 2 × 107 CFU of acid-pretreated M. avium subsp. paratuberculosis in 100 μl of ANT (pH 6) and allowed to react with the cells for 90 min at room temperature. Bound FN was detected as described above. Percent inhibition was determined by dividing counts from peptide-treated wells by that from positive control wells (containing biotinylated FN only), subtracting this quotient from 1, and multiplying by 100. Data were compared by Student's t test.
Antibody blocking assays.
For one set of experiments, 2 × 107 CFU of acid-pretreated M. avium subsp. paratuberculosis was reacted in microplate wells with various concentrations of either anti-FAP IgG or normal rabbit (NRb) IgG in ANT (pH 6) for 1 h prior to the addition of biotinylated FN. In other experiments, biotinylated FN was added to acid-pretreated cell suspensions immediately following the addition of blocking antibody in ANT (pH 6). FN binding was assayed as described above. Percent inhibition was determined as above. An inhibition index was calculated by dividing counts from anti-FAP IgG-treated wells by that from NRb IgG-treated wells and substracting the quotient from 1. Data were evaluated by one-way analysis of variance and Tukey's multiple comparison test.
Immunofluorescence analysis.
Cell wall lipids were removed from bacteria by extracting PBST-washed bacterial cells with 2:1 chloroform-methanol for 5 min. The extraction mixture was centrifuged for 20 min at 2,000 × g. The aqueous and organic phases were carefully collected and discarded, and the remaining cellular interface was heated to 50°C for 20 min. The interface was resuspended in PBST and spotted on individual wells of treated microscope slides (Erie Scientific, Portsmouth, N.H.). Washed acid-treated or untreated cells were spotted in parallel wells. Suspensions were air dried and then fixed for 10 min in cold acetone. Anti-FAP antiserum, NRS, and anti-BCG IgG were diluted 1:100 in PBST and added separately to both extracted and untreated cells. Negative control wells received PBST only. After incubation in a humidity chamber for 30 min at room temperature, slides were rinsed and washed for 10 min in PBS. Wells were probed with a 1:100 dilution of FITC-conjugated goat anti-rabbit IgG for 30 min at room temperature, and slides were washed as before. After brief air drying, coverslips were mounted and slides were examined with an Optiphot-2 epifluorescence microscope (Nikon, Melville, N.Y.).
Western blotting of cell lysate fractions.
Thirty milliliters of bacterial culture was centrifuged 20 min at 1,200 × g. Cell pellets were resuspended in 35 ml of PBS and centrifuged again. Pellets were resuspended in 1 ml of PBS, and phenylmethylsulfonyl fluoride (Sigma) was added to a final concentration of 1 mM. Suspensions were sonicated in three 30-s bursts with 20-s cooling intervals. Sonicates were fractionated essentially as described elsewhere (22). Briefly, a portion of the sonicate was retained as the whole cell lysate, while the bulk of the sonicate was centrifuged for 10 min at 13,000 × g. The supernatant was regarded as the crude cytoplasmic fraction (including cytoplasm, cytoplasmic membrane, and soluble material between the cytoplasmic membrane and mycolate layer of the cell wall), and the pellet was considered the cell wall fraction. Triton X-114 (Sigma) extraction was used to partition cell wall proteins into aqueous and detergent fractions. A portion of the crude cytoplasmic fraction was centrifuged at 100,000 × g for 2 h. The resulting pellet was resuspended in a minimal volume of PBST and considered the cytoplasmic membrane fraction. Components of each fraction were separated on sodium dodecyl sulfate–12% polyacrylamide gels and electroblotted to nitrocellulose membranes. After blocking membranes in PBST with 5% BSA (PBSTB), blots were incubated with anti-FAP antiserum diluted 1:2,000 in PBSTB. Bound antibodies were detected with a 1:400 dilution of goat anti-rabbit IgG in PBSTB and developed using diaminobenzidine (Sigma).
Immunoelectron microscopy.
Bacterial cells were fixed in 4% paraformaldehyde and 0.1% glutaraldehyde in 0.15 M phosphate buffer (Millonig's buffer) for 3 h at 4°C. Fixed cells were washed twice in Millonig's buffer two times for 10 min each and pelleted in 2% low-gelling-temperature agarose (Sigma). Following dehydration with increasing concentrations of ethanol, the cell bead was embedded in LR white (Electron Microscopy Sciences, Fort Washington, Pa.). After blocking briefly with Tris-buffered saline with 0.3% Tween 20 and 1% BSA, thin sections were incubated overnight at 4°C with a 1:750 dilution of anti-FAP antiserum. Bound anti-FAP antibody was labeled with goat anti-rabbit IgG conjugated to 10-nm gold particles. Sections were stained with uranyl acetate and lead citrate and examined with a JEOL (Palo Alto, Calif.) 100 CX transmission electron microscope.
Nucleotide sequence accession number.
