Highlights
-
•
High expression of circ_0101692 is identified in ccRCC.
-
•
Knockdown of circ_0101692 suppresses ccRCC cell proliferation and migration, and induced cell apoptosis in vitro, and reduced tumor xenograft growth in vivo.
-
•
MiR-384 is a potential downstream target of circ_0101692.
-
•
Downregulation of miR-384 partially eliminates the suppressive effects of circ_0101692 knockdown on ccRCC cell malignant phenotypes.
-
•
FN1 was a target gene of miR-384.
-
•
MiR-384 represses ccRCC cell progression through targeting FN1.
Keywords: Clear cell renal cell carcinoma, Circ_0101692, miR-384, FN1, Proliferation
Abstract
Purpose
Circular RNA_0101692 (circ_0101692) is overexpressed in clear cell renal cell carcinoma (ccRCC) by microarray analyses. However, its function and action mechanism in ccRCC tumorigenesis is still elusive.
Methods
Western blotting and qRT-PCR were executed to assess the circ_0101692, miR-384 and FN1 expression in ccRCC cells and tissues. Target relationships among them were determined via dual luciferase reporter and/or RNA immunoprecipitation assays. Cell proliferation was evaluated by CCK-8 assay. Caspase-3 activity assay was utilized to analyze cell apoptosis. To find out whether ccRCC cells might migrate, a transwell assay was performed. To assess the effects of circ_0101692 on tumor development in vivo, a mouse xenograft model was used.
Results
High expression of circ_0101692 and FN1, and decreased miR-384 were determined in ccRCC. Cell growth, migration and viability were decreased whereas cell apoptosis was stimulated when circ_0101692 was knockdown. miR-384 inhibitor transfection attenuated the inhibiting impacts of circ_0101692 silencing on ccRCC cell progression. FN1 deletion further inverted the cancer-promoting effect of miR-384 downregulation on cell viability and migration. In addition, circ_0101692 could sponge miR-384 to relieve the inhibition of miR-384 on FN1 in ccRCC.
Conclusions
Circ_0101692 targeted miR-384/FN1 axis to facilitate cell proliferation, migration and repress apoptosis, thereby accelerating the development of ccRCC. This points out that circ_0101692/miR-384/FN1 axis might be a prospective target implemented for the future treatment of ccRCC.
Introduction
Renal cell carcinoma (RCC) is thought to be the most common type of urinary system cancer in the world [1] and the mortality is still increasing annually [2]. Histological features show that clear cell RCC (ccRCC) is the predominant type, making up around 80% of RCC [3]. Generally speaking, ccRCC patients at an early stage are curable but unfortunately, the 5-year survival rate is only 10% for the advanced patients due to the frequent relapse and high metastasis [4], [5], [6]. Therefore, exploring the early diagnostic marker and occurrence mechanisms of ccRCC progression are useful for the advance of ccRCC diagnosis and prognosis.
A unique kind of non-coding RNA known as circular RNAs (circRNAs) is found throughout in the cytoplasm [7,8]. Distinguished from linear RNAs, circRNAs have no 5ʹ caps and 3ʹ tails, and therefore exhibit a unique and stable ring structure [9,10]. Emerging evidences have uncovered the crucial role of circRNAs in cancer progression, including ccRCC. Certain highly-expressed circRNAs represents relatively poor prognosis and identified as a prognostic biomarker in ccRCC patients, and regulates malignant behaviors of ccRCC cells, such as hyperproliferation and rapid metastasis [11], [12], [13]. Additionally, circRNAs have been shown to serve as the microRNAs' (miRNAs') sponges in recent years, allowing for competitive regulation of miRNA production, therefore modulating the occurrence and development of ccRCC together [11,[13], [14], [15]], such as circ_001895-miR-296-5p-SOX12 [11], circ_101341-miR-411-EGLN3 [13], and circ_RPL23A-miR-1233-ACAT2 [15]. However, it is well known that competitive endogenous RNA (ceRNA) networks are complex systems, not just composed of a few circRNA-miRNA-mRNA. Therefore, we expect to continue to characterize the ceRNA mechanisms in ccRC and to determine their regulatory role in ccRC.
A tumor xenograft model and ccRCC were used to evaluate a circRNA expression and function. Furthermore, this research is unique in uncovered the action mechanism of circRNA-miRNA-mRNA axis in ccRCC progression at a cellular level. Some of these results may indicate promising treatment targets for ccRCC.
Methods
Clinical specimens
A total of 45 pairs of ccRCC patients were registered in our hospital from June 2019 to July 2020. Each subject underwent a histology investigation to determine their diagnosis; they were all untreated prior to arrival. ccRCC and the relative normal tissues (five centimeters distant from the tumor edge) were obtained by surgery. All the subjects signed the written informed consent, and this research has been approved by the ethics committee of our hospital (IRB number: WHSHIRB-K-2021079).
Cell culture
The American Type Culture Collection (Manassas, VA, USA) provided human embryonic kidney cells HEK293 and two kinds of ccRCC cell lines (786-O and 769-P). The other two types of ccRCC cell lines (KMRC-1, and KMRC-3) were obtained from Japanese Collection of Research Bioresources Cell Bank. All cells were grown at 37°C with 5% CO2 in DMEM (Invitrogen, Carlsbad, CA, USA) with 10% FBS (Invitrogen). Cells at passages 2-5 were used for the subsequent experiments.
Cell transfection
Short interfering RNA (SiRNA)-mediated circ_0101692 (si-circ), siRNA-FN1 (si-FN1) and their negative control si-NC, and miR-NC, miR-384 mimic (miR-384) as well as miR-384 inhibitor (inhibitor) and inhibitor-NC were from Sangon Biotech (Shanghai, China). Using Lipofectamine 3000 (Invitrogen), the aforementioned agents were transfected into KMRC-1 or KMRC-3 cells. Transfection efficiency was evaluated via quantitative real-time PCR (qRT-PCR). Two days later, the transfected cells were collected for functional experiments. The siRNA sequences of this study are shown in Supplementary Table 1.
qRT-PCR
From ccRCC tissues or cells, total RNA was extracted using an RNA extraction kit (Cwbio, Beijing, China). Afterwards, RNA was purified using phenol–chloroform extraction method. After reversely transcribed using Evo m-mlv reverse transcription reagent (Accyrate Biology, Changsha, China), qRT-PCR was conducted employing SYBR green kit (Accyrate Biology) under the Real-time PCR System (APPlied Biosystems, Foster City, CA, USA). The used primers were shown in Table 1. GAPDH (circ_0101692 and FN1) and U6 (miR-384) were used for normalization and the 2−ΔΔCt technique was utilized to analyze the target gene expression.
