Abstract
Environmental factors, including temperature, can modulate an animal’s lifespan. However, their underlying mechanisms remain largely undefined. We observed a profound effect of temperature on the aging of Caenorhabditis elegans (C. elegans) by performing proteomic analysis at different time points (young adult, middle age, and old age) and temperature conditions (20 °C and 25 °C). Importantly, although at the higher temperature, animals had short life spans, the shift from 20 °C to 25 °C for one day during early adulthood was beneficial for protein homeostasis since; it decreased protein synthesis and increased degradation. Consistent with our findings, animals who lived longer in the 25 °C shift were also more resistant to high temperatures along with oxidative and UV stresses. Furthermore, the lifespan extension by the 25 °C shift was mediated by three important transcription factors, namely FOXO/DAF-16, HSF-1, and HIF-1. We revealed an unexpected and complicated mechanism underlying the effects of temperature on aging, which could potentially aid in developing strategies to treat age-related diseases. Our data are available in ProteomeXchange with the identifier PXD024916.
Keywords: Lifespan, Temperature, C. elegans, Proteostasis, DAF-16, HSF-1, HIF-1
Graphical Abstract
1. Introduction
Aging is a complex phenomenon and is associated with various diseases. To address the underlying mechanisms, intensive work has been done with different model organisms, including the nematode C. elegans. Studies have identified many conserved genes/pathways, including the insulin signaling pathway (daf-2, daf-16) [1], [2], oxidative stress factors (hif-1) [3], dietary restriction signaling pathway (pha-4, skn-1) [4], or heat shock response transcription factors (hsf-1) that regulate aging processes [5]. Besides genetic factors, aging is also regulated by environmental factors, including temperature [6], [7], [8], [9].
Animals generally live shorter at higher temperatures and longer at lower temperatures. Previous studies have shown that temperature could regulate the lifespan through the nervous system in C. elegans [10], [11], [12], [13], while low temperatures extended the lifespan of Drosophila through 4E-BP protein synthesis and/or post-translation modifications [14]. Contrastingly, Miller et al. suggested that temperature regulate the lifespan of C. elegans independently of any known genetic pathways [9]. The inconsistent findings could be due to the complicated effect of temperature on aging than known to us [15], [16]. Considering aging as a complex and systematic phenomenon, it may be interesting to understand how temperature affects aging at system levels.
Recently, the advancement in proteomics methodology has made it possible to identify and quantify thousands of proteins involved in the aging process. Walther et al. used stable-isotope labeling of amino acids in a cell culture (SILAC)-based experiment, and profiled more than 5000 proteins in aging C. elegans [17]. A deep proteomics analysis by Narayan et al. provided a profile of over 10,000 proteins [18]. Although proteomics strategy in aging C. elegans has made a lot of gratifying discoveries, little is known about the mechanism underlying the effect of temperature on lifespan modulation at the proteomic level.
In this study, we systematically analyzed C. elegans aging at 20 °C and 25 °C using an isobaric tag for relative and absolute quantitation (iTRAQ)-based proteomics strategy [19] and identified 4955 proteins in aging worms. Of which, 1557 were differentially expressed at different ages or temperature conditions. Additionally, we unexpectedly found that switching from 20 °C to 25 °C for one day during early adulthood stages could maintain protein homeostasis, extend the lifespan and increase stress resistance. These results suggested that temperature played a complex role in animals’ aging and that an optimal shift of high temperature in the early adulthood stage may be beneficial.
2. Materials and methods
2.1. Strains and maintenance
Bristol N2, daf-16(mu86), daf-12(rh257), daf-12(rh61rh441), hif-1(ia4), hsf-1(sy441), and drIs20(Pvha-6::Q44::YFP) strains were obtained from the Caenorhabditis Genetics Center (CGC). For lifespan analysis, all these mutant strains were back-crossed at least four times with the Bristol N2 strain. The worms were cultivated on NGM plates with E. coli OP50 or HT115 as the food source [20].
2.2. Sampling
Age-synchronized L1 populations were obtained initially by isolating eggs from gravid adult nematodes using alkaline hypochlorite treatment, then the eggs were hatched overnight in the M9 buffer. Next, these synchronized L1 worms were cultivated on NGM plates at 20 °C until the late L4 stage and then divided into two groups and maintained at 20 °C and 25 °C, separately. Furthermore, the worms at late L4 (Day 0), 1 day post L4 (Day 1), 5 days post L4 (Day 5), and 10 days post L4 (Day 10) stages were purified using a 25 µm filter membrane [21] and then immediately frozen in liquid nitrogen and stored at − 80 °C.
2.3. Proteomic analysis
Briefly, worms were resuspended in lysis buffer (4 M Guanidine hydrochloride, 100 mM TEAB, pH 7.0, and proteases inhibitor) and sonicated for 6 min (2 s sonication with 5 s intervals) on ice. Next, the sample was centrifuged at 20,000 g for 20 min at 4 °C and the supernatant was collected. The protein concentration was determined using the Bradford assay. Later, proteins were reduced, alkylated, and digested using Lys-C and trypsin, as previously described [22], [23]. Peptide mixtures were labeled with the 8-plex iTRAQ reagent according to the manufacturer's protocol. Three biological replicates, including late L4 (Day 0), Day 1 at 20 °C, Day 1 at 25 °C, Day 5 at 20 °C, Day 5 at 25 °C, Day 10 at 20 °C, Day 10 at 25 °C, and their mixture were labeled with 113, 114, 115, 116, 117, 118, 119, and 121 iTRAQ reagents, respectively.
2.4. Basic RPLC fractionation
Each labeled peptide was pooled in the same amount and fractionated using the Dionex UltiMate-3000 HPLC system (Thermo Fisher Scientific, USA). The first-dimensional separation was performed using a high pH reverse phase separation. Peptides were fractionated using Dionex UltiMate-3000 HPLC system (Thermo Fisher Scientific, USA) on a Kinetex EVO C18 column (5 µm, 100 Å, 3 mm i.d. ×150 mm (Phenomenex, USA)). The solvent consisted of 10 mM NH4COOH (pH 10) as mobile phase A, and 10 mM NH4COOH in 80% ACN (pH 10) as mobile phase B. Next, 300 µg of the generated peptides was loaded onto the column using mobile phase A, at the following flow rate: 0.1 mL/min for 10 min, 0.1–0.2 mL/min for 1 min, and 0.2 mL/min for 34 min. The separation gradient was as follows: 0% B for 11 min, 0–40% B for 25 min, 40–100% B for 2 min, 100% B for 2 min, 100–0% B for 2 min, and 0% B for 2 min. In the next 40 min gradient, fractions from the sixth minute were collected at 1-min intervals. These 40 fractions were pooled and mixed into 12 fractions of 7–19–31, 8–20–32, and so on. Finally, 12 fractions were obtained.
