Abstract
Highly pathogenic avian H5N1 influenza virus (H5N1) infection in humans causes acute respiratory distress syndrome, leading to multiple organ failure. Five fatal cases of H5N1 infection in Vietnam were analyzed pathologically to reveal virus distribution, and local proinflammatory cytokine and chemokine expression profiles in formalin-fixed, paraffin-embedded lung tissues. Our main histopathological findings showed diffuse alveolar damage in the lungs. The infiltration of myeloperoxidase-positive and/or CD68 (clone KP-1)-positive neutrophils and monocytes/macrophages was remarkable in the alveolar septa and alveolar spaces. Immunohistochemistry revealed that H5N1 mainly infected alveolar epithelial cells and monocytes/macrophages in lungs. H5N1 replication was confirmed by detecting H5N1 mRNA in epithelial cells using in situ hybridization. Quantitation of H5N1 RNA using quantitative reverse transcription PCR assays revealed that the level of H5N1 RNA was increased in cases during early phases of the disease. We quantified the expression of tumor necrosis factor-alpha (TNF-α), interleukin (IL)-6, IL-8, regulated on activation normal T-cell expressed and secreted (commonly known as RANTES), and interferon-gamma-inducible protein of 10 kDa (IP-10) in formalin-fixed, paraffin-embedded lung sections. Their expression levels correlated with H5N1 RNA copy numbers detected in the same lung region. Double immunofluorescence staining revealed that TNF-α, IL-6, IL-8 and IP-10 were expressed in epithelial cells and/or monocytes/macrophages. In particular, IL-6 was also expressed in endothelial cells. The dissemination of H5N1 beyond respiratory organs was not confirmed in two cases examined in this study.
Keywords: autopsy, cytokine, chemokine, H5N1, influenza, immunohistochemistry, lung
Main
Since the outbreak of highly pathogenic avian H5N1 influenza virus infection in humans in 2003, the WHO has reported 608 confirmed cases from 15 countries, with a mortality rate of about 60% by August 2012 (http://www.who.int/influenza/human_animal_interface/EN_GIP_20120810CumulativeNumberH5N1cases.pdf). The potential for mutation and reassortment of the viral genome, which may be responsible for human-to-human transmission, has increased the threat of an influenza pandemic.
Many H5N1-infected patients develop acute respiratory distress syndrome and die because of respiratory failure or multiple organ failure.1, 2 Pathological study of autopsied cases revealed that the H5N1 virus infected type I and type II pneumocytes and caused primary viral pneumonia, which further developed into acute respiratory distress syndrome.3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18 Although seasonal influenza virus infection is also associated with pneumonia, the virus mainly infects epithelial cells of the upper respiratory tract.19, 20, 21 Complications involving pneumonia are often due to bacterial infection. In the case of the influenza pandemic that occurred in 2009, the virus, designated A(H1N1)pdm09, infected pneumocytes in some patients. It also caused severe respiratory failure, similar to that seen for H5N1 infection, during the first year of the outbreak.22, 23, 24, 25, 26 The analysis of 20 autopsied A(H1N1)pdm09 infection cases in Japan revealed that diffuse alveolar damage, similar to that seen in a case of H5N1 infection, was present in the lungs of 5 cases (25%).27
The pathological study of human tissues can be restricted because of the difficulty in obtaining samples, but it is crucial for elucidating pathogenesis of the disease. There have been only 16 reports of pathological studies carried out on fatal cases of H5N1 infection. These have been reported from Hong Kong,3, 4, 5, 6 Thailand,7, 8, 9, 10, 11, 12, 13 China14, 15, 16, 17 and Vietnam.18 Lungs infected with H5N1 presented with diffuse alveolar damage in all cases. In most cases, H5N1 antigens and RNA were detected in extrapulmonary organs, suggesting disseminated systemic H5N1 infection and viremia.
Nearly 77% (10/13 patients) of H5N1 infection cases have resulted in death at the National Hospital of Pediatrics Hanoi, since December 2003. In this study, post-mortem biopsied or autopsied tissues from five fatal cases were analyzed pathologically to reveal virus distribution and the expression levels of cytokines and chemokines in lungs. Immunohistochemistry, in situ hybridization, double immunofluorescence staining and quantitative reverse transcription PCR (qRT-PCR) methods were used in our studies.
Materials and methods
Patients and Formalin-Fixed, Paraffin-Embedded Tissues
H5N1 infection of the five cases was confirmed by detecting viral RNA in bronchiolar aspirate using RT-PCR. The numbering of the case was in order of the shortness of the disease duration. In three cases (cases 1, 4 and 5; Table 1 ), post-mortem biopsied lung tissues were examined; these cases have been reported previously in a short communication.18 In the other two cases, patients were autopsied at National Hospital of Pediatrics; in addition to lung tissue, liver, heart, kidney, intestine, spleen and pancreas were examined in case 2. For case 3, liver, heart and kidney tissue were also examined in addition to lung tissue (Table 1). Only formalin-fixed, paraffin-embedded tissues were available for this study. This study was approved by the institutional medical ethical committee of the National Institute of Infectious Diseases, Japan (approval no. 320) and National Hospital of Pediatrics Hanoi.
Table 1.