Table 1.
Real-time PCR primer synthesis list.
| Gene | Sequences | |
|---|---|---|
| circ_0101692 | Forward | 5, -CAGAAATTCCTGCAGACATCC-3 |
| Reverse | 5′-TGATGCAGCGACCATCAC-3′ | |
| miR-384 | Forward | 5, -TGTTAAATCAGGAATTTTAA-3 |
| Reverse | 5′-TGTTACAGGCATTATGAA-3′ | |
| FN1 | Forward | 5, -ACCCTGGGTATGACACTGGA-3, |
| Reverse | 5, -TGCCTCTGCTGGTCTTTCAG-3, | |
| U6 | Forward | 5, -CTCGCTTCGGCAGCACA-3, |
| Reverse | 5, -AACGCTTCACGAATTTGCGT-3, | |
| GAPDH | Forward | 5, -AGAAAAACCTGCCAAATATGATGAC-3, |
| Reverse | 5, -TGGGTGTCGCTGTTGAAGTC-3, | |
RNase R treatment
We incubated total RNA (4 µg) with 3U/µg of RNase R from Geneseed Biotech company at Guangzhou, China at 37°C for one hour. The control group received no RNase R treatment. circ_0101692 and its linear gene EGLN3 expression levels were calculated via qRT-PCR.
Assessment for subcellular location
RNA from cytoplasm or nucleus of KMRC-1/KMRC-3 cells was isolated using a Cytoplasmic/Nuclear Extraction Kit acquired from Thermo Fisher Scientific (Waltham, USA). A qRT-PCR was used to determine the expression level. For the nucleus, U6 was used, while for the cytoplasm, GAPDH was used.
Cell viability assay
KMRC-1/KMRC-3 cells were cultured for 0, 24, 48 and 72 h, respectively, before being plated onto 96-well plates (4 × 103 cells/well). Thereafter, CCK-8 (10 μL, Sangon Biotech) was supplemented to each well to incubate for another 2 h. Using a microplate reader obtained from DR-3518G, Hiwell Diatek, China, the optical density at 450 nm was calculated.
Apoptosis detection assay
The apoptosis of ccRCC cells was determined by caspase-3 activity assay kit (QCbio Science & Technologies) located at Shanghai, China. In brief, cells were digested via trypsin treatment, and centrifugation for 15 min at 600 g at 4°C is then performed. The cell lysate was further lysed for 15 min on ice before being centrifuged at 20,000 g for 5 min at 4°C. Thereupon, Ac-DEVD-pNA (2 mM) was added to incubate 2 h at 37°C. Caspase-3 activity at 405 nm was assessed via a microplate reader from Hiwell Diatek.
Transwell migration assay
In upper chamber of Transwell (pore size, 8 µm), 500 µl cell suspension in FBS-free DMEM (2 × 105) was added, while the lower chamber received 750 µl of DMEM comprising FBS (10%). The migratory cells were fixed with 4 percent methanol for 15 min and stained for 30 min with 0.2 percent crystal violet after being cultured for 24 h. A light microscope from Olympus located at Japan was applied to count the stained cells at a magnification of 200x.
DLR (Dual luciferase reporter) assay
circInteractome was used to estimate the target linkage among circ_0101692 and miR-384, while TargetScan was applied to determine the binding site among miR-384 and FN1. circ_0101692/FN1 sequence comprising miR-384 binding site were inserted into pGL3 vector (Promega) as circ_0101692-wild type (WT)/FN1-WT or circ_0101692-mutant type (MUT)/FN1-MUT, which were co-transfected into KMRC-1 or KMRC-3 cells with the help of Lipofectamine 3000 from Thermo Fisher Scientific for around 48 h at 37°C. A Dual-Glo Luciferase assay system from Promega, USA was used to measure the luciferase activity.
RNA immunoprecipitation (RIP) assay
The Magna RIP RNA-Binding Protein Immunoprecipitation Kit from Millipore, Bedford, USA, was used to carry out the RIP assay. After being lysed in RIP lysis solution, magnetic beads were used to attach KMRC-1 or KMRC-3 cells and treated with anti-Ago2 or anti-IgG antibodies (1:3000; Millipore). RNA enrichment was determined using qRT-PCR.
Western blotting analysis
A RIPA lysis buffer (BOSTER, Wuhan, China) was used to extract total proteins from KMRC-1 or KMRC-3 cells. The proteins were then transferred onto PVDF membranes after being electrophoretically segregated on a SDS polyacrylamide gel (10%). 5% nonfat milk was used to block the membranes for 1 h at room temperature, and incubated with corresponding primary antibodies FN1 (Abcam, Cambridge, UK; cat.no. ab2413; 1:1000) and GAPDH (Abcam; cat.no. 9485; 1:1000) at 4°C overnight, and further incubated with specific secondary antibody (Abcam; cat.no. ab6721; 1:5000) at 25°C for 30 min. Internal control was performed using GAPDH. The protein bands were visualized using ECL kit (Tanon, Shanghai, China) on a Tanon3500 gel imaging system (Tanon), and quantified using Alpha Innotech software.
Xenograft tumor model
Prior to testing, BALB/c nude mice acquired from Esebio located at Shanghai, China were given time to acclimate to the lab setting. The mice (n = 5) were divided ad libitum into the short hairpin RNA-circ_0101692 (sh-circ) and sh-NC groups a week later. Prior to transfection into KMRC-1 cells, the sh-NC or sh-circ was first incorporated into the lentiviral vector. Mice were injected subcutaneously with the transfected cells (1 × 105 cells/100 µL). Tumor width and length were measured each week, with the formula of (width × length)2 × 1/2 to estimate tumor volume. After 5th week, mice were euthanatized via pentobarbital sodium and the weight of tumor xenograft was evaluated. The hospital's ethical committee also allowed the use of animals in tests.