2.5. Nanoflow LC-ESI-MS/MS
A nanoflow EASY-nLC 1000 system (Thermo Fisher Scientific, Odense, Denmark) coupled with an LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) was used to perform LC-ESI-MS/MS analysis. We adopted a two-column system for all analyses. First, samples were loaded onto an Acclaim PepMap100 C18 Nano Trap Column (5 µm, 100 Å, 100 µm i.d. ×2 cm, (Thermo Fisher Scientific, Sunnyvale, CA)) and then analyzed on an Acclaim PepMap RSLC C18 column (2 µm, 100 Å, 75 µm i.d. ×25 cm (Thermo Fisher Scientific, Sunnyvale, CA)). The mobile phases included Solvent A (0.1% formic acid) and Solvent B (0.1% formic acid in ACN). The derivatized peptides were eluted in the following gradients: 2–5% B for 2 min, 5–28% B for 98 min, 28–35% B for 5 min, 35–90% B for 2 min, and 90% B for 13 min at a flow rate of 200nL/min. Data acquisition mode was set to obtain one MS scan at the resolution of 60,000 (400–1600 m/z; automatic gain control (AGC) target of 1e6; maximum ion injection time (IIT) of 100 ms), followed by 15 MS/MS scans with the most intense ions using HCD (normalized collision energy (NCE) of 35; isolation width of 2 m/z; resolution of 15,000; AGC target of 5e4; maximum IIT of 100 ms). A 60 s dynamic exclusion was applied with a precursor mass tolerance of 10 ppm.
2.6. Proteomic data analysis
Raw data were processed using Proteome Discover (Version 1.4, Thermo Fisher Scientific, Germany), matched against the C. elegans database (20161228, 17,392 sequences), and scored using the Mascot server (Version 2.3, Matrix Science, London, UK). Parameters were set as follows: up to two missed cleavage sites allowed; a mass tolerance of 10 ppm for MS and 0.05 Da for MS/MS fragment ions; carbamidomethylation on cysteine and iTRAQ 8-plex on lysine; N-term as fixed modifications; oxidation on methionine as variable modifications. The search results were validated using the incorporated Target Decoy PSM Validator in Proteome Discoverer (FDR ≤ 0.01).
Based on G. Dennis et al., functional annotation and KEGG pathway analysis were performed using DAVID bioinformatics resource [24]. PCA analysis and volcano plots of proteins were performed using Omicbean tools (www.omicsbean.cn), while the heat map analysis was performed using Omicshare tools (www.omicshare.com/tools). Box plots of expressional changes in proteins were displayed using R.
Unbiased hierarchical clustering analysis was performed for the altered proteins at different temperatures, separately. Compared to L4 (Day 0), the bias caused by the large differences in abundance was avoided by scaling all change ratios between –1 (maximum decrease) and + 1 (maximum increase) [18]. Altered proteins in different groups were clustered using Mfuzz analysis [25]. The time course expression pattern was drawn using Omicshare tools.
2.7. Pharyngeal pumping assays
The age-synchronized L1 worms were cultivated at 20 °C on NGM plates until the late L4 stage, and then divided into two groups and maintained at 20 °C and 25 °C, separately. The number of contractions per minute was calculated by counting the pumping 3 times at 30 s intervals (n = 10). At least three biological replicates were performed.
2.8. Eggs-hatching assays
On Day 1 (24 h post L4), ten worms were transferred to fresh NGM/OP50 plates (one worm/plate). After 24 h culture, worms were transferred to fresh NGM/OP50 plates (one worm/plate), and the process was repeated every 24 h until no more eggs were laid. After the eggs were laid, we counted the animals hatching from eggs three days later. Data were presented as the number of progenies ± SEM. At least three independent biological replicates were performed.
2.9. Protein aggregation assays
OG412 (Q44::YFP) animals were cultured at 21 °C until they reached the L4 stage and then divided into groups and exposed to 21 °C and 25 °C temperatures for one day. Next, both groups were treated with 500 mM NaCl for 4 h and the percentage of animals with Q44::YFP aggregation puncta was calculated. Statistical analyses were performed using the Chi-Square test.
2.10. Fluorescence microscope and Confocal imaging
Worms were anesthetized using 50 mM muscimol on 3% agarose pads. Images were captured using Andor Dragonfly Spinning Disc Confocal Microscope with a 10x objective and a 488 nm laser. The rotation and brightness/contrast of the images were processed using Adobe Photoshop CC.
2.11. Lifespan assays
Lifespan assays were performed at 20 °C unless otherwise indicated. In this study, the first day of adulthood (24 h post L4) was considered day 1. Animals were considered dead if the pharyngeal pumping and response to gentle touching by a platinum wire were absent. For each lifespan experiment, we transferred 80–100 worms in their first six days of adulthood, into fresh NGM plates every day. The worms were then transferred to new plates every three days. To test the temperature effects on their longevity, the worms at different ages (late L4, Day 1, Day 2, Day 3, Day 4, and Day 5) were subjected to one-day 25 °C treatment, followed by their transfer back to 20 °C. Moreover, to determine the minimum/maximum duration of high-temperature treatment extending the lifespan, we shifted the late L4 worms to 25 °C for different time periods (3 h, 6 h, 12 h, 18 h, 1 day, 2 days, 3 days, 4 days, and 5 days), and then transferred them back to 20 °C. For RNAi experiments, we used the standard protocol as described previously [26]. RNAi plasmids (skn-1 and pha-4) from the Ahringer library were used [27]. The lifespan experiments were performed in more than three replicates. GraphPad Prism 5 (GraphPad Software, Inc.) and IBM SPSS Statistics 26 (IBM) were used to generate plot survival curves and perform statistical analyses. The log-rank (Kaplan-Meier) test was used to calculate the p-values.
2.12. Stress assays
For stress assays, day 1 adults were either incubated at 35 °C (heat stress), in an S-basal buffer containing 10 mM hydrogen peroxide (oxidative stress), or under 1200 J/m2 UV light (UV stress) [28], [29], [30]. All assays were carried out in triplicates, with 60–70 worms in each replicate. The log-rank (Kaplan-Meier) test was used to calculate p-values.
2.13. Statistics
Protein aggregation assays were calculated using the Chi-Square test while the lifespan and stress assays were calculated using the log-rank (Kaplan-Meier) test.
3. Results
3.1. Quantitative proteomic data is highly reproducible
To understand how temperature regulated lifespan at the system level, we applied an isobaric tag for relative and absolute quantitation (iTRAQ)-based proteomics strategy [19] in the model organism C. elegans at 20 °C and 25 °C. First, we validated aging-related phenotypes at 20 °C and 25 °C and then collected the samples for proteomic analysis. Consistent with previous reports [31], high temperature accelerated aging-related phenotypes, including the decline of pharyngeal pumping rate, reproduction age, and lifespan (Supplementary Fig. S1a-c).
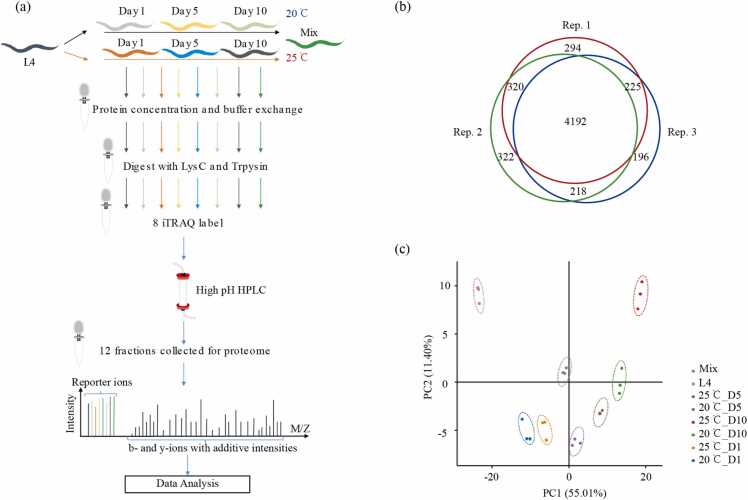
Sample preparation and processing are illustrated in Fig. 1a. Briefly, nematodes at 20 °C and 25 °C were collected at the late larval 4 (L4, Day 0, only at 20 °C), 1 day post L4 (Day 1), 5 days post L4, and 10 days post L4 (Day 10) stages and then digested using FASP method. The resulting tryptic peptides and the mixture of all seven samples in an equal amount were labeled with iTRAQ 8-plex reagent. The labeled peptides were pooled and fractionated using basic RPLC. For the LC-MS/MS analysis, 12 fractions were collected from each biological replicate.