Summary of the five H5N1 infection cases examined in this study
| Case | Date of admission (month, year) | Age (year)/sex | Duration of illness (days) | Day of admission | Sample examined | Lung histology |
Laboratory data (on admission) |
Treatment |
Detection of viral-RNA or antigen |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
qRT-PCR |
||||||||||||||||
| WBC (/mm3) | Plt (/mm3) | CRP (mg/dl) | Antibiotics | Oseltamivir | MSL | Bronchial aspirate | FFPE lung copies per cell | IHC | ISH | |||||||
| 1 | December, 2003 | 12/F | 8 | Day 6 | Lung | DAD (exudative phase) | 2100 | 45 000 | 3.1 | Yes | No | Yes | + | 256 | + | + |
| 2 | March, 2008 | 11/M | 10 | Day 7 | Lung, liver, heart, kidney, spleen, intestine, pancreas | DAD (exudative phase) | 1700 | 207 000 | 6.8 | Yes | Yes | Yes | + | 4.89 | + | + |
| 3 | December, 2007 | 4/M | 13 | Day 11 | Lung, liver, heart, kidney | DAD (exudative phase) | 2300 | 154 000 | 2.4 | Yes | Yes | Yes | + | 23.1 | + | + |
| 4 | July, 2004 | 4/M | 16 | Day 8 | Lung | DAD (proliferation phase) | 2300 | 150 000 | 0.59 | Yes | Yes | Yes | + | 1.05 | − | − |
| 5 | December, 2003 | 5/M | 18 | Day 8 | Lung | DAD (proliferation phase) | 3400 | 174 000 | 0.59 | Yes | No | Yes | + | UDL | − | − |
Abbreviations: CRP, C-reactive protein; DAD, diffuse alveolar damage; FFPE, formalin-fixed, paraffin-embedded; IHC, immunohistochemistry; ISH, in situ hybridization; MSL, methylpredonisolone(10 mg/kg/day); Plt, Platelet; qRT-PCR, quantitaive real-time RT-PCR; UDL, under detection limit; WBC, white blood cell; +, positive; −, negative.
Histopathological Studies and Immunohistochemistry
The formalin-fixed, paraffin-embedded tissues were cut (3-μm thick sections) and mounted on silane-coated glass slides. Histopathological studies were performed on all samples using hematoxylin–eosin (HE) staining. For lung sections, Elastica–Masson Goldner staining was also done. Immunohistochemistry for influenza A nucleoprotein antigen (InfA-NP) was performed to evaluate the distribution of H5N1 antigens with a mouse monoclonal antibody against InfA-NP28 as previously described.18, 23, 27
Immunohistochemistry for cell type-specific marker proteins, cytokines and chemokines was performed using primary antibodies against the following proteins: myeloperoxidase (MPO; Nichirei Bioscience, Tokyo, Japan); neutrophil elastase (DAKO Cytomation, Copenhagen, Denmark); CD68 (KP-1 or PGM-1; DAKO); CD8 (Novocastra Laboratories, Newcastle, UK); tumor necrosis factor-alpha (TNF-α; BD Pharmingen, San Diego, CA, USA); interleukin (IL)-6 (R&D Systems, Minneapolis, MN, USA); IL-8 (R&D Systems); regulated on activation normal T-cell expressed and secreted (RANTES; R&D Systems); and interferon-gamma-inducible protein of 10 kDa (IP-10; R&D Systems). After incubation with these primary antibodies, signals were detected using the avidin–biotin complex immunoperoxidase method (LSAB2, DAKO). A biotin-free catalyzed signal amplification system (CSAII, DAKO) was used for the detection of TNF-α, IL-6 and CD8. The EnVision system (DAKO) was used for the detection of neutrophil elastase. As a negative control, an irrelevant antibody was used in place of the primary antibody.
In Situ Hybridization
Influenza virus genomic RNA and mRNA were detected in formalin-fixed, paraffin-embedded lung sections by in situ hybridization AT tailing combined with a catalyzed signal amplification method (ISH-AT) as described previously.29, 30 Anti-sense and sense ISH-AT probes against H5N1 nucleoprotein (NP) genes were prepared and used to detect H5N1 mRNA and genomic RNA. The anti-sense ISH-AT probe against the rabies virus NP gene was used as an irrelevant negative control (Table 2 ).
Table 2.
Primers and probes for quantitative real-time RT-PCR and in situ hybridization AT tailing
| Target | Primer and probe | Sequence | Gene bank or reference |
|---|---|---|---|
| Influenza virus-matrix | Forward primer Reverse primer Probe |
5′-AGCAAAAGCAGGTAGATRTT-3′ 5′-TCGGCTTTGAGGGGG-3′ 5′-FAM-AMCCGAGGTCGAAACGTAYG-TAMRA-3′ |
CY006300 Ref. Ng et al31 |
| TNF-α | Forward primer Reverse primer Probe |
5′-CAGAGGGAAGAGTTCCCCAGG-3′ 5′-GGCTACAGGCTTGTCACTGG-3′ 5′-FAM-TGGCCCAGGCAGTCAGATCATCTTCTCG-TAMRA-3′ |
NM_000594.2 |
| IL-6 | Forward primer Reverse primer Probe |
5′-GAAGCTCTATCTCCCCTCCAGG-3′ 5′-GCAACACCAGGAGCAGCC-3′ 5′-FAM-ACTCCTTCTCCACAAGCGCCTTCGGT-TAMRA-3′ |
NM_000600.3 |
| IL-8 | Forward primer Reverse primer Probe |
5′-CTTGGCAGCCTTCCTGATTTC-3′ 5′-GCACTGACATCTAAGTTCTTTAGCA-3′ 5′-FAM-GCTCTGTGTGAAGGTGCAGTTTTGCCA-TAMRA-3′ |
NM_000584.2 |
| RANTES | Forward primer Reverse primer Probe |
5′-GCATCTGCCTCCCCATATTCC-3′ 5′-CCACTGGTGTAGAAATACTCCTTGA-3′ 5′-FAM-CTGCTTTGCCTACATTGCCCGCCCA-TAMRA-3′ |
NM_002985.2 |
| IP-10 | Forward primer Reverse primer Probe |
5′-GCCATTCTGATTTGCTGCCTTA-3′ 5′-TGCAGGTACAGCGTACAGTTC-3′ 5′-FAM-AGTGGCATTCAAGGAGTACCTCTCTCT-TAMRA-3′ |
NM_001565 |
| β-Actin | Forward primer Reverse primer Probe |
5′-TGAGCGCGGCTACAGCTT-3′ 5′-TCCTTAATGTTCACGCACGATTT-3′ 5′-FAM-ACCACCACGGCCGAGCGG-TAMRA-3′ |
NM_00110 Ref. Krafft et al32 |
| H5N1 virus -NP | Sense probe Antisense probe |
5′-GCAAGGGTCAACTCTCCCGAGGAGATCTGGAGCTGCTGGT-(AT)10-3′ 5′-ACCAGCAGCTCCAGATCTCCTCGGGAGAGTTGACCCTTGC-(AT)10-3′ |
AY651530 |
| Rabies virus -NP | Antisense probe | 5′-CAGTGGGGTCCCTTGTCAGCTCCATACCTCCCGTCAGAGC-(AT)10-3′ | AB573762.1 |
Double Immunofluorescence Staining
Double immunofluorescence staining for InfA-NP, cytokines, chemokines or for cell type-specific marker proteins was performed. We used antibodies against the following marker proteins to determine cell type: MPO (DAKO); surfactant apoprotein D (SP-D; Chemicon, Temecula, CA, USA); CD68 (KP-1 or PGM-1; DAKO); cytokeratin AE1/AE3 (DAKO); epithelial membrane antigen (EMA; DAKO); and CD34 (Novocastra). AlexaFluor 488-conjugated anti-rabbit or anti-mouse IgG (Molecular Probes) and AlexaFluor 568-conjugated anti-mouse or anti-rabbit IgG (Molecular Probes) were used as secondary antibodies. Nuclei were stained with TO-PRO-3, a specific nucleic acid stain (Molecular Probes). Confocal laser scanning microscopy was used to visualize double immunofluorescence staining as described previously.18, 23, 27
RNA Extraction
RNA was extracted from formalin-fixed, paraffin-embedded tissue sections (10μm × 3) using a PureLink FFPE total RNA isolation kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. Each sample was treated with DNase I to eliminate DNA contamination using a Turbo DNA-free kit (Ambion, Austin, TX, USA). The total RNA concentration was determined from spectrophotometric optical density measurements.