The sequence shown in Fig. 2 has been assigned GenBank accession no. AY007557.
FIG. 2.
Nucleotide and predicted amino acid sequences of FAP-P. The putative Shine-Dalgarno sequence is boxed, and the N-terminal signal sequence is underlined. Regions of FAP-P that correspond to the FN-binding peptides of FAP-A (27, 37) are in boldface.
RESULTS
Effect of pH on FN binding.
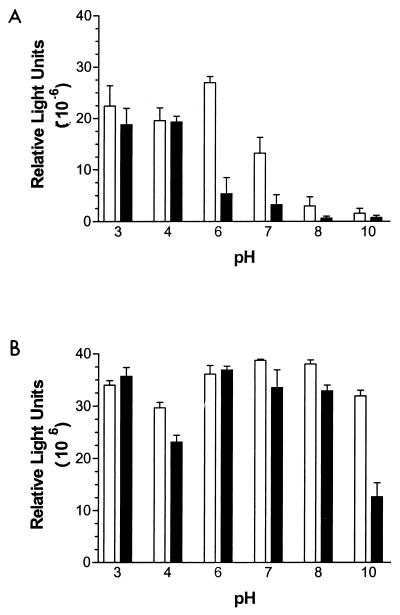
To examine the effect of pH on FN binding by M. avium subsp. paratuberculosis, we exposed bacteria to a range of FN solutions at different pH. In general, FN binding was highest at acidic pH and was low to negligible at pH above 7 (Fig. 1A). The two M. avium subsp. paratuberculosis strains differed with respect to their pH optima. Strain 5781 bound FN equally well at pH 3 to 6 (P = 0.2186), whereas FN binding by strain 6594 was significantly lower at pH 6 than at pH 3 or 4 (P < 0.05).
FIG. 1.
Effect of pH on FN binding by M. avium subsp. paratuberculosis. Bacteria (2 × 107 CFU) were resuspended in pH 3, 4, 6, 7, 8, or 10 buffer, and 200 ng of biotinylated FN, prepared in each buffer, was added to the appropriate wells. After 90 min, bound FN was determined by a chemiluminescence assay. □, strain 5781; ■, strain 6594. (A) No pretreatment. (B) M. avium subsp. paratuberculosis were treated for 5 mins with pH 3 buffer at room temperature and then incubated with FN prepared in pH 3, 4, 6, 7, 8, or 10 buffer. Data shown represent the mean and standard error of the mean of four replicates for each treatment.
It was not clear from these experiments whether acidophilic binding of FN by M. avium subsp. paratuberculosis reflected a favored conformation assumed by FN at acidic pH or if the FN-binding activity of the organism was enhanced at low pH. To determine this, M. avium subsp. paratuberculosis cells were briefly pretreated with pH 3 buffer before addition of FN. As shown in Fig. 1B, pretreatment enhanced FN binding by M. avium subsp. paratuberculosis, particularly at pH above 7. The enhancement effect was significantly lower for binding at pH 4 (strain 5781, P < 0.05; strain 6594, P < 0.01), such that the pH-dependent binding profiles were bimodal for acid-pretreated cells of both strains. The original acidophilic FN binding profile was essentially restored when acid-pretreated cells were neutralized prior to the addition of FN (data not shown). These observations demonstrate that FN binding by M. avium subsp. paratuberculosis is favored at low pH and that this effect results primarily from acid-mediated alteration of the binding capability of the organism. Acid-pretreated M. avium subsp. paratuberculosis was used in each subsequent FN binding assay.
Detection of an open reading frame (ORF) encoding a FAP homologue.
To identify sequences within the M. avium subsp. paratuberculosis genome that encode a FAP homologue, PCR was performed with oligonucleotide primers that flank a region that is highly conserved among FAPs (27). Sequencing of the cloned PCR product (pTS011) revealed a 474-bp fragment that showed 98% sequence identity with the corresponding region of M. avium subsp. avium fap (data not shown). A 415-bp probe generated by PCR from pTS011 hybridized with a 4.6-kbp fragment on Southern blots of EcoRI-digested M. avium subsp. paratuberculosis 5781 and 6594 DNAs (data not shown). These observations suggested that M. avium subsp. paratuberculosis contained coding information for a FAP homologue and that this sequence was present as a single copy in the M. avium subsp. paratuberculosis genome.
We constructed a λ ZAP II library of EcoRI-digested M. avium subsp. paratuberculosis 5781 DNA to recover the entire coding sequence of the M. avium subsp. paratuberculosis FAP homologue. Approximately 15 to 20 probe-reactive plaques were observed among every 2 × 104 recombinant plaques upon primary screening of the amplified library. After plaque purification and excision, all phagemids produced a 415-bp band when amplified in the presence of the FAPA-1B–FAPA-2B primer pair. The insert contained in one of these phagemids (pTS036) was sequenced bidirectionally using primer walking from both internal sequences (obtained from pTS011 sequencing) and vector DNA sequences, revealing an 1,107-bp ORF that showed 97% sequence identity with M. avium subsp. avium fap (Fig. 2). This ORF and its putative translation product, designated FAP-P, did not show any significant identity with nucleotide or amino acid sequences other than those of mycobacterial FAPs, as determined by BLAST searches (3).