Statistical analysis
Data were presented as means ± SD. Based on SPSS 23.0 software (SPSS, Chicago, IL, USA), the differences between two groups were compared using Student's t-test and those among multiple groups were compared using one-way ANOVA followed by Tukey's post-hoc test or two-way ANOVA after that Sidak's multiple comparisons test. An analysis of Pearson associations was conducted to determine the significance of the correlation. If P < 0.05, a difference was considered significant.
Results
High expression of circ_0101692 is identified in ccRCC
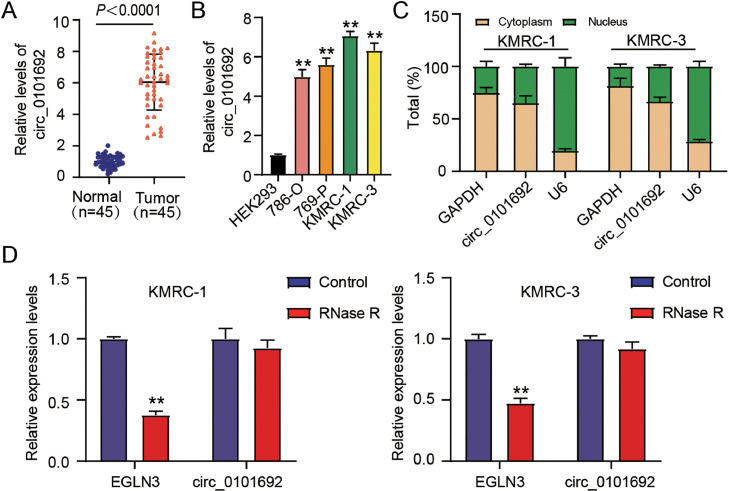
We first determined circ_0101692 expression in ccRCC cell lines as well as in tissues. As featured in Fig. 1A and 1B, amplified circ_0101692 expression was recognized in ccRCC cell lines (769-P, KMRC-1, 786-O, and KMRC-3) and tumor tissues compared to their corresponding controls, respectively. Subcellular localization analysis demonstrated that circ_0101692 is predominantly distributed in cytoplasm and the cytoplasm/nucleus distribution ratio is approximately 65%/35% (Fig. 1C). Circ_0101692 was resistant to RNase R, but RNase R treatment dramatically lowered the expression level of EGLN3 (Fig. 1D).
Fig. 1.
High expression of circ_0101692 is identified in ccRCC. (A) The expression of circ_0101692 in ccRCC tissues and normal tissues was detected by qRT-PCR. P < 0.0001 vs. Normal. (B) circ_0101692 expression in ccRCC cell lines (786-O, SK-RC-42, KMRC-1 and KMRC-3) and HEK293 cells was detected by qRT-PCR. **P < 0.01 vs. HEK293. (C) The expression of circ_0101692 in cytoplasm and nucleus of KMRC-1 and KMRC-3 cells via qRT-PCR. (D) The levels of circ-0101692 and corresponding linear transcript (EGLN3) in total cellular RNA incubated with RNase R. **P < 0.01 vs. Control.
Knockdown of circ_0101692 suppresses ccRCC cell proliferation and migration, and induced cell apoptosis in vitro, and reduced tumor xenograft growth in vivo
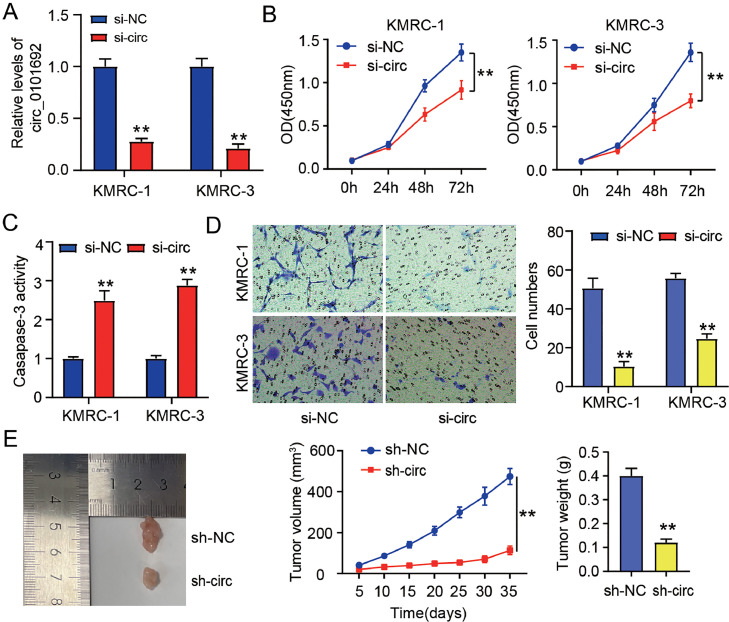
Then, the influences of circ_0101692 on ccRCC progression in vitro were explored. After transfecting si-circ into KMRC-1 and KMRC-3 cells, the result showed that circ_0101692 expression was decreased by about 75%, suggesting that si-circ was transfected successfully (Fig. 2A). As demonstrated in Fig. 2B, the vitality of ccRCC cells was remarkably repressed by circ_0101692 knockdown. Additionally, we discovered that si-circ transfected KMRC-1 or KMRC-3 cells had higher caspase-3 activity than si-NC-transfected cells (Fig. 2C), indicating that knocking-down circ_0101692 significantly stimulated cell apoptosis in ccRCC. Comparing the si-NC group with si-circ group, it was evident that the si-circ group's ability to inhibit KMRC-1 and KMRC-3 cell migration was stronger than si-NC group (Fig. 2D). As shown in Fig. 2E, injection of KMRC-1 cells that transfected with sh-circ significantly reduced tumor volume and weight. Based on the above results, silencing of circ_0101692 can inhibit ccRCC cell growth in vitro and tumor xenografts in vivo.
Fig. 2.
Knockdown of circ_0101692 suppresses ccRCC cell proliferation and migration, and induced cell apoptosis in vitro, and reduced tumor xenograft growth in vivo. (A) circ_0101692 expression in KMRC-1 and KMRC-3 cells after transfection with si-circ_0101692 (si-circ) or si-NC was determined using qRT-PCR. **P < 0.01 vs. si-NC. (B) The viability of KMRC-1 and KMRC-3 cells transfected with si-circ/si-NC was detected by CCK-8 assay. **P < 0.01 vs. si-NC. (C) Apoptosis in KMRC-1 and KMRC-3 cells transfected with si-circ/si-NC was determined by caspase-3 activity assay. **P < 0.01 vs. si-NC. (D) The numbers of migratory in transfected KMRC-1 and KMRC-3 cells with si-circ/si-NC were assessed by Transwell assay. **P < 0.01 vs. the si-NC. (E) The representative solid image, tumor volume (5, 10, 15, 20, 25, 30 and 35 days), and tumor weight (at the end of 35 days) in mouse xenograft model after injection of transfected KMRC-1 cells that was stably with sh-circ/NC. **P < 0.01 vs. sh-NC.
