Abstract
Lipase is a very important digestive enzyme for triglyceride absorption in vivo. The inhibitory activities of 26 dietary flavonoids, including flavone, flavanone, isoflavone and flavanol, on lipase were determined. Flavone exhibited stronger inhibitory activity than other types of flavonoids. Among them, luteolin exhibited the strongest inhibitory activity with IC50 value of 99 ± 11 μM, followed by quercetin and baicalein. The binding affinity of these flavonoids with lipase was investigated by fluorescence titration method. The binding affinity of flavones was stronger than flavanones, and was linearly positively correlated with their inhibitory activity. The binding of flavones on lipase caused the blue-shift of fluorescence, while flavanones caused red-shift. The analysis of structure-activity relationship of flavonoids on lipase revealed that the structure of C ring is very crucial. The hydrogenation of C2=C3 bond and the absence of C=O group in C ring both caused significant decrease of inhibitory activity. Besides, the hydroxylation on ring A and B of flavones increased the activity, while glycosylation weakened the activity. Molecular docking analysis confirmed that C2=C3 bond in C ring of flavones increases the π-conjugation and contributes to maintaining the planarity of flavonoid structure, which favour its Pi-Pi interaction with lipase.
Keywords: Flavonoids, Lipase, Inhibitory activity, Binding affinity, Structure-activity relationship
Graphical abstract
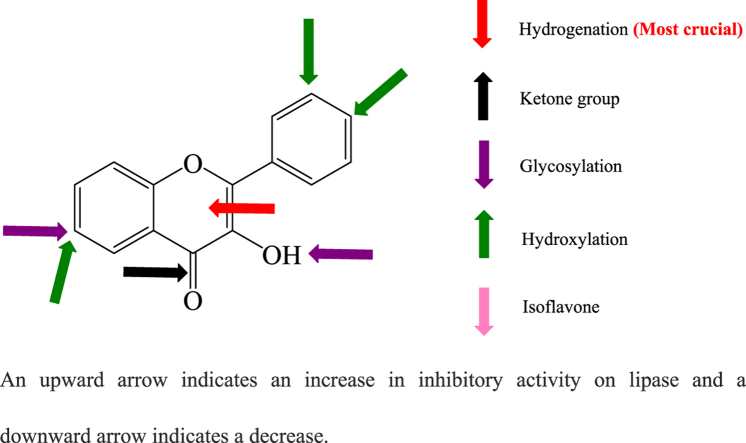
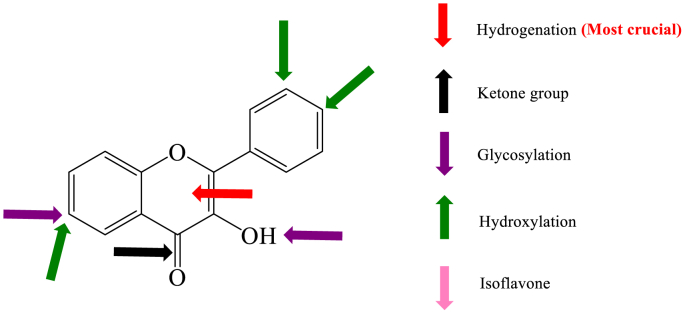
An upward arrow indicates an increase in inhibitory activity on lipase and a downward arrow indicates a decrease.
Highlights
-
•
The inhibitory activity of flavonoids on lipase was structure dependent.
-
•
The unsaturation of C2=C3 bond in C ring is very crucial.
-
•
Flavone exhibited stronger inhibitory activity than other types of flavonoids.
-
•
The binding affinity of flavones on lipase determined their inhibitory activity.
1. Introduction
Triglyceride (TG) is the nutrient with highest calorie in food. Excessive TG intake is the main cause of obesity. However, after ingestion, TG is not absorbed directly in human gastrointestinal tract. It should be hydrolyzed by lipase to form free fatty acids, monoacylglycerol and diacylglycerol before absorption by the enterocyte. Pancreatic lipase is the most important lipase, which responds for the hydrolysis of more than 70% food TG (Liu et al., 2020). Hence, the inhibition of pancreatic lipase can effectively reduce calorie intake without reducing food intake. Lipase inhibitor is a promising candidate in controlling obesity. Orlistat, a very powerful lipase inhibitor derived from natural product (lipstatin), is approved by the FDA for long-term obesity treatment (Birari and Bhutani, 2007). Besides, Orlistat is poorly absorbed in circulatory blood (Zhi et al., 1999). Hence, Orlistat is safe without accumulation and affecting the central nervous system. However, its complete inhibition of lipase may also cause side effects such as oily stools, flatulence, etc.
Compared with drug administration for obesity treatment, natural lipase inhibitors contained in food maybe more convenient for obesity prevention. Plant-based food contains many secondary metabolites, such as polyphenols, saponins, terpenes, etc. Flavonoids are the most commonly found polyphenols in human diet. Flavonoids are composed of three rings, in which benzene ring (A) and pyranone ring (C) are condensed together, and benzene ring B is connected to C ring through 2 or 3 position. According to the structure, dietary flavonoids could be subdivided into flavones (including flavonols), flavanone (including dihydroflavonols), isoflavone, flavan-3-ols and anthocyanin (Crozier et al., 2009). Studies showed that some flavonoids possess noteworthy lipase inhibition activity (Buchholz and Melzig, 2015). For example, quercetin exhibited inhibitory effect on pancreatic lipase with IC50 value of 70 μg/mL. After oral administration in rats, quercetin significantly reduced fat absorption in the gastrointestinal tract, but increased fat excretion in feces (Zhou et al., 2021). Galangin showed dose-dependent inhibition on pancreatic lipase activity and exhibited anti-obesity effects in rats (Kumar and Alagawadi, 2013). Tea polyphenols, epigallocatechin and epigallocatechin gallate (EGCG) also exhibited promising effects on lipase (Nakai et al., 2005; Rahim et al., 2015).