The FAP-P ORF encodes a protein comprised of 368 amino acids that contains a 39-residue putative N-terminal signal peptide (34, 35) (Fig. 2). The predicted molecular mass of the precursor protein is 36.1 kDa, whereas that of the mature FAP-P is expected to be 32.2 kDa. Proline residues are predicted to comprise 22.5% of the mature protein. The FAP-P precursor shows 91% sequence identity with FAP-A. Most of the discrepancies between aligned protein sequences arise from a shift in reading frame caused by four single-base deletions in the N-terminal third of the FAP-P coding sequence. The high degree of amino acid sequence identity extends into regions of FAP-A that have been demonstrated to have FN-binding capacity, namely, residues 177 to 201 and 269 to 292 (27). Within each of the corresponding positions for FAP-P (residues 173 to 197 and 265 to 288, respectively), there are single amino acid substitutions: a minor substitution at position 185 (Leu for Val) and a major substitution at position 267 (Ala for Arg). The latter substitution does not affect FN binding by FAP-A (37).
Blocking by FAP-specific peptide and antibodies.
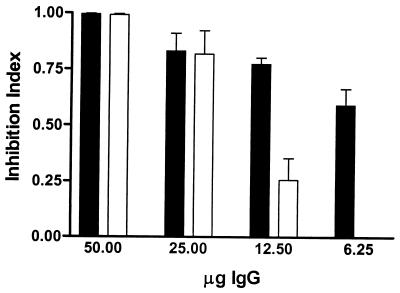
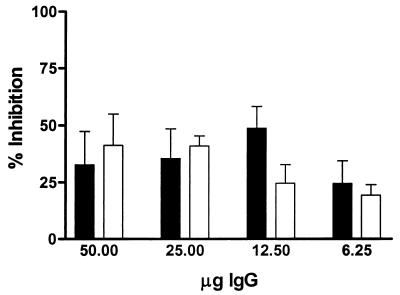
To determine if FN binding by M. avium subsp. paratuberculosis is mediated by FAP-P, we first sought to block binding by pretreating FN with FAP-A peptide FAPA 269–292, which has been shown to block binding of M. avium subsp. avium, M. bovis BCG, and M. smegmatis to FN-coated surfaces. FAP-A 269-292 significantly inhibited FN binding by both M. avium subsp. paratuberculosis strains studied (67.59% ± 11.38% [5781] and 89.71% ± 6.54% [6594] of the peptide-free control level) relative to the scrambled peptide (14.05% ± 14.05% [5781] and 31.59% ± 18.39% [6594] of the control level) (5781, P = 0.0253; 6594, P = 0.0247). This indicated that M. avium subsp. paratuberculosis binds FN in a FAP-dependent manner. To support this conclusion, we added either anti-FAP IgG or NRb IgG to acid-treated cell suspensions immediately prior to the addition of FN. NRb IgG blocked FN binding in a nonspecific manner; i.e., blocking by NRb IgG was dose independent (P = 0.227). In contrast, treatment with anti-FAP IgG resulted in a dose-dependent abrogation of FN binding by M. avium subsp. paratuberculosis (P < 0.0001); at higher concentrations, the inhibitory effect was significantly greater for anti-FAP IgG than for NRb IgG (P < 0.01 at both 50 and 25 μg of IgG). To evaluate specific inhibition of FN binding by anti-FAP, raw data were corrected for nonspecific (NRb IgG) blocking to create an inhibition index (Fig. 3). This clearly showed that specific blocking by anti-FAP IgG almost entirely abolished FN binding, confirming that this capability is a FAP-dependent process in M. avium subsp. paratuberculosis.
FIG. 3.
Sequential addition of anti-FAP IgG and FN results in specific blocking of FN binding by M. avium subsp. paratuberculosis. FN (in pH 6 buffer) was added immediately following the addition of graded concentrations of either NRb IgG or anti-FAP IgG to acid-treated M. avium subsp. paratuberculosis. To factor out nonspecific inhibition of FN binding by M. avium subsp. paratuberculosis, an inhibition index was calculated by dividing counts from anti-FAP-treated wells by companion NRb- IgG-treated wells and subtracting this quotient from unity. ■, strain 5781; □, strain 6594. For both panels, data shown represent the mean and standard error of the mean of four replicates for each treatment.
Subcellular localization of FAP-P.