qRT-PCR Assays
Copy numbers for each RNA of interest and human β-actin mRNA from each sample were determined by qRT-PCR performed in a Mx3005P (Stratagene, La Jolla, CA, USA) using a QuantiTect Probe RT-PCR kit (Qiagen GmbH, Hilden, Germany). Human β-actin mRNA was used as an internal reference gene that provided a normalization factor for the amount of RNA isolated from a specimen. The copy numbers of H5N1 per cell were calculated using β-actin mRNA copy number, which was estimated to be 1500 copies per cell. For H5N1 RNA, we used a primer-probe set that amplified a segment within the matrix protein (M) region of H5N1 RNA. The RT-PCR thermal cycling conditions were 50 °C for 30 min, then 95 °C for 15 min, followed by 40 cycles of 94 °C for 15 s, and 60 °C for 1 min. All samples were run in triplicate. Primers and probes were synthesized by Sigma Genosys (Sigma-Aldrich, St Louis, MO, USA; Table 2).
Results
Patients
The characteristics of the five patients are presented in Table 1. H5N1 infection was confirmed by detecting H5N1 RNA in bronchial aspirate by RT-PCR. The durations of illness were 8, 10, 13, 16 and 18 days for cases 1, 2, 3, 4 and 5, respectively. The patients did not have significant medical histories or underlying diseases. All of them had a chance to be infected because of direct contact with sick poultry. Clinical symptoms exhibited by all patients included fever, cough and dyspnea. The average day of admission, which corresponded to when patients complained of dyspnea, was day 8 of the illness. Soon after admission, they were placed on a ventilator because of reduced oxygen saturation. Laboratory data on admission showed that white blood cell count was low for all cases (1700–3400/mm3). Antibiotics had been prescribed in all cases before admission. Antiviral treatment with oseltamivir was used in cases 2, 3 and 4 after admission. Methylpredonisolone was administered in all cases except for case 5. Hemophilus influenza was cultured from the sputa of case 5 on admission.
Histopathological Findings in the Lung
Acute intra-alveolar edema, congestion and/or hemorrhage, desquamation of pneumocytes, interstitial and intra-alveolar inflammatory cell infiltration, fibrosis and type II pneumocyte hyperplasia were observed. Three cases died before day 13 of the illness and presented with the exudative phase of diffuse alveolar damage (Figures 1a and c ). The other two cases died after day 16 of the illness and presented with the proliferative phase of diffuse alveolar damage. The numbers of myofibroblasts and fibroblasts were increased in case 4 (Figure 1e). Hyaline membrane formation was shown by Elastica–Masson Goldner staining (Figure 1d).
Figure 1.
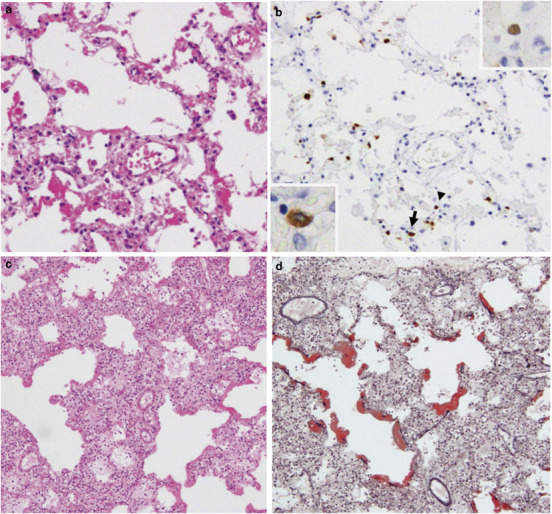
Representative histopathological findings in lung sections following hematoxylin–eosin (HE) (a, c, e) or Elastica–Masson Goldner staining (d). Immunohistochemistry was conducted to detect type A influenza virus nucleoprotein antigen (InfA-NP) (b). In situ hybridization AT-tailing (ISH-AT) (f–h) was also used. (a) Exudative phase of diffuse alveolar damage in Case 1. (b) InfA-NP antigen (brown) was detected in the nucleus (arrow head, upper inset) or cytoplasm (arrow, lower inset) of alveolar epithelial cells for Case 1. (c) Exudative phase of diffuse alveolar damage in case 3. (d) Elastica–Masson Goldner staining showed hyaline membrane formation (red) for case 3. (e) Proliferative phase of diffuse alveolar damage. The numbers of fibroblasts and myofibroblasts were increased in the alveolar septa and alveolar space in case 4. (f) ISH-AT using an antisense H5N1 nucleoprotein (NP) probe was able to detect H5N1 mRNA. (g) ISH-AT with a sense H5N1 NP probe detected H5N1 genomic RNA. (h) ISH-AT using an antisense rabies virus nucleocapsid probe as a negative control. Original magnifications, × 10 (c, d), × 20 (a, b), × 40 (b, inset, f–h).
