Abstract
Legionella pneumophila, the etiologic agent of Legionnaires' disease, contains a single, monopolar flagellum which is composed of one major subunit, the FlaA protein. To evaluate the role of the flagellum in the pathogenesis and ecology of Legionella, the flaA gene of L. pneumophila Corby was mutagenized by introduction of a kanamycin resistance cassette. Immunoblots with antiflagellin-specific polyclonal antiserum, electron microscopy, and motility assays confirmed that the specific flagellar mutant L. pneumophila Corby KH3 was nonflagellated. The redelivery of the intact flaA gene into the chromosome (L. pneumophila Corby CD10) completely restored flagellation and motility. Coculture studies showed that the invasion efficiency of the flaA mutant was moderately reduced in amoebae and severely reduced in HL-60 cells. In contrast, adhesion and the intracellular rate of replication remained unaffected. Taking these results together, we have demonstrated that the flagellum of L. pneumophila positively affects the establishment of infection by facilitating the encounter of the host cell as well as by enhancing the invasion capacity.
Legionella pneumophila, the etiologic agent of Legionnaires' disease, is a ubiquitous microorganism inhabiting natural and man-made freshwater biotopes (5). In these environments, the gram-negative, rod-shaped bacteria survive as intracellular pathogens of protozoan organisms such as Acanthamoeba castellanii, Hartmannella vermiformis, and Naegleria spp. (15). Upon transmission to individuals via L. pneumophila-containing aerosols generated by showerheads and air-conditioning systems, the bacteria invade and multiply within alveolar macrophages (1, 2, 7) and nonphagocytic cells (17). The infection which mainly affects immunocompromised patients results in a life-threatening atypical pneumonia (7).
Detailed ultrastructural and molecular studies of the intracellular fate of the bacterium revealed that human macrophages and protozoan cells infected with L. pneumophila exhibit remarkable similarities concerning the establishment of a replicative phagosome (3, 16, 22, 42, 45). However, significant differences were observed during early stages of infection (21). Uptake by Hartmannella is accomplished by a microfilament-independent mechanism that is sensitive to methylamine, which is an inhibitor of receptor-mediated endocytosis (28). So far, one receptor of Hartmanella vermiformis, a Gal/GalNAc lectin, could be identified (46). Attachment of L. pneumophila to this lectin results in tyrosine dephosphorylation of multiple host cell proteins. However, depending on the type of amoeba, different receptors might be involved (22). In contrast, the uptake by human macrophages occurs following binding of complement receptors CR1 and CR3 via microfilament-dependent phagocytosis (26). In addition to this cytochalasin D-sensitive mechanism, complement-independent mechanisms for uptake by nonphagocytic cells have been described (39).
The influence of bacterial motility on infection processes or on survival of legionellae in aquatic habitats is not well understood, but motility has been associated with the growth phase of L. pneumophila (40). Bacteria which actively multiply within the host cell vacuole are nonmotile, whereas bacteria in the later stages of infection and cell lysis are flagellated and highly motile (8, 38). This trait during the postexponential phase suggests that motility enables Legionella to escape from a spent host and facilitates its attempt to find a new host by dispersion into the environment.
The single, monopolar flagellum of L. pneumophila is composed of one major subunit, the flagellin, encoded by the flaA gene (23). Previous reports suggest that the expression of flagella is temperature regulated, since it is repressed at temperatures higher than 37°C (36). Recently, we demonstrated that the expression of the flaA gene is regulated at the transcriptional level (24) by the alternative ς28 factor (FliA) and probably by FlaR, a regulator of the LysR family (25). Moreover, the complex flagellum expression and assembly seems to be coordinatively regulated with other virulence-associated traits (6, 16, 38, 40). These traits include thickening of the cell envelope, sensitivity to NaCl, contact-dependent cytotoxicity, osmotic resistance, and evasion of macrophage lysosomes (8). However, experiments with an insertion mutation in the fliI gene of L. pneumophila indicated that intracellular growth in macrophage-like U937 cells does not require flagellar assembly (32). Therefore, it has been proposed that the flagellum might be a virulence-associated factor in the infection process of L. pneumophila. In addition, it has been proposed that proteins involved in the assembly of flagella may be required for export of factors involved in intracellular growth (32).
To determine the exact role of the flagellum for the pathogenicity, we constructed and phenotypically characterized a specific flaA mutant of L. pneumophila Corby. The results of this study provide evidence for the importance of the flagellum during the early stage of infection of eukaryotic host cells.
MATERIALS AND METHODS
Bacterial strains and plasmids.
L. pneumophila Corby (serogroup 1) (27) was used for the construction of the flaA mutant strain KH3 carrying the chromosomal flaA::kam gene fusion (47). Escherichia coli DH5α was used for propagation of recombinant plasmid DNA, and E. coli K-12 (SM10λpir) (43) was used for propagation of pCVD442 and pMSS704. Plasmid pUC18 (Pharmacia LKB, Freiburg, Germany) was used for the construction of pKH106, and plasmid pKS (Stratagene, Heidelberg, Germany) was used for the generation of pKH2 and pKH3. The suicide vectors pCVD442 and pMSS704 (11) were used to generate plasmids pKH4 and pCD10, respectively.