Fig. 1.
Proteomic analysis of aging C. elegans at different temperatures (a) Schematic overview of proteome analysis. Synchronized worms at different stages were lysed and mixed with iTRAQ-labeled peptides. After separation into 12 fractions, the peptides were analyzed by a nano HPLC system coupled to ESI-MS. (b) Venn diagrams of the shared and distinctive proteins identified from three biological replicates. (c) The Principal Component Analysis (PCA) map of three replicates.
Overall, 4955 proteins were identified and quantified from at least two biological replicates at a false discovery rate (FDR) of 0.05% (Fig. 1b and Supplementary Table 1). The reliability of experiments was evaluated using the principal component analysis (PCA) (Fig. 1c), which revealed that the three biological replicates in each sample displayed a tendency to be clustered together, indicating high reproducibility [32].
3.2. High temperature accelerated aging at the proteomic levels
To understand the dynamics of each protein, we generated a heat map using the Omicshare tools. The heat map showed the same tendency of expression changes in the majority of proteins at both 20 °C and 25 °C conditions (Fig. 2a). These results indicated that as the animal aged, the global changes in protein expression did not differ at both temperatures; however, the rate of change was generally faster at 25 °C than at 20 °C.
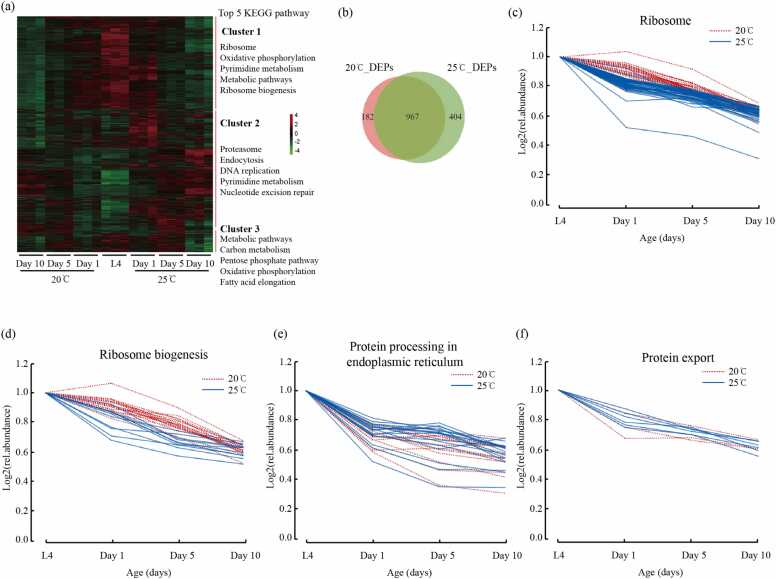
Fig. 2.
High temperature accelerated expression changes in proteome but played a beneficial role in proteostasis (a) Heatmap of the abundance profiles of three major clusters listed with the top 5 KEGG pathways. (b) Venn diagrams of the shared and distinctive differentially expressed proteins (DEPs) between 20 °C (red) and 25 °C (green). (c) The relative abundance of ribosomal proteins overtime at 20 °C (red) and 25 °C (blue). (d) The relative abundance of ribosomal biogenesis proteins over time at 20 °C (red) and 25 °C (blue). (e) The relative abundance of proteins in the endoplasmic reticulum over time at 20 °C (red) and 25 °C (blue). (f) The relative abundance of proteins related to protein export overtime at 20 °C (red) and 25 °C (blue).
The proteomic dynamics during the aging process were investigated by generating volcano plots, which determined the differentially expressed protein (DEPs) profiles of samples on L4, Day 1, Day 5, and Day 10 stages (Supplementary Fig. S2 and Supplementary Table 2). Here, we identified 1575 DEPs (criteria of fold-change >1.5 or <0.67 and p-value <0.05). Of these, 1149 belonged to the 20 °C group and 1371 to the 25 °C group (Fig. 2b). The expression of more altered proteins within the same period also indicated that the proteome change rate was higher at a high temperature, which was consistent with the hypothesis of animals aging faster at a higher temperature.
To closely explore the function of DEPs, we performed GO term analysis using Database for Annotation, Visualization, and Integrated Discovery (DAVID) [33] (Supplementary Table 2). Since the data from day 10 revealed more DEPs, we focused our analysis on the same day. We found that at a higher temperature, expression of proteins involved in lipid transporting and trafficking (VIT-1, VIT-2, VIT-3, VIT-5, and VIT-6), heat stress response (HSP-1, HSP-2, HSP-48, and VIT-6), protein metabolic processing (TAG-225, CPI-1, SRP-1, TEP-1, and FBF-1), and proteolysis inhibition (TAG-225, CPI-1, SRP-1, and TEP-1) were high (Supplementary Table 3). Of these, many were shown to negatively regulate lifespan [34], [35], [36]. Contrastingly, we found lower expression levels of proteins involved in defense response (SPP-5, GST-5, CELE_F55G11.4, GSTK-1, LEC-8, IRG-3, DCT-17, LIPL-5, CPR-3), fatty acid biosynthetic process (ELO-1, HPO-8, ELO-2), and metabolic processing (UGT-22, UGT-23, UGT-62, ELO-1, HPO-8, ELO-2, CYP-3C1, CELE_T05E7.1) (Supplementary Table 3), some of which were considered positive lifespan regulators [37], [38]. These data suggested that high temperatures accelerated the changes in aging-associated proteins and promoted aging.
To further reveal the differences in proteomic changes during aging at different temperatures, we performed an unbiased hierarchical clustering analysis [39]. We used a six-cluster mode to group the age-variant proteins at two temperature conditions (Supplementary Fig. S3a and 3b; Supplementary Table 4). Clusters 1 and 2 comprised the proteins in the aging process that increased sharply and gradually in abundance, respectively, while clusters 3 and 4 comprised those that decreased sharply and gradually, respectively. Clusters 5 and 6 included in the remaining small fraction. Cluster 1 and cluster 3 proteins were significantly more at 25 °C than at 20 °C (776 vs. 448), which was consistent with the result of the volcano plots analysis (Fig. S2). Our data further supported the model that high temperature accelerated proteomic changes and promoted aging.
KEGG pathway analysis was performed using DAVID, which provided the altered protein profile in each cluster changes [33]. Our data showed that while clusters 1 and 2 components were similar between animals at 20 °C and 25 °C conditions, clusters 3 and 4 were quite different. Notably, the ribosome-related proteins decreased rapidly at 25 °C (in cluster 3) but slowly at 20 °C (in cluster 4) (Supplementary Fig. S3a and 3b). Surprisingly, we noticed that proteostasis-related proteins were regulated differently between 20 °C and 25 °C conditions. Specifically, more DEPs in the components of the ribosome, endoplasmic reticulum, and protein exporting components were downregulated (Figs. 2c, 2e, 2 f). Also, a faster reduction in the expression of ribosomal biogenesis components (Fig. 2d) was observed at 25 °C than at 20 °C. These data suggested that a higher temperature may be beneficial for maintaining proteostasis by reducing protein synthesis.
Overall, our proteomic data indicated that the high temperature was a double-edged sword. Although it accelerated changes in aging-related proteins, it also maintained proteostasis by reducing protein synthesis.