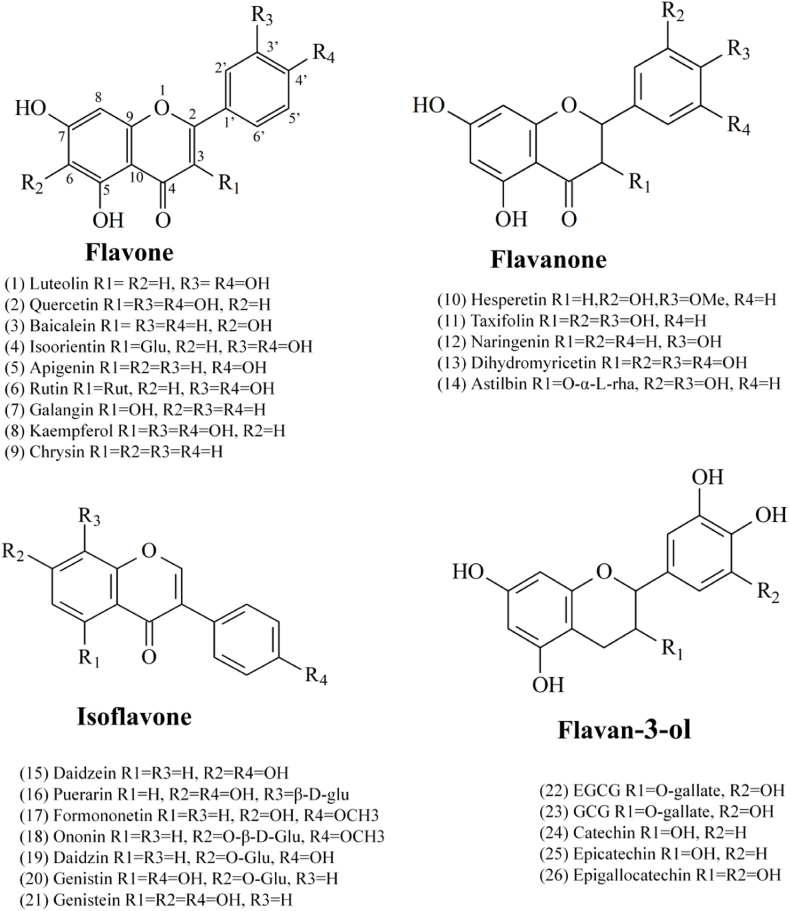
However, there are thousands of flavonoids found in nature. Because many flavonoids are market unavailable, as well as the huge labor intensity, it is impossible to investigate the inhibitory activity of flavonoids on lipase one by one. Hence, illuminating the structure-activity relationship of flavonoids on lipase is of great significance for guiding the discovery of more active flavonoids in diet. Hence, in the present study, the inhibitory activities of 26 flavonoids on lipase were compared simultaneously. The molecular structure of tested flavonoids was shown in Fig. 1. Their binding affinity with the enzyme was calculated by fluorescence titration and simulated by molecular docking analysis. To the best of our knowledge, the structure-activity relationship of flavonoids on lipase was discussed for the first time.
Fig. 1.
The molecular structure of 26 tested flavonoids.
2. Materials and methods
2.1. Chemicals and reagents
Taxifolin (Su et al., 2020), astilbin (Zheng and Zhang, 2019) and dihydromyricetin (Sun et al., 2021) (all>98%) were purified in our laboratory. EGCG, sodium deoxycholate and porcine pancreatic lipase were purchased from Yuanye Bio-Technology Co., Ltd. (Shanghai, China). Quercetin, luteolin, kaempferol, puerarin, apigenin, hesperetin, baicalein, chrysin, genistein, epigallocatechin, daidzein, rutin, epicatechin, naringenin and p-nitrophenyl pal-mitate (pNPP) were purchased from Shanghai Aladdin Biochemical Technology Co., Ltd. Galangin, formononetin, ononin, daidzin, genistin, catechin and gallocatechin 3-O-gallate (GCG) were purchased from Chengdu Push Bio-Technology Co., Ltd. Tris and gum acacia were purchased from Beijing Solarbio Science & Technology Co., Ltd. Milli-Q water was used throughout the study.
2.2. Inhibitory activity of flavonoids on pancreatic lipase
The inhibitory activity of flavonoids on pancreatic lipase was determined by using a Thermo Microplate Spectrophotometer (Multiskan FC, USA) according to the method of Zhou et al. (2021). Pancreatic lipase (20 mg) was suspended in 10 ml Tris-HCl buffer (50 mM, pH 8.0, containing 0.1% gum Arabic powder and 0.2% sodium deoxycholate). After gently shaking, the mixture was centrifuged at 2000 g for 10 min to obtain the enzyme supernatant. In 96-well plates, 80 μL Tris-HCl buffer, 100 μL enzyme supernatant and 10 μL flavonoids solution (dissolved in 50% ethanol with different concentration) were mixed together. After incubation at 37 °C for 20 min, 10 μL pNPP (10 mM, dissolved in absolute ethanol) was added to start the reaction. The absorbance at 405 nm was recorded every minute for 20 min. Then, the absorbance growth slope (V), which represents the enzyme activity was calculated. The inhibitory activity of flavonoids was calculated by the following equation (1):
| (1) |
where V0 and V were the enzyme activity in the absence and presence of inhibitor.