Several attempts were made to block FN binding by preincubating acid-treated M. avium subsp. paratuberculosis with anti-FAP IgG, washing the cells, and then exposing the cells to FN. No specific blocking was observed, indicating that the antibodies failed to bind FAP-P (Fig. 4). This suggested that FAP-P is not exposed on the surface of M. avium subsp. paratuberculosis. To determine the location of FAP-P within the cell, we first performed an indirect immunofluorescence assay on intact and envelope lipid-extracted bacteria. Anti-FAP-specific fluorescence was observed only from lipid-extracted bacteria. Acid-treated M. avium subsp. paratuberculosis fluoresced strongly when reacted with anti-M. bovis BCG IgG, as did M. avium subsp. paratuberculosis held at neutral pH, but neither yielded fluorescence greater than that from secondary antibody-only controls when reacted with anti-FAP antiserum (data not shown).
FIG. 4.
Pretreatment with anti-FAP antiserum fails to block FN binding by M. avium subsp. paratuberculosis. Graded concentrations of NRb IgG (□) or anti-FAP IgG (■) in pH 6 buffer were incubated with acid-treated M. avium subsp. paratuberculosis for 1 h. After unbound antibodies were separated from cells by centrifugation, FN was added to the microplate wells, and the binding assay proceeded as described in Materials and Methods. Data are shown for strain 6594 and represent the mean and standard error of the mean of four replicates for each treatment.
Western blotting of whole cell lysates yielded two anti-FAP-reactive bands, at 54.2 and 49.1 kDa (Fig. 5). These immunoreactive bands migrated much slower than predicted above. This anomalous migration has been reported for other FAPs and was attributed to the high Pro content of these proteins (14, 27). The faster-migrating band is speculated to have resulted from C-terminal proteolytic cleavage of the secreted protein (11). Both bands were present in the crude cytoplasmic fraction. No anti-FAP reactivity was observed in the aqueous or detergent fractions of the cell wall or the cytoplasmic membrane fraction. Because the faster-migrating form was also present in the crude cytoplasmic fraction, these results suggested that the mature form of FAP resided between the cytoplasmic membrane and the mycolate layer, either as a soluble protein or weakly associated with a membrane- or cell wall-bound moiety.
FIG. 5.
Western blotting reveals that the M. avium subsp. paratuberculosis FAP homologue is not tightly associated with the cell wall. Washed M. avium subsp. paratuberculosis were sonicated and separated into subcellular fractions. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transfer to nitrocellulose, blots were probed with polyclonal anti-FAP antiserum. Positions of molecular weight markers are indicated in kilodaltons at the left. Two bands migrating to 54.2 and 49.1 kDa can be seen in the whole cell lysate (WL) and the crude cytoplasmic fraction (C/P). No immunoreactivity was seen in the aqueous (CWA) or detergent (CWD) phase of the cell wall fraction or in the cytoplasmic membrane fraction (CM).
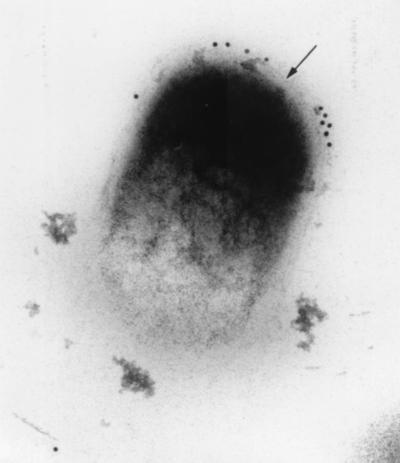
Immunoelectron microscopy revealed that FAP-P was predominantly located immediately exterior to the peptidoglycan layer of the cell envelope but well beneath the surface of the cell (Fig. 6). Immunogold labeling was also seen occasionally within the cytoplasm. These observations essentially corroborated the results of Western blot analysis, indicating that mature FAP-P is located between the peptidoglycan layer and inner face of the mycolate layer in M. avium subsp. paratuberculosis.
FIG. 6.
FAP-P is located within the cell envelope. Uranyl acetate-stained thin sections of M. avium subsp. paratuberculosis prepared from an M. avium subsp. paratuberculosis pellet were reacted with polyclonal anti-FAP rabbit serum. After washing, retained antibodies were detected by 10-nm gold-labeled anti-rabbit antibody. Most of the labeling is confined to an area immediately exterior to the peptidoglycan layer of the cell envelope, visible as a thin line (arrow) bounding the cytoplasm. Magnification, ×119,000.
DISCUSSION
The previously identified FN-binding activity of M. avium subsp. paratuberculosis was found to be strongly influenced by pH in the present study. Both M. avium subsp. paratuberculosis strains studied bound FN much more readily at acidic pH, and binding was negligible at pH above 7. Pretreatment of M. avium subsp. paratuberculosis with an acidic buffer greatly enhanced FN binding across the pH range studied. When acid-treated cells were held in neutral buffer prior to the addition of FN, binding reverted to the original pH profile. An 1,107-bp ORF that bore very high sequence identity with M. avium subsp. avium gene encoding FAP-A was detected in M. avium subsp. paratuberculosis genomic DNA. FN binding was specifically blocked by a FAP-A FN-binding peptide as well as anti-FAP IgG, indicating that FN binding by M. avium subsp. paratuberculosis was mediated by a FAP homologue, FAP-P, which was not surface expressed.