3.2.1. High temperature at the early adult stage beneficial for proteostasis maintenance
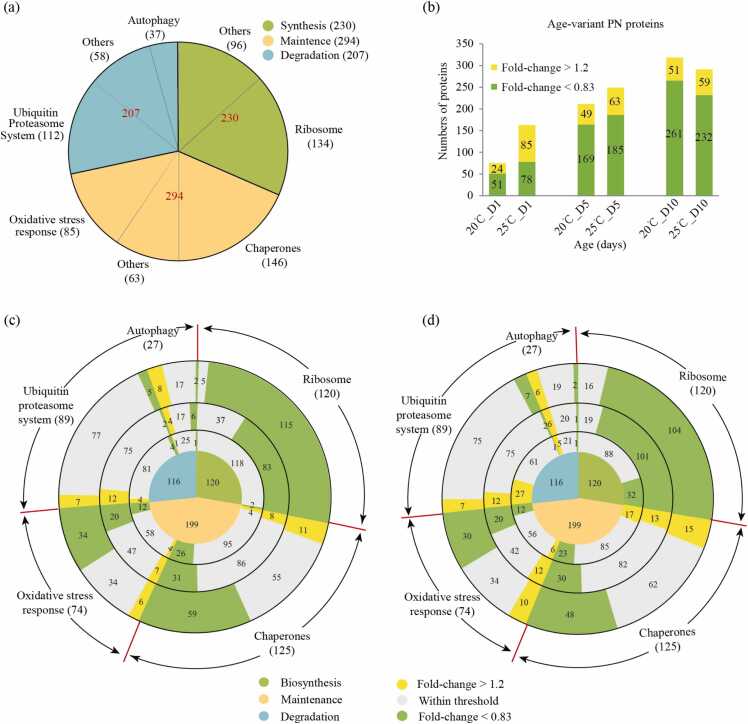
The above data showed that high temperature played a contradictory role in aging. To further analyze the proteostasis net (PN) profile in aging at 20 °C and 25 °C, we specifically analyzed 731 altered PN proteins involved in protein synthesis, maintenance, and degradation during aging (Fig. 3a). Although the number of altered PN proteins at both temperatures increased as animals age, they were generally more abundant at 25 °C than at 20 °C (Fig. 3b and Supplementary Table 5), which was consistent with the fact that high temperatures accelerated the aging-related protein changes.
Fig. 3.
High temperature was beneficial for the proteostasis network (PN). (a) The identified number of PN components classified based on their functions. (b) The number of PN proteins altered across different time points (day 1, 5, and 10 versus L4; fold-change >1.2 or <0.83; p < 0.05) at 20 °C and 25 °C. (c–d) The change in the relative abundance of proteostasis network components at 20 °C (c) or 25 °C (d). The number in the innermost solid circle represents the total PN components. Three functional categories i.e., protein biosynthesis (green), protein degradation (orange), and protein maintenance (blue) are displayed at the center. The three concentric circles from center to periphery represent days 1, 5, and 10. The relative abundance of components compared to the L4 stage (yellow, fold-change >1.2 and green, fold-change <0.83) are indicated in the bars.
Next, we classified the PN proteins based on DAVID [33] (Figs. 3c and 3d). Interestingly, we found more down-regulated ribosomal proteins (32 vs 2) along with upregulated chaperones (17 vs 4) and protein degradation regulators (32 vs 5) in young adult animals on day 1 at 25 °C than at 20 °C. However, the difference was less obvious on day 5 and disappeared on day 10, indicating that animals treated at 25 °C for one day showed robust benefits on proteostasis.
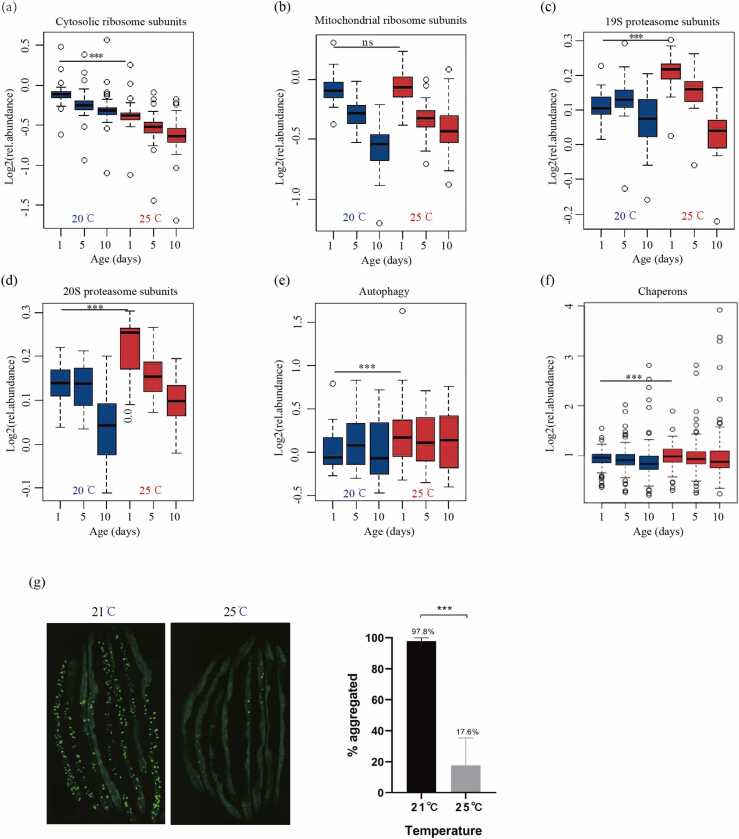
To further understand the effect of temperature on the aging process, we analyzed the expression levels of 456 PN components across different functional categories and found that the cytosolic and mitochondrial ribosomal proteins during aging were down-regulated at both temperatures (Fig. 4a-b). However, the median level of cytosolic ribosomal proteins was lower at 25 °C than at 20 °C. Furthermore, we found that the protein degradation components, including the 19 S and 20 S proteasome subunits, were higher at 25 °C than at 20 °C (Fig. 4c-d). Moreover, the expressions of proteins involved in autophagy, chaperons, and oxidative stress defense were much higher at 25 °C than at 20 °C (Fig. 4e-f and Supplementary Fig. S4). Interestingly, although the beneficial effect of high temperatures on PN was more robust on day 1, it lasted till day 10. Besides PN, various other DNA repair proteins were upregulated at 25 °C (Supplementary Fig. S4). Overall, these data further suggested that high temperatures played a beneficial role in the aging process.
Fig. 4.
Temperature affected the abundance of PN components and protein homeostasis. (a-b) The relative abundance of ribosomal proteins overtime at 20 °C and 25 °C. Quantification of 75 cytosolic (a) and 54 mitochondrial (b) ribosomal subunits. Box plots representing log2 values of fold-changes. Horizontal solid lines showing median values. ***p < 0.001 for cytosolic ribosomal proteins and p > 0.05 for mitochondrial ribosomal proteins based on Wilcoxon signed-rank test. (c-d) The relative abundance of proteasome subunits over time at 20 °C and 25 °C. Quantification of proteasome components of 17 subunits of the 19 S (c) and 14 subunits of the 20 S (d). ***p < 0.001 for 20 S subunits and 19 S subunits based on Wilcoxon signed-rank test. (e) The relative abundance of autophagy components over time at 20 °C and 25 °C, with quantification of 14 autophagy components. ***p < 0.001 based on Wilcoxon signed-rank test. (f) The relative abundance of chaperon components overtime at 20 °C and 25 °C, with quantification of 125 proteins. * **p < 0.001 based on Wilcoxon signed-rank test. (g) Protein aggregation analysis using a YFP-tagged polyQ marker at 21 °C and 25 °C, showing a 25 °C shift for 24 h robustly suppressing the aggregation induced by high salt concentration. Three independent biological replicates were performed with > 80 animals under each condition in each replicate. ***p < 0.001 based on the Chi-Square test.
