Abstract
As important drug targets, G protein-coupled receptors (GPCRs) play pivotal roles in a wide range of physiological processes. Extensive efforts of structural biology have been made on the study of GPCRs. However, a large portion of GPCR structures remain unsolved due to structural instability. Recently, AlphaFold2 has been developed to predict structure models of many functionally important proteins including all members of the GPCR family. Herein we evaluated the accuracy of GPCR structure models predicted by AlphaFold2. We revealed that AlphaFold2 could capture the overall backbone features of the receptors. However, the predicted models and experimental structures were different in many aspects including the assembly of the extracellular and transmembrane domains, the shape of the ligand-binding pockets, and the conformation of the transducer-binding interfaces. These differences impeded the use of predicted structure models in the functional study and structure-based drug design of GPCRs, which required reliable high-resolution structural information.
Keywords: G protein-coupled receptors, AlphaFold2, protein structure prediction, drug design, structural biology
Introduction
As an extensive membrane receptor family, G protein-coupled receptors (GPCRs) have over 800 members in humans. They are involved in diverse physiological and pathological processes, including sense of light, neural signal transmission, and endocrine system homeostasis [1–4]. As such, GPCRs have a rich history as drug targets, which account for nearly one-third of all approved drugs [5]. In the field of structural biology, it is an important research object to elucidate activation mechanism of GPCRs. The progress of X-ray-based methods used to be slow [6, 7]. The development of new techniques, such as cryo-electron microscopy, leads to the enormous surge of GPCR structures from 2017 [8, 9]. However, due to the considerable workload and internal receptor flexibility, only 757 GPCR structures have been released to date [6, 10, 11]. The high cost and difficulty of obtaining GPCR structures hinder the function research and drug development of GPCRs, which calls for an accurate efficient structure prediction algorithm.
Developed by DeepMind company in 2021, AlphaFold2 shows the best-in-class performance of protein structure prediction in the Critical Assessment of protein Structure Prediction 14 competition, which has reached near experimental accuracy in a lot of cases [12]. The success of AlphaFold2 comes from its understanding of protein folding problems and powerful deep learning network composition, including a combination of multiple sequence alignment and template information, a new Evoformer structure, and an equivariant module to produce structures [13, 14]. With the powerful algorithm, the AlphaFold2 developers predicted structures for most human proteome and publicly released the predicted models [15].
Considering the structures of hundreds of important GPCRs remained unsolved, structure models predicted by AlphaFold2 may provide information for GPCR research, such as structure-based drug design, template build in structure biology, and GPCR functional study [16, 17]. Thus, it is necessary and valuable to comprehensively evaluate the reliability of GPCR structure models predicted by AlphaFold2. In this work, we compared the experimental structures of GPCRs with the corresponding structure models predicted by AlphaFold2. To exclude the influence of the training data, we collected the experimental structures released after the emergence of the AlphaFold2 database. Computational analysis including deviation and distance calculations was performed on all available structures. The comprehensive comparison reflects the performance of AlphaFold2 in addressing sub-domain assembly, ligand-binding, and activation of GPCRs.
Evaluation for G-protein coupled receptors
Evaluation for overall receptor structures
In order to evaluate the performance of AlphaFold2 on GPCRs, we collected 29 GPCR structures released after the publication of the AlphaFold2 database, thereby confirming that the prediction of these GPCR structures did not involve the experimental structural information. The experimental information of 29 structures is listed in Supplementary Table S1. According to the traditional classification of GPCRs, these 29 structures are categorized in classes A, B1, and F [18, 19]. Most of them are functionally active structures in complex with G proteins (Supplementary Table S1) [20].
During GPCR activation, the conserved seven transmembrane (TM) helices show different movement modes [21–23]. TM1–TM4 are relatively stable, while TM5–TM7 conformations are distinct from inactive to active states to provide space for transducers [23, 24]. Three extracellular loops (ECLs) and three intracellular loops (ICLs) between TMs are traditionally flexible due to their intrinsic unstable secondary structure [25]. We measured the Cα root mean square deviation (Cα RMSD) between predicted models and experimental structures for entire receptors, TM1–TM4, TM5–TM7, and TM domain (TMD) in Supplementary Table S2. The predicted models show an average global Cα RMSD of 1.64 ± 1.08 Å from the experimental structures, suggesting that the global structure properties have been captured by AlphaFold2 (Supplementary Fig. S1). Notably, the prediction of models in TMD is accurate, especially for stable TM1–TM4 with an average Cα RMSD of 0.79 ± 0.19 Å. TM5–TM7 shows slightly different conformations (1.26 ± 0.45 Å) between the predicted models and experimental structures. In particular, prostaglandin D2 receptor 2 (CRTH2), an inactive-state class A structure in our collection, shows a small Cα RMSD of 0.63 Å (Supplementary Fig. S1a and Table S2). As an active-state structure in class B1 GPCRs, glucagon-like peptide 1 receptor (GLP1R) also shows small deviation between the experimental structure and predicted model in TMD (Cα RMSD of 1.04 Å, Supplementary Fig. S1b and Table S2). These findings suggested that AlphaFold2 maintains its performance in predicting overall TMD structures of GPCRs.