The pH response curve for soluble FN binding by M. avium subsp. paratuberculosis was remarkably similar to that observed for BCG (5). Because of the brief time (5 min) in which organisms were exposed to acidic conditions, the enhancement observed from acid pretreatment is believed to reflect activation or augmentation of existing FN-binding capacity rather that up-regulation of gene expression. The acidophic binding profile would support the significance of FN binding as a virulence factor in M. avium subsp. paratuberculosis. Because Johne's disease is spread primarily by a fecal-oral transmission cycle, M. avium subsp. paratuberculosis will encounter a relatively wide range of pH environments as it passes through the digestive tract of the ruminant host. The pHs of the rumen and omasum do not vary far from neutral, whereas that of the abomasum, or true stomach, is maintained at about 3. The pH remains below 4.5 within the duodenum and increases from approximately neutral in the jejunum to pH 8 in the terminal ileum, wherein paratuberculosis lesions usually first develop (24, 33). Our data suggest that M. avium subsp. paratuberculosis may become activated to bind FN by passage through the abomasum and become opsonized by FN present in bile secretions in the duodenum (36). Once bound to M. avium subsp. paratuberculosis, FN could bridge the organisms to integrins on the surface of M cells in the terminal ileum and facilitate their translocation across the epithelial barrier into Peyer's patches. Organisms that are not opsonized as they enter the jejunum would likely remain so and would be less likely to attach to and invade M cells.
Given the very high degree of genomic relatedness between M. avium subsp. paratuberculosis and M. avium subsp. avium, we expected to find a FAP homologue in M. avium subsp. paratuberculosis. We did not anticipate that FAP-P would be located well beneath the cell surface. The results of our Western analysis, which were corroborated by immunofluorescence and immunoelectron microscopy, contradict results obtained from M. bovis BCG, in which FAP was located in the cell wall fraction (25). Cultures in the previous study were sonicated much more vigorously than was done in this investigation. Thus, FAP may occupy different locations these species, which may in turn reflect the different routes of infection used by these organisms. The subsurface location of FAP-P may represent an adaptation in M. avium subsp. paratuberculosis that prevents exposure of this protein to proteolytic enzymes present in the small intestine.
Scatchard analysis indicated that a single protein was responsible for FN binding by M. avium subsp. paratuberculosis (31). Nonetheless, it is difficult to reconcile the requirement for FAP-P in FN binding with its cellular location if one maintains that this protein is solely responsible for interacting with FN. It is possible that low pH may alter the conformation of FAP-P such that it becomes more exposed and therefore available for direct interaction with FN. However, we were not able to block acid-treated M. avium subsp. paratuberculosis with anti-FAP IgG in the absence of FN. Moreover, the essential FN-binding residues 273RWFV276 within FAP-A 269–292 (37) and, by extrapolation, residues 192KLYA195 of FAP-A 177–201 (27) have a net positive charge at neutral pH and most probably remain so at low pH. These tetrapeptides are identical in sequence and relative position in FAP-P. Hence, it is doubtful that acid enhancement of FN binding in M. avium subsp. paratuberculosis is mediated through FAP-P.
How then does FN gain access to a buried binding protein? It is possible that low pH effects a change in cell wall permeability, either through a pH-sensitive porin or by rearranging the organization of cell wall structural components. Although immunoelectron microscopy was not performed on acid-treated cells, the data presented herein provide indirect evidence that this possibility is unlikely. Since a permeability change sufficient to accommodate the passage of FN through the cell wall would also permit entry of much smaller IgG molecules, one would expect that pretreatment of acid-activated M. avium subsp. paratuberculosis with anti-FAP antibodies would block FN binding. However, antibody pretreatment failed to abrogate binding. Similarly, acid-treated M. avium subsp. paratuberculosis did not demonstrate anti-FAP reactivity in immunofluorescence experiments. FAP-specific fluorescence was observed only when cell wall lipids were extracted from the organisms. We propose a model for FN binding by M. avium subsp. paratuberculosis in which at least one component at or near the cell surface interacts with FN and shuttles it inward toward FAP-P in a coordinated fashion. Alternatively, interaction of cell surface components of M. avium subsp. paratuberculosis with FN may transmit a signal that leads to the translocation of FAP-P to the cell surface. For either of these to occur, two conditions would have to be met: first, the initial interaction with FN must occur with a lower affinity than that of FAP-P; second, it must occur at a site on FN other than that used by FAP-P.