To confirm the beneficial role of the high-temperature shift in the early adult stage on protein homeostasis, we examined the polyQ::YFP aggregation [40] and found that the Q44::YFP aggregation was robustly suppressed in animals exposed to 25 °C for one day compared to the control animals (Fig. 4g), which was consistent with the proteomic data, further supporting the model that optimal high-temperature shift was beneficial for proteostasis maintenance.
These observations collectively demonstrated that the 25 °C shift during the early adult stage was potentially beneficial to animals, which was most likely through proteostasis maintenance.
3.2.2. Temporally restricted beneficial effect of high temperature on lifespan extension
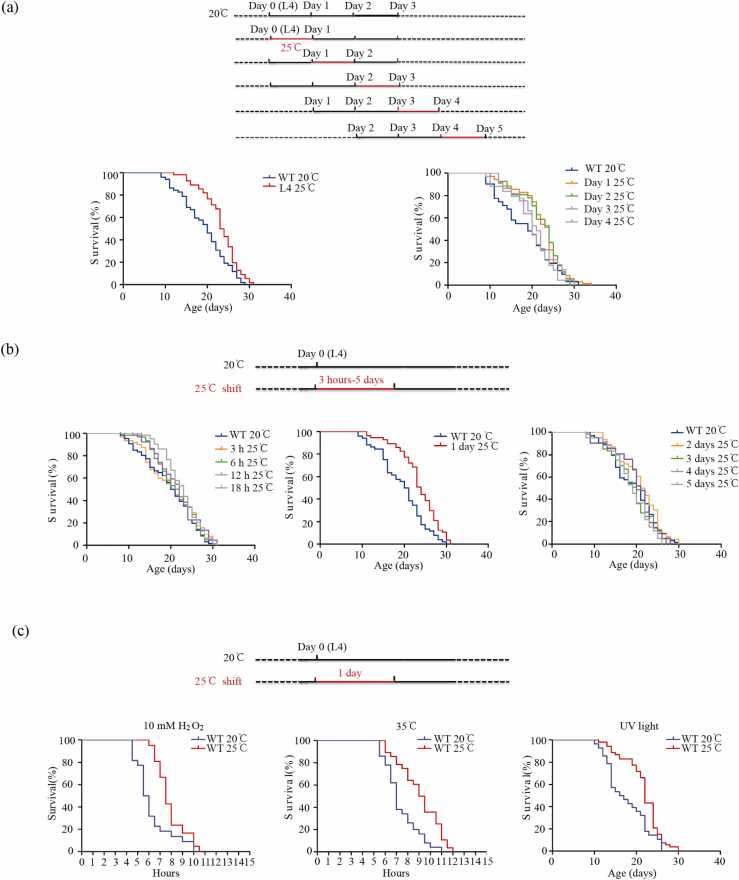
Our proteomic data suggested that a high temperature during the early adult stage played beneficial roles in proteostasis. To investigate this further, we shifted L4 stage animals from 20 °C to 25 °C for one day and tested the effects on their lifespan. Interestingly, theses animals lived significantly longer than those kept at a constant 20 °C (Fig. 5a and Supplementary Table 6), which supported the model that a high-temperature shift for one day was beneficial in increasing the lifespan.
Fig. 5.
The regime of high temperature that extends the lifespan was temporally restricted (a) The temporal effects of high-temperature shift on the lifespan. Animals were shifted from 20 °C to 25 °C at specific stages (late-L4, Day 1, Day 2, Day 3, Day 4) for 24 h then back to 20 °C, followed by the performance of adult lifespan assay (late-L4: p < 0.001; Day 1: p = 0.01; Day 2: p = 0.023; Day 3: p = 0.369; Day 4: p = 0.826; log-rank test). (b) The optimal high-temperature duration is required for the lifespan extension. Animals were shifted from 20 °C to 25 °C for different period between 3 h and 5 days starting from the L4 stage ( 3 h from L4: p = 0.268; 6 h from L4: p = 0.292; 12 h from L4: p = 0.085; 18 h from L4: p = 0.032; 1 day from L4: p < 0.001; 2 days from L4: p = 0.233; 3 days from L4: p = 0.354; 4 days from L4: p = 0.848; 5 days from L4: p = 0.151; log-rank test). (c) High-temperature regime promotes stress resistance. Animals in the high-temperature regime were more resistant to oxidative stress (p < 0.001; log-rank test), heat stress (p < 0.001; log-rank test), and UV light (p < 0.001; log-rank test).
Next, we determined the time window when one-day high-temperature treatment was beneficial. For this, we shifted animals at different stages (from the pre-adult L4 stage to the day 5 adult stage) from 20 °C to 25 °C for 24 h at different time points and found that one-day 25 °C treatment extended lifespan only when the shifting was done before the day 3-adult stage. During this time window, the earlier the animals were shifted, the more robust the lifespan showed extension (Fig. 5a and Supplementary Table 6). When the shift happened after day 3 of the adult stage, the lifespan no longer showed extension, indicating that the beneficial effect of the high-temperature shift was restricted to a critical time window, which was before day 3 of adulthood.
To determine the minimum and maximum duration of high temperature during which the shift extended the lifespan, we shifted animals from 20 °C to 25 °C starting from the L4 stage for different periods between 3 h and 5 days. Our results showed that only 18 h or 24 h shifts extended the lifespan while the rest did not (Fig. 5b). Although the duration of high temperature may depend on the starting time point, we speculate that its beneficial effect requires maintenance of high temperature for about one day.
Lifespan extension is usually accompanied by various stress resistance [28], [29], [30]. We investigated if high-temperature shift during the pre-adult stage affected stress resistance and found that shifted animals were significantly more resistant to various stresses, including heat shock, ultraviolet (UV), and oxidative stresses (Fig. 5c). This indicated that a high-temperature shift for a day starting from the L4 stage was beneficial for stress resistance as well.
