Summary
Background
Global estimates suggest millions of deaths annually are associated with antimicrobial resistance (AMR) but these are generated from scarce data on the relative risk of death attributable to drug-resistant versus drug-sensitive infections.
Methods
We examined all episodes of E. coli bloodstream infection in Ontario, Canada between 2017 and 2020, and measured 90 day mortality among those with resistant versus sensitive isolates for each of 8 commonly used antibiotic classes and a category of difficult to treat resistance (DTTR). We used multivariable logistic regression to calculate an adjusted odds of mortality associated with AMR, after accounting for patient demographics, comorbidities, and prior healthcare exposure.
Findings
Among 14,548 eligible episodes of E. coli bloodstream infection, resistance was most common to aminopenicillins (46.8%), followed by first generation cephalosporins (38.8%), fluoroquinolones (26.5%), sulfonamides (24.1%), third generation cephalosporins (13.8%), aminoglycosides (11.7%), beta-lactam-beta-lactamase-inhibitors (9.1%) and carbapenems (0.2%). Only 18 (0.1%) episodes exhibited DTTR. For each antibiotic class, the unadjusted odds of mortality (OR) were higher among resistant isolates, but after accounting for patient characteristics the adjusted odds (aOR) of mortality were attenuated: aminopenicillins (OR 1.22, 95% CI 1.12–1.33; aOR 1.09, 95% CI 0.99–1.20), first generation cephalosporins (OR 1.24, 95% CI 1.14–1.35; aOR 1.07, 95% CI 0.97–1.18), third generation cephalosporins (OR 1.64, 95% CI 1.47–1.82; aOR 1.29, 95% CI 1.15–1.46), beta-lactam-beta-lactamase-inhibitors (OR 1.69, 95% CI 1.52–1.89, aOR 1.28, 95% CI 1.13–1.45), carbapenems (OR 3.11, 95% CI 1.52–6.34; aOR 2.06, 95% CI 0.91–4.66), sulfonamides (OR 1.19, 95% CI 1.07–1.31, aOR 1.06, 95% CI 0.95–1.18), fluoroquinolones (OR 1.49, 95% CI 1.36–1.64, aOR 1.16, 95% CI 1.05–1.29), aminoglycosides (OR 1.43, 95% CI 1.27–1.62; aOR 1.27, 95% CI 1.11–1.46), and DTTR (OR 3.71, 95% CI 1.46–9.41; aOR 2.58, 95% CI 0.87–7.66).
Interpretation
AMR is associated with substantial increased mortality among patients with E. coli bloodstream infection, particularly for resistance to classes commonly used as empiric treatment. Surveillance for AMR-associated mortality should incorporate adjustment for patient characteristics and prior healthcare utilization.
Funding
This work was supported by a project grant from CIHR (grant number 159503). This study was also supported by ICES, which is funded by an annual grant from Ontario Ministry of Health and Long-Term Care (MOHLTC).
Keywords: Antimicrobial resistance, Bloodstream infections, Bacteremia, Mortality, Escherichia coli
Research in context.
Evidence before this study
The Antimicrobial Resistance Collaboration recently published in the Lancet a comprehensive global estimate of the burden of antimicrobial resistance (AMR) across 23 pathogens, 88 pathogen-drug combinations and 204 countries. This work estimated that there are currently 4.95 million (3.62–6.57) annual deaths associated with bacterial AMR, including 1.27 million (95% UI 0.911–1.71) deaths attributable to bacterial AMR. The immense source data included 471 million individual bacterial isolates, making it possible to precisely estimate the prevalence of pathogens causing different infections and the prevalence of resistance among those pathogens. However, the global team of investigators learned that data were relatively scarce for determining the relative risk (RR) of death for drug-resistant compared with drug-susceptible infection. Where possible the collaboration used a modelling approach which accounted for age, admission diagnosis, site of culture and hospital versus community onset, but in some cases the primary literature provided only a crude RR without adjustment for patient characteristics.
Seeking additional evidence to fill this gap, we searched OVID MEDLINE from database inception until August 2022 using the terms “Drug Resistance, Bacterial” and “Mortality” with a filter for systematic reviews. Through that search we uncovered one systematic review, which included 16 studies assessing the impact of resistance on patient outcomes, but was focused only on multi-drug resistance and healthcare-associated infections. Among a subset of 14 eligible studies, the pooled mortality was higher among patients infected with multi-drug versus non-multi-drug resistant bacteria (RR 1.61, 95% CI 1.36–1.90), but this result was obtained by pooling crude mortality rates without any adjustment for patient characteristics.
Added value of this study
With access to linked microbiology test results and healthcare administrative datasets across a large region, we were able to study the adjusted odds of death among patients infected with drug-resistant versus drug-susceptible pathogens, allowing for a more accurate estimate of mortality associated with bacterial drug resistance. We focused on bloodstream infections with E. coli, the most common bloodstream pathogen and the leading pathogen among AMR-associated deaths, and detected substantial increased odds of mortality with resistance to each common class of antimicrobial. After accounting for patient characteristics, and in particular extent of prior healthcare exposure, the adjusted odds of mortality were greatly attenuated. However, there was a persistent and large association between AMR and mortality for almost all individual classes, and the association was strongest for resistance to antibiotics commonly used as empiric treatment.
Implications of all the available evidence
The mortality following E. coli bloodstream infections is significantly higher for strains resistant to each class of common antimicrobial agents, and in particular those used commonly in empiric treatment. Some of the current excess mortality with AMR E. coli is explained by patient characteristics, including extent of prior healthcare exposure and so surveillance for AMR should incorporate adjustment for these confounders to prevent over-estimation.
