Abstract
Amyotrophic lateral sclerosis (ALS) is an incurable neurodegenerative disease characterized by the degeneration of motor neurons in the spinal cord. Main symptoms are manifested as weakness, muscle loss, and muscle atrophy. Some studies have reported that alterations in sphingolipid metabolism may be intimately related to neurodegenerative diseases, including ALS. Acid sphingomyelinase (ASM), a sphingolipid-metabolizing enzyme, is considered an important mediator of neurodegenerative diseases. Herein, we show that ASM activity increases in samples from patients with ALS and in a mouse model. Moreover, genetic inhibition of ASM improves motor function impairment and spinal neuronal loss in an ALS mouse model. Therefore, these results suggest the role of ASM as a potentially effective target and ASM inhibition may be a possible therapeutic approach for ALS.
Keywords: Acid sphingomyelinase, Amyotrophic lateral sclerosis, FUS, Motor behavioral dysfunction, Motor neuronal loss
INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disease that influences motor neurons in the brain, brainstem, and spinal cord. Common symptoms of ALS include weakness, weight loss, and muscle atrophy, which ultimately lead to muscle paralysis (1-3). To date, mutations in more than 50 genes have been associated with ALS pathogenesis. Among these, fused in sarcoma (FUS), superoxide dismutase 1 (SOD1), TANK-binding kinase 1 (TBK1), TAR DNA-binding protein (TARDBP), and guanine nucleotide exchange C9orf72 (C9ORF72) are the most common genes associated with ALS (4-6). Although the exact mechanism of these mutations in ALS pathogenesis remains unclear, several studies have shown that these genes are associated with RNA processing, oxidative stress, cytoskeletal stability, protein aggregation and degradation, and autophagy (6-12).
Acid sphingomyelinase (ASM) encoded by Smpd1 is a significant sphingolipid-metabolizing enzyme (13). Previous studies have shown that the activity or expression of ASM increases in several neurodegenerative diseases, including Alzheimer’s disease, Parkinson’s disease, and ALS (14-17). The primary role of ASM is to catalyze the conversion of sphingomyelin, a significant component of membranes, into ceramide and phosphocholine (18). Increased ASM activity is involved in various pathological processes, such as neuronal cell death and differentiation, oxidative stress, senescence, disorganization of the cytoskeleton, and defective autophagy (15, 17, 18-21). However, the correlation between altered ASM levels and ALS pathology has not yet been fully characterized.
Herein, our study showed an increased ASM activity in the plasma or spinal cord of ALS patients and in a mouse model. Moreover, we found that genetic inhibition of ASM improved motor behavioral dysfunction and motor neuronal loss in the spinal cord of an ALS mouse model. Thus, our study provided evidence that increased ASM may influence ALS pathology, suggesting the possibility of ASM as a therapeutic target for ALS.
RESULTS
Increased ASM activity in ALS patients and mice
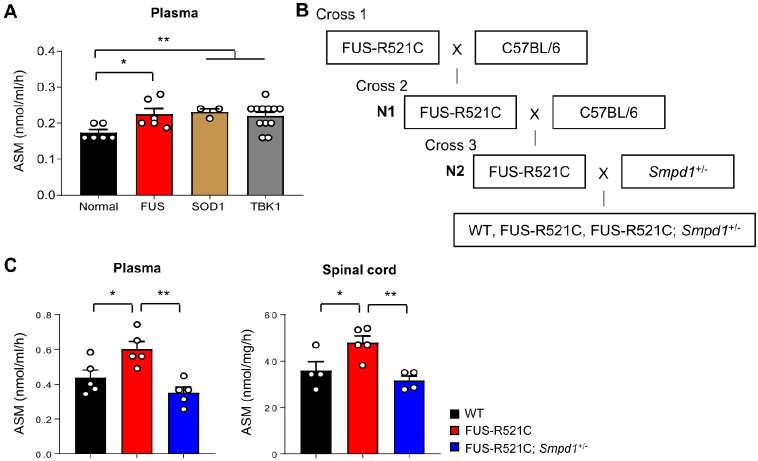