2.3. Fluorescence titration
In a tube, 4 mL Tris-HCl buffer (pH = 8.0), 1 mL lipase supernatant and 5 μL flavonoid solution (dissolved in 50% ethanol with different concentration) were mixed together. The Fluorescence spectra of the mixture was measured using a 970CRT spectrofluorophotometer (Shanghai Scientific Instruments Limited Company, Shanghai, China) with excitation wavelength at 280 nm. The binding constants (Ka) and number of binding sites (n) are calculated according to the logarithmic equation (2) (Hua et al., 2018):
| (2) |
where F0 and F represent the fluorescence intensity of pancreatic lipase in the absence and presence of flavonoids, [Q] is the concentration of flavonoids, respectively.
2.4. Molecular docking analysis
The molecular docking analysis between lipase and flavonoids was performed using software of Discovery Studio 2019 (Accelrys Co., Ltd., US). The crystal structure of pancreatic lipase (PDB ID: 1ETH) was downloaded from RCSB Protein Data Bank (http://www.rcsb.org/pdb/home.do), while the 3D structures of flavonoids were generated by Chem3D 19.0 software (CambridgeSoft Corporation, USA). A sphere centered on the active site cavities of lipase with a diameter of 10 Å was used as the docking site.
2.5. Statistic analysis
Data were expressed as means ± standard deviation of triplicate trials. Data analysis and plotting were performed with software of Origin 9.0 (Origin Lab Co., Northampton, MA, USA).
3. Results and discussion
3.1. Lipase inhibitory activity of 26 flavonoids
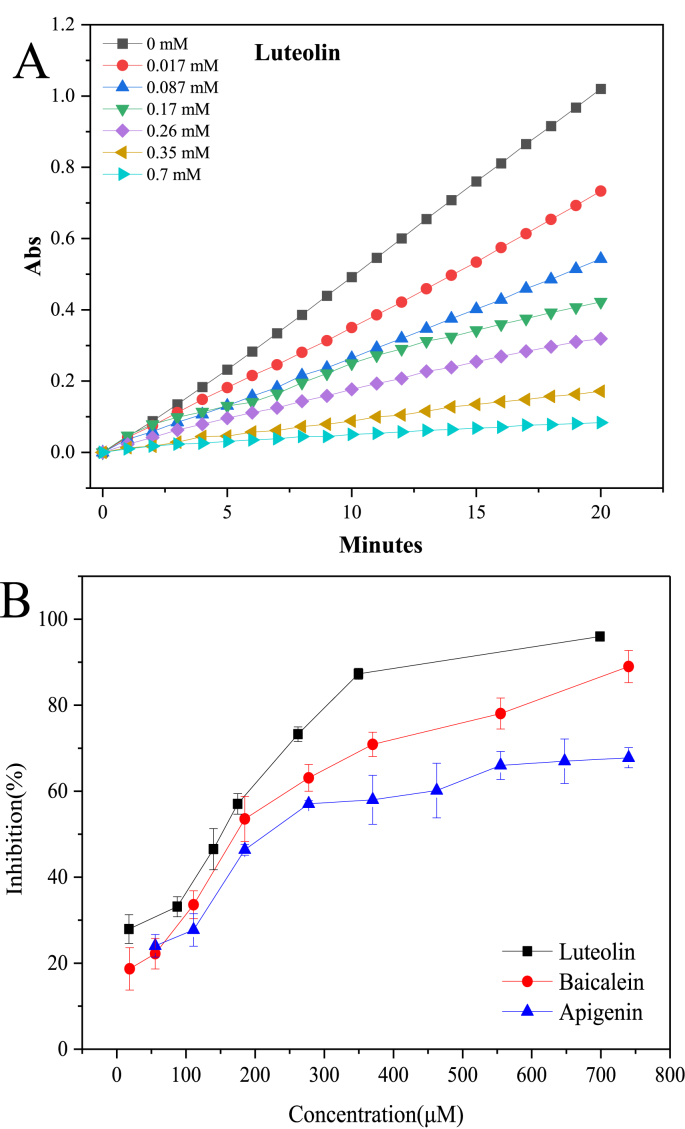
In the present study, pNPP was used as the substrate for pancreatic lipase. It can be hydrolyzed by pancreatic lipase to produce colored p-nitrophenol, which has maximum absorption at about 405 nm. Hence, the absorbance of the reaction mixture should linearly grow with the reaction time until the substrate is insufficient. The initial growth slope can represent the enzyme activity. When lipase inhibitor is present, the growth slope should decline. As shown in Fig. 2A, the slope declined gradually with the increase of luteolin concentration in the mixture. The concentration-inhibitory activity plots of all flavonoids were shown in Fig. 2B and Fig. S1. The IC50 value is defined as the concentration of inhibitor reaching 50% inhibition on enzyme activity. According to the definition, the IC50 values of the 26 flavonoids were simulated by SPSS software with the experimental data and were listed in Table 1. Orlistat was used as the positive control, which exhibited very strong lipase inhibitory activity with IC50 value of only 0.092 μM. The inhibitory activity of flavonoids was molecular structure dependent, but all far weaker than Orlistat. Among the 26 flavonoids, luteolin exhibited strongest inhibitory activity with IC50 of 99 ± 11 μM, followed by quercetin (IC50 = 128 ± 22 μM) and baicalein (IC50 = 156 ± 22 μM), while epigallocatechin and epicatechin were the weakest. Because of poor solubility combined with weak inhibitory activity, the IC50 values of some flavonoids were not obtained, e.g., chrysin, kaempferol and genistein. According to the results in Table 1, generally, flavones exhibited stronger inhibitory activity than other types of flavonoids. The IC50 values of most flavones were only about one tenth that of flavanones. Quercetin (or its glycoside) is one of the most abundant flavones in plants, and is almost found in all vegetables and fruits. For instance, consuming 100 g onion could obtain about 35–120 mg quercetin, and 200 g Curly kale equals about 60–120 mg (Manach et al., 2004). Luteolin and apigenin are also abundant in Parsley, Celery, etc (Manach et al., 2004). Because of the strong inhibitory activity of these flavones on lipase, the anti-obesity effects of such plant foods deserve further attention.