Evaluation for receptors with large extracellular domains (ECDs)
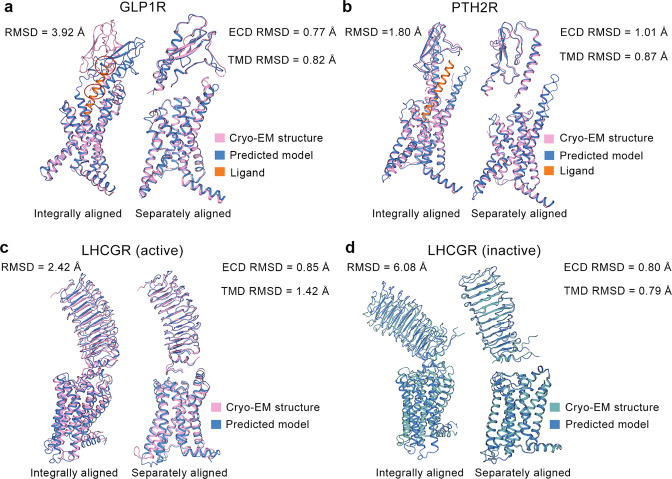
Several structures of GPCRs with large ECDs show higher overall Cα RMSDs (≥1.80 Å). In the case of semaglutide-bound GLP1R structure with ECD, an integral alignment of the predicted model to the experimental structure leads to a large Cα RMSD of 3.92 Å, while the ECD or TMD in the predicted model is similar to those in the experimental structure (Cα RMSD of 0.77 Å for ECD and Cα RMSD of 0.82 Å for TMD). As shown in Fig. 1a, the ECD of the predicted model has a different orientation with respect to the TMD, compared with that of the experimental structure. Structurally aligning the predicted model to the ligand-bound experimental one, we observed that the ligand from the experimental structure has two conflicts with the predicted model in the ECD (Fig. 1a). Due to these conflicts, the ligand cannot be placed in the predicted model in the same way as which in the experimental complex structure of GLP1R. In another class B1 GPCR, i.e., parathyroid hormone 2 receptor (PTH2R), the assembly of ECD and TMD in the predicted model is also distinct from the experimental structure (Fig. 1b). Its overall Cα RMSD is 1.80 Å, while its ECD Cα RMSD and TMD Cα RMSD are 1.01 Å and 0.87 Å, respectively. Luteinizing hormone and choriogonadotropin receptor (LHCGR) with a large ECD also appears large Cα RMSD values in active (2.42 Å) and inactive states (6.08 Å) (Fig. 1c, d). Similarly, separate alignments of the ECD or TMD of LHCGR decrease the Cα RMSD to a value less than 1.5 Å (Cα RMSD of 0.85 Å for active ECD, Cα RMSD of 1.42 Å for active TMD, Cα RMSD of 0.80 Å for inactive ECD, Cα RMSD of 0.79 Å for inactive TMD). It suggests that the sub-domain prediction of LHCGR is mostly accurate. However, the predicted models present different assemblies of ECD and TMD, compared with those of experimental structures.
Fig. 1. Comparison between predicted models and experimental structures with large ECD.
Experimental structures and predicted models for glucagon-like peptide 1 receptor (GLP1R) (a), parathyroid hormone 2 receptor (PTH2R) (b), active (c) and inactive (d) luteinizing hormone and choriogonadotropin receptor (LHCGR) in different ways of alignments. Cryo-EM structures of GLP1R (PDB ID: 7KI0), PTH2R (PDB ID: 7F16), LHCGR (PDB IDs: 7FIG and 7FIJ) are selected for calculation. Pink (or cyan) and blue cartoons represent experimental structures and predicted models, respectively. Two conflicts between the predicted model of GLP1R and the peptide ligand from the experimental structure are highlighted in red circles.
Evaluation for the orthosteric ligand-binding sites
An endogenous modulator of a GPCR binds to the center of an extracellular pocket consisting of TM helices, which is known as the orthosteric site [26, 27]. From this site, ligands could induce receptor conformational changes and alter the functional states of GPCRs [22, 28, 29]. Thus, the pocket information is valuable in structure-based drug design and functional research [30–32].