We first confirmed whether ASM activity was altered in plasma samples of ALS patients with FUS, SOD1, and TBK1 mutations (Table 1). The results showed a significant increase in ASM activity in the plasma of patients with ALS compared to that in controls (Fig. 1A). To investigate the influence of ASM on ALS pathology in a moue model, we used transgenic mice expressing FUS-R521C, which is a common mutation in familial ALS-FUS patients. These transgenic mice show severe dendritic and synaptic defects in spinal motor neurons, reduced muscle mass, and motor behavioral abnormalities. Moreover, most mice die within 4-6 weeks after symptom onset, and a minority can survive up to 4-6 months (22). To maintain the FUS-R521C colony, FUS-R521C mice were mated with C57BL/6 mice to generate N1 progeny, and N1 transgenic mice were further mated with C57BL/6 mice. FUS-R521C; Smpd1+/− mice were produced by mating N2 transgenic and Smpd1+/− mice (Fig. 1B). Similar to patients with ALS, ASM activity was elevated in the plasma and spinal cord of 12-week-old FUS-R521C mice (Fig. 1C). Importantly, ASM activity in age-matched FUS-R521C; Smpd1+/− mice was significantly lower than that in the FUS-R521C mice, with levels within the normal range (Fig. 1C). These results suggest that the elevation of ASM activity may influence the pathogenesis of ALS.
Table 1.
Human plasma information for control subjects and subjects with amyotrophic lateral sclerosis (ALS)
| Gene | Mutation | Number | Age (y) |
|---|---|---|---|
| Control | - | 6 | >60 |
| FUS | FUS (R495X) | 6 | >60 |
| FUS (G504WfsX12) | |||
| FUS (Q519E) | |||
| SOD1 | SOD1 (G11V) | 3 | >60 |
| SOD1 (I105T) | |||
| TBK1 | TBK1 (R384W) | 12 | >60 |
| TBK1 (I475T) | |||
| TBK1 (E476K) | |||
| TBK1 (I472Sfs*8) |
Fig. 1.
ASM activity increases in the plasma of ALS patients and both the plasma and spinal cords of ALS model mice; its activity decreases after genetic inhibition of ASM in ALS mice. (A) ASM activity in plasma of normal and ALS patients (n = 3-12 per group). (B) A schematic diagram showing the strategy used for generating FUS-R521C; Smpd1+/− mice. (C) ASM activity in the plasma and spinal cord of wild type (WT), FUS-R521C, and FUS-R521C; Smpd1+/− mice (n = 4-5 per group). One-way analysis of variance, Tukey’s post hoc test. *P < 0.05, **P < 0.01. All error bars indicate S.E.M. All data analysis was done on 12-week-old mice.
Genetic ASM inhibition improves motor behavioral dysfunction in FUS-R521C mice
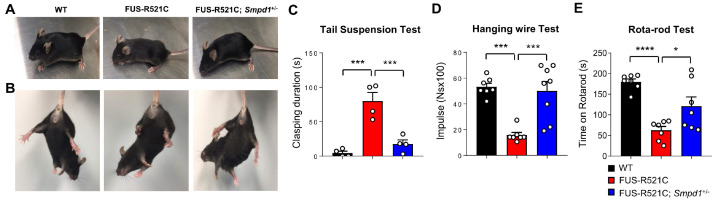
Next, to assess the potential effect of genetic ASM inhibition on motor function in FUS-R521C mice, we performed the tail suspension, hanging wire, and rotarod tests. Fig. 2A shows that FUS-R521C mice exhibited growth retardation, spastic paraplegia, and severe muscle wasting, but not in FUS-R521C; Smpd1+/− mice. In addition, we observed prolonged hind limb clasping in FUS-R521C mice during the tail-suspension test. However, FUS-R521C; Smpd1+/− mice showed hind limb clasping similar to that in wild type (WT) mice (Fig. 2B). In a 120-s period, the FUS-R521C mice spent approximately 80 s with hind limbs clasped together, whereas the FUS-R521C; Smpd1+/− mice exhibited a clasping phenotype for less than 20 s (Fig. 2C). The hanging wire test was used to evaluate the neuromuscular strength of the paws of experimental mice. The FUS-R521C mice showed a significant decrease compared to the WT mice regarding the time duration for which they could hang on to the wire. Compared to the FUS-R521C mice, the hanging wire test performance was better in the FUS-R521C; Smpd1+/− mice (Fig. 2D). We also confirmed a significant decrease in motor function/coordination in the FUS-R521C mice compared to that in WT mice through the rotarod test, the performance of which was better in the FUS-R521C; Smpd1+/− mice (Fig. 2E). Therefore, these results indicate that restoration of ASM activity to the normal range can improve motor behavioral dysfunction in FUS-R521C mice.