Fig. 2.
(A) The inhibitory effects of luteolin on pancreatic lipase; (B) The concentration-inhibitory activity plots of typical flavonoids.
Table 1.
The inhibitory activity and binding property of 26 tested flavonoids on pancreatic lipase.
| Class | Name | Substitution | IC50(μM) | lgKa | n |
|---|---|---|---|---|---|
| Control | Orlistat | 0.092 | |||
| Flavone |
Luteolin | 5,7,3′,4′-OH | 99 ± 11 | 6.808 | 1.435 |
| Quercetin | 3,5,7,3′,4′-OH | 128 ± 22 | 6.586 | 1.419 | |
| Baicalein | 5,6,7-OH | 156 ± 22 | 6.173 | 1.270 | |
| Isoorientin | 5,7,3′,4′-OH, 6-Glu | 201 ± 23 | 5.332 | 1.278 | |
| Apigenin | 5,7,4′-OH | 256 ± 54 | 5.413 | 1.156 | |
| Rutin | 3-rut,5,7,3′,4′-OH | 367 ± 182 | 5.269 | 1.115 | |
| Galangin | 3,5,7-OH | 574 ± 132 | 4.864 | 1.097 | |
| Kaempferol | 3,5,7,4′-OH | >983 | 5.391 | 1.163 | |
| Chrysin |
5,7-OH |
>593 |
4.789 |
0.964 |
|
| Flavanone |
Hesperetin | 5,7,3′-OH,4′-OMe | 3033 ± 829 | 4.383 | 0.998 |
| Taxifolin | 3,5,7,3′,4′-OH | 3868 ± 939 | 4.250 | 0.956 | |
| Naringenin | 5,7,4′-OH | 6032 ± 41 | 4.477 | 1.013 | |
| Dihydromyricetin | 3,5,7,3′,4′,5′-OH | 7175 ± 418 | 3.979 | 0.911 | |
| Astilbin |
5,7,3′4′-OH,3-O-α-L-Rha |
7316 ± 66 |
3.822 |
0.850 |
|
| Isoflavone |
Daidzein | 7,4′-OH | 852 ± 667 | 4.937 | 1.101 |
| Puerarin | 7,4′-OH, 8-β-D-Glu | 16840 ± 692 | 5.846 | 1.232 | |
| Formononetin | 7-OH, 4′-OCH3 | >1863 | 4.08 | 0.926 | |
| Ononin | 7-O-β-D-Glu,4′-OCH3 | >1161 | 4.964 | 1.086 | |
| Daidzin | 7-O-Glu, 4′-OH | >1152 | 3.850 | 0.877 | |
| Genistin | 5,4′-OH, 7-O-Glu | >925 | 4.966 | 1.086 | |
| Genistein |
5,7,4′-OH |
>2960 |
5.306 |
1.129 |
|
| Flavan-3-ol | EGCG | 5,7,3′4′,5′-OH, 3-O-gallate | 863 ± 107 | 4.444 | 0.984 |
| GCG | 5,7,3′4′,5′-OH, 3-O-gallate | 858 ± 17 | 4.143 | 0.937 | |
| Catechin | 3,5,7,3′4′-OH | >12919 | 2.131 | 0.691 | |
| Epicatechin | 3,5,7,3′4′-OH | >20670 | 2.501 | 0.734 | |
| Epigallocatechin | 3,5,7,3′,4′,5′-OH | >48977 | 2.235 | 0.670 |
3.2. Binding affinity between flavonoids and lipase determined by fluorescence titration
To exhibit lipase inhibitory activity, the flavonoids should bind on the protein first. Because of containing fluorescent groups such as tryptophan, tyrosine and phenylalanine, most proteins can emit endogenous fluorescence under exciting light. The binding of flavonoids on protein usually causes fluorescence quenching. Hence, the fluorescence titration method was used to determine the binding constant in the present study.
As shown in Fig. 3, under the exciting wavelength at 280 nm, lipase emits maximum fluorescence at about 350 nm. With the addition of flavonoids, its endogenous fluorescence was gradually quenched, which implied the interaction between flavonoids and the protein. Accordingly, the binding constant and number of binding sites can be calculated. The results were listed in Table 1. The values of lgKa of most flavonoids were in the range of 4–7, and the number of binding sites was all around one. However, epigallocatechin and epicatechin showed very weak binding affinity with lipase, which was in accordance with their inhibitory performance. Xiao et al. (2011b) investigated the structure-affinity relationship of dietary polyphenols (most are flavonoids) binding on bovine hemoglobin. Our results are similar to their finding that the saturation of C2-C3 bond in C ring is crucial to the binding affinity. Flavones with unsaturated C2=C3 bond exhibited stronger binding affinity with lipase than flavanones with saturated C2-C3 bond. Hydrophobic interaction is the main driving force of flavonoid-protein binding (Xiao et al., 2011b). Zhang et al. (2017) showed that the unsaturated C2=C3 bond significantly decreases the water solubility of flavonoids. Besides, the saturation of C2-C3 bond may change the connection conformation between C ring and B ring in the molecule. A saturated C2-C3 bond permits the twisting of the B-ring, while C2=C3 bond increases the π-conjugation and favors near-planarity of the two rings. Molecules with near-planar structure more easily enter the hydrophobic pockets in proteins (Xiao et al., 2011b).