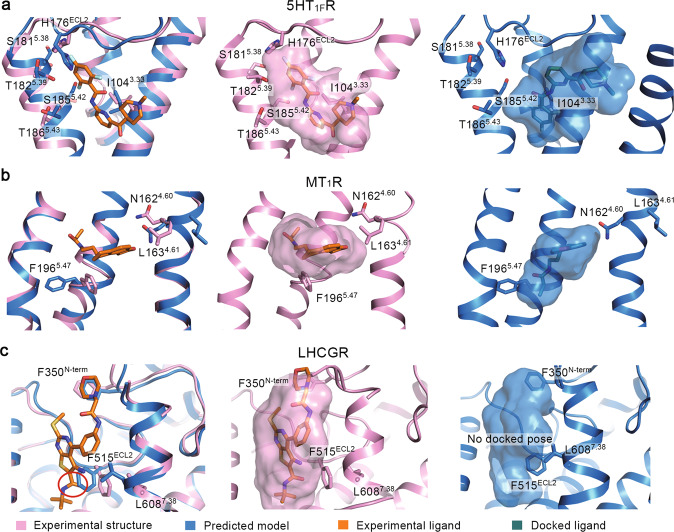
In the full set of 29 experimental structures, only four are GPCR-small molecule complexes (Supplementary Table S1). We performed molecular docking against the orthosteric sites in their predicted models and experimental structures to evaluate the feasibility of predicted models in drug development and functional study (Fig. 2). In the orthosteric ligand-binding sites (Fig. 2 and Supplementary Fig. S2), the TMD backbones are highly similar between predicted models and experimental structures (Supplementary Table S3, average backbone RMSD is 0.89 ± 0.34 Å). Nevertheless, some residues at these sites show different sidechain conformations, leading to higher sidechain RMSD (1.90 ± 0.58 Å) and all-atom RMSD (1.52 ± 0.42 Å). As a result, different binding poses of ligands were observed in predicted models and experimental structures (Fig. 2). In 5-hydroxytryptamine 1F receptor (5HT1FR), backbone and sidechain conformations of the ligand-binding sites are different between the predicted model and experimental structure; its all-atom RMSD is 1.63 Å, backbone RMSD is 1.29 Å and sidechain RMSD is 1.87 Å (Fig. 2a). The sidechain of H176ECL2 was predicted to insert between the extracellular region of TM4 and TM5. Additionally, I1043.33, S1815.38, T1825.39, S1855.42, and T1865.43 rotate their sidechains to the center of TMD in the predicted model. Due to these distinct sidechain conformations, the predicted model has a narrower pocket compared with that of the experimental structure. The interaction between the trifluorobenzene ring of ligand and H176ECL2 is also blocked in the predicted model (Fig. 2a). In the molecular docking, the narrow ligand-binding pocket determines a binding pose of ligand toward TM2 of the predicted model (Fig. 2a). The RMSD of the ligand docked to the predicted model from the ligand in the experimental complex structure (PDB ID: 7EXD) is 7.15 Å. For the predicted orthosteric pocket of melatonin receptor 1A (MT1R), its all-atom RMSD is 2.13 Å, backbone RMSD is 0.99 Å and sidechain RMSD is 2.82 Å from experimental structure (Fig. 2b). In the predicted model, the sidechain of F1965.47 moves out of the receptor core to form a cavity that favors a deep insertion of the ligand. Also, the sidechain rotations of N1624.60 and L1634.61 contribute to movement of the ligand toward the center of TM helix bundle. As a result, the docked ligand shows a deep-inserted pose close to the backbone of F1965.47. The RMSD of the ligand docked to the predicted model from the one of the experimental complex structure (PDB ID: 7DB6) is 4.79 Å. In the positive allosteric modulator-binding pocket of LHCGR, though the backbone difference between the predicted model and experimental structure is small (all-atom RMSD is 1.34 Å and backbone RMSD is 0.94 Å), the sidechain distinctions are critical (sidechain RMSD is 1.66 Å) (Fig. 2c). The sidechain of F515ECL2 is predicted to insert into the cavity between the TM5 and TM7, and L6087.38 turns its sidechain to the receptor core in the predicted model. The N-terminal loop conformation adjacent to F350N-term alters the environment at the top of the pocket. As a consequence, the ligand fails to bind to the pocket of the predicted model in molecular docking (Fig. 2c). For an antagonist bound CRTH2, its ligand-binding pocket is highly similar between the predicted model and experimental structure with respect to backbone and sidechain (all-atom RMSD is 0.97 Å, backbone RMSD is 0.34 Å and sidechain RMSD is 1.23 Å, Supplementary Fig. S2). Consequentially, the ligand-binding pose to the predicted model and the one in the experimental complex structure (PDB ID: 7M8W) are highly similar (ligand RMSD is 0.90 Å). Collectively, in three out of four cases, the predicted orthosteric ligand-binding sites present different sidechain conformations from those of the experimental structures, which alters the docking results of the active compounds.
Fig. 2. Evaluation for the orthosteric ligand-binding sites.
Ligands binding to the orthosteric sites in experimental structures and predicted models of 5-hydroxytryptamine receptor 1F receptor (5HT1FR) (a), melatonin receptor 1A (MT1R) (b), and luteinizing hormone and choriogonadotropin receptor (LHCGR) (c). Cryo-EM structures of 5HT1FR (PDB ID: 7EXD), MT1R (PDB ID: 7DB6), LHCGR (PDB ID: 7FIH) are selected for analysis. Key residues and ligands are shown as sticks. Pink, blue, orange, and green represent experimental structures, predicted models, ligands from experimental complex structures, and docked ligands, respectively. The ligand-binding pockets are shown as surfaces.