The antigen 85 complex was the first family of mycobacterial proteins to be identified as having FN-binding activity (1, 2, 30). Members of this complex are 30- to 31-kDa mycolyltransferase isozymes found within the outer envelope and culture supernatants of mycobacteria. Previous investigations have found that whereas antigen 85 proteins present in M. bovis BCG and M. tuberculosis culture supernatant bound labeled FN bound in Western blots, cell-bound forms did not (2). However, polyclonal antibodies against the antigen 85 complex of M. bovis BCG as well as purified antigens 85A and 85B blocked attachment of this organism to FN-coated surfaces (2). Members of the antigen 85 complex contain a well-conserved 11-residue motif, FEWYYQSSGLSSV, that that comprises the minimal FN binding domain (19). This motif is present on the surface of antigen 85C of M. bovis BCG (26). The Glu residue within this motif is likely to be protonated at pH 3 to 4. Thus, it is tempting to speculate that FN binding by M. avium subsp. paratuberculosis involves the participation of antigen 85 components and that the observed effect of pH on FN binding may be mediated through these proteins. There are conflicting reports regarding the region of FN bound by antigen 85. It was concluded from an earlier investigation that antigen 85 recognized the collagen binding site near the N terminus of FN (23). M. bovis BCG, M. leprae, and M. avium subsp. avium are able to bind the C-terminal heparin-binding fragment of FN (27, 28). This fragment, which is recognized by FAP, is sufficient to promote attachment and ingestion of M. leprae by epithelial cells in vitro (28, 37). These observations would thus call the contribution of the antigen 85 complex to FN binding by mycobacteria into question in view of the previous binding site localization study. However, a more recent report found that antigen 85 B bound to recombinant FN fragments containing the central integrin-binding and C-terminal heparin binding domains. Further, it was observed that heparin and not gelatin inhibited binding of antigen 85B to FN (18). In light of these findings, it remains quite possible that the antigen 85 complex participates in the binding of FN by mycobacteria. M. tuberculosis mutants deficient in the expression of antigen 85 components have been recently described (4). Development of similar mutants of M. avium subsp. paratuberculosis as well as antibody and peptide blocking studies, may help to clarify the role of these proteins in FAP-P-dependent binding.
One member of the PE-PGRS (proline-glutamine polymorphic GC-repetitive sequence) protein family of M. tuberculosis (8) has recently been found to possess FN-binding activity (10). This protein, Rv1759c, does not show any significant identity with the amino acid sequences of the FAP or antigen 85 family. Further study is needed to determine whether the ability to bind FN extends to other members of the PE-PGRS family, where these proteins are located within the cell, and whether a homologue is present in M. avium subsp. paratuberculosis.
Attachment of M. avium subsp. paratuberculosis to M cells and the subsequent translocation of organisms by these cells to the underlying gut-associated immune tissue are the necessary initial steps in the development of paratuberculosis. By gaining an understanding of the mechanisms and macromolecules involved in these processes, one can begin to develop herd management and immunization strategies that may limit the spread of infection within herds. Further work is needed to confirm the existence of coordinated FN binding in M. avium subsp. paratuberculosis as well as the significance of this process in establishing infections in vivo. Nonetheless, the participation of several proteins in the process of FN binding may offer multiple targets for therapeutic and immunologic intervention against Johne's disease.
ACKNOWLEDGMENTS
We thank Jeffrey Schorey for providing anti-FAP antiserum and Deborah Van Horn for performing the electron microscopy work described herein.
This work was supported by a Purdue School of Veterinary Medicine Internal Grant and the Purdue President's Distinguished Fellowship.