3.2.3. High temperature promoted lifespan extension through transcription factors including FOXO/DAF-16, HSF-1, and HIF-1
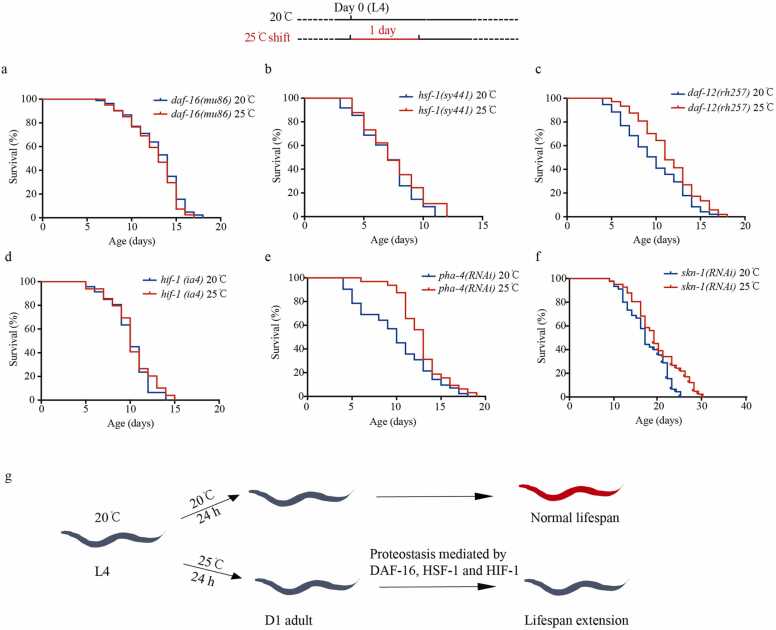
The molecular mechanisms underlying the lifespan extension by high-temperature shifts during the early adult stage were investigated by studying the interaction between high-temperature shifts and genetic longevity signaling pathways, including daf-16(mu86), daf-12(rh257), daf-12(rh61rh441), hif-1(ia4), hsf-1(sy441), skn-1(RNAi), and pha-4(RNAi). These are considered the major longevity signaling pathway molecules [41]. Interestingly, the mutations in daf-16, hsf-1, and hif-1 showed complete suppression of the lifespan extension by the high-temperature shift, while the mutations in daf-12 or RNAi treatment for skn-1 and pha-4 exhibited minimum effects (Fig. 6a-e, Supplementary Table 6). These results suggested that daf-16, hsf-1, and hif-1 played major roles in the lifespan extension induced by high-temperature shifts (Fig. 6f).
Fig. 6.
Lifespan extension induced by high-temperature shift required daf-16, hsf-1, and hif-1. (a-e) Lifespan analysis with daf-16(mu86), hsf-1(sy441), and hif-1(ia4) mutants and pha-4 and skn-1 RNAi animals at constant 20 °C and 25 °C shift for a day at the L4 stage. daf-16(mu86) (p = 0.20; log-rank test) (a), hsf-1(sy441) (p = 0.07; log-rank test) (b), hif-1(ia4) (p = 0.24; log-rank test), (c), pha-4 (p < 0.001; log-rank test) (d), and skn-1 (p < 0.001; log-rank test) (e). (f) A model illustrating a one-day thermal regime to extend the lifespan of animals.
4. Discussion
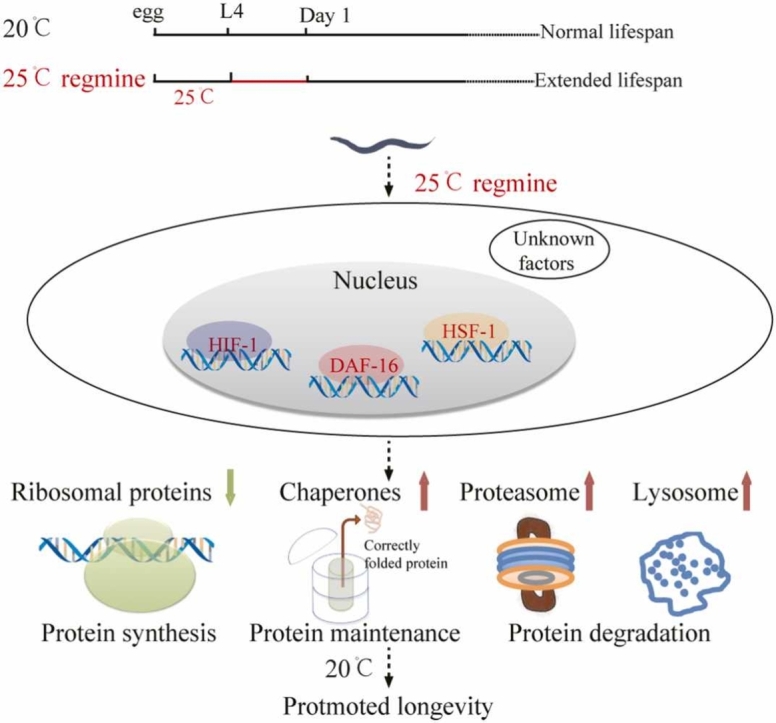
This study revealed six major findings. 1. A similar proteome remodeling trend was observed during animal aging at both low and high temperatures; 2. Animals showed more rapid changes at proteomic expression level at 25 °C than at 20 °C, which was consistent with the fact that animals aged faster at higher temperatures; 3. Higher temperature shift on the day 1-adult stage was found beneficial for proteostasis maintenance; 4. Consistent with the proteomic data, high-temperature shifts for one day at the early adult stage extended animals’ lifespan along with improving stress resistance; 5. The temporal window of high temperature was essential to exert the beneficial effect; 6. High temperature-induced lifespan extension required several transcription factors, including FOXO/DAF-16, HSF-1, and HIF-1. These data provided important insights into the molecular mechanisms regulated by high temperatures.
Aging is a systematic and complex phenomenon with proteostasis loss considered as its indicator [42]. Proteostasis maintenance relies on the PN, which can be classified into the following three categories: synthesis (ribosomes), maintenance (molecular chaperones), and degradation (ubiquitin-proteasome and lysosome-autophagy proteolytic system) [43]. With the increase in animals’ age, we found that cytosolic and mitochondrial ribosomal proteins decreased, which indicated a decrease in protein synthesis and a beneficial effect on proteostasis. Previously, several other groups have reported the same results independently [17], [44]. During aging, the reduction in global translation may redirect the energy resources toward optimal maintenance and repair [45]. Additionally, the number of down-regulated chaperons and oxidative response components showed an increase with an increase in animals' age, which was consistent with the hypothesis that the accumulation of damaged proteins is one of the main causalities of aging.
Generally, increasing the temperature of cultivation can speed up the aging process. However, the expression of cytosolic ribosomal proteins in animals cultured at a higher temperature was found to be significantly lower, while the expression of the proteasome components was found to be much higher, suggesting its beneficial effect on proteostasis on day 1. The decrease in protein synthesis or the increase in protein degradation was usually correlated to lifespan extension [46], [47], which was consistent with this study.
One explanation of how the high-temperature conditions benefitted animals’ lifespan lies in the induction of hormesis by the short-time high-temperature treatment. Hormesis is defined as a moderate stress-induced benefit effect, for example, thermotolerance and lifespan extension induced by heat shock [48]. In the laboratory, C. elegans is normally maintained between 15 and 25 °C with 32 °C or above being the heat shock temperatures [49]. The response to such a condition is generally exhibited within two hours or earlier. However, our proteomic data were obtained from animals treated with high temperatures for 24 h. Also, in our lifespan assay, no lifespan extension was observed upon treatment for 12 h or lesser. Furthermore, very few heat shock proteins were upregulated in the proteomic result. Therefore, the beneficial effect in our study was different from traditional heat shock responses. Consistently, Pispa et al. recently reported that L4 stage-C. elegans exposed to 25 °C temperature for one day affected selective protein degradation via the UPS but not the heat shock or UPRmt responses [50]. Overall, these studies suggested that a 25 °C shift from a constant 20 °C temperature during the early adult stage may induce a response different from traditional heat shock hormesis. However, further studies are needed to address the detailed mechanisms.