Evaluation for functionally important TM6–TM7 helices
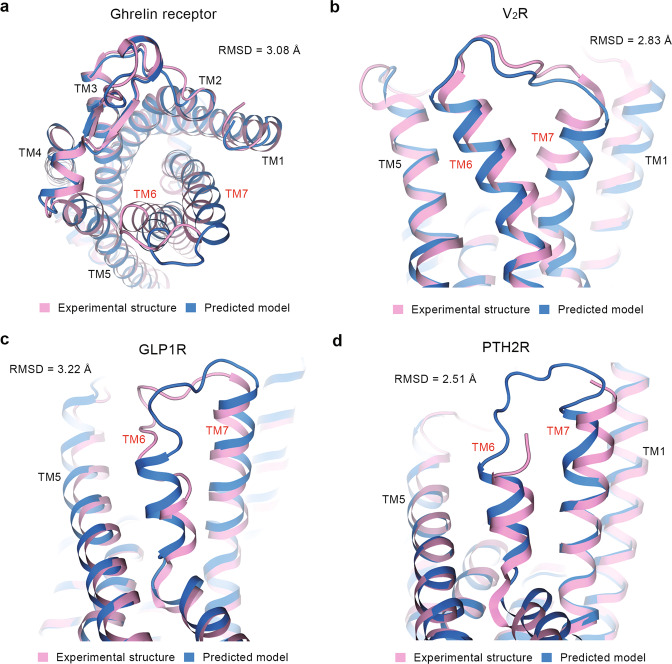
A critical activation property of a GPCR is the conformational adjustment of TM6–TM7 helices, which provides space for the binding of transducers [21, 28, 33–35]. Compared with the experimental structures, several predicted models of GPCRs show different extracellular TM6–TM7 conformations (TM6–TM7 heavy-atom RMSDs are larger than 2 Å), though aligned with respect to TMD (Supplementary Table S4). As representative class A GPCRs, ghrelin receptor and vasopressin V2 receptor (V2R) show large values of TM6–TM7 heavy-atom RMSD (3.08 Å for ghrelin receptor and 2.83 Å for V2R). The TM6 and TM7 helices of the ghrelin receptor are predicted to move out from the TM bundle center compared with the experimental structure (PDB ID: 7F9Y) (Fig. 3a). In the predicted model of V2R (Fig. 3b), TM6 stays far from TM7 and TM7 moves downward compared with the experimental structure (PDB ID: 7DW9). For class B1 GPCRs, GLP1R and PTH2R have large TM6–TM7 heavy-atom RMSDs of 3.22 Å and 2.51 Å, respectively. As shown in Fig. 4c, d, GLP1R and PTH2R are predicted to have TM6 and TM7 move upward to the extracellular side compared with the experimental structures (PDB IDs: 7KI1 and 7F16). Of note, the different TM6–TM7 conformations at the extracellular side mostly exist in the GPCR complexed with peptide ligands (Supplementary Table S4).
Fig. 3. Comparison of TMD conformations at extracellular side.
TM6 and TM7 helices in experimental structures and predicted models of ghrelin receptor (a), vasopressin V2 receptor (V2R) (b), glucagon-like peptide 1 receptor (GLP1R) (c), and parathyroid hormone 2 receptor (PTH2R) (d). Cryo-EM structures of ghrelin receptor (PDB ID: 7F9Y), V2R (PDB ID: 7DW9), GLP1R (PDB ID: 7KI1) and PTH2R (PDB ID: 7F16) are selected for analysis. Pink and blue cartoons represent experimental structures and predicted models, respectively. TM6–TM7 heavy-atom RMSD values are shown in each plot.
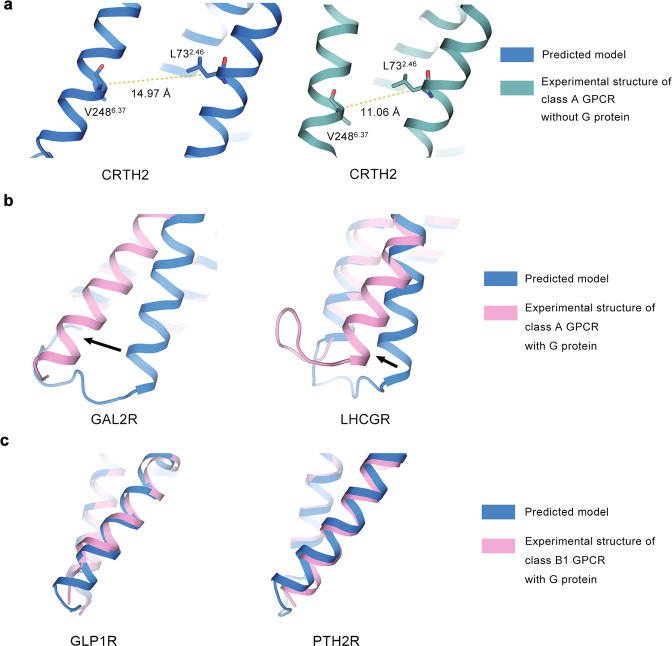
Fig. 4. Comparison of TM6 conformations at intracellular side.