Histopathological Findings in Extrapulmonary Organs
Several extrapulmonary organs were examined in cases 2 and 3 (Table 1). Inflammatory cell infiltration was not observed in any sections from extrapulmonary tissues. In the liver, we observed focal mild fatty changes around a lobule with a ballooning change of hepatocytes in both cases. Reactive hemophagocytosis3, 5, 6 and lymphoid depletion,5, 17 which are sometimes associated with hypercytokinemea, were not remarkable in the spleen. Renal tubular necrosis and congestion in the kidney17 were not observed. The pathological findings in extrapulmonary organs from both cases were limited and appeared to be caused by hypoxic changes rather than H5N1 infection.
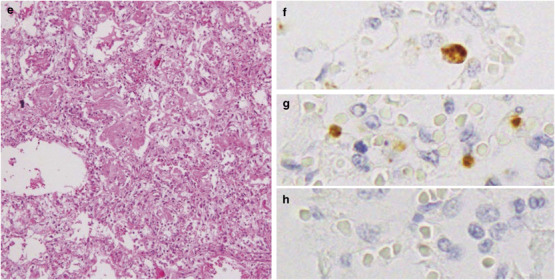
Characterization of Infiltrating Cells
The infiltrating cells in the alveolar septa and alveolar space (Figure 2a ) were characterized using immunohistochemistry on serial lung sections from the exudative phase of diffuse alveolar damage. Most infiltrating cells were MPO-positive (Figure 2b) and/or CD68 clone KP-1-positive (Figure 2c). MPO was expressed in neutrophils, monocytes and their precursors. CD68 clone KP-1 was expressed in not only monocytes/macrophages but also myeloid precursors and neutrophils. Neutrophil elastase was expressed in some of the infiltrated cells, suggesting they were neutrophils or precursor cells (Figure 2d). CD68 clone PGM-1 is more specific for mature macrophages in comparison with CD68 clone KP-1. CD68 clone PGM-1-positive alveolar macrophages were also detected (Figure 2e). Taken together, the results indicate that the infiltrating cells were mainly neutrophils and monocytes/macrophages, and their precursors. CD8-positive cytotoxic T lymphocytes were rarely detected in this region (Figure 2f). Several CD8-positive T lymphocytes were detected in other areas of the same lung section, but there were few of these in total.
Figure 2.
Hematoxylin–eosin (HE) staining (a) and immunohistochemistry for cell type-specific marker proteins (b–f) in serial lung sections from case 3. (a) Inflammatory cells infiltrated the alveolar septa and alveolar spaces. The phenotype of infiltrating cells were characterized using immunostaining for the detection of myeloperoxidase (MPO) (b), CD68 clone KP-1 (c), neutrophil elastase (d), CD68 clone PGM-1 (e) and CD8 T cells (f). Positive signals are indicated by brown staining. Original magnification, × 20 (a–f), × 40 (a–f, inset).
Viral Load
The relative levels of H5N1 viral RNA (genomic RNA and mRNA) in formalin-fixed, paraffin-embedded sections were quantified by qRT-PCR using primer-probe sets for the H5N1 M protein and β-actin gene sequences (Table 2). The relative H5N1 viral copy number per cell was calculated as described in the Materials and methods section. H5N1 RNA was under the detection limit (UDL) in all extrapulmonary tissue sections. The viral load varied among the lung regions from the same case (Figure 4a). The highest copy number of H5N1 RNA was seen from the lung in case 1, who had the shortest duration of illness. The lung in the proliferative phase of diffuse alveolar damage (cases 4 and 5) presented with a low viral load. In case 5, who died on day 18 of the disease, the amount of viral RNA was UDL in all lung sections (Figure 4a).
H5N1-Infected Cells Detected by Immunohistochemistry and In Situ Hybridization
The distribution of InfA-NP was examined by immunohistochemistry using monoclonal antibodies against the protein. It was detected in the lung tissue sections from cases 1, 2 and 3 (Table 1, Figure 1b). H5N1 mRNA and genomic RNA were detected separately in the lung sections of cases 1, 2 and 3 using ISH-AT with anti-sense and sense probes (Figures 1f and g). The detection of influenza mRNA was an indicator of virus proliferation in the lung. The InfA-NP-positive cells (red) were identified using double immunofluorescence staining for cell type-specific marker proteins (green) and TO-PRO-3 nucleic acid staining (blue). Positive signals were visualized by confocal laser scanning microscopy (Figure 3 ). The InfA-NP signals (Figures 3a, e and i) were mainly detected in the SP-D-positive type II pneumocytes (Figure 3b), and the CD68-positive monocytes/macrophages (Figure 3f). They were also detected in AE1/AE3-positive bronchiolar epithelial cells (Figure 3j). InfA-NP signals were detected in the nuclei (Figures 3d, h).
Figure 3.
Phenotype of influenza A nucleoprotein antigen (InfA-NP)-positive cells. (a, e, i) InfA-NP immunoreactivity can be seen in red. (b) Immunoreactivity for surfactant apoprotein D (SP-D) in type II pneumocytes. (f) CD68 clone KP-1 in monocytes. (j) Cytokeratin (AE1/AE3) in bronchiolar epithelial cells. (c, g, k) Colocalization for each type is also presented. (d, h, l) TO-PRO-3 nucleic acid staining (blue) revealed the InfA-NP in nuclei. Original magnification, × 400.
Cytokines and Chemokines
The elevation of cytokine and chemokine levels occurred in H5N1-infected lungs. Their expression levels were examined by quantifying the mRNA copy number of TNF-α, IL-6, IL8, RANTES and IP-10 in the five cases. The extracted RNA from 3 to 5 lung regions from each case was analyzed separately. The expression levels were variable from every region of the same case. Case 1 presented with the highest titers of cytokines and chemokines of all. In case 5, only IL-8 mRNA was detected. The expression level of every cytokine and chemokine correlated with the copy number of H5N1 RNA (Figure 4g ). This suggested that the local elevation of cytokines and chemokines in the lung were possibly caused by H5N1 infection in the same region.
Figure 4.