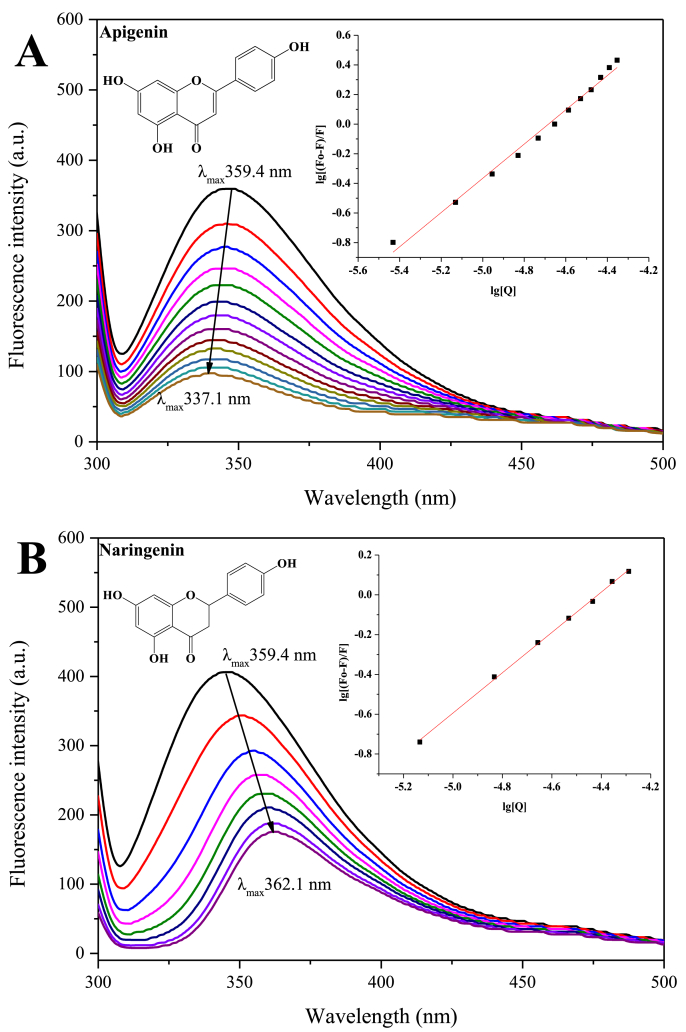
Fig. 3.
The fluorescence titration results of apigenin (A) and naringenin (B) on lipase. The concentration of apigenin from 1 to 13 were 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11 and 12 μg/mL, respectively; The concentration of naringenin from 1 to 8 were 0, 2, 4, 6, 8, 10, 12 and 14 μg/mL, respectively.
In the present study, an interesting phenomenon was found that the binding of flavones on lipase all caused the blue-shift of fluorescence emission, while flavanones all caused red-shift. The fluorescence titration results of apigenin and naringenin, which have identical molecular structure besides C2-C3 bond, were shown in Fig. 3. The fluorescence changes of other flavonoids on lipase were presented in Fig. S2 and Fig. S3. The results may imply that the binding location of flavones on lipase is different from flavanones. The binding location of flavones may be closer to the active pockets of lipase, which cause stronger inhibitory activity. In our previous study, the binding location of quercetin on lipase was simulated by molecular docking method. The results revealed that quercetin had a binding site near the active pocket of lipase. It can form Pi-Cation interaction with His264, which is one of the three key residues involved in the catalytic process (Zhou et al., 2021).
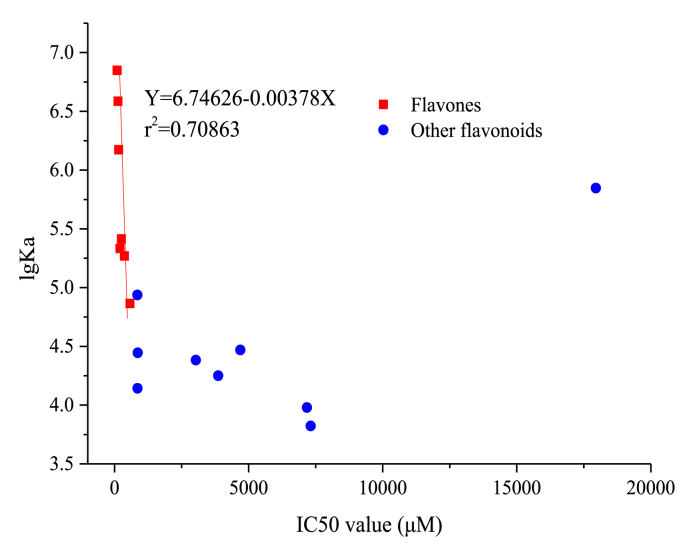
The relationship between the binding affinity and inhibitory activity of flavonoids on lipase was shown in Fig. 4. As shown, a linear relationship for flavones was found (r2 = 0.708), while other types of flavonoids didn't show a close relationship. The result supported our deduction that the binding location of flavones and flavanones on lipase may be different.
Fig. 4.
Correlation between inhibitory activity (IC50) and binding affinity (lgKa) of flavonoids on lipase.