TM6 helices of predicted models and experimental structures for a class A GPCR without G protein (a), a class A GPCR with G protein (b), and a class B1 GPCR with G protein (c). Structures without G protein, structures with G protein, and predicted models are shown in green, pink, and blue cartoons, respectively. PDB IDs of experimental structures are labeled in each plot. Dashed lines indicate the TM6 tilt distance (TD). Arrows indicate the orientational differences of TM6 between predicted models and experimental structures. All structures are aligned with respect to TMD.
Considering that the outward movement of TM6 provides space for transducer binding, the orientation of TM6 is a significant symbol for GPCR activation [36–38]. Thus, we measured the TM6 tilt distance (TD) in predicted models (TDpred) and experimental structures (TDexp) to characterize TM6 orientation (Supplementary Table S5) [39, 40]. The difference of TM6 tilt distance between TDexp and TDpred was also calculated as ΔTD. A positive value of ΔTD indicates that the experimental structure has a larger TD and a more spacious intracellular protein-binding pocket compared with the predicted model, or vice versa.
Different patterns of ΔTD exist among class A GPCRs without G proteins, class A GPCRs with G proteins, and class B1 GPCRs with G proteins (Fig. 4). Class A GPCRs have an average ΔTD of −1.85 ± 2.06 Å, suggesting that they are predicted to provide more space for protein binding at the intracellular side compared with the experimental structures. For example, the TM6 helix of CRTH2 moves outward in the predicted model (TDpred is 14.97 Å) relative to the experimental structure (TDexp is 11.06 Å) (Fig. 4a). G protein-bound class A GPCRs have an average ΔTD of 3.61 ± 1.59 Å, suggesting that they are predicted to provide less space for protein binding than the experimental structures. As shown in Fig. 4b, the predicted TM6 helices stay closer to the TM bundle at the intracellular side compared with experimental structures. The values of TDpred for GAL2R and LHCGR are 11.72 Å and 15.61 Å, respectively, but their TDexp are 16.46 Å and 17.09 Å. Class B1 GPCRs have the average ΔTD of 0.05 ± 0.46 Å, suggesting that the predicted models are highly consistent with the experimental structures. As shown in Fig. 4c, the predicted TM6 helices of GLP1R and PTH2R overlap with those of experimental structures; the values of ΔTD for GLP1R and PTH2R are −0.02 Å and −0.70 Å, respectively. To sum up, for class A GPCRs, the predicted models tend to adopt an “averaged” TM6 conformation (TDpred is 13.78 ± 1.69 Å) between G protein-bound state (TDexp is 17.54 ± 0.82 Å) and non-G protein-bound state (TDexp is 12.74 ± 1.84 Å). For class B1 GPCRs, the predicted models (TDpred is 22.92 ± 0.04 Å) tend to adopt G protein-bound state conformations (TDexp is 22.62 ± 0.37 Å).
Notably, the ICL3 between TM5 and TM6 is predicted as a long-extended helix in some cases of class A GPCRs (Supplementary Fig. S3). In the experimental structure of 5HT1FR, 26 residues (from L211 to K236) adjacent to the C-terminus of TM5 and 32 residues (from D257 to E288) adjacent to the C-terminus of TM6 should participate in forming the ICL3 of the receptor. But they are predicted to be a part of the TM helix in the predicted model (Supplementary Fig. S3a). Similarly, for cholecystokinin receptor 1 (CCKAR), 8 residues (from Q246 to R253) linked to the C-terminus of TM5 and 14 residues (from G288 to S301) linked to N-terminus of TM6 are predicted as long helices, while they belong to ICL3 according to the experimental structure (Supplementary Fig. S3b).
The possible reasons for deviations of predictions from experiments
The predicted models by AlphaFold2 show ECD-TMD assemblies that are different from those of experimental structures. Considering that GPCRs are highly dynamic proteins, more than one ECD-TMD assemblies may exist for a particular receptor [23, 29]. Even though a predicted ECD-TMD assembly of a GPCR is distinct from the experimental one, we could not rule out the possibility that this predicted model captures an unveiled metastable state of the receptor. In case of GLP1R, the predicted model shows conflicts that obstruct the peptide binding due to its distinct assembly of ECD and TMD (Fig. 1a). Since an active ligand may induce conformational changes of its receptor, the loss of ligand information in training may be the reason why AlphaFold2 fails to construct an ECD-TMD assembly of GLP1R, which favors peptide binding. As functionally important membrane proteins, many GPCRs receive activation signals via ECD upon peptide binding [29, 41] and the ligand-binding process is determined by the relative orientation between ECD and TMD [42]. Therefore, it needs to be cautious to use predicted models by AlphaFold2 to study the peptide-binding process of GPCRs.