Fig. 2.
Genetic inhibition of ASM improves motor behavioral deficits in FUS-R521C mice. (A) Top, the majority of FUS-R521C mice show growth retardation, spastic paraplegia, and severe muscle wasting, but this is not evident in FUS-R521C; Smpd1+/− mice. Bottom, the FUS-R521C mice also exhibit prolonged hind limb clasping on the tail-suspension test, but this is not evident in the FUS-R521C; Smpd1+/− mice. (B, C) Images of hind limb clasping in mice and the quantification of clasping duration on the tail suspension test (n = 4 per group). (D) Holding impulse of each group on the hanging wire test (n = 7-8 per group). (E) Rotarod scores of each group on the rotarod test (n = 7 per group). One-way analysis of variance, Tukey’s post hoc test. *P < 0.05, ***P < 0.001, ****P < 0.0001. All error bars indicate S.E.M. All data analysis was done on 12-week-old mice.
Genetic ASM inhibition protects motor neuronal loss in FUS-R521C mice
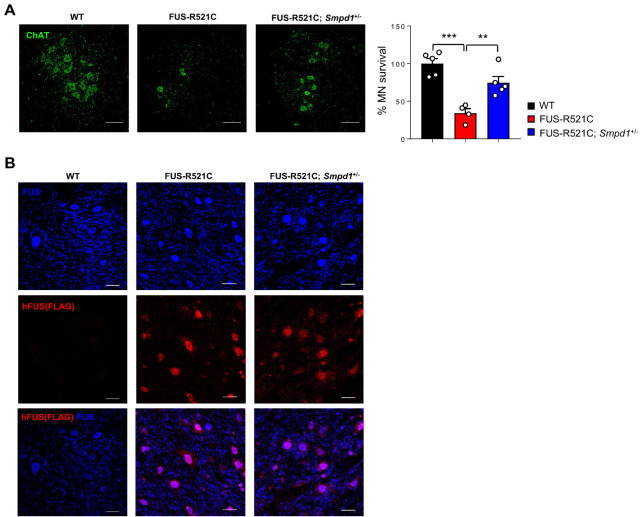
Motor neuron loss in the spinal cord is generally observed in ALS pathologies, and the FUS-R521C mutant is associated with motor neuron degeneration (23, 24). Based on this, we confirmed motor neuron survival using an antibody against choline acetyl transferase (ChAT) to visualize and quantitate spinal motor neurons. The number of ChAT+ motor neurons was significantly reduced in the spinal cord of the FUS-R521C mice. However, the FUS-R521C; Smpd1+/− mice showed an increase in the number of ChAT+ motor neurons (Fig. 3A), indicating that a decrease in ASM activity had a positive effect on motor neuron survival in FUS-R521C mice. The FUS protein is predominantly found in the nucleus than in the cytoplasm of neurons. However, in ALS patients with mutations in FUS, this protein is mislocalized in the cytoplasm, leading to motor neuron death (24, 25). To determine whether the FUS protein related to motor neuron survival is affected by ASM inhibition, the spinal cords of WT, FUS-R521C, and FUS-R521C; Smpd1+/− mice were immunostained with anti-FLAG and anti-FUS antibodies. The anti-FLAG antibody was used to detect FLAG-tagged FUS-R521C mutant proteins, and the anti-FUS antibody was used to detect endogenous FUS proteins. The confocal images showed that endogenous FUS protein remained localized to the nucleus and was less abundant in the cytoplasm for all groups. In contrast, FLAG-tagged FUS-R521C mutant proteins were found in both the nucleus and cytoplasm of the spinal cords of FUS-R521C mice. Moreover, we observed no significant differences in the expression of FLAG-tagged FUS-R521C mutant proteins between FUS-R521C; Smpd1+/− mice and FUS-R521C mice (Fig. 3B). These results suggest that the increase in spinal motor neuron survival by ASM inhibition in FUS-R521C mice is independent of the mislocalization of FUS-R521C mutant proteins.