Introduction
The Antimicrobial Resistance Collaboration recently provided the most comprehensive global estimate of the burden of antimicrobial resistance (AMR) across 23 pathogens, 88 pathogen-drug combinations and 204 countries.1 The collaboration amassed data from systematic literature reviews, healthcare systems, and surveillance programs, and through a 10 step process estimated that there were 4.95 million deaths in 2019 associated with bacterial AMR, including 1.27 million deaths attributable to bacterial AMR. Escherichia coli (E. coli) was the number one culprit pathogen, with 829,000 AMR associated deaths and 219,000 AMR attributable deaths.1
The source data was massive and included 471 million individual bacterial isolates, making it possible to precisely estimate the prevalence of pathogens causing different infections and the prevalence of resistance among those pathogens. However, the investigators learned that data were relatively scarce for determining the relative risk of death for drug-resistant compared with drug-sensitive infection.1 This information is crucial towards understanding the impact of AMR on mortality, but is lacking in most regions of the world because there are very few healthcare systems in which microbiology results can be linked to patient characteristics and outcomes.
In Ontario, Canada's most populous province with more than 14 million residents, we have recently linked microbiology test results to healthcare administrative datasets,2 such that it is now possible to study the relative risk of death among drug-resistant versus drug-susceptible pathogens, after accounting for patient risk factors allowing for a more accurate estimate of mortality associated with bacterial drug resistance. In this study, we focused on bloodstream infections with E. coli, the most common bloodstream pathogen and the leading pathogen among AMR-associated deaths,1,2 to determine the extent to which AMR is associated with an increased odds of death in the context of a well-resourced healthcare system.
Methods
General study design and setting
We conducted a retrospective cohort study of all people with E. coli bacteremia in Ontario, Canada between January 1, 2017 and December 31, 2020, and examined the odds of mortality associated with resistant versus susceptible isolates after accounting for clinically relevant patient characteristics.
Data sources
The study was made possible by the availability of province-wide microbiology results from the Ontario Laboratories Information System (OLIS), combining data form more than 100 hospital, community and public health laboratories into a single repository.2,3 OLIS is linkable to administrative datasets from Ontario's universal healthcare system, via a unique and confidential identifier held at ICES (formerly, the Institute for Clinical Evaluative Sciences). ICES is an independent, non-profit research institute whose legal status under Ontario's health information privacy law allows it to collect and analyze health care and demographic data, without consent, for health system evaluation and improvement. The relevant linked databases for this study included hospital information from the Canadian Institute for Health Information Discharge Abstract Database (CIHI-DAD), emergency department data from the National Ambulatory Care Reporting System (NACRS), physician claims data from the Ontario Health Insurance Plan (OHIP), and vital statistics and demographic data from the Registered Persons Database (RPDB).4,5
Index event and inclusion/exclusion criteria
We included all unique episodes of E. coli bacteremia from 2017 to 2020. A person could be included more than once during the four year study period, but all E. coli positive blood cultures within 7 days of an initial blood culture were considered part of the same episode.2 The index date was the date of collection of the first E. coli positive blood culture in the episode. We excluded non-Ontario residents, the rare patients with missing age, sex, or postal code, and episodes with polymicrobial bacteremia, or no antibiotic susceptibility testing.
Primary predictor variables: Antimicrobial resistance
The primary predictor in this study was antimicrobial resistant as compared to antimicrobial susceptible E. coli. We studied eight individual classes of agents including: aminopenicillins (represented by ampicillin), first generation cephalosporins (cefazolin), third generation cephalosporins (ceftriaxone, ceftazidime), beta-lactam beta-lactamase inhibitors (piperacillin-tazobactam), carbapenems (ertapenem, meropenem), fluoroquinolones (ciprofloxacin, levofloxacin, moxifloxacin), sulfonamides (trimethoprim-sulfamethoxazole), and commonly used aminoglycosides (gentamicin, tobramycin). In addition, we examined a special category of difficult to treat resistance (DTTR) defined as resistance to carbapenems, fluoroquinolones, and at least one of third generation cephalosporins or beta-lactam beta-lactamase inhibitors.6
Measurement of antimicrobial resistance
OLIS includes data from many laboratories, and not all test and report the same panel of antibiotics.3 Even within a laboratory, resistance reporting for E. coli could be variably suppressed or released based on results of other agents. Therefore, in addition to reported susceptibility results, we applied rule-based logic to impute missing susceptibility results where they could be reliably inferred from available results. For example, an ampicillin susceptible isolate could be imputed to be piperacillin-tazobactam susceptible, but not vice versa. After rule-based imputation, we applied a logistic regression model-based imputation approach for remaining antibiotics. The multivariable model accounted for the overall rate of susceptibility to that agent (intercept) as well as patient age, sex, location at time of culture collection, and the results of all other classes (including those recorded as ‘missing’). In sensitivity analyses we examined other approaches to imputing these missing susceptibility results, including: imputing all missing results as susceptible, imputing all missing results as the proportion of susceptible results among available data for that class, or deleting all organisms with missing results.
Primary outcome
The outcome of interest was 90 day mortality, defined from the date of collection of the initial blood culture yielding E. coli.
Patient characteristics for multivariable risk adjustment
From the ICES databases we extracted patient characteristics that could potentially be associated with AMR and patient outcomes, including: age, sex, setting at the time of blood culture draw (community, hospital ward, intensive care unit (ICU), long term care), total days spent in hospital in prior 12 months, total days spent in ICU in prior 12 months, total days spent in long term care in prior 12 months, total physician visits in prior 12 months, source of bacteremia (urinary tract versus other/unknown), immunosuppressive illnesses, and 18 individual comorbidities (osteoarthritis, cancer, arrhythmias, mood disorders, mental health disorders, osteoporosis, renal disease, stroke, coronary artery disease, myocardial infarction, asthma, congestive heart failure, chronic obstructive pulmonary disease, dementia, diabetes mellitus, hypertension, rheumatoid arthritis, and chronic liver disease).