In three out of four small-molecule bound GPCRs, the predicted models by AlphaFold2 show different ligand-binding pocket shapes compared with the experimental structures (Fig. 2). Even though it achieves a Cα backbone RMSD accuracy of ~1 Å, AlphaFold2 can hardly predict the pocket side chains of GPCRs with high accuracies. This has also been revealed for the cases of the other proteins [43]. As a result, the docking poses of active compounds to the predicted models are very different from the one observed in the experimental complex structures in most cases (Fig. 2a, b). In the case of LHCGR, AlphaFold2 even fails to produce suitable small molecule pockets for docking (Fig. 2c). Meanwhile, the shapes of ligand-binding pockets could be determined by the chemical and physical characteristics of ligands [44, 45]. Although trained with the PDB database containing numerous small molecular ligand-bound proteins, AlphaFold2 is still unable to capture sufficient features of a specific ligand-binding site of a GPCR, whose ligand information is unknown for the AI system. Therefore, AlphaFold2 alone cannot provide sufficient information for structure-based drug design. The application of its predicted models in the drug development requires great efforts to validate, remodel or refine the ligand-binding site.
The conformational changes of TM6–TM7 alter the intracellular protein-binding interfaces for different transducers to determine the functional states of GPCRs [36–38]. For the functionally important TM6 helix, AlphaFold2 tends to produce an “average” conformation in class A GPCRs and an active-like conformation in class B1 GPCRs. We analyze the composition of the published GPCR structures before the release of the AlphaFold2 database in Supplementary Table S6. The structures were classified as active, intermediate, and inactive ones in the GPCRdb database (https://gpcrdb.org/structure/) according to the open degree of the TM6 helix [39, 40]. In class A GPCRs, 55% of structures are inactive and 37% are active. However, in class B1 GPCRs, 70% of structures are active, completely dominating the population. This finding indicates that the data bias of AlphaFold2 contributes to the biased predictions of the TM6 conformations. In addition, AlphaFold2 produces long helices in the region of ICL3 in some cases. The ICL3 itself is flexible in the GPCR family [46]. In the experimental structures, the fully solved ICL3 shows irregular conformation [3]. To construct a stable structure in the experiments, an unstable ICL3 is always replaced by thermostabilized apocytochrome b562RIL (BRIL) to link TM5 and TM6, which yields to an artificial long helix instead of ICL3 [47–49]. AlphaFold2 may learn from these unnatural structures of GPCRs containing BRIL to produce a long helix instead of ICL3. Due to the training data bias regarding TM6 and ICL3, AlphaFold2 may produce biased or unnatural models of GPCRs. These models need to be excluded in the biological mechanism study of GPCRs. In application of predicted models, homolog structures may be referred.
Conclusion
As a state-of-the-art protein structure prediction tool, AlphaFold2 has provided structure models for more than nine hundred thousand proteins. Among these proteins, GPCR is one of the most important classes in structural biology and drug development [17, 50]. To provide insights into the usage of these predicted models for drug design and function exploration, we systematically evaluated AlphaFold2 by comparing predicted models and experimental structures of GPCRs regarding sub-domain assembly, ligand-binding, and functional state of the receptors. Even though AlphaFold2 achieves a Cα RMSD accuracy of ~1 Å in protein structure prediction [12], it alone cannot manage the structure-based drug design of GPCRs, which require high-resolution information of protein side chains. AlphaFold2 also shows limitations in predicting the ECD-TMD assembly and the transducer-binding interface of a GPCR. Due to data bias, AlphaFold2 may produce biased or artificial predicted models of GPCRs, which need to be excluded by experimental validation. Thus, experimental structural determination is still an indispensable tool for obtaining the structure of GPCRs and providing credentials for structure-based drug design and GPCR function exploration.
Supplementary information
Acknowledgements
This work was partially supported by Lingang Laboratory grant (LG202102-01-01 to XC); Ministry of Science and Technology (China) grants (2018YFA0507002 to HEX); Shanghai Municipal Science and Technology Major Project (2019SHZDZX02 to HEX); Shanghai Municipal Science and Technology Major Project (HEX); CAS Strategic Priority Research Program (XDB37030103 to HEX); the National Natural Science Foundation of China (32130022 to HEX, 32171187 to YJ, 82121005 to HEX and YJ); and Shanghai Municipal Science and Technology Major Project.
Competing interests
The authors declare no competing interests.
Contributor Information
Yi Jiang, Email: yijiang@simm.ac.cn.
H. Eric Xu, Email: Eric.Xu@simm.ac.cn.
Xi Cheng, Email: xicheng@simm.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41401-022-00938-y.