Media and chemicals.
E. coli was cultivated in Luria-Bertani broth. Solid medium for growth of L. pneumophila was ACES [N-(2-acetamido)-2-aminoethanesulfonic acid]-buffered charcoal yeast extract medium (ABCYE) (pH 6.9), essentially as described previously (13). Strains were incubated at 37°C for 48 h before being harvested. For broth culture, L. pneumophila was grown in ACES-buffered yeast extract broth (YEB; 1% yeast), supplemented with 0.025% ferric pyrophosphate and 0.04% l-cysteine, to stationary growth phase at 37°C unless indicated otherwise. Where indicated, drugs were included in bacteriological media at the following concentrations: ampicillin, 100 mg ml−1; kanamycin, 12.5 to 25 mg ml−1; and chloramphenicol, 5 to 25 mg ml−1. Enzymes were purchased from Pharmacia LKB, Boehringer GmbH (Mannheim, Germany), and GIBCO BRL (Eggenstein, Germany). All other chemicals were supplied by Merck (Darmstadt, Germany), Oxoid (Wesel, Germany), Roth (Karlsruhe, Germany), and Sigma (Deisenhofen, Germany).
DNA techniques.
Preparation of genomic DNA and plasmid DNA, as well as DNA-cloning procedures and Southern blot analysis, were performed by standard methods (41). Several oligonucleotide primer sets (MWG, Ebersberg, Germany) were designed and synthesized for amplification of DNA fragments encompassing the chromosomal flaA region or the flaA region with the inserted antibiotic resistance marker. The nucleotide sequences of the primers were as follows: KHACCD, 5′-TCCAACCGTCGCTCCCATGGAGCCACCCA-3′; FLA1, 5′-GTAATCAACACTAATGTGGC-3′; and FLA5, 5′-GTTGCAGAATTTGGTTTTTGGTC-3′. PCR was carried out using a Thermocycler 60 apparatus from Biomed (Theres, Germany) and GoldStar DNA polymerase (Eurogentec, Seraing, Belgium). Amplification was performed at 95°C for 3 min followed by 30 cycles of denaturation (95°C for 1 min), annealing (55°C for 1.5 min), and extension (72°C for 5 min).
Construction of plasmids.
To construct a disrupted flaA allele replacement vehicle, plasmid pKH106 was isolated from an expression library of L. pneumophila Corby (23). The 2,580-bp Sau3AI chromosomal fragment of this plasmid comprises the flaA gene, the putative ς28 promoter region, and the beginning of an open reading frame which shows significant homology to the accD gene of E. coli. Subsequent cloning of a 2,170-bp XbaI-KpnI fragment containing the flaA gene into pKS resulted in pKH2. The cloned flaA gene was disrupted by replacement of an internal 109-bp EcoRI-HindIII fragment with a Kmr cassette, generating plasmid pKH3. To construct the suicide delivery vector, the 4,000-bp XbaI-KpnI fragment of pKH3 was ligated into pCVD442, resulting in pKH4. For complementation, the SacI-XbaI fragment (2,170 bp) from pKH106 was cloned into suicide plasmid pMSS704, resulting in pCD10.
Electrotransformation of L. pneumophila.
For transformation of L. pneumophila, bacteria were grown on ABCYE agar plates at 37°C for 24 h. They were then suspended in 200 ml of chilled (4°C) water containing 10% glycerol, and pelleted by centrifugation. This procedure was repeated three times, and the final pellet was suspended in 1 ml of 10% glycerol. Aliquots of 80 μl were stored at −80°C. Samples were prepared for electroporation by mixing competent cells and plasmid DNA on ice. The samples were placed into prechilled 0.1-cm electroporation cuvettes (Bio-Rad) and pulsed with the pulse controller set at 2.3 kV, 25 mF, and 100 Ω of resistance. Immediately following electric discharge, 1 ml of prewarmed YEB was added to each cuvette. Phenotypic expression was allowed to occur overnight in 4 ml of YEB. For selection of recombinants, cultures were plated onto ABCYE agar plates containing the appropriate antibiotic, with or without 5% sucrose. After 4 to 5 days of incubation at 37°C, colonies were isolated and streaked again on selective ABCYE medium. All strains were stored at −80°C to avoid serial-passage effects.
SDS-PAGE and immunoblotting.
Total-cell extracts of L. pneumophila strains were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting. SDS-PAGE was performed by use of the discontinuous buffer system of Laemmli (30). Legionella cells were grown in overnight cultures at 37°C to the stationary phase unless indicated otherwise. A 1-ml volume was pelleted by centrifugation, the cells were suspended in 100 μl of SDS sample lysis buffer, and a 5-μl volume was then loaded onto an SDS–13% polyacrylamide gel. Purified flagellar extracts were prepared from cultures grown on agar plates for 5 days at 30°C. Flagella were isolated by differential centrifugation as described elsewhere (34). Western blot analyses were carried out as described elsewhere by using a polyclonal monospecific antibody against L. pneumophila Corby flagellin (23).