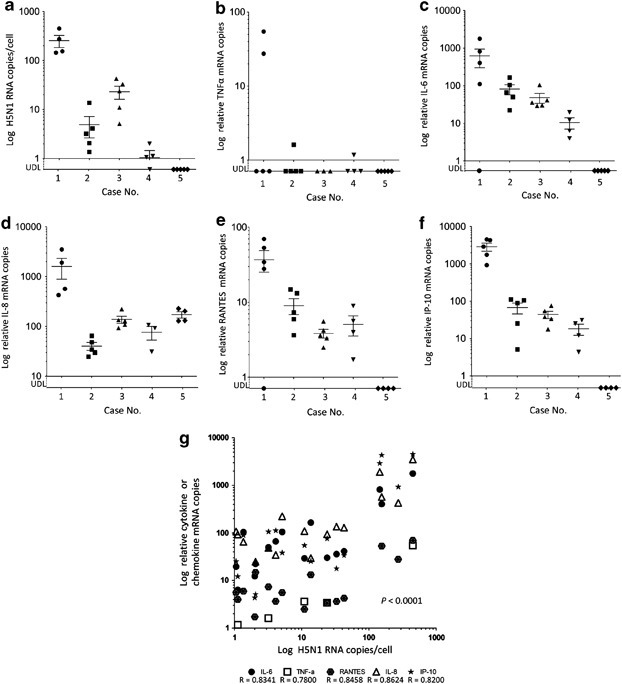
H5N1 RNA and proinflammatory cytokine or chemokine expression levels in several lung regions. The x axis indicates the case number. The durations of disease for cases 1, 2, 3, 4 and 5 were 8, 10, 13, 16 and 18 days, respectively. The y axis indicates H5N1 RNA (a), tumor necrosis factor-alpha (TNF-α) (b), interleukin (IL)-6 (c), IL-8 (d), regulated on activation normal T-cell expressed and secreted (RANTES) (e) and interferon-gamma-inducible protein of 10 kDa (IP-10) (f) mRNA copy numbers on a logarithmic scale. •, case 1; ▪, case 2; ▴, case 3; ▾, case 4; and ♦, case 5. (g) Correlation between copy numbers of H5N1 RNA and cytokine or chemokine mRNA in lung tissue. Data from the five cases (a–f) were combined, with any values below the limit of detection (UDL) excluded. The horizontal lines indicate the mean, and vertical error bars indicate the mean±s.d., P<0.0001.
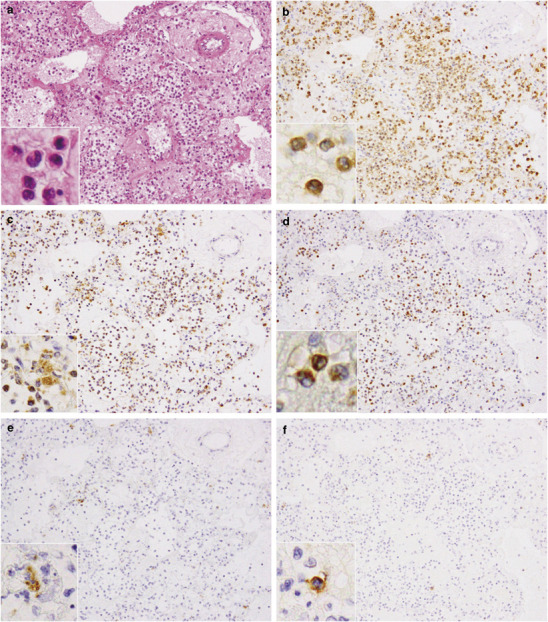
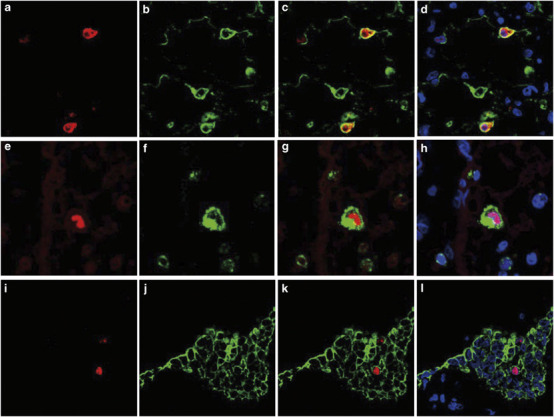
Next, we tried to detect these cytokines and chemokines in the lungs of cases 1 and 3 using immunohistochemistry (Figure 5 ). TNF-α, IL-6, IL-8, RANTES and IP-10 were detected at much higher levels in case 1 compared with case 3, which was compatible with the copy number of each mRNA shown in Figure 4g. The phenotype of each cytokine/chemokine-positive cell was determined using double immunofluorescence staining. Cells expressing RANTES could not be identified. TNF-α was detected in a MPO-positive monocytes (Figure 6a-3 ) and SPD-positive type II pneumocytes (Figure 6b-3). IL-6 was detected in PGM-1-positive monocytes/macrophages (Figure 6c-3) but also in an EMA-positive alveolar epithelial cells (Figure 6d-3) and in CD34-positive endothelial cells (Figure 6e-3). IL-8 was detected in PGM-1-positive monocytes/macrophages (Figure 6f-3). IP-10 was detected in PGM-1-positive monocytes/macrophages (Figure 6g-3) and EMA-positive bronchiolar epithelial cells (Figure 6h-3).
Figure 5.
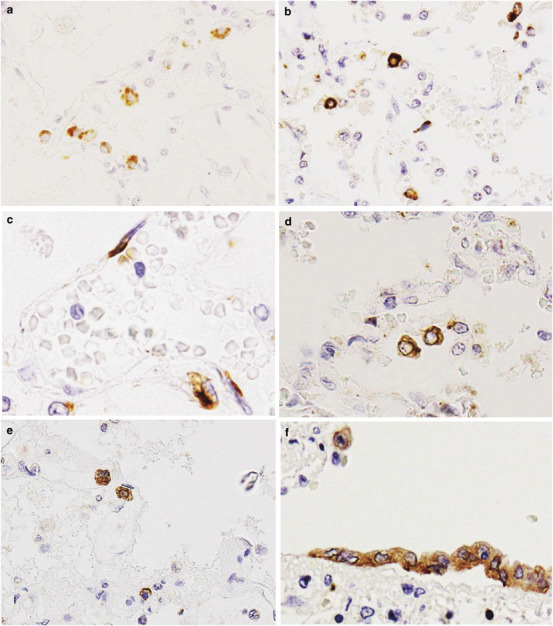
Immunohistochemistry on lung tissue sections for the detection of cytokines and chemokines. (a) Tumor necrosis factor-alpha (TNF-α), (b, c) interleukin (IL)-6, (d) IL-8, (e) regulated on activation normal T-cell expressed and secreted (RANTES) and (f) interferon-gamma-inducible protein of 10 kDa (IP-10) were detected in the cytoplasm of several cells. (c) IL-6 immunoreactivity was observed in endothelial cells. (f) IP-10 immunoreactivity was seen in bronchiolar epithelial cells. A positive signal is indicated by brown staining. Original magnification, × 40.