MiR-384 is a possible downstream circ_0101692 target
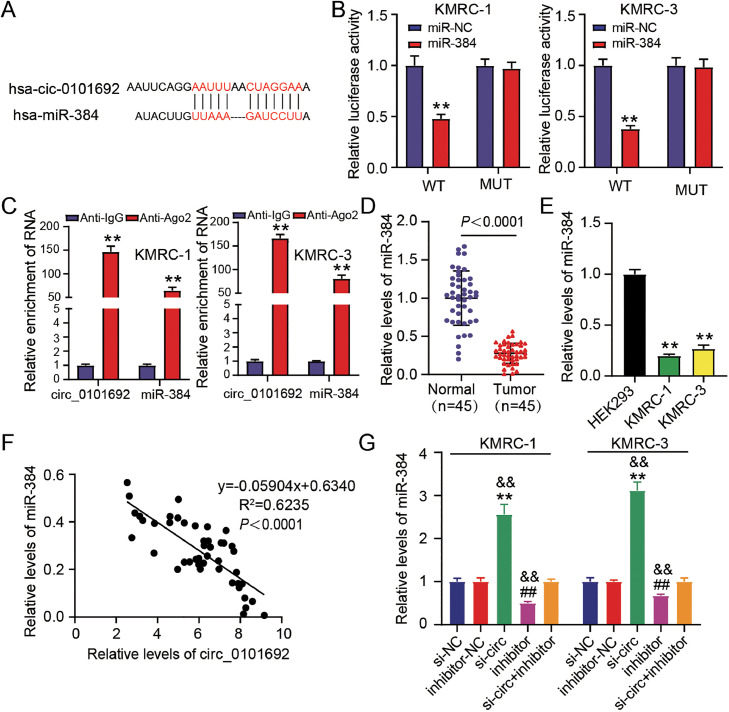
Based on the predicted results of circinteractome software (https://circinteractome.nia.nih.gov/index.html), there is an underlying binding site among circ_0101692 and miR-384 (Fig. 3A). When comparing with the circ 0101692 WT/miR-NC group, the DLR assay revealed that the luciferase activity in circ_0101692 WT/miR-384 group was significantly reduced, but there appeared to be no changes between the circ_0101692 MUT/miR-384 and circ_0101692 MUT/miR-NC groups (Fig. 3B). Meanwhile, RIP assay was performed to further validate this target relationship, indicating that both circ_0101692 and miR-384 were enriched in Ago2-containing beads by contrast to the IgG immunoprecipitate controls (Fig. 3C). Then, we noticed that the expression of miR-384 in ccRCC tissues and cell lines was lower than it was in the matching controls (Fig. 3D and 3E). Interestingly, circ_0101692 expression was correlated negatively with miR-384 expression in ccRCCs (Fig. 3F). In addition, in both KMRC-1 and KMRC-3 cells, we further revealed that miR-384 expression level was significantly suppressed by transfection of inhibitor, whereas miR-384 expression was elevated after si-circ transfection. Meanwhile, inhibitor transfection attenuated the promoting impacts of circ_0101692 silencing on miR-384 expression (Fig. 3G). As a result, we assumed that miR-384 and circ 0101692 were targets for each other. Furthermore, circ_0101692 lowered miR-384 levels in ccRCCs.
Fig. 3.
MiR-384 is a potential downstream target of circ_0101692. (A) The predicted binding site of circ_0101692 and miR-384 via circInteractome (https://circinteractome.nia.nih.gov/index.html). (B) The luciferase activity in co-transfected KMRC-1 and KMRC-3 cells with circ_0101692 WT (WT) or circ_0101692 MUT (MUT) and miR-384 mimic (miR-384) or miR-NC was ascertained by dual luciferase reporter assay. **P < 0.01 vs. miR-NC. (C) RIP assay was performed to validate the association among circ_0101692 and miR-384. **P < 0.01 vs. Anti-IgG. (D) The expression of miR-384 in ccRCC tissues and normal tissues was detected by qRT-PCR. P < 0.0001 vs. Normal. (E) The expression of miR-384 in ccRCC cell lines (KMRC-1 and KMRC-3) and HEK293 cells was ascertained with the help of qRT-PCR. **P < 0.01 vs. HEK293. (F) Correlation between the expression of circ_0101692 and miR-384 in ccRCC tissues was assessed by Pearson correlation analysis. (G) The expression of miR-384 in KMRC-1 and KMRC-3 cells after transfection si-circ, si-NC, miR-384 inhibitor (inhibitor), inhibitor-NC, or si-circ + inhibitor was detected by qRT-PCR. **P < 0.01 vs. the si-NC group; ##P < 0.01 vs. inhibitor-NC; &&P < 0.01 vs. si-circ + inhibitor.
Downregulation of miR-384 partially eliminates the suppressive effects of circ_0101692 knockdown on ccRCC cell malignant phenotypes
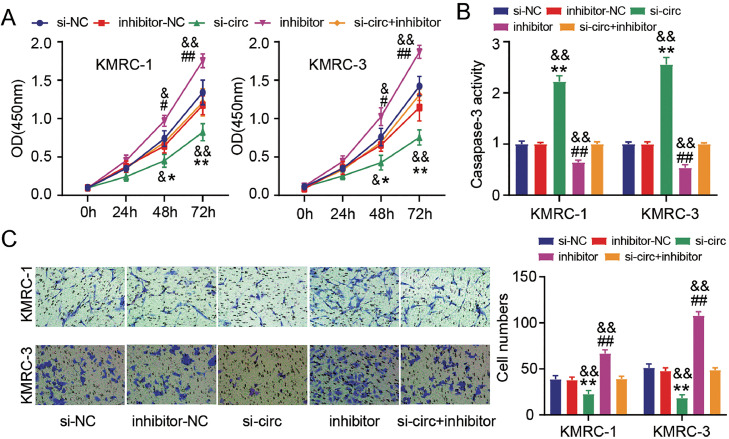
Then, the interaction between circ_0101692 with miR-384 was further confirmed through functional experiments. Inhibition of miR-384 could dramatically increase the proliferative potentials (Fig. 4A), decline caspase-3 activity (Fig. 4B), and potentiate the migrated capacities of KMRC-1 and KMRC-3 cells (Fig. 4C). As shown in Fig. 4A–C, we further demonstrated that transfection of inhibitor significantly inverted the inhibiting effects of si-circ transfection on cell viability and migration, and the promoting effect on caspase-3 activity. These results provided additional evidence that miR-384 is circ_0101692′s direct target and is associated in the growth of ccRCC.