Statistical analysis
We used univariable logistic regression to examine the crude association between AMR and 90 day mortality following E. coli bloodstream infection, with separate models for each of the 8 antibiotic classes, as well as DTTR; results are reported as odds ratios and 95% confidence intervals. Next, we used multivariable logistic regression, adjusting for all patient characteristics (listed above), again with separate models for each resistance measure. In sensitivity analyses, we used Generalized Estimating Equations to account for multiple E. coli bloodstream infections within patients, and random effects to account for potential clustering by treating facility. For facility random effects we used unique institution numbers for each acute and long term care facility; community onset infections were categorized separately.
We conducted pre-specified sensitivity analyses in which we used alternative approaches for imputing missing susceptibility results (described above), and a post-hoc sensitivity analysis examining 30 day rather than 90 day mortality. In an additional post-hoc sensitivity analysis we incorporated healthcare utilization as non-linear covariates using cubic splines with 4 knots.
Lastly, we conducted subgroup analyses based on setting of specimen collection (community, long term care, hospital ward, intensive care). All analyses were conducted in SAS version 9.4 (Cary, NC).
Role of the funding source
The sponsor of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
Between 2017 and 2020 there were 15,843 episodes of E. coli bloodstream infection among Ontario residents; after excluding patients with missing age, sex or postal code information (n = 14), non-Ontario residents (n = 8), and patients with polymicrobial bloodstream infection (n = 1273), there were 14,548 episodes of E. coli bloodstream infection eligible for analysis, among 13,706 unique patients. These bloodstream infection episodes were distributed across community (2382, 16.4%), acute care hospital wards (10,233, 70.3%), intensive care units (1784, 12.3%), and long term care facilities (149, 1.0%). The treating facilities included 143 acute care hospitals and 107 long term care facilities.
The median age of people across episodes of E. coli bloodstream infections was 74 years and women outnumbered men (8102; 55.7%) (Table 1, column 1). Patients had spent an average of 11.95 days in hospital in the preceding year (median 1 day, interquartile range 0–11 days). Nearly half of the E. coli bloodstream infections originated from a urinary tract source (6913; 47.5%). Overall, 2585 (17.8%) of episodes were associated with death within 90 days.
Table 1.
Characteristics among patients with E. coli bloodstream infection, overall and by isolate resistance to individual antibiotic classes.
| Characteristic | Total |
Aminopenicillin resistant |
First gen. cephalosporin resistant |
Third gen. cephalosporin resistant |
Beta-lactam beta-lactamase inhibitor resistant |
Carbapenem resistant |
Sulfonamide resistant |
Fluoroquinolone resistant |
Aminoglycosidea resistant |
Difficult to treat resistanceb |
|---|---|---|---|---|---|---|---|---|---|---|
| N = 14,548 | N = 6807 | N = 5647 | N = 2012 | N = 1323 | N = 28 | N = 3501 | N = 3856 | N = 1700 | N = 18 | |
| Sex (n, %) | ||||||||||
| Female | 8102 (55.7%) | 3688 (54.2%) | 2953 (52.3%) | 971 (48.3%) | 622 (47.0%) | 11 (39.3%) | 1877 (53.6%) | 1779 (46.1%) | 875 (51.5%) | 6 (33.3%) |
| Male | 6446 (44.3%) | 3119 (45.8%) | 2694 (47.7%) | 1041 (51.7%) | 701 (53.0%) | 17 (60.7%) | 1624 (46.4%) | 2077 (53.9%) | 825 (48.5%) | 12 (66.7%) |
| Age | ||||||||||
| Mean (SD) | 70.57 (18.01) | 69.97 (17.83) | 70.12 (17.72) | 70.81 (16.47) | 70.35 (16.09) | 64.21 (18.50) | 69.67 (17.63) | 72.09 (15.37) | 70.67 (17.35) | 66.61 (15.93) |
| Median (Q1-Q3) | 74 (62–84) | 73 (62–83) | 73 (62–83) | 74 (63–82) | 72 (62–82) | 70 (52–79) | 72 (61–82) | 74 (64–83) | 73 (63–83) | 68 (55–79) |