3.3. Molecular docking analysis
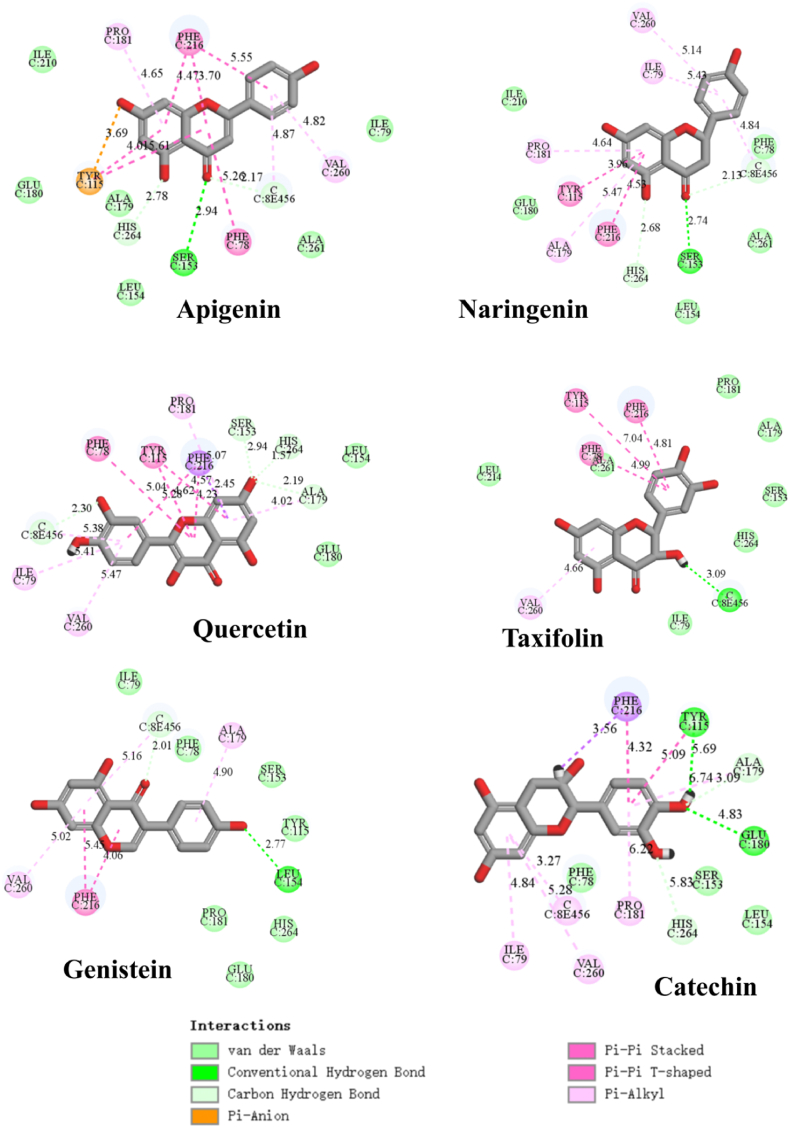
Computational molecular docking can predict the interactions and orientation of ligand with macromolecular receptor, which is a complement to the experimental data. The molecular docking results of six typical flavonoids among four types in Table 1 with lipase were shown in Fig. 5 and Fig. S4. As shown, flavonoids can interact with the amino acid residues in lipase through many kinds of secondary interactions, including hydrogen bond, van der Waals, Pi-Pi stacked, etc. It is clearly showed that the secondary bonds of flavones (quercetin and apigenin) with lipase simulated by the software were much more than other types of flavonoids, particularly at C ring. The C2=C3 bond in C ring of flavones increases the π-conjugation and favours the Pi-Pi interaction with enzyme. The twisting of the B-ring in flavanones (apigenin and naringenin) during docking in the enzyme was also found. The results revealed that the C2=C3 bond also contributes to maintaining the planarity of flavonoid structure. The position of B ring also affects the sencondary interaction. Genistein (isoflavone) with B ring at C3 position has less secondary bonds with lipase than apigenin with B ring at C2 position. Ser153, Asp 177 and His264 were the three key residues involved in the catalytic process of lipase (Du et al., 2018). The secondary interaction of flavonoids with these residues will undoubtedly reduce the catalytic activity of the enzyme. The molecular docking results confirmed the crucial role of C2=C3 bond in flavonoids in maintaining its secondary interactions with lipase, which was in accordance with the enzyme activity inhibitory test and fluorescence titration results.
Fig. 5.
The molecular interactions of typical flavonoids with lipase simulated by molecular docking.
3.4. Structure-activity relationship analysis
Hydrogenation of C2 and C3 on C Ring The hydrogenation of C2 and C3 on C ring of flavonoids is the difference between flavones and flavanones. According to our results, the saturation of C2-C3 bond is crucial to the inhibitory activity of flavonoids on lipase. All flavones exhibited much stronger inhibitory activity than flavanones. Taxifolin and quercetin are two flavonoids with identical substituted groups besides C2-C3 bond on C ring, as are apigenin and naringenin. Quercetin (IC50 = 128 ± 22 μM) and apigenin (IC50 = 256 ± 54 μM) exhibited much stronger inhibitory activity on lipase than taxifolin (IC50 = 3868 ± 939 μM) and naringenin (IC50 = 6032 ± 41 μM), respectively. According to the fluorescence titration study, the binding affinity of quercetin and apigenin with lipase was also much stronger than taxifolin and naringenin, respectively. The result may be explained by two reasons. First, the hydrogenation of C2 and C3 increases the solubility of flavonoids (Zhang et al., 2017), which decrease its binding affinity with liapse through weakening the hydrophobic interaction. Second, the hydrogenation of C2 and C3 permits more twisting of B ring when connected to C ring (Edenharder et al., 1993). It is deduced that the nonplanarity of molecular structure of flavonoid may weak its binding affinity on lipase. Molecular docking analysis confirmed that C2=C3 bond in C ring of flavones increases the π-conjugation and contributes to maintaining the planarity of flavonoid structure, which favour its Pi-Pi interaction with lipase.
Hydroxylation According to the IC50 values of flavonoids listed in Table 1, the hydroxylation had a significant effect on their inhibitory activity on lipase. Generally, the hydroxylation on ring A and B of flavones increased the activity. For instance, the IC50 of baicalein with three hydroxyl groups (5,6,7-OH) in A ring was much smaller than chrysin (5,7-OH); The number hydroxyl group of luteolin, apigenin and chrysin in B ring was positive correlated with their inhibitory activity. However, the effect of hydroxylation on C3 position in C ring is uncertain. For instance, Galangin with C3 hydroxy was stronger than chrysin, while kaempferol with C3 hydroxy was weaker than apigenin.