Fig. 4.
Downregulation of miR-384 partially eliminates the suppressive effects of circ_0101692 knockdown on ccRCC cell malignant phenotypes. (A) The viability of KMRC-1 and KMRC-3 cells transfected with si-circ, si-NC, inhibitor, inhibitor-NC, or si-circ + inhibitor was measured by CCK-8 assay. (B) Apoptosis in these above transfected ccRCC cells was measured by the caspase-3 activity assay. (C) The numbers of migratory in these above transfected ccRCC cells were assessed by Transwell assay. *P < 0.05, **P < 0.01 vs. si-NC; #P < 0.05, ##P < 0.01 vs. inhibitor-NC; &P < 0.05, &&P < 0.01 vs. si-circ + inhibitor.
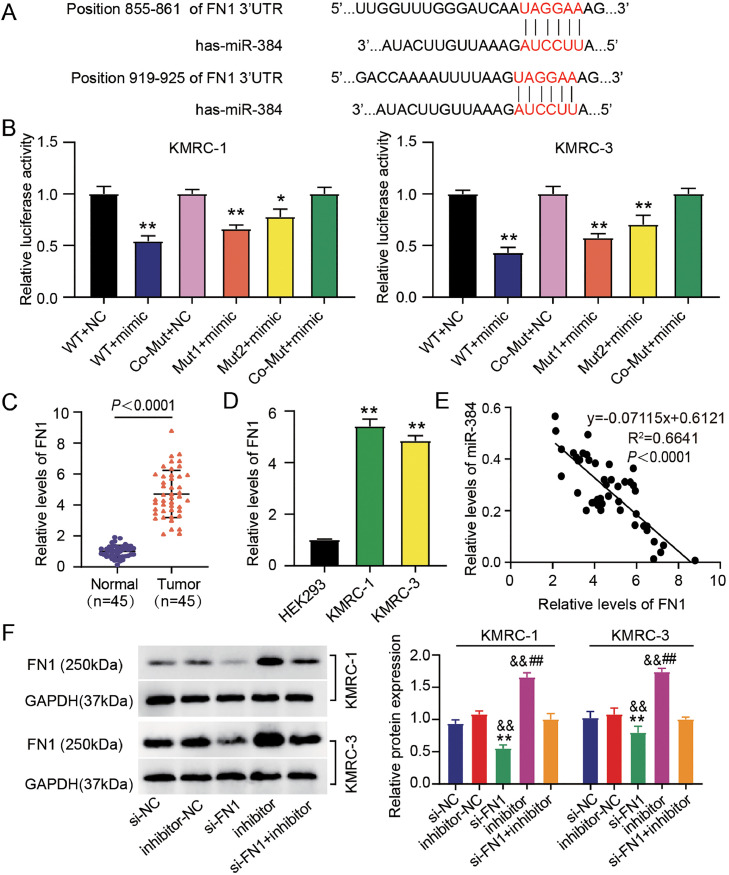
FN1 was a target gene of miR-384
The underlying binding site of miR-384 with FN1 was predicted by the TargetScan software (https://www.targetscan.org/vert_80/) (Fig. 5A), and further confirmed by DLR assay. As illustrated in Fig. 5B, there seemed no significant impact of co-MUT on luciferase activity. However, one site mutation (MUT 1 or MUT 2) reduced approximately 30-40% luciferase activity, whereas vector including two binding sites (WT) decreased luciferase activity approximately 50-60%. Besides, increased expression of FN1 was identified in ccRCC tissues and cell lines (Fig. 5C and 5D). The expression of miR-384 was negatively associated with FN1 in ccRCC tissues (Fig. 5E). After transfection, the protein level of FN1 was then measured. As presented in Fig. 5F, western blotting analysis exhibited that FN1 protein level was decreased by si-FN1 transfection, while was elevated after transfection of inhibitor. More intriguingly, miR-384 downregulation stimulated FN1 protein production when FN1 was lost, although only to a limited extent. FN1 is a miR-384 target gene in ccRCC when taken collectively.
Fig. 5.
FN1 was a target gene of miR-384. (A) TargetScan (https://www.targetscan.org/vert 80/) was used to estimate the binding site among miR-384 and FN1. (B) The target association among miR-384 and FN1 was verified by dual luciferase reporter assay. *P < 0.05, **P < 0.01 vs. WT +NC. (C) FN1 level in ccRCC and normal tissues was measured by qRT-PCR. P < 0.0001 vs. Normal. (D) FN1 level in ccRCC cell lines (KMRC-1 and KMRC-3) and HEK293 cells was detected by qRT-PCR. **P < 0.01 vs. HEK293. (E) Correlation between the expression of FN1 and miR-384 in ccRCC tissues was assessed by Pearson correlation analysis. (F) The protein levels of FN1 in KMRC-1 and KMRC-3 cells after transfection of si-FN1, si-NC, inhibitor, inhibitor-NC, or si-FN1 + inhibitor were measured by western blotting. ##P < 0.01 vs. inhibitor-NC; **P < 0.01 vs. the si-NC group; &&P < 0.01 vs. si-FN1 + inhibitor.