| Setting (n, %) | ||||||||||
| Community | 2382 (16.4%) | 1038 (15.2%) | 842 (14.9%) | 231 (11.5%) | 131 (9.9%) | 0 (0.0%) | 544 (15.5%) | 471 (12.2%) | 230 (13.5%) | 0 (0.0%) |
| Hospital | 10,233 (70.3%) | 4804 (70.6%) | 3991 (70.7%) | 1470 (73.1%) | 991 (74.9%) | 21 (75.0%) | 2481 (70.9%) | 2857 (74.1%) | 1240 (72.9%) | 13–17a |
| ICU | 1784 (12.3%) | 891 (13.1%) | 746 (13.2%) | 289 (14.4%) | 195 (14.7%) | 7 (25.0%) | 432 (12.3%) | 475 (12.3%) | 209 (12.3%) | 1–5a |
| LTC | 149 (1.0%) | 74 (1.1%) | 68 (1.2%) | 22 (1.1%) | 6 (0.5%) | 0 (0.0%) | 44 (1.3%) | 53 (1.4%) | 21 (1.2%) | 0 (0.0%) |
| Days spent in hospital in prior 12 months | ||||||||||
| Mean (SD) | 11.95 (30.20) | 14.80 (34.66) | 15.97 (35.96) | 23.80 (44.75) | 24.38 (44.59) | 45.43 (52.65) | 16.46 (37.17) | 19.56 (42.01) | 19.65 (39.91) | 45.28 (60.76) |
| Median (Q1-Q3) | 1 (0–11) | 2 (0–15) | 3 (0–16) | 7 (0–27) | 8 (0–28) | 29 (8–63) | 3 (0–17) | 4 (0–20) | 4 (0–22) | 24 (5–64) |
| Days spent in ICU in prior 12 months | ||||||||||
| Mean (SD) | 1.03 (8.26) | 1.40 (10.51) | 1.56 (11.48) | 2.72 (17.35) | 3.26 (19.68) | 5.86 (14.23) | 1.38 (10.41) | 1.98 (13.70) | 1.37 (6.97) | 2.67 (9.26) |
| Median (Q1-Q3) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–3) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–0) |
| Number of physician visits in prior 12 months | ||||||||||
| Mean (SD) | 6.26 (6.30) | 6.65 (6.65) | 6.71 (6.69) | 7.58 (7.30) | 8.04 (7.94) | 11.39 (9.81) | 6.94 (6.90) | 6.91 (7.05) | 7.06 (6.77) | 11.22 (8.17) |
| Median (Q1-Q3) | 5 (2–9) | 5 (2–9) | 5 (2–9) | 6 (2–11) | 6 (3–11) | 9 (4–16) | 5 (2–9) | 5 (2–9) | 5 (2–10) | 9 (4–16) |
| Days spent in LTC in prior 12 months | ||||||||||
| Mean (SD) | 24.20 (86.44) | 27.89 (91.67) | 30.43 (95.51) | 39.10 (105.53) | 32.32 (96.62) | 0.00 (0.00) | 29.43 (94.26) | 44.62 (113.48) | 36.55 (102.28) | 0.00 (0.00) |
| Median (Q1-Q3) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–0) |
| Source of bacteremia (n, %) | ||||||||||
| Other | 7635 (52.5%) | 3526 (51.8%) | 2942 (52.1%) | 1095 (54.4%) | 680 (51.4%) | 19 (67.9%) | 1740 (49.7%) | 2001 (51.9%) | 843 (49.6%) | 11 (61 |
| Urinary tract | 6913 (47.5%) | 3281 (48.2%) | 2705 (47.9%) | 917 (45.6%) | 643 (48.6%) | 9 (32.1%) | 1761 (50.3%) | 1855 (48.1%) | 857 (50.4%) | 7 (38.9%) |
| Immunosuppressive illness (n, %) | 1267 (8.7%) | 691 (10.2%) | 570 (10.1%) | 224 (11.1%) | 163 (12.3%) | 1–5a | 473 (13.5%) | 425 (11.0%) | 180 (10.6%) | 1–5a |
| Comorbidities (n, %) | ||||||||||
| Osteoarthritis | 9776 (67.2%) | 4625 (67.9%) | 3835 (67.9%) | 1388 (69.0%) | 908 (68.6%) | 17 (60.7%) | 2355 (67.3%) | 2655 (68.9%) | 1147 (67.5%) | 12 (66.7%) |
| Cancer | 8344 (57.4%) | 3855 (56.6%) | 3213 (56.9%) | 1101 (54.7%) | 741 (56.0%) | 15 (53.6%) | 2039 (58.2%) | 2227 (57.8%) | 969 (57.0%) | 10 (55.6%) |
| Arrhythmia | 2733 (18.8%) | 1291 (19.0%) | 1073 (19.0%) | 385 (19.1%) | 246 (18.6%) | 1–5a | 653 (18.7%) | 738 (19.1%) | 311 (18.3%) | 0 (0.0%) |
| Mood disorder | 8036 (55.2%) | 3724 (54.7%) | 3062 (54.2%) | 1056 (52.5%) | 690 (52.2%) | 9 (32.1%) | 1922 (54.9%) | 2084 (54.0%) | 932 (54.8%) | 1–5a |
| Mental health disorder | 5198 (35.7%) | 2542 (37.3%) | 2124 (37.6%) | 841 (41.8%) | 537 (40.6%) | 10 (35.7%) | 1335 (38.1%) | 1533 (39.8%) | 644 (37.9%) | 1–5a |
| Osteoporosis | 1766 (12.1%) | 825 (12.1%) | 673 (11.9%) | 244 (12.1%) | 164 (12.4%) | 1–5a | 427 (12.2%) | 468 (12.1%) | 210 (12.4%) | 1–5a |
| Renal disease | 4410 (30.3%) | 2293 (33.7%) | 1959 (34.7%) | 872 (43.3%) | 551 (41.6%) | 13 (46.4%) | 1310 (37.4%) | 1514 (39.3%) | 657 (38.6%) | 8 (44.4%) |
| Stroke | 1631 (11.2%) | 780 (11.5%) | 680 (12.0%) | 272 (13.5%) | 184 (13.9%) | 1–5a | 418 (11.9%) | 518 (13.4%) | 207 (12.2%) | 1–5a |
| Coronary artery disease | 4278 (29.4%) | 2057 (30.2%) | 1733 (30.7%) | 672 (33.4%) | 413 (31.2%) | 1–5a | 1042 (29.8%) | 1236 (32.1%) | 520 (30.6%) | 1–5a |