References
- 1.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 2.Kang Y, Zhou XE, Gao X, He Y, Liu W, Ishchenko A, et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–7. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu P, Huang S, Zhang H, Mao C, Zhou XE, Cheng X, et al. Structural insights into the lipid and ligand regulation of serotonin receptors. Nature. 2021;592:469–73. doi: 10.1038/s41586-021-03376-8. [DOI] [PubMed] [Google Scholar]
- 4.Duan J, Xu P, Cheng X, Mao C, Croll T, He X, et al. Structures of full-length glycoprotein hormone receptor signalling complexes. Nature. 2021;598:688–92. doi: 10.1038/s41586-021-03924-2. [DOI] [PubMed] [Google Scholar]
- 5.Hauser AS, Attwood MM, Rask-Andersen M, Schiöth HB, Gloriam DE. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov. 2017;16:829–42. doi: 10.1038/nrd.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kooistra AJ, Mordalski S, Pándy-Szekeres G, Esguerra M, Mamyrbekov A, Munk C, et al. GPCRdb in 2021: integrating GPCR sequence, structure and function. Nucleic Acids Res. 2021;49:D335–43.. doi: 10.1093/nar/gkaa1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thal DM, Vuckovic Z, Draper-Joyce CJ, Liang Y-L, Glukhova A, Christopoulos A, et al. Recent advances in the determination of G protein-coupled receptor structures. Curr Opin Struct Biol. 2018;51:28–34. doi: 10.1016/j.sbi.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Safdari HA, Pandey S, Shukla AK, Dutta S. Illuminating GPCR signaling by Cryo-EM. Trends Cell Biol. 2018;28:591–4. doi: 10.1016/j.tcb.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Duan J, Shen D-D, Zhou XE, Bi P, Liu Q-F, Tan Y-X, et al. Cryo-EM structure of an activated VIP1 receptor-G protein complex revealed by a NanoBiT tethering strategy. Nat Commun. 2020;11:4121. doi: 10.1038/s41467-020-17933-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rose PW, Prlić A, Altunkaya A, Bi C, Bradley AR, Christie CH, et al. The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2017;45:D271–81.. doi: 10.1093/nar/gkw1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jonić S. Cryo-electron microscopy analysis of structurally heterogeneous macromolecular complexes. Comput Struct Biotechnol J. 2016;14:385–90. doi: 10.1016/j.csbj.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. Advances in Neural Information Processing Systems 30. Long Beach, CA: USA; 2017. p 5998–6008.
- 14.Thomas N, Smidt T, Kearnes S, Yang L, Li L, Kohlhoff K, et al. Tensor field networks: rotation-and translation-equivariant neural networks for 3d point clouds. Computing Research Repository. 2018;abs/1802.08219:1–19.
- 15.Tunyasuvunakool K, Adler J, Wu Z, Green T, Zielinski M, Žídek A, et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–6. doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masrati G, Landau M, Ben-Tal N, Lupas A, Kosloff M, Kosinski J. Integrative structural biology in the era of accurate structure prediction. J Mol Biol. 2021;433:167127. doi: 10.1016/j.jmb.2021.167127. [DOI] [PubMed] [Google Scholar]
- 17.Cramer P. AlphaFold2 and the future of structural biology. Nat Struct Mol Biol. 2021;28:704–5. doi: 10.1038/s41594-021-00650-1. [DOI] [PubMed] [Google Scholar]
- 18.Schiöth HB, Fredriksson R. The GRAFS classification system of G-protein coupled receptors in comparative perspective. Gen Comp Endocrinol. 2005;142:94–101. doi: 10.1016/j.ygcen.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 19.Isberg V, de Graaf C, Bortolato A, Cherezov V, Katritch V, Marshall FH, et al. Generic GPCR residue numbers—aligning topology maps while minding the gaps. Trends Pharmacol Sci. 2015;36:22–31. doi: 10.1016/j.tips.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–50. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 21.Lu S, He X, Yang Z, Chai Z, Zhou S, Wang J, et al. Activation pathway of a G protein-coupled receptor uncovers conformational intermediates as targets for allosteric drug design. Nat Commun. 2021;12:4721. doi: 10.1038/s41467-021-25020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dror RO, Arlow DH, Maragakis P, Mildorf TJ, Pan AC, Xu H, et al. Activation mechanism of the β2-adrenergic receptor. Proc Natl Acad Sci USA. 2011;108:18684–9. doi: 10.1073/pnas.1110499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Latorraca NR, Venkatakrishnan AJ, Dror RO. GPCR dynamics: structures in motion. Chem Rev. 2016;117:139–55. doi: 10.1021/acs.chemrev.6b00177. [DOI] [PubMed] [Google Scholar]
- 24.Ulloa-Aguirre A, Zariñán T, Jardón-Valadez E, Gutiérrez-Sagal R, Dias JA. Structure-function relationships of the follicle-stimulating hormone receptor. Front Endocrinol. 2018;29:707. doi: 10.3389/fendo.2018.00707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wingler LM, Lefkowitz RJ. Conformational basis of G protein-coupled receptor signaling versatility. Trends Cell Biol. 2020;30:736–47. doi: 10.1016/j.tcb.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu S, Zhang J. Small molecule allosteric modulators of G-protein-coupled receptors: drug–target Interactions. J Med Chem. 2019;62:24–45. doi: 10.1021/acs.jmedchem.7b01844. [DOI] [PubMed] [Google Scholar]