Figure 6.
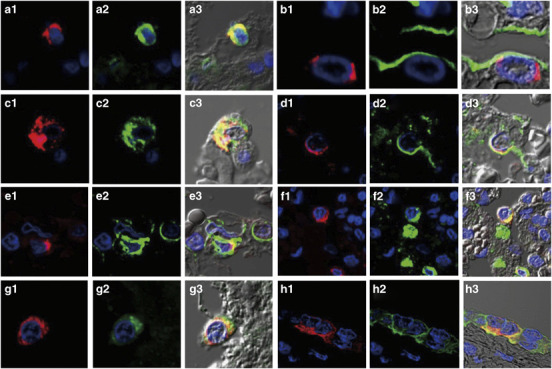
Double immunofluorescence staining for phenotype determination of tumor necrosis factor-alpha (TNF-α), interleukin (IL)-6-, IL-8- or IP-10-positive cells. (a-1, b-1) TNF-α, (c-1, d-1, e-1) IL-6, (f-1) IL-8 and (g-1, h-1) interferon-gamma-inducible protein of 10 kDa (IP-10) immunoreactivity (red) and cell type-specific marker protein immunoreactivity (green). (a-3, b-3, c-3, d-3, e-3, f-3, g-3, h-3) Colocalization of immunostaining. (a-2) Myeloperoxidase (MPO) in monocytes. (b-2) Surfactant apoprotein D (SP-D) in type II pneumocytes. (c-2, f-2, g-2) CD68 clone PGM-1 in monocytes/macrophages. (d-2) Epithelial membrane antigen (EMA) in alveolar epithelial cells. (e-2) CD34 in endothelial cells. (h-2) EMA in bronchiolar epithelial cells. TO-PRO-3 nucleic acid staining (blue) is shown. Differential interference contrast (DIC) images are shown in a-3, b-3, c-3, d-3, e-3, f-3, g-3 and h-3. Original magnification, × 600.
Discussion
All post-mortem biopsied or autopsied cases with H5N1 infection reported to date have shown acute respiratory distress syndrome clinically and diffuse alveolar damage in lung histopathology.3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18 The five cases analyzed in this study also suffered from viral pneumonia, which led to acute respiratory distress syndrome. H5N1 antigens and RNA were detected in pneumocytes and monocytes/macrophages in cases 1, 2 and 3 (Figures 1e–g). In particular, these three cases presented with the exudative phase of diffuse alveolar damage via histopathology, with infiltration of inflammatory cells in the alveolar septa and alveolar space remarkably high. Immunohistochemistry revealed that the inflammatory cells were not lymphocytes, but mostly MPO- and/or CD68 clone KP-1-positive neutrophils or monocytes/macrophages (Figure 2). A more detailed study is necessary to elucidate the mechanism of infiltration for these cells in the H5N1-infected lung.
H5N1 patients presented with dysregulation of cytokine and chemokine levels, which is often referred to as a ‘cytokine storm'. This is thought to be one of the key mechanisms in the pathogenesis of H5N1 infection.3, 33 According to in vitro experiments, H5N1 infection of primary human macrophages, along with alveolar and bronchial epithelial cells induced proinflammatory cytokines and chemokines more potently than seasonal influenza virus infection.34, 35, 36 Animal experiments also showed induction of proinflammatory cytokines and chemokines occurred because of H5N1 infection.37, 38 In H5N1-infected human lungs, the local expression of cytokines and chemokines was investigated using RT-PCR9, 10, 11, 13, 17 or immunohistochemistry.4, 15, 16 The cytokines and chemokines reported to be upregulated in H5N1-infected lungs were: TNF-α,4, 9, 10, 13, 16, 17 IFN-α/β,11 IP-10,11, 13, 16, 17 RANTES,16, 17 MIP-3β,17 IL-6,16 IFN-γ,16 IFN-β,16 IL-8,16 MCP-1,16 and MIP-1α.15, 16 In this study, based on these findings, we quantified the expression of five proinflammatory cytokines and chemokines in formalin-fixed, paraffin-embedded H5N1-infected lung tissues. We examined 3–5 lung regions per case. It was impossible to examine the time course of expression for each mediator in the post-mortem biopsied or autopsied lungs. The five patients in this study had no significant medical histories or underlying diseases, which could possibly affect expression levels of cytokines and chemokines. Furthermore, all formalin-fixed, paraffin-embedded samples were prepared using the same procedures at the same hospital. We have presented the expression levels of each mediator in the lungs of the five cases in order of shortness of disease duration (Figure 4). The local induction of proinflammatory cytokines and chemokines were confirmed in several H5N1-infected lung tissues. The expression levels of cytokines and chemokines in case 1, with the shortest duration of disease, were the highest among all cases (Figures 4b–f). In addition, the expression levels of cytokines and chemokines correlated with viral load in every lung region, suggesting that H5N1-induced upregulation of cytokines and chemokines in the lung (Figure 4g). Double immunofluorescence staining revealed that cells expressing the cytokines and chemokines were mainly monocytes/macrophages or epithelial cells (Figure 6). Our results were consistent with those previously reported.4, 16 We also noticed the new finding that IL-6 was expressed in EMA-positive alveolar epithelial cells (Figure 6d-3), CD34-positive endothelial cells (Figure 6e-3), and PGM-1-positive monocytes/macrophages (Figure 6c-3). It should be noted that IL-6 was also detected in endothelial cells, which may be related to local vascular injury in H5N1-infected lung (Figure 6e-3).