| Acute myocardial infarction | 1148 (7.9%) | 547 (8.0%) | 447 (7.9%) | 175 (8.7%) | 108 (8.2%) | 1–5a | 277 (7.9%) | 343 (8.9%) | 140 (8.2%) | 1–5a |
| Asthma | 2616 (18.0%) | 1307 (19.2%) | 1069 (18.9%) | 421 (20.9%) | 280 (21.2%) | 1–5a | 700 (20.0%) | 729 (18.9%) | 347 (20.4%) | 1–5a |
| Congestive heart failure | 3105 (21.3%) | 1596 (23.4%) | 1327 (23.5%) | 597 (29.7%) | 384 (29.0%) | 7 (25.0%) | 899 (25.7%) | 1007 (26.1%) | 462 (27.2%) | 1–5a |
| Chronic obstructive pulmonary disease | 1937 (13.3%) | 978 (14.4%) | 787 (13.9%) | 312 (15.5%) | 196 (14.8%) | 1–5a | 517 (14.8%) | 582 (15.1%) | 267 (15.7%) | 0 (0.0%) |
| Dementia | 2471 (17.0%) | 1180 (17.3%) | 1024 (18.1%) | 410 (20.4%) | 240 (18.1%) | 1–5a | 603 (17.2%) | 822 (21.3%) | 335 (19.7%) | 0 (0.0%) |
| Diabetes mellitus | 6174 (42.4%) | 3062 (45.0%) | 2560 (45.3%) | 1050 (52.2%) | 677 (51.2%) | 13 (46.4%) | 1610 (46.0%) | 1936 (50.2%) | 803 (47.2%) | 9 (50.0%) |
| Hypertension | 10,532 (72.4%) | 5010 (73.6%) | 4166 (73.8%) | 1556 (77.3%) | 1015 (76.7%) | 18 (64.3%) | 2572 (73.5%) | 2979 (77.3%) | 1294 (76.1%) | 12 (66.7%) |
| Rheumatoid arthritis | 236 (1.6%) | 138 (2.0%) | 107 (1.9%) | 44 (2.2%) | 26 (2.0%) | 0 (0.0%) | 73 (2.1%) | 72 (1.9%) | 42 (2.5%) | 0 (0.0%) |
| Chronic liver disease | 862 (5.9%) | 448 (6.6%) | 392 (6.9%) | 166 (8.3%) | 122 (9.2%) | 6 (21.4%) | 236 (6.7%) | 257 (6.7%) | 113 (6.6%) | 1–5a |
| 30-day mortality (n, %) | 1822 (12.5%) | 944 (13.9%) | 796 (14.1%) | 356 (17.7%) | 240 (18.1%) | 10 (35.7%) | 497 (14.2%) | 604 (15.7%) | 265 (15.6%) | 8 (44.4%) |
| 90-day mortality (n, %) | 2585 (17.8%) | 1312 (19.3%) | 1111 (19.7%) | 500 (24.9%) | 334 (25.2%) | 11 (39.3%) | 687 (19.6%) | 856 (22.2%) | 387 (22.8%) | 8 (44.4%) |
ICES privacy/confidentiality safeguards require suppression of small cell sizes.
Difficult to treat resistance is defined as resistance to carbapenems, fluoroquinolones, and at least one of third generation cephalosporins or beta-lactam beta-lactamase inhibitors.
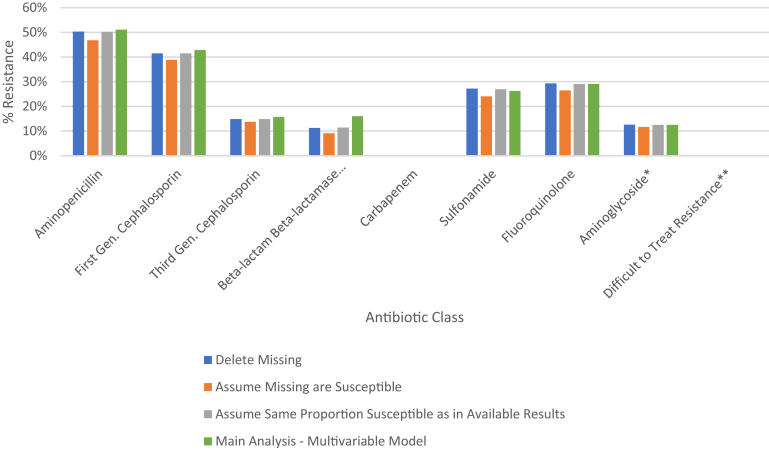
Among the 8 antibiotic classes of interest, resistance was most common to aminopenicillins (6807; 46.8%), followed by first generation cephalosporins (5647; 38.8%), fluoroquinolones (3856; 26.5%), sulfonamides (3501; 24.1%), third generation cephalosporins (2012; 13.8%), aminoglycosides (1700; 11.7%), beta-lactam beta-lactamase inhibitors (1323; 9.1%) and carbapenems (28; 0.2%). There were only (18; 0.1%) episodes exhibiting a DTTR profile. Only a minority of susceptibility results were missing after rule-based imputation (ranging from 4.8% for carbapenems results to 19.8% for beta-lactam beta-lactamase inhibitor results). Furthermore, final resistance rates were robust across multiple methods for imputing missing susceptibilities (Fig. 1).
Fig. 1.
Antibiotic class-specific resistance rates among E. coli bloodstream infections (similar across different imputation methods). ∗Aminoglycosides refers to gentamicin and tobramycin. ∗∗Difficult to treat resistance is defined as resistance to carbapenems, fluoroquinolones, and at least one of third generation cephalosporins or beta-lactam beta-lactamase inhibitors.
Patient demographic characteristics, healthcare exposure histories, and comorbidities differed substantially among resistant versus susceptible isolates (Table 1). For example, the average (SD) number of days admitted to hospital in the prior 12 months was 14.8 (34.7) for those with aminopenicillin resistance, 16.0 (36.0) for those with first generation cephalosporin resistance, 23.8 (44.8) with third generation cephalosporin resistance, 24.4 (44.6) for beta-lactamase inhibitor resistance, 45.4 (52.7) for carbapenem resistance, 16.5 (37.2) for sulfonamide resistance, 19.6 (42.0) for fluoroquinolone resistance, 19.7 (39.9) for aminoglycoside resistance, and 45.3 (60.8) days for DTTR (Table 1).