REFERENCES
- 1.Abou-Zeid C, Garbe T, Lathigra R, Wiker H G, Harboe M, Rook G A, Young D B. Genetic and immunological analysis of Mycobacterium tuberculosis fibronectin-binding proteins. Infect Immun. 1991;59:2712–2718. doi: 10.1128/iai.59.8.2712-2718.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abou-Zeid C, Ratliff T L, Wiker H G, Harboe M, Bennedsen J, Rook G A. Characterization of fibronectin-binding antigens released by Mycobacterium tuberculosis and Mycobacterium bovis BCG. Infect Immun. 1988;56:3046–3051. doi: 10.1128/iai.56.12.3046-3051.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armitige L Y, Jagannath C, Wanger A R, Norris S J. Disruption of the genes encoding antigen 85A and antigen 85B of Mycobacterium tuberculosis H37Rv: effect on growth in culture and in macrophages. Infect Immun. 2000;68:767–778. doi: 10.1128/iai.68.2.767-778.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aslanzadeh J, Brown E J, Quillin S P, Ritchey J K, Ratliff T L. Characterization of soluble fibronectin binding to Bacille Calmette- Guerin. J Gen Microbiol. 1989;135:2735–2741. doi: 10.1099/00221287-135-10-2735. [DOI] [PubMed] [Google Scholar]
- 6.Belisle J T, Sonnenberg M G. Isolation of genomic DNA from mycobacteria. Methods Mol Biol. 1998;101:31–44. doi: 10.1385/0-89603-471-2:31. [DOI] [PubMed] [Google Scholar]
- 7.Cheung A L, Yeaman M R, Sullam P M, Witt M D, Bayer A S. Role of the sar locus of Staphylococcus aureus in induction of endocarditis in rabbits. Infect Immun. 1994;62:1719–1725. doi: 10.1128/iai.62.5.1719-1725.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 9.De Strooper B, Van der Schueren B, Jaspers M, Saison M, Spaepen M, Van Leuven F, Van den Berghe H, Cassiman J J. Distribution of the beta 1 subgroup of the integrins in human cells and tissues. J Histochem Cytochem. 1989;37:299–307. doi: 10.1177/37.3.2645360. [DOI] [PubMed] [Google Scholar]
- 10.Espitia C, Laclette J P, Mondragon-Palomino M, Amador A, Campuzano J, Martens A, Singh M, Cicero R, Zhang Y, Moreno C. The PE-PGRS glycine-rich proteins of Mycobacterium tuberculosis: a new family of fibronectin-binding proteins? Microbiology. 1999;145:3487–3495. doi: 10.1099/00221287-145-12-3487. [DOI] [PubMed] [Google Scholar]
- 11.Horn C, Namane A, Pescher P, Riviere M, Romain F, Puzo G, Barzu O, Marchal G. Decreased capacity of recombinant 45/47-kDa molecules (Apa) of Mycobacterium tuberculosis to stimulate T lymphocyte responses related to changes in their mannosylation pattern. J Biol Chem. 1999;274:32023–32030. doi: 10.1074/jbc.274.45.32023. [DOI] [PubMed] [Google Scholar]
- 12.Kirschner P, Meier A, Bottiger E C. Genotypic identification and detection of mycobacteria—facing novel and uncultured pathogens. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology. Washington, D.C.: ASM Press; 1993. pp. 173–190. [Google Scholar]
- 13.Kuroda K, Brown E J, Telle W B, Russell D G, Ratliff T L. Characterization of the internalization of bacillus Calmette-Guerin by human bladder tumor cells. J Clin Investig. 1993;91:69–76. doi: 10.1172/JCI116202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laqueyrerie A, Militzer P, Romain F, Eiglmeier K, Cole S, Marchal G. Cloning, sequencing, and expression of the apa gene coding for the Mycobacterium tuberculosis 45/47-kilodalton secreted antigen complex. Infect Immun. 1995;63:4003–4010. doi: 10.1128/iai.63.10.4003-4010.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manning J E, Hume E B, Hunter N, Knox K W. An appraisal of the virulence factors associated with streptococcal endocarditis. J Med Microbiol. 1994;40:110–114. doi: 10.1099/00222615-40-2-110. [DOI] [PubMed] [Google Scholar]
- 16.Middleton A M, Chadwick M V, Nicholson A G, Dewar A, Groger R K, Brown E J, Wilson R. The role of Mycobacterium avium complex fibronectin attachment protein in adherence to the human respiratory mucosa. Mol Microbiol. 2000;38:381–391. doi: 10.1046/j.1365-2958.2000.02137.x. [DOI] [PubMed] [Google Scholar]
- 17.Momotani E, Whipple D L, Thiermann A B, Cheville N F. Role of M cells and macrophages in the entrance of Mycobacterium paratuberculosis into domes of ileal Peyer's patches in calves. Vet Pathol. 1988;25:131–137. doi: 10.1177/030098588802500205. [DOI] [PubMed] [Google Scholar]
- 18.Naito M, Fukuda T, Sekiguchi K, Yamada T. The domains of human fibronectin mediating the binding of alpha antigen, the most immunopotent antigen of mycobacteria that induces protective immunity against mycobacterial infection. Biochem J. 2000;347:725–731. [PMC free article] [PubMed] [Google Scholar]