Cell culture and growth of L. pneumophila in HL-60 cells.
The human leukemia cell line HL-60 (Deutsche Sammlung von Mikroorganismen und Zellkultuven, Braunschweig, Germany) was maintained at 37°C under 5% CO2 in RPMI 1640 medium (PAA, Cölbe, Germany) supplemented with 2 mM l-glutamine (Gln) and 10% fetal calf serum (FCS) (Sigma). HL-60 cells were differentiated into macrophages by incubation for 2 days with 10 ng of phorbol 12-myristate 13-acetate per ml in RPMI–2 mM Gln–10% FCS in 24-well plates (Falcon, Schwandorf, Germany). Adherent cells were washed three times and then incubated with RPMI–2 mM Gln–10% FCS prior to infection.
The ability of L. pneumophila strains to grow in macrophage-like cells was determined in coculture assays. Bacterial strains were cultivated overnight at 37°C in YEB to the beginning of the stationary phase. They were adjusted in RPMI–2 mM Gln–10% FCS medium to a concentration of 2 × 103 cells per ml prior to infection. Differentiated HL-60 cells (2 × 105 cells per well) in 24-well plates were infected with 1 ml of bacterial suspension (multiplicity of infection (MOI) 0.01) and incubated for 4 days. Macrophages were lysed daily with 1 ml of cold H2O and combined with the culture supernatant, and serial dilutions were spread on ABCYE plates to determine the number of bacterial CFU.
To analyze the intracellular growth rates during the first 24 h, a gentamicin assay was used. HL-60 cells were infected at an MOI of 10 for 2 h. After extracellular bacteria were killed by addition of gentamicin (80 μg/ml) for 1 h, CFU were determined immediately and after 24 h.
In situ hybridization of infected host cells was performed as described elsewhere (19). Briefly, HL-60 cells were infected with bacteria at a MOI of 0.01 in Permanox chamber slides (Nunc, Wiesbaden, Germany). After 72 h, the bacteria were labeled with the Legionella-specific probe LEG705. Fluorescence was detected using a Zeiss (Oberkochen, Germany) Axiolab microscope and the Zeiss filter set 00 and 10.
To determine the number of intracellular bacteria at the beginning of infection, HL-60 cells were infected with 2 × 106 bacteria, (MOI, 10), incubated for different time intervals (15, 30, 60, and 120 min), treated with gentamicin (80 μg/ml) for 1 h, washed twice, lysed as described above, and plated on agar plates in serial dilutions. To minimize uptake during the attachment assay, HL-60 cells were pretreated with 1 μg of cytochalasin D per ml for 1 h (44). The number of adherent bacteria was determined after sedimentation of bacteria by centrifugation (1,000 × g for 5 min) and a 20-min incubation in the presence of cytochalasin D, followed by vigorous washing. All assays were performed independently in triplicate.
Amoeba culture and growth of L. pneumophila in A. castellanii.
Acanthamoeba castellanii was obtained from the American Type Culture Collection (ATCC 30234). Axenic cultures of A. castellanii were prepared in 20 ml of Acanthamoeba medium PYG 712 (4) at room temperature. Subculture of the amoebae was performed at intervals of 4 days. The axenic culture was adjusted to a titer of 2 × 105 cells per ml, and 1 ml of culture was pipetted into each well of 24-well plates. Following overnight incubation, the medium was replaced with Acanthamoeba buffer (i.e., PYG 712 medium without proteose peptone and yeast extract), and the next day the amoeba cultures were infected with bacteria as described for HL-60 cells (see above). To determine the number of adherent bacteria, amoebae were incubated with cycloheximide (100 μg/ml) and cytochalasin D (5 μg/ml) for 2 h prior to and throughout the infection (29).
Electron microscopy.
Bacteria were grown to stationary phase in supplemented YEB at 37°C. They were then carefully resuspended in distilled water, and a drop of the suspension was directly applied to Formvar-coated copper grids. After sedimentation of the bacteria and removal of the remaining fluid, the samples were stained with 2% uranyl acetate and examined with a transmission electron microscope (EM10; Zeiss) at 60 kV.
RESULTS
Construction of an L. pneumophila flaA mutant and a flaA-positive complementant.
Plasmid pKH4 was used to inactivate the targeted flaA locus of L. pneumophila strain Corby (Fig. 1). We obtained three putative mutants (KH1, KH2, and KH3) on ABCYE plates plus kanamycin and sucrose, where the allelic exchange was possibly due to a double crossover. An additional three recombinants (KH4, KH5, and KH6) were isolated from ABCYE plates plus kanamycin as a result of single crossover.