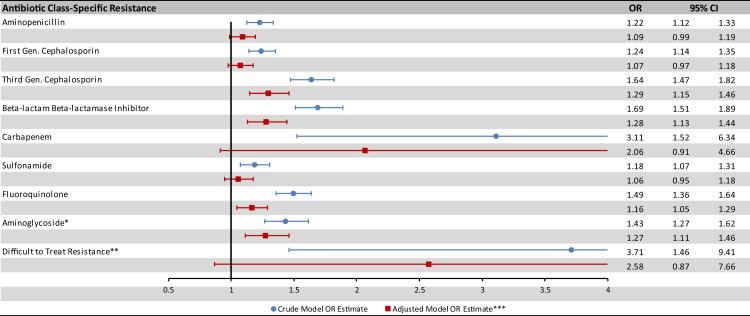
The 90 day mortality rate among resistant isolates was higher than the mortality rate among overall isolates (17.8%), including isolates with aminopenicillin resistance (19.3%), first generation cephalosporin resistance (19.7%), third generation cephalosporin resistance (24.9%), beta-lactamase inhibitor resistance (25.2%), carbapenem resistance (39.3%), sulfonamide resistance (19.6%), fluoroquinolone resistance (22.2%), aminoglycoside resistance (22.8%) and DTTR (44.4%). The unadjusted odds ratio for 90 day mortality was significantly greater than 1 for each antibiotic class (Fig. 2, blue bars).
Fig. 2.
Association of antibiotic resistance and 90-day mortality among patients with E. coli bacteremia (crude and adjusted OR estimates). ∗Aminoglycosides refers to gentamicin and tobramycin. ∗∗Difficult to treat resistance is defined as resistance to carbapenems, fluoroquinolones, and at least one of third generation cephalosporins or beta-lactam beta-lactamase inhibitors. ∗∗∗The adjusted model accounts for patient age, sex, setting, healthcare utilization, 18 individual comorbidities, and source of bacteremia.
After adjusting for patient demographics, health care exposure and comorbidities the association between AMR and mortality was attenuated, but remained statistically significant for many individual antibiotic classes (Fig. 2, red bars). The point estimates for the adjusted odds ratio (aOR) of AMR associated mortality were highest for DTTR (aOR 2.58, 95% CI 0.87–7.66), carbapenems (aOR 2.06, 95% CI 0.91–4.66), third generation cephalosporins (aOR 1.29, 95% CI 1.15–1.46), and beta-lactam beta-lactamase inhibitors (aOR 1.28, 95% CI 1.13–1.45).
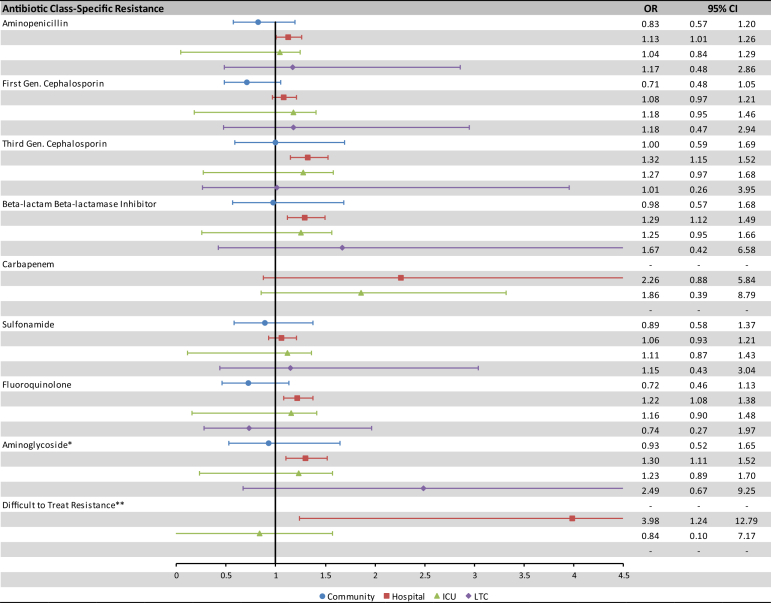
Fig. 3 displays subgroup analyses of blood culture draws occurring in hospitals, ICUs, long term care facilities and the community. In post-hoc sensitivity analyses, there was a substantial attenuation in the association of AMR and mortality when healthcare utilization variables were added sequentially into the models (Supplemental Fig. S1), a further strengthened association of AMR with an earlier 30 day mortality time-point (Supplemental Fig. S2), and minimal impact of accounting for clustering at the patient or facility level (Supplemental Fig. S3).
Fig. 3.
Association of antibiotic resistance and 90-day mortality among patients with E. coli bacteremia by care setting (adjusted∗∗∗ OR estimates only). ∗Aminoglycosides refers to gentamicin and tobramycin. ∗∗Difficult to treat resistance is defined as resistance to carbapenems, fluoroquinolones, and at least one of third generation cephalosporins or beta-lactam beta-lactamase inhibitors. ∗∗∗The adjusted model accounts for patient age, sex, setting, healthcare utilization, 18 individual comorbidities, and source of bacteremia.
Discussion
Through population-wide linkable microbiology and clinical databases we were able to study the association of resistance and mortality among more than 14,000 episodes of E. coli bloodstream infection. For each class of antimicrobial agents, we detected much higher crude mortality associated with resistant versus susceptible E. coli. After accounting for patient characteristics including age, sex, comorbidities, test location, and especially healthcare exposure, the associations between resistance and mortality were greatly attenuated but many remained statistically significant. The point estimates for the adjusted odds ratio of AMR associated mortality remained highest for DTTR and individual antibiotic classes most commonly used in empiric treatment (third generation cephalosporins, beta-lactam-beta-lactamase inhibitors and carbapenems).