Glycosylation Most dietary flavonoids exist in the form of glycoside in plants (Manach et al., 2004). The glycosylation could improve its solubility. However, in vitro study revealed that glycosylation significantly weakened binding affinity of flavonoids on protein (Xiao et al., 2011a). Accordingly, their inhibitory activity on the enzyme (e.g. α-glucosidase) was also weakened (Xiao et al., 2013). For instance, Li et al. showed that glucosidylation of quercetin significantly weakened its inhibitory activity on α-glucosidase (Li et al., 2009). Similar to these studies, our results also confirmed that the glycosylation of flavonoids weakens their inhibitory activity on lipase, e.g. the IC50 values comparison between quercetin and rutin, taxifolin and astilbin, daidzein and daidzin, etc. However, according to the metabolism pathway of flavonoids in vivo, glycosylated flavonoids are usually hydrolyzed by lactase phlorizin hydrolase in the brush border of enterocyte in small intestine to form their aglycones in the lumen (Williamson et al., 2018). Digestive enzymes such as pancreatic lipase and α-amylase are extracellular enzymes. When the flavonoid aglycones are formed, they can immediately bind with the enzymes in the lumen. Hence, we believe that the glycosylation of flavonoids in plants may not seriously affect their digestive enzyme inhibitory activity in vivo.
Isoflavone Because of containing C2=C3 bond in C ring, some isoflavones (e.g. daidzein) also exhibited stronger binding affinity and inhibition on lipase than flavanones. However, its location of B ring at C3 position weakens its inhibitory activity. For example, the IC50 values of apigenin and genistein were 256 μM ± 54 and >2960 μM, respectively. The weaker binding affinity of genistein with lipase than apigenin was also found in molecular docking analysis (Fig. 5).
Ketone group (C = O) at C ring Catechins, including catechin, epicatechin, epigallocatechin, epicatechin gallate, EGCG are the main flavan-3-ols found in tea leaves. Compared with other flavonoids, flavan-3-ols lack the ketone group (C=O) at C ring, which causes the nonplanarity of their molecular conformation (Crozier et al., 2009). The disruption of molecular planarity of flavonoids could dramatically increase their solubility (Lewin et al., 2013). Hence, in the present study, epigallocatechin and epicatechin had weak binding affinity with lipase, which was similar to their binding property with other proteins, e.g. bovine hemoglobin (Xiao et al., 2011b), rat plasma proteins (Xiao et al., 2011a). Correspondingly, very weak lipase inhibitory activity was found. The gallicacylation of epigallocatechin to form EGCG increased its affinity with lipase and enhanced its inhibitory activity. Similar results were found for the inhibitory activities of catechins on α-glucosidase (Xiao et al., 2013).
4. Conclusion
Among the 26 dietary flavonoids tested in the present study, their inhibitory activity on lipase was structure dependent and the details were listed in Fig. 6 and Table S1. Flavones exhibited much stronger inhibitory activity than other types of flavonoids. Structure-activity relationship analysis showed that the features of C ring are very crucial. The hydrogenation of C2=C3 bond and the absence of C=O group in C ring both cause significant decrease of inhibitory activity. According to the fluorescence titration and molecular docking results, the structure of C ring may also determine the binding affinity and binding location of flavonoids on lipase. These features further determine their inhibitory activity. A positive linear correlation between the inhibitory activity and binding affinity of flavones on lipase was found. Besides, the hydroxylation on ring A and B of flavones increases the inhibitory activity, while glycosylation weakens the activity.
Fig. 6.
Potential sites where flavonoids affect pancreatic lipase inhibition. The upward arrow indicates an increase in inhibitory activity and a downward arrow indicates a decrease.
CRediT authorship contribution statement
Mang-Mang Li: Investigation, Methodology, Data curation, Formal analysis, Writing – original draft. Yi-Ting Chen: Investigation, Methodology, Data curation. Jin-Cang Ruan: Formal analysis, Validation. Wen-Jun Wang: Methodology, Supervision. Ji-Guang Chen: Data curation, Formal analysis. Qing-Feng Zhang: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant 32060541).
