Summary
The pilosebaceous unit (PSU) is composed of multiple compartments and the self-renewal of PSU depends on distinct hair follicle stem cell (HFSC) populations. However, the differential roles of the HFSCs in sebaceous gland (SG) renewal have not been completely understood. Here, we performed multiple lineage tracing analysis to unveil the contribution of different HFSC populations to PSU regeneration during the hair cycle and wound healing. Our results indicated that the upper bulge stem cells contributed extensively to the SG replenishment during hair cycling, while HFSCs in the lower bugle did not. During skin wound healing, all HFSC populations participated in the SG replenishment. Moreover, β-catenin activation promoted the contribution of HFSCs to SG replenishment, whereas β-catenin deletion substantially repressed the event. Thus, our findings indicated that HFSCs contributed to SG replenishment in a β-catenin-dependent manner.
Subject areas: Cell biology, Stem cells research
Graphical abstract
Highlights
-
•
Hair follicle stem cells (HFSCs) lineage tracing unveils contributions to sebocytes
-
•
HFSCs in the upper bulge contribute to sebaceous gland (SG) renewal during cycling
-
•
All HFSCs in the hair follicle adjacent to wounds contribute to SG replenishment
-
•
β-catenin signaling activate HFSCs to sebocytes replenishment
Cell biology; Stem cells research
Introduction
The hair follicle (HF) undergoes periodic cycling in mice. In each cycle there are anagen, catagen and telogen phases (Joost et al., 2018; Paus et al., 1999; Saxena et al., 2019). Multiple populations of HF stem cells (HFSCs) reside in the HF to maintain distinct pilosebaceous unit (PSU) compartments in mice. The PSU contains the HF and sebaceous gland (SG) (Brownell et al., 2011; Füllgrabe et al., 2015; Horsley et al., 2006; Jaks et al., 2008; Morris et al., 2004). HF bulge and other regions of the PSU are reconstituted by mitotically active stem cells (Solanas and Benitah, 2013). Regional HFSC heterogeneity and compartmentalization are crucial for tissue homeostasis (Page et al., 2013). The upper part of the PSU, which comprises the isthmus, junctional zone, infundibulum, and SG, contains SCs expressing Lgr6, Plet1/Mts24, and Lrig1 (Jensen et al., 2009; Nijhof et al., 2006; Page et al., 2013; Snippert et al., 2010). SCs in the bulge express a series of marker genes including Lgr5, CD34, K15, and Gli1 (Brownell et al., 2011; Cheng et al., 2018; Jaks et al., 2008; Morris et al., 2004; Trempus et al., 2003) and are considered as multipotent SCs (Horsley et al., 2006). In addition, Lrig1 defines the HF junctional zone adjacent to the SGs. And Lrig1-expressing cells contribute to the sebaceous gland and interfollicular epidermis during homeostasis (Jensen et al., 2009).
Quiescent and active cyclical SCs are located in separate yet adjacent locations in HFs (Lavker et al., 2003; Li and Clevers, 2010; Wang et al., 2017, 2022). These SCs are categorized into two types: hair germ SCs and bulge SCs. Although the hair germ and bulge are two adjacent compartmentalized structures, they share many common cellular biomarkers such as Gli1, K15, and Lgr5 (Brownell et al., 2011; Cheng et al., 2018; Jaks et al., 2008; Morris et al., 2004; Wang et al., 2017). A two-step activation mechanism of hair germ and bulge SCs during hair regeneration further clarifies this point (Greco et al., 2009). During the resting phase (telogen), hair germ SCs are quiescent because of the net balance of bone morphogenetic protein (BMP) signaling (major inhibitor) and Wnt signaling (major activator), which is recognized for its critical role in multiple stages of the hair cycle (Calvo-Sánchez et al., 2019; Geyfman et al., 2015). At the transition from the telogen to anagen, hair germ SCs initially proliferate rapidly prior to bulge SCs activation and then progress to become transit-amplifying cells (TACs) in the matrix (Greco et al., 2009). Subsequently, TACs further differentiate into various epithelial lineages along the HF longitudinal axis, such as the inner root sheath (IRS) and hair shaft (Greco et al., 2009; Xin et al., 2018; Yang et al., 2017). Meanwhile, bulge SCs receive morphogen Sonic hedgehog (Shh) signals that are known for their critical role in regulating HF epithelial growth and maintenance of TAC-derived tissue polarity, and thus are activated, with the progenies of bulge SCs migrating downward and differentiating into the outer root sheath (ORS) (Hsu et al., 2014b; Rompolas et al., 2013). ORS cells can differentiate into matrix cells, followed by differentiation into the IRS and hair shaft (Xin et al., 2018). During the transition phase (catagen), the lower ORS cells undergo apoptosis, whereas the upper ORS cells survive and contribute to the new bulge and hair germ (Hsu et al., 2011, 2014a, 2011; Rompolas et al., 2013). Available evidence has proven that follicular epithelial SCs reside in the bulge and that such SCs can give rise to not only all components of the hair shaft but also the epidermis and SG (Lavker et al., 2003). The bulge SC population itself is heterogeneous (Krieger and Simons, 2015; Rompolas et al., 2013). Live imaging of individual bulge or hair germ SCs revealed that HFSCs accomplish different functions depending on their locations. Upper bulge SCs are the principle cellular source of the upper PSU; these SCs are usually quiescent and can undergo self-renewal. Lower bulge SCs instead differentiate downward into the ORS (Rompolas et al., 2013).
Lineage tracing and functional genetics studies have elegantly demonstrated that bulge SCs continuously produce other SCs or progenitor cells that differentiate and replenish sebocytes (Morris et al., 2004; Petersson et al., 2011, 2015). HFSCs and their progenies traced to SGs in the K15-CrePGR;LacZ mouse (Morris et al., 2004), and manipulation of TCF/Lef1 signaling in K15-CreERT2;R26RYFP mice resulted in abnormal SG formation (Petersson et al., 2011). Furthermore, ΔNLef1 mutation in K15 mice gives rise to SG tumorigenesis (Petersson et al., 2015). All these studies indicate an association of HFSCs with SG renewal and aberrant growth. However, the contribution of different HFSC subpopulations in SG renewal remains unclear.
This study elucidated the ability of different HFSCs to give rise to the whole PSU in the normal hair cycle and after wounding to the skin. Our data also demonstrated that β-catenin signaling has a significant role in SG replenishment.
Results
Bulge progeny can repopulate the entire pilosebaceous unit during hair follicle cycling
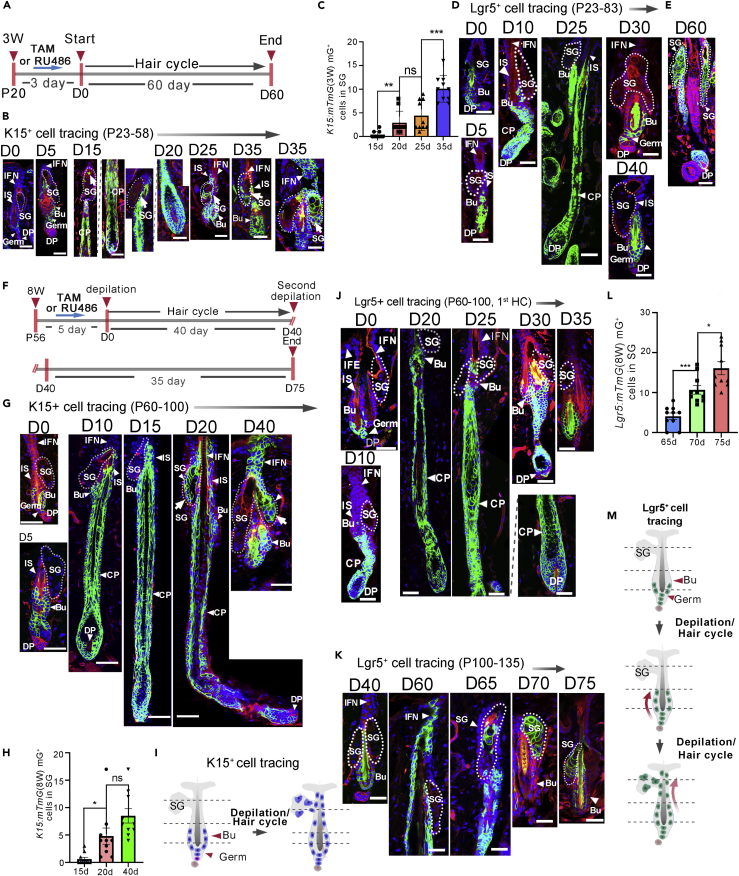
To assay the role of bulge cells in replenishing the SG, an inducible lineage tracing system for tracking Keratin (K) 15+ bulge SCs, K15CrePGR:Rosa26-mTmG (referred to as K15:mTmG), was developed, as K15 is a well-defined HFSC marker. Firstly, K15+ cells were traced during the first HF cycle (postnatal day 20–days 40, P20–40) (Figure 1A). After 3 consecutive days of mifepristone (RU486) administration, the bulge SCs were efficiently labeled with the membrane GFP (mGFP). By contrast, no mGFP signal was detected in SGs. As the HF cycle continued, mGFP fluorescence could be detected in the SG as early as the 15th tracing day. As the HFs proceeded to catagen and telogen phases, the number of mGFP+ cells in the SGs significantly increased (Figures 1B and 1C). These data indicated that bulge SCs could dynamically replenish SG cells during the HF cycle.
Figure 1.
The participation of K15+ and Lgr5+ HFSCs in SG renewal in the depilation-induced hair cycle
(A) Experimental timeline of tamoxifen/RU486 injection in 3-weeks old (3W) Lgr5:mTmG and K15:mTmG mice. P, postpartum; D, day.
(B) Representative images showing the fate of K15-lineage cells (green) during the hair cycle in 3W K15:mTmG mice.
(C) Quantitation of mGFP+ cells within SGs in K15:mTmG mice in the first hair cycle. Data are represented as mean ± SEM, ns, not significant, ∗p < 0.05, ∗∗∗p < 0.005, t-test, n = 3.
(D and E) Tracing of progenies of Lgr5+ HFSCs in Lgr5:mTmG mice after tamoxifen induction from P23 to P58 (D), and in the second hair cycle (E).
(F) Experimental scheme for RU486/tamoxifen injection and analysis of depilation-induced HF activation in K15:mTmG or Lgr5:mTmG mice.
(G) Tracing of K15+ HFSCs in K15:mTmG mice from P60 to P80 after RU486 induction.
(H) Quantitation of mGFP+ cells within SGs in 8W K15:mTmG mice at the first cycle. Data are represented as mean ± SEM, ns, not significant, ∗p < 0.05, t-test, n = 3.
(I) A schematic diagram showing the progeny of K15+ SCs in the depilation-induced hair cycle.
(J and K) Tracing the fate of Lgr5+ HFSCs in Lgr5:mTmG mice in the first (0–35 days) (J) and the second (40–75 days) (K) hair cycle following tamoxifen treatment.
(L) Quantitation of mGFP+ cells within SGs in 8W Lgr5:mTmG mice at the second cycle. Data are represented as mean ± SEM, ∗p < 0.05, ∗∗∗p < 0.005, t-test, n = 3.
(M) A schematic portraying the fate of Lgr5+ HFSCs in two consecutive hair cycles induced by depilation. Bu, Bulge; DP, dermal papilla; SG, sebaceous gland. Scale bars: 50 μm.
To further explore the differential capacities of various HFSC populations in contributing to the SG lineage, we employed Lgr5CreER:Rosa26-mTmG (referred to as Lgr5:mTmG) mice for tracing cells located in the lower bulge and hair germ. Lgr5+ cell tracing during the first HF cycle (P20–40) showed that the progenies of Lgr5+ cells repopulated the upper bulge and isthmus in the newly formed HF; however, no mGFP+ cells were detected in the junctional zone or SGs (Figure 1D). In the second cycle (anagen; P60), mGFP+ cells were detected in SGs (Figure 1E). The combined results of K15+ and Lgr5+ cell lineage tracings suggested that different HFSC populations exhibit different capacities in the formation of SG cells.
To further analyze the relationship between bulge SCs and SG, telogen (P56) HFs were forced into cycling through depilation (Figures 1F and 1G). The tracing of K15+ cells showed that the progenies of bulge SCs could extensively contribute to sebocytes, with some SGs being completely replenished by mGFP+ sebocytes (Figures 1G–1I). Similar experiments were conducted to trace Lgr5+ cells during HF cycling in adult mice. Consistent with the lineage tracing results of the first HF cycle, during the depilation-induced HF cycle (P56–100) of adult mice, the progenies of Lgr5+ cells were found in the upper bulge and isthmus but not in the SG (Figure 1J). To further explore the contribution of Lgr5+ cell progeny to SG cells, telogen (P100) HFs of the Lgr5:mTmG mice were induced into another HF cycle (P100–140) through depilation. Interestingly, during the second depilation-induced HF cycle, Lgr5+ cell progenies continued to move upward and efficiently contributed to SG cells, and part of the SG was completely replenished by mGFP+ cells (Figure 1K–1M). These data indicated that in depilation-induced successive HF cycles, Lgr5+ SCs in the lower bulge could first repopulate the upper bulge and isthmus and then further moved upward and replenished the SG.
Germ cells do not contribute to sebaceous gland cells during hair follicle cycling
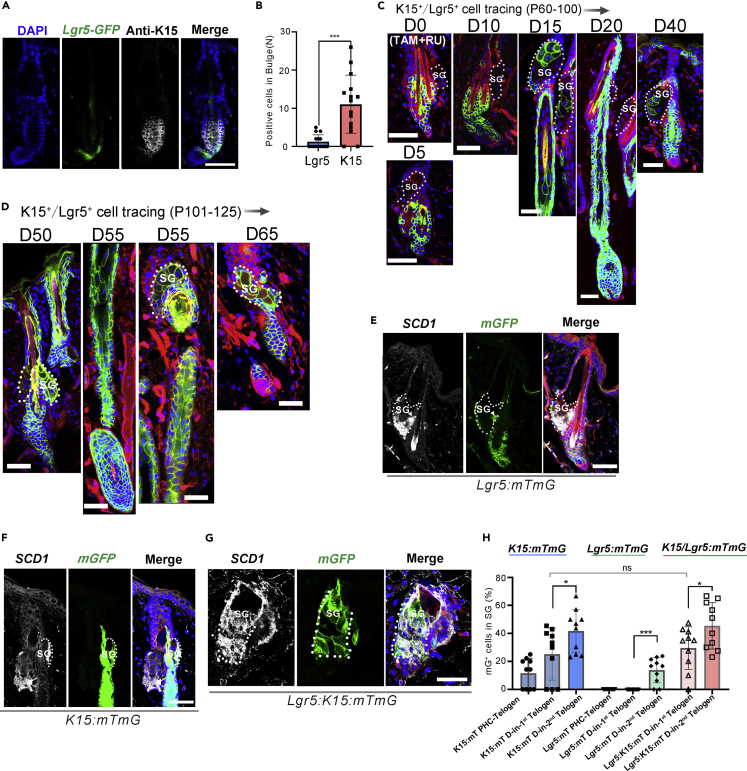
Previous studies indicated that germ SCs were derived from the bulge and triggered the initial step of the HF cycle (Hsu et al., 2011, 2014b, 2011). To further evaluate the role of germ SCs in HF cycling and its contribution to the PSU, we performed more lineage tracing analysis. Since all germ SCs expressed Lgr5 but a fraction of them did not express K15 (Figures 2A and 2B), mice that could trace both germ and bulge SCs, namely Lgr5CreER:K15CrePGR:Rosa26-mTmG (referred to as Lgr5:K15:mTmG), were developed. Histological analysis showed that both germ and bulge cells were efficiently labeled in the mice in both the first and second depilation-induced HF cycles (Figures 2C and 2D).
Figure 2.
Lineage tracing of HFSCs (Lgr5+K15+) in SG renewal during the depilation-induced hair cycle
(A) Immunofluorescence staining for K15 expression in telogen HFs of Lgr5-CreERT2-EGFP mice showed that K15 expression was present in the whole bulge and some hair germ cells, while Lgr5 expression marked cells in the lower bulge and hair germ.
(B) Quantitation of cells negative for K15 or Lgr5 in the germ (Telogen) (Data are represented as mean ± SEM, ∗∗∗p < 0.005, t-test, n = 15 mice).
(C and D) Tracing of Lgr5+ and K15+ HFSCs in the first (C) and second (D) depilation-induced hair cycles in Lgr5:K15:mTmG mice after RU486/tamoxifen induction (n = 5 mice).
(E–G) After depilation, immunostaining for SCD1+ sebocytes in Lgr5:mTmG mice in second cycle anagen (E), in K15:mTmG mice after the first cycle (F), and in Lgr5:K15:mTmG mice in the first cycle telogen (G), co-stained mGFP+ progenies of the SCs in the SG (n = 5 mice).
(H) The progenies which were mGFP+ and found in the SG in two consecutive hair cycles were quantified (Data are represented as mean ± SEM, ns, not significant, ∗p < 0.05, ∗∗∗p < 0.005, t-test, n = 5 mice). PHC, postnatal hair cycle. Scale bars: 50 μm.
To confirm that the progenies of HFSCs present in the SG were sebocytes, we performed immunostaining for stearoyl coenzyme A desaturase1 (SCD1), which had been known to be exclusively expressed in sebocytes in the skin (Niemann and Horsley, 2012), and found that the mGFP+ cells in the SG also expressed SCD1 (Figures 2E–2G). The results indicated that the progenies of HFSCs in the SG have adopted a sebocyte phenotype. Quantitative analysis of mGFP+ cells in SGs displayed no significant difference between the Lgr5:K15:mTmG and K15:mTmG mice after the first and second depilation-induced HF cycles (Figure 2H), indicating a negligible contribution of germ SCs to SG cells.
In summary, our results indicated that in depilation-induced HF cycles, K15+ HFSCs, but not Lgr5+ HFSCs, contributed to sebocytes during the first hair cycle; the progeny of Lgr5+ HFSCs were detected in the SG in the second hair cycle and in a much lower level, compared to cells derived from K15+ HFSCs.
Sebaceous gland cells were extensively replenished by the upper bulge SCs during the hair cycle
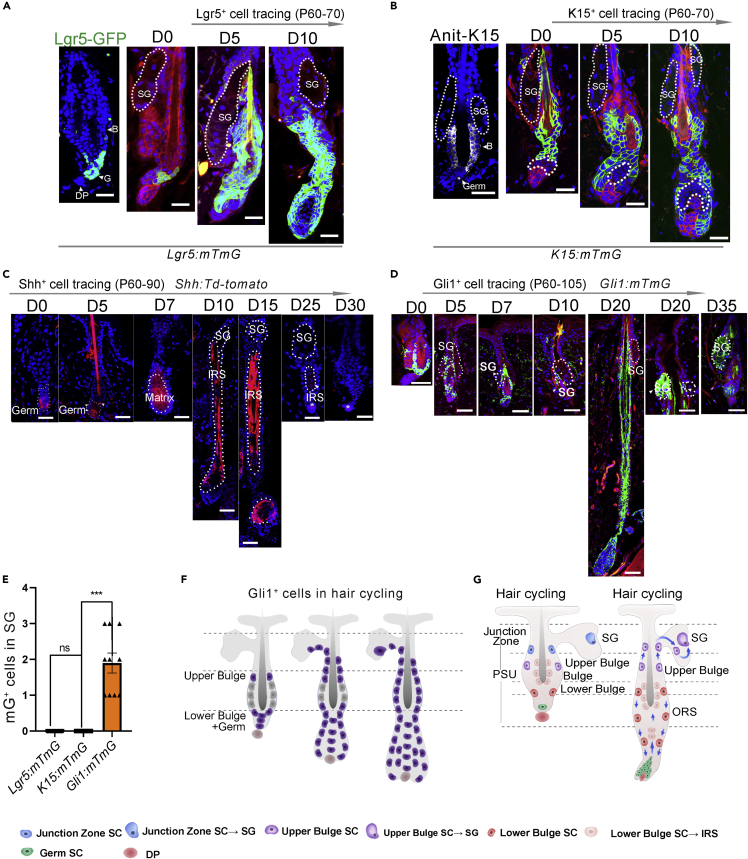
Based on all the aforementioned data, bulge SCs, instead of hair germ SCs, are assumed to generate differentiated progenies to replenish the SG. To determine which bulge SC population acts as the cellular source that contributes to SG renewal, we performed the lineage tracking of different SC populations. Four transgenic mice (Lgr5+, K15+, Gli1+, Shh+) were selected to specifically track HFSCs located in different anatomical sites of the HF. Lgr5 was expressed in SCs in the lower bulge and germ of telogen HFs. During anagen, Lgr5+ lower bulge and germ SCs differentiated into the ORS, IRS, and hair shaft (Figure 3A). K15 immunostaining showed that K15+ cells were located in the inner and outer layer bulge cells. In K15:mTmG mice, a small fraction of hair germ SCs, which were not labeled by mGFP, expanded at the early stage of anagen (Figure 3B), indicating that hair germ SCs divided into TACs. In both Lgr5:mTmG and K15:mTmG mouse lines, no mGFP+ cells were detected within the SGs in the telogen HF before hair cycling.
Figure 3.
Lineage tracing of multiple HFSCs for their contribution to sebocytes
(A) Lgr5 expression in telogen HF from Lgr5-GFP mouse line (left panel). The tracing of Lgr5-labeled cells in the early and mid-anagen phase (right panel) (n = 5).
(B) Immunofluorescence staining showing K15 expression in the telogen HF (left panel). In K15:mTmG mice, the progeny of K15+ HFSCs at early and mid-anagen were shown (right panel).
(C) Tracing of Gli1-lineage cells in 8-week-old Gli1:mTmG mice (n = 5).
(D) Tracing of Shh+ matrix progenitors in 8-week-old Shh:tdTomato mice after tamoxifen induction (n = 5).
(E) Counts of progenies of Lgr5+, K15+, and Gli1+ SCs in the SG, which were detected by the expression of mG, Data are represented as mean ± SEM, ns, not significant, ∗∗∗p < 0.005, t-test, n = 5.
(F) A schematic diagram showing the tracing of Gli1+ SCs during the first depilation-induced hair cycle.
(G) A schematic diagram showing the tracing of the lower bulge and upper bulge progenitors during the first depilation-induced hair cycle. Bu, bulge; LB, lower bulge; UB, upper bulge; SG: sebaceous gland; ORS, outer root sheath. Scale bars: 50 μm.
Shh marks matrix progenitors at early anagen (Figure S1A). We performed lineage tracing experiments of Shh+ matrix progenitors in 8-week-old ShhCreER:Rosa26-tdTomato (referred to as Shh:tdTomato) mice. During anagen, the progenitors differentiated into the IRS and hair shaft but not into SGs, and their progenies gradually disappeared during the catagen and telogen phases (Figure 3D). Thus, no SG cells were marked during the entire HF cycle. This indicated that the Shh+ matrix progenitors derived from hair germ were not the source of cells for SG renewal.
To further evaluate the role of upper bulge cells in SG renewal, we developed another transgenic reporter mice Gli1CreER:Rosa26-mTmG (commonly referred to as Gli1:mTmG), as Gli1 is known to be expressed in the upper bulge cells (Brownell et al., 2011), and was also detected in our analysis (Figure S1B). In hair depilation experiments of 8-week-old Gli1:mTmG mice after tamoxifen administration, a small number of mGFP+ cells were observed in the SGs in the early anagen phase (Figure 3D). By contrast, no mGFP+ cells was detected in early anagen HFs of Lgr5:mTmG mice, as well as in K15:mTmG mice (Figures 3A, 3B, and 3E). Gli1+ upper bulge SCs differentiated into sebaceous lineages early probably due to their proximity to the SG (Figures 3D–3G and S1B). When Gli1:mTmG HFs progressed to the next telogen phase, SGs contained higher labeling efficiency of mGFP+ cells (Figures S1C and S1D). Results in Gli1:mTmG mice strongly support the notion that the upper bulge is the principal cellular source of SG cells (Figures 3G and S1E).
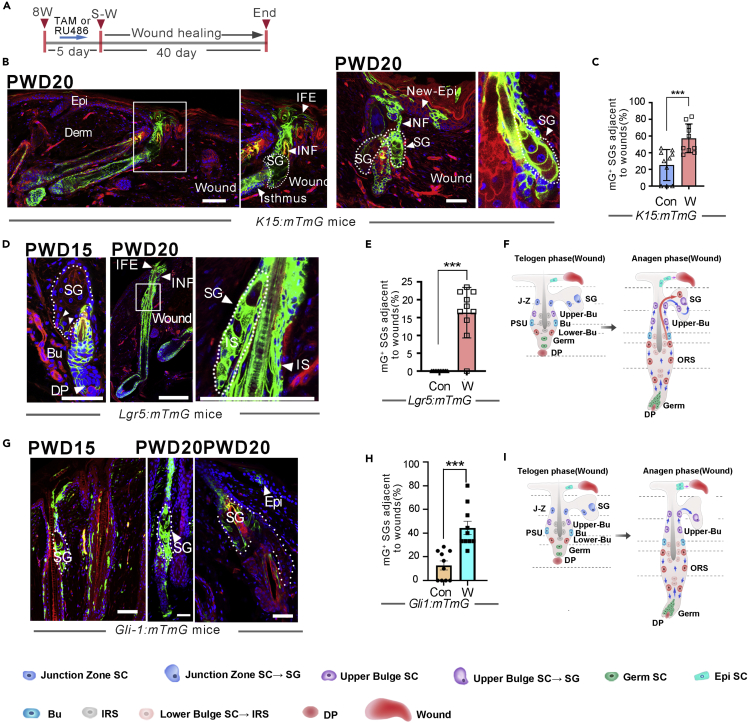
Bulge and hair germ SCs substantially reconstitute the sebaceous gland after the skin wound formation
Previous studies indicate that Lgr5+ and K15+ HFSCs contribute to the interfollicular epidermis during wound healing (Ito et al., 2007). To determine whether wounding enhanced SG renewal by recruiting HFSC progenies, we introduced four full-thickness, circular wounds (diameter: 0.5 cm) into the back skin of K15:mTmG and Lgr5:mTmG mice (Figure 4A). In the K15:mTmG mice, at 20 days after wound formation, K15-derived progenies traced to the SG adjacent to wounds and exhibited a high labeling efficiency during wound repair (Figure 4B). K15-derived descendants were no longer confined to the PSU but extended into the interfollicular epidermis and infundibulum (Figure 4B). The number of SGs containing mGFP-labeled cells adjacent to the wounds in K15:mTmG mice was counted. Compared to unwounded mice, wounding to the skin significantly accelerated SG repopulation by K15+ SCs progenies (Figure 4C). Similarly, after wounding to the skin, Lgr5-derived progenies also replenished the SGs with a high labeling efficiency and then migrated into the interfollicular epidermis and infundibulum (Figure 4D), and the percentage of SGs containing mGFP-labeled cells was much greater than that in the unwounded skin (Figures 4E and 4F).
Figure 4.
Wounding enhances SG renewal by recruiting progenies of bulge stem cells
(A) Experimental timeline for fate mapping experiments using Lgr5:mTmG and K15:mTmG mice after wounding the back skin of 8-week-old mice.
(B and C) Tracing of K15+ SCs in K15:mTmG mice 20 days post-wounding. Note that the vast majority of sebocytes in the HF adjacent to the wound and some interfollicular epidermal cells were labeled (B), and mGFP+ cells within the SGs were counted (B and C, Data are represented as mean ± SEM, ∗∗∗p < 0.005, t-test, n = 3).
(D and E) Tracing of Lgr5+ SCs in Lgr5:mTmG mice at 15 and 20 days post-wounding. The vast majority of SG cells adjacent to wounds were labeled (D), and mGFP+ cells within the SGs were counted (D and E), Data are represented as mean ± SEM, ∗∗∗p < 0.005, t-test, n = 3).
(F) A schematic summary shows that wounding to the skin accelerated the differentiation of bulge and lower bulge SCs into SG cells.
(G and H) Tracing of Gli1+ SCs in Gli1:mTmG mice at 15 and 20 days post-wounding (G), and mGFP+ cells within the SGs were counted (H, Data are represented as mean ± SEM, ∗∗∗p < 0.005, t-test, n = 3 mice).
(I) A schematic summary showing the contribution of upper bulge SCs to SG cells after wounding to the skin. PWD, post-wound day; Derm: dermis; Epi, epidermis; New-Epi, new-epidermis; IFE, interfollicular epidermis; IFN, infundibulum; SG, sebaceous glands; W: wound. Scale bars: 50 μm.
To determine whether hair germ SCs contribute to SG renewal after wounding to the skin, we performed the same wounding experiments in Gli1:mTmG mice. The results showed that wounding accelerated the differentiation of Gli1+ upper bulge and hair germ SCs into SG cells (Figure 4G).
Together, the results indicated that the wound environment broke the lineage commitment of the bulge and hair germ SCs, and accelerated the recruitment of their progenies to SGs and the interfollicular epidermis (Figures 4H and 4I).
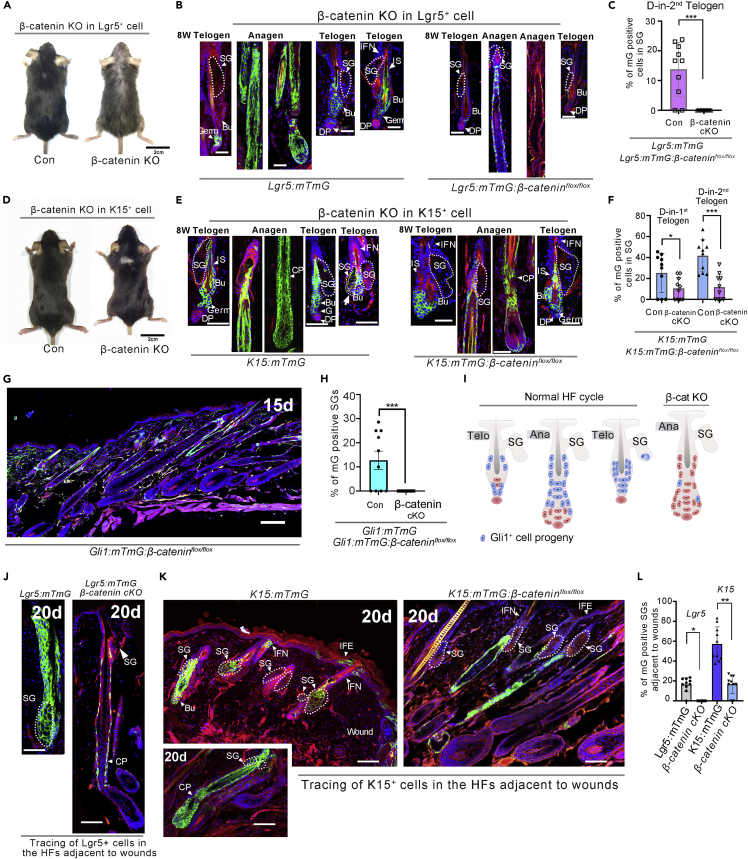
Loss of β-catenin in Lgr5+ and K15+ cells blocks their contribution to sebaceous gland renewal
To determine whether altering Wnt/β-catenin signaling affects the degree of participation of K15+ and Lgr5+ SCs in SG renewal, we specifically deleted β-catenin in K15- and Lgr5-expressing cells by crossing Ctnnb1flox/flox mice with Lgr5:mTmG, K15:mTmG, or Lgr5:K15:mTmG mice. Indeed, we observed that β-catenin deletion in Lgr5-and Lgr5:K15-expressing cells led to a reduction of their contribution to sebocytes associated with aberrant hair shaft shape (Figures S2A–SF) and hair shaft density (Figures 5A and S2G), illustrating that β-catenin loss in bulge and hair germ SCs affected hair growth. Genetic deletion of β-catenin in Lgr5 cells led to reduced β-catenin protein in the cells (Figures S4C and SD). Notably, no mGFP+ cells existed in the SGs after the second depilation in Lgr5CreER:Rosa26-mTmG:Ctnnb1flox/flox (referred to as Lgr5:mTmG:β-cateninflox/flox) mice (Figures 5A–5C). In K15CrePGR:Rosa26-mTmG:Ctnnb1flox/flox (referred to as K15:mTmG:β-cateninflox/flox) mice, genetic deletion of β-catenin in K15 cells resulted in reduced β-catenin protein in the cells (Figures S5A and S5B), and the mice showed reduced involvement of K15+ SCs in SG renewal during the first depilation-induced hair cycle (Figures 5D and 5E). Notably, at subsequent depilation-induced hair regeneration in the mice, the number of K15-traced cells was dramatically reduced within SGs (Figure 5E). Similarly, the loss of β-catenin in Lgr5:K15:mTmG mice considerably reduced the number of mGFP-labeled cells within SGs (Figures S2G and S2H). In addition, knockout of β-catenin in Gli1 cells in Gli1:mTmG:β-cateninflox/flox mice inhibited the differentiation of Gli1+ HFSCs into sebocytes (Figure 5G–5I and S3A–S3E), and mGFP-labeled cells were not detected even during the second depilation-induced hair cycle.
Figure 5.
β-catenin deletion in Lgr5+ and K15+ stem cells inhibits their differentiation into sebocytes
(A) Representative images of Lgr5:mTmG and Lgr5: mTmG:β-cateninflox/flox mice at the second telogen.
(B and C) Lineage tracing of Lgr5+ SCs in Lgr5:mTmG and Lgr5: mTmG:β-cateninflox/flox mice. The inhibition of β-catenin signaling in Lgr5+ cells blocked their differentiation into SG cells (B); mGFP+ cells within the SGs were counted (A–C), Data are represented as mean ± SEM, ∗∗∗p < 0.005, t-test, n = 3).
(D) Representative images of K15:mTmG and K15: mTmG:β-cateninflox/flox mice at the second telogen.
(E and F) Lineage tracing of K15+ SCs in K15:mTmG and K15: mTmG:β-cateninflox/flox mice showed a decrease in the number of mGFP-labeled cells within the SGs upon β-catenin deletion (E–F, Data are represented as mean ± SEM, ∗p < 0.05, ∗∗∗p < 0.005, t-test, n = 3).
(G and H) Representative images of HFs close to wounds in Lgr5:mTmG and Lgr5:mTmG:β-cateninflox/flox mice (G) Tracing of Gli1-expressing cells in Gli1:mTmG:β-cateninflox/flox mice (8W) at 15 days post-injections of TAM. (H) Quantitation of mGFP+ cells within the SGs in Gli1:mTmG and Gli1:mTmG: β-cateninflox/flox mice (G-H), Data are represented as mean ± SEM, ∗p < 0.05, ∗∗∗p < 0.005, t-test, n = 3).
(I) Schematic illustrations of the fate of Gli1+ SCs during normal hair cycle in wild-type mice, and in Gli1:mTmG: β-cateninflox/flox mice with loss of β-catenin expression (J and k) and K15:mTmG and K15:mTmG:β-cateninflox/flox mice (K) at 20 days post-wounding.
(L) Quantitation of mGFP+ cells within SGs in Lgr5:mTmG, Lgr5:mTmG:β-cateninflox/flox, K15:mTmG, and K15:mTmG:β-cateninflox/flox mice adjacent to wounds (J–L), Data are represented as mean ± SEM ∗p < 0.05 and ∗∗∗p < 0.005, n = 3). Bu, bulge; DP, dermal papilla; IFN, infundibulum; IS, isthmus; SG, sebaceous gland. Scale bars: 2 cm in A and D; 50 μm in B, E, G, and H.
To examine the influence of β-catenin loss in HFSCs on their contribution to SG renewal after wounding, full-thickness circular skin wounds in the K15:mTmG: β-cateninflox/flox and Lgr5:mTmG:β-catenin flox/flox mice were induced. At day 20 after wounding, β-catenin deletion in Lgr5+ SCs dramatically reduced their involvement in SG renewal at the wound edge (Figures 5J and S2I–S2N). In addition, the loss of β-catenin in K15-expressing cells in K15:mTmG: β-cateninflox/flox mice also significantly decreased their contribution to sebocytes at the wound margin (Figures 5K–5L). These data suggest a crucial role of β-catenin signal in the replenishment of SG by bugle cells during HF cycling and wound healing.
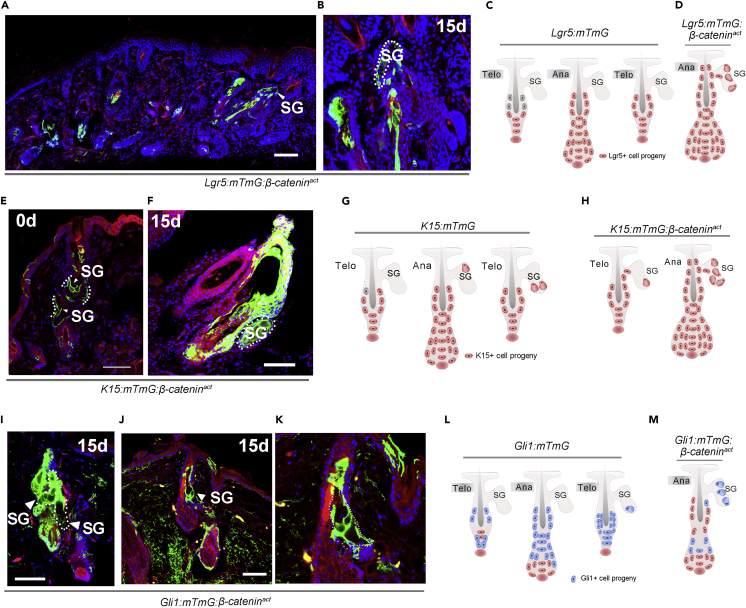
β-catenin activation promotes Lgr5+ cells to replenish the sebaceous gland
To further assess the effect of β-catenin on the recruitment of HFSC progenies to the SG, β-catenin was constitutively activated in Lgr5+ SCs in Lgr5CreERT2-GFP:Rosa26-mTmG:Ctnnb1tm1Mmt/+ (referred to as Lgr5:mTmG: β-cateninact) and Lgr5CreERT2-GFP:Rosa26-EYFP:Ctnnb1tm1Mmt/+ (referred to as Lgr5:YFP:β-cateninact) mice after tamoxifen treatment. Eight-week-old mice were depilated after injection of tamoxifen. In contrast to Lgr5:mTmG (Figure 1C) or Lgr5CreER-GFP:Rosa26-EYFP (referred to as to Lgr5:YFP) (Figures S4A and S4B) mice, by 15 days after depilation, GFP+ cells exited from the base of HFs and streamed into the upper PSU to replenish the SG (Figures 6A–6C) and the infundibulum in Lgr5:YFP: β-cateninact mice (Figures S4A and S4B). Immunofluorescence staining for β-catenin in Lgr5:mTmG:β-cateninact and Lgr5:mTmG mice showed upregulated levels of β-catenin protein in the HF of Lgr5:mTmG:β-cateninact mice (Figures S4C and S4F). However, probably due to the rapid differentiation of β-catenin-activated Lgr5+ progeny, very few HFSCs were labeled. Then, we conducted lineage tracing experiments in K15CrePGR:Rosa26-mTmG:Ctnnb1tm1Mmt/+ (referred to as K15:mTmG: β-cateninact). Similarly, increased levels of β-catenin were found in the HF in K15:mTmG:β-cateninact mice (Figures S5A and S5C), and the number of SGs containing mGFP-labeled cells increased rapidly (Figures 6E–6H). Moreover, in Gli1CreERT2:Rosa26-mTmG:Ctnnb1tm1Mmt/+ (referred to as Gli1:mTmG: β-cateninact) mice, the number of SGs containing mGFP-labeled cells also increased rapidly (Figures 6I–6M).
Figure 6.
Overexpression of β-catenin increases the contribution of bulge SCs to SG cells
(A–D) Tracing of Lgr5+ HFSCs with β-catenin activation. Representative microscopic images in low- (A) and high (B)-magnification of skin tissue sections of 8-weeks-old Lgr5:mTmG:β-cateninact mice after hair depilation showing mGFP-labeled cells in the SG (A-B, n = 5); the contribution of Lgr5+ SCs to sebocytes in Lgr5:mTmG mice with normal β-catenin gene (C) or in Lgr5:mTmG:β-cateninact mice (D) was illustrated.
(E–H) Tracing of K15+ HFSCs with activated β-catenin. Images showing the progenies of K15+ HFSCs in the SG in K15:mTmG:β-cateninact mice at 0 days (E) and 15 days (F) after depilation (E–F, n = 5).Schematic summary of the fate of K15-lineage cells (red) in K15:mTmG (G) and K15:mTmG: β-cateninact mice (H).
(I–K) Tracing the fate of Gli1-lineage in Gli1:mTmG:β-cateninact mice at 15 days post-depilation. Note that most SG cells were mGFP-labeled (I-K, n = 5).
(L and M) Schematic illustrations of the fate of Gli1+ SCs during normal hair cycle in wild-type mice (L), and in Gli1:mTmG: β-cateninact mice with altered expression of β-catenin (M). Telo, telogen; Ana, anagen; SG, sebaceous gland. Scale bars: 50 μm.
In summary, forced β-catenin activation in HFSCs accelerated their capacity in replenishing SG cells, and Lgr5+ SC progenies were even present in the SG during the first depilation-induced hair cycle (this was not found in mice with normal β-catenin). Conversely, β-catenin inhibition blocked Lgr5+ SC differentiation into SG cells (Figures 7A–7F, Figures S4C, S4E, and S5A–S5C). Thus, our data suggested a key role of Wnt/β-catenin signaling in directing the sebaceous lineage fate of HFSCs.
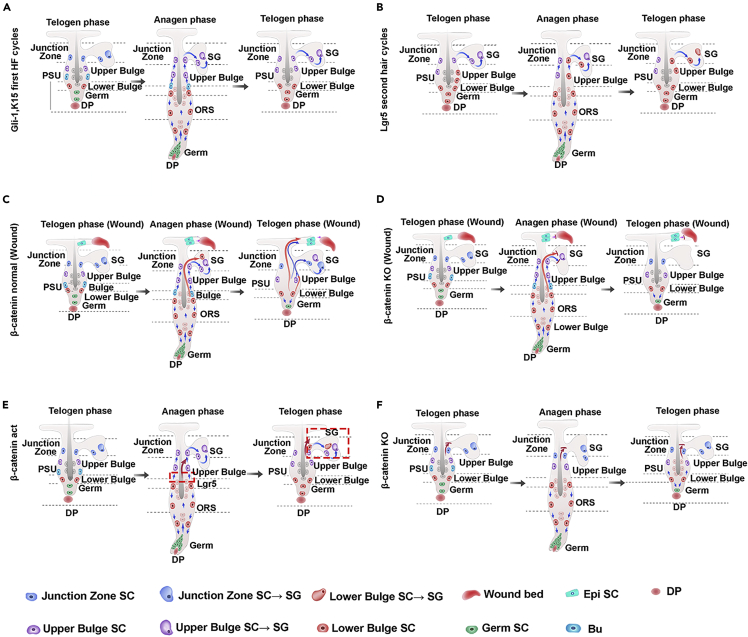
Figure 7.
Proposed models showing migration and differentiation of HFSCs to sebocytes in normal and altered conditions
(A) A schematic presentation depicting the progenies of Gli1+ and K15+ HFSCs in the HF and SG during the first hair cycle.
(B) A schematic illustration of the fate of Lgr5+ HFSCs in the second hair cycle. (C) A schematic illustration showing the fate of Lgr5+ and K15+ HFSCs in Lgr5:mTmG and K15:mTmG mice, respectively.
(C and D) A schematic summarizes that wounding to the skin accelerates the differentiation of bulge SCs into SG cells (C), and the process is abolished by β-catenin inhibition (D).
(E and F) Schematic diagrams depicting the activation (E) and inhibition of β-catenin on the differentiation of bulge SCs into SG cells. J-Z, junctional zone; PWD, post-wound day; Derm, dermis; Epi, epidermis; IFE, interfollicular epidermis; IFN: infundibulum; SG: sebaceous gland; W, wound; PSU, the pilosebaceous unit.
Discussion
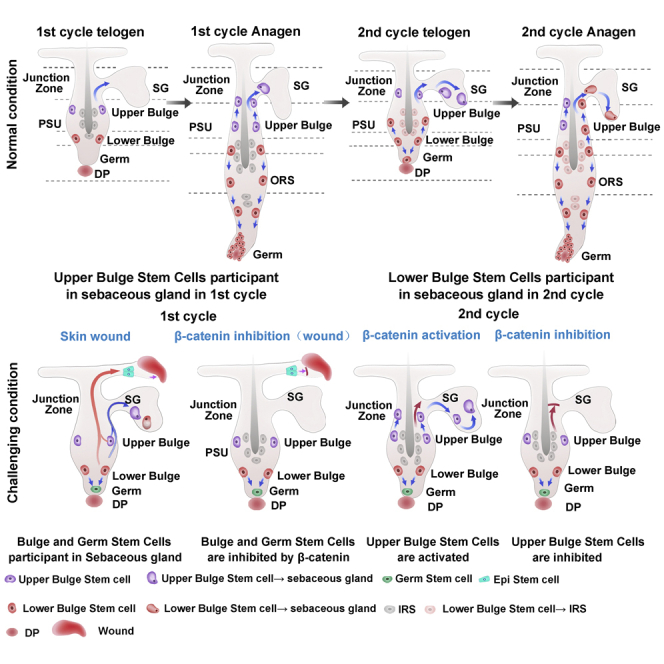
HFs undergo cyclical regeneration lifelong, progressing through stages of anagen, catagen, and telogen (Greco et al., 2009; Joost et al., 2016; Wang et al., 2017). HFSCs located in different parts of the HFs perform different functions and exhibit different differentiation potential (Hsu et al., 2014a, 2011, 2014b; Rompolas et al., 2013; Xin et al., 2018). Bulge SCs are a heterogeneous cell population (Jaks et al., 2010; Krieger and Simons, 2015; Rompolas et al., 2013; Wells and Watt, 2018). Previous studies about lineage tracing of bulge SCs have largely focused on their contribution to cells within the HF (Jaks et al., 2010; Krieger and Simons, 2015; Rompolas et al., 2013; Wells and Watt, 2018). In one study, using K15;R26RYFP mice, the progenies K15+ SCs were found in the SG (Petersson et al., 2011), but the contribution of other HFSC populations to sebocytes along with the mechanisms underlying HFSC differentiation into sebocytes have not been completely understood. We here traced the fates of different HFSCs and elucidated their distinctive roles in sebocyte replenishment during hair cycling and after skin wound formation. The results indicated that the progeny of K15+, Lgr5+, and Gli1+ SCs in the upper bulge contributed to sebocytes in a β-catenin-dependent manner, whereas Shh+ matrix progenitors in the lower HF region did not. As an important source of SG units, SCs in the upper bulge migrated upward and differentiated into SG cells in successive HF cycles to renew SGs.
Our lineage tracing analysis of multiple HFSCs showed differential contributions of these SCs to sebocytes. While the progenies of HFSCs in the upper bulge including Gli1+ and K15+ SCs were traced to the SG early in the first hair cycle, sebocytes derived from Lgr5+ HFSCs located in the lower bulge and germ appeared in the SG during the second hair cycle. A similar fashion was found in these HFSCs in hair depilation-induced hair cycle. These results indicated that HFSCs anatomically adjacent to the SG were prone to differentiation into sebocytes. Our findings are in line with several previous studies. With live imaging tracing of bulge or hair germ SC populations, earlier findings revealed that HFSCs accomplish different functions depending on their spatial locations (Krieger and Simons, 2015; Rompolas et al., 2013). Notably, several recent studies revealed a multipotent HFSC population located in the upper part of the HF adjacent to the SG, named HF-associated pluripotent (HAP) SCs. The cells were defined as nestin+CD34+K15-, and their progenies were found in the outer root sheath of the HF (Amoh and Hoffman, 2017; Li et al., 2003). In addition, the cells were found capable of differentiating into several cell lineages outside of the HF, such as nerve cells, endothelial cells, and adipocytes (Amoh and Hoffman, 2017; Amoh et al., 2005a, 2005b, 2005b; Hoffman, 2006; Li et al., 2003), suggesting a possibility of these cells to differentiate to SG cells.
We further explored the mechanisms underlying HFSC differentiation into sebocytes. We found that wounding to the skin greatly promoted the recruitment of the progenies of all HFSCs to the SG, regardless of their location in the HF, suggesting that wounding mediated a potent activating signal to the SCs for SG renewal. It has been known that wounding activates Wnt/β-catenin in the HFSCs adjacent to the wound site (Ito et al., 2007; Wang et al., 2017). We performed experiments to examine the loss and gain of β-catenin in HFSCs, and found that the genetic ablation of β-catenin in the HFSCs markedly suppressed their differentiation into sebocytes; conversely, the activation of β-catenin in the SCs greatly promoted their differentiation into sebocytes. Thus, our results indicated a crucial role of Wnt/β-catenin signaling in HFSC differentiation into sebocytes during physiological HF cycling and after skin wounding. Our results are in line with several previous studies regarding the role of β-catenin in the skin, where β-catenin activation has been shown to play a central role in HF formation, and be involved in the development of skin tumors (Baker et al., 2010; Brown et al., 2017; Deschene et al., 2014; Kretzschmar et al., 2016). We anticipate that our findings would inspire and establish a solid foundation for future skin research development.
Limitations of the study
In this study, we elucidated the distinct bulge stem cell populations maintain the pilosebaceous unit during normal homeostasis and post-wounding in a β-catenin-dependent manner. Undoubtedly, continual efforts to unravel HFSC function and mechanism have promised to shed the light on mechanisms governing HF physiology and pathophysiology. A crucial question regarding the relationship between multilineage- and lineage-restricted progenitors such as for the SG also need to be investigated.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-CD34(1:50) | Invitrogen | Cat#MA5-32059; RRID: AB_2809353 |
| SCD-1(1:150) | Cell signaling | Cat#2794S |
| Shh Polyclonal antibody(1:200) | Proteintech | Cat#20697-1-AP; RRID: AB_10694828 |
| Cytokeratin 15 Polyclonal antibody(1:50) | Proteintech | Cat#10137-1-AP; RRID: AB_2234359 |
| Anti-Gli1 antibody(1:50) | Abcam | Cat#ab217326 |
| Cytokeratin 15 Polyclonal antibody(1:50) | Proteintech | Cat#10137-1-AP |
| Experimental models: Organisms/strains | ||
| B6; SJL-Tg(Krt1-15-cre/PGR)22Cot/J(K15-CrePR1) | 005249 (Jax mice) | N/A |
| Gli1tm3(cre/ERT2)Alj/J | 007913 (Jax mice) | N/A |
| Ctnnb1tm1Mmt | (Southern University of Science and Technology) | N/A |
| B6.129-Ctnnb1tm2Kem/KnwJ | 004152(Jax mice ) | N/A |
| Gt(ROSA)26Sortm4(ACTB-tdTomato,EGFP)Luo/J | 007576 (Jax mice) | N/A |
| B6.129S6-Shhtm2(cre/ERT2)Cjt/J | 005623 (Jax mice) | N/A |
| B6.129P2-Lgr5tm1(cre/ERT2)Cle/J | 008875 (Jax mice) | N/A |
| B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | 007909 (Jax mice) | N/A |
| B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J | 006148 (Jax mice) | N/A |
| Software and algorithms | ||
| Prism, Version 8.30 | GraphPad | https://www.graphpad.com/scientific-software/prism |
| NLS Element Viewer 4.50 | Nikon | https://www.microscope.healthcare.nikon.com |
| ZEISS 2.30 | Carl Zeiss | https://www.zeiss.com/microscopy/us/products/microscope-software |
| R 3.62 | R | https://www.r-project.org/ |
| Bioconductor | Bioconductor | https://bioconductor.org/packages/ |
| FV1000 | Olympus | https://olympus-fv1000-viewer.updatestar.com/ |
| SPSS 26.0 | IBM | https://spss.en.softonic.com/ |
Resource availability
Lead contact
Further information and requests for resources, reagents, and protocols should be directed to and will be fulfilled by the lead contact, Wu Yaojiong (wu.yaojiong@sz.tsinghua.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Mice
Lgr5-GFP-Cre-ERT2 (Lgr5-Cre) (JAX mice, stock no.: 008875), Gli1CreERT2 (JAX mice, stock no.: 007913), β-catenin flox/flox mice(JAX mice, stock no.:004152), and Rosa mTmG (JAX mice, Stock no.: 007576) mice were obtained from the Jackson Laboratory β-catenin flox/flox mice were a gift from Dr. Zhenge Luo (Institute of Neuroscience, CAS). Ctnnb1tm1Mmt(β-cateninact) mice were a gift fromLiang Fang(Southern University of Science and Technology). TdTomato (JAX mice, stock no.: 007909) and ShhCreERT2 (Jax mice no.: 005623) were gifts fromYing Xiao(Run Run Shaw Hospital affiliated with Zhejiang University Medical College). Gli1CreERT2 was a gift from Sun Yat-sen University, Guanghua School of Stomatology. K15-CrePR1 was obtained from Jackson Laboratories (JAX mice, Stock no.: 005249). K15-CrePR1, Gli1CreERT2, and Lgr5-CreERT2 mice were crossed with β-catenin flox/flox mice, and Rosa mTmG mice to acquire K15:mTmG: β-catenin flox/flox, Gli1:mTmG: β-catenin flox/flox, Lgr5:mTmG:β-catenin flox/flox, K15:mTmG, Gli1:mTmG, and Lgr5:mTmG mice. K15-CrePR1, Gli1CreERT2, and Lgr5-Cre:Rosa-mTmG were crossed with Ctnnb1tm1Mmt to acquire K15:mTmG:β-cateninact, Gli1:mTmG:Ctnnb1act and Lgr5:mTmG:β-cateninact mice. Shh-CreER mice were crossed with Rosa mTmG mice to acquire Shh:mTmG mice. K15:mTmG mice were crossed with Lgr5: mTmG to acquire Lgr5:K15:mTmG mice. K15:mTmG:β-cateninflox/flox mice were crossed with Lgr5:mTmG:β-cateninflox/flox mice to acquire Lgr5:K15:mTmG:β-cateninflox/flox mice. Lgr5:mTmG mice were crossed with Ctnnb1tm1Mmt/+ mice to acquire Lgr5:mTmG:Ctnnb1tm1Mmt/+. Lgr5 mice were crossed with Rosa26-EYFP:Ctnnb1tm1Mmt/+ mice to acquire Lgr5: YFP:Ctnnb1tm1Mmt/+ mice. ERT-Cre recombinase was activated in mice by injecting tamoxifen (16.7 mg/mL dissolved in sunflower oil) at 24-h intervals, and PGR-Cre recombinase was activated in mice by injecting mifepristone (10 mg/mL dissolved in sunflower oil) at 24-h intervals. Mice were maintained in a temperature-controlled environment (20 ± 1°C) with a 12 h light/dark cycle and free access to food and water. Mice in normal body weights (average 18 g in female and 20 g in male) and mental and physical conditions were used for experiments. See Table for more information of mice. For preparation of the mice at time when mice were 3-weeks and 8-weeks old. 8-week-old transgenic mice and 3-weeks old Lgr5:mTmG and K15:mTmG mice.
Ethical considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors. All procedures were performed with the approval of the Animal Ethics Committee of Medical Key Laboratory of Health Toxicology of Shenzhen, Shenzhen Center for Disease Control and Prevention, and the Animal Ethics Committee of Tsinghua Shenzhen International Graduate School. The committees approving the experiments and confirming that all experiments conform to the relevant regulatory standards.
Method details
Excisional wounds in mice
Mice (body weight 18-20 g) were anesthetized with an intraperitoneal injection of sodium pentobarbital (50 mg/kg). Symmetrical 5-mm-diameter full thickness skin wounds were created on the back mice with a skin biopsy punch. We create the skin wounds with bandages to prevent infection (Wang et al., 2013). Full thickness skin samples of the wound and surrounding tissues were harvested with a 10-mm skin biopsy punch 20 days after surgery.
DNA extraction
Tissues were cut and completely soaked in digestion buffer. Protease Plus (Vazyme PD101 AC, 2 μL) and Buffer L (100 μL) tissue digestion buffer was mixed evenly before use. Digestion was performed in a 55°C water bath for 30 min. Samples were then centrifuged at 12,000 rpm for 5 min, the supernatant was used as the PCR template.
PCR amplification
The configuration of the reaction system was performed at low temperatures to ensure PCR amplification efficiency and specificity. The PCR components in 20 μL were as follows: template 1.3–1.4 μL, forward primer 1 μL, reverse primer 1 μL, 2 × Mix (Vazyme PD101 or Accurate Taq Master Mix (dye plus)) 10 μL, with ddH2O added for the remaining liquid volume. PCR conditions followed the Jackson mice genotyping protocol.
Immunohistochemistry
For immunostaining, fixed samples were cryoprotected through a graded treatment process. Tissues were first fixed in 4% paraformaldehyde overnight and then transferred to 30% sucrose (w/v) for another 24 h. Tissue was then embedded on cryomolds in tissue-freezing medium (OCT), then snap-frozen at −80 °C and sectioned to a thickness of 12 or 15 μm on a Leica cryostat. Slides were washed in blocking buffer phosphate-buffered saline (PBS) for 10 min and cryosections were blocked in 10% goat serum, 3% bovine serum albumin (BSA) and 0.5 TritonX-100 for an hour at room temperature. Then frozen tissue sections were incubated with different primary antibodies (see Table for antibody information) at 4°C overnight, incubated with fluorescent-dye conjugated secondary antibodies (Alex Fluor 647) at room temperature for 2 h and visualized under confocal microscope (ZEISS, see Table for information of software and algorithms).
Quantification and statistical analysis
The number of male and female animals was approximately equal, with no significant phenotypic differences between mice in different genders. All experiments and presented results were replicated from at least three times independently treated animals, and genotyping was performed before and after animal treatment for conformation purposes. There was no exclusion of animals or data.
All values were expressed as means ± SD. Comparison between two groups was performed with two-tailed Student’s t test. Comparisons among more than two groups were performed using one-way analysis of variance (ANOVA). Differences were considered statistically significant when p < 0.05.
Acknowledgments
This work was supported by grants from the The National Natural Science Foundation of China (NSFC) (31961160702), Shenzhen Science and Technology Innovation Committee (WDZC20200820173710001, KCXFZ20201221173207022, JCYJ20190809180217220), State Key Laboratory of Chemical Oncogenomics fund, and Tsinghua Shenzhen International Graduate School Overseas Cooperation Fund (HW2021003).
Author contributions
J.H. and K.L. performed experiments and analyzed data. X.W. and Y.W. designed the project, provided resources, and wrote the article. Other authors assisted in the experiments. All authors provided scientific input and approved the final article.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: January 20, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105805.
Contributor Information
Xusheng Wang, Email: Wangxsh27@mail.sysu.edu.cn.
Yaojiong Wu, Email: wu.yaojiong@sz.tsinghua.edu.cn.
Supplemental information
Data and code availability
-
•
Data: The data that support the findings of this study are available from the corresponding author upon reasonable request.
-
•
Code: This research paper does not list code.
-
•
Availability statement: Any additional information in this study are available from the lead contact upon request. upon reasonable request.
References
- Amoh Y., Hoffman R.M. Hair follicle-associated-pluripotent (HAP) stem cells. Cell Cycle. 2017;16:2169–2175. doi: 10.1080/15384101.2017.1356513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoh Y., Li L., Campillo R., Kawahara K., Katsuoka K., Penman S., Hoffman R.M. Implanted hair follicle stem cells form Schwann cells that support repair of severed peripheral nerves. Proc. Natl. Acad. Sci. USA. 2005;102:17734–17738. doi: 10.1073/pnas.0508440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoh Y., Li L., Katsuoka K., Penman S., Hoffman R.M. Multipotent nestin-positive, keratin-negative hair-follicle bulge stem cells can form neurons. Proc. Natl. Acad. Sci. USA. 2005;102:5530–5534. doi: 10.1073/pnas.0501263102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C.M., Verstuyf A., Jensen K.B., Watt F.M. Differential sensitivity of epidermal cell subpopulations to beta-catenin-induced ectopic hair follicle formation. Dev. Biol. 2010;343:40–50. doi: 10.1016/j.ydbio.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S., Pineda C.M., Xin T., Boucher J., Suozzi K.C., Park S., Matte-Martone C., Gonzalez D.G., Rytlewski J., Beronja S., Greco V. Correction of aberrant growth preserves tissue homeostasis. Nature. 2017;548:334–337. doi: 10.1038/nature23304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell I., Guevara E., Bai C.B., Loomis C.A., Joyner A.L. Nerve-derived sonic hedgehog defines a niche for hair follicle stem cells capable of becoming epidermal stem cells. Cell Stem Cell. 2011;8:552–565. doi: 10.1016/j.stem.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo-Sánchez M.I., Fernández-Martos S., Carrasco E., Moreno-Bueno G., Bernabéu C., Quintanilla M., Espada J. A role for the Tgf-β/Bmp co-receptor Endoglin in the molecular oscillator that regulates the hair follicle cycle. J. Mol. Cell Biol. 2019;11:39–52. doi: 10.1093/jmcb/mjy051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.C., Tsutsui K., Taguchi T., Sanzen N., Nakagawa A., Kakiguchi K., Yonemura S., Tanegashima C., Keeley S.D., Kiyonari H., et al. Hair follicle epidermal stem cells define a niche for tactile sensation. Elife. 2018;7:e38883. doi: 10.7554/eLife.38883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschene E.R., Myung P., Rompolas P., Zito G., Sun T.Y., Taketo M.M., Saotome I., Greco V. β-Catenin activation regulates tissue growth non-cell autonomously in the hair stem cell niche. Science. 2014;343:1353–1356. doi: 10.1126/science.1248373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füllgrabe A., Joost S., Are A., Jacob T., Sivan U., Haegebarth A., Linnarsson S., Simons B.D., Clevers H., Toftgård R., Kasper M. Dynamics of Lgr6⁺ progenitor cells in the hair follicle, sebaceous gland, and interfollicular epidermis. Stem Cell Rep. 2015;5:843–855. doi: 10.1016/j.stemcr.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyfman M., Plikus M.V., Treffeisen E., Andersen B., Paus R. Resting no more: re-defining telogen, the maintenance stage of the hair growth cycle. Biol. Rev. Camb. Phil. Soc. 2015;90:1179–1196. doi: 10.1111/brv.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco V., Chen T., Rendl M., Schober M., Pasolli H.A., Stokes N., Dela Cruz-Racelis J., Fuchs E. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 2009;4:155–169. doi: 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman R.M. The pluripotency of hair follicle stem cells. Cell Cycle. 2006;5:232–233. doi: 10.4161/cc.5.3.2397. [DOI] [PubMed] [Google Scholar]
- Horsley V., O'Carroll D., Tooze R., Ohinata Y., Saitou M., Obukhanych T., Nussenzweig M., Tarakhovsky A., Fuchs E. Blimp1 defines a progenitor population that governs cellular input to the sebaceous gland. Cell. 2006;126:597–609. doi: 10.1016/j.cell.2006.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y.C., Li L., Fuchs E. Emerging interactions between skin stem cells and their niches. Nat. Med. 2014;20:847–856. doi: 10.1038/nm.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y.C., Li L., Fuchs E. Transit-amplifying cells orchestrate stem cell activity and tissue regeneration. Cell. 2014;157:935–949. doi: 10.1016/j.cell.2014.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y.C., Pasolli H.A., Fuchs E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell. 2011;144:92–105. doi: 10.1016/j.cell.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Yang Z., Andl T., Cui C., Kim N., Millar S.E., Cotsarelis G. Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature. 2007;447:316–320. doi: 10.1038/nature05766. [DOI] [PubMed] [Google Scholar]
- Jaks V., Barker N., Kasper M., van Es J.H., Snippert H.J., Clevers H., Toftgård R. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- Jaks V., Kasper M., Toftgård R. The hair follicle-a stem cell zoo. Exp. Cell Res. 2010;316:1422–1428. doi: 10.1016/j.yexcr.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Jensen K.B., Collins C.A., Nascimento E., Tan D.W., Frye M., Itami S., Watt F.M. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell. 2009;4:427–439. doi: 10.1016/j.stem.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joost S., Jacob T., Sun X., Annusver K., La Manno G., Sur I., Kasper M. Single-cell transcriptomics of traced epidermal and hair follicle stem cells reveals rapid adaptations during wound healing. Cell Rep. 2018;25:585–597.e7. doi: 10.1016/j.celrep.2018.09.059. [DOI] [PubMed] [Google Scholar]
- Joost S., Zeisel A., Jacob T., Sun X., La Manno G., Lönnerberg P., Linnarsson S., Kasper M. Single-cell transcriptomics reveals that differentiation and spatial signatures shape epidermal and hair follicle heterogeneity. Cell Syst. 2016;3:221–237.e9. doi: 10.1016/j.cels.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar K., Weber C., Driskell R.R., Calonje E., Watt F.M. Compartmentalized epidermal activation of β-catenin differentially affects lineage reprogramming and underlies tumor heterogeneity. Cell Rep. 2016;14:269–281. doi: 10.1016/j.celrep.2015.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger T., Simons B.D. Dynamic stem cell heterogeneity. Development. 2015;142:1396–1406. doi: 10.1242/dev.101063. [DOI] [PubMed] [Google Scholar]
- Lavker R.M., Sun T.T., Oshima H., Barrandon Y., Akiyama M., Ferraris C., Chevalier G., Favier B., Jahoda C.A.B., Dhouailly D., et al. Hair follicle stem cells. J. Invest. Dermatol. Symp. Proc. 2003;8:28–38. doi: 10.1046/j.1523-1747.2003.12169.x. [DOI] [PubMed] [Google Scholar]
- Li L., Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Mignone J., Yang M., Matic M., Penman S., Enikolopov G., Hoffman R.M. Nestin expression in hair follicle sheath progenitor cells. Proc. Natl. Acad. Sci. USA. 2003;100:9958–9961. doi: 10.1073/pnas.1733025100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris R.J., Liu Y., Marles L., Yang Z., Trempus C., Li S., Lin J.S., Sawicki J.A., Cotsarelis G. Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- Niemann C., Horsley V. Development and homeostasis of the sebaceous gland. Semin. Cell Dev. Biol. 2012;23:928–936. doi: 10.1016/j.semcdb.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhof J.G.W., Braun K.M., Giangreco A., van Pelt C., Kawamoto H., Boyd R.L., Willemze R., Mullenders L.H.F., Watt F.M., de Gruijl F.R., van Ewijk W. The cell-surface marker MTS24 identifies a novel population of follicular keratinocytes with characteristics of progenitor cells. Development. 2006;133:3027–3037. doi: 10.1242/dev.02443. [DOI] [PubMed] [Google Scholar]
- Page M.E., Lombard P., Ng F., Göttgens B., Jensen K.B. The epidermis comprises autonomous compartments maintained by distinct stem cell populations. Cell Stem Cell. 2013;13:471–482. doi: 10.1016/j.stem.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus R., Müller-Röver S., van der Veen C., Maurer M., Eichmüller S., Ling G., Hofmann U., Foitzik K., Mecklenburg L., Handjiski B. A comprehensive guide for the recognition and classification of distinct stages of hair follicle morphogenesis. J. Invest. Dermatol. 1999;113:523–532. doi: 10.1046/j.1523-1747.1999.00740.x. [DOI] [PubMed] [Google Scholar]
- Petersson M., Brylka H., Kraus A., John S., Rappl G., Schettina P., Niemann C. TCF/Lef1 activity controls establishment of diverse stem and progenitor cell compartments in mouse epidermis. EMBO J. 2011;30:3004–3018. doi: 10.1038/emboj.2011.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson M., Reuter K., Brylka H., Kraus A., Schettina P., Niemann C. Interfering with stem cell-specific gatekeeper functions controls tumour initiation and malignant progression of skin tumours. Nat. Commun. 2015;6:5874. doi: 10.1038/ncomms6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P., Mesa K.R., Greco V. Spatial organization within a niche as a determinant of stem-cell fate. Nature. 2013;502:513–518. doi: 10.1038/nature12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena N., Mok K.W., Rendl M. An updated classification of hair follicle morphogenesis. Exp. Dermatol. 2019;28:332–344. doi: 10.1111/exd.13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert H.J., Haegebarth A., Kasper M., Jaks V., van Es J.H., Barker N., van de Wetering M., van den Born M., Begthel H., Vries R.G., et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327:1385–1389. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- Solanas G., Benitah S.A. Regenerating the skin: a task for the heterogeneous stem cell pool and surrounding niche. Nat. Rev. Mol. Cell Biol. 2013;14:737–748. doi: 10.1038/nrm3675. [DOI] [PubMed] [Google Scholar]
- Trempus C.S., Morris R.J., Bortner C.D., Cotsarelis G., Faircloth R.S., Reece J.M., Tennant R.W. Enrichment for living murine keratinocytes from the hair follicle bulge with the cell surface marker CD34. J. Invest. Dermatol. 2003;120:501–511. doi: 10.1046/j.1523-1747.2003.12088.x. [DOI] [PubMed] [Google Scholar]
- Wang J., He J., Zhu M., Han Y., Yang R., Liu H., Xu X., Chen X. Cellular heterogeneity and plasticity of skin epithelial cells in wound healing and tumorigenesis. Stem Cell Rev. Rep. 2022;18:1912–1925. doi: 10.1007/s12015-021-10295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chen H., Tian R., Zhang Y., Drutskaya M.S., Wang C., Ge J., Fan Z., Kong D., Wang X., et al. Macrophages induce AKT/β-catenin-dependent Lgr5+ stem cell activation and hair follicle regeneration through TNF. Nat. Commun. 2017;8 doi: 10.1038/ncomms14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Ge J., Tredget E.E., Wu Y. The mouse excisional wound splinting model, including applications for stem cell transplantation. Nat. Protoc. 2013;8:302–309. doi: 10.1038/nprot.2013.002. [DOI] [PubMed] [Google Scholar]
- Wells J.M., Watt F.M. Diverse mechanisms for endogenous regeneration and repair in mammalian organs. Nature. 2018;557:322–328. doi: 10.1038/s41586-018-0073-7. [DOI] [PubMed] [Google Scholar]
- Xin T., Gonzalez D., Rompolas P., Greco V. Flexible fate determination ensures robust differentiation in the hair follicle. Nat. Cell Biol. 2018;20:1361–1369. doi: 10.1038/s41556-018-0232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Adam R.C., Ge Y., Hua Z.L., Fuchs E. Epithelial-mesenchymal micro-niches govern stem cell lineage choices. Cell. 2017;169:483–496.e13. doi: 10.1016/j.cell.2017.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data: The data that support the findings of this study are available from the corresponding author upon reasonable request.
-
•
Code: This research paper does not list code.
-
•
Availability statement: Any additional information in this study are available from the lead contact upon request. upon reasonable request.