Our study findings bolster the Antimicrobial Resistance Collaboration's current estimate of 1 million deaths per year attributable to AMR,1 by demonstrating a strong association between AMR and mortality. There are a number of prior large studies and systematic reviews which have documented substantial increased mortality associated with resistant isolates, but most of these are crude estimates. For example, a nationwide study of more than 11,000 E. coli bloodstream infections in Israel, detected a higher case fatality rate with multi-drug resistant (MDR) versus non-MDR E. coli (47% versus 28%), but these were crude rates derived from submissions to a national death registry.7 A systematic review of 16 studies involving MDR bacteria in hospitalized patients, detected increased mortality compared to non-MDR bacteria (RR 1.61, 95% CI 1.36–1.90), but this involved pooling of crude mortality rates without any adjustment for patient characeristics.8 Similarly, a systematic review pooling 15 studies of carbapenem-resistant versus sensitive Klebsiella pneumoniae infections calculated increased mortality in unadjusted analyses (OR 2.2, 95% CI 1.8–2.6), but was unable to generate a pooled adjusted odds ratio because too few studies undertook multivariable adjustment.9 Our study builds upon this work by establishing an association between AMR and mortality, after adjusting for key patient characteristics available in linked data sources.
Increases in mortality with AMR are unlikely to be driven by increased virulence among resistant pathogens,10 but rather because it is well established that delays in adequate antibiotic coverage are associated with increased mortality among patients with serious bacterial infections.11, 12, 13, 14 In a systematic review of seventy studies evaluating the importance of adequate empiric coverage for sepsis, among the subset of 26 studies accounting for comorbidities and severity of illness, inadequate coverage was associated with a pooled aOR of 1.60 (95% CI 1.37–1.86) for mortality.11 This translated to a number needed to harm of 10 – one additional fatality for each 10 patients receiving inadequate coverage.11 Although antibiotic treatments are not available in this cohort, a prior Canadian survey indicated that empiric treatment recommendations for sepsis are variable, but most commonly include ceftriaxone, piperacillin-tazobactam or meropenem.15 This likely explains why resistance to these three classes of beta-lactam agents exhibited the strongest associations with 90 day mortality. DTTR, although uncommon in Ontario, was even more strongly associated with 90 day mortality because it portends a lower likelihood of adequate empiric coverage, and even after susceptibility results become available DTTR treatment often requires use of toxic agents such as aminoglycosides.6
The attenuation in odds ratios on multivariable adjustment in our study suggests that many prior studies over-estimate AMR attributable mortality by failing to adjust for potential confounding patient characteristics that could be associated with both the acquisition of AMR and the likelihood of death – including age, comorbidities and healthcare exposure history. Incorporating unadjusted odds of death with resistant versus susceptible organisms can lead to unrealistic projections and predictions of the population burden of AMR.16,17
Our study is potentially limited by the fact that methods of antibiotic susceptibility testing and reporting, including the specific panels of antibiotics, are not standardized across all laboratories. However, our results were robust across multiple imputation techniques for missing susceptibilities. Our analyses are potentially underpowered for carbapenem resistance and DTTR because of low local rates of these resistance profiles. Similarly, we are unable to study the impact of resistance to reserve use agents (including older agents such as amikacin and colistin, and newer agents such as ceftolozane-tazobactam) given that these are only rarely tested and reported. A major limitation of this study is lack of access to information on the empiric and targeted antibiotic agents used to treat these bloodstream infections. This information would have been helpful to determine the extent to which selection of inadequate treatment regimens mediates the impact of resistance on outcomes. Ontario has a well-resourced healthcare system with both easy access to therapeutics and low prevalence of pan-drug resistance, and so the generalizability of our findings is unclear to low- and middle-income countries as well as regions with high prevalence of resistance. Although we accounted for a rich array of patient characteristics, there is still potential for unmeasured confounding by unavailable measures.
E. coli bloodstream infections are associated with a high 90 day mortality rate, and the crude mortality is significantly higher for strains resistant to each class of common antimicrobial agents. Some of the current elevated mortality with AMR E. coli is explained by patient characteristics, and in particular confounding by extent of prior healthcare exposure. Therefore many estimates of the current mortality associated with AMR may be over-estimates, and ongoing surveillance for AMR-associated mortality should incorporate adjustment for patient characteristics. However, our data detected strong signals of associated increased mortality with AMR, even after multivariable adjustment. The association was strongest for the most important empiric and therapeutic classes of agents, and so future estimates will also need to take into account antibiotic availability and treatment practices in different regions. Ongoing global and local efforts are essential to help curtail AMR, and thereby minimize mortality among patients with E. coli bloodstream infection and other common bacterial pathogens and syndromes.
Contributors
Conceptualization: N.D., K.A.B. Data Curation: D.F. Formal Analysis: D.F. Funding Acquisition: N.D., K.A.B., K.L.S., J.J., D.M.M., S.N.P. Investigation: N.D., D.F., J.J., B.J.L., S.M.L., D.M.M., K.M., S.N.P., K.L.S., K.A.B. Methodology: N.D., D.F., J.J., B.J.L., S.M.L., D.M.M., K.M., S.N.P., K.L.S., K.A.B. Project Administration: S.M.L. Writing Original Draft: N.D., S.M.L. Writing-Reviewing/Editing: N.D., D.F., J.J., B.J.L., S.M.L., D.M.M., K.M., S.N.P., K.L.S., K.A.B. Accessed and Verified all Data: D.F., N.D., K.A.B.