Furthermore, our data showed that the shift from 20 °C to 25 °C for 24 h extended lifespan only when the treatment was given before day 3 of the adult stage. Starting from the L4 stage, the lifespan was extended only when the shifting duration was between 18 h and 24 h. The data describe a temporal window that exerted a beneficial effect of the moderate high-temperature shift. High-temperature treatment at the developmental stage has been reported to extend lifespan [11], [51]. Our study showed that a one-day high-temperature shift at the early adult stage extended the lifespan, broadening our knowledge of the high-temperature effect. Moreover, we observed that the effect of high-temperature shifts on longevity was mainly regulated by DAF-16, HIF-1, and HSF-1. DAF-16 is required for the high-temperature shift-induced lifespan extension since previous studies indicated the critical role of DAF-16 in proteostasis. DAF-16 could enhance proteasome activity through up-regulation of the proteasomal subunit RPN-6, which was responsible for the stabilization of the interaction between the 20 S core and the 19 S cap of the proteasome [52]. Furthermore, the overexpression of DAF-16 helped to maintain proteostasis and enhance longevity by activating multiple stress response pathways [53]. Hypoxia-inducible factor 1 (HIF-1) is also required for the high-temperature shift-induced lifespan extension, which was consistent with the previous finding that HIF-1 was required for 25 °C induced heat acclimatization [54]. It is surprising that HSF-1 is also required for the high-temperature shift-induced lifespan extension since heat shock responses were not observed under this condition [50]. Besides DAF-16, HSF-1, and HIF-1, other factors, including DAF-12 and SKN-1 may be involved in the lifespan extension since their mutations or RNAi knowckdown partially suppressed the lifespan extension. These data demonstrated that the beneficial role of high-temperature treatment during the early adult stage required multiple transcription factors.
In summary, our work not only provided valuable insights into the aging mechanisms regulated by temperature, but also uncovered a novel phenomenon of moderate high-temperature shifts at the early adult stage, extending lifespan and promoting stress resistance. Therefore, our study suggested that high temperatures played both detrimental and beneficial roles in aging. The beneficial effect exerted during the specific temporal window can be explored to provide potential aid in developing strategies for treating aging-related disorders.
CRediT authorship contribution statement
Jichang Huang: Conceptualization, Methodology, Investigation, Formal analysis, Writing – original draft. Kai Wang: Investigation, Data curation. Mengqing Wang: Investigation, Validation;, Zhen Wu: Methodology, Formal analysis. Guangjie Xie: Investigation. Yuling Peng: Investigation. Yan Zhang: Resources, Supervision, Xumin Zhang: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing. Zhiyong Shao: Conceptualization, Funding Acquisition, Resources, Supervision, Writing – review & editing.
Conflict of Interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work.
Acknowledgements
We thank Miss Lin Huang for the kind help with MS analysis. This work was supported by the Ministry of Science and Technology, People’s Republic of China (2021YFA0909300 to ZS), National Natural Science Foundation of China (No. 31870822 and 31470806 to XZ, 31872762 and 32170828 to ZS), the Shanghai Municipal Science and Technology Major Project (2018SHZDZX01), ZJLab, the Research Fund of the State Key Laboratory of Genetic Engineering, Fudan University, the Foundation of Chengdu Medical College (CYZZD21–04 and 2021LHPJ-02 to JH), National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University (Z2021JC006 to YZ). We thank CGC for strains.
Author contributions
J.H. performed proteomic experiments and functional assays. K.W. performed RNAi experiments. M.W. performed part of lifespan assays. J.H. and Z.W. performed proteomic data analysis. G.X. did the protein aggregation assays. J.H., X.Z. and Z.S. designed the experiments and wrote the manuscript. Y.Z., X.Z. and Z.S. supervised experiments. All the authors revised the manuscript.
Supplementary materials
The data that supports the findings of this study are available in the supplementary materials.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2022.12.017.
Contributor Information
Xumin Zhang, Email: xumin_zhang@fudan.edu.cn.
Zhiyong Shao, Email: shaozy@fudan.edu.cn.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Ogg S., Paradis S., Gottlieb S., Patterson G.I., Lee L., Tissenbaum H.A., et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389(6654):994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 2.Tissenbaum H.A., Ruvkun G. An insulin-like signaling pathway affects both longevity and reproduction in Caenorhabditis elegans. Genetics. 1998;148(2):703–717. doi: 10.1093/genetics/148.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y., Shao Z., Zhai Z., Shen C., Powell-Coffman J.A. The HIF-1 hypoxia-inducible factor modulates lifespan in C. elegans. PLoS One. 2009;4(7) doi: 10.1371/journal.pone.0006348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith-Vikos T., de Lencastre A., Inukai S., Shlomchik M., Holtrup B., Slack F.J. MicroRNAs mediate dietary-restriction-induced longevity through PHA-4/FOXA and SKN-1/Nrf transcription factors. Curr Biol. 2014;24(19):2238–2246. doi: 10.1016/j.cub.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsu A.L., Murphy C.T., Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300(5622):1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 6.Conti B. Considerations on temperature, longevity and aging. Cell Mol Life Sci. 2008;65(11):1626–1630. doi: 10.1007/s00018-008-7536-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farmer K.J., Sohal R.S. Effects of ambient temperature on free radical generation, antioxidant defenses and life span in the adult housefly, Musca domestica. Exp Gerontol. 1987;22(1):59–65. doi: 10.1016/0531-5565(87)90015-5. [DOI] [PubMed] [Google Scholar]
- 8.Liu R.K., Walford R.L. The effect of lowered body temperature on lifespan and immune and non-immune processes. Gerontologia. 1972;18(5–6):363–388. doi: 10.1159/000211944. [DOI] [PubMed] [Google Scholar]
- 9.Miller H., Fletcher M., Primitivo M., Leonard A., Sutphin G.L., Rintala N., et al. Genetic interaction with temperature is an important determinant of nematode longevity. Aging Cell. 2017;16(6):1425–1429. doi: 10.1111/acel.12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao R., Zhang B., Dong Y., Gong J., Xu T., Liu J., et al. A genetic program promotes C. elegans longevity at cold temperatures via a thermosensitive TRP channel. Cell. 2013;152(4):806–817. doi: 10.1016/j.cell.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang B., Xiao R., Ronan E.A., He Y., Hsu A.L., Liu J., et al. Environmental Temperature Differentially Modulates C. elegans Longevity through a Thermosensitive TRP Channel. Cell Rep. 2015;11(9):1414–1424. doi: 10.1016/j.celrep.2015.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang B., Gong J., Zhang W., Xiao R., Liu J., Xu X.Z.S. Brain-gut communications via distinct neuroendocrine signals bidirectionally regulate longevity in C. elegans. Genes Dev. 2018;32(3–4) doi: 10.1101/gad.309625.117. 258-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y.C., Chen H.J., Tseng W.C., Hsu J.M., Huang T.T., Chen C.H., et al. A C. elegans Thermosensory Circuit Regulates Longevity through crh-1/CREB-Dependent flp-6 Neuropeptide Signaling. Dev Cell. 2016;39(2) doi: 10.1016/j.devcel.2016.08.021. 209-23. [DOI] [PubMed] [Google Scholar]