Handling Editor: Aiqian Ye
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crfs.2022.100424.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- Birari R.B., Bhutani K.K. Pancreatic lipase inhibitors from natural sources: unexplored potential. Drug Discov. Today. 2007;12:879–889. doi: 10.1016/j.drudis.2007.07.024. [DOI] [PubMed] [Google Scholar]
- Buchholz T., Melzig M.F. Polyphenolic compounds as pancreatic lipase inhibitors. Planta Med. 2015;81:771–783. doi: 10.1055/s-0035-1546173. [DOI] [PubMed] [Google Scholar]
- Crozier A., Jaganath I.B., Clifford M.N. Dietary phenolics: chemistry, bioavailability and effects on health. Nat. Prod. Rep. 2009;26:1001–1043. doi: 10.1039/B802662A. [DOI] [PubMed] [Google Scholar]
- Du X., Bai M., Huang Y., Jiang Z., Chen F., Ni H., Li Q. Inhibitory effect of astaxanthin on pancreatic lipase with inhibition kinetics integrating molecular docking simulation. J. Funct.Foods. 2018;48:551–557. doi: 10.1016/j.jff.2018.07.045. [DOI] [Google Scholar]
- Edenharder R.V., Von Petersdorff I., Rauscher R. Antimutagenic effects of flavoniods, chalcones and structurally related compounds on the activity of 2-amino-3-methylinidazo [4, 5-ƒ] quinoline (IQ) and other heterocyclic amine mutagens from cooked food. Mutat. Res.-Fund Mol. M. 1993;287:261–274. doi: 10.1016/0027-5107(93)90019-C. [DOI] [PubMed] [Google Scholar]
- Hua F., Zhou P., Wu H., Chu G., Xie Z., Bao G. Inhibition of α-glucosidase and α-amylase by flavonoid glycosides from Lu'an GuaPian tea: molecular docking and interaction mechanism. Food Funct. 2018;9:4173–4183. doi: 10.1039/C8FO00562A. [DOI] [PubMed] [Google Scholar]
- Kumar S., Alagawadi K.R. Anti-obesity effects of galangin, a pancreatic lipase inhibitor in cafeteria diet fed female rats. Pharm. Biol. 2013;51:607–613. doi: 10.3109/13880209.2012.757327. [DOI] [PubMed] [Google Scholar]
- Lewin G., Maciuk A., Moncomble A., Cornard J.P. Enhancement of the water solubility of flavone glycosides by disruption of molecular planarity of the aglycone moiety. J. Nat. Prod. 2013;76:8–12. doi: 10.1021/np300460a. [DOI] [PubMed] [Google Scholar]
- Li Y.Q., Zhou F.C., Gao F., Bian J.S., Shan F. Comparative evaluation of quercetin, isoquercetin and rutin as inhibitors of α-glucosidase. J. Agric. Food Chem. 2009;57:11463–11468. doi: 10.1021/jf903083h. [DOI] [PubMed] [Google Scholar]
- Liu T.T., Liu X.T., Chen Q.X., Shi Y. Lipase inhibitors for obesity: a review. Biomed. Pharmacother. 2020;128 doi: 10.1016/j.biopha.2020.110314. [DOI] [PubMed] [Google Scholar]
- Manach C., Scalbert A., Morand C., Rémésy C., Jiménez L. Polyphenols: food sources and bioavailability. Am. J. Clin. Nutr. 2004;79:727–747. doi: 10.1093/ajcn/79.5.727. [DOI] [PubMed] [Google Scholar]
- Nakai M., Fukui Y., Asami S., Toyoda-Ono Y., Iwashita T., Shibata H., Kiso Y. Inhibitory effects of oolong tea polyphenols on pancreatic lipase in vitro. J. Agric. Food Chem. 2005;53:4593–4598. doi: 10.1021/jf047814+. [DOI] [PubMed] [Google Scholar]
- Rahim A.T.M., Takahashi Y., Yamaki K. Mode of pancreatic lipase inhibition activity in vitro by some flavonoids and non-flavonoid polyphenols. Food Res. Int. 2015;75:289–294. doi: 10.1016/j.foodres.2015.05.017. [DOI] [PubMed] [Google Scholar]
- Su H., Ruan Y.T., Li Y., Chen J.G., Yin Z.P., Zhang Q.F. In vitro and in vivo inhibitory activity of taxifolin on three digestive enzymes. Int. J. Biol. Macromol. 2020;150:31–37. doi: 10.1016/j.ijbiomac.2020.02.027. [DOI] [PubMed] [Google Scholar]
- Sun C.C., Li Y., Yin Z.P., Zhang Q.F. Physicochemical properties of dihydromyricetin and the effects of ascorbic acid on its stability and bioavailability. J. Sci. Food Agric. 2021;101:3862–3869. doi: 10.1002/jsfa.11022. [DOI] [PubMed] [Google Scholar]
- Williamson G., Colin D.K., Crozier A. The bioavailability, transport, and bioactivity of dietary flavonoids: a review from a historical perspective. Compr. Rev. Food Sci. Food Saf. 2018;17:1054–1112. doi: 10.1111/1541-4337.12351. [DOI] [PubMed] [Google Scholar]
- Xiao J., Cao H., Chen T., Yang F., Liu C., Xu X. Molecular property–binding affinity relationship of flavonoids for common rat plasma proteins in vitro. Biochimie. 2011;93:134–140. doi: 10.1016/j.biochi.2010.08.013. [DOI] [PubMed] [Google Scholar]
- Xiao J.B., Huo J.L., Yang F., Chen X.Q. Noncovalent interaction of dietary polyphenols with bovine hemoglobin in vitro: molecular structure/property–affinity relationship aspects. J. Agric. Food Chem. 2011;59:8484–8490. doi: 10.1021/jf201536v. [DOI] [PubMed] [Google Scholar]
- Xiao J., Kai G., Yamamoto K., Chen X. Advance in dietary polyphenols as α-glucosidases inhibitors: a review on structure-activity relationship aspect. Crit. Rev. Food Sci. Nutr. 2013;53:818–836. doi: 10.1080/10408398.2011.561379. [DOI] [PubMed] [Google Scholar]
- Zhang H., Wang M., Chen L., Liu Y., Liu H., Huo H., Qi A. Structure-solubility relationships and thermodynamic aspects of solubility of some flavonoids in the solvents modeling biological media. J. Mol. Liq. 2017;225:439–445. doi: 10.1016/j.molliq.2016.11.036. [DOI] [Google Scholar]
- Zheng D., Zhang Q.F. Bioavailability enhancement of astilbin in rats through zein–caseinate nanoparticles. J. Agric. Food Chem. 2019;67:5746–5753. doi: 10.1021/acs.jafc.9b00018. [DOI] [PubMed] [Google Scholar]
- Zhi J., Mulligan T.E., Hauptman J.B. Long‐term systemic exposure of orlistat, a lipase inhibitor, and its metabolites in obese patients. J. Clin. Pharmacol. 1999;39:41–46. doi: 10.1177/00912709922007543. [DOI] [PubMed] [Google Scholar]
- Zhou J.F., Wang W.J., Yin Z.P., Zheng G.D., Chen J.G., Li J.E., Zhang Q.F. Quercetin is a promising pancreatic lipase inhibitor in reducing fat absorption in vivo. Food Biosci. 2021;43 doi: 10.1016/j.fbio.2021.101248. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.