- 19.Naito M, Ohara N, Matsumoto S, Yamada T. The novel fibronectin-binding motif and key residues of mycobacteria. J Biol Chem. 1998;273:2905–2909. doi: 10.1074/jbc.273.5.2905. [DOI] [PubMed] [Google Scholar]
- 20.Neeman R, Keller N, Barzilai A, Korenman Z, Sela S. Prevalence of internalisation-associated gene, prtF1, among persisting group-A streptococcus strains isolated from asymptomatic carriers. Lancet. 1998;352:1974–1977. doi: 10.1016/S0140-6736(97)12452-7. [DOI] [PubMed] [Google Scholar]
- 21.Ott S L, Wells S J, Wagner B A. Herd-level economic losses associated with Johne's disease on US dairy operations. Prev Vet Med. 1999;40:179–192. doi: 10.1016/s0167-5877(99)00037-9. [DOI] [PubMed] [Google Scholar]
- 22.Parish T, Wheeler P R. Preparation of cell-free extracts from mycobacteria. Methods Mol Biol. 1998;101:77–89. doi: 10.1385/0-89603-471-2:77. [DOI] [PubMed] [Google Scholar]
- 23.Peake P, Gooley A, Britton W J. Mechanism of interaction of the 85B secreted protein of Mycobacterium bovis with fibronectin. Infect Immun. 1993;61:4828–4834. doi: 10.1128/iai.61.11.4828-4834.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillipson A T. Ruminant digestion. In: Swenson M J, editor. Dukes' physiology of domestic animals. 9th ed. Ithaca, N.Y: Comstock Publishing Associates; 1977. pp. 250–286. [Google Scholar]
- 25.Ratliff T L, McCarthy R, Telle W B, Brown E J. Purification of a mycobacterial adhesin for fibronectin. Infect Immun. 1993;61:1889–1894. doi: 10.1128/iai.61.5.1889-1894.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ronning D R, Klabunde T, Besra G S, Vissa V D, Belisle J T, Sacchettini J C. Crystal structure of the secreted form of antigen 85C reveals potential targets for mycobacterial drugs and vaccines. Nat Struct Biol. 2000;7:141–146. doi: 10.1038/72413. [DOI] [PubMed] [Google Scholar]
- 27.Schorey J S, Holsti M A, Ratliff T L, Allen P M, Brown E J. Characterization of the fibronectin-attachment protein of Mycobacterium avium reveals a fibronectin-binding motif conserved among mycobacteria. Mol Microbiol. 1996;21:321–329. doi: 10.1046/j.1365-2958.1996.6381353.x. [DOI] [PubMed] [Google Scholar]
- 28.Schorey J S, Li Q, McCourt D W, Bong-Mastek M, Clark-Curtiss J E, Ratliff T L, Brown E J. A Mycobacterium leprae gene encoding a fibronectin binding protein is used for efficient invasion of epithelial cells and Schwann cells. Infect, Immun. 1995;63:2652–2657. doi: 10.1128/iai.63.7.2652-2657.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stabel J R. Johne's disease: a hidden threat. J Dairy Sci. 1998;81:283–288. doi: 10.3168/jds.S0022-0302(98)75577-8. [DOI] [PubMed] [Google Scholar]
- 30.Thole J E, Schoningh R, Janson A A, Garbe T, Cornelisse Y E, Clark-Curtiss J E, Kolk A H, Ottenhoff T H, De Vries R R, Abou-Zeid C. Molecular and immunological analysis of a fibronectin-binding protein antigen secreted by Mycobacterium leprae. Mol Microbiol. 1992;6:153–163. doi: 10.1111/j.1365-2958.1992.tb01996.x. [DOI] [PubMed] [Google Scholar]
- 31.Valentin-Weigand P, Moriarty K M. Mycobacterium paratuberculosis binds fibronectin. Res Microbiol. 1992;143:75–79. doi: 10.1016/0923-2508(92)90036-n. [DOI] [PubMed] [Google Scholar]
- 32.Westerlund B, Korhonen T K. Bacterial proteins binding to the mammalian extracellular matrix. Mol Microbiol. 1993;9:687–694. doi: 10.1111/j.1365-2958.1993.tb01729.x. [DOI] [PubMed] [Google Scholar]
- 33.Whitlock R H, Buergelt C. Preclinical and clinical manifestations of paratuberculosis (including pathology) Vet Clin North Am Food Anim Pract. 1996;12:345–356. doi: 10.1016/s0749-0720(15)30410-2. [DOI] [PubMed] [Google Scholar]
- 34.Wieles B, van Agterveld M, Janson A, Clark-Curtiss J, Rinke de Wit T, Harboe M, Thole J. Characterization of a Mycobacterium leprae antigen related to the secreted Mycobacterium tuberculosis protein MPT32. Infect Immun. 1994;62:252–258. doi: 10.1128/iai.62.1.252-258.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiker H G, Wilson M A, Schoolnik G K. Extracytoplasmic proteins of Mycobacterium tuberculosis—mature secreted proteins often start with aspartic acid and proline. Microbiology. 2000;146:1525–1533. doi: 10.1099/00221287-146-7-1525. [DOI] [PubMed] [Google Scholar]
- 36.Yu J L, Andersson R, Ljungh A. Protein adsorption and bacterial adhesion to biliary stent materials. J Surg Res. 1996;62:69–73. doi: 10.1006/jsre.1996.0175. [DOI] [PubMed] [Google Scholar]
- 37.Zhao W, Schorey J S, Groger R, Allen P M, Brown E J, Ratliff T L. Characterization of the fibronectin binding motif for a unique mycobacterial fibronectin attachment protein, FAP. J Biol Chem. 1999;274:4521–4526. doi: 10.1074/jbc.274.8.4521. [DOI] [PubMed] [Google Scholar]