The most important characteristics of H5N1 infection that distinguish it from other subtypes of influenza virus infection, are that H5N1 disseminates beyond the respiratory system.14 For disseminated infection, H5N1 should have been in the bloodstream at some point. Virus isolation from peripheral blood is considered evidence of viremia.33, 39 Actually, virus antigens and H5N1 RNA have been reported to be detected in extrapulmonary tissues from several fatal cases.6, 9, 10, 12, 13, 14, 15, 17 The positive strand of H5N1 mRNA, indicative of viral replication, was detected by strand-specific RT-PCR in the intestines,6, 13, 14 liver,9, 13 heart,13, 14 lymph node,13 placenta14 and brain.14 On the other hand, histopathological findings were mostly nonspecific for H5N1 infection, such as hemophagocytotic activity, depletion of lymphoid cells, acute tubular necrosis, fatty changes in the liver and brain edema. More specific pathological changes, such as inflammatory cell infiltrations associated with the detection of viral antigen would be needed to show disseminate H5N1 infection. In extrapulmonary organs of two autopsied cases in this study, we were unable to obtain evidence of H5N1 dissemination. For both cases, illness lasted >10 days and the titers of H5N1 RNA were low, even in lung sections (Table 1, Figure 4a). Therefore, it might be reasonable to suggest that H5N1 RNA in extrapulmonary organs were below the level of detection. In addition, the histopathological findings of the extrapulmonary tissues were limited to nonspecific ischemic changes. The inconsistent results regarding the extent of H5N1 distribution in fatal cases are likely due to several factors, including the conditions of samples, the duration of disease and the medication given to patients. In addition to this, the differences in permissivity and immunological reactivity to H5N1 among patients should be also considered.
We investigated formalin-fixed, paraffin-embedded tissues from five fatal H5N1 cases. H5N1 viral load was highest in the lung of the case with the shortest duration of disease. Proinflammatory cytokine and chemokine mRNA copy numbers correlated with H5N1 RNA copy numbers in each lung region. In H5N1-infected lungs, monocytes/macrophages, epithelial cells and endothelial cells produced several cytokines and chemokines. We were unable to determine any dissemination of H5N1 beyond the respiratory organs in two autopsied cases. Further investigation is necessary to elucidate the pathogenesis of H5N1 infection in humans.
Acknowledgments
We thank Dr Thuy TB Phung for clinical information of patients and Ms K Shimonohara for technical assistance. This work was supported by the Health and Labor Sciences Research Grants on Emerging and Re-emerging Infectious Diseases (H22 Shinko-Ippan-014), from the Ministry of Health, Labour and Welfare, Japan.
Competing interests
The authors declare no conflict of interest.
References
- 1.Beigel JH, Farrar J, Han AM. Avian influenza A (H5N1) infection in humans. N Engl J Med. 2005;353:1374–1385. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- 2.Yuen KY, Chan PK, Peiris M. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998;351:467–471. doi: 10.1016/S0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
- 3.To KF, Chan PKS, Chan KF. Pathology of fatal human infection associated with avian influenza A H5N1 virus. J Med Virol. 2001;63:242–246. doi: 10.1002/1096-9071(200103)63:3. [DOI] [PubMed] [Google Scholar]
- 4.Peiris JSM, Yu WC, Leung CW. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet. 2004;363:617–619. doi: 10.1016/S0140-6736(04)15595-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng WF, To KF, Lam WWL. The comparative pathology of severe acute respiratory syndrome and avian influenza A subtype H5N1-a review. Hum Pathol. 2006;37:381–390. doi: 10.1016/j.humpath.2006.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z, Zhang J, Huang K. Systemic infection of avian influenza A virus H5N1 subtype in human. Hum Pathol. 2009;40:735–739. doi: 10.1016/j.humpath.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ungchusak K, Aucwarakul P, Dowell SF. Probable person-to-person transmission of avian influenza (H5N1) N Engl J Med. 2005;352:333–340. doi: 10.1056/NEJMoa044021. [DOI] [PubMed] [Google Scholar]
- 8.Chotpitayasunondh T, Ungchusak K, Hanshaoworakul W. Human disease from influenza A (H5N1), Thailand, 2004. Emerg Infect Dis. 2005;11:201–209. doi: 10.3201/eid1102.041061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uiprasertkul M, Puthavathana P, Sangsiriwut K. Influenza A H5N1 replication sites in humans. Emerg Infect Dis. 2005;11:1036–1041. doi: 10.3201/eid1107.041313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uiprasertkul M, Kitphati R, Puthavathana P. Apoptosis and pathogenesis of avian influenza A (H5N1) virus in Humans. Emerg Infect Dis. 2007;13:708–712. doi: 10.3201/eid1305.060572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thitithanyanont A, Engering A, Uiprasertkul M. Antiviral immune response in H5N1-infected human lung tissue and possible mechanisms underlying the hyper production of interferon-inducible IP-10. Biochem Biophys Res Commun. 2010;398:752–758. doi: 10.1016/j.bbrc.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piwpankaew Y, Monteerarat Y, Suptawiwat O. Distribution of viral RNA, sialic acid receptor, and pathology in H5N1 avian influenza patients. Acta Pathol Microbiol Scand. 2010;118:895–902. doi: 10.1111/j.1600-0463.2010.02676.x. [DOI] [PubMed] [Google Scholar]
- 13.Sirinonthanawech N, Uiprasertkul M, Suptawiwat O. Viral load of the highly pathogenic avian influenza H5N1 virus in infected human tissues. J Med Virol. 2011;83:1418–1423. doi: 10.1002/jmv.22146. [DOI] [PubMed] [Google Scholar]