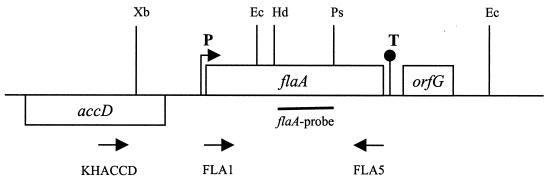
FIG. 1.
flaA region of L. pneumophila. Bars represent open reading frames; flaA is the flagellin-encoding gene, orfG is the homolog to flaG of P. aeruginosa, and accD is the homolog to accD of E. coli. Deduced promoters (P), terminators (T), binding sites for primers (arrows), and the flaA-specific probe are indicated. Restriction sites: Ec, EcoRI; Hd, HindIII; Ps, PstI; Xb, XbaI.
Candidate flagellar mutants were screened by PCR with a primer pair specific for the 5′ (FLA1) and 3′ (FLA5) region of the flaA gene. Amplification products of the predicted length of 1,300 bp were observed for the wild type and the single-crossover mutants. In contrast, strains KH1, KH2, and KH3 revealed amplification products with a predicted length of 3,000 bp, indicating the integration of the neo gene (1,800 bp) and the deletion of 109 bp within the flaA locus. During a second screen using primer pair KHACCD and FLA5, we excluded the possibility of extrachromosomal copies or a single-crossover event for these mutants, since the binding site of KHACCD is lacking in construct pKH4. The desired recombination event was confirmed by Southern blot analysis using a flaA specific probe as well as a Kmr gene probe (data not shown). Strain KH3 was chosen for complementation and further characterization.
Successful reintegration of the intact flaA gene into the chromosome was achieved after cloning the 2.2-kb SacI-XbaI fragment of pKH106H4 into the suicide vector pMSS704. After delivery of the complete vector into the chromosome by a single crossover, the presence of the intact flaA gene was proven by PCR and Southern blotting with flaA-specific, Kmr-specific, and pMSS704-specific probes (data not shown). The insertion was shown to be upstream of the disrupted flaA gene, and the wild-type phenotype was completely restored.
Flagellar expression and motility.
The effect of targeted mutagenesis on the expression of the L. pneumophila FlaA protein was assessed by SDS-PAGE and Western blot analysis by probing whole-cell lysates with a polyclonal monospecific antibody against L. pneumophila Corby flagellin (reference 23 and data not shown). The flaA mutant strain KH3 was shown to be devoid of the 48-kDa FlaA protein, while wild-type strain Corby and the complemented flaA mutant CD10 clearly expressed the protein. The formation of intact flagella correlated with the flagellin expression, as demonstrated by electron microscopy (Fig. 2). Light microscopy was used to monitor the motility of the wild-type strain and the complemented mutant. Consistent with the results obtained by Western blotting and electron microscopy, this motile phenotype was absent in mutant KH3.
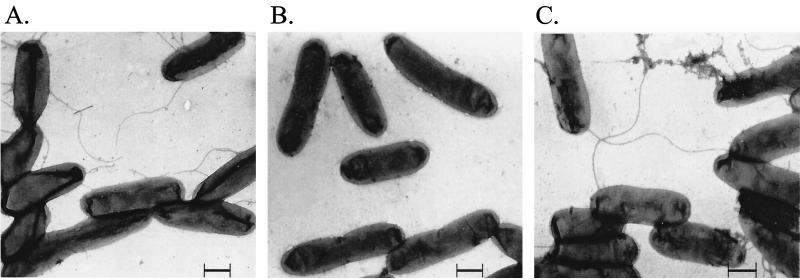
FIG. 2.
Electron micrographs of the flagellated wild-type strain (A), the nonflagellated mutant KH3 (B), and the complemented mutant strain CD10 (C). Bacteria were grown to stationary phase, suspended in H2O, applied to Formvar-coated copper grids, shadowed with 2% uranyl acetate, and examined with a Zeiss EM10 transmission electron microscope. Bars, 0.5 μm.
Adherence to host cells.
To investigate the effects of the disruption of the flaA gene on the life cycle of Legionella, we evaluated the ability of the L. pneumophila wild-type strain, the mutant strain KH3, and the complemented strain CD10 to adhere to host cells. In the presence of phagocytosis inhibitors, the bacteria were centrifuged onto A. castellanii and differentiated HL-60 cells, and the number of adherent bacteria was determined after 20 min of coincubation. No significant differences in adherence among the three strains and their host cells were observed (data not shown).
Early stages of invasion.
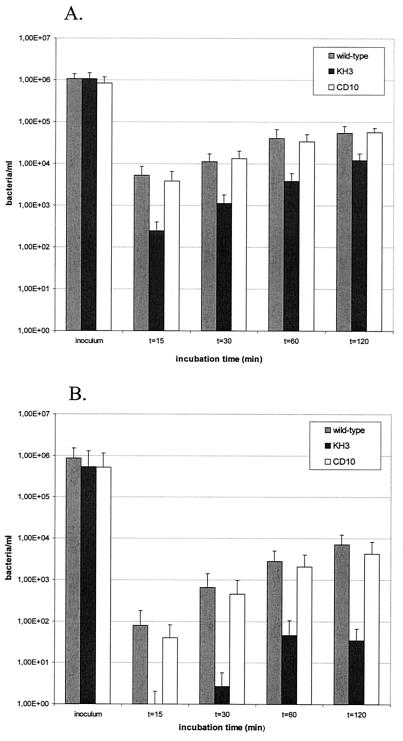
Gentamicin infection assays revealed a clear difference in the number of intracellular bacteria at the onset of multiplication (Fig. 3). After 30 min of coincubation with A. castellanii, 10-fold fewer bacteria of strain KH3 than of the wild-type strain were found inside the host cells. In HL-60 cells, the rate of invasion of strain KH3 was 150-fold lower, while the complemented strain CD10 exhibited the wild-type phenotype in both host cell systems. After 120 min of coincubation, the flagellated and nonflagellated bacteria still showed a 5-fold difference for amoebae and a 130-fold difference for HL-60 cells.
FIG. 3.
Invasion efficiency of L. pneumophila Corby wild type, the flaA mutant KH3, and the complemented mutant CD10 into A. castellanii cells (A) and HL-60 cells (B). Adherent host cells were incubated with legionellae at an MOI of 10 for different time intervals and treated with gentamicin (80 μg/ml) for 1 h, and the CFU were determined by plating on ABCYE agar plates. Error bars indicate the standard deviations obtained from three independent experiments.
Intracellular multiplication in amoebae and macrophage-like HL-60 cells.
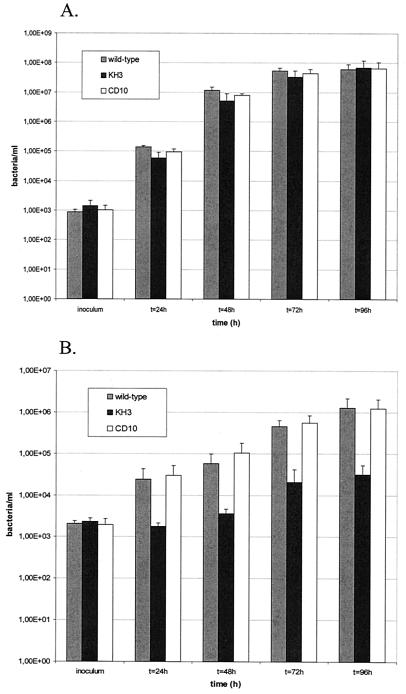
To determine whether the disruption of the flaA gene influences the intracellular multiplication of the bacteria in host cells, A. castellanii and HL-60 cells were infected with the same set of flagellated and nonflagellated strains. The results of the intracellular growth assays are shown in Fig. 4. In A. castellanii, no difference between flagellated and nonflagellated bacteria was observed. All strains showed a 50,000 to 70,000-fold increase in numbers by 4 days postinfection (Fig. 4A). However, a significant difference in infection by flagellated bacteria and the nonflagellated mutant KH3 was observed in HL-60 cells 24 to 96 h postinfection (Fig. 4B). While the wild-type strain Corby had multiplied 600-fold by 4 days postinfection, the flaA mutant had only multiplied 14-fold. The defect of growth of mutant KH3 was completely restored in the complemented strain CD10. To exclude variable growth kinetics of the wild-type and mutant strains, growth curves for the growth of both strains in YEB were established. No difference in growth could be observed. To distinguish between invasion and intracellular growth, an intracellular growth assay (MOI, 10) with gentamicin treatment (2 h postinfection) to kill extracellular bacteria was performed. With this method, the intracellular growth rate of the wild-type strain can be calculated with respect to that of the mutant strain. The number of intracellular bacteria was determined directly after the gentamicin treatment and again after 24 h. The two strains showed comparable intracellular growth rates. The wild-type and CD10 strains replicated 75- to 125-fold, and the KH3 strain replicated 150-fold. To determine the number of infected host cells and the number of bacteria per host cell, in situ hybridization was performed at the later stages of infection (72 and 96 h). These experiments showed that the percentage of infected HL-60 cells was higher for the wild-type strain than for the flaA mutant. Taken together, these results demonstrate that the expression of the FlaA protein is especially relevant for the invasion efficiency of macrophage-like HL-60 cells but not for intracellular growth.
FIG. 4.
Multiplication of L. pneumophila Corby wild type, the flaA mutant KH3, and the complemented mutant CD10 in A. castellanii and HL-60 cells. (A) A. castellanii cells (2 × 105/ml) were infected with 2 × 103 bacteria, and the CFU per well were determined daily in duplicate by plating on ABCYE plates. (B) Differentiated HL-60 cells (2 × 105 cells/ml) were infected and treated as described for A. castellanii. Error bars indicate the standard deviations obtained from three independent experiments.
DISCUSSION
Surface structures, such as capsules, lipopolysaccarides, pili, and flagella, play an important role in bacterial pathogenicity and in the ability of bacteria to survive in the environment. Motility-defective bacteria of some species have been reported to be affected in virulence because of aberrations in host cell adherence, invasion mechanisms, or other unknown factors (10, 14, 35, 36). Previous studies utilizing insertional mutagenesis have linked the expression of flagella by L. pneumophila and the ability of this pathogen to infect A. castellanii, H. vermiformis, and human U937 cells (6, 38). Moreover, flagellated legionellae have been found in lung alveolar spaces of legionellosis patients (9). In contrast, other reports do not support a direct link between flagella and virulence in Legionella. The insertion mutation in the fliI open reading frame, an essential component of flagellum assembly, had no effect on intracellular growth in cultured cells (32). Also, the finding that flagellar expression is not required for the intraperitoneal infection of guinea pigs argues against the involvment of flagella in virulence (12). Due to this conflicting results, other authors have suggested that Legionella virulence factors may be coregulated with flagellar expression (6).
To elucidate the exact role of the flagellum of L. pneumophila during the infection process of amoebae and human macrophages, we constructed a specific flagellum-negative mutant of L. pneumophila Corby. Construction of this flagellar mutant, KH3, was accomplished by using a kanamycin gene cassette for targeted disruption of flaA. The allelic exchange completely abolished the assembly of the flagellar filament and, accordingly, revealed that only one copy of flaA exists in the L. pneumophila genome. The redelivery of the intact flaA gene into the chromosome completely restored the flagellated motile wild-type phenotype.
Earlier reports have shown that pili improve the adherence of L. pneumophila to mammalian and protozoan cells (44). By using the nonflagellated mutant KH3, we found that the flaA mutation does not additionally affect the adhesion of Legionella organisms to their respective host cells but clearly reduces the capacity to invade macrophage-like HL-60 cells and A. castellanii. After 30 min, the motile wild type shows a 150-fold-higher invasion rate in HL-60 cells and a 10-fold higher invasion rate in A. castellanii cells than the nonflagellated mutant does. Therefore, we conclude that the flagella of Legionella facilitate the initial encounter of the host cell and somehow enhance the process of uptake whereas intracellular replication is not affected. Similar effects have also been shown for other organisms. For example, the flagella of Agrobacterium tumefaciens are required for efficient encounter of the root surface and possibly aid in orientating the bacterial cells at various sites for infection (10). The construction of a nonmotile, nonflagellated Campylobacter jejuni mutant resulted in a decrease of internalization by a factor of 30 to 40 compared to the parent strain, while attachment appeared to be unaffected (18). Pseudomonas aeruginosa nonflagellated mutants exhibited a lower rate of uptake by murine macrophages, whereas the attachment via flagella did not play a major role (14). Similar results were also obtained for Proteus mirabilis (33). Adherence studies with concanavalin-pretreated A. castellanii demonstrated that the flagella of Pseudomonas fluorescens and Proteus mirabilis interact with carbohydrates on the host cell surface (37). Moreover, in P. aeruginosa the presence of the bacterial flagellum is required for nonopsonic phagocytosis by macrophages (31). Accordingly, similar opsonin-independent adherence and uptake mechanisms have been described for L. pneumophila (39). However, further studies are needed to determine whether the flagellum of Legionella is involved in this specific mode of uptake.
The negative effect of the flaA mutation decreases during the course of infection in A. castellanii but not in HL-60 cells. This indicates that the severely reduced invasion efficiency results in smaller intracellular numbers of bacteria in HL-60 cells even at later stages of infection. In amoebae, this effect of the flaA mutant is not observed due to a higher invasion efficiency compared to that in HL-60 cells. This may also be the explanation for the observed replication of the nonflagellated fliI mutant in U937 cells (32). Differences between human macrophages and amoebae have also been observed on infection with mutants with mutations in the macrophage-specific infectivity loci (mil) of Legionella (20).
Earlier reports that flagella are not required for the intracellular growth of legionellae correlate with our results and the finding that flagellin is not expressed during the replicative phase of infection (8, 24, 32). Our current view holds that the expression of the flagellum is especially relevant for the initial encounter of the host. This may be important in the environment, where the availability of host cells may be limited. Additionally, our results show that flagellation improves the invasion capacity of Legionella organisms into their respective host cells. The observation that intracellular legionellae are highly motile after the multiplicative phase of the bacteria raises the question whether flagellation also contributes to the lysis of the host cell. Further studies with our nonflagellated mutant KH3 and with motility-defective flagellated strains will help to elucidate whether chemotaxis, host cell lysis, or anchorage to biofilm components in the environment are responsible for the widespread dissemination of L. pneumophila.
ACKNOWLEDGMENTS
We thank Ute Hentschel for critically reading the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG HA 1434/12-1), by the “Graduiertenkolleg Infektiologie,” and by the Fonds der Chemischen Industrie.
REFERENCES
- 1.Abu Kwaik Y, Gao L Y, Stone B J, Venkataraman C, Harb O S. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl Environ Microbiol. 1998;64:3127–3133. doi: 10.1128/aem.64.9.3127-3133.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu Kwaik Y A. The use of differential display-PCR to isolate and characterize a Legionella pneumophila locus induced during the intracellular infection of macrophages. Mol Microbiol. 1996;21:543–556. doi: 10.1111/j.1365-2958.1996.tb02563.x. [DOI] [PubMed] [Google Scholar]
- 3.Abu Kwaik Y A. The phagosome containing Legionella pneumophila within the protozoan Hartmannella vermiformis is surrounded by the rough endoplasmatic reticulum. Appl Environ Microbiol. 1996;62:2022–2028. doi: 10.1128/aem.62.6.2022-2028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.American Type Culture Collection. Cataloge of protists—algae and protozoa. 16th ed. 1985. Supplement: media formulations. American Type Culture Collection, Rockville, Md. [Google Scholar]
- 5.Barker J, Brown M R W. Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology. 1994;140:1253–1259. doi: 10.1099/00221287-140-6-1253. [DOI] [PubMed] [Google Scholar]
- 6.Bosshardt S C, Benson R F, Fields B S. Flagella are a positive predictor for virulence in Legionella. Microb Pathog. 1997;23:107–112. doi: 10.1006/mpat.1997.0134. [DOI] [PubMed] [Google Scholar]
- 7.Brand B C, Hacker J. The biology of Legionella infection. In: Kaufmann S H E, editor. Host response to intracellular pathogens. London, United Kingdom: Chapman & Hall, Ltd.; 1997. pp. 291–312. [Google Scholar]
- 8.Byrne B, Swanson M S. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun. 1998;66:3029–3034. doi: 10.1128/iai.66.7.3029-3034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandler F W, Thomason B M, Hebert G A. Flagella on Legionnaires' disease bacteria in the human lung. Ann Intern Med. 1980;93:715–716. doi: 10.7326/0003-4819-93-5-715. [DOI] [PubMed] [Google Scholar]
- 10.Chesnokova O, Coutinho J B, Khan I H, Mikhail M S, Kado C I. Characterization of flagella genes of Agrobacterium tumefaciens, and the effect of a bald strain on virulence. Mol Microbiol. 1997;23:579–590. doi: 10.1046/j.1365-2958.1997.d01-1875.x. [DOI] [PubMed] [Google Scholar]
- 11.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliot J A, Johnson W. Virulence conversion of Legionella pneumophila serogroup 1 by passage in guinea pigs and embryontaed eggs. Infect Immun. 1982;35:943–947. doi: 10.1128/iai.35.3.943-946.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feeley J C, Gibson R J, Gorman G W, Langford N C, Rasheed J K, Makel D C, Baine W B. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J Clin Microbiol. 1979;10:437–441. doi: 10.1128/jcm.10.4.437-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feldman M, Bryan R, Rajan S, Scheffler L, Brunnert S, Tang H, Prince A. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect Immun. 1998;66:43–51. doi: 10.1128/iai.66.1.43-51.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fields B S. The molecular ecology of Legionellae. Trends Microbiol. 1996;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 16.Gao L Y, Harb O S, Abu Kwaik Y. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant host cells, mammalian macrophages and protozoa. Infect Immun. 1997;65:4738–4746. doi: 10.1128/iai.65.11.4738-4746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garduno R A, Garduno E, Hoffman P S. Surface-associated hsp60 chaperonin of Legionella pneumophila mediates invasion in a HeLa cell model. Infect Immun. 1998;66:4602–4610. doi: 10.1128/iai.66.10.4602-4610.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grant C C R, Konkel M E, Cieplak W, Jr, Tompkins L S. Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect Immun. 1993;61:1764–1771. doi: 10.1128/iai.61.5.1764-1771.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimm D, Merkert H, Ludwig W, Schleifer K-H, Hacker J, Brand B C. Specific detection of Legionella pneumophila: Construction of a new 16S rRNA-targeted oligonucleotide probe. Appl Environ Microbiol. 1998;64:2686–2690. doi: 10.1128/aem.64.7.2686-2690.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harb O S, Venkataraman C, Haack B J, Gao L Y, Abu Kwaik Y. Heterogeneity in the attachment and uptake mechanisms of the Legionnaires' disease bacterium, Legionella pneumophila, by protozoan hosts. Appl Environ Microbiol. 1998;64:126–132. doi: 10.1128/aem.64.1.126-132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harb O S, Abu Kwaik Y. Characterization of a macrophage-specific infectivity locus (milA) of Legionella pneumophila. Infect Immun. 2000;68:368–376. doi: 10.1128/iai.68.1.368-376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hentschel U, Steinert M, Hacker J. Common molecular mechanisms of symbiosis and pathogenesis. Trends Microbiol. 2000;8:226–231. doi: 10.1016/s0966-842x(00)01758-3. [DOI] [PubMed] [Google Scholar]
- 23.Heuner K, Bender-Beck L, Brand B C, Lück P C, Mann K-H, Marre R, Ott M, Hacker J. Cloning and genetic characterization of the flagellum subunit gene (flaA) of Legionella pneumophila serogroup 1. Infect Immun. 1995;63:2499–2507. doi: 10.1128/iai.63.7.2499-2507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heuner K, Hacker J, Brand B C. The alternative sigma factor ς28 of Legionella pneumophila restores flagellation and motility to an Escherichia coli fliA mutant. J Bacteriol. 1997;179:17–23. doi: 10.1128/jb.179.1.17-23.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heuner K, Dietrich C, Steinert M, Göbel U B, Hacker J. Cloning and characterization of a Legionella pneumophila specific gene encoding a member of the LysR family of transcriptional regulators. Mol Gen Genet. 2000;264:204–211. doi: 10.1007/s004380000310. [DOI] [PubMed] [Google Scholar]
- 26.Horwitz M A. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med. 1983;158:1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jepras R I, Fitzgeorge R B, Baskerville A. A comparison of virulence of two strains of Legionella pneumophila based on experimental aerosol infection of guinea pigs. J Hyg. 1985;95:29–38. doi: 10.1017/s0022172400062252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King C H, Fields B S, Shotts E B, Jr, White E H. Effects of cytochalasin D and methylamine on intracellular growth of Legionella pneumophila in amoebae and human monocyte-like cells. Infect Immun. 1991;59:758–763. doi: 10.1128/iai.59.3.758-763.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Köhler R, Bubert A, Goebel W, Steinert M, Hacker J, Bubert B. Expression and use of the green fluorescent protein as a reporter system in Legionella pneumophila. Mol Gen Genet. 2000;262:1060–1069. doi: 10.1007/pl00008649. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Mahenthiralingam E, Speert D P. Nonopsonic phagocytosis of Pseudomonas aeruginosa by macrophages and polymorphonuclear leukocytes requires the presence of the bacterial flagellum. Infect Immun. 1995;63:4519–4523. doi: 10.1128/iai.63.11.4519-4523.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merriam J J, Mathur R, Maxfield-Boumil R, Isberg R. Analysis of the Legionella pneumophila fliI gene: intracellular growth of defined mutant defective for flagellum biosynthesis. Infect Immun. 1997;65:2497–2501. doi: 10.1128/iai.65.6.2497-2501.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mobley H L, Belas R, Lockatell V, Chippendale G, Trifillis A L, Johnson D E, Warren J W. Construction of a flagellum-negative mutant of Proteus mirabilis: effect on internalization by human renal epithelial cells and virulence in a mouse model of ascending urinary tract infection. Infect Immun. 1996;64:5332–5340. doi: 10.1128/iai.64.12.5332-5340.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montie T C, Craven R C, Holder I A. Flagellar preparations from Pseudomonas aeruginosa: isolation and characterization. Infect Immun. 1982;35:281–288. doi: 10.1128/iai.35.1.281-288.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ormonde P, Horstedt P, O'Toole R, Milton D L. Role of motility in adherence to and invasion of a fish cell line by Vibrio anguillarum. J Bacteriol. 2000;182:2326–2328. doi: 10.1128/jb.182.8.2326-2328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ott M, Messner P, Heesemann J, Marre R, Hacker J. Temperature-dependent expression of flagella in Legionella. J Gen Microbiol. 1991;137:1955–1961. doi: 10.1099/00221287-137-8-1955. [DOI] [PubMed] [Google Scholar]
- 37.Preston T M, King C A. Binding sites for bacterial flagella at the surface of the soil amoeba Acanthamoeba. J Gen Microbiol. 1984;130:1449–1458. [Google Scholar]
- 38.Pruckler J M, Benson R F, Moyenuddin M, Martin W T, Fields B S. Association of flagellum expression and intracellular growth of Legionella pneumophila. Infect Immun. 1995;63:4928–4932. doi: 10.1128/iai.63.12.4928-4932.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodgers F G, Gibson F C. Opsonin-independent adherence and intracellular development of Legionella pneumophila within U937 cells. Can J Microbiol. 1993;39:718–722. doi: 10.1139/m93-103. [DOI] [PubMed] [Google Scholar]
- 40.Rowbotham T J. Current views on the relationships between amoebae, legionellae and man. Isr J Med Sci. 1986;22:678–689. [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Segal G, Shuman H A. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect Immun. 1999;67:2117–2124. doi: 10.1128/iai.67.5.2117-2124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon R, O'Connell M, Labes M, Pühler A. Plasmid vectors for the genetic analysis and manipulation of Rhizobia and other gram-negative bacteria. Methods Enzymol. 1986;118:640–658. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- 44.Stone B J, Abu Kwaik Y. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect Immun. 1998;66:1768–1775. doi: 10.1128/iai.66.4.1768-1775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swanson M S, Isberg R R. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun. 1995;63:3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Venkataraman C, Haack B J, Bondada S, Abu Kwaik Y. Identification of a Gal/GalNAc lectin in the protozoan Hartmannella vermiformis as a potential receptor for attachment and invasion by the Legionnaires' disease bacterium. J Exp Med. 1997;18:537–547. doi: 10.1084/jem.186.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woodcock D M, Crowther P J, Doherty J, Jefferson S, De Cruz E, Nayer-Weidner M, Smith S S, Michael M Z, Graham M W. Quantification evaluation of Escherichia coli for tolerance of cytosine methylation in plasmid and phage recombination. Nucleic Acids Res. 1989;9:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]