- 14.Carvalho G.B., Drago I., Hoxha S., Yamada R., Mahneva O., Bruce K.D., et al. The 4E-BP growth pathway regulates the effect of ambient temperature on Drosophila metabolism and lifespan. Proc Natl Acad Sci U S A. 2017;114(36) doi: 10.1073/pnas.1618994114. 9737-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grumiaux C., Andersen M.K., Colinet H., Overgaard J. Fluctuating thermal regime preserves physiological homeostasis and reproductive capacity in Drosophila suzukii. J Insect Physiol. 2019;113:33–41. doi: 10.1016/j.jinsphys.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Torson A.S., Yocum G.D., Rinehart J.P., Kemp W.P., Bowsher J.H. Transcriptional responses to fluctuating thermal regimes underpinning differences in survival in the solitary bee Megachile rotundata. J Exp Biol. 2015;218(Pt 7):1060–1068. doi: 10.1242/jeb.113829. [DOI] [PubMed] [Google Scholar]
- 17.Walther D.M., Kasturi P., Zheng M., Pinkert S., Vecchi G., Ciryam P., et al. Widespread proteome remodeling and aggregation in aging C. elegans. Cell. 2015;161(4):919–932. doi: 10.1016/j.cell.2015.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narayan V., Ly T., Pourkarimi E., Murillo A.B., Gartner A., Lamond A.I., et al. Deep proteome analysis identifies age-related processes in C. elegans. Cell Syst. 2016;3(2) doi: 10.1016/j.cels.2016.06.011. 144-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross P.L., Huang Y.N., Marchese J.N., Williamson B., Parker K., Hattan S., et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3(12):1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larance M., Bailly A.P., Pourkarimi E., Hay R.T., Buchanan G., Coulthurst S., et al. Stable-isotope labeling with amino acids in nematodes. Nat Methods. 2011;8(10):849–851. doi: 10.1038/nmeth.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang J., Wu Z., Wang J., Zhang X. Quantitative phosphoproteomics reveals GTBP-1 regulating C.elegans lifespan at different environmental temperatures. Biochem Biophys Res Commun. 2018;503(3):1962–1967. doi: 10.1016/j.bbrc.2018.07.142. [DOI] [PubMed] [Google Scholar]
- 23.Huang J., Wu Z., Zhang X. Short-term mild temperature-stress-induced alterations in the C. elegans phosphoproteome. Int J Mol Sci. 2020;21:17. doi: 10.3390/ijms21176409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dennis G., Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., et al. Vol. 4. Database for Annotation, Visualization, and Integrated Discovery; DAVID: 2003. p. P3. (Genome Biol). [PubMed] [Google Scholar]
- 25.Kumar L., M EF. Mfuzz: a software package for soft clustering of microarray data. Bioinformation 2007; 2(1): 5–7. [DOI] [PMC free article] [PubMed]
- 26.Fraser A.G., Kamath R.S., Zipperlen P., Martinez-Campos M., Sohrmann M., Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408(6810):325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 27.Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., et al. Systematic functional analysis of the C. elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 28.Vilchez D., Morantte I., Liu Z., Douglas P.M., Merkwirth C., Rodrigues A.P., et al. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489(7415):263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 29.Lithgow G.J., White T.M., Hinerfeld D.A., Johnson T.E. Thermotolerance of a long-lived mutant of Caenorhabditis elegans. J Gerontol. 1994;49(6):B270–B276. doi: 10.1093/geronj/49.6.b270. [DOI] [PubMed] [Google Scholar]
- 30.Hirota K., Shigekawa C., Araoi S., Sha L., Inagawa T., Kanou A., et al. Simultaneous ablation of prmt-1 and prmt-5 abolishes asymmetric and symmetric arginine dimethylations in Caenorhabditis elegans. J Biochem. 2017;161(6):521–527. doi: 10.1093/jb/mvw101. [DOI] [PubMed] [Google Scholar]
- 31.Huang C., Xiong C., Kornfeld K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2004;101(21):8084–8089. doi: 10.1073/pnas.0400848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan M., Breitkopf S.B., Yang X., Asara J.M. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7(5):872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Halaschek-Wiener J., Khattra J.S., McKay S., Pouzyrev A., Stott J.M., Yang G.S., et al. Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res. 2005;15(5):603–615. doi: 10.1101/gr.3274805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu H., Fu W., Mattson M.P. The catalytic subunit of telomerase protects neurons against amyloid beta-peptide-induced apoptosis. J Neurochem. 2000;75(1):117–124. doi: 10.1046/j.1471-4159.2000.0750117.x. [DOI] [PubMed] [Google Scholar]
- 36.Murphy C.T., McCarroll S.A., Bargmann C.I., Fraser A., Kamath R.S., Ahringer J., et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424(6946):277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 37.Ayyadevara S., Dandapat A., Singh S.P., Siegel E.R., Shmookler Reis R.J., Zimniak L., et al. Life span and stress resistance of Caenorhabditis elegans are differentially affected by glutathione transferases metabolizing 4-hydroxynon-2-enal. Mech Aging Dev. 2007;128(2):196–205. doi: 10.1016/j.mad.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinkston-Gosse J., Kenyon C. DAF-16/FOXO targets genes that regulate tumor growth in Caenorhabditis elegans. Nat Genet. 2007;39(11):1403–1409. doi: 10.1038/ng.2007.1. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki R., Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22(12):1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 40.Mohri-Shiomi A., Garsin D.A. Insulin signaling and the heat shock response modulate protein homeostasis in the Caenorhabditis elegans intestine during infection. J Biol Chem. 2008;283:194–201. doi: 10.1074/jbc.M707956200. [DOI] [PubMed] [Google Scholar]
- 41.Kenyon C.J. The genetics of aging. Nature. 2010;464(7288):504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 42.Basisty N., Meyer J.G., Schilling B. Protein turnover in aging and longevity. Proteomics. 2018;18(5–6) doi: 10.1002/pmic.201700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartl F.U., Bracher A., Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 44.Kirstein-Miles J., Scior A., Deuerling E., Morimoto R.I. The nascent polypeptide-associated complex is a key regulator of proteostasis. EMBO J. 2013;32(10):1451–1468. doi: 10.1038/emboj.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Syntichaki P., Troulinaki K., Tavernarakis N. eIF4E function in somatic cells modulates aging in Caenorhabditis elegans. Nature. 2007;445(7130):922–926. doi: 10.1038/nature05603. [DOI] [PubMed] [Google Scholar]
- 46.Pan K.Z., Palter J.E., Rogers A.N., Olsen A., Chen D., Lithgow G.J., et al. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6(1):111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Low P. The role of ubiquitin-proteasome system in aging. Gen Comp Endocrinol. 2011;172(1):39–43. doi: 10.1016/j.ygcen.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 48.Lithgow G.J., White T.M., Melov S., Johnson T.E. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci U S A. 1995;92(16):7540–7544. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zevian S.C., Yanowitz J.L. Methodological considerations for heat shock of the nematode Caenorhabditis elegans. Methods. 2014;68(3):450–457. doi: 10.1016/j.ymeth.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pispa J., Matilainen O., Holmberg C.I. Tissue-specific effects of temperature on proteasome function. Cell Stress Chaperones. 2020;25(3):563–572. doi: 10.1007/s12192-020-01107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou L., He B., Deng J., Pang S., Tang H. Histone acetylation promotes long-lasting defense responses and longevity following early life heat stress. PLoS Genet. 2019;15(4) doi: 10.1371/journal.pgen.1008122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shemesh N., Shai N., Ben-Zvi A. Germline stem cell arrest inhibits the collapse of somatic proteostasis early in Caenorhabditis elegans adulthood. Aging Cell. 2013;12(5):814–822. doi: 10.1111/acel.12110. [DOI] [PubMed] [Google Scholar]
- 53.Shore D.E., Carr C.E., Ruvkun G. Induction of cytoprotective pathways is central to the extension of lifespan conferred by multiple longevity pathways. Plos Genetics. 2012;8(7) doi: 10.1371/journal.pgen.1002792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Treinin M., Shliar J., Jiang H., Powell-Coffman J.A., Bromberg Z., Horowitz M. HIF-1 is required for heat acclimation in the nematode Caenorhabditis elegans. Physiol Genomics. 2003;14(1):17–24. doi: 10.1152/physiolgenomics.00179.2002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material