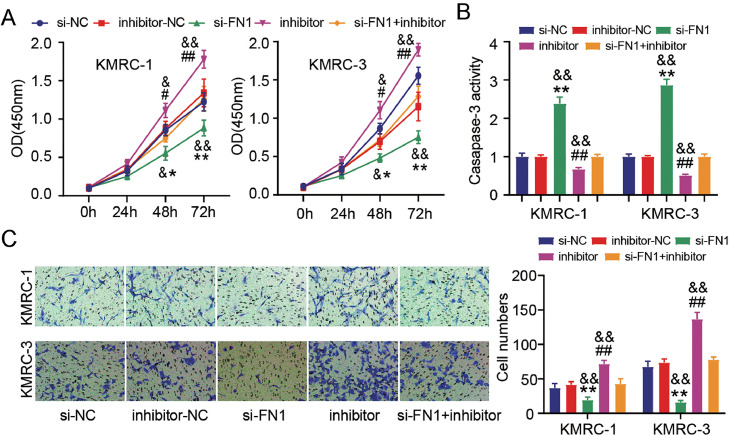
MiR-384 represses ccRCC cell progression through targeting FN1
Finally, the action mechanism of miR-384/FN1 axis in the progression of ccRCC in vitro was also evaluated. We found that FN1 deficiency repressed cell proliferation (Fig. 6A) and accelerated apoptosis (Fig. 6B). Not only that, it also distinctly decreased the migrated abilities of KMRC-1 and KMRC-3 cells (Fig. 6C). As mentioned above, inhibition of miR-384 had obviously enhanced impacts on cell viability and migration, and suppressive effect on caspase-3 activity. However, FN1 deletion partially reversed the effects of miR-384 suppression on ccRCC cells as shown above (Fig. 6A–C). We speculated miR-384 may interact with FN1 to modulate ccRCC tumorigenesis.
Fig. 6.
MiR-384 represses ccRCC cell progression through targeting FN1. (A) The viability of KMRC-1 and KMRC-3 cells transfected with si-FN1, si-NC, inhibitor, inhibitor-NC, or si-FN1 + inhibitor was measured by CCK-8 assay. (B) Apoptosis in these above transfected ccRCC cells 2was determined by the caspase-3 activity assay. (C) The numbers of migratory in these above transfected ccRCC cells were assessed by Transwell assay. *P < 0.05, **P < 0.01 vs. si-NC group; #P < 0.05, ##P < 0.01 vs. inhibitor-NC; &P < 0.05, &&P < 0.01 vs. si-FN1 + inhibitor.
Discussion
As a high metastasis malignant urinary system tumor, ccRCC generally originates from the dysfunction of proximal renal tubule epithelial cells [3]. It is reported that there are approximately 60,000 people newly diagnosed as ccRCC in USA each year, while 20% of them is died [16], suggesting a high mortality of ccRCC. Therefore, exploring underlying molecular targets for ccRCC therapy is a problem to be solved urgently. Our study revealed that silencing of circ_0101692 can repress cell proliferation, migration and induce apoptosis in ccRCC through miR-384/FN1 pathway.
Past researches have revealed that circ_0101692 is highly expressed in pancreatic cancer and breast cancer [17,18]. In this investigation, we also discovered that circ_0101692 was expressed more often in ccRCC tissues and cell lines. Similarly, Franz et al. performed microarray analysis in 7 ccRCC samples, and demonstrated that circ_0101692 expression is upregulated [19]. Therefore, we speculated that circ_0101692 may be related to ccRCC oncogenicity. Interestingly, we demonstrated that knockdown of circ_0101692 exerted the anti-cancer effect in ccRCC through inhibiting cell migration, cell proliferation and promoting apoptosis in vitro. We speculated that circ_0101692 deletion may function as tumor inhibitor in ccRCC. Our in vivo research showing that circ_0101692 knockdown also inhibited tumor xenograft development further supported our hypothesis. Collectively, these findings suggested that circ_0101692 silencing may repress the development of ccRCC via controlling cancer cell proliferation and metastasis.
A microRNA (miRNA) is a 16–22 nucleotide RNA that modulates gene expression [20]. With the in-depth researches of miRNAs in recent years, their regulatory functions in ccRCC tumorigenesis have been greatly demonstrated [21], [22], [23], [24]. The anti-tumor function of miR-384 in different kinds of human malignancies has been revealed by several research, that is in colorectal cancer [25], gastric cancer [26], and non-small cell lung cancer [27]. In the tumorigenesis of these cancers, miR-384 is found to be downregulated to affect cancer progression. In line with theses previous data, we found that ccRCC cell lines and tissues also had a low expression of miR-384. Meanwhile, a recent study conducted by Guo et al. has indicated that miR-384 expression is reduced in ccRCC tissues and cells [28]. The results suggested that high miR-384 expression may represent a good prognosis for ccRCC patients. Additionally, some circRNAs generally serve as miR-384′s sponges, playing a significant role in the emergence of cancers, featuring circ_0020123/circ_PRKCA/circ_0000376-miR-384 in non-small cell lung cancer [29], [30], [31], circ_0000670-miR-384 in gastric cancer [32], circ_0092276-miR-384 in breast cancer [33]. We speculated that there may be also some interactions of miR-384 with circ_0101692 in ccRCC. In line with expectations, our study found that circ_0101692 directly targets miR-384. In addition, we showed that transfection of the miR-384 inhibitor partially overturned the inhibitory impacts of si-circ_0101692 transfection on cell survival and migration as well as the enhancing impact on apoptosis. In general, we believed that silencing of circ_0101692 suppressive the progression of ccRCC via sponging miR-384.
FN1, belongings to the FN family, is generally overexpressed and serves as an oncogene in tumorigenesis [34], such as in cervical cancer [35], diffuse large B-cell lymphoma [36], and medullary thyroid cancer [37]. Increased expression of FN1 was seen in ccRCC tissues and cell lines in the current investigation, indicating that FN1 may also have an oncogenic role in the formation of ccRCC. Recently, Bai et al. revealed that circRNA/miRNA/mRNA network may be an effective therapy option for ccRCC patients [38]. Given that the regulating effects of circ_0101692/miR-384 axis in ccRCC progression, we further hypothesized a circ_0101692/miR-384/mRNA network in ccRCC. In the current investigation, we discovered that miR-384 directly targeted FN1 and speculated FN1 may involve in ccRCC progression regulated by miR-384. As anticipated, FN1 silencing reversed the miR-384 downregulation's boosting effects on cell survival and migration as well as its inhibitory impact on apoptosis. Taken together, we concluded that silencing of circ_0101692 interacted with miR-384/FN1 axis to affect ccRCC progression.