Data sharing statement
ICES is an independent, non-profit research institute whose legal status under Ontario's health information privacy law allows it to collect and analyze health care and demographic data, without consent, for health system evaluation and improvement. This data is subject to extensive privacy and confidentiality safeguards, and so cannot be released for open use. However, ICES can enable data access to external researchers through ICES Data & Analytic Services (ICES DAS) (https://www.ices.on.ca/DAS).
Declaration of interests
We declare that we have no conflicts of interest.
Acknowledgement
This work was supported by a project grant from CIHR (grant number 159503 to K.A.B. and N.D.). This study was supported by ICES, which is funded by an annual grant from the Ontario Ministry of Health (MOH) and the Ministry of Long-Term Care (MLTC). Parts of this material are based on data and information compiled and provided by: MOH, OLIS and CIHI. The analyses, conclusions, opinions and statements expressed herein are solely those of the authors and do not reflect those of the funding or data sources; no endorsement is intended or should be inferred. Parts of this material are based on data and/or information compiled and provided by CIHI. However, the analyses, conclusions, opinions and statements expressed in the material are those of the author(s), and not necessarily those of CIHI.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2022.101781.
Appendix A. Supplementary data
References
- 1.Antimicrobial Resistance Collaborators Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verway M., Brown K.A., Marchand-Austin A., et al. Prevalence and mortality associated with bloodstream organisms: a population-wide retrospective cohort study. J Clin Microbiol. 2022;6(4) doi: 10.1128/jcm.02429-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langford B.J., Daneman N., Diong C., et al. Antibiotic susceptibility reporting and association with antibiotic prescribing: a cohort study. Clin Microbiol Infect. 2021;27(4):568–575. doi: 10.1016/j.cmi.2020.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Juurlink D.P., Preyra C., Croxford R., et al. Canadian Institute for Health Information discharge abstract database: a validation study. Toronto: Institute for Clinical Evaluative Sciences; June, 2006. https://www.ices.on.ca/Publications/Atlases-and-Reports/2006/Canadian-Institute-for-Health-Information Available from:
- 5.Williams J.I., Young W. In: Patterns of health care in Ontario: the ICES practice atlas. 2 ed. Goel V., Williams J.I., Anderson G.M., Blackstein-Hirsch P., Fooks C., Naylor C.D., editors. Canadian Medical Association; Ottawa: 1996. A summary of the quality of healthcare adminstrative databases in Canada; pp. 339–346. [Google Scholar]
- 6.Kadri S.S., Adjemian J., Lai Y.L., et al. Difficult-to-treat resistance in gram-negative bacteremia at 173 US hospitals: retrospective cohort analysis of prevalence, predictors, and outcome of resistance to all first-line agents. Clin Infect Dis. 2018;67(12):1803–1814. doi: 10.1093/cid/ciy378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldman S.F., Temkin E., Wullfhart L., et al. A nationwide population-based study of Escherichia coli bloodstream infections: incidence, antimicrobial resistance and mortality. Clin Microbiol Infect. 2022;28(6):879.e1–879.e7. doi: 10.1016/j.cmi.2021.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Serra-Burriel M., Keys M., Campillo-Artero C., et al. Impact of multi-drug resistant bacteria on economic and clinical outcomes of healthcare-associated infections in adults: systematic review and meta-analysis. PLoS One. 2020;15(1) doi: 10.1371/journal.pone.0227139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohler P.P., Volling C., Green K., Uleryk E.M., Shah P.S., McGeer A. Carbapenem resistance, initial antibiotic therapy, and mortality in Klebsiella pneumoniae Bacteremia: a systematic review and meta-analysis. Infect Control Hosp Epidemiol. 2017;38(11):1319–1328. doi: 10.1017/ice.2017.197. [DOI] [PubMed] [Google Scholar]
- 10.Da Silva G.J., Mendonca N. Association between antimicrobial resistance and virulence in Escherichia coli. Virulence. 2012;3(1):18–28. doi: 10.4161/viru.3.1.18382. [DOI] [PubMed] [Google Scholar]
- 11.Paul M., Shani V., Muchtar E., Kariv G., Robenshtok E., Leibovici L. Systematic review and meta-analysis of the efficacy of appropriate empiric antibiotic therapy for sepsis. Antimicrob Agents Chemother. 2010;54(11):4851–4863. doi: 10.1128/AAC.00627-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar A., Roberts D., Wood K.E., et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006;34(6):1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9. [DOI] [PubMed] [Google Scholar]
- 13.Kadri S.S., Lai Y.L., Warner S., et al. forming the National Institutes of Health Antimicrobial Resistance Outcomes Research Initiative (NIH-ARORI). Inappropriate empirical antibiotic therapy for bloodstream infections based on discordant in-vitro susceptibilities: a retrospective cohort analysis of prevalence, predictors, and mortality risk in US hospitals. Lancet Infect Dis. 2021;21(2):241–251. doi: 10.1016/S1473-3099(20)30477-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ibrahim E.H., Sherman G., Ward S., Fraser V.J., Kollef M.H. The influence of inadequate antimicrobial treatment of bloodstream infections on patient outcomes in the ICU setting. Chest. 2000;118(1):145–155. doi: 10.1378/chest.118.1.146. [DOI] [PubMed] [Google Scholar]
- 15.Cressman A.M., MacFadden D.R., Verma A.A., Fahad R., Daneman N. Empiric antibiotic treatment thresholds for serious bacterial infections: a scenario-based survey study. Clin Infect Dis. 2019;69(6):930–937. doi: 10.1093/cid/ciy1031. [DOI] [PubMed] [Google Scholar]
- 16.de Kraker M.E., Stewardson A.J., Harbarth S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 2016;13(11) doi: 10.1371/journal.pmed.1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Neill J. Tackling drug-resistant infections globally: final report and recommendations. London, England. 2016. https://wellcomecollection.org/works/thvwsuba Available from:
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.