Fig. 3.
Genetic inhibition of ASM prevents motor neuronal loss in the spinal cord of FUS-R521C mice. (A) Left, representative immunofluorescence images of motor neuron (ChAT, green) in the spinal cord of wild type (WT), FUS-R521C, and FUS-R521C; Smpd1+/− mice. Scale bars, 50 μm. Right, quantification of motor neuron survival in each group (n = 4-5 per group). (B) Representative immunofluorescence images of FUS and FUS-R521C protein detected by FUS antibody (blue) and hFUS (Flag) antibody (red). Scale bars, 20 μm. One-way analysis of variance, Tukey’s post hoc test. **P < 0.01, ***P < 0.001. All error bars indicate S.E.M. All data analysis was done on 12-week-old mice.
DISCUSSION
Although the causative pathogenic mechanisms of ALS remain unclear, the complex interactions between genetic and environmental factors contribute to the development and progression of the disease. ASM is increased by environmental stress and several neurodegenerative diseases (14). Previous studies have shown that patients with ALS and mouse models have high levels of ceramide in the spinal cord, which affects inflammation, the loss of neuromuscular junctions, and the degeneration of motor neurons (26). In this study, we revealed that ASM activity increased in samples from ALS patients and FUS-R521C mice, indicating that increased ASM correlated with high levels of ceramide in ALS. Moreover, this finding suggests that elevation of ASM activity could influence the pathogenesis of ALS through ceramide.
Genetic inhibition of ASM improved motor behavioral function and spinal neuronal loss in FUS-R521C mice without affecting the expression of FUS-R521C proteins in the nucleus and cytoplasm. Mutations in FUS cause impaired neuronal autophagy and abnormal protein accumulation in motor neurons, leading to motor neuronal death (27, 28). A previous study has also shown that ASM increase is involved in autophagy dysfunction and contributes to defective degradation of abnormal proteins in neurons (15). These reports suggest that the protective effect of ASM inhibition on spinal neuronal loss may be related to the correction of defective autophagy in FUS-R521C mice. Moreover, it was possibly a direct improvement effect of ASM inhibition, independent of FUS-R521C protein expression. We will conduct further studies on more detailed therapeutic mechanisms of ASM inhibition in the future.
The most widely used drugs for ALS are those that manage symptoms, prevent complications, and slow down progression. However, since the mechanisms of action are likely to be varied and complex, they have failed to show significant efficacy in ALS (6, 7). Another treatment strategy is RNA-targeting therapeutics, such as short interfering RNA (siRNA). siRNA is used to decrease the expression of target genes by interacting with the RNA-induced silencing complex. This treatment has also been delayed by inefficient and poorly targeted delivery (6, 7). Small molecules have been investigated as alternatives and potential therapeutics for ALS. These small molecules are compounds with anti-inflammatory, anti-oxidative, anti-apoptotic, autophagy-inducing, and neuroprotective properties (29). Although the vast majority of compounds have limitations in the exact mode of action and their efficacy, the concomitant use of some compounds is allowed for the treatment of ALS.
Previous studies have suggested that several antidepressant drugs affect ASM inhibition through action as functional inhibitors (17, 30). However, these inhibitors lack specificity, have off-target effects, and have unclear mechanisms of therapeutic action in neurodegenerative diseases. Based on this, a new small compound was recently identified as a selective and direct ASM inhibitor without off-target effects (31). This ASM inhibitor is significantly effective in improving neuroinflammation, autophagy dysfunction, synapse loss, neuronal survival, and activity in neurodegenerative diseases (31). Moreover, this inhibitor exhibits excellent bioavailability, central nervous system distribution, and microsomal stability. Although ASM activity and its inhibitory efficacy should be further investigated in various ALS mouse models, this ASM inhibitor could be a potential therapeutic agent for ALS. Furthermore, we believe that the combination of ASM inhibitors with current therapeutic agents might present synergetic effects for the improvement of ALS pathology.