- 27.He X, Ni D, Lu S, Zhang J. Characteristics of allosteric proteins, sites, and modulators. In: Zhang J, Nussinov R, editors. Advances in experimental medicine and biology; v 1163. Protein Allostery in Drug Discovery. Singapore: Springer; 2019. p. 107–39. [DOI] [PubMed]
- 28.Zhou Q, Yang D, Wu M, Guo Y, Guo W, Zhong L, et al. Common activation mechanism of class A GPCRs. Elife. 2019;8:e50279. doi: 10.7554/eLife.50279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mattedi G, Acosta-Gutiérrez S, Clark T, Gervasio FL. A combined activation mechanism for the glucagon receptor. Proc Natl Acad Sci USA. 2020;117:15414–22. doi: 10.1073/pnas.1921851117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischer A, Smieško M, Sellner M, Lill MA. Decision making in structure-based drug discovery: visual inspection of docking results. J Med Chem. 2021;64:2489–500. doi: 10.1021/acs.jmedchem.0c02227. [DOI] [PubMed] [Google Scholar]
- 31.Jacobson KA. New paradigms in GPCR drug discovery. Biochem Pharmacol. 2015;98:541–55. doi: 10.1016/j.bcp.2015.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roth BL, Irwin JJ, Shoichet BK. Discovery of new GPCR ligands to illuminate new biology. Nat Chem Biol. 2017;13:1143–51. doi: 10.1038/nchembio.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang YL, Belousoff MJ, Zhao P, Koole C, Fletcher MM, Truong TT, et al. Toward a structural understanding of class B GPCR peptide binding and activation. Mol Cell. 2020;77:656–68. doi: 10.1016/j.molcel.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 34.DeVree BT, Mahoney JP, Vélez-Ruiz GA, Rasmussen SGF, Kuszak AJ, Edwald E, et al. Allosteric coupling from G protein to the agonist-binding pocket in GPCRs. Nature. 2016;535:182–6. doi: 10.1038/nature18324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weis WI, Kobilka BK. The molecular basis of G protein–coupled receptor activation. Annu Rev Biochem. 2018;87:897–919. doi: 10.1146/annurev-biochem-060614-033910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Madan BM. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–94. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 37.Josephs TM, Belousoff MJ, Liang Y-L, Piper SJ, Cao J, Garama DJ, et al. Structure and dynamics of the CGRP receptor in apo and peptide-bound forms. Science. 2021;372:eabf7258. doi: 10.1126/science.abf7258. [DOI] [PubMed] [Google Scholar]
- 38.Tehan BG, Bortolato A, Blaney FE, Weir MP, Mason JS. Unifying family A GPCR theories of activation. Pharmacol Ther. 2014;143:51–60. doi: 10.1016/j.pharmthera.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Munk C, Mutt E, Isberg V, Nikolajsen LF, Bibbe JM, Flock T, et al. An online resource for GPCR structure determination and analysis. Nat Methods. 2019;16:151–62. doi: 10.1038/s41592-018-0302-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pándy-Szekeres G, Munk C, Tsonkov TM, Mordalski S, Harpsøe K, Hauser AS, et al. GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res. 2018;46:D440–6.. doi: 10.1093/nar/gkx1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karageorgos V, Venihaki M, Sakellaris S, Pardalos M, Kontakis G, Matsoukas M-T, et al. Current understanding of the structure and function of family B GPCRs to design novel drugs. Hormones. 2018;17:45–59. doi: 10.1007/s42000-018-0009-5. [DOI] [PubMed] [Google Scholar]
- 42.Yang D, Zhou Q, Labroska V, Qin S, Darbalaei S, Wu Y, et al. G protein-coupled receptors: structure- and function-based drug discovery. Signal Transduct Target Ther. 2021;6:7. doi: 10.1038/s41392-020-00435-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore PB, Hendrickson WA, Henderson R, Brunger AT. The protein-folding problem: not yet solved. Science. 2022;375:507. doi: 10.1126/science.abn9422. [DOI] [PubMed] [Google Scholar]
- 44.Zhao S, Wu B, Stevens RC. Advancing chemokine GPCR structure based drug discovery. Structure. 2019;27:405–8. doi: 10.1016/j.str.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 45.Massink A, Amelia T, Karamychev A, IJzerman AP. Allosteric modulation of G protein-coupled receptors by amiloride and its derivatives. Perspectives for drug discovery? Med Res Rev. 2020;40:683–708. doi: 10.1002/med.21633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chun E, Thompson AA, Liu W, Roth CB, Griffith MT, Katritch V, et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure. 2012;20:967–76. doi: 10.1016/j.str.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X, He C, Wang M, Zhou Q, Yang D, Zhu Y, et al. Structures of the human cholecystokinin receptors bound to agonists and antagonists. Nat Chem Biol. 2021;17:1230–7. doi: 10.1038/s41589-021-00866-8. [DOI] [PubMed] [Google Scholar]
- 48.McCorvy JD, Wacker D, Wang S, Agegnehu B, Liu J, Lansu K, et al. Structural determinants of 5-HT2B receptor activation and biased agonism. Nat Struct Mol Biol. 2018;25:787–96. doi: 10.1038/s41594-018-0116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim K, Che T, Panova O, DiBerto JF, Lyu J, Krumm BE, et al. Structure of a hallucinogen-activated Gq-coupled 5-HT2A serotonin receptor. Cell. 2020;182:1574–88. doi: 10.1016/j.cell.2020.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rossmann MG. Molecular replacement-historical background. Acta Crystallogr D Biol Crystallogr. 2001;57:1360–6. doi: 10.1107/S0907444901009386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.