- 14.Gu J, Xie Z, Gao Z. H5N1 infection of the respiratory tract and beyond: a molecular pathology study. Lancet. 2007;370:1137–1145. doi: 10.1016/S0140-6736(07)61515-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korteweg C, Gu J. Pathology, molecular biology, and pathogenesis of avian influenza A (H5N1) infection in humans. Am J Pathol. 2008;172:1155–1170. doi: 10.2353/ajpath.2008.070791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng R, Lu M, Korteweg C. Distinctly different expression of cytokines and chemokines in the lung of two H5N1 avian influenza patients. J Pathol. 2008;216:328–336. doi: 10.1002/path.2417. [DOI] [PubMed] [Google Scholar]
- 17.Gao R, Dong L, Dong J. A systemic molecular pathology study of a laboratory confirmed H5N1 human case. PLoS ONE. 2010;5:e13315. doi: 10.1371/journal.pone.0013315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liem NT, Nakajima N, Phat LP. H5N1-infected cells in lung with diffuse alveolar damage in exudative phase from a fatal case in Vietnam. Jpn J Infect Dis. 2008;61:157–160. PubMed PMID: 18362412. [PubMed] [Google Scholar]
- 19.Taubenberger JK, Morens DM. The pathology of influenza virus infections. Annu Rev Pathol. 2008;3:499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guarner J, Shieh WJ, Dawson J. Immunohistochemical and in situ hybridization studies of influenza A virus infection in human lungs. Am J Clin Pathol. 2000;114:227–233. doi: 10.1309/HV74-N24T-2K2C-3E8Q. [DOI] [PubMed] [Google Scholar]
- 21.Guarner J, Pddock CD, Shieh WJ. Histopathologic and immunohistochemical features of fatal influenza virus infection in children during the 2003-2004 season. Clin Infect Dis. 2006;43:132–140. doi: 10.1086/505122. [DOI] [PubMed] [Google Scholar]
- 22.Soto-Abraham MV, Soriano-Rosas J, Diaz-Quinonez A. Pathological changes associated with 2009 H1N1 virus. N Engl J Med. 2009;361:2001–2003. doi: 10.1056/NEJMc0907171. [DOI] [PubMed] [Google Scholar]
- 23.Nakajima N, Hata S, Sato Y. The first case of pandemic influenza (A/H1N1) virus infection in Japan: Detection of a high copy number of the virus in type II alveolar epithelial cells by pathological and virological examination. Jpn J Infect Dis. 2010;63:67–71. PubMed PMID: 20093768. [PubMed] [Google Scholar]
- 24.Maud T, Hajjar LA, Callegari GD. Lung pathology in fatal novel human influenza A (H1N1) infection. Am J Respir Crit Care Med. 2010;181:72–79. doi: 10.1164/rccm.200909-1420OC. [DOI] [PubMed] [Google Scholar]
- 25.Gill JR, Sheng ZM, Ely SF. Pulmonary pathologic findings of fatal 2009 pandemic influenza A/H1N1 viral infections. Arch Pathol Lab Med. 2010;134:235–243. doi: 10.5858/134.2.235. PMCID 2819217; PubMed PMID: 20121613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shieh WJ, Blau DM, Denison AM. 2009 Pandemic influenza A (H1N1): pathology and pathogenesis of 100 fatal cases in the United States. Am J Pathol. 2010;177:166–175. doi: 10.2353/ajpath.2010.100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakajima N, Sato Y, Katano H. Histopathological and immunohistochemical findings of 20 autopsy cases with 2009 H1N1 virus infection. Modern Pathol. 2012;25:1–13. doi: 10.1038/modpathol.2011.125. [DOI] [PubMed] [Google Scholar]
- 28.Chen Z, Sahashi Y, Matsuo K. Comparison of the ability of viral protein- expressing plasmid DNAs to protect against influenza. Vaccine. 1998;16:1544–1549. doi: 10.1016/S0264-410X(98)00043-7. [DOI] [PubMed] [Google Scholar]
- 29.Nakajima N, Petronela Ionescu, Sato Y. In situ hybridization AT-tailing with catalyzed signal amplification for sensitive and specific in situ detection of human immunodeficiency virus -1 mRNA in formaline-fixed and paraffin-embedded tissues. Am J Pathol. 2003;2:381–389. doi: 10.1016/S0002-9440(10)63833-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakajima N, Ozaki YA, Nagata N. SARS Coronavirus-infected cells in lung detected by new in situ hybridization technique. Jpn J Infect Dis. 2003;56:139–141. PubMed PMID: 12944688. [PubMed] [Google Scholar]
- 31.Ng EK, Cheng PK, Ng AY. Influenza A H5N1 detection. Emerg Infect Dis. 2005;11:21303–21305. doi: 10.3201/eid1108.041317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krafft AE, Russell KL, Hawksworth AW. Evaluation of PCR testing of ethanol-fixed nasal swab specimens as an augumented surveillance strategy for influenza virus and adenovirus identification. J Clin Microbiol. 2005;43:1768–1775. doi: 10.1128/JCM.43.4.1768-1775.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Jong MD, Simmons CP, Thanh TT. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheung CY, Poon LLM, Lau AS. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet. 2002;360:1831–1837. doi: 10.1016/S0140-6736(02)11772-7. [DOI] [PubMed] [Google Scholar]
- 35.Chan MC, Cheung CY, Chui WH. Proinflammatory cytokine reponses induced by influenza A (H5N1) viruses in primary human alveolar and bronchial epithelial cells. Respir Res. 2005;6:135. doi: 10.1186/1465-9921-6-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam WY, Yeung AC, Chu IM. Profiles of cytokine and chemokine gene expression in human pulmonary epithelial cells induced by human and avian influenza viruses. Virol J. 2010;7:344. doi: 10.1186/1743-422X-7-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baskin CR, Bielefeldt-Ohmann H, Tumpey TM. Early and sustained innate immune response defines pathology and death in nonhuman primates infected by highly pathogenic influenza virus. Proc Natl Acad Sci USA. 2009;9:3455–3460. doi: 10.1073/pnas.0813234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maines TR, Belser JA, Gustin KM. Local innate immune responses and influenza virus transmission and virulence in ferrets. J Infect Dis. 2012;205:474–485. doi: 10.1093/infdis/jir768. [DOI] [PubMed] [Google Scholar]
- 39.Chutinimithul S, Bhattarakosol P, Srisuratanon S. H5N1 influenza A virus and infected human plasma. Emerg Infect Dis. 2006;12:1041–1043. doi: 10.3201/eid1206.060227. [DOI] [PMC free article] [PubMed] [Google Scholar]