MATERIALS AND METHODS
Mice
The following mouse lines were used: C57BL/6 WT mice (The Jackson Laboratory), FUS-R521C transgenic mice (22) (C57BL/6 background), and Smpd1-/- mice (32) (C57BL/6 background). The block randomization method was used to allocate animals to the experimental groups. To eliminate bias, all investigators were blinded to the experimental groups during data collection and data analysis. Mice were housed under a 12 h day/12 h night cycle with adlibitum access to water and food pellets. All protocols were approved by the Kyungpook National University Institutional Animal Care and Use Committee (IACUC).
Plasma collection
Blood was collected into sodium heparin-coated tubes via intracardial bleeding at the time of death. Plasma was generated by centrifugation of freshly collected blood, and aliquots were stored at −80°C until use. Human plasma samples were obtained from individuals with ALS and age-matched controls from Hanyang University Hospital. Informed consent was obtained from all subjects according to the ethics committee guidelines of Hanyang University Hospital.
ASM activity assays
Enzymatic activity was measured as previously described (15, 31) using a UPLC system (Waters). Briefly, the spinal cord was lysed in a homogenization buffer containing 50 mM HEPES (Sigma-Aldrich, H3375), 150 mM NaCl (Sigma-Aldrich, S3014), 0.2% Igepal CA-630 (Sigma-Aldrich, I8896), and protease inhibitors (Calbiochem, 539131). Three microliters of the samples (plasma or spinal cord) were mixed with 3 μl of 200 μm BODIPY-C12-sphingomyelin (Invitrogen, D7711) diluted in 0.2 M of sodium acetate buffer, pH 5.0, 0.2 mM ZnCl2, and 0.2% Igepal CA-630, and incubated at 37°C for 1 h. Hydrolysis reactions were stopped by adding 114 μl and centrifuged at 13,000 rpm for 5 min. Thirty microliters of the supernatant was then transferred to a sampling glass vial, and 5 μl was applied to a UPLC system for analysis. Quantification was achieved by comparison with BODIPY-C12-ceramide using the Waters Millennium software.
Histological analysis
For immunofluorescence staining, the spinal cord was cut using a vibratome (30 μm). ChAT antibody (1:100, Millipore, AB144p), FLAG-M2 (1:100, Sigma-Aldrich, F3165), and FUS (1:100, Sigma-Aldrich, HPA008784) were used to stain the neurons and FUS proteins in the spinal cord. Sections were analyzed using a laser-scanning confocal microscope (FV3000; Olympus). The MetaMorph software (Molecular Devices) was used for quantification.
Behavioral studies
We performed behavioral studies to assess motor function as previously described (22, 33). For the tail suspension test, each mouse was hung from its tail at a height of 80 cm for 120 s, and the clasping duration was scored. For the hanging wire test, each mouse was placed on the wire lid of a conventional housing cage, and the lid was turned upside down. The latency from the beginning of the test until the mouse stood with at least two limbs on the lid was recorded. The animals made three attempts to stand for a maximum of 180 s per trial, and the longest latency was recorded. The rotarod apparatus (accelerating model 47600; Ugo Basile) was set to an initial speed of 4 rpm, and the acceleration was increased by 32 rpm every 25-30 s. Scores were registered every two days, and three independent tests were performed at each measurement. Uniform conditions were carefully maintained for each test, with a rest time of 1 h between trials. Each test was limited to 300 s.
Statistical analysis
Sample sizes were determined using the G*Power software (with α = 0.05 and a power of 0.8). In general, statistical methods were not used to recalculate or pre-determine sample sizes. The variance was similar within comparable experimental groups. Individuals performing the experiments were blinded to the identity of the experimental groups until the end of the data collection and analysis for at least one independent experiment. In cases where more than two groups were compared, one-way analysis of variance (ANOVA) was used, followed by Tukey’s HSD test. All statistical analyses were performed using the GraphPad Prism software (version 7.0). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 were considered to be significant.
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2020R1A2C3006875, 2020R1A2C3006734). This research was also supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare and MSIT, Republic of Korea (HU20C0345).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Rowland LP, Shneider NA. Amyotrophic lateral sclerosis. New Engl J Med. 2001;344:1688–1700. doi: 10.1056/NEJM200105313442207. [DOI] [PubMed] [Google Scholar]
- 2.Chio A, Logroscino G, Hardiman O, et al. Prognostic factors in ALS: a critical review. Amyotroph Lateral Scler. 2009;10:310–323. doi: 10.3109/17482960802566824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardiman O, Al-Chalabi A, Chio A, et al. Amyotrophic lateral sclerosis. Nat Rev Dis Primers. 2017;3:17071. doi: 10.1038/nrdp.2017.71. [DOI] [PubMed] [Google Scholar]
- 4.Zou ZY, Zhou ZR, Che CH, Liu CY, He RL, Huang HP. Genetic epidemiology of amyotrophic lateral sclerosis: a systematic review and meta-analysis. J Neurol Neurosurg Psychiatr. 2017;88:540–549. doi: 10.1136/jnnp-2016-315018. [DOI] [PubMed] [Google Scholar]
- 5.Boylan K. Familial ALS. Neurol Clin. 2015;33:807–830. doi: 10.1016/j.ncl.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mejzini R, Flynn LL, Pitout IL, Fletcher S, Wilton SD, Akkari PA. ALS genetics, mechanisms, and therapeutics: where are we now? Front Neurosci. 2019;13:1310. doi: 10.3389/fnins.2019.01310.097cd21b39bf48b0950c0db564d5c9d4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim G, Gautier O, Tassoni-Tsuchida E, Ma XR, Gitler AD. ALS genetics: gains, losses, and implications for future therapies. Neuron. 2020;108:822–842. doi: 10.1016/j.neuron.2020.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vance C, Rogelj B, Hortobágyi T, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosen DR, Siddique T, Patterson D, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 10.Oakes JA, Davies MC, Collins MO. TBK1: a new player in ALS linking autophagy and neuroinflammation. Mol Brain. 2017;10:5. doi: 10.1186/s13041-017-0287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.eedharan J, Sr, Blair IP, Tripathi VB, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schissel SL, Schuchman EH, Williams KJ, Tabas I. Zn2+-stimulated sphingomyelinase is secreted by many cell types and is a product of the acid sphingomyelinase gene. J Biol Chem. 1996;271:18431–18436. doi: 10.1074/jbc.271.31.18431. [DOI] [PubMed] [Google Scholar]
- 14.Kornhuber J, Rhein C, Muller CP, Muhle C. Secretory sphingomyelinase in health and disease. Biol Chem. 2015;396:707–736. doi: 10.1515/hsz-2015-0109. [DOI] [PubMed] [Google Scholar]
- 15.Lee JK, Jin HK, Park MH, et al. Acid sphingomyelinase modulates the autophagic process by controlling lysosomal biogenesis in Alzheimer's disease. J Exp Med. 2014;211:1551–1570. doi: 10.1084/jem.20132451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ong WY, Herr DR, Farooqui T, Ling EA, Farooqui AA. Role of sphingomyelinases in neurological disorders. Expert Opin Ther Targets. 2015;19:1725–1742. doi: 10.1517/14728222.2015.1071794. [DOI] [PubMed] [Google Scholar]
- 17.Park MH, Jin HK, Bae JS. Potential therapeutic target for aging and age-related neurodegenerative diseases: the role of acid sphingomyelinase. Exp Mol Med. 2020;52:380–389. doi: 10.1038/s12276-020-0399-8.c83739d0e4604ace88f605bf29c188de [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenkins RW, Canals D, Hannun YA. Roles and regulation of secretory and lysosomal acid sphingomyelinase. Cell Signal. 2009;21:836–846. doi: 10.1016/j.cellsig.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Xiang, Gulbins Erich, Zhang Yang. Oxidative stress triggers Ca-dependent lysosome trafficking and activation of acid sphingomyelinase. Cell Physiol Biochem. 2012;30:815–826. doi: 10.1159/000341460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith EL, Schuchman EH. The unexpected role of acid sphingomyelinase in cell death and the pathophysiology of common diseases. FASEB J. 2008;22:3419–3431. doi: 10.1096/fj.08-108043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park MH, Lee JY, Park KH, et al. Vascular and neurogenic rejuvenation in aging mice by modulation of ASM. Neuron. 2018;100:167–182. doi: 10.1016/j.neuron.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 22.Qiu H, Lee S, Shang Y, et al. ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J Clin Invest. 2014;124:981–999. doi: 10.1172/JCI72723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell JC, McGoldrick P, Vance C, et al. Overexpression of human wild-type FUS causes progressive motor neuron degeneration in an age- and dose-dependent fashion. Acta Neuropathol. 2013;125:273–288. doi: 10.1007/s00401-012-1043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma A, Lyashchenko AK, Lu L, et al. ALS-associated mutant FUS induces selective motor neuron degeneration through toxic gain of function. Nat Commun. 2016;7:10465. doi: 10.1038/ncomms10465.320664c72a834d799df983f0633c7d56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hewitt C, Kirby J, Highley JR, et al. Novel FUS/TLS mutations and pathology in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2010;67:455–461. doi: 10.1001/archneurol.2010.52. [DOI] [PubMed] [Google Scholar]
- 26.Cutler RG, Pedersen WA, Camandola S, Rothstein JD, Mattson MP. Evidence that accumulation of ceramides and cholesterol esters mediates oxidative stress-induced death of motor neurons in amyotrophic lateral sclerosis. Ann Neurol. 2002;52:448–457. doi: 10.1002/ana.10312. [DOI] [PubMed] [Google Scholar]
- 27.Arenas A, Kuang L, Zhang J, Kingren MS, Zhu H. FUS regulates autophagy by mediating the transcription of genes critical to the autophagosome formation. J Neurochem. 2021;157:752–763. doi: 10.1111/jnc.15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baskoylu SN, Chapkis N, Unsal B, et al. Disrupted autophagy and neuronal dysfunction in C. elegans knockin models of FUS amyotrophic lateral sclerosis. Cell Rep. 2022;38:110195. doi: 10.1016/j.celrep.2021.110195. [DOI] [PubMed] [Google Scholar]
- 29.Petrov D, Moussy A, Hermine O Mansfield C, author. ALS clinical trials review: 20 years of failure. Are we any closer to registering a new treatment? Front Aging Neurosci. 2017;9:68. doi: 10.3389/fnagi.2017.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kornhuber J, Tripal P, Reichel M, et al. Functional Inhibitors of Acid Sphingomyelinase (FIASMAs): a novel pharmacological group of drugs with broad clinical applications. Cell Physiol Biochem. 2010;26:9–20. doi: 10.1159/000315101. [DOI] [PubMed] [Google Scholar]
- 31.Park MH, Park KH, Choi BJ, et al. Discovery of a dual-action small molecule that improves neuropathological features of Alzheimer's disease mice. Proc Natl Acad Sci U S A. 2022;119:e2115082119. doi: 10.1073/pnas.2115082119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horinouchi K, Erlich S, Perl DP, et al. Acid sphingomyelinase deficient mice: a model of types A and B Niemann-Pick disease. Nat Genet. 1995;10:288–293. doi: 10.1038/ng0795-288. [DOI] [PubMed] [Google Scholar]
- 33.Shiihashi G, Ito D, Yagi T, Nihei Y, Ebine T, Suzuki N. Mislocated FUS is sufficient for gain-of-toxic-function amyotrophic lateral sclerosis phenotypes in mice. Brain. 2016;139:2380–2394. doi: 10.1093/brain/aww161. [DOI] [PubMed] [Google Scholar]