The research does have certain restrictions. Firstly, the underlying regulatory functions of circ_0101692/miR-384/FN1 axis were not validated in vivo. We attempt to elucidate this issue in future investigation. In addition, the correlation of circ_0101692/miR-384/FN1 axis with clinical practice was not been explored. In the future, we will collect more clinical samples to explore this. In addition, FN1 regulation of carcinogenesis is a complex process involving changes in multiple signaling pathways. For example, FN1 promotes proliferation and metastasis of thyroid cancer cells by activating the NF-κB pathway [39]. FN1 knockdown inhibited the proliferation, migration and invasion of breast cancer cells by blocking the activation of PI3K/AKT [40]. However, the downstream regulatory mechanism of FN1 in this study is still unclear, so we will continue to study the downstream pathway of circ_0101692/miR-384/FN1 in the future.
In a word, the current study uncovers that circ_0101692 aggravates ccRCC through regulating miR-384/FN1 axis. These discoveries shed light on how circ_0101692 affects the growth of ccRCC and may be a useful molecular target for ccRCC treatment.
Declarations
Funding: This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflicts of interest: The authors affirm that they do not have any competing interests.
Ethics approval
The Ethics Committee of The Sixth Hospital of Wuhan, Affiliated Hospital of Jianghan University, gave its approval to the current study (Wuhan, China). The Declaration of Helsinki's ethical guidelines are strictly followed when processing clinical tissue samples. Each patient completed an informed consent form in writing.
This animal experiment was conducted in accordance with the ARRIVE guidelines and was authorized by the Ethics Committee of The Sixth Hospital of Wuhan, Affiliated Hospital of Jianghan University's Ethics Committee.
Consent to participate
All patients signed written informed consent.
Consent for publication
Participants gave their permission for their names to be published.
Authors' contributions
The tests and data analysis were carried out by HZ. The study was created and planned by MM. The data was acquired by MM. The data were analyzed and interpreted by HZ. The article was reviewed and approved by all authors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
None.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101612.
Appendix. Supplementary materials
Availability of data and material
All data generated or analyzed during this study are included in this article.
References
- 1.Capitanio U., Bensalah K., Bex A., Boorjian S.A., Bray F., Coleman J., et al. Epidemiology of renal cell carcinoma. Eur. Urol. 2019;75(1):74–84. doi: 10.1016/j.eururo.2018.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Zhai W., Sun Y., Jiang M., Wang M., Gasiewicz T.A., Zheng J., et al. Differential regulation of LncRNA-SARCC suppresses VHL-mutant RCC cell proliferation yet promotes VHL-normal RCC cell proliferation via modulating androgen receptor/HIF-2α/C-MYC axis under hypoxia. Oncogene. 2017;36(31) doi: 10.1038/onc.2016.514. [DOI] [PubMed] [Google Scholar]
- 4.Motzer R.J., Molina A.M. Targeting renal cell carcinoma. J. Clin. Oncol. 2009;27(20):3274–3276. doi: 10.1200/JCO.2009.21.8461. [DOI] [PubMed] [Google Scholar]
- 5.Dutcher J.P. Recent developments in the treatment of renal cell carcinoma. Ther Adv Urol. 2013;5(6):338–353. doi: 10.1177/1756287213505672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eggener S.E., Yossepowitch O., Pettus J.A., Snyder M.E., Motzer R.J., Russo P. Renal cell carcinoma recurrence after nephrectomy for localized disease: predicting survival from time of recurrence. J. Clin. Oncol. 2006;24(19):3101–3106. doi: 10.1200/JCO.2005.04.8280. [DOI] [PubMed] [Google Scholar]
- 7.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Kulcheski F.R., Christoff A.P., Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J. Biotechnol. 2016;238:42–51. doi: 10.1016/j.jbiotec.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 9.Vicens Q., Westhof E. Biogenesis of circular RNAs. Cell. 2014;159(1):13–14. doi: 10.1016/j.cell.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 11.Chen Z., Xiao K., Chen S., Huang Z., Ye Y., Chen T. Circular RNA hsa_circ_001895 serves as a sponge of microRNA-296-5p to promote clear cell renal cell carcinoma progression by regulating SOX12. Cancer Sci. 2020;111(2):713–726. doi: 10.1111/cas.14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mo J., Zhao Y., Ao Z., Chen L., Lin S., Zeng W., et al. Circ-APBB1IP as a prognostic biomarker promotes clear cell renal cell carcinoma progression through the ERK1/2 Signaling pathway. Int. J. Med. Sci. 2020;17(9):1177–1186. doi: 10.7150/ijms.44550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yue Y., Cui J., Zhao Y., Liu S., Niu W. Circ_101341 deteriorates the progression of clear cell renal cell carcinoma through the miR- 411/EGLN3 Axis. Cancer Manag. Res. 2020;12:13513–13525. doi: 10.2147/CMAR.S272287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holdt L.M., Stahringer A., Sass K., Pichler G., Kulak N.A., Wilfert W., et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016;7 doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng L., Cao H., Xu J., Xu M., He W., Zhang W., et al. Circ_RPL23A acts as a miR-1233 sponge to suppress the progression of clear cell renal cell carcinoma by promoting ACAT2. J. Bioenerg. Biomembr. 2021;53(4):415–428. doi: 10.1007/s10863-021-09901-8. [DOI] [PubMed] [Google Scholar]
- 16.Siegel R., Ma J., Zou Z., Jemal A. Cancer statistics, 2014. CA Cancer J. Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 17.Zhao B., Li Z., Qin C., Li T., Wang Y., Cao H., et al. Mobius strip in pancreatic cancer: biogenesis, function and clinical significance of circular RNAs. Cell. Mol. Life Sci. CMLS. 2021;78(17-18):6201–6213. doi: 10.1007/s00018-021-03908-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.L. Wang, X. Li, D. Wang, H. Feng, F. Ma, J. Cao, et al. Identification of circular RNAs hsa-circ-000696 as a novel biomarker for breast cancer. 2019.
- 19.Franz A., Ralla B., Weickmann S., Jung M., Rochow H., Stephan C., et al. Circular RNAs in clear cell renal cell carcinoma: their microarray-based identification, analytical validation, and potential use in a Clinico-Genomic model to improve prognostic accuracy. Cancers. 2019;11(10) doi: 10.3390/cancers11101473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H., Lei C., He Q., Pan Z., Xiao D., Tao Y. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol. Cancer. 2018;17(1) doi: 10.1186/s12943-018-0765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren Y., Zhang L., Zhang W., Gao Y. MiR-30a suppresses clear cell renal cell carcinoma proliferation and metastasis by targeting LRP6. Hum. Cell. 2021;34(2):598–606. doi: 10.1007/s13577-020-00472-1. [DOI] [PubMed] [Google Scholar]
- 22.Wang X.G., Zhu Y.W., Wang T., Chen B., Xing J.C., Xiao W. MiR-483-5p downregulation contributed to cell proliferation, metastasis, and inflammation of clear cell renal cell carcinoma. Kaohsiung J. Med. Sci. 2021;37(3):192–199. doi: 10.1002/kjm2.12320. [DOI] [PubMed] [Google Scholar]
- 23.Deng J.H., Zheng G.Y., Li H.Z., Ji ZG. MiR-212-5p inhibits the malignant behavior of clear cell renal cell carcinoma cells by targeting TBX15. Eur. Rev. Med. Pharmacol. Sci. 2019;23(24):10699–10707. doi: 10.26355/eurrev_201912_19770. [DOI] [PubMed] [Google Scholar]
- 24.Wang J., Wang C., Li Q., Guo C., Sun W., Zhao D., et al. miR-429-CRKL axis regulates clear cell renal cell carcinoma malignant progression through SOS1/MEK/ERK/MMP2/MMP9 pathway. Biomed. Pharmacother. 2020;127 doi: 10.1016/j.biopha.2020.110215. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y.X., Zhu H.F., Zhang Z.Y., Ren F., Hu Y.H. MiR-384 inhibits the proliferation of colorectal cancer by targeting AKT3. Cancer Cell Int. 2018;18 doi: 10.1186/s12935-018-0628-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang F. miR-384 targets metadherin gene to suppress growth, migration, and invasion of gastric cancer cells. J. Int. Med. Res. 2019;47(2):926–935. doi: 10.1177/0300060518817171. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Guo Q., Zheng M., Xu Y., Wang N., Zhao W. MiR-384 induces apoptosis and autophagy of non-small cell lung cancer cells through the negative regulation of Collagen α-1(X) chain gene. Biosci. Rep. 2019;39(2) doi: 10.1042/BSR20181523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo L., Wang D., Zhang Z. MiR-384 represses tumorigenesis by regulating CDK6 and predicts prognosis of clear cell renal cell carcinoma. J. BUON. 2018;23(3):787–794. official journal of the Balkan Union of Oncology. [PubMed] [Google Scholar]
- 29.Ma Q., Huai B., Liu Y., Jia Z., Zhao Q. Circular RNA circ_0020123 promotes non-small cell lung cancer progression through miR-384/TRIM44 Axis. Cancer Manag. Res. 2021;13:75–87. doi: 10.2147/CMAR.S278913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu X., Zhang X., Zhang Y., Wang Z. Curcumin suppresses the malignancy of non-small cell lung cancer by modulating the circ-PRKCA/miR-384/ITGB1 pathway. Biomed. Pharmacother. 2021;138 doi: 10.1016/j.biopha.2021.111439. [DOI] [PubMed] [Google Scholar]
- 31.Sun H., Chen Y., Fang Y.Y., Cui T.Y., Qiao X., Jiang C.Y., et al. Circ_0000376 enhances the proliferation, metastasis, and chemoresistance of NSCLC cells via repressing miR-384. Cancer Biomark. Sect. A Dis. Markers. 2020;29(4):463–473. doi: 10.3233/CBM-190912. [DOI] [PubMed] [Google Scholar]
- 32.Liu P., Cai S., Li N. Circular RNA-hsa-circ-0000670 promotes gastric cancer progression through the microRNA-384/SIX4 axis. Exp. Cell. Res. 2020;394(2) doi: 10.1016/j.yexcr.2020.112141. [DOI] [PubMed] [Google Scholar]
- 33.Wang Q., Liang D., Shen P., Yu Y., Yan Y., You W. Hsa_circ_0092276 promotes doxorubicin resistance in breast cancer cells by regulating autophagy via miR-348/ATG7 axis. Transl. Oncol. 2021;14(8) doi: 10.1016/j.tranon.2021.101045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stehling-Sun S., Dade J., Nutt S.L., DeKoter R.P., Camargo FD. Regulation of lymphoid versus myeloid fate 'choice' by the transcription factor Mef2c. Nat. Immunol. 2009;10(3):289–296. doi: 10.1038/ni.1694. [DOI] [PubMed] [Google Scholar]
- 35.Wang S., Gao B., Yang H., Liu X., Wu X., Wang W. MicroRNA-432 is downregulated in cervical cancer and directly targets FN1 to inhibit cell proliferation and invasion. Oncol. Lett. 2019;18(2):1475–1482. doi: 10.3892/ol.2019.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song Y., Gao F., Peng Y., Yang X. Long non-coding RNA DBH-AS1 promotes cancer progression in diffuse large B-cell lymphoma by targeting FN1 via RNA-binding protein BUD13. Cell Biol. Int. 2020;44(6):1331–1340. doi: 10.1002/cbin.11327. [DOI] [PubMed] [Google Scholar]
- 37.Zhan S., Li J., Wang T., Ge W. Quantitative proteomics analysis of sporadic medullary thyroid cancer reveals FN1 as a potential novel candidate prognostic biomarker. Oncologist. 2018;23(12):1415–1425. doi: 10.1634/theoncologist.2017-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bai S., Wu Y., Yan Y., Shao S., Zhang J., Liu J., et al. Construct a circRNA/miRNA/mRNA regulatory network to explore potential pathogenesis and therapy options of clear cell renal cell carcinoma. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-70484-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen C., Shen Z. FN1 promotes thyroid carcinoma cell proliferation and metastasis by activating the NF-κB pathway. Protein Pept. Lett. 2022 doi: 10.2174/0929866530666221019162943. [DOI] [PubMed] [Google Scholar]
- 40.Huang S., Huang P., Wu H., Wang S., Liu G. LINC02381 aggravates breast cancer through the miR-1271-5p/FN1 axis to activate PI3K/AKT pathway. Mol. Carcinog. 2022;61(3):346–358. doi: 10.1002/